Please be patient as the page loads
|
ZNHI4_HUMAN
|
||||||
| SwissProt Accessions | Q9C086 | Gene names | ZNHIT4, HMGA1L4, PAPA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger HIT domain-containing protein 4 (PAP-1-associated protein 1) (PAPA-1) (High mobility group AT-hook 1-like 4). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ZNHI4_MOUSE
|
||||||
| θ value | 1.53401e-116 (rank : 1) | NC score | 0.983677 (rank : 2) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99PT3, Q9CY38 | Gene names | Znhit4, Hmga1l4, Papa1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger HIT domain-containing protein 4 (PAP-1-associated protein 1) (PAPA-1) (High mobility group AT-hook 1-like 4). | |||||
|
ZNHI4_HUMAN
|
||||||
| θ value | 7.37397e-111 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9C086 | Gene names | ZNHIT4, HMGA1L4, PAPA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger HIT domain-containing protein 4 (PAP-1-associated protein 1) (PAPA-1) (High mobility group AT-hook 1-like 4). | |||||
|
VPS72_HUMAN
|
||||||
| θ value | 0.125558 (rank : 3) | NC score | 0.068852 (rank : 5) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15906, Q53GJ2, Q5U0R4 | Gene names | VPS72, TCFL1, YL1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 72 homolog (Transcription factor-like 1) (Protein YL-1). | |||||
|
ZN580_MOUSE
|
||||||
| θ value | 0.125558 (rank : 4) | NC score | 0.026368 (rank : 12) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9DB38 | Gene names | Znf580, Zfp580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 580. | |||||
|
ZN580_HUMAN
|
||||||
| θ value | 0.21417 (rank : 5) | NC score | 0.024465 (rank : 14) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UK33, Q9NPP7 | Gene names | ZNF580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 580 (LDL-induced EC protein). | |||||
|
ZNHI2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 6) | NC score | 0.110921 (rank : 3) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UHR6, Q3SY14, Q8IUV0 | Gene names | ZNHIT2, C11orf5 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger HIT domain-containing protein 2 (Protein FON). | |||||
|
IF4G1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 7) | NC score | 0.032288 (rank : 10) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
SYNG_HUMAN
|
||||||
| θ value | 1.06291 (rank : 8) | NC score | 0.064473 (rank : 7) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UMZ2, Q6ZT17 | Gene names | AP1GBP1, SYNG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP1 subunit gamma-binding protein 1 (Gamma-synergin). | |||||
|
ZN518_HUMAN
|
||||||
| θ value | 1.81305 (rank : 9) | NC score | 0.016644 (rank : 19) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6AHZ1, O15044, Q32MP4 | Gene names | ZNF518, KIAA0335 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 518. | |||||
|
PANK1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 10) | NC score | 0.025038 (rank : 13) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TE04, Q7RTX6, Q7Z495, Q8TBQ8 | Gene names | PANK1, PANK | |||
|
Domain Architecture |
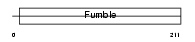
|
|||||
| Description | Pantothenate kinase 1 (EC 2.7.1.33) (Pantothenic acid kinase 1) (hPanK1) (hPanK). | |||||
|
MYPN_MOUSE
|
||||||
| θ value | 3.0926 (rank : 11) | NC score | 0.013681 (rank : 20) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5DTJ9, Q7TPW5, Q8BZ76 | Gene names | Mypn, Kiaa4170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myopalladin. | |||||
|
FREA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 12) | NC score | 0.008563 (rank : 22) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43638, Q96D28 | Gene names | FKHL18, FREAC10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Forkhead-related transcription factor 10 (FREAC-10) (Forkhead-like 18 protein). | |||||
|
PS1C2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 13) | NC score | 0.068650 (rank : 6) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80ZC9 | Gene names | Psors1c2, Spr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Psoriasis susceptibility 1 candidate gene 2 protein homolog precursor (SPR1 protein). | |||||
|
TP53B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 14) | NC score | 0.018789 (rank : 17) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
ZBT17_HUMAN
|
||||||
| θ value | 5.27518 (rank : 15) | NC score | 0.006018 (rank : 24) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 925 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13105, Q15932, Q5JYB2, Q9NUC9 | Gene names | ZBTB17, MIZ1, ZNF151 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 17 (Zinc finger protein 151) (Myc-interacting zinc finger protein) (Miz-1 protein). | |||||
|
PKHA4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 16) | NC score | 0.040492 (rank : 9) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VC98, Q8R3N3 | Gene names | Plekha4, Pepp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
SYNG_MOUSE
|
||||||
| θ value | 6.88961 (rank : 17) | NC score | 0.045848 (rank : 8) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5SV85, Q5SV84, Q6PHT6 | Gene names | Ap1gbp1, Syng | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP1 subunit gamma-binding protein 1 (Gamma-synergin). | |||||
|
TAF6L_MOUSE
|
||||||
| θ value | 6.88961 (rank : 18) | NC score | 0.018912 (rank : 16) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R2K4, Q8C5T2 | Gene names | Taf6l, Paf65a | |||
|
Domain Architecture |
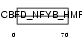
|
|||||
| Description | TAF6-like RNA polymerase II p300/CBP-associated factor-associated factor 65 kDa subunit 6L (PCAF-associated factor 65 alpha) (PAF65- alpha). | |||||
|
ZNHI2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 19) | NC score | 0.069952 (rank : 4) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QY66, Q8C356, Q8VCQ7 | Gene names | Znhit2, ORF6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger HIT domain-containing protein 2 (Protein FON). | |||||
|
CRTC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 20) | NC score | 0.018068 (rank : 18) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6UUV9, O75114, Q6Y3A3, Q7LDZ2, Q8IUL3, Q8IZ34, Q8IZL1, Q8N6W3, Q96AI8, Q9H801 | Gene names | CRTC1, KIAA0616, MECT1, TORC1, WAMTP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Transducer of regulated cAMP response element-binding protein 1) (Transducer of CREB protein 1) (Mucoepidermoid carcinoma translocated protein 1). | |||||
|
HIS3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 21) | NC score | 0.032170 (rank : 11) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P15516, Q16243, Q502Z1 | Gene names | HTN3, HIS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histatin-3 precursor (Histidine-rich protein 3) (PB) (Basic histidine- rich protein) (Hst) [Contains: Histatin-3; Histatin-3 1/25 (Histatin- 6); Histatin-3 1/24 (Histatin-5); Histatin-3 1/13; Histatin-3 1/12; Histatin-3 1/11; Histatin-3 5/13; Histatin-3 5/12 (Histatin-11); Histatin-3 5/11 (Histatin-12); Histatin-3 6/13; Histatin-3 6/11; Histatin-3 7/13; Histatin-3 7/12; Histatin-3 7/11; Histatin-3 12/32 (Histatin-4); Histatin-3 12/25 (Histatin-9); Histatin-3 12/24 (Histatin-7); Histatin-3 13/25 (Histatin-10); Histatin-3 13/24 (Histatin-8); Histatin-3 14/25; Histatin-3 14/24; Histatin-3 15/25; Histatin-3 15/24; Histatin-3 26/32; Histatin-3 28/32; Histatin-3 29/32]. | |||||
|
KCNQ1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 22) | NC score | 0.022845 (rank : 15) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97414, O88702, Q7TNZ1, Q7TPL7, Q80VR7 | Gene names | Kcnq1, Kcna9, Kvlqt1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
P3H1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 23) | NC score | 0.010728 (rank : 21) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q32P28, Q7KZR4, Q96BR8, Q96SK8, Q96SL5, Q96SN3, Q9H6K3, Q9HC86, Q9HC87 | Gene names | LEPRE1, GROS1, P3H1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prolyl 3-hydroxylase 1 precursor (EC 1.14.11.7) (Leucine- and proline- enriched proteoglycan 1) (Leprecan-1) (Growth suppressor 1). | |||||
|
PRD13_HUMAN
|
||||||
| θ value | 8.99809 (rank : 24) | NC score | 0.007204 (rank : 23) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 716 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H4Q3, Q5TGC1 | Gene names | PRDM13, PFM10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PR domain zinc finger protein 13 (PR domain-containing protein 13). | |||||
|
ZNHI4_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 7.37397e-111 (rank : 2) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9C086 | Gene names | ZNHIT4, HMGA1L4, PAPA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger HIT domain-containing protein 4 (PAP-1-associated protein 1) (PAPA-1) (High mobility group AT-hook 1-like 4). | |||||
|
ZNHI4_MOUSE
|
||||||
| NC score | 0.983677 (rank : 2) | θ value | 1.53401e-116 (rank : 1) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99PT3, Q9CY38 | Gene names | Znhit4, Hmga1l4, Papa1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger HIT domain-containing protein 4 (PAP-1-associated protein 1) (PAPA-1) (High mobility group AT-hook 1-like 4). | |||||
|
ZNHI2_HUMAN
|
||||||
| NC score | 0.110921 (rank : 3) | θ value | 0.62314 (rank : 6) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UHR6, Q3SY14, Q8IUV0 | Gene names | ZNHIT2, C11orf5 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger HIT domain-containing protein 2 (Protein FON). | |||||
|
ZNHI2_MOUSE
|
||||||
| NC score | 0.069952 (rank : 4) | θ value | 6.88961 (rank : 19) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QY66, Q8C356, Q8VCQ7 | Gene names | Znhit2, ORF6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger HIT domain-containing protein 2 (Protein FON). | |||||
|
VPS72_HUMAN
|
||||||
| NC score | 0.068852 (rank : 5) | θ value | 0.125558 (rank : 3) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15906, Q53GJ2, Q5U0R4 | Gene names | VPS72, TCFL1, YL1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 72 homolog (Transcription factor-like 1) (Protein YL-1). | |||||
|
PS1C2_MOUSE
|
||||||
| NC score | 0.068650 (rank : 6) | θ value | 5.27518 (rank : 13) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80ZC9 | Gene names | Psors1c2, Spr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Psoriasis susceptibility 1 candidate gene 2 protein homolog precursor (SPR1 protein). | |||||
|
SYNG_HUMAN
|
||||||
| NC score | 0.064473 (rank : 7) | θ value | 1.06291 (rank : 8) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UMZ2, Q6ZT17 | Gene names | AP1GBP1, SYNG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP1 subunit gamma-binding protein 1 (Gamma-synergin). | |||||
|
SYNG_MOUSE
|
||||||
| NC score | 0.045848 (rank : 8) | θ value | 6.88961 (rank : 17) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5SV85, Q5SV84, Q6PHT6 | Gene names | Ap1gbp1, Syng | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP1 subunit gamma-binding protein 1 (Gamma-synergin). | |||||
|
PKHA4_MOUSE
|
||||||
| NC score | 0.040492 (rank : 9) | θ value | 6.88961 (rank : 16) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VC98, Q8R3N3 | Gene names | Plekha4, Pepp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
IF4G1_MOUSE
|
||||||
| NC score | 0.032288 (rank : 10) | θ value | 0.813845 (rank : 7) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
HIS3_HUMAN
|
||||||
| NC score | 0.032170 (rank : 11) | θ value | 8.99809 (rank : 21) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P15516, Q16243, Q502Z1 | Gene names | HTN3, HIS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histatin-3 precursor (Histidine-rich protein 3) (PB) (Basic histidine- rich protein) (Hst) [Contains: Histatin-3; Histatin-3 1/25 (Histatin- 6); Histatin-3 1/24 (Histatin-5); Histatin-3 1/13; Histatin-3 1/12; Histatin-3 1/11; Histatin-3 5/13; Histatin-3 5/12 (Histatin-11); Histatin-3 5/11 (Histatin-12); Histatin-3 6/13; Histatin-3 6/11; Histatin-3 7/13; Histatin-3 7/12; Histatin-3 7/11; Histatin-3 12/32 (Histatin-4); Histatin-3 12/25 (Histatin-9); Histatin-3 12/24 (Histatin-7); Histatin-3 13/25 (Histatin-10); Histatin-3 13/24 (Histatin-8); Histatin-3 14/25; Histatin-3 14/24; Histatin-3 15/25; Histatin-3 15/24; Histatin-3 26/32; Histatin-3 28/32; Histatin-3 29/32]. | |||||
|
ZN580_MOUSE
|
||||||
| NC score | 0.026368 (rank : 12) | θ value | 0.125558 (rank : 4) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9DB38 | Gene names | Znf580, Zfp580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 580. | |||||
|
PANK1_HUMAN
|
||||||
| NC score | 0.025038 (rank : 13) | θ value | 2.36792 (rank : 10) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TE04, Q7RTX6, Q7Z495, Q8TBQ8 | Gene names | PANK1, PANK | |||
|
Domain Architecture |

|
|||||
| Description | Pantothenate kinase 1 (EC 2.7.1.33) (Pantothenic acid kinase 1) (hPanK1) (hPanK). | |||||
|
ZN580_HUMAN
|
||||||
| NC score | 0.024465 (rank : 14) | θ value | 0.21417 (rank : 5) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UK33, Q9NPP7 | Gene names | ZNF580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 580 (LDL-induced EC protein). | |||||
|
KCNQ1_MOUSE
|
||||||
| NC score | 0.022845 (rank : 15) | θ value | 8.99809 (rank : 22) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97414, O88702, Q7TNZ1, Q7TPL7, Q80VR7 | Gene names | Kcnq1, Kcna9, Kvlqt1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
TAF6L_MOUSE
|
||||||
| NC score | 0.018912 (rank : 16) | θ value | 6.88961 (rank : 18) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R2K4, Q8C5T2 | Gene names | Taf6l, Paf65a | |||
|
Domain Architecture |

|
|||||
| Description | TAF6-like RNA polymerase II p300/CBP-associated factor-associated factor 65 kDa subunit 6L (PCAF-associated factor 65 alpha) (PAF65- alpha). | |||||
|
TP53B_MOUSE
|
||||||
| NC score | 0.018789 (rank : 17) | θ value | 5.27518 (rank : 14) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
CRTC1_HUMAN
|
||||||
| NC score | 0.018068 (rank : 18) | θ value | 8.99809 (rank : 20) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6UUV9, O75114, Q6Y3A3, Q7LDZ2, Q8IUL3, Q8IZ34, Q8IZL1, Q8N6W3, Q96AI8, Q9H801 | Gene names | CRTC1, KIAA0616, MECT1, TORC1, WAMTP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Transducer of regulated cAMP response element-binding protein 1) (Transducer of CREB protein 1) (Mucoepidermoid carcinoma translocated protein 1). | |||||
|
ZN518_HUMAN
|
||||||
| NC score | 0.016644 (rank : 19) | θ value | 1.81305 (rank : 9) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6AHZ1, O15044, Q32MP4 | Gene names | ZNF518, KIAA0335 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 518. | |||||
|
MYPN_MOUSE
|
||||||
| NC score | 0.013681 (rank : 20) | θ value | 3.0926 (rank : 11) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5DTJ9, Q7TPW5, Q8BZ76 | Gene names | Mypn, Kiaa4170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myopalladin. | |||||
|
P3H1_HUMAN
|
||||||
| NC score | 0.010728 (rank : 21) | θ value | 8.99809 (rank : 23) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q32P28, Q7KZR4, Q96BR8, Q96SK8, Q96SL5, Q96SN3, Q9H6K3, Q9HC86, Q9HC87 | Gene names | LEPRE1, GROS1, P3H1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prolyl 3-hydroxylase 1 precursor (EC 1.14.11.7) (Leucine- and proline- enriched proteoglycan 1) (Leprecan-1) (Growth suppressor 1). | |||||
|
FREA_HUMAN
|
||||||
| NC score | 0.008563 (rank : 22) | θ value | 5.27518 (rank : 12) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43638, Q96D28 | Gene names | FKHL18, FREAC10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Forkhead-related transcription factor 10 (FREAC-10) (Forkhead-like 18 protein). | |||||
|
PRD13_HUMAN
|
||||||
| NC score | 0.007204 (rank : 23) | θ value | 8.99809 (rank : 24) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 716 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H4Q3, Q5TGC1 | Gene names | PRDM13, PFM10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PR domain zinc finger protein 13 (PR domain-containing protein 13). | |||||
|
ZBT17_HUMAN
|
||||||
| NC score | 0.006018 (rank : 24) | θ value | 5.27518 (rank : 15) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 925 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13105, Q15932, Q5JYB2, Q9NUC9 | Gene names | ZBTB17, MIZ1, ZNF151 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 17 (Zinc finger protein 151) (Myc-interacting zinc finger protein) (Miz-1 protein). | |||||