Please be patient as the page loads
|
TMEPA_HUMAN
|
||||||
| SwissProt Accessions | Q969W9, Q5TDR6, Q96B72, Q9UJD3 | Gene names | TMEPAI, PMEPA1, STAG1 | |||
|
Domain Architecture |
|
|||||
| Description | Transmembrane prostate androgen-induced protein (Solid tumor- associated 1 protein). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TMEPA_HUMAN
|
||||||
| θ value | 5.94893e-161 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q969W9, Q5TDR6, Q96B72, Q9UJD3 | Gene names | TMEPAI, PMEPA1, STAG1 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane prostate androgen-induced protein (Solid tumor- associated 1 protein). | |||||
|
TMEPA_MOUSE
|
||||||
| θ value | 1.29561e-123 (rank : 2) | NC score | 0.987021 (rank : 2) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9D7R2, Q9EQH9 | Gene names | Tmepai, N4wbp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane prostate androgen-induced protein (NEDD4 WW domain- binding protein 4). | |||||
|
CR001_HUMAN
|
||||||
| θ value | 3.56943e-81 (rank : 3) | NC score | 0.886250 (rank : 4) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O15165, O15166, O15167, O15168, Q6NXP3 | Gene names | C18orf1 | |||
|
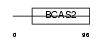
Domain Architecture |
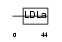
|
|||||
| Description | Protein C18orf1. | |||||
|
CR001_MOUSE
|
||||||
| θ value | 1.14825e-79 (rank : 4) | NC score | 0.887366 (rank : 3) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BWJ4 | Gene names | D18Ertd653e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C18orf1 homolog. | |||||
|
CRTC1_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 5) | NC score | 0.134398 (rank : 5) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6UUV9, O75114, Q6Y3A3, Q7LDZ2, Q8IUL3, Q8IZ34, Q8IZL1, Q8N6W3, Q96AI8, Q9H801 | Gene names | CRTC1, KIAA0616, MECT1, TORC1, WAMTP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Transducer of regulated cAMP response element-binding protein 1) (Transducer of CREB protein 1) (Mucoepidermoid carcinoma translocated protein 1). | |||||
|
ATN1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 6) | NC score | 0.084801 (rank : 9) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
CRTC1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 7) | NC score | 0.112518 (rank : 6) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q68ED7, Q6ZQ85 | Gene names | Crtc1, Kiaa0616, Mect1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Mucoepidermoid carcinoma translocated protein 1 homolog). | |||||
|
CPSF7_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 8) | NC score | 0.104507 (rank : 7) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N684, Q7Z3H9, Q9H025, Q9H9V1 | Gene names | CPSF7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 7 (Cleavage and polyadenylation specificity factor 59 kDa subunit) (CPSF 59 kDa subunit) (Pre-mRNA cleavage factor Im 59 kDa subunit). | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 9) | NC score | 0.074772 (rank : 11) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
CPSF7_MOUSE
|
||||||
| θ value | 0.279714 (rank : 10) | NC score | 0.095740 (rank : 8) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BTV2, Q3TNF1, Q8BKE7, Q8CFS8 | Gene names | Cpsf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 7. | |||||
|
CAC1B_HUMAN
|
||||||
| θ value | 0.365318 (rank : 11) | NC score | 0.019655 (rank : 29) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
GAB1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 12) | NC score | 0.076973 (rank : 10) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9QYY0, Q91VW7 | Gene names | Gab1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
PDC6I_HUMAN
|
||||||
| θ value | 0.365318 (rank : 13) | NC score | 0.072561 (rank : 12) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8WUM4, Q9BX86, Q9NUN0, Q9P2H2, Q9UKL5 | Gene names | PDCD6IP, AIP1, ALIX, KIAA1375 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death 6-interacting protein (PDCD6-interacting protein) (ALG-2-interacting protein 1) (Hp95). | |||||
|
TULP4_HUMAN
|
||||||
| θ value | 0.365318 (rank : 14) | NC score | 0.064009 (rank : 14) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NRJ4, Q9HD22, Q9P2F0 | Gene names | TULP4, KIAA1397, TUSP | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-like protein 4 (Tubby superfamily protein). | |||||
|
PDC6I_MOUSE
|
||||||
| θ value | 0.47712 (rank : 15) | NC score | 0.068181 (rank : 13) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9WU78, O88695, O89014, Q8BSL8, Q8R0H5, Q99LR3, Q9QZN8 | Gene names | Pdcd6ip, Aip1, Alix | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death 6-interacting protein (ALG-2-interacting protein X) (ALG-2-interacting protein 1) (E2F1-inducible protein) (Eig2). | |||||
|
TULP4_MOUSE
|
||||||
| θ value | 0.47712 (rank : 16) | NC score | 0.057535 (rank : 18) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JIL5, Q8CA75, Q922C2 | Gene names | Tulp4, Tusp | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-like protein 4 (Tubby superfamily protein). | |||||
|
TROAP_HUMAN
|
||||||
| θ value | 0.62314 (rank : 17) | NC score | 0.057833 (rank : 17) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q12815 | Gene names | TROAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trophinin-associated protein (Tastin) (Trophinin-assisting protein). | |||||
|
BL1S3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 18) | NC score | 0.056619 (rank : 19) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5U5M8, Q8C6R4, Q8R0Z3 | Gene names | Bloc1s3, Blos3, Rp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Biogenesis of lysosome-related organelles complex-1 subunit 3 (Reduced pigmentation protein) (BLOC subunit 3). | |||||
|
CASC3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 19) | NC score | 0.050839 (rank : 21) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8K3W3, Q8K219, Q99NF0 | Gene names | Casc3, Mln51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC3 (Cancer susceptibility candidate gene 3 protein homolog) (Metastatic lymph node protein 51 homolog) (Protein MLN 51 homolog) (Protein barentsz) (Btz). | |||||
|
NFAC4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 20) | NC score | 0.025623 (rank : 24) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14934 | Gene names | NFATC4, NFAT3 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 4 (NF-ATc4) (NFATc4) (T cell transcription factor NFAT3) (NF-AT3). | |||||
|
CELR2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 21) | NC score | 0.009680 (rank : 41) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9HCU4, Q92566 | Gene names | CELSR2, CDHF10, EGFL2, KIAA0279, MEGF3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 2 precursor (Epidermal growth factor-like 2) (Multiple epidermal growth factor-like domains 3) (Flamingo 1). | |||||
|
IL3RB_MOUSE
|
||||||
| θ value | 2.36792 (rank : 22) | NC score | 0.026662 (rank : 23) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P26955 | Gene names | Csf2rb, Aic2b, Csf2rb1, Il3rb1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytokine receptor common beta chain precursor (GM-CSF/IL-3/IL-5 receptor common beta-chain) (CD131 antigen). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 23) | NC score | 0.019368 (rank : 30) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
SMR3B_HUMAN
|
||||||
| θ value | 2.36792 (rank : 24) | NC score | 0.055229 (rank : 20) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P02814, Q9UBN0, Q9UCT0 | Gene names | SMR3B, PBII, PROL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 3 homolog B precursor (Proline-rich protein 3) (Proline-rich peptide P-B) [Contains: Peptide P-A; Peptide D1A]. | |||||
|
CNOT3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 25) | NC score | 0.022889 (rank : 27) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O75175, Q9NZN7, Q9UF76 | Gene names | CNOT3, KIAA0691, NOT3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
PDZD8_HUMAN
|
||||||
| θ value | 3.0926 (rank : 26) | NC score | 0.060684 (rank : 15) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NEN9, Q86WE0, Q86WE5, Q9UFF1 | Gene names | PDZD8, PDZK8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 8 (Sarcoma antigen NY-SAR-84/NY-SAR- 104). | |||||
|
AMOL2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 27) | NC score | 0.010388 (rank : 40) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K371, Q3TPM1, Q7TPE4, Q8BP84, Q8BS08, Q9QUS0 | Gene names | Amotl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin-like protein 2. | |||||
|
ANKZ1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 28) | NC score | 0.020160 (rank : 28) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80UU1, Q9CZF5 | Gene names | Ankzf1, D1Ertd161e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and zinc finger domain-containing protein 1. | |||||
|
BSN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 29) | NC score | 0.024344 (rank : 25) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
FOXN1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 30) | NC score | 0.015194 (rank : 35) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O15353, O15352 | Gene names | FOXN1, WHN | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein N1 (Transcription factor winged-helix nude). | |||||
|
NCOR2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 31) | NC score | 0.016381 (rank : 32) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
SFPQ_HUMAN
|
||||||
| θ value | 4.03905 (rank : 32) | NC score | 0.015881 (rank : 33) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
SHAN1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 33) | NC score | 0.012314 (rank : 38) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
SYNJ2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 34) | NC score | 0.013102 (rank : 36) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15056, Q5TA13, Q5TA16, Q5TA19, Q86XK0, Q8IZA8, Q9H226 | Gene names | SYNJ2, KIAA0348 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptojanin-2 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 2). | |||||
|
HES1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 35) | NC score | 0.029236 (rank : 22) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14469 | Gene names | HES1, HL, HRY | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1) (Hairy- like) (HHL) (Hairy homolog). | |||||
|
PHC2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 36) | NC score | 0.023302 (rank : 26) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IXK0, Q5T0C1, Q6NUJ6, Q6ZQR1, Q8N306, Q8TAG8, Q96BL4, Q9Y4Y7 | Gene names | PHC2, EDR2, PH2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 2 (hPH2) (Early development regulatory protein 2). | |||||
|
FRS2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 37) | NC score | 0.012265 (rank : 39) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8C180 | Gene names | Frs2, Frs2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibroblast growth factor receptor substrate 2 (FGFR substrate 2) (Suc1-associated neurotrophic factor target 1) (SNT-1) (FGFR signaling adaptor SNT) (FRS2 alpha). | |||||
|
GNAS3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 38) | NC score | 0.058195 (rank : 16) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z0F1, Q9QXW5, Q9Z0H2 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroendocrine secretory protein 55 (NESP55) [Contains: LHAL tetrapeptide; GPIPIRRH peptide]. | |||||
|
IRF5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 39) | NC score | 0.008945 (rank : 42) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P56477 | Gene names | Irf5 | |||
|
Domain Architecture |
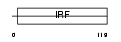
|
|||||
| Description | Interferon regulatory factor 5 (IRF-5). | |||||
|
LPIN3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 40) | NC score | 0.017839 (rank : 31) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BQK8, Q5TDB9, Q9NPY8, Q9UJE5 | Gene names | LPIN3, LIPN3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipin-3 (Lipin 3-like). | |||||
|
PDZD2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 41) | NC score | 0.015756 (rank : 34) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
ZN516_MOUSE
|
||||||
| θ value | 6.88961 (rank : 42) | NC score | 0.004160 (rank : 44) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7TSH3 | Gene names | Znf516, Kiaa0222, Zfp516 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 516. | |||||
|
CELR2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 43) | NC score | 0.007117 (rank : 43) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9R0M0, Q99K26, Q9Z2R4 | Gene names | Celsr2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 2 precursor (Flamingo 1) (mFmi1). | |||||
|
KRHB2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 44) | NC score | 0.003625 (rank : 45) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NSB4 | Gene names | KRTHB2 | |||
|
Domain Architecture |
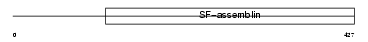
|
|||||
| Description | Keratin, type II cuticular Hb2 (Hair keratin, type II Hb2). | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 45) | NC score | 0.012662 (rank : 37) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 46) | NC score | 0.000411 (rank : 46) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
TMEPA_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 5.94893e-161 (rank : 1) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q969W9, Q5TDR6, Q96B72, Q9UJD3 | Gene names | TMEPAI, PMEPA1, STAG1 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane prostate androgen-induced protein (Solid tumor- associated 1 protein). | |||||
|
TMEPA_MOUSE
|
||||||
| NC score | 0.987021 (rank : 2) | θ value | 1.29561e-123 (rank : 2) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9D7R2, Q9EQH9 | Gene names | Tmepai, N4wbp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane prostate androgen-induced protein (NEDD4 WW domain- binding protein 4). | |||||
|
CR001_MOUSE
|
||||||
| NC score | 0.887366 (rank : 3) | θ value | 1.14825e-79 (rank : 4) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BWJ4 | Gene names | D18Ertd653e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C18orf1 homolog. | |||||
|
CR001_HUMAN
|
||||||
| NC score | 0.886250 (rank : 4) | θ value | 3.56943e-81 (rank : 3) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O15165, O15166, O15167, O15168, Q6NXP3 | Gene names | C18orf1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C18orf1. | |||||
|
CRTC1_HUMAN
|
||||||
| NC score | 0.134398 (rank : 5) | θ value | 0.00665767 (rank : 5) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6UUV9, O75114, Q6Y3A3, Q7LDZ2, Q8IUL3, Q8IZ34, Q8IZL1, Q8N6W3, Q96AI8, Q9H801 | Gene names | CRTC1, KIAA0616, MECT1, TORC1, WAMTP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Transducer of regulated cAMP response element-binding protein 1) (Transducer of CREB protein 1) (Mucoepidermoid carcinoma translocated protein 1). | |||||
|
CRTC1_MOUSE
|
||||||
| NC score | 0.112518 (rank : 6) | θ value | 0.0563607 (rank : 7) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q68ED7, Q6ZQ85 | Gene names | Crtc1, Kiaa0616, Mect1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Mucoepidermoid carcinoma translocated protein 1 homolog). | |||||
|
CPSF7_HUMAN
|
||||||
| NC score | 0.104507 (rank : 7) | θ value | 0.0961366 (rank : 8) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N684, Q7Z3H9, Q9H025, Q9H9V1 | Gene names | CPSF7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 7 (Cleavage and polyadenylation specificity factor 59 kDa subunit) (CPSF 59 kDa subunit) (Pre-mRNA cleavage factor Im 59 kDa subunit). | |||||
|
CPSF7_MOUSE
|
||||||
| NC score | 0.095740 (rank : 8) | θ value | 0.279714 (rank : 10) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BTV2, Q3TNF1, Q8BKE7, Q8CFS8 | Gene names | Cpsf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 7. | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.084801 (rank : 9) | θ value | 0.0563607 (rank : 6) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
GAB1_MOUSE
|
||||||
| NC score | 0.076973 (rank : 10) | θ value | 0.365318 (rank : 12) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9QYY0, Q91VW7 | Gene names | Gab1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.074772 (rank : 11) | θ value | 0.0961366 (rank : 9) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
PDC6I_HUMAN
|
||||||
| NC score | 0.072561 (rank : 12) | θ value | 0.365318 (rank : 13) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8WUM4, Q9BX86, Q9NUN0, Q9P2H2, Q9UKL5 | Gene names | PDCD6IP, AIP1, ALIX, KIAA1375 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death 6-interacting protein (PDCD6-interacting protein) (ALG-2-interacting protein 1) (Hp95). | |||||
|
PDC6I_MOUSE
|
||||||
| NC score | 0.068181 (rank : 13) | θ value | 0.47712 (rank : 15) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9WU78, O88695, O89014, Q8BSL8, Q8R0H5, Q99LR3, Q9QZN8 | Gene names | Pdcd6ip, Aip1, Alix | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death 6-interacting protein (ALG-2-interacting protein X) (ALG-2-interacting protein 1) (E2F1-inducible protein) (Eig2). | |||||
|
TULP4_HUMAN
|
||||||
| NC score | 0.064009 (rank : 14) | θ value | 0.365318 (rank : 14) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NRJ4, Q9HD22, Q9P2F0 | Gene names | TULP4, KIAA1397, TUSP | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-like protein 4 (Tubby superfamily protein). | |||||
|
PDZD8_HUMAN
|
||||||
| NC score | 0.060684 (rank : 15) | θ value | 3.0926 (rank : 26) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NEN9, Q86WE0, Q86WE5, Q9UFF1 | Gene names | PDZD8, PDZK8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 8 (Sarcoma antigen NY-SAR-84/NY-SAR- 104). | |||||
|
GNAS3_MOUSE
|
||||||
| NC score | 0.058195 (rank : 16) | θ value | 6.88961 (rank : 38) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z0F1, Q9QXW5, Q9Z0H2 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroendocrine secretory protein 55 (NESP55) [Contains: LHAL tetrapeptide; GPIPIRRH peptide]. | |||||
|
TROAP_HUMAN
|
||||||
| NC score | 0.057833 (rank : 17) | θ value | 0.62314 (rank : 17) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q12815 | Gene names | TROAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trophinin-associated protein (Tastin) (Trophinin-assisting protein). | |||||
|
TULP4_MOUSE
|
||||||
| NC score | 0.057535 (rank : 18) | θ value | 0.47712 (rank : 16) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JIL5, Q8CA75, Q922C2 | Gene names | Tulp4, Tusp | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-like protein 4 (Tubby superfamily protein). | |||||
|
BL1S3_MOUSE
|
||||||
| NC score | 0.056619 (rank : 19) | θ value | 0.813845 (rank : 18) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5U5M8, Q8C6R4, Q8R0Z3 | Gene names | Bloc1s3, Blos3, Rp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Biogenesis of lysosome-related organelles complex-1 subunit 3 (Reduced pigmentation protein) (BLOC subunit 3). | |||||
|
SMR3B_HUMAN
|
||||||
| NC score | 0.055229 (rank : 20) | θ value | 2.36792 (rank : 24) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P02814, Q9UBN0, Q9UCT0 | Gene names | SMR3B, PBII, PROL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 3 homolog B precursor (Proline-rich protein 3) (Proline-rich peptide P-B) [Contains: Peptide P-A; Peptide D1A]. | |||||
|
CASC3_MOUSE
|
||||||
| NC score | 0.050839 (rank : 21) | θ value | 0.813845 (rank : 19) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8K3W3, Q8K219, Q99NF0 | Gene names | Casc3, Mln51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC3 (Cancer susceptibility candidate gene 3 protein homolog) (Metastatic lymph node protein 51 homolog) (Protein MLN 51 homolog) (Protein barentsz) (Btz). | |||||
|
HES1_HUMAN
|
||||||
| NC score | 0.029236 (rank : 22) | θ value | 5.27518 (rank : 35) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14469 | Gene names | HES1, HL, HRY | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1) (Hairy- like) (HHL) (Hairy homolog). | |||||
|
IL3RB_MOUSE
|
||||||
| NC score | 0.026662 (rank : 23) | θ value | 2.36792 (rank : 22) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P26955 | Gene names | Csf2rb, Aic2b, Csf2rb1, Il3rb1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytokine receptor common beta chain precursor (GM-CSF/IL-3/IL-5 receptor common beta-chain) (CD131 antigen). | |||||
|
NFAC4_HUMAN
|
||||||
| NC score | 0.025623 (rank : 24) | θ value | 0.813845 (rank : 20) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14934 | Gene names | NFATC4, NFAT3 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 4 (NF-ATc4) (NFATc4) (T cell transcription factor NFAT3) (NF-AT3). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.024344 (rank : 25) | θ value | 4.03905 (rank : 29) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
PHC2_HUMAN
|
||||||
| NC score | 0.023302 (rank : 26) | θ value | 5.27518 (rank : 36) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IXK0, Q5T0C1, Q6NUJ6, Q6ZQR1, Q8N306, Q8TAG8, Q96BL4, Q9Y4Y7 | Gene names | PHC2, EDR2, PH2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 2 (hPH2) (Early development regulatory protein 2). | |||||
|
CNOT3_HUMAN
|
||||||
| NC score | 0.022889 (rank : 27) | θ value | 3.0926 (rank : 25) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O75175, Q9NZN7, Q9UF76 | Gene names | CNOT3, KIAA0691, NOT3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
ANKZ1_MOUSE
|
||||||
| NC score | 0.020160 (rank : 28) | θ value | 4.03905 (rank : 28) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80UU1, Q9CZF5 | Gene names | Ankzf1, D1Ertd161e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and zinc finger domain-containing protein 1. | |||||
|
CAC1B_HUMAN
|
||||||
| NC score | 0.019655 (rank : 29) | θ value | 0.365318 (rank : 11) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.019368 (rank : 30) | θ value | 2.36792 (rank : 23) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
LPIN3_HUMAN
|
||||||
| NC score | 0.017839 (rank : 31) | θ value | 6.88961 (rank : 40) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BQK8, Q5TDB9, Q9NPY8, Q9UJE5 | Gene names | LPIN3, LIPN3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipin-3 (Lipin 3-like). | |||||
|
NCOR2_HUMAN
|
||||||
| NC score | 0.016381 (rank : 32) | θ value | 4.03905 (rank : 31) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
SFPQ_HUMAN
|
||||||
| NC score | 0.015881 (rank : 33) | θ value | 4.03905 (rank : 32) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
PDZD2_HUMAN
|
||||||
| NC score | 0.015756 (rank : 34) | θ value | 6.88961 (rank : 41) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
FOXN1_HUMAN
|
||||||
| NC score | 0.015194 (rank : 35) | θ value | 4.03905 (rank : 30) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O15353, O15352 | Gene names | FOXN1, WHN | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein N1 (Transcription factor winged-helix nude). | |||||
|
SYNJ2_HUMAN
|
||||||
| NC score | 0.013102 (rank : 36) | θ value | 4.03905 (rank : 34) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15056, Q5TA13, Q5TA16, Q5TA19, Q86XK0, Q8IZA8, Q9H226 | Gene names | SYNJ2, KIAA0348 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptojanin-2 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 2). | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.012662 (rank : 37) | θ value | 8.99809 (rank : 45) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
SHAN1_HUMAN
|
||||||
| NC score | 0.012314 (rank : 38) | θ value | 4.03905 (rank : 33) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
FRS2_MOUSE
|
||||||
| NC score | 0.012265 (rank : 39) | θ value | 6.88961 (rank : 37) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8C180 | Gene names | Frs2, Frs2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibroblast growth factor receptor substrate 2 (FGFR substrate 2) (Suc1-associated neurotrophic factor target 1) (SNT-1) (FGFR signaling adaptor SNT) (FRS2 alpha). | |||||
|
AMOL2_MOUSE
|
||||||
| NC score | 0.010388 (rank : 40) | θ value | 4.03905 (rank : 27) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K371, Q3TPM1, Q7TPE4, Q8BP84, Q8BS08, Q9QUS0 | Gene names | Amotl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin-like protein 2. | |||||
|
CELR2_HUMAN
|
||||||
| NC score | 0.009680 (rank : 41) | θ value | 2.36792 (rank : 21) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9HCU4, Q92566 | Gene names | CELSR2, CDHF10, EGFL2, KIAA0279, MEGF3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 2 precursor (Epidermal growth factor-like 2) (Multiple epidermal growth factor-like domains 3) (Flamingo 1). | |||||
|
IRF5_MOUSE
|
||||||
| NC score | 0.008945 (rank : 42) | θ value | 6.88961 (rank : 39) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P56477 | Gene names | Irf5 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 5 (IRF-5). | |||||
|
CELR2_MOUSE
|
||||||
| NC score | 0.007117 (rank : 43) | θ value | 8.99809 (rank : 43) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9R0M0, Q99K26, Q9Z2R4 | Gene names | Celsr2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 2 precursor (Flamingo 1) (mFmi1). | |||||
|
ZN516_MOUSE
|
||||||
| NC score | 0.004160 (rank : 44) | θ value | 6.88961 (rank : 42) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7TSH3 | Gene names | Znf516, Kiaa0222, Zfp516 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 516. | |||||
|
KRHB2_HUMAN
|
||||||
| NC score | 0.003625 (rank : 45) | θ value | 8.99809 (rank : 44) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NSB4 | Gene names | KRTHB2 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cuticular Hb2 (Hair keratin, type II Hb2). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.000411 (rank : 46) | θ value | 8.99809 (rank : 46) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||