Please be patient as the page loads
|
SYNG_HUMAN
|
||||||
| SwissProt Accessions | Q9UMZ2, Q6ZT17 | Gene names | AP1GBP1, SYNG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP1 subunit gamma-binding protein 1 (Gamma-synergin). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SYNG_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 160 | |
| SwissProt Accessions | Q9UMZ2, Q6ZT17 | Gene names | AP1GBP1, SYNG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP1 subunit gamma-binding protein 1 (Gamma-synergin). | |||||
|
SYNG_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.946481 (rank : 2) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q5SV85, Q5SV84, Q6PHT6 | Gene names | Ap1gbp1, Syng | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP1 subunit gamma-binding protein 1 (Gamma-synergin). | |||||
|
ITSN1_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 3) | NC score | 0.125338 (rank : 7) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
DSPP_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 4) | NC score | 0.113618 (rank : 12) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
ITSN1_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 5) | NC score | 0.121606 (rank : 8) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
EP15_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 6) | NC score | 0.150630 (rank : 3) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P42567 | Gene names | Eps15 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
EP15_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 7) | NC score | 0.146834 (rank : 4) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1021 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P42566 | Gene names | EPS15, AF1P | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
GORS2_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 8) | NC score | 0.110548 (rank : 13) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99JX3, Q3U9D2, Q8CCD0, Q91W68 | Gene names | Gorasp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 2 (GRS2) (Golgi reassembly-stacking protein of 55 kDa) (GRASP55). | |||||
|
ITSN2_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 9) | NC score | 0.119107 (rank : 10) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1173 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9NZM3, O95062, Q15812, Q9HAK4, Q9NXE6, Q9NYG0, Q9NZM2, Q9ULG4 | Gene names | ITSN2, KIAA1256, SH3D1B, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (SH3P18) (SH3P18-like WASP-associated protein). | |||||
|
ITSN2_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 10) | NC score | 0.114094 (rank : 11) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1371 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9Z0R6, Q9Z0R5 | Gene names | Itsn2, Ese2, Sh3d1B | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (EH and SH3 domains protein 2) (EH domain and SH3 domain regulator of endocytosis 2). | |||||
|
EP15R_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 11) | NC score | 0.130109 (rank : 5) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q60902, Q8CB60, Q8CB70, Q91WH8 | Gene names | Eps15l1, Eps15-rs, Eps15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R) (Epidermal growth factor receptor pathway substrate 15-related sequence) (Eps15-rs). | |||||
|
PTN13_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 12) | NC score | 0.028843 (rank : 112) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q64512, Q61494, Q62135, Q64499 | Gene names | Ptpn13, Ptp14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein tyrosine phosphatase PTP-BL) (Protein-tyrosine phosphatase RIP) (protein tyrosine phosphatase DPZPTP) (PTP36). | |||||
|
EP15R_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 13) | NC score | 0.127224 (rank : 6) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1083 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UBC2 | Gene names | EPS15L1, EPS15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R). | |||||
|
AKA12_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 14) | NC score | 0.070345 (rank : 37) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
IL4RA_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 15) | NC score | 0.081009 (rank : 24) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P16382, O54690, Q60583, Q8CBW5 | Gene names | Il4r, Il4ra | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-4 receptor alpha chain precursor (IL-4R-alpha) (CD124 antigen) [Contains: Soluble interleukin-4 receptor alpha chain (IL-4- binding protein) (IL4-BP)]. | |||||
|
CSTFT_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 16) | NC score | 0.061300 (rank : 47) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8C7E9, Q6A016, Q8BHH7, Q8C3W7, Q8R2Y1, Q9EPU3 | Gene names | Cstf2t, Kiaa0689 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage stimulation factor 64 kDa subunit, tau variant (CSTF 64 kDa subunit, tau variant) (CF-1 64 kDa subunit, tau variant) (TauCstF-64). | |||||
|
LHX3_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 17) | NC score | 0.023630 (rank : 129) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P50481, Q61800, Q61801 | Gene names | Lhx3, Lim-3, Lim3, Plim | |||
|
Domain Architecture |
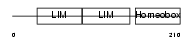
|
|||||
| Description | LIM/homeobox protein Lhx3 (Homeobox protein LIM-3) (Homeobox protein P-LIM) (LIM homeobox protein 3). | |||||
|
ASPP1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 18) | NC score | 0.051192 (rank : 62) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 956 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96KQ4, O94870 | Gene names | PPP1R13B, ASPP1, KIAA0771 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
ATN1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 19) | NC score | 0.073441 (rank : 33) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
ALMS1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 20) | NC score | 0.064125 (rank : 43) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8TCU4, Q53S05, Q580Q8, Q86VP9, Q9Y4G4 | Gene names | ALMS1, KIAA0328 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alstrom syndrome protein 1. | |||||
|
S23IP_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 21) | NC score | 0.080663 (rank : 26) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y6Y8, Q8IXH5, Q9BUK5 | Gene names | SEC23IP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SEC23-interacting protein (p125). | |||||
|
HECD1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 22) | NC score | 0.030219 (rank : 109) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 371 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ULT8, Q6P445, Q86VJ1, Q96F34, Q9UFZ7 | Gene names | HECTD1, KIAA1131 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase HECTD1 (HECT domain-containing protein 1) (E3 ligase for inhibin receptor) (EULIR). | |||||
|
ROBO2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 23) | NC score | 0.020281 (rank : 138) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9HCK4, O43608 | Gene names | ROBO2, KIAA1568 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 2 precursor. | |||||
|
GPSM2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 24) | NC score | 0.057948 (rank : 54) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P81274, Q8N0Z5 | Gene names | GPSM2, LGN | |||
|
Domain Architecture |

|
|||||
| Description | G-protein signaling modulator 2 (Mosaic protein LGN). | |||||
|
GPSM2_MOUSE
|
||||||
| θ value | 0.125558 (rank : 25) | NC score | 0.057707 (rank : 55) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VDU0, Q8BLX3 | Gene names | Gpsm2, Lgn, Pins | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein signaling modulator 2 (Pins homolog). | |||||
|
ROBO3_HUMAN
|
||||||
| θ value | 0.125558 (rank : 26) | NC score | 0.030717 (rank : 107) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96MS0 | Gene names | ROBO3 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 3 precursor (Roundabout-like protein 3). | |||||
|
YLPM1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 27) | NC score | 0.090204 (rank : 17) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
HGS_MOUSE
|
||||||
| θ value | 0.163984 (rank : 28) | NC score | 0.067504 (rank : 40) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
PDC6I_MOUSE
|
||||||
| θ value | 0.163984 (rank : 29) | NC score | 0.080356 (rank : 27) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9WU78, O88695, O89014, Q8BSL8, Q8R0H5, Q99LR3, Q9QZN8 | Gene names | Pdcd6ip, Aip1, Alix | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death 6-interacting protein (ALG-2-interacting protein X) (ALG-2-interacting protein 1) (E2F1-inducible protein) (Eig2). | |||||
|
TRIP6_HUMAN
|
||||||
| θ value | 0.163984 (rank : 30) | NC score | 0.029235 (rank : 110) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15654, O15170, O15275, Q9BTB2, Q9UNT4 | Gene names | TRIP6, OIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 6 (TRIP6) (OPA-interacting protein 1) (Zyxin-related protein 1) (ZRP-1). | |||||
|
BCL9_MOUSE
|
||||||
| θ value | 0.21417 (rank : 31) | NC score | 0.071102 (rank : 36) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
CSTFT_HUMAN
|
||||||
| θ value | 0.21417 (rank : 32) | NC score | 0.040350 (rank : 81) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H0L4, O75174, Q53HK6, Q7LGE8, Q8N6T1 | Gene names | CSTF2T, KIAA0689 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage stimulation factor 64 kDa subunit, tau variant (CSTF 64 kDa subunit, tau variant) (CF-1 64 kDa subunit, tau variant) (TauCstF-64). | |||||
|
FA54A_MOUSE
|
||||||
| θ value | 0.21417 (rank : 33) | NC score | 0.072215 (rank : 34) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VED8 | Gene names | Fam54a, Dufd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM54A (DUF729 domain-containing protein 1). | |||||
|
TCF20_HUMAN
|
||||||
| θ value | 0.21417 (rank : 34) | NC score | 0.064054 (rank : 44) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UGU0, O14528, Q13078, Q4V353, Q9H4M0 | Gene names | TCF20, KIAA0292, SPBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP) (AR1). | |||||
|
AFF1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 35) | NC score | 0.046341 (rank : 68) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
GRIN1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 36) | NC score | 0.073963 (rank : 32) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
NUPL_MOUSE
|
||||||
| θ value | 0.279714 (rank : 37) | NC score | 0.060960 (rank : 48) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8K2K6, O70448, Q8BQL5, Q8CDK9 | Gene names | Hrb, Rip | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoporin-like protein RIP (HIV-1 Rev-binding protein homolog). | |||||
|
LMNB1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 38) | NC score | 0.034015 (rank : 96) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 719 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P14733, Q61791 | Gene names | Lmnb1 | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-B1. | |||||
|
MAVS_MOUSE
|
||||||
| θ value | 0.365318 (rank : 39) | NC score | 0.078193 (rank : 29) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8VCF0 | Gene names | Mavs, Ips1, Visa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial antiviral signaling protein (Interferon-beta promoter stimulator protein 1) (IPS-1) (Virus-induced signaling adapter) (CARD adapter inducing interferon-beta) (Cardif). | |||||
|
PTN22_MOUSE
|
||||||
| θ value | 0.365318 (rank : 40) | NC score | 0.026497 (rank : 120) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P29352, Q7TMP9 | Gene names | Ptpn22, Ptpn8 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 22 (EC 3.1.3.48) (Hematopoietic cell protein-tyrosine phosphatase 70Z-PEP). | |||||
|
SMC3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 41) | NC score | 0.058378 (rank : 53) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1186 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9UQE7, O60464 | Gene names | CSPG6, BAM, BMH, SMC3, SMC3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome-associated polypeptide) (hCAP) (Bamacan) (Basement membrane-associated chondroitin proteoglycan). | |||||
|
SMC3_MOUSE
|
||||||
| θ value | 0.365318 (rank : 42) | NC score | 0.058465 (rank : 52) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1187 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9CW03, O35667, Q9QUS3 | Gene names | Cspg6, Bam, Bmh, Mmip1, Smc3, Smc3l1, Smcd | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome segregation protein SmcD) (Bamacan) (Basement membrane-associated chondroitin proteoglycan) (Mad member- interacting protein 1). | |||||
|
AF10_HUMAN
|
||||||
| θ value | 0.47712 (rank : 43) | NC score | 0.027906 (rank : 114) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P55197 | Gene names | MLLT10, AF10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-10. | |||||
|
TCP1L_HUMAN
|
||||||
| θ value | 0.47712 (rank : 44) | NC score | 0.084817 (rank : 19) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TDR4, Q96LN5 | Gene names | TCP10L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-complex protein 10A homolog 2 (T-complex protein 10A-2) (TCP10A-2) (TCP10-like). | |||||
|
EPN2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 45) | NC score | 0.042601 (rank : 74) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95208, O95207, Q9H7Z2, Q9UPT7 | Gene names | EPN2, KIAA1065 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-2 (EPS-15-interacting protein 2). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 46) | NC score | 0.054037 (rank : 59) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
TCF20_MOUSE
|
||||||
| θ value | 0.62314 (rank : 47) | NC score | 0.066521 (rank : 41) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 0.813845 (rank : 48) | NC score | 0.055277 (rank : 58) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 0.813845 (rank : 49) | NC score | 0.069495 (rank : 38) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 0.813845 (rank : 50) | NC score | 0.061346 (rank : 46) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TCOF_HUMAN
|
||||||
| θ value | 0.813845 (rank : 51) | NC score | 0.097095 (rank : 15) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
TXND2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 52) | NC score | 0.040738 (rank : 79) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
CCD91_HUMAN
|
||||||
| θ value | 1.06291 (rank : 53) | NC score | 0.049531 (rank : 64) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q7Z6B0, Q68D43, Q6IA78, Q8NEN7, Q9NUW9 | Gene names | CCDC91, GGABP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner) (p56 accessory protein). | |||||
|
DHB4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 54) | NC score | 0.040856 (rank : 77) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51659 | Gene names | HSD17B4, EDH17B4 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal multifunctional enzyme type 2 (MFE-2) (D-bifunctional protein) (DBP) (17-beta-hydroxysteroid dehydrogenase 4) (17-beta-HSD 4) (D-3-hydroxyacyl-CoA dehydratase) (EC 4.2.1.107) (3-alpha,7- alpha,12-alpha-trihydroxy-5-beta-cholest-24-enoyl-CoA hydratase) (3- hydroxyacyl-CoA dehydrogenase) (EC 1.1.1.35). | |||||
|
LMNB1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 55) | NC score | 0.033200 (rank : 99) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P20700, Q3SYN7, Q96EI6 | Gene names | LMNB1, LMN2, LMNB | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-B1. | |||||
|
NRG2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 56) | NC score | 0.032597 (rank : 103) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P56974 | Gene names | Nrg2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pro-neuregulin-2, membrane-bound isoform precursor (Pro-NRG2) [Contains: Neuregulin-2 (NRG-2) (Divergent of neuregulin 1) (DON-1)]. | |||||
|
REPS2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 57) | NC score | 0.120107 (rank : 9) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8NFH8, O43428, Q8NFI5 | Gene names | REPS2, POB1 | |||
|
Domain Architecture |

|
|||||
| Description | RalBP1-associated Eps domain-containing protein 2 (RalBP1-interacting protein 2) (Partner of RalBP1). | |||||
|
ZNHI4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 58) | NC score | 0.064473 (rank : 42) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9C086 | Gene names | ZNHIT4, HMGA1L4, PAPA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger HIT domain-containing protein 4 (PAP-1-associated protein 1) (PAPA-1) (High mobility group AT-hook 1-like 4). | |||||
|
ARMX2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 59) | NC score | 0.032833 (rank : 100) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6A058, Q9CXI9 | Gene names | Armcx2, Kiaa0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2. | |||||
|
CN032_MOUSE
|
||||||
| θ value | 1.38821 (rank : 60) | NC score | 0.075407 (rank : 30) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 1.38821 (rank : 61) | NC score | 0.062680 (rank : 45) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
SYM_MOUSE
|
||||||
| θ value | 1.38821 (rank : 62) | NC score | 0.045680 (rank : 69) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q68FL6 | Gene names | Mars | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methionyl-tRNA synthetase (EC 6.1.1.10) (Methionine--tRNA ligase) (MetRS). | |||||
|
SYNJ1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 63) | NC score | 0.031248 (rank : 105) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
TR10B_HUMAN
|
||||||
| θ value | 1.38821 (rank : 64) | NC score | 0.026917 (rank : 119) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O14763, O14720, O15508, O15517, O15531, Q9BVE0 | Gene names | TNFRSF10B, DR5, KILLER, TRAILR2, TRICK2, ZTNFR9 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 10B precursor (Death receptor 5) (TNF-related apoptosis-inducing ligand receptor 2) (TRAIL receptor 2) (TRAIL-R2) (CD262 antigen). | |||||
|
UACA_MOUSE
|
||||||
| θ value | 1.38821 (rank : 65) | NC score | 0.030981 (rank : 106) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8CGB3, Q69ZG3, Q7TN77, Q8BJC8 | Gene names | Uaca, Kiaa1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein (Nucling) (Nuclear membrane-binding protein). | |||||
|
UBP42_HUMAN
|
||||||
| θ value | 1.38821 (rank : 66) | NC score | 0.024355 (rank : 127) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
XRN1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 67) | NC score | 0.026365 (rank : 121) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P97789, O35651, P97790, Q3TE44, Q3TRE9, Q3UQL5 | Gene names | Xrn1, Dhm2, Exo, Sep1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5'-3' exoribonuclease 1 (EC 3.1.11.-) (mXRN1) (Strand-exchange protein 1 homolog) (Protein Dhm2). | |||||
|
CK024_HUMAN
|
||||||
| θ value | 1.81305 (rank : 68) | NC score | 0.052699 (rank : 61) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
NCOR2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 69) | NC score | 0.034263 (rank : 95) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
SELPL_MOUSE
|
||||||
| θ value | 1.81305 (rank : 70) | NC score | 0.048843 (rank : 66) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 2.36792 (rank : 71) | NC score | 0.059383 (rank : 51) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
CDN1C_MOUSE
|
||||||
| θ value | 2.36792 (rank : 72) | NC score | 0.036910 (rank : 88) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P49919 | Gene names | Cdkn1c, Kip2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
CSPG5_MOUSE
|
||||||
| θ value | 2.36792 (rank : 73) | NC score | 0.032828 (rank : 101) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q71M36, Q71M37, Q7TNT8, Q8BPJ5, Q9QY32 | Gene names | Cspg5, Caleb, Ngc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate proteoglycan 5 precursor (Neuroglycan C) (Acidic leucine-rich EGF-like domain-containing brain protein). | |||||
|
HGS_HUMAN
|
||||||
| θ value | 2.36792 (rank : 74) | NC score | 0.068445 (rank : 39) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 517 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O14964, Q9NR36 | Gene names | HGS, HRS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate (Protein pp110) (Hrs). | |||||
|
LACTB_MOUSE
|
||||||
| θ value | 2.36792 (rank : 75) | NC score | 0.029067 (rank : 111) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9EP89 | Gene names | Lactb, Lact1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine beta-lactamase-like protein LACTB. | |||||
|
NUCB2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 76) | NC score | 0.053038 (rank : 60) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P81117 | Gene names | Nucb2, Nefa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleobindin-2 precursor (DNA-binding protein NEFA). | |||||
|
TIM44_MOUSE
|
||||||
| θ value | 2.36792 (rank : 77) | NC score | 0.037457 (rank : 84) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O35857 | Gene names | Timm44, Mimt44, Tim44 | |||
|
Domain Architecture |

|
|||||
| Description | Import inner membrane translocase subunit TIM44, mitochondrial precursor. | |||||
|
UPK3B_HUMAN
|
||||||
| θ value | 2.36792 (rank : 78) | NC score | 0.071936 (rank : 35) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BT76, Q86W06 | Gene names | UPK3B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uroplakin-3B precursor (Uroplakin IIIb) (UPIIIb) (p35). | |||||
|
ARI1B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 79) | NC score | 0.037457 (rank : 85) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
ARI5B_MOUSE
|
||||||
| θ value | 3.0926 (rank : 80) | NC score | 0.024972 (rank : 125) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BM75, Q8C0E0, Q8K4G8, Q8K4L9, Q9CU78, Q9JIX4 | Gene names | Arid5b, Desrt, Mrf2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (MRF1-like) (Modulator recognition factor protein 2) (MRF-2) (Developmentally and sexually retarded with transient immune abnormalities protein). | |||||
|
BRDT_MOUSE
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.027462 (rank : 115) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q91Y44, Q59HJ4 | Gene names | Brdt, Fsrg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain testis-specific protein (RING3-like protein) (Bromodomain- containing female sterile homeotic-like protein). | |||||
|
CEND3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | 0.023394 (rank : 131) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R5G7, Q5DTN4, Q6NVF1, Q8R5G6 | Gene names | Centd3, Arap3, Drag1, Kiaa4097 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3) (Dual specificity Rho- and Arf-GTPase-activating protein 1). | |||||
|
CT2NL_MOUSE
|
||||||
| θ value | 3.0926 (rank : 83) | NC score | 0.041246 (rank : 75) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 902 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q99LJ0, Q6ZPR0, Q8BSV1 | Gene names | Cttnbp2nl, Kiaa1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
EP400_HUMAN
|
||||||
| θ value | 3.0926 (rank : 84) | NC score | 0.018357 (rank : 141) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
HCN2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 85) | NC score | 0.022784 (rank : 133) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O88703, O70506 | Gene names | Hcn2, Bcng2, Hac1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2) (Hyperpolarization-activated cation channel 1) (HAC-1). | |||||
|
KI67_HUMAN
|
||||||
| θ value | 3.0926 (rank : 86) | NC score | 0.039912 (rank : 82) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 3.0926 (rank : 87) | NC score | 0.040839 (rank : 78) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 88) | NC score | 0.045062 (rank : 71) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
TMM28_HUMAN
|
||||||
| θ value | 3.0926 (rank : 89) | NC score | 0.039799 (rank : 83) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75949 | Gene names | TMEM28, TED | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 28 (TED protein). | |||||
|
WNK2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 90) | NC score | 0.009859 (rank : 158) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
CENPF_HUMAN
|
||||||
| θ value | 4.03905 (rank : 91) | NC score | 0.032073 (rank : 104) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
CIC_MOUSE
|
||||||
| θ value | 4.03905 (rank : 92) | NC score | 0.023502 (rank : 130) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CK5P2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 93) | NC score | 0.036687 (rank : 90) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
CN032_HUMAN
|
||||||
| θ value | 4.03905 (rank : 94) | NC score | 0.059533 (rank : 50) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
DHB13_MOUSE
|
||||||
| θ value | 4.03905 (rank : 95) | NC score | 0.022554 (rank : 134) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VCR2, Q8CIU2 | Gene names | Hsd17b13, Scdr9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 17-beta hydroxysteroid dehydrogenase 13 precursor (EC 1.1.-.-) (Short- chain dehydrogenase/reductase 9) (Alcohol dehydrogenase PAN1B-like). | |||||
|
ING3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 96) | NC score | 0.033366 (rank : 98) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VEK6, Q99JS6, Q9ERB2 | Gene names | Ing3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 3 (p47ING3 protein). | |||||
|
MINT_MOUSE
|
||||||
| θ value | 4.03905 (rank : 97) | NC score | 0.056853 (rank : 56) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
NSD1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 98) | NC score | 0.025244 (rank : 124) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
NUMBL_HUMAN
|
||||||
| θ value | 4.03905 (rank : 99) | NC score | 0.035168 (rank : 93) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Y6R0, Q7Z4J9 | Gene names | NUMBL | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein (Numb-R). | |||||
|
PARM1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 100) | NC score | 0.036133 (rank : 92) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6UWI2, Q96DV8, Q9Y4S1 | Gene names | ames=UNQ1879/PRO4322 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein PARM-1 precursor. | |||||
|
RTN1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 101) | NC score | 0.018863 (rank : 140) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q16799, Q16800, Q16801 | Gene names | RTN1, NSP | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-1 (Neuroendocrine-specific protein). | |||||
|
SRBP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 102) | NC score | 0.036496 (rank : 91) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
TTK_MOUSE
|
||||||
| θ value | 4.03905 (rank : 103) | NC score | 0.002810 (rank : 164) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35761 | Gene names | Ttk, Esk | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein kinase TTK (EC 2.7.12.1) (ESK) (PYT). | |||||
|
UBQL2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 104) | NC score | 0.033862 (rank : 97) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9QZM0, Q7TSJ8, Q8VDH9 | Gene names | Ubqln2, Plic2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-2 (Protein linking IAP with cytoskeleton 2) (PLIC-2) (Ubiquitin-like product Chap1/Dsk2) (DSK2 homolog) (Chap1). | |||||
|
CRTC1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 105) | NC score | 0.041083 (rank : 76) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6UUV9, O75114, Q6Y3A3, Q7LDZ2, Q8IUL3, Q8IZ34, Q8IZL1, Q8N6W3, Q96AI8, Q9H801 | Gene names | CRTC1, KIAA0616, MECT1, TORC1, WAMTP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Transducer of regulated cAMP response element-binding protein 1) (Transducer of CREB protein 1) (Mucoepidermoid carcinoma translocated protein 1). | |||||
|
EHD4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 106) | NC score | 0.082173 (rank : 23) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H223, Q9HAR1, Q9NZN2 | Gene names | EHD4, HCA10, HCA11 | |||
|
Domain Architecture |

|
|||||
| Description | EH-domain-containing protein 4 (Hepatocellular carcinoma-associated protein 10/11). | |||||
|
HHEX_HUMAN
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.013652 (rank : 152) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q03014 | Gene names | HHEX, HEX, PRH, PRHX | |||
|
Domain Architecture |
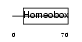
|
|||||
| Description | Homeobox protein PRH (Hematopoietically expressed homeobox) (Homeobox protein HEX). | |||||
|
KLC2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 108) | NC score | 0.020659 (rank : 137) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O88448 | Gene names | Klc2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 2 (KLC 2). | |||||
|
M3K4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 109) | NC score | 0.001584 (rank : 165) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O08648, O08649, O70124 | Gene names | Map3k4, Mekk4 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 4 (EC 2.7.11.25) (MAPK/ERK kinase kinase 4) (MEK kinase 4) (MEKK 4). | |||||
|
MN1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 110) | NC score | 0.042714 (rank : 73) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q10571 | Gene names | MN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable tumor suppressor protein MN1. | |||||
|
MNAB_HUMAN
|
||||||
| θ value | 5.27518 (rank : 111) | NC score | 0.013645 (rank : 153) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9HBD1, Q5JPD7, Q86ST6, Q8N3D6, Q96F27, Q9H5J2, Q9HBD2, Q9NWN9, Q9NXE1 | Gene names | MNAB, RNF164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated nucleic acid-binding protein (RING finger protein 164). | |||||
|
MYCD_MOUSE
|
||||||
| θ value | 5.27518 (rank : 112) | NC score | 0.028527 (rank : 113) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8VIM5, Q8C3W6, Q8VIL4 | Gene names | Myocd, Bsac2, Mycd, Srfcp | |||
|
Domain Architecture |

|
|||||
| Description | Myocardin (SRF cofactor protein) (Basic SAP coiled-coil transcription activator 2). | |||||
|
NU155_MOUSE
|
||||||
| θ value | 5.27518 (rank : 113) | NC score | 0.027342 (rank : 116) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99P88 | Gene names | Nup155 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup155 (Nucleoporin Nup155) (155 kDa nucleoporin). | |||||
|
PDZD6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 114) | NC score | 0.027201 (rank : 118) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ULD6, Q4W5I8, Q86V55 | Gene names | PDZD6, KIAA1284, PDZK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 6. | |||||
|
PO121_HUMAN
|
||||||
| θ value | 5.27518 (rank : 115) | NC score | 0.060032 (rank : 49) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
PTN12_MOUSE
|
||||||
| θ value | 5.27518 (rank : 116) | NC score | 0.017782 (rank : 145) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35831 | Gene names | Ptpn12 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 12 (EC 3.1.3.48) (Protein-tyrosine phosphatase P19) (P19-PTP) (MPTP-PEST). | |||||
|
RD23B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 117) | NC score | 0.025675 (rank : 123) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P54727, Q8WUB0 | Gene names | RAD23B | |||
|
Domain Architecture |
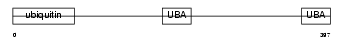
|
|||||
| Description | UV excision repair protein RAD23 homolog B (hHR23B) (XP-C repair- complementing complex 58 kDa protein) (p58). | |||||
|
ROCK2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 118) | NC score | 0.016007 (rank : 151) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 2073 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O75116, Q9UQN5 | Gene names | ROCK2, KIAA0619 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2) (Rho kinase 2). | |||||
|
SQSTM_HUMAN
|
||||||
| θ value | 5.27518 (rank : 119) | NC score | 0.019227 (rank : 139) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13501, Q13446, Q9BUV7, Q9BVS6, Q9UEU1 | Gene names | SQSTM1, ORCA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sequestosome-1 (Phosphotyrosine-independent ligand for the Lck SH2 domain of 62 kDa) (Ubiquitin-binding protein p62) (EBI3-associated protein of 60 kDa) (p60) (EBIAP). | |||||
|
TCRG1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 120) | NC score | 0.048495 (rank : 67) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
TCRG1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 121) | NC score | 0.051014 (rank : 63) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
TM16H_MOUSE
|
||||||
| θ value | 5.27518 (rank : 122) | NC score | 0.016889 (rank : 147) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PB70, Q69ZE4 | Gene names | Tmem16h, Kiaa1623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 16H. | |||||
|
ZNHI4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 123) | NC score | 0.040685 (rank : 80) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99PT3, Q9CY38 | Gene names | Znhit4, Hmga1l4, Papa1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger HIT domain-containing protein 4 (PAP-1-associated protein 1) (PAPA-1) (High mobility group AT-hook 1-like 4). | |||||
|
3BP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 124) | NC score | 0.017442 (rank : 146) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y3L3, Q6IBZ2, Q6ZVL9, Q96HQ5, Q9NSQ9 | Gene names | SH3BP1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
ARHGG_HUMAN
|
||||||
| θ value | 6.88961 (rank : 125) | NC score | 0.044990 (rank : 72) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5VV41, Q86TF0, Q99434 | Gene names | ARHGEF16, NBR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 16. | |||||
|
BPA1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 126) | NC score | 0.022500 (rank : 136) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
CCD82_HUMAN
|
||||||
| θ value | 6.88961 (rank : 127) | NC score | 0.024970 (rank : 126) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8N4S0, Q9H2Q5, Q9H5E3 | Gene names | CCDC82 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 82. | |||||
|
CT2NL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 128) | NC score | 0.049062 (rank : 65) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9P2B4, Q96B40 | Gene names | CTTNBP2NL, KIAA1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
DCR1C_HUMAN
|
||||||
| θ value | 6.88961 (rank : 129) | NC score | 0.017897 (rank : 144) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96SD1, Q5JSR4, Q5JSR5, Q5JSR7, Q5JSR8, Q5JSR9, Q5JSS0, Q5JSS7, Q6PK14, Q8N101, Q8N132, Q8TBW9, Q9BVW9, Q9HAM4 | Gene names | DCLRE1C, ARTEMIS, ASCID, SCIDA, SNM1C | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Artemis protein (EC 3.1.-.-) (DNA cross-link repair 1C protein) (SNM1- like protein) (A-SCID protein) (hSNM1C). | |||||
|
EHD1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 130) | NC score | 0.084153 (rank : 20) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H4M9, O14611, Q9UNR3 | Gene names | EHD1, PAST, PAST1 | |||
|
Domain Architecture |

|
|||||
| Description | EH-domain-containing protein 1 (Testilin) (hPAST1). | |||||
|
EHD1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 131) | NC score | 0.084125 (rank : 21) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9WVK4 | Gene names | Ehd1, Past1 | |||
|
Domain Architecture |

|
|||||
| Description | EH-domain-containing protein 1 (mPAST1). | |||||
|
EHD4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 132) | NC score | 0.083592 (rank : 22) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9EQP2 | Gene names | Ehd4, Past2 | |||
|
Domain Architecture |

|
|||||
| Description | EH-domain-containing protein 4 (mPAST2). | |||||
|
KCNH2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 133) | NC score | 0.009433 (rank : 159) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O35219, O35220, O35221, O35989 | Gene names | Kcnh2, Erg, Merg1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (ERG1) (MERG) (Merg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1). | |||||
|
NBEAL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 134) | NC score | 0.012964 (rank : 155) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZS30, Q6Y876, Q6ZQY5, Q96Q30 | Gene names | NBEAL1, ALS2CR17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurobeachin-like 1 (Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 17 protein). | |||||
|
NR4A1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 135) | NC score | 0.003980 (rank : 163) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P12813 | Gene names | Nr4a1, Gfrp, Hmr, N10, Nur77 | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR4A1 (Orphan nuclear receptor HMR) (Nuclear hormone receptor NUR/77) (N10 nuclear protein). | |||||
|
PXK_MOUSE
|
||||||
| θ value | 6.88961 (rank : 136) | NC score | 0.016338 (rank : 150) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 320 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BX57, Q3TD60, Q3TEL7, Q3TZZ1, Q3U197, Q3U773, Q3UCS5, Q4FBH7, Q91WB6 | Gene names | Pxk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PX domain-containing protein kinase-like protein (Modulator of Na,K- ATPase) (MONaKA). | |||||
|
ROCK2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 137) | NC score | 0.016527 (rank : 149) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 2072 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P70336, Q8BR64, Q8CBR0 | Gene names | Rock2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2). | |||||
|
ROD1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 138) | NC score | 0.012996 (rank : 154) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95758, Q86YB3, Q86YH9 | Gene names | ROD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of differentiation 1 (Rod1). | |||||
|
ROD1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 139) | NC score | 0.012956 (rank : 156) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BHD7, Q923C3 | Gene names | Rod1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of differentiation 1 (Rod1). | |||||
|
WNK1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 140) | NC score | 0.005505 (rank : 162) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1158 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9H4A3, O15052, Q86WL5, Q8N673, Q9P1S9 | Gene names | WNK1, KDP, KIAA0344, PRKWNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1) (Kinase deficient protein). | |||||
|
ZN318_HUMAN
|
||||||
| θ value | 6.88961 (rank : 141) | NC score | 0.045540 (rank : 70) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q5VUA4, O94796, Q4G0E4, Q8NEM6, Q9UNU8, Q9Y2W9 | Gene names | ZNF318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Endocrine regulatory protein). | |||||
|
AFTIN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 142) | NC score | 0.017954 (rank : 143) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6ULP2, Q6ZM66, Q86VW3, Q8TCF3, Q9H7E3, Q9HAB9, Q9NXS4 | Gene names | AFTPH, AFTH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Aftiphilin. | |||||
|
EHD3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 143) | NC score | 0.080856 (rank : 25) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NZN3, Q8N514, Q9NZB3 | Gene names | EHD3, EHD2 | |||
|
Domain Architecture |

|
|||||
| Description | EH-domain-containing protein 3. | |||||
|
F8I2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 144) | NC score | 0.034994 (rank : 94) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q00558 | Gene names | F8a1, F8a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Factor VIII intron 22 protein (CpG island protein). | |||||
|
FMN2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 145) | NC score | 0.016576 (rank : 148) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9JL04 | Gene names | Fmn2 | |||
|
Domain Architecture |

|
|||||
| Description | Formin-2. | |||||
|
IRF5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 146) | NC score | 0.007059 (rank : 161) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P56477 | Gene names | Irf5 | |||
|
Domain Architecture |
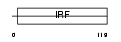
|
|||||
| Description | Interferon regulatory factor 5 (IRF-5). | |||||
|
K0460_HUMAN
|
||||||
| θ value | 8.99809 (rank : 147) | NC score | 0.023944 (rank : 128) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q5VT52, O75048, Q5VT53, Q6MZL4, Q86XD2 | Gene names | KIAA0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
LEUK_MOUSE
|
||||||
| θ value | 8.99809 (rank : 148) | NC score | 0.027277 (rank : 117) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P15702 | Gene names | Spn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukosialin precursor (Leukocyte sialoglycoprotein) (Sialophorin) (Ly- 48) (B cell differentiation antigen LP-3) (CD43 antigen). | |||||
|
MAP4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 149) | NC score | 0.025750 (rank : 122) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
MYT1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 150) | NC score | 0.012933 (rank : 157) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8CFC2, O08995, Q8CFH1 | Gene names | Myt1, Kiaa0835, Nzf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1 (MyT1) (Neural zinc finger factor 2) (NZF-2). | |||||
|
NALDL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 151) | NC score | 0.007440 (rank : 160) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7M758 | Gene names | Naaladl1, Naaladasel | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylated-alpha-linked acidic dipeptidase-like protein (EC 3.4.17.21) (NAALADase L). | |||||
|
NRG2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 152) | NC score | 0.022553 (rank : 135) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O14511 | Gene names | NRG2, NTAK | |||
|
Domain Architecture |

|
|||||
| Description | Pro-neuregulin-2, membrane-bound isoform precursor (Pro-NRG2) [Contains: Neuregulin-2 (NRG-2) (Neural- and thymus-derived activator for ERBB kinases) (NTAK) (Divergent of neuregulin-1) (DON-1)]. | |||||
|
PEPL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 153) | NC score | 0.030358 (rank : 108) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9R269, O70231, Q9CUT1, Q9JLZ7 | Gene names | Ppl | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin. | |||||
|
PRG4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 154) | NC score | 0.037432 (rank : 86) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
R3HD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 155) | NC score | 0.036829 (rank : 89) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15032, Q8IW32 | Gene names | R3HDM1, KIAA0029, R3HDM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R3H domain-containing protein 1. | |||||
|
SEPT7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 156) | NC score | 0.018093 (rank : 142) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q16181, Q6NX50 | Gene names | SEPT7, CDC10 | |||
|
Domain Architecture |

|
|||||
| Description | Septin-7 (CDC10 protein homolog). | |||||
|
TPR_HUMAN
|
||||||
| θ value | 8.99809 (rank : 157) | NC score | 0.032685 (rank : 102) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
UBQL2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 158) | NC score | 0.023155 (rank : 132) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UHD9, O94798, Q9H3W6, Q9HAZ4 | Gene names | UBQLN2, PLIC2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-2 (Protein linking IAP with cytoskeleton 2) (PLIC-2) (hPLIC- 2) (Ubiquitin-like product Chap1/Dsk2) (DSK2 homolog) (Chap1). | |||||
|
ZN318_MOUSE
|
||||||
| θ value | 8.99809 (rank : 159) | NC score | 0.036987 (rank : 87) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99PP2, Q3TZL5, Q9JJ01 | Gene names | Znf318, Tzf, Zfp318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Testicular zinc finger protein). | |||||
|
ZN710_MOUSE
|
||||||
| θ value | 8.99809 (rank : 160) | NC score | -0.003054 (rank : 166) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3U288, Q3U341, Q6P9R3, Q8BJH0, Q8BTL6 | Gene names | Znf710, Zfp710 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 710. | |||||
|
EHD2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.086335 (rank : 18) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NZN4, Q96CB6 | Gene names | EHD2 | |||
|
Domain Architecture |

|
|||||
| Description | EH-domain-containing protein 2. | |||||
|
EHD3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.074723 (rank : 31) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9QXY6 | Gene names | Ehd3, Ehd2 | |||
|
Domain Architecture |

|
|||||
| Description | EH-domain-containing protein 3. | |||||
|
REPS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.078349 (rank : 28) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96D71, Q8NDR7, Q8WU62, Q9BXY9 | Gene names | REPS1 | |||
|
Domain Architecture |

|
|||||
| Description | RalBP1-associated Eps domain-containing protein 1 (RalBP1-interacting protein 1). | |||||
|
REPS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.097372 (rank : 14) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O54916, Q8C9J9, Q99LR8 | Gene names | Reps1 | |||
|
Domain Architecture |

|
|||||
| Description | RalBP1-associated Eps domain-containing protein 1 (RalBP1-interacting protein 1). | |||||
|
REPS2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.090871 (rank : 16) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q80XA6 | Gene names | Reps2, Pob1 | |||
|
Domain Architecture |
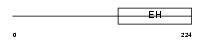
|
|||||
| Description | RalBP1-associated Eps domain-containing protein 2 (RalBP1-interacting protein 2) (Partner of RalBP1). | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.056140 (rank : 57) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
SYNG_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 160 | |
| SwissProt Accessions | Q9UMZ2, Q6ZT17 | Gene names | AP1GBP1, SYNG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP1 subunit gamma-binding protein 1 (Gamma-synergin). | |||||
|
SYNG_MOUSE
|
||||||
| NC score | 0.946481 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q5SV85, Q5SV84, Q6PHT6 | Gene names | Ap1gbp1, Syng | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP1 subunit gamma-binding protein 1 (Gamma-synergin). | |||||
|
EP15_MOUSE
|
||||||
| NC score | 0.150630 (rank : 3) | θ value | 0.000786445 (rank : 6) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P42567 | Gene names | Eps15 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
EP15_HUMAN
|
||||||
| NC score | 0.146834 (rank : 4) | θ value | 0.00102713 (rank : 7) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1021 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P42566 | Gene names | EPS15, AF1P | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
EP15R_MOUSE
|
||||||
| NC score | 0.130109 (rank : 5) | θ value | 0.00509761 (rank : 11) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q60902, Q8CB60, Q8CB70, Q91WH8 | Gene names | Eps15l1, Eps15-rs, Eps15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R) (Epidermal growth factor receptor pathway substrate 15-related sequence) (Eps15-rs). | |||||
|
EP15R_HUMAN
|
||||||
| NC score | 0.127224 (rank : 6) | θ value | 0.0113563 (rank : 13) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1083 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UBC2 | Gene names | EPS15L1, EPS15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R). | |||||
|
ITSN1_HUMAN
|
||||||
| NC score | 0.125338 (rank : 7) | θ value | 0.000158464 (rank : 3) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
ITSN1_MOUSE
|
||||||
| NC score | 0.121606 (rank : 8) | θ value | 0.000270298 (rank : 5) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
REPS2_HUMAN
|
||||||
| NC score | 0.120107 (rank : 9) | θ value | 1.06291 (rank : 57) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8NFH8, O43428, Q8NFI5 | Gene names | REPS2, POB1 | |||
|
Domain Architecture |

|
|||||
| Description | RalBP1-associated Eps domain-containing protein 2 (RalBP1-interacting protein 2) (Partner of RalBP1). | |||||
|
ITSN2_HUMAN
|
||||||
| NC score | 0.119107 (rank : 10) | θ value | 0.00175202 (rank : 9) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1173 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9NZM3, O95062, Q15812, Q9HAK4, Q9NXE6, Q9NYG0, Q9NZM2, Q9ULG4 | Gene names | ITSN2, KIAA1256, SH3D1B, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (SH3P18) (SH3P18-like WASP-associated protein). | |||||
|
ITSN2_MOUSE
|
||||||
| NC score | 0.114094 (rank : 11) | θ value | 0.00175202 (rank : 10) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1371 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9Z0R6, Q9Z0R5 | Gene names | Itsn2, Ese2, Sh3d1B | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (EH and SH3 domains protein 2) (EH domain and SH3 domain regulator of endocytosis 2). | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.113618 (rank : 12) | θ value | 0.000270298 (rank : 4) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
GORS2_MOUSE
|
||||||
| NC score | 0.110548 (rank : 13) | θ value | 0.00134147 (rank : 8) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99JX3, Q3U9D2, Q8CCD0, Q91W68 | Gene names | Gorasp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 2 (GRS2) (Golgi reassembly-stacking protein of 55 kDa) (GRASP55). | |||||
|
REPS1_MOUSE
|
||||||
| NC score | 0.097372 (rank : 14) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O54916, Q8C9J9, Q99LR8 | Gene names | Reps1 | |||
|
Domain Architecture |

|
|||||
| Description | RalBP1-associated Eps domain-containing protein 1 (RalBP1-interacting protein 1). | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.097095 (rank : 15) | θ value | 0.813845 (rank : 51) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
REPS2_MOUSE
|
||||||
| NC score | 0.090871 (rank : 16) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q80XA6 | Gene names | Reps2, Pob1 | |||
|
Domain Architecture |

|
|||||
| Description | RalBP1-associated Eps domain-containing protein 2 (RalBP1-interacting protein 2) (Partner of RalBP1). | |||||
|
YLPM1_MOUSE
|
||||||
| NC score | 0.090204 (rank : 17) | θ value | 0.125558 (rank : 27) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
EHD2_HUMAN
|
||||||
| NC score | 0.086335 (rank : 18) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NZN4, Q96CB6 | Gene names | EHD2 | |||
|
Domain Architecture |

|
|||||
| Description | EH-domain-containing protein 2. | |||||
|
TCP1L_HUMAN
|
||||||
| NC score | 0.084817 (rank : 19) | θ value | 0.47712 (rank : 44) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TDR4, Q96LN5 | Gene names | TCP10L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-complex protein 10A homolog 2 (T-complex protein 10A-2) (TCP10A-2) (TCP10-like). | |||||
|
EHD1_HUMAN
|
||||||
| NC score | 0.084153 (rank : 20) | θ value | 6.88961 (rank : 130) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H4M9, O14611, Q9UNR3 | Gene names | EHD1, PAST, PAST1 | |||
|
Domain Architecture |

|
|||||
| Description | EH-domain-containing protein 1 (Testilin) (hPAST1). | |||||
|
EHD1_MOUSE
|
||||||
| NC score | 0.084125 (rank : 21) | θ value | 6.88961 (rank : 131) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9WVK4 | Gene names | Ehd1, Past1 | |||
|
Domain Architecture |

|
|||||
| Description | EH-domain-containing protein 1 (mPAST1). | |||||
|
EHD4_MOUSE
|
||||||
| NC score | 0.083592 (rank : 22) | θ value | 6.88961 (rank : 132) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9EQP2 | Gene names | Ehd4, Past2 | |||
|
Domain Architecture |

|
|||||
| Description | EH-domain-containing protein 4 (mPAST2). | |||||
|
EHD4_HUMAN
|
||||||
| NC score | 0.082173 (rank : 23) | θ value | 5.27518 (rank : 106) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H223, Q9HAR1, Q9NZN2 | Gene names | EHD4, HCA10, HCA11 | |||
|
Domain Architecture |

|
|||||
| Description | EH-domain-containing protein 4 (Hepatocellular carcinoma-associated protein 10/11). | |||||
|
IL4RA_MOUSE
|
||||||
| NC score | 0.081009 (rank : 24) | θ value | 0.0193708 (rank : 15) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P16382, O54690, Q60583, Q8CBW5 | Gene names | Il4r, Il4ra | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-4 receptor alpha chain precursor (IL-4R-alpha) (CD124 antigen) [Contains: Soluble interleukin-4 receptor alpha chain (IL-4- binding protein) (IL4-BP)]. | |||||
|
EHD3_HUMAN
|
||||||
| NC score | 0.080856 (rank : 25) | θ value | 8.99809 (rank : 143) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NZN3, Q8N514, Q9NZB3 | Gene names | EHD3, EHD2 | |||
|
Domain Architecture |

|
|||||
| Description | EH-domain-containing protein 3. | |||||
|
S23IP_HUMAN
|
||||||
| NC score | 0.080663 (rank : 26) | θ value | 0.0736092 (rank : 21) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y6Y8, Q8IXH5, Q9BUK5 | Gene names | SEC23IP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SEC23-interacting protein (p125). | |||||
|
PDC6I_MOUSE
|
||||||
| NC score | 0.080356 (rank : 27) | θ value | 0.163984 (rank : 29) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9WU78, O88695, O89014, Q8BSL8, Q8R0H5, Q99LR3, Q9QZN8 | Gene names | Pdcd6ip, Aip1, Alix | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death 6-interacting protein (ALG-2-interacting protein X) (ALG-2-interacting protein 1) (E2F1-inducible protein) (Eig2). | |||||
|
REPS1_HUMAN
|
||||||
| NC score | 0.078349 (rank : 28) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96D71, Q8NDR7, Q8WU62, Q9BXY9 | Gene names | REPS1 | |||
|
Domain Architecture |

|
|||||
| Description | RalBP1-associated Eps domain-containing protein 1 (RalBP1-interacting protein 1). | |||||
|
MAVS_MOUSE
|
||||||
| NC score | 0.078193 (rank : 29) | θ value | 0.365318 (rank : 39) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8VCF0 | Gene names | Mavs, Ips1, Visa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial antiviral signaling protein (Interferon-beta promoter stimulator protein 1) (IPS-1) (Virus-induced signaling adapter) (CARD adapter inducing interferon-beta) (Cardif). | |||||
|
CN032_MOUSE
|
||||||
| NC score | 0.075407 (rank : 30) | θ value | 1.38821 (rank : 60) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
EHD3_MOUSE
|
||||||
| NC score | 0.074723 (rank : 31) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9QXY6 | Gene names | Ehd3, Ehd2 | |||
|
Domain Architecture |

|
|||||
| Description | EH-domain-containing protein 3. | |||||
|
GRIN1_HUMAN
|
||||||
| NC score | 0.073963 (rank : 32) | θ value | 0.279714 (rank : 36) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.073441 (rank : 33) | θ value | 0.0563607 (rank : 19) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
FA54A_MOUSE
|
||||||
| NC score | 0.072215 (rank : 34) | θ value | 0.21417 (rank : 33) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VED8 | Gene names | Fam54a, Dufd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM54A (DUF729 domain-containing protein 1). | |||||
|
UPK3B_HUMAN
|
||||||
| NC score | 0.071936 (rank : 35) | θ value | 2.36792 (rank : 78) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BT76, Q86W06 | Gene names | UPK3B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uroplakin-3B precursor (Uroplakin IIIb) (UPIIIb) (p35). | |||||
|
BCL9_MOUSE
|
||||||
| NC score | 0.071102 (rank : 36) | θ value | 0.21417 (rank : 31) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.070345 (rank : 37) | θ value | 0.0193708 (rank : 14) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.069495 (rank : 38) | θ value | 0.813845 (rank : 49) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
HGS_HUMAN
|
||||||
| NC score | 0.068445 (rank : 39) | θ value | 2.36792 (rank : 74) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 517 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O14964, Q9NR36 | Gene names | HGS, HRS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate (Protein pp110) (Hrs). | |||||
|
HGS_MOUSE
|
||||||
| NC score | 0.067504 (rank : 40) | θ value | 0.163984 (rank : 28) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
TCF20_MOUSE
|
||||||
| NC score | 0.066521 (rank : 41) | θ value | 0.62314 (rank : 47) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
ZNHI4_HUMAN
|
||||||
| NC score | 0.064473 (rank : 42) | θ value | 1.06291 (rank : 58) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9C086 | Gene names | ZNHIT4, HMGA1L4, PAPA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger HIT domain-containing protein 4 (PAP-1-associated protein 1) (PAPA-1) (High mobility group AT-hook 1-like 4). | |||||
|
ALMS1_HUMAN
|
||||||
| NC score | 0.064125 (rank : 43) | θ value | 0.0736092 (rank : 20) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8TCU4, Q53S05, Q580Q8, Q86VP9, Q9Y4G4 | Gene names | ALMS1, KIAA0328 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alstrom syndrome protein 1. | |||||
|
TCF20_HUMAN
|
||||||
| NC score | 0.064054 (rank : 44) | θ value | 0.21417 (rank : 34) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UGU0, O14528, Q13078, Q4V353, Q9H4M0 | Gene names | TCF20, KIAA0292, SPBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP) (AR1). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.062680 (rank : 45) | θ value | 1.38821 (rank : 61) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.061346 (rank : 46) | θ value | 0.813845 (rank : 50) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
CSTFT_MOUSE
|
||||||
| NC score | 0.061300 (rank : 47) | θ value | 0.0252991 (rank : 16) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8C7E9, Q6A016, Q8BHH7, Q8C3W7, Q8R2Y1, Q9EPU3 | Gene names | Cstf2t, Kiaa0689 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage stimulation factor 64 kDa subunit, tau variant (CSTF 64 kDa subunit, tau variant) (CF-1 64 kDa subunit, tau variant) (TauCstF-64). | |||||
|
NUPL_MOUSE
|
||||||
| NC score | 0.060960 (rank : 48) | θ value | 0.279714 (rank : 37) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8K2K6, O70448, Q8BQL5, Q8CDK9 | Gene names | Hrb, Rip | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoporin-like protein RIP (HIV-1 Rev-binding protein homolog). | |||||
|
PO121_HUMAN
|
||||||
| NC score | 0.060032 (rank : 49) | θ value | 5.27518 (rank : 115) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
CN032_HUMAN
|
||||||
| NC score | 0.059533 (rank : 50) | θ value | 4.03905 (rank : 94) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.059383 (rank : 51) | θ value | 2.36792 (rank : 71) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
SMC3_MOUSE
|
||||||
| NC score | 0.058465 (rank : 52) | θ value | 0.365318 (rank : 42) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1187 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9CW03, O35667, Q9QUS3 | Gene names | Cspg6, Bam, Bmh, Mmip1, Smc3, Smc3l1, Smcd | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome segregation protein SmcD) (Bamacan) (Basement membrane-associated chondroitin proteoglycan) (Mad member- interacting protein 1). | |||||
|
SMC3_HUMAN
|
||||||
| NC score | 0.058378 (rank : 53) | θ value | 0.365318 (rank : 41) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1186 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9UQE7, O60464 | Gene names | CSPG6, BAM, BMH, SMC3, SMC3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome-associated polypeptide) (hCAP) (Bamacan) (Basement membrane-associated chondroitin proteoglycan). | |||||
|
GPSM2_HUMAN
|
||||||
| NC score | 0.057948 (rank : 54) | θ value | 0.125558 (rank : 24) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P81274, Q8N0Z5 | Gene names | GPSM2, LGN | |||
|
Domain Architecture |

|
|||||
| Description | G-protein signaling modulator 2 (Mosaic protein LGN). | |||||
|
GPSM2_MOUSE
|
||||||
| NC score | 0.057707 (rank : 55) | θ value | 0.125558 (rank : 25) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VDU0, Q8BLX3 | Gene names | Gpsm2, Lgn, Pins | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein signaling modulator 2 (Pins homolog). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.056853 (rank : 56) | θ value | 4.03905 (rank : 97) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.056140 (rank : 57) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.055277 (rank : 58) | θ value | 0.813845 (rank : 48) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.054037 (rank : 59) | θ value | 0.62314 (rank : 46) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
NUCB2_MOUSE
|
||||||
| NC score | 0.053038 (rank : 60) | θ value | 2.36792 (rank : 76) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P81117 | Gene names | Nucb2, Nefa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleobindin-2 precursor (DNA-binding protein NEFA). | |||||
|
CK024_HUMAN
|
||||||
| NC score | 0.052699 (rank : 61) | θ value | 1.81305 (rank : 68) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
ASPP1_HUMAN
|
||||||
| NC score | 0.051192 (rank : 62) | θ value | 0.0563607 (rank : 18) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 956 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96KQ4, O94870 | Gene names | PPP1R13B, ASPP1, KIAA0771 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
TCRG1_MOUSE
|
||||||
| NC score | 0.051014 (rank : 63) | θ value | 5.27518 (rank : 121) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
CCD91_HUMAN
|
||||||
| NC score | 0.049531 (rank : 64) | θ value | 1.06291 (rank : 53) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q7Z6B0, Q68D43, Q6IA78, Q8NEN7, Q9NUW9 | Gene names | CCDC91, GGABP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner) (p56 accessory protein). | |||||
|
CT2NL_HUMAN
|
||||||
| NC score | 0.049062 (rank : 65) | θ value | 6.88961 (rank : 128) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9P2B4, Q96B40 | Gene names | CTTNBP2NL, KIAA1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
SELPL_MOUSE
|
||||||
| NC score | 0.048843 (rank : 66) | θ value | 1.81305 (rank : 70) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
TCRG1_HUMAN
|
||||||
| NC score | 0.048495 (rank : 67) | θ value | 5.27518 (rank : 120) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
AFF1_MOUSE
|
||||||
| NC score | 0.046341 (rank : 68) | θ value | 0.279714 (rank : 35) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
SYM_MOUSE
|
||||||
| NC score | 0.045680 (rank : 69) | θ value | 1.38821 (rank : 62) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q68FL6 | Gene names | Mars | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methionyl-tRNA synthetase (EC 6.1.1.10) (Methionine--tRNA ligase) (MetRS). | |||||
|
ZN318_HUMAN
|
||||||
| NC score | 0.045540 (rank : 70) | θ value | 6.88961 (rank : 141) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q5VUA4, O94796, Q4G0E4, Q8NEM6, Q9UNU8, Q9Y2W9 | Gene names | ZNF318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Endocrine regulatory protein). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.045062 (rank : 71) | θ value | 3.0926 (rank : 88) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
ARHGG_HUMAN
|
||||||
| NC score | 0.044990 (rank : 72) | θ value | 6.88961 (rank : 125) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5VV41, Q86TF0, Q99434 | Gene names | ARHGEF16, NBR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 16. | |||||
|
MN1_HUMAN
|
||||||
| NC score | 0.042714 (rank : 73) | θ value | 5.27518 (rank : 110) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q10571 | Gene names | MN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable tumor suppressor protein MN1. | |||||
|
EPN2_HUMAN
|
||||||
| NC score | 0.042601 (rank : 74) | θ value | 0.62314 (rank : 45) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95208, O95207, Q9H7Z2, Q9UPT7 | Gene names | EPN2, KIAA1065 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-2 (EPS-15-interacting protein 2). | |||||
|
CT2NL_MOUSE
|
||||||
| NC score | 0.041246 (rank : 75) | θ value | 3.0926 (rank : 83) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 902 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q99LJ0, Q6ZPR0, Q8BSV1 | Gene names | Cttnbp2nl, Kiaa1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
CRTC1_HUMAN
|
||||||
| NC score | 0.041083 (rank : 76) | θ value | 5.27518 (rank : 105) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6UUV9, O75114, Q6Y3A3, Q7LDZ2, Q8IUL3, Q8IZ34, Q8IZL1, Q8N6W3, Q96AI8, Q9H801 | Gene names | CRTC1, KIAA0616, MECT1, TORC1, WAMTP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Transducer of regulated cAMP response element-binding protein 1) (Transducer of CREB protein 1) (Mucoepidermoid carcinoma translocated protein 1). | |||||
|
DHB4_HUMAN
|
||||||
| NC score | 0.040856 (rank : 77) | θ value | 1.06291 (rank : 54) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51659 | Gene names | HSD17B4, EDH17B4 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal multifunctional enzyme type 2 (MFE-2) (D-bifunctional protein) (DBP) (17-beta-hydroxysteroid dehydrogenase 4) (17-beta-HSD 4) (D-3-hydroxyacyl-CoA dehydratase) (EC 4.2.1.107) (3-alpha,7- alpha,12-alpha-trihydroxy-5-beta-cholest-24-enoyl-CoA hydratase) (3- hydroxyacyl-CoA dehydrogenase) (EC 1.1.1.35). | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.040839 (rank : 78) | θ value | 3.0926 (rank : 87) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.040738 (rank : 79) | θ value | 0.813845 (rank : 52) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
ZNHI4_MOUSE
|
||||||
| NC score | 0.040685 (rank : 80) | θ value | 5.27518 (rank : 123) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99PT3, Q9CY38 | Gene names | Znhit4, Hmga1l4, Papa1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger HIT domain-containing protein 4 (PAP-1-associated protein 1) (PAPA-1) (High mobility group AT-hook 1-like 4). | |||||
|
CSTFT_HUMAN
|
||||||
| NC score | 0.040350 (rank : 81) | θ value | 0.21417 (rank : 32) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H0L4, O75174, Q53HK6, Q7LGE8, Q8N6T1 | Gene names | CSTF2T, KIAA0689 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage stimulation factor 64 kDa subunit, tau variant (CSTF 64 kDa subunit, tau variant) (CF-1 64 kDa subunit, tau variant) (TauCstF-64). | |||||
|
KI67_HUMAN
|
||||||
| NC score | 0.039912 (rank : 82) | θ value | 3.0926 (rank : 86) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
TMM28_HUMAN
|
||||||
| NC score | 0.039799 (rank : 83) | θ value | 3.0926 (rank : 89) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75949 | Gene names | TMEM28, TED | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 28 (TED protein). | |||||
|
TIM44_MOUSE
|
||||||
| NC score | 0.037457 (rank : 84) | θ value | 2.36792 (rank : 77) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O35857 | Gene names | Timm44, Mimt44, Tim44 | |||
|
Domain Architecture |

|
|||||
| Description | Import inner membrane translocase subunit TIM44, mitochondrial precursor. | |||||
|
ARI1B_HUMAN
|
||||||
| NC score | 0.037457 (rank : 85) | θ value | 3.0926 (rank : 79) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.037432 (rank : 86) | θ value | 8.99809 (rank : 154) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
ZN318_MOUSE
|
||||||
| NC score | 0.036987 (rank : 87) | θ value | 8.99809 (rank : 159) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99PP2, Q3TZL5, Q9JJ01 | Gene names | Znf318, Tzf, Zfp318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Testicular zinc finger protein). | |||||
|
CDN1C_MOUSE
|
||||||
| NC score | 0.036910 (rank : 88) | θ value | 2.36792 (rank : 72) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P49919 | Gene names | Cdkn1c, Kip2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
R3HD1_HUMAN
|
||||||
| NC score | 0.036829 (rank : 89) | θ value | 8.99809 (rank : 155) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15032, Q8IW32 | Gene names | R3HDM1, KIAA0029, R3HDM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R3H domain-containing protein 1. | |||||
|
CK5P2_HUMAN
|
||||||
| NC score | 0.036687 (rank : 90) | θ value | 4.03905 (rank : 93) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
SRBP1_MOUSE
|
||||||
| NC score | 0.036496 (rank : 91) | θ value | 4.03905 (rank : 102) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
PARM1_HUMAN
|
||||||
| NC score | 0.036133 (rank : 92) | θ value | 4.03905 (rank : 100) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6UWI2, Q96DV8, Q9Y4S1 | Gene names | ames=UNQ1879/PRO4322 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein PARM-1 precursor. | |||||
|
NUMBL_HUMAN
|
||||||
| NC score | 0.035168 (rank : 93) | θ value | 4.03905 (rank : 99) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Y6R0, Q7Z4J9 | Gene names | NUMBL | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein (Numb-R). | |||||
|
F8I2_MOUSE
|
||||||
| NC score | 0.034994 (rank : 94) | θ value | 8.99809 (rank : 144) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q00558 | Gene names | F8a1, F8a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Factor VIII intron 22 protein (CpG island protein). | |||||
|
NCOR2_HUMAN
|
||||||
| NC score | 0.034263 (rank : 95) | θ value | 1.81305 (rank : 69) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
LMNB1_MOUSE
|
||||||
| NC score | 0.034015 (rank : 96) | θ value | 0.365318 (rank : 38) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 719 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P14733, Q61791 | Gene names | Lmnb1 | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-B1. | |||||
|
UBQL2_MOUSE
|
||||||
| NC score | 0.033862 (rank : 97) | θ value | 4.03905 (rank : 104) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9QZM0, Q7TSJ8, Q8VDH9 | Gene names | Ubqln2, Plic2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-2 (Protein linking IAP with cytoskeleton 2) (PLIC-2) (Ubiquitin-like product Chap1/Dsk2) (DSK2 homolog) (Chap1). | |||||
|
ING3_MOUSE
|
||||||
| NC score | 0.033366 (rank : 98) | θ value | 4.03905 (rank : 96) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VEK6, Q99JS6, Q9ERB2 | Gene names | Ing3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 3 (p47ING3 protein). | |||||
|
LMNB1_HUMAN
|
||||||
| NC score | 0.033200 (rank : 99) | θ value | 1.06291 (rank : 55) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P20700, Q3SYN7, Q96EI6 | Gene names | LMNB1, LMN2, LMNB | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-B1. | |||||
|
ARMX2_MOUSE
|
||||||
| NC score | 0.032833 (rank : 100) | θ value | 1.38821 (rank : 59) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6A058, Q9CXI9 | Gene names | Armcx2, Kiaa0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2. | |||||
|
CSPG5_MOUSE
|
||||||
| NC score | 0.032828 (rank : 101) | θ value | 2.36792 (rank : 73) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q71M36, Q71M37, Q7TNT8, Q8BPJ5, Q9QY32 | Gene names | Cspg5, Caleb, Ngc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate proteoglycan 5 precursor (Neuroglycan C) (Acidic leucine-rich EGF-like domain-containing brain protein). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.032685 (rank : 102) | θ value | 8.99809 (rank : 157) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
NRG2_MOUSE
|
||||||
| NC score | 0.032597 (rank : 103) | θ value | 1.06291 (rank : 56) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P56974 | Gene names | Nrg2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pro-neuregulin-2, membrane-bound isoform precursor (Pro-NRG2) [Contains: Neuregulin-2 (NRG-2) (Divergent of neuregulin 1) (DON-1)]. | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.032073 (rank : 104) | θ value | 4.03905 (rank : 91) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
SYNJ1_MOUSE
|
||||||
| NC score | 0.031248 (rank : 105) | θ value | 1.38821 (rank : 63) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
UACA_MOUSE
|
||||||
| NC score | 0.030981 (rank : 106) | θ value | 1.38821 (rank : 65) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8CGB3, Q69ZG3, Q7TN77, Q8BJC8 | Gene names | Uaca, Kiaa1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein (Nucling) (Nuclear membrane-binding protein). | |||||
|
ROBO3_HUMAN
|
||||||
| NC score | 0.030717 (rank : 107) | θ value | 0.125558 (rank : 26) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96MS0 | Gene names | ROBO3 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 3 precursor (Roundabout-like protein 3). | |||||
|
PEPL_MOUSE
|
||||||
| NC score | 0.030358 (rank : 108) | θ value | 8.99809 (rank : 153) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9R269, O70231, Q9CUT1, Q9JLZ7 | Gene names | Ppl | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin. | |||||
|
HECD1_HUMAN
|
||||||
| NC score | 0.030219 (rank : 109) | θ value | 0.0961366 (rank : 22) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 371 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ULT8, Q6P445, Q86VJ1, Q96F34, Q9UFZ7 | Gene names | HECTD1, KIAA1131 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase HECTD1 (HECT domain-containing protein 1) (E3 ligase for inhibin receptor) (EULIR). | |||||
|
TRIP6_HUMAN
|
||||||
| NC score | 0.029235 (rank : 110) | θ value | 0.163984 (rank : 30) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15654, O15170, O15275, Q9BTB2, Q9UNT4 | Gene names | TRIP6, OIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 6 (TRIP6) (OPA-interacting protein 1) (Zyxin-related protein 1) (ZRP-1). | |||||
|
LACTB_MOUSE
|
||||||
| NC score | 0.029067 (rank : 111) | θ value | 2.36792 (rank : 75) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9EP89 | Gene names | Lactb, Lact1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine beta-lactamase-like protein LACTB. | |||||
|
PTN13_MOUSE
|
||||||
| NC score | 0.028843 (rank : 112) | θ value | 0.00665767 (rank : 12) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q64512, Q61494, Q62135, Q64499 | Gene names | Ptpn13, Ptp14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein tyrosine phosphatase PTP-BL) (Protein-tyrosine phosphatase RIP) (protein tyrosine phosphatase DPZPTP) (PTP36). | |||||
|
MYCD_MOUSE
|
||||||
| NC score | 0.028527 (rank : 113) | θ value | 5.27518 (rank : 112) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8VIM5, Q8C3W6, Q8VIL4 | Gene names | Myocd, Bsac2, Mycd, Srfcp | |||
|
Domain Architecture |

|
|||||
| Description | Myocardin (SRF cofactor protein) (Basic SAP coiled-coil transcription activator 2). | |||||
|
AF10_HUMAN
|
||||||
| NC score | 0.027906 (rank : 114) | θ value | 0.47712 (rank : 43) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P55197 | Gene names | MLLT10, AF10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-10. | |||||
|
BRDT_MOUSE
|
||||||
| NC score | 0.027462 (rank : 115) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q91Y44, Q59HJ4 | Gene names | Brdt, Fsrg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain testis-specific protein (RING3-like protein) (Bromodomain- containing female sterile homeotic-like protein). | |||||
|
NU155_MOUSE
|
||||||
| NC score | 0.027342 (rank : 116) | θ value | 5.27518 (rank : 113) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99P88 | Gene names | Nup155 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup155 (Nucleoporin Nup155) (155 kDa nucleoporin). | |||||
|
LEUK_MOUSE
|
||||||
| NC score | 0.027277 (rank : 117) | θ value | 8.99809 (rank : 148) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P15702 | Gene names | Spn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukosialin precursor (Leukocyte sialoglycoprotein) (Sialophorin) (Ly- 48) (B cell differentiation antigen LP-3) (CD43 antigen). | |||||
|
PDZD6_HUMAN
|
||||||
| NC score | 0.027201 (rank : 118) | θ value | 5.27518 (rank : 114) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ULD6, Q4W5I8, Q86V55 | Gene names | PDZD6, KIAA1284, PDZK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 6. | |||||
|
TR10B_HUMAN
|
||||||
| NC score | 0.026917 (rank : 119) | θ value | 1.38821 (rank : 64) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O14763, O14720, O15508, O15517, O15531, Q9BVE0 | Gene names | TNFRSF10B, DR5, KILLER, TRAILR2, TRICK2, ZTNFR9 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 10B precursor (Death receptor 5) (TNF-related apoptosis-inducing ligand receptor 2) (TRAIL receptor 2) (TRAIL-R2) (CD262 antigen). | |||||
|
PTN22_MOUSE
|
||||||
| NC score | 0.026497 (rank : 120) | θ value | 0.365318 (rank : 40) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P29352, Q7TMP9 | Gene names | Ptpn22, Ptpn8 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 22 (EC 3.1.3.48) (Hematopoietic cell protein-tyrosine phosphatase 70Z-PEP). | |||||
|
XRN1_MOUSE
|
||||||
| NC score | 0.026365 (rank : 121) | θ value | 1.38821 (rank : 67) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P97789, O35651, P97790, Q3TE44, Q3TRE9, Q3UQL5 | Gene names | Xrn1, Dhm2, Exo, Sep1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5'-3' exoribonuclease 1 (EC 3.1.11.-) (mXRN1) (Strand-exchange protein 1 homolog) (Protein Dhm2). | |||||
|
MAP4_MOUSE
|
||||||
| NC score | 0.025750 (rank : 122) | θ value | 8.99809 (rank : 149) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
RD23B_HUMAN
|
||||||
| NC score | 0.025675 (rank : 123) | θ value | 5.27518 (rank : 117) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P54727, Q8WUB0 | Gene names | RAD23B | |||
|
Domain Architecture |

|
|||||
| Description | UV excision repair protein RAD23 homolog B (hHR23B) (XP-C repair- complementing complex 58 kDa protein) (p58). | |||||
|
NSD1_MOUSE
|
||||||
| NC score | 0.025244 (rank : 124) | θ value | 4.03905 (rank : 98) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
ARI5B_MOUSE
|
||||||
| NC score | 0.024972 (rank : 125) | θ value | 3.0926 (rank : 80) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BM75, Q8C0E0, Q8K4G8, Q8K4L9, Q9CU78, Q9JIX4 | Gene names | Arid5b, Desrt, Mrf2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (MRF1-like) (Modulator recognition factor protein 2) (MRF-2) (Developmentally and sexually retarded with transient immune abnormalities protein). | |||||
|
CCD82_HUMAN
|
||||||
| NC score | 0.024970 (rank : 126) | θ value | 6.88961 (rank : 127) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8N4S0, Q9H2Q5, Q9H5E3 | Gene names | CCDC82 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 82. | |||||
|
UBP42_HUMAN
|
||||||
| NC score | 0.024355 (rank : 127) | θ value | 1.38821 (rank : 66) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
K0460_HUMAN
|
||||||
| NC score | 0.023944 (rank : 128) | θ value | 8.99809 (rank : 147) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q5VT52, O75048, Q5VT53, Q6MZL4, Q86XD2 | Gene names | KIAA0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
LHX3_MOUSE
|
||||||
| NC score | 0.023630 (rank : 129) | θ value | 0.0330416 (rank : 17) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P50481, Q61800, Q61801 | Gene names | Lhx3, Lim-3, Lim3, Plim | |||
|
Domain Architecture |

|
|||||
| Description | LIM/homeobox protein Lhx3 (Homeobox protein LIM-3) (Homeobox protein P-LIM) (LIM homeobox protein 3). | |||||
|
CIC_MOUSE
|
||||||
| NC score | 0.023502 (rank : 130) | θ value | 4.03905 (rank : 92) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CEND3_MOUSE
|
||||||
| NC score | 0.023394 (rank : 131) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R5G7, Q5DTN4, Q6NVF1, Q8R5G6 | Gene names | Centd3, Arap3, Drag1, Kiaa4097 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3) (Dual specificity Rho- and Arf-GTPase-activating protein 1). | |||||
|
UBQL2_HUMAN
|
||||||
| NC score | 0.023155 (rank : 132) | θ value | 8.99809 (rank : 158) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UHD9, O94798, Q9H3W6, Q9HAZ4 | Gene names | UBQLN2, PLIC2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-2 (Protein linking IAP with cytoskeleton 2) (PLIC-2) (hPLIC- 2) (Ubiquitin-like product Chap1/Dsk2) (DSK2 homolog) (Chap1). | |||||
|
HCN2_MOUSE
|
||||||
| NC score | 0.022784 (rank : 133) | θ value | 3.0926 (rank : 85) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O88703, O70506 | Gene names | Hcn2, Bcng2, Hac1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2) (Hyperpolarization-activated cation channel 1) (HAC-1). | |||||
|
DHB13_MOUSE
|
||||||
| NC score | 0.022554 (rank : 134) | θ value | 4.03905 (rank : 95) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VCR2, Q8CIU2 | Gene names | Hsd17b13, Scdr9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 17-beta hydroxysteroid dehydrogenase 13 precursor (EC 1.1.-.-) (Short- chain dehydrogenase/reductase 9) (Alcohol dehydrogenase PAN1B-like). | |||||
|
NRG2_HUMAN
|
||||||
| NC score | 0.022553 (rank : 135) | θ value | 8.99809 (rank : 152) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O14511 | Gene names | NRG2, NTAK | |||
|
Domain Architecture |

|
|||||
| Description | Pro-neuregulin-2, membrane-bound isoform precursor (Pro-NRG2) [Contains: Neuregulin-2 (NRG-2) (Neural- and thymus-derived activator for ERBB kinases) (NTAK) (Divergent of neuregulin-1) (DON-1)]. | |||||
|
BPA1_MOUSE
|
||||||
| NC score | 0.022500 (rank : 136) | θ value | 6.88961 (rank : 126) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
KLC2_MOUSE
|
||||||
| NC score | 0.020659 (rank : 137) | θ value | 5.27518 (rank : 108) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O88448 | Gene names | Klc2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 2 (KLC 2). | |||||
|
ROBO2_HUMAN
|
||||||
| NC score | 0.020281 (rank : 138) | θ value | 0.0961366 (rank : 23) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9HCK4, O43608 | Gene names | ROBO2, KIAA1568 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 2 precursor. | |||||
|
SQSTM_HUMAN
|
||||||
| NC score | 0.019227 (rank : 139) | θ value | 5.27518 (rank : 119) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13501, Q13446, Q9BUV7, Q9BVS6, Q9UEU1 | Gene names | SQSTM1, ORCA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sequestosome-1 (Phosphotyrosine-independent ligand for the Lck SH2 domain of 62 kDa) (Ubiquitin-binding protein p62) (EBI3-associated protein of 60 kDa) (p60) (EBIAP). | |||||
|
RTN1_HUMAN
|
||||||
| NC score | 0.018863 (rank : 140) | θ value | 4.03905 (rank : 101) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q16799, Q16800, Q16801 | Gene names | RTN1, NSP | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-1 (Neuroendocrine-specific protein). | |||||
|
EP400_HUMAN
|
||||||
| NC score | 0.018357 (rank : 141) | θ value | 3.0926 (rank : 84) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
SEPT7_HUMAN
|
||||||
| NC score | 0.018093 (rank : 142) | θ value | 8.99809 (rank : 156) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q16181, Q6NX50 | Gene names | SEPT7, CDC10 | |||
|
Domain Architecture |

|
|||||
| Description | Septin-7 (CDC10 protein homolog). | |||||
|
AFTIN_HUMAN
|
||||||
| NC score | 0.017954 (rank : 143) | θ value | 8.99809 (rank : 142) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6ULP2, Q6ZM66, Q86VW3, Q8TCF3, Q9H7E3, Q9HAB9, Q9NXS4 | Gene names | AFTPH, AFTH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Aftiphilin. | |||||
|
DCR1C_HUMAN
|
||||||
| NC score | 0.017897 (rank : 144) | θ value | 6.88961 (rank : 129) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96SD1, Q5JSR4, Q5JSR5, Q5JSR7, Q5JSR8, Q5JSR9, Q5JSS0, Q5JSS7, Q6PK14, Q8N101, Q8N132, Q8TBW9, Q9BVW9, Q9HAM4 | Gene names | DCLRE1C, ARTEMIS, ASCID, SCIDA, SNM1C | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Artemis protein (EC 3.1.-.-) (DNA cross-link repair 1C protein) (SNM1- like protein) (A-SCID protein) (hSNM1C). | |||||
|
PTN12_MOUSE
|
||||||
| NC score | 0.017782 (rank : 145) | θ value | 5.27518 (rank : 116) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35831 | Gene names | Ptpn12 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 12 (EC 3.1.3.48) (Protein-tyrosine phosphatase P19) (P19-PTP) (MPTP-PEST). | |||||
|
3BP1_HUMAN
|
||||||
| NC score | 0.017442 (rank : 146) | θ value | 6.88961 (rank : 124) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y3L3, Q6IBZ2, Q6ZVL9, Q96HQ5, Q9NSQ9 | Gene names | SH3BP1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
TM16H_MOUSE
|
||||||
| NC score | 0.016889 (rank : 147) | θ value | 5.27518 (rank : 122) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PB70, Q69ZE4 | Gene names | Tmem16h, Kiaa1623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 16H. | |||||
|
FMN2_MOUSE
|
||||||
| NC score | 0.016576 (rank : 148) | θ value | 8.99809 (rank : 145) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9JL04 | Gene names | Fmn2 | |||
|
Domain Architecture |

|
|||||
| Description | Formin-2. | |||||
|
ROCK2_MOUSE
|
||||||
| NC score | 0.016527 (rank : 149) | θ value | 6.88961 (rank : 137) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 2072 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P70336, Q8BR64, Q8CBR0 | Gene names | Rock2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2). | |||||
|
PXK_MOUSE
|
||||||
| NC score | 0.016338 (rank : 150) | θ value | 6.88961 (rank : 136) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 320 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BX57, Q3TD60, Q3TEL7, Q3TZZ1, Q3U197, Q3U773, Q3UCS5, Q4FBH7, Q91WB6 | Gene names | Pxk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PX domain-containing protein kinase-like protein (Modulator of Na,K- ATPase) (MONaKA). | |||||
|
ROCK2_HUMAN
|
||||||
| NC score | 0.016007 (rank : 151) | θ value | 5.27518 (rank : 118) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 2073 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O75116, Q9UQN5 | Gene names | ROCK2, KIAA0619 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2) (Rho kinase 2). | |||||
|
HHEX_HUMAN
|
||||||
| NC score | 0.013652 (rank : 152) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q03014 | Gene names | HHEX, HEX, PRH, PRHX | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein PRH (Hematopoietically expressed homeobox) (Homeobox protein HEX). | |||||
|
MNAB_HUMAN
|
||||||
| NC score | 0.013645 (rank : 153) | θ value | 5.27518 (rank : 111) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9HBD1, Q5JPD7, Q86ST6, Q8N3D6, Q96F27, Q9H5J2, Q9HBD2, Q9NWN9, Q9NXE1 | Gene names | MNAB, RNF164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated nucleic acid-binding protein (RING finger protein 164). | |||||
|
ROD1_HUMAN
|
||||||
| NC score | 0.012996 (rank : 154) | θ value | 6.88961 (rank : 138) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95758, Q86YB3, Q86YH9 | Gene names | ROD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of differentiation 1 (Rod1). | |||||
|
NBEAL_HUMAN
|
||||||
| NC score | 0.012964 (rank : 155) | θ value | 6.88961 (rank : 134) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZS30, Q6Y876, Q6ZQY5, Q96Q30 | Gene names | NBEAL1, ALS2CR17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurobeachin-like 1 (Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 17 protein). | |||||
|
ROD1_MOUSE
|
||||||
| NC score | 0.012956 (rank : 156) | θ value | 6.88961 (rank : 139) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BHD7, Q923C3 | Gene names | Rod1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of differentiation 1 (Rod1). | |||||
|
MYT1_MOUSE
|
||||||
| NC score | 0.012933 (rank : 157) | θ value | 8.99809 (rank : 150) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8CFC2, O08995, Q8CFH1 | Gene names | Myt1, Kiaa0835, Nzf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1 (MyT1) (Neural zinc finger factor 2) (NZF-2). | |||||
|
WNK2_HUMAN
|
||||||
| NC score | 0.009859 (rank : 158) | θ value | 3.0926 (rank : 90) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
KCNH2_MOUSE
|
||||||
| NC score | 0.009433 (rank : 159) | θ value | 6.88961 (rank : 133) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O35219, O35220, O35221, O35989 | Gene names | Kcnh2, Erg, Merg1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (ERG1) (MERG) (Merg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1). | |||||
|
NALDL_MOUSE
|
||||||
| NC score | 0.007440 (rank : 160) | θ value | 8.99809 (rank : 151) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7M758 | Gene names | Naaladl1, Naaladasel | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylated-alpha-linked acidic dipeptidase-like protein (EC 3.4.17.21) (NAALADase L). | |||||
|
IRF5_MOUSE
|
||||||
| NC score | 0.007059 (rank : 161) | θ value | 8.99809 (rank : 146) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P56477 | Gene names | Irf5 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 5 (IRF-5). | |||||
|
WNK1_HUMAN
|
||||||
| NC score | 0.005505 (rank : 162) | θ value | 6.88961 (rank : 140) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1158 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9H4A3, O15052, Q86WL5, Q8N673, Q9P1S9 | Gene names | WNK1, KDP, KIAA0344, PRKWNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1) (Kinase deficient protein). | |||||
|
NR4A1_MOUSE
|
||||||
| NC score | 0.003980 (rank : 163) | θ value | 6.88961 (rank : 135) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P12813 | Gene names | Nr4a1, Gfrp, Hmr, N10, Nur77 | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR4A1 (Orphan nuclear receptor HMR) (Nuclear hormone receptor NUR/77) (N10 nuclear protein). | |||||
|
TTK_MOUSE
|
||||||
| NC score | 0.002810 (rank : 164) | θ value | 4.03905 (rank : 103) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35761 | Gene names | Ttk, Esk | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein kinase TTK (EC 2.7.12.1) (ESK) (PYT). | |||||
|
M3K4_MOUSE
|
||||||
| NC score | 0.001584 (rank : 165) | θ value | 5.27518 (rank : 109) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O08648, O08649, O70124 | Gene names | Map3k4, Mekk4 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 4 (EC 2.7.11.25) (MAPK/ERK kinase kinase 4) (MEK kinase 4) (MEKK 4). | |||||
|
ZN710_MOUSE
|
||||||
| NC score | -0.003054 (rank : 166) | θ value | 8.99809 (rank : 160) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3U288, Q3U341, Q6P9R3, Q8BJH0, Q8BTL6 | Gene names | Znf710, Zfp710 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 710. | |||||