Please be patient as the page loads
|
IRF5_MOUSE
|
||||||
| SwissProt Accessions | P56477 | Gene names | Irf5 | |||
|
Domain Architecture |
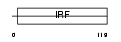
|
|||||
| Description | Interferon regulatory factor 5 (IRF-5). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
IRF5_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.995672 (rank : 2) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q13568, Q9BQF0 | Gene names | IRF5 | |||
|
Domain Architecture |
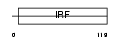
|
|||||
| Description | Interferon regulatory factor 5 (IRF-5). | |||||
|
IRF5_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P56477 | Gene names | Irf5 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 5 (IRF-5). | |||||
|
IRF6_MOUSE
|
||||||
| θ value | 1.63515e-126 (rank : 3) | NC score | 0.983668 (rank : 3) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P97431 | Gene names | Irf6 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 6 (IRF-6). | |||||
|
IRF6_HUMAN
|
||||||
| θ value | 3.08376e-125 (rank : 4) | NC score | 0.983283 (rank : 4) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O14896 | Gene names | IRF6 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 6 (IRF-6). | |||||
|
IRF4_HUMAN
|
||||||
| θ value | 2.7521e-57 (rank : 5) | NC score | 0.930214 (rank : 6) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q15306, Q99660 | Gene names | IRF4, MUM1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 4 (IRF-4) (Lymphocyte-specific interferon regulatory factor) (LSIRF) (NF-EM5) (Multiple myeloma oncogene 1). | |||||
|
IRF4_MOUSE
|
||||||
| θ value | 3.974e-56 (rank : 6) | NC score | 0.930930 (rank : 5) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q64287, Q60802 | Gene names | Irf4, Spip | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 4 (IRF-4) (Lymphocyte-specific interferon regulatory factor) (LSIRF) (NF-EM5) (PU.1 interaction partner) (Transcriptional activator PIP). | |||||
|
IRF8_HUMAN
|
||||||
| θ value | 3.71957e-54 (rank : 7) | NC score | 0.926846 (rank : 7) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q02556 | Gene names | IRF8, ICSBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 8 (IRF-8) (Interferon consensus sequence- binding protein) (ICSBP). | |||||
|
IRF8_MOUSE
|
||||||
| θ value | 3.1488e-53 (rank : 8) | NC score | 0.924206 (rank : 8) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P23611 | Gene names | Irf8, Icsbp, Icsbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 8 (IRF-8) (Interferon consensus sequence- binding protein) (ICSBP). | |||||
|
IRF3_HUMAN
|
||||||
| θ value | 5.38352e-45 (rank : 9) | NC score | 0.900261 (rank : 10) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q14653 | Gene names | IRF3 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 3 (IRF-3). | |||||
|
IRF3_MOUSE
|
||||||
| θ value | 4.88048e-38 (rank : 10) | NC score | 0.902641 (rank : 9) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P70671 | Gene names | Irf3 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 3 (IRF-3). | |||||
|
IRF7_HUMAN
|
||||||
| θ value | 2.50655e-34 (rank : 11) | NC score | 0.881091 (rank : 13) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q92985, O00331, O00332, O00333, O75924 | Gene names | IRF7 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 7 (IRF-7). | |||||
|
IRF7_MOUSE
|
||||||
| θ value | 3.27365e-34 (rank : 12) | NC score | 0.894929 (rank : 11) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P70434 | Gene names | Irf7 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 7 (IRF-7). | |||||
|
IRTF_HUMAN
|
||||||
| θ value | 2.12192e-33 (rank : 13) | NC score | 0.891904 (rank : 12) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q00978 | Gene names | ISGF3G, IRF9 | |||
|
Domain Architecture |
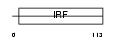
|
|||||
| Description | Transcriptional regulator ISGF3 subunit gamma (Interferon regulatory factor 9) (IRF-9) (IFN-alpha-responsive transcription factor subunit) (Interferon-stimulated gene factor 3 gamma) (ISGF3 p48 subunit) (ISGF- 3 gamma). | |||||
|
IRF2_HUMAN
|
||||||
| θ value | 8.08199e-25 (rank : 14) | NC score | 0.838899 (rank : 17) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P14316, Q96B99 | Gene names | IRF2 | |||
|
Domain Architecture |
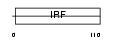
|
|||||
| Description | Interferon regulatory factor 2 (IRF-2). | |||||
|
IRF2_MOUSE
|
||||||
| θ value | 1.05554e-24 (rank : 15) | NC score | 0.835749 (rank : 18) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P23906 | Gene names | Irf2 | |||
|
Domain Architecture |
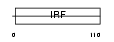
|
|||||
| Description | Interferon regulatory factor 2 (IRF-2). | |||||
|
IRF1_HUMAN
|
||||||
| θ value | 8.93572e-24 (rank : 16) | NC score | 0.842592 (rank : 15) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P10914, Q96GG7 | Gene names | IRF1 | |||
|
Domain Architecture |
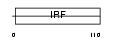
|
|||||
| Description | Interferon regulatory factor 1 (IRF-1). | |||||
|
IRF1_MOUSE
|
||||||
| θ value | 8.93572e-24 (rank : 17) | NC score | 0.839181 (rank : 16) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P15314 | Gene names | Irf1 | |||
|
Domain Architecture |
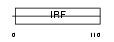
|
|||||
| Description | Interferon regulatory factor 1 (IRF-1). | |||||
|
IRTF_MOUSE
|
||||||
| θ value | 4.58923e-20 (rank : 18) | NC score | 0.861097 (rank : 14) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q61179 | Gene names | Isgf3g | |||
|
Domain Architecture |
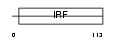
|
|||||
| Description | Transcriptional regulator ISGF3 subunit gamma (IFN-alpha-responsive transcription factor subunit) (Interferon-stimulated gene factor 3 gamma) (ISGF3 p48 subunit) (ISGF-3 gamma). | |||||
|
GNAS1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 19) | NC score | 0.012329 (rank : 28) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6R0H7, Q6R0H4, Q6R0H5, Q6R2J5, Q9JJX0, Q9Z1N8 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
PLXB3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 20) | NC score | 0.011288 (rank : 31) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ULL4, Q9HDA4 | Gene names | PLXNB3, KIAA1206 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B3 precursor. | |||||
|
RPGF3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 21) | NC score | 0.024673 (rank : 19) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95398, O95634, Q8WVN0 | Gene names | RAPGEF3, CGEF1, EPAC, EPAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 3 (cAMP-regulated guanine nucleotide exchange factor I) (cAMP-GEFI) (Exchange factor directly activated by cAMP 1) (Epac 1) (Rap1 guanine-nucleotide-exchange factor directly activated by cAMP). | |||||
|
RPGF3_MOUSE
|
||||||
| θ value | 0.62314 (rank : 22) | NC score | 0.024491 (rank : 20) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VCC8, Q8BZK9, Q8R1R1 | Gene names | Rapgef3, Epac, Epac1 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 3 (cAMP-regulated guanine nucleotide exchange factor I) (cAMP-GEFI) (Exchange factor directly activated by cAMP 1) (Epac 1). | |||||
|
ZN692_HUMAN
|
||||||
| θ value | 0.813845 (rank : 23) | NC score | 0.000313 (rank : 47) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BU19, Q5SRA5, Q5SRA6, Q9HBC9, Q9NW93, Q9NWY6, Q9UF97 | Gene names | ZNF692 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 692. | |||||
|
SSBP4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 24) | NC score | 0.012210 (rank : 29) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
SON_HUMAN
|
||||||
| θ value | 1.38821 (rank : 25) | NC score | 0.012441 (rank : 27) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
MNT_HUMAN
|
||||||
| θ value | 1.81305 (rank : 26) | NC score | 0.015682 (rank : 24) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
PIGT_HUMAN
|
||||||
| θ value | 2.36792 (rank : 27) | NC score | 0.018335 (rank : 21) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q969N2, Q7Z3N7, Q9BQY7, Q9BQY8, Q9UJG6, Q9Y2Z5 | Gene names | PIGT | |||
|
Domain Architecture |

|
|||||
| Description | GPI transamidase component PIG-T precursor (Phosphatidylinositol- glycan biosynthesis class T protein). | |||||
|
PO121_HUMAN
|
||||||
| θ value | 2.36792 (rank : 28) | NC score | 0.018012 (rank : 22) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
ACK1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 29) | NC score | -0.000963 (rank : 49) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1080 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O54967, Q8C2U0, Q8K0K4 | Gene names | Tnk2, Ack1 | |||
|
Domain Architecture |
|
|||||
| Description | Activated CDC42 kinase 1 (EC 2.7.10.2) (ACK-1) (Non-receptor protein tyrosine kinase Ack) (Tyrosine kinase non-receptor protein 2). | |||||
|
DEN1A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 30) | NC score | 0.013866 (rank : 25) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TEH3, Q5VWF0, Q8IVD6, Q9H796 | Gene names | DENND1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 1A. | |||||
|
TLE1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 31) | NC score | 0.009553 (rank : 33) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q04724, Q5T3G4, Q969V9 | Gene names | TLE1 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 1 (ESG1) (E(Sp1) homolog). | |||||
|
TLE1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 32) | NC score | 0.009252 (rank : 34) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q62440 | Gene names | Tle1, Grg1 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 1 (Groucho-related protein 1) (Grg- 1). | |||||
|
MTSS1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 33) | NC score | 0.016516 (rank : 23) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R1S4, Q8BMM3, Q99LB3 | Gene names | Mtss1, Mim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein). | |||||
|
DPOE2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 34) | NC score | 0.012593 (rank : 26) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P56282, O43560 | Gene names | POLE2, DPE2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase epsilon subunit 2 (EC 2.7.7.7) (DNA polymerase II subunit 2) (DNA polymerase epsilon subunit B). | |||||
|
NALP6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 35) | NC score | 0.005679 (rank : 44) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P59044 | Gene names | NALP6, PYPAF5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 6 (PYRIN-containing APAF1-like protein 5). | |||||
|
SEZ6L_HUMAN
|
||||||
| θ value | 5.27518 (rank : 36) | NC score | 0.005939 (rank : 43) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BYH1, O95917, Q5THY5, Q6IBZ4, Q6UXD4, Q9NUI3, Q9NUI4, Q9NUI5, Q9Y2E1, Q9Y3J6 | Gene names | SEZ6L, KIAA0927 | |||
|
Domain Architecture |

|
|||||
| Description | Seizure 6-like protein precursor. | |||||
|
XYLT1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 37) | NC score | 0.008307 (rank : 39) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86Y38, Q9H1B6 | Gene names | XYLT1, XT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Xylosyltransferase 1 (EC 2.4.2.26) (Xylosyltransferase I) (XylT-I) (XT-I) (Peptide O-xylosyltransferase 1). | |||||
|
ZMYM3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 38) | NC score | 0.008439 (rank : 37) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JLM4, Q80U17 | Gene names | Zmym3, Kiaa0385, Zfp261, Znf261 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 3 (Zinc finger protein 261) (DXHXS6673E protein). | |||||
|
BIM_MOUSE
|
||||||
| θ value | 6.88961 (rank : 39) | NC score | 0.011481 (rank : 30) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O54918, O54919, O54920 | Gene names | Bcl2l11, Bim | |||
|
Domain Architecture |

|
|||||
| Description | Bcl-2-like protein 11 (Bcl2-interacting mediator of cell death). | |||||
|
ENPP7_HUMAN
|
||||||
| θ value | 6.88961 (rank : 40) | NC score | 0.007238 (rank : 40) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6UWV6, Q6ZTS5, Q8IUS8 | Gene names | ENPP7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 7 precursor (EC 3.1.4.12) (E-NPP7) (NPP-7) (Alkaline sphingomyelin phosphodiesterase) (Intestinal alkaline sphingomyelinase) (Alk-SMase). | |||||
|
SAPS1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 41) | NC score | 0.008316 (rank : 38) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
TMEPA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 42) | NC score | 0.008945 (rank : 36) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q969W9, Q5TDR6, Q96B72, Q9UJD3 | Gene names | TMEPAI, PMEPA1, STAG1 | |||
|
Domain Architecture |
|
|||||
| Description | Transmembrane prostate androgen-induced protein (Solid tumor- associated 1 protein). | |||||
|
CNTRB_MOUSE
|
||||||
| θ value | 8.99809 (rank : 43) | NC score | 0.000067 (rank : 48) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CB62, Q5NCF3 | Gene names | Cntrob, Lip8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
ECM1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 44) | NC score | 0.009164 (rank : 35) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61508 | Gene names | Ecm1 | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular matrix protein 1 precursor (Secretory component p85). | |||||
|
MDFI_HUMAN
|
||||||
| θ value | 8.99809 (rank : 45) | NC score | 0.011140 (rank : 32) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99750 | Gene names | MDFI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MyoD family inhibitor (Myogenic repressor I-mf). | |||||
|
PHC2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 46) | NC score | 0.006636 (rank : 42) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QWH1, O88463, Q8K5D9 | Gene names | Phc2, Edr2, Ph2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 2 (mPH2) (Early development regulatory protein 2) (p36). | |||||
|
SOX21_HUMAN
|
||||||
| θ value | 8.99809 (rank : 47) | NC score | 0.003991 (rank : 45) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y651, P35715, Q15504 | Gene names | SOX21, SOX25, SOXA | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-21 (SOX-A). | |||||
|
SOX21_MOUSE
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.003989 (rank : 46) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q811W0, Q8VH35 | Gene names | Sox21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor SOX-21. | |||||
|
SYNG_HUMAN
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | 0.007059 (rank : 41) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UMZ2, Q6ZT17 | Gene names | AP1GBP1, SYNG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP1 subunit gamma-binding protein 1 (Gamma-synergin). | |||||
|
IRF5_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P56477 | Gene names | Irf5 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 5 (IRF-5). | |||||
|
IRF5_HUMAN
|
||||||
| NC score | 0.995672 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q13568, Q9BQF0 | Gene names | IRF5 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 5 (IRF-5). | |||||
|
IRF6_MOUSE
|
||||||
| NC score | 0.983668 (rank : 3) | θ value | 1.63515e-126 (rank : 3) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P97431 | Gene names | Irf6 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 6 (IRF-6). | |||||
|
IRF6_HUMAN
|
||||||
| NC score | 0.983283 (rank : 4) | θ value | 3.08376e-125 (rank : 4) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O14896 | Gene names | IRF6 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 6 (IRF-6). | |||||
|
IRF4_MOUSE
|
||||||
| NC score | 0.930930 (rank : 5) | θ value | 3.974e-56 (rank : 6) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q64287, Q60802 | Gene names | Irf4, Spip | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 4 (IRF-4) (Lymphocyte-specific interferon regulatory factor) (LSIRF) (NF-EM5) (PU.1 interaction partner) (Transcriptional activator PIP). | |||||
|
IRF4_HUMAN
|
||||||
| NC score | 0.930214 (rank : 6) | θ value | 2.7521e-57 (rank : 5) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q15306, Q99660 | Gene names | IRF4, MUM1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 4 (IRF-4) (Lymphocyte-specific interferon regulatory factor) (LSIRF) (NF-EM5) (Multiple myeloma oncogene 1). | |||||
|
IRF8_HUMAN
|
||||||
| NC score | 0.926846 (rank : 7) | θ value | 3.71957e-54 (rank : 7) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q02556 | Gene names | IRF8, ICSBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 8 (IRF-8) (Interferon consensus sequence- binding protein) (ICSBP). | |||||
|
IRF8_MOUSE
|
||||||
| NC score | 0.924206 (rank : 8) | θ value | 3.1488e-53 (rank : 8) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P23611 | Gene names | Irf8, Icsbp, Icsbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 8 (IRF-8) (Interferon consensus sequence- binding protein) (ICSBP). | |||||
|
IRF3_MOUSE
|
||||||
| NC score | 0.902641 (rank : 9) | θ value | 4.88048e-38 (rank : 10) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P70671 | Gene names | Irf3 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 3 (IRF-3). | |||||
|
IRF3_HUMAN
|
||||||
| NC score | 0.900261 (rank : 10) | θ value | 5.38352e-45 (rank : 9) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q14653 | Gene names | IRF3 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 3 (IRF-3). | |||||
|
IRF7_MOUSE
|
||||||
| NC score | 0.894929 (rank : 11) | θ value | 3.27365e-34 (rank : 12) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P70434 | Gene names | Irf7 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 7 (IRF-7). | |||||
|
IRTF_HUMAN
|
||||||
| NC score | 0.891904 (rank : 12) | θ value | 2.12192e-33 (rank : 13) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q00978 | Gene names | ISGF3G, IRF9 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ISGF3 subunit gamma (Interferon regulatory factor 9) (IRF-9) (IFN-alpha-responsive transcription factor subunit) (Interferon-stimulated gene factor 3 gamma) (ISGF3 p48 subunit) (ISGF- 3 gamma). | |||||
|
IRF7_HUMAN
|
||||||
| NC score | 0.881091 (rank : 13) | θ value | 2.50655e-34 (rank : 11) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q92985, O00331, O00332, O00333, O75924 | Gene names | IRF7 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 7 (IRF-7). | |||||
|
IRTF_MOUSE
|
||||||
| NC score | 0.861097 (rank : 14) | θ value | 4.58923e-20 (rank : 18) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q61179 | Gene names | Isgf3g | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ISGF3 subunit gamma (IFN-alpha-responsive transcription factor subunit) (Interferon-stimulated gene factor 3 gamma) (ISGF3 p48 subunit) (ISGF-3 gamma). | |||||
|
IRF1_HUMAN
|
||||||
| NC score | 0.842592 (rank : 15) | θ value | 8.93572e-24 (rank : 16) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P10914, Q96GG7 | Gene names | IRF1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 1 (IRF-1). | |||||
|
IRF1_MOUSE
|
||||||
| NC score | 0.839181 (rank : 16) | θ value | 8.93572e-24 (rank : 17) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P15314 | Gene names | Irf1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 1 (IRF-1). | |||||
|
IRF2_HUMAN
|
||||||
| NC score | 0.838899 (rank : 17) | θ value | 8.08199e-25 (rank : 14) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P14316, Q96B99 | Gene names | IRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 2 (IRF-2). | |||||
|
IRF2_MOUSE
|
||||||
| NC score | 0.835749 (rank : 18) | θ value | 1.05554e-24 (rank : 15) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P23906 | Gene names | Irf2 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 2 (IRF-2). | |||||
|
RPGF3_HUMAN
|
||||||
| NC score | 0.024673 (rank : 19) | θ value | 0.47712 (rank : 21) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95398, O95634, Q8WVN0 | Gene names | RAPGEF3, CGEF1, EPAC, EPAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 3 (cAMP-regulated guanine nucleotide exchange factor I) (cAMP-GEFI) (Exchange factor directly activated by cAMP 1) (Epac 1) (Rap1 guanine-nucleotide-exchange factor directly activated by cAMP). | |||||
|
RPGF3_MOUSE
|
||||||
| NC score | 0.024491 (rank : 20) | θ value | 0.62314 (rank : 22) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VCC8, Q8BZK9, Q8R1R1 | Gene names | Rapgef3, Epac, Epac1 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 3 (cAMP-regulated guanine nucleotide exchange factor I) (cAMP-GEFI) (Exchange factor directly activated by cAMP 1) (Epac 1). | |||||
|
PIGT_HUMAN
|
||||||
| NC score | 0.018335 (rank : 21) | θ value | 2.36792 (rank : 27) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q969N2, Q7Z3N7, Q9BQY7, Q9BQY8, Q9UJG6, Q9Y2Z5 | Gene names | PIGT | |||
|
Domain Architecture |

|
|||||
| Description | GPI transamidase component PIG-T precursor (Phosphatidylinositol- glycan biosynthesis class T protein). | |||||
|
PO121_HUMAN
|
||||||
| NC score | 0.018012 (rank : 22) | θ value | 2.36792 (rank : 28) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
MTSS1_MOUSE
|
||||||
| NC score | 0.016516 (rank : 23) | θ value | 4.03905 (rank : 33) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R1S4, Q8BMM3, Q99LB3 | Gene names | Mtss1, Mim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein). | |||||
|
MNT_HUMAN
|
||||||
| NC score | 0.015682 (rank : 24) | θ value | 1.81305 (rank : 26) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
DEN1A_HUMAN
|
||||||
| NC score | 0.013866 (rank : 25) | θ value | 3.0926 (rank : 30) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TEH3, Q5VWF0, Q8IVD6, Q9H796 | Gene names | DENND1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 1A. | |||||
|
DPOE2_HUMAN
|
||||||
| NC score | 0.012593 (rank : 26) | θ value | 5.27518 (rank : 34) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P56282, O43560 | Gene names | POLE2, DPE2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase epsilon subunit 2 (EC 2.7.7.7) (DNA polymerase II subunit 2) (DNA polymerase epsilon subunit B). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.012441 (rank : 27) | θ value | 1.38821 (rank : 25) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
GNAS1_MOUSE
|
||||||
| NC score | 0.012329 (rank : 28) | θ value | 0.163984 (rank : 19) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6R0H7, Q6R0H4, Q6R0H5, Q6R2J5, Q9JJX0, Q9Z1N8 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
SSBP4_HUMAN
|
||||||
| NC score | 0.012210 (rank : 29) | θ value | 1.06291 (rank : 24) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
BIM_MOUSE
|
||||||
| NC score | 0.011481 (rank : 30) | θ value | 6.88961 (rank : 39) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O54918, O54919, O54920 | Gene names | Bcl2l11, Bim | |||
|
Domain Architecture |

|
|||||
| Description | Bcl-2-like protein 11 (Bcl2-interacting mediator of cell death). | |||||
|
PLXB3_HUMAN
|
||||||
| NC score | 0.011288 (rank : 31) | θ value | 0.47712 (rank : 20) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ULL4, Q9HDA4 | Gene names | PLXNB3, KIAA1206 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B3 precursor. | |||||
|
MDFI_HUMAN
|
||||||
| NC score | 0.011140 (rank : 32) | θ value | 8.99809 (rank : 45) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99750 | Gene names | MDFI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MyoD family inhibitor (Myogenic repressor I-mf). | |||||
|
TLE1_HUMAN
|
||||||
| NC score | 0.009553 (rank : 33) | θ value | 3.0926 (rank : 31) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q04724, Q5T3G4, Q969V9 | Gene names | TLE1 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 1 (ESG1) (E(Sp1) homolog). | |||||
|
TLE1_MOUSE
|
||||||
| NC score | 0.009252 (rank : 34) | θ value | 3.0926 (rank : 32) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q62440 | Gene names | Tle1, Grg1 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 1 (Groucho-related protein 1) (Grg- 1). | |||||
|
ECM1_MOUSE
|
||||||
| NC score | 0.009164 (rank : 35) | θ value | 8.99809 (rank : 44) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61508 | Gene names | Ecm1 | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular matrix protein 1 precursor (Secretory component p85). | |||||
|
TMEPA_HUMAN
|
||||||
| NC score | 0.008945 (rank : 36) | θ value | 6.88961 (rank : 42) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q969W9, Q5TDR6, Q96B72, Q9UJD3 | Gene names | TMEPAI, PMEPA1, STAG1 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane prostate androgen-induced protein (Solid tumor- associated 1 protein). | |||||
|
ZMYM3_MOUSE
|
||||||
| NC score | 0.008439 (rank : 37) | θ value | 5.27518 (rank : 38) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JLM4, Q80U17 | Gene names | Zmym3, Kiaa0385, Zfp261, Znf261 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 3 (Zinc finger protein 261) (DXHXS6673E protein). | |||||
|
SAPS1_HUMAN
|
||||||
| NC score | 0.008316 (rank : 38) | θ value | 6.88961 (rank : 41) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
XYLT1_HUMAN
|
||||||
| NC score | 0.008307 (rank : 39) | θ value | 5.27518 (rank : 37) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86Y38, Q9H1B6 | Gene names | XYLT1, XT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Xylosyltransferase 1 (EC 2.4.2.26) (Xylosyltransferase I) (XylT-I) (XT-I) (Peptide O-xylosyltransferase 1). | |||||
|
ENPP7_HUMAN
|
||||||
| NC score | 0.007238 (rank : 40) | θ value | 6.88961 (rank : 40) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6UWV6, Q6ZTS5, Q8IUS8 | Gene names | ENPP7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 7 precursor (EC 3.1.4.12) (E-NPP7) (NPP-7) (Alkaline sphingomyelin phosphodiesterase) (Intestinal alkaline sphingomyelinase) (Alk-SMase). | |||||
|
SYNG_HUMAN
|
||||||
| NC score | 0.007059 (rank : 41) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UMZ2, Q6ZT17 | Gene names | AP1GBP1, SYNG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP1 subunit gamma-binding protein 1 (Gamma-synergin). | |||||
|
PHC2_MOUSE
|
||||||
| NC score | 0.006636 (rank : 42) | θ value | 8.99809 (rank : 46) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QWH1, O88463, Q8K5D9 | Gene names | Phc2, Edr2, Ph2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 2 (mPH2) (Early development regulatory protein 2) (p36). | |||||
|
SEZ6L_HUMAN
|
||||||
| NC score | 0.005939 (rank : 43) | θ value | 5.27518 (rank : 36) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BYH1, O95917, Q5THY5, Q6IBZ4, Q6UXD4, Q9NUI3, Q9NUI4, Q9NUI5, Q9Y2E1, Q9Y3J6 | Gene names | SEZ6L, KIAA0927 | |||
|
Domain Architecture |

|
|||||
| Description | Seizure 6-like protein precursor. | |||||
|
NALP6_HUMAN
|
||||||
| NC score | 0.005679 (rank : 44) | θ value | 5.27518 (rank : 35) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P59044 | Gene names | NALP6, PYPAF5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 6 (PYRIN-containing APAF1-like protein 5). | |||||
|
SOX21_HUMAN
|
||||||
| NC score | 0.003991 (rank : 45) | θ value | 8.99809 (rank : 47) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y651, P35715, Q15504 | Gene names | SOX21, SOX25, SOXA | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-21 (SOX-A). | |||||
|
SOX21_MOUSE
|
||||||
| NC score | 0.003989 (rank : 46) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q811W0, Q8VH35 | Gene names | Sox21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor SOX-21. | |||||
|
ZN692_HUMAN
|
||||||
| NC score | 0.000313 (rank : 47) | θ value | 0.813845 (rank : 23) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BU19, Q5SRA5, Q5SRA6, Q9HBC9, Q9NW93, Q9NWY6, Q9UF97 | Gene names | ZNF692 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 692. | |||||
|
CNTRB_MOUSE
|
||||||
| NC score | 0.000067 (rank : 48) | θ value | 8.99809 (rank : 43) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CB62, Q5NCF3 | Gene names | Cntrob, Lip8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
ACK1_MOUSE
|
||||||
| NC score | -0.000963 (rank : 49) | θ value | 3.0926 (rank : 29) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1080 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O54967, Q8C2U0, Q8K0K4 | Gene names | Tnk2, Ack1 | |||
|
Domain Architecture |

|
|||||
| Description | Activated CDC42 kinase 1 (EC 2.7.10.2) (ACK-1) (Non-receptor protein tyrosine kinase Ack) (Tyrosine kinase non-receptor protein 2). | |||||