Please be patient as the page loads
|
CR001_HUMAN
|
||||||
| SwissProt Accessions | O15165, O15166, O15167, O15168, Q6NXP3 | Gene names | C18orf1 | |||
|
Domain Architecture |
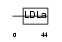
|
|||||
| Description | Protein C18orf1. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CR001_HUMAN
|
||||||
| θ value | 6.35588e-163 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O15165, O15166, O15167, O15168, Q6NXP3 | Gene names | C18orf1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C18orf1. | |||||
|
CR001_MOUSE
|
||||||
| θ value | 4.26331e-159 (rank : 2) | NC score | 0.994703 (rank : 2) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8BWJ4 | Gene names | D18Ertd653e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C18orf1 homolog. | |||||
|
TMEPA_HUMAN
|
||||||
| θ value | 3.56943e-81 (rank : 3) | NC score | 0.886250 (rank : 4) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q969W9, Q5TDR6, Q96B72, Q9UJD3 | Gene names | TMEPAI, PMEPA1, STAG1 | |||
|
Domain Architecture |
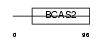
|
|||||
| Description | Transmembrane prostate androgen-induced protein (Solid tumor- associated 1 protein). | |||||
|
TMEPA_MOUSE
|
||||||
| θ value | 1.95861e-79 (rank : 4) | NC score | 0.890232 (rank : 3) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9D7R2, Q9EQH9 | Gene names | Tmepai, N4wbp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane prostate androgen-induced protein (NEDD4 WW domain- binding protein 4). | |||||
|
SSPO_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 5) | NC score | 0.219731 (rank : 35) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
CO6_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 6) | NC score | 0.245451 (rank : 31) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P13671 | Gene names | C6 | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C6 precursor. | |||||
|
LRP2_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 7) | NC score | 0.284353 (rank : 22) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P98164, O00711, Q16215 | Gene names | LRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 2 precursor (Megalin) (Glycoprotein 330) (gp330). | |||||
|
LRP4_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 8) | NC score | 0.289603 (rank : 16) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8VI56, Q8BPX5, Q8CBB3, Q8CCP5 | Gene names | Lrp4, Kiaa0816 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 4 precursor (LDLR dan). | |||||
|
LRP1_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 9) | NC score | 0.262814 (rank : 29) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q07954, Q2PP12, Q8IVG8 | Gene names | LRP1, A2MR | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1 precursor (LRP) (Alpha-2-macroglobulin receptor) (A2MR) (Apolipoprotein E receptor) (APOER) (CD91 antigen). | |||||
|
LRP4_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 10) | NC score | 0.289933 (rank : 15) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O75096 | Gene names | LRP4, KIAA0816, MEGF7 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 4 precursor (Multiple epidermal growth factor-like domains 7). | |||||
|
CORIN_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 11) | NC score | 0.174344 (rank : 39) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9Z319 | Gene names | Corin, Crn, Lrp4 | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuteric peptide-converting enzyme (EC 3.4.21.-) (pro-ANP- converting enzyme) (Corin) (Low density lipoprotein receptor-related protein 4). | |||||
|
LRP12_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 12) | NC score | 0.289346 (rank : 18) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Y561 | Gene names | LRP12, ST7 | |||
|
Domain Architecture |
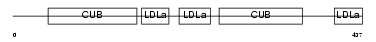
|
|||||
| Description | Low-density lipoprotein receptor-related protein 12 precursor (Suppressor of tumorigenicity protein 7). | |||||
|
LRP12_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 13) | NC score | 0.289407 (rank : 17) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8BUJ9, Q8BWM9 | Gene names | Lrp12 | |||
|
Domain Architecture |
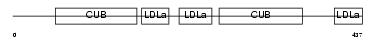
|
|||||
| Description | Low-density lipoprotein receptor-related protein 12 precursor. | |||||
|
CORIN_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 14) | NC score | 0.174376 (rank : 38) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9Y5Q5, Q9UHY2 | Gene names | CORIN, CRN, TMPRSS10 | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuteric peptide-converting enzyme (EC 3.4.21.-) (pro-ANP- converting enzyme) (Corin) (Heart-specific serine proteinase ATC2) (Transmembrane protease, serine 10). | |||||
|
LRP3_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 15) | NC score | 0.312450 (rank : 8) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O75074 | Gene names | LRP3 | |||
|
Domain Architecture |
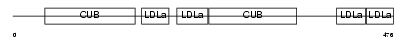
|
|||||
| Description | Low-density lipoprotein receptor-related protein 3 precursor (hLRp105). | |||||
|
LRP1B_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 16) | NC score | 0.262403 (rank : 30) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 602 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9NZR2, Q8WY29, Q8WY30, Q8WY31 | Gene names | LRP1B, LRPDIT | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1B precursor (Low- density lipoprotein receptor-related protein-deleted in tumor) (LRP- DIT). | |||||
|
LRP1B_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 17) | NC score | 0.262981 (rank : 28) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 606 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9JI18, Q8BZD3, Q8BZM7 | Gene names | Lrp1b, Lrpdit | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1B precursor (Low- density lipoprotein receptor-related protein-deleted in tumor) (LRP- DIT). | |||||
|
LRP8_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 18) | NC score | 0.282565 (rank : 23) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q924X6, Q8CAK9, Q8CDF5, Q921B6 | Gene names | Lrp8, Apoer2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 8 precursor (Apolipoprotein E receptor 2). | |||||
|
VLDLR_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 19) | NC score | 0.288996 (rank : 19) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P98155, Q5VVF6 | Gene names | VLDLR | |||
|
Domain Architecture |

|
|||||
| Description | Very low-density lipoprotein receptor precursor (VLDL receptor) (VLDL- R). | |||||
|
CO9_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 20) | NC score | 0.314248 (rank : 7) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P02748 | Gene names | C9 | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C9 precursor [Contains: Complement component C9a; Complement component C9b]. | |||||
|
ST14_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 21) | NC score | 0.168330 (rank : 41) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P56677 | Gene names | St14, Prss14 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of tumorigenicity protein 14 (EC 3.4.21.-) (Serine protease 14) (Epithin). | |||||
|
VLDLR_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 22) | NC score | 0.286376 (rank : 21) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P98156, Q64022 | Gene names | Vldlr | |||
|
Domain Architecture |

|
|||||
| Description | Very low-density lipoprotein receptor precursor (VLDL receptor) (VLDL- R). | |||||
|
LDLR_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 23) | NC score | 0.287315 (rank : 20) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P35951 | Gene names | Ldlr | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor precursor (LDL receptor). | |||||
|
LRP6_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 24) | NC score | 0.304261 (rank : 11) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O75581 | Gene names | LRP6 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 6 precursor. | |||||
|
LRP6_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 25) | NC score | 0.304713 (rank : 10) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O88572 | Gene names | Lrp6 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 6 precursor. | |||||
|
CO9_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 26) | NC score | 0.295447 (rank : 12) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P06683, Q91XA7 | Gene names | C9 | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C9 precursor. | |||||
|
LDLR_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 27) | NC score | 0.279598 (rank : 25) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P01130, Q53ZD9 | Gene names | LDLR | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor precursor (LDL receptor). | |||||
|
ST14_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 28) | NC score | 0.167090 (rank : 42) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9Y5Y6, Q9BS01, Q9H3S0, Q9HB36, Q9HCA3 | Gene names | ST14, PRSS14, SNC19, TADG15 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of tumorigenicity protein 14 (EC 3.4.21.-) (Serine protease 14) (Matriptase) (Membrane-type serine protease 1) (MT-SP1) (Prostamin) (Serine protease TADG-15) (Tumor-associated differentially-expressed gene 15 protein). | |||||
|
LRP8_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 29) | NC score | 0.282025 (rank : 24) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q14114, O14968, Q86V27, Q99876, Q9BR78 | Gene names | LRP8, APOER2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 8 precursor (Apolipoprotein E receptor 2). | |||||
|
SORL_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 30) | NC score | 0.271863 (rank : 26) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O88307, O54711, O70581 | Gene names | Sorl1 | |||
|
Domain Architecture |

|
|||||
| Description | Sortilin-related receptor precursor (Sorting protein-related receptor containing LDLR class A repeats) (mSorLA) (SorLA-1) (Low-density lipoprotein receptor relative with 11 ligand-binding repeats) (LDLR relative with 11 ligand-binding repeats) (LR11) (Gp250). | |||||
|
LRP10_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 31) | NC score | 0.319069 (rank : 6) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q7Z4F1, O95882, Q86T02, Q8NCZ4, Q9HC42, Q9UG33 | Gene names | LRP10 | |||
|
Domain Architecture |
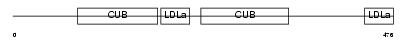
|
|||||
| Description | Low-density lipoprotein receptor-related protein 10 precursor. | |||||
|
CO8B_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 32) | NC score | 0.234655 (rank : 32) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P07358 | Gene names | C8B | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C8 beta chain precursor (Complement component 8 subunit beta). | |||||
|
CO8B_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 33) | NC score | 0.227941 (rank : 33) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8BH35, Q4VAH1, Q8CHJ5, Q8CHJ6, Q8VC14 | Gene names | C8b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement component C8 beta chain precursor (Complement component 8 subunit beta). | |||||
|
SORL_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 34) | NC score | 0.270490 (rank : 27) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q92673, Q92856 | Gene names | SORL1 | |||
|
Domain Architecture |

|
|||||
| Description | Sortilin-related receptor precursor (Sorting protein-related receptor containing LDLR class A repeats) (SorLA) (SorLA-1) (Low-density lipoprotein receptor relative with 11 ligand-binding repeats) (LDLR relative with 11 ligand-binding repeats) (LR11). | |||||
|
LRP10_MOUSE
|
||||||
| θ value | 0.125558 (rank : 35) | NC score | 0.310001 (rank : 9) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q7TQH7, Q7TS95, Q921T0, Q9EPE8 | Gene names | Lrp10, Lrp9 | |||
|
Domain Architecture |
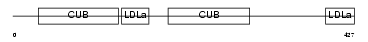
|
|||||
| Description | Low-density lipoprotein receptor-related protein 10 precursor. | |||||
|
CD320_HUMAN
|
||||||
| θ value | 0.21417 (rank : 36) | NC score | 0.348844 (rank : 5) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9NPF0, Q53HF7 | Gene names | CD320, 8D6A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CD320 antigen precursor (8D6 antigen) (FDC-signaling molecule 8D6) (FDC-SM-8D6). | |||||
|
CO7_HUMAN
|
||||||
| θ value | 0.21417 (rank : 37) | NC score | 0.196091 (rank : 37) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P10643, Q6P3T5, Q92489 | Gene names | C7 | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C7 precursor. | |||||
|
LRP5_MOUSE
|
||||||
| θ value | 0.47712 (rank : 38) | NC score | 0.294081 (rank : 13) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q91VN0, O88883, Q9R208 | Gene names | Lrp5, Lrp7 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 5 precursor (LRP7) (Lr3). | |||||
|
MINT_HUMAN
|
||||||
| θ value | 0.47712 (rank : 39) | NC score | 0.036312 (rank : 164) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MINT_MOUSE
|
||||||
| θ value | 0.47712 (rank : 40) | NC score | 0.035067 (rank : 166) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
CO8A_MOUSE
|
||||||
| θ value | 0.62314 (rank : 41) | NC score | 0.226259 (rank : 34) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8K182, Q8CHJ9, Q8CHK1 | Gene names | C8a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement component C8 alpha chain precursor (Complement component 8 subunit alpha). | |||||
|
ENTK_MOUSE
|
||||||
| θ value | 0.62314 (rank : 42) | NC score | 0.093224 (rank : 66) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P97435 | Gene names | Prss7, Entk | |||
|
Domain Architecture |

|
|||||
| Description | Enteropeptidase (EC 3.4.21.9) (Enterokinase) [Contains: Enteropeptidase non-catalytic heavy chain; Enteropeptidase catalytic light chain]. | |||||
|
GAB1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 43) | NC score | 0.070299 (rank : 73) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9QYY0, Q91VW7 | Gene names | Gab1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
TMPS9_HUMAN
|
||||||
| θ value | 0.62314 (rank : 44) | NC score | 0.085393 (rank : 67) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q7Z410, Q6ZND6, Q7Z411 | Gene names | TMPRSS9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 9 (EC 3.4.21.-) (Polyserase-1) (Polyserase-I) (Polyserine protease 1) [Contains: Serase-1; Serase-2; Serase-3]. | |||||
|
TMPS9_MOUSE
|
||||||
| θ value | 0.62314 (rank : 45) | NC score | 0.083327 (rank : 69) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P69525 | Gene names | Tmprss9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 9 (EC 3.4.21.-) (Polyserase-1) (Polyserine protease 1) (Polyserase-I) [Contains: Serase-1; Serase-2; Serase-3]. | |||||
|
CO8A_HUMAN
|
||||||
| θ value | 1.06291 (rank : 46) | NC score | 0.217326 (rank : 36) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P07357, Q13668, Q9H130 | Gene names | C8A | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C8 alpha chain precursor (Complement component 8 subunit alpha). | |||||
|
PGBM_HUMAN
|
||||||
| θ value | 1.06291 (rank : 47) | NC score | 0.114647 (rank : 60) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P98160, Q16287, Q9H3V5 | Gene names | HSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
CFAI_MOUSE
|
||||||
| θ value | 1.38821 (rank : 48) | NC score | 0.118964 (rank : 56) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q61129, Q9WU07 | Gene names | Cfi, If | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor I precursor (EC 3.4.21.45) (C3B/C4B inactivator) [Contains: Complement factor I heavy chain; Complement factor I light chain]. | |||||
|
LRP5_HUMAN
|
||||||
| θ value | 1.81305 (rank : 49) | NC score | 0.290574 (rank : 14) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O75197, Q96TD6, Q9UP66 | Gene names | LRP5, LRP7 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 5 precursor. | |||||
|
PGBM_MOUSE
|
||||||
| θ value | 1.81305 (rank : 50) | NC score | 0.119887 (rank : 55) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q05793 | Gene names | Hspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
TMPS6_MOUSE
|
||||||
| θ value | 1.81305 (rank : 51) | NC score | 0.118582 (rank : 57) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9DBI0, Q6PF94 | Gene names | Tmprss6 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 6 (EC 3.4.21.-) (Matriptase-2). | |||||
|
TMPS7_MOUSE
|
||||||
| θ value | 1.81305 (rank : 52) | NC score | 0.131370 (rank : 50) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8BIK6 | Gene names | Tmprss7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 7 precursor (EC 3.4.21.-). | |||||
|
PDZD8_HUMAN
|
||||||
| θ value | 2.36792 (rank : 53) | NC score | 0.057634 (rank : 112) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NEN9, Q86WE0, Q86WE5, Q9UFF1 | Gene names | PDZD8, PDZK8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 8 (Sarcoma antigen NY-SAR-84/NY-SAR- 104). | |||||
|
5NT1B_MOUSE
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.021115 (rank : 168) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91YE9, Q91Y48 | Gene names | Nt5c1b, Airp | |||
|
Domain Architecture |

|
|||||
| Description | Cytosolic 5'-nucleotidase 1B (EC 3.1.3.5) (Cytosolic 5'-nucleotidase IB) (cN1B) (cN-IB) (Autoimmune infertility-related protein). | |||||
|
CASR_MOUSE
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.009621 (rank : 174) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QY96, O08968, O88519, Q9QY95, Q9QZU8, Q9R1D6, Q9R1Y2 | Gene names | Casr, Gprc2a | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular calcium-sensing receptor precursor (CaSR) (Parathyroid Cell calcium-sensing receptor). | |||||
|
CFAI_HUMAN
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.122698 (rank : 53) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P05156, O60442 | Gene names | CFI, IF | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor I precursor (EC 3.4.21.45) (C3B/C4B inactivator) [Contains: Complement factor I heavy chain; Complement factor I light chain]. | |||||
|
GPR63_MOUSE
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.001264 (rank : 175) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9EQQ3 | Gene names | Gpr63 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 63 (PSP24-beta) (PSP24-2). | |||||
|
SPIT1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.145940 (rank : 47) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9R097 | Gene names | Spint1, Hai1 | |||
|
Domain Architecture |

|
|||||
| Description | Kunitz-type protease inhibitor 1 precursor (Hepatocyte growth factor activator inhibitor type 1) (HAI-1). | |||||
|
TMPS6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | 0.121621 (rank : 54) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8IU80, Q5TI06, Q6UXD8, Q8IUE2, Q8IXV8 | Gene names | TMPRSS6 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 6 (EC 3.4.21.-) (Matriptase-2). | |||||
|
ATS9_HUMAN
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.025839 (rank : 167) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9P2N4, Q9NR29 | Gene names | ADAMTS9, KIAA1312 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-9 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 9) (ADAM-TS 9) (ADAM-TS9). | |||||
|
GNAS3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.061009 (rank : 95) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z0F1, Q9QXW5, Q9Z0H2 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroendocrine secretory protein 55 (NESP55) [Contains: LHAL tetrapeptide; GPIPIRRH peptide]. | |||||
|
MCP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.017796 (rank : 170) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88174, Q9R0R9 | Gene names | Cd46, Mcp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane cofactor protein precursor (CD46 antigen). | |||||
|
SMAD4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.009630 (rank : 173) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P97471, Q9CW56 | Gene names | Smad4, Dpc4, Madh4 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 4 (SMAD 4) (Mothers against DPP homolog 4) (Deletion target in pancreatic carcinoma 4 homolog) (Smad4). | |||||
|
CELR2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.013648 (rank : 172) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9R0M0, Q99K26, Q9Z2R4 | Gene names | Celsr2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 2 precursor (Flamingo 1) (mFmi1). | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.035836 (rank : 165) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
LRP11_MOUSE
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.168885 (rank : 40) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8CB67, Q8C7Y7 | Gene names | Lrp11 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 11 precursor. | |||||
|
CELR2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 67) | NC score | 0.018162 (rank : 169) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9HCU4, Q92566 | Gene names | CELSR2, CDHF10, EGFL2, KIAA0279, MEGF3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 2 precursor (Epidermal growth factor-like 2) (Multiple epidermal growth factor-like domains 3) (Flamingo 1). | |||||
|
LPIN3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 68) | NC score | 0.014259 (rank : 171) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BQK8, Q5TDB9, Q9NPY8, Q9UJE5 | Gene names | LPIN3, LIPN3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipin-3 (Lipin 3-like). | |||||
|
BMP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.055692 (rank : 121) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P13497, Q13292, Q13872, Q14874, Q99421, Q99422, Q99423, Q9UL38 | Gene names | BMP1, PCOLC | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein 1 precursor (EC 3.4.24.19) (BMP-1) (Procollagen C-proteinase) (PCP) (Mammalian tolloid protein) (mTld). | |||||
|
BMP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.055971 (rank : 117) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P98063 | Gene names | Bmp1 | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein 1 precursor (EC 3.4.24.19) (BMP-1) (Procollagen C-proteinase) (PCP) (Mammalian tolloid protein) (mTld). | |||||
|
C1QR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.060293 (rank : 97) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NPY3, O00274 | Gene names | CD93, C1QR1, MXRA4 | |||
|
Domain Architecture |
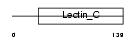
|
|||||
| Description | Complement component C1q receptor precursor (Complement component 1 q subcomponent receptor 1) (C1qR) (C1qRp) (C1qR(p)) (C1q/MBL/SPA receptor) (CD93 antigen) (CDw93). | |||||
|
C1QR1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.056638 (rank : 114) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O89103, Q3U2X0 | Gene names | Cd93, Aa4, C1qr1, C1qrp, Ly68 | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C1q receptor precursor (Complement component 1 q subcomponent receptor 1) (C1qRp) (C1qR(p)) (C1q/MBL/SPA receptor) (CD93 antigen) (Cell surface antigen AA4) (Lymphocyte antigen 68). | |||||
|
C1RA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.052393 (rank : 144) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8CG16, Q99KI6, Q9ET60 | Gene names | C1ra, C1r | |||
|
Domain Architecture |

|
|||||
| Description | Complement C1r-A subcomponent precursor (EC 3.4.21.41) (Complement component 1, r-A subcomponent) [Contains: Complement C1r-A subcomponent heavy chain; Complement C1r-A subcomponent light chain]. | |||||
|
C1RB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.051664 (rank : 150) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8CFG9 | Gene names | C1rb, C1r | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement C1r-B subcomponent precursor (EC 3.4.21.41) (Complement component 1, r-B subcomponent) [Contains: Complement C1r-B subcomponent heavy chain; Complement C1r-B subcomponent light chain]. | |||||
|
C1R_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.051555 (rank : 152) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P00736, Q8J012 | Gene names | C1R | |||
|
Domain Architecture |

|
|||||
| Description | Complement C1r subcomponent precursor (EC 3.4.21.41) (Complement component 1, r subcomponent) [Contains: Complement C1r subcomponent heavy chain; Complement C1r subcomponent light chain]. | |||||
|
CD248_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.066665 (rank : 78) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9HCU0, Q3SX55, Q96KB6 | Gene names | CD248, CD164L1, TEM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
CD248_MOUSE
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.067599 (rank : 77) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q91V98, Q3UMV6, Q91ZV1 | Gene names | Cd248, Tem1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
CREL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.052835 (rank : 141) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96HD1, Q6I9X5, Q8NFT4, Q9Y409 | Gene names | CRELD1, CIRRIN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich with EGF-like domain protein 1 precursor. | |||||
|
CREL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.053218 (rank : 135) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q91XD7, Q8BGJ8 | Gene names | Creld1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich with EGF-like domain protein 1 precursor. | |||||
|
CRUM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.050373 (rank : 162) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P82279, Q5K3A6, Q5TC28, Q5VUT1, Q6N027, Q8WWY0 | Gene names | CRB1 | |||
|
Domain Architecture |

|
|||||
| Description | Crumbs homolog 1 precursor. | |||||
|
CUBN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.055788 (rank : 119) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9JLB4 | Gene names | Cubn, Ifcr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cubilin precursor (Intrinsic factor-cobalamin receptor). | |||||
|
DLL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.052916 (rank : 140) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O00548, Q9NU41, Q9UJV2 | Gene names | DLL1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 1 precursor (Drosophila Delta homolog 1) (Delta1) (H-Delta-1). | |||||
|
DLL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.053143 (rank : 138) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61483 | Gene names | Dll1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 1 precursor (Drosophila Delta homolog 1) (Delta1). | |||||
|
EGFL7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.051005 (rank : 158) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UHF1, Q5M7Y5, Q5VUD5, Q96EG0 | Gene names | EGFL7, MEGF7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EGF-like domain-containing protein 7 precursor (Multiple EGF-like domain protein 7) (Multiple epidermal growth factor-like domain protein 7) (Vascular endothelial statin) (VE-statin) (NOTCH4-like protein) (ZNEU1). | |||||
|
EGF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.140414 (rank : 48) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P01133 | Gene names | EGF | |||
|
Domain Architecture |

|
|||||
| Description | Pro-epidermal growth factor precursor (EGF) [Contains: Epidermal growth factor (Urogastrone)]. | |||||
|
EGF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.136178 (rank : 49) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P01132, Q6P9J2 | Gene names | Egf | |||
|
Domain Architecture |

|
|||||
| Description | Pro-epidermal growth factor precursor (EGF) [Contains: Epidermal growth factor]. | |||||
|
ENTK_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.084135 (rank : 68) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P98073 | Gene names | PRSS7, ENTK | |||
|
Domain Architecture |

|
|||||
| Description | Enteropeptidase precursor (EC 3.4.21.9) (Enterokinase) [Contains: Enteropeptidase non-catalytic heavy chain; Enteropeptidase catalytic light chain]. | |||||
|
FA10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.051310 (rank : 155) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O88947, O54740, Q99L32 | Gene names | F10 | |||
|
Domain Architecture |
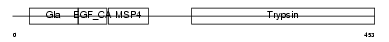
|
|||||
| Description | Coagulation factor X precursor (EC 3.4.21.6) (Stuart factor) [Contains: Factor X light chain; Factor X heavy chain; Activated factor Xa heavy chain]. | |||||
|
FA9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.052286 (rank : 146) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P00740 | Gene names | F9 | |||
|
Domain Architecture |
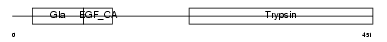
|
|||||
| Description | Coagulation factor IX precursor (EC 3.4.21.22) (Christmas factor) (Plasma thromboplastin component) (PTC) [Contains: Coagulation factor IXa light chain; Coagulation factor IXa heavy chain]. | |||||
|
FBLN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.061364 (rank : 93) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P23142, P23143, P23144, P37888, Q5TIC4, Q8TBH8, Q9HBQ5, Q9UC21, Q9UGR4, Q9UH41 | Gene names | FBLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-1 precursor. | |||||
|
FBLN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.064690 (rank : 81) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q08879, Q08878, Q8C3B1, Q91ZC9, Q922K8 | Gene names | Fbln1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-1 precursor (Basement-membrane protein 90) (BM-90). | |||||
|
FBLN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.059840 (rank : 99) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P98095 | Gene names | FBLN2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-2 precursor. | |||||
|
FBLN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.062957 (rank : 89) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P37889, Q9WUI2 | Gene names | Fbln2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-2 precursor. | |||||
|
FBLN3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.053880 (rank : 133) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q12805 | Gene names | EFEMP1, FBLN3, FBNL | |||
|
Domain Architecture |
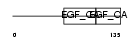
|
|||||
| Description | EGF-containing fibulin-like extracellular matrix protein 1 precursor (Fibulin-3) (FIBL-3) (Fibrillin-like protein) (Extracellular protein S1-5). | |||||
|
FBLN3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.051388 (rank : 154) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BPB5, Q6Y3N6 | Gene names | Efemp1, Fbln3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EGF-containing fibulin-like extracellular matrix protein 1 precursor (Fibulin-3) (FIBL-3). | |||||
|
FBLN4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.054122 (rank : 131) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O95967, O75967 | Gene names | EFEMP2, FBLN4 | |||
|
Domain Architecture |

|
|||||
| Description | EGF-containing fibulin-like extracellular matrix protein 2 precursor (Fibulin-4) (FIBL-4) (UPH1 protein). | |||||
|
FBLN4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.054198 (rank : 129) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9WVJ9 | Gene names | Efemp2, Fbln4, Mbp1 | |||
|
Domain Architecture |

|
|||||
| Description | EGF-containing fibulin-like extracellular matrix protein 2 precursor (Fibulin-4) (FIBL-4) (Mutant p53-binding protein 1). | |||||
|
FBLN5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.057928 (rank : 108) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UBX5, O75966, Q6UWA3 | Gene names | FBLN5, DANCE | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-5 precursor (FIBL-5) (Developmental arteries and neural crest EGF-like protein) (Dance) (Urine p50 protein) (UP50). | |||||
|
FBLN5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.057392 (rank : 113) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9WVH9, Q541Z7 | Gene names | Fbln5, Dance | |||
|
Domain Architecture |
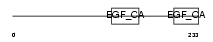
|
|||||
| Description | Fibulin-5 precursor (FIBL-5) (Developmental arteries and neural crest EGF-like protein) (Dance). | |||||
|
FBN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.059655 (rank : 101) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P35555, Q15972, Q75N87 | Gene names | FBN1, FBN | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-1 precursor. | |||||
|
FBN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.059800 (rank : 100) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 563 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61554, Q60826 | Gene names | Fbn1, Fbn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-1 precursor. | |||||
|
FBN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.058474 (rank : 106) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P35556 | Gene names | FBN2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-2 precursor. | |||||
|
FBN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.057924 (rank : 109) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61555, Q63957 | Gene names | Fbn2, Fbn-2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-2 precursor. | |||||
|
FBN3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.059205 (rank : 104) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q75N90, Q75N91, Q75N92, Q75N93, Q86SJ5, Q96JP8 | Gene names | FBN3, KIAA1776 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibrillin-3 precursor. | |||||
|
GAS6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.063975 (rank : 83) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q14393, Q7Z7N3 | Gene names | GAS6 | |||
|
Domain Architecture |

|
|||||
| Description | Growth-arrest-specific protein 6 precursor (GAS-6). | |||||
|
GAS6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.064924 (rank : 79) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q61592, Q99K57 | Gene names | Gas6 | |||
|
Domain Architecture |

|
|||||
| Description | Growth-arrest-specific protein 6 precursor (GAS-6). | |||||
|
GLU2B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.110362 (rank : 62) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P14314, Q96BU9, Q96D06, Q9P0W9 | Gene names | PRKCSH, G19P1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glucosidase 2 subunit beta precursor (Glucosidase II subunit beta) (Protein kinase C substrate, 60.1 kDa protein, heavy chain) (PKCSH) (80K-H protein). | |||||
|
GLU2B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.104247 (rank : 65) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O08795, Q921X2 | Gene names | Prkcsh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glucosidase 2 subunit beta precursor (Glucosidase II subunit beta) (Protein kinase C substrate, 60.1 kDa protein, heavy chain) (PKCSH) (80K-H protein). | |||||
|
GRN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.082583 (rank : 70) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P28799, P23781, P23782, P23783, P23784, Q9BWE7, Q9UCH0 | Gene names | GRN | |||
|
Domain Architecture |

|
|||||
| Description | Granulins precursor (Proepithelin) (PEPI) [Contains: Acrogranin; Paragranulin; Granulin-1 (Granulin G); Granulin-2 (Granulin F); Granulin-3 (Granulin B); Granulin-4 (Granulin A); Granulin-5 (Granulin C); Granulin-6 (Granulin D); Granulin-7 (Granulin E)]. | |||||
|
GRN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.059449 (rank : 103) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P28798 | Gene names | Grn | |||
|
Domain Architecture |

|
|||||
| Description | Granulins precursor (Proepithelin) (PEPI) (PC cell-derived growth factor) (PCDGF) [Contains: Acrogranin; Granulin-1; Granulin-2; Granulin-3; Granulin-4; Granulin-5; Granulin-6; Granulin-7]. | |||||
|
IDD_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.162637 (rank : 43) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P98153 | Gene names | DGCR2, IDD, KIAA0163 | |||
|
Domain Architecture |
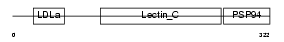
|
|||||
| Description | Integral membrane protein DGCR2/IDD precursor. | |||||
|
IDD_MOUSE
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.147889 (rank : 45) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P98154, Q61844 | Gene names | Dgcr2, Dgsc, Idd, Sez-12, Sez12 | |||
|
Domain Architecture |
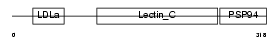
|
|||||
| Description | Integral membrane protein DGCR2/IDD precursor (Seizure-related membrane-bound adhesion protein). | |||||
|
JAG1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.050711 (rank : 160) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 514 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P78504, O14902, O15122, Q15816 | Gene names | JAG1, JAGL1 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-1 precursor (Jagged1) (hJ1) (CD339 antigen). | |||||
|
JAG1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.051646 (rank : 151) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9QXX0 | Gene names | Jag1 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-1 precursor (Jagged1) (CD339 antigen). | |||||
|
JAG2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.054500 (rank : 128) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y219, Q9UE17, Q9UE99, Q9UNK8, Q9Y6P9, Q9Y6Q0 | Gene names | JAG2 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-2 precursor (Jagged2) (HJ2). | |||||
|
JAG2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.054720 (rank : 126) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9QYE5, O55139, O70219 | Gene names | Jag2 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-2 precursor (Jagged2). | |||||
|
KR107_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.050635 (rank : 161) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P60409, Q70LJ2 | Gene names | KRTAP10-7, KAP10.7, KAP18-7, KRTAP10.7, KRTAP18-7, KRTAP18.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-7 (Keratin-associated protein 10.7) (High sulfur keratin-associated protein 10.7) (Keratin-associated protein 18-7) (Keratin-associated protein 18.7). | |||||
|
LCE3B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.053163 (rank : 136) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5TA77 | Gene names | LCE3B, LEP14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 3B (Late envelope protein 14). | |||||
|
LRP11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.151255 (rank : 44) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q86VZ4, Q5VYC0, Q96SN6 | Gene names | LRP11 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 11 precursor. | |||||
|
LTB1L_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.063125 (rank : 88) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q14766 | Gene names | LTBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1L precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
LTB1L_MOUSE
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.058945 (rank : 105) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 514 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8CG19, O88349, Q8BNW7, Q8C7F5, Q8CIR0 | Gene names | Ltbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1L precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
LTB1S_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.061311 (rank : 94) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P22064, Q8TD95 | Gene names | LTBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1S precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
LTB1S_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.057857 (rank : 111) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8CG18, Q8BNW7, Q8C7F5, Q8CIR0 | Gene names | Ltbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1S precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
LTBP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.063624 (rank : 84) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 552 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q14767, Q99907, Q9NS51 | Gene names | LTBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
LTBP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.052091 (rank : 147) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O08999, Q8C6W9 | Gene names | Ltbp2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
LTBP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.051967 (rank : 149) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9NS15, O15107, Q96HB9, Q9H7K2, Q9UFN4 | Gene names | LTBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 3 precursor (LTBP-3). | |||||
|
LTBP3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.052581 (rank : 143) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61810, Q8BNQ6 | Gene names | Ltbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 3 precursor (LTBP-3). | |||||
|
LTBP4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.053976 (rank : 132) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8K4G1, Q8K4G0 | Gene names | Ltbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 4 precursor (LTBP-4). | |||||
|
MASP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.054133 (rank : 130) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P48740, O95570, Q9UF09 | Gene names | MASP1, CRARF, CRARF1, PRSS5 | |||
|
Domain Architecture |

|
|||||
| Description | Complement-activating component of Ra-reactive factor precursor (EC 3.4.21.-) (Ra-reactive factor serine protease p100) (RaRF) (Mannan-binding lectin serine protease 1) (Mannose-binding protein- associated serine protease) (MASP-1) [Contains: Complement-activating component of Ra-reactive factor heavy chain; Complement-activating component of Ra-reactive factor light chain]. | |||||
|
MASP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.052316 (rank : 145) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P98064 | Gene names | Masp1, Crarf | |||
|
Domain Architecture |

|
|||||
| Description | Complement-activating component of Ra-reactive factor precursor (EC 3.4.21.-) (Ra-reactive factor serine protease p100) (RaRF) (Mannan-binding lectin serine protease 1) [Contains: Complement- activating component of Ra-reactive factor heavy chain; Complement- activating component of Ra-reactive factor light chain]. | |||||
|
MASP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.053303 (rank : 134) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O00187, O75754, Q5TEQ5, Q5TER0, Q96QG4, Q9BZH0, Q9H498, Q9H499, Q9UBP3, Q9ULC7, Q9UMV3, Q9Y270 | Gene names | MASP2 | |||
|
Domain Architecture |

|
|||||
| Description | Mannan-binding lectin serine protease 2 precursor (EC 3.4.21.104) (Mannose-binding protein-associated serine protease 2) (MASP-2) (MBL- associated serine protease 2) [Contains: Mannan-binding lectin serine protease 2 A chain; Mannan-binding lectin serine protease 2 B chain]. | |||||
|
MASP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.051097 (rank : 157) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q91WP0, Q9QXA4, Q9QXD2, Q9QXD5, Q9Z338 | Gene names | Masp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mannan-binding lectin serine protease 2 precursor (EC 3.4.21.104) (Mannose-binding protein-associated serine protease 2) (MASP-2) (MBL- associated serine protease 2) [Contains: Mannan-binding lectin serine protease 2 A chain; Mannan-binding lectin serine protease 2 B chain]. | |||||
|
MATN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.056512 (rank : 116) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O00339, Q6UWA5, Q7Z5X1, Q8NDE6, Q96FT5, Q9NSZ1 | Gene names | MATN2 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-2 precursor. | |||||
|
MATN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.050739 (rank : 159) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O08746 | Gene names | Matn2 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-2 precursor. | |||||
|
MATN4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.053147 (rank : 137) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O95460, Q5QPU2, Q5QPU3, Q5QPU4, Q8N2M5, Q8N2M7, Q9H1F8, Q9H1F9 | Gene names | MATN4 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-4 precursor. | |||||
|
MATN4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.052815 (rank : 142) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O89029, O89030, Q9QWS3 | Gene names | Matn4 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-4 precursor (MAT-4). | |||||
|
MEGF6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.055650 (rank : 122) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O75095, Q5VV39 | Gene names | MEGF6, EGFL3, KIAA0815 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple epidermal growth factor-like domains 6 precursor (EGF-like domain-containing protein 3) (Multiple EGF-like domain protein 3). | |||||
|
MFRP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.127706 (rank : 51) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9BY79, Q335M3, Q96DQ9 | Gene names | MFRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane frizzled-related protein (Membrane-type frizzled-related protein). | |||||
|
MFRP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.116974 (rank : 58) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8K480, Q8BPP4 | Gene names | Mfrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane frizzled-related protein (Membrane-type frizzled-related protein). | |||||
|
NELL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.050089 (rank : 163) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q92832, Q9Y472 | Gene names | NELL1, NRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein NELL1 precursor (NEL-like protein 1) (Nel-related protein 1). | |||||
|
NELL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.052032 (rank : 148) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 449 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99435 | Gene names | NELL2, NRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein NELL2 precursor (NEL-like protein 2) (Nel-related protein 2). | |||||
|
NELL2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.051148 (rank : 156) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q61220 | Gene names | Nell2, Mel91 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein NELL2 precursor (NEL-like protein 2) (MEL91 protein). | |||||
|
NETO1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.067940 (rank : 75) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8TDF5, Q86W85, Q8ND78, Q8TDF4 | Gene names | NETO1, BTCL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 1 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 1). | |||||
|
NETO1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.067649 (rank : 76) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8R4I7, Q80X39, Q8C4S3, Q8CCM2 | Gene names | Neto1, Btcl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 1 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 1). | |||||
|
NETO2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.064712 (rank : 80) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8NC67, Q7Z381, Q8ND51, Q96SP4, Q9NVY8 | Gene names | NETO2, BTCL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 2 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 2). | |||||
|
NETO2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.064484 (rank : 82) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8BNJ6, Q5VM49, Q8C4Q8 | Gene names | Neto2, Btcl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 2 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 2). | |||||
|
NID1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.113492 (rank : 61) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P14543, Q14942, Q59FL2, Q5TAF2, Q5TAF3, Q86XD7 | Gene names | NID1, NID | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-1 precursor (Entactin). | |||||
|
NID1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.114722 (rank : 59) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P10493, Q8BQI3, Q8C3U8, Q8C9P6 | Gene names | Nid1, Ent | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-1 precursor (Entactin). | |||||
|
NID2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.105986 (rank : 64) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q14112, O43710 | Gene names | NID2 | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-2 precursor (NID-2) (Osteonidogen). | |||||
|
NID2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.107593 (rank : 63) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O88322 | Gene names | Nid2 | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-2 precursor (NID-2) (Entactin-2). | |||||
|
NOTC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.051458 (rank : 153) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q01705, Q06007, Q61905, Q99JC2, Q9QW58, Q9R0X7 | Gene names | Notch1, Motch | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (Motch A) (mT14) (p300) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
PERF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.055599 (rank : 123) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P10820 | Gene names | Prf1, Pfp | |||
|
Domain Architecture |

|
|||||
| Description | Perforin-1 precursor (P1) (Lymphocyte pore-forming protein) (Cytolysin). | |||||
|
PROC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.053024 (rank : 139) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P33587, O35498, Q91WN8, Q99PC6 | Gene names | Proc | |||
|
Domain Architecture |
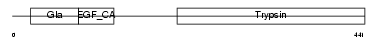
|
|||||
| Description | Vitamin K-dependent protein C precursor (EC 3.4.21.69) (Autoprothrombin IIA) (Anticoagulant protein C) (Blood coagulation factor XIV) [Contains: Vitamin K-dependent protein C light chain; Vitamin K-dependent protein C heavy chain; Activation peptide]. | |||||
|
PROS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.061963 (rank : 92) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P07225, Q15518, Q7Z715 | Gene names | PROS1, PROS | |||
|
Domain Architecture |

|
|||||
| Description | Vitamin K-dependent protein S precursor. | |||||
|
PROS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.059993 (rank : 98) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q08761, P43483 | Gene names | Pros1, Pros | |||
|
Domain Architecture |

|
|||||
| Description | Vitamin K-dependent protein S precursor. | |||||
|
PROZ_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.061986 (rank : 91) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P22891, Q15213 | Gene names | PROZ | |||
|
Domain Architecture |
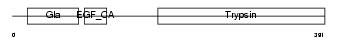
|
|||||
| Description | Vitamin K-dependent protein Z precursor. | |||||
|
PROZ_MOUSE
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.055853 (rank : 118) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9CQW3 | Gene names | Proz | |||
|
Domain Architecture |
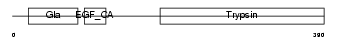
|
|||||
| Description | Vitamin K-dependent protein Z precursor. | |||||
|
RXFP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.055524 (rank : 124) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8WXD0 | Gene names | RXFP2, GPR106, GREAT, LGR8 | |||
|
Domain Architecture |

|
|||||
| Description | Relaxin receptor 2 (Relaxin family peptide receptor 2) (Leucine-rich repeat-containing G-protein coupled receptor 8) (G-protein coupled receptor affecting testicular descent) (G-protein coupled receptor 106). | |||||
|
SCUB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.074447 (rank : 71) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8IX30, Q5CZB3, Q86UZ9, Q8NAU9 | Gene names | SCUBE3, CEGF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal peptide, CUB and EGF-like domain-containing protein 3 precursor. | |||||
|
SCUB3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.074125 (rank : 72) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q66PY1, Q68FG9 | Gene names | Scube3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal peptide, CUB and EGF-like domain-containing protein 3 precursor. | |||||
|
SPIT1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.146332 (rank : 46) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O43278, Q7Z7D2 | Gene names | SPINT1, HAI1 | |||
|
Domain Architecture |

|
|||||
| Description | Kunitz-type protease inhibitor 1 precursor (Hepatocyte growth factor activator inhibitor type 1) (HAI-1). | |||||
|
TENA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.063506 (rank : 85) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 631 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P24821, Q14583, Q15567 | Gene names | TNC, HXB | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin precursor (TN) (Tenascin-C) (TN-C) (Hexabrachion) (Cytotactin) (Neuronectin) (GMEM) (JI) (Myotendinous antigen) (Glioma- associated-extracellular matrix antigen) (GP 150-225). | |||||
|
TENA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.063476 (rank : 86) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q80YX1, Q64706, Q80YX2 | Gene names | Tnc, Hxb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tenascin precursor (TN) (Tenascin-C) (TN-C) (Hexabrachion). | |||||
|
TLL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.057882 (rank : 110) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O43897, Q96AN3, Q9NQS4 | Gene names | TLL1, TLL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 1 precursor (EC 3.4.24.-). | |||||
|
TLL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.058048 (rank : 107) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q62381, Q3UTT9, Q8BNP5 | Gene names | Tll1, Tll | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 1 precursor (EC 3.4.24.-) (mTll). | |||||
|
TLL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.054709 (rank : 127) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y6L7, Q6PJN5, Q9UQ00 | Gene names | TLL2, KIAA0932 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 2 precursor (EC 3.4.24.-). | |||||
|
TLL2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.055782 (rank : 120) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9WVM6 | Gene names | Tll2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 2 precursor (EC 3.4.24.-). | |||||
|
TMPS2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.056634 (rank : 115) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O15393, Q9BXX1 | Gene names | TMPRSS2, PRSS10 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 2 precursor (EC 3.4.21.-) [Contains: Transmembrane protease, serine 2 non-catalytic chain; Transmembrane protease, serine 2 catalytic chain]. | |||||
|
TMPS2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.063399 (rank : 87) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9JIQ8, Q9JKC4, Q9QY82 | Gene names | Tmprss2 | |||
|
Domain Architecture |
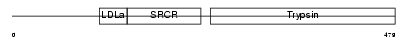
|
|||||
| Description | Transmembrane protease, serine 2 (EC 3.4.21.-) (Epitheliasin) (Plasmic transmembrane protein X) [Contains: Transmembrane protease, serine 2 non-catalytic chain; Transmembrane protease, serine 2 catalytic chain]. | |||||
|
TMPS3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.059637 (rank : 102) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P57727, Q6ZMC3 | Gene names | TMPRSS3, ECHOS1, TADG12 | |||
|
Domain Architecture |
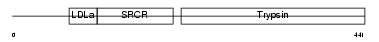
|
|||||
| Description | Transmembrane protease, serine 3 (EC 3.4.21.-) (Serine protease TADG- 12) (Tumor-associated differentially-expressed gene 12 protein). | |||||
|
TMPS3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.054814 (rank : 125) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8K1T0, Q8VDE0 | Gene names | Tmprss3 | |||
|
Domain Architecture |
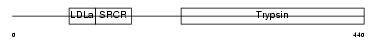
|
|||||
| Description | Transmembrane protease, serine 3 (EC 3.4.21.-). | |||||
|
TMPS7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.125001 (rank : 52) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q7RTY8 | Gene names | TMPRSS7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 7 precursor (EC 3.4.21.-). | |||||
|
TRBM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.060963 (rank : 96) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P07204, Q9UC32 | Gene names | THBD, THRM | |||
|
Domain Architecture |
|
|||||
| Description | Thrombomodulin precursor (TM) (Fetomodulin) (CD141 antigen). | |||||
|
TRBM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.062590 (rank : 90) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P15306 | Gene names | Thbd | |||
|
Domain Architecture |
|
|||||
| Description | Thrombomodulin precursor (TM) (Fetomodulin) (CD141 antigen). | |||||
|
ZAN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.069270 (rank : 74) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
CR001_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 6.35588e-163 (rank : 1) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O15165, O15166, O15167, O15168, Q6NXP3 | Gene names | C18orf1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C18orf1. | |||||
|
CR001_MOUSE
|
||||||
| NC score | 0.994703 (rank : 2) | θ value | 4.26331e-159 (rank : 2) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8BWJ4 | Gene names | D18Ertd653e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C18orf1 homolog. | |||||
|
TMEPA_MOUSE
|
||||||
| NC score | 0.890232 (rank : 3) | θ value | 1.95861e-79 (rank : 4) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9D7R2, Q9EQH9 | Gene names | Tmepai, N4wbp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane prostate androgen-induced protein (NEDD4 WW domain- binding protein 4). | |||||
|
TMEPA_HUMAN
|
||||||
| NC score | 0.886250 (rank : 4) | θ value | 3.56943e-81 (rank : 3) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q969W9, Q5TDR6, Q96B72, Q9UJD3 | Gene names | TMEPAI, PMEPA1, STAG1 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane prostate androgen-induced protein (Solid tumor- associated 1 protein). | |||||
|
CD320_HUMAN
|
||||||
| NC score | 0.348844 (rank : 5) | θ value | 0.21417 (rank : 36) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9NPF0, Q53HF7 | Gene names | CD320, 8D6A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CD320 antigen precursor (8D6 antigen) (FDC-signaling molecule 8D6) (FDC-SM-8D6). | |||||
|
LRP10_HUMAN
|
||||||
| NC score | 0.319069 (rank : 6) | θ value | 0.0252991 (rank : 31) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q7Z4F1, O95882, Q86T02, Q8NCZ4, Q9HC42, Q9UG33 | Gene names | LRP10 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 10 precursor. | |||||
|
CO9_HUMAN
|
||||||
| NC score | 0.314248 (rank : 7) | θ value | 0.00390308 (rank : 20) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P02748 | Gene names | C9 | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C9 precursor [Contains: Complement component C9a; Complement component C9b]. | |||||
|
LRP3_HUMAN
|
||||||
| NC score | 0.312450 (rank : 8) | θ value | 0.00134147 (rank : 15) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O75074 | Gene names | LRP3 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 3 precursor (hLRp105). | |||||
|
LRP10_MOUSE
|
||||||
| NC score | 0.310001 (rank : 9) | θ value | 0.125558 (rank : 35) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q7TQH7, Q7TS95, Q921T0, Q9EPE8 | Gene names | Lrp10, Lrp9 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 10 precursor. | |||||
|
LRP6_MOUSE
|
||||||
| NC score | 0.304713 (rank : 10) | θ value | 0.00869519 (rank : 25) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O88572 | Gene names | Lrp6 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 6 precursor. | |||||
|
LRP6_HUMAN
|
||||||
| NC score | 0.304261 (rank : 11) | θ value | 0.00869519 (rank : 24) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O75581 | Gene names | LRP6 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 6 precursor. | |||||
|
CO9_MOUSE
|
||||||
| NC score | 0.295447 (rank : 12) | θ value | 0.0113563 (rank : 26) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P06683, Q91XA7 | Gene names | C9 | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C9 precursor. | |||||
|
LRP5_MOUSE
|
||||||
| NC score | 0.294081 (rank : 13) | θ value | 0.47712 (rank : 38) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q91VN0, O88883, Q9R208 | Gene names | Lrp5, Lrp7 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 5 precursor (LRP7) (Lr3). | |||||
|
LRP5_HUMAN
|
||||||
| NC score | 0.290574 (rank : 14) | θ value | 1.81305 (rank : 49) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O75197, Q96TD6, Q9UP66 | Gene names | LRP5, LRP7 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 5 precursor. | |||||
|
LRP4_HUMAN
|
||||||
| NC score | 0.289933 (rank : 15) | θ value | 0.000786445 (rank : 10) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O75096 | Gene names | LRP4, KIAA0816, MEGF7 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 4 precursor (Multiple epidermal growth factor-like domains 7). | |||||
|
LRP4_MOUSE
|
||||||
| NC score | 0.289603 (rank : 16) | θ value | 0.000602161 (rank : 8) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8VI56, Q8BPX5, Q8CBB3, Q8CCP5 | Gene names | Lrp4, Kiaa0816 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 4 precursor (LDLR dan). | |||||
|
LRP12_MOUSE
|
||||||
| NC score | 0.289407 (rank : 17) | θ value | 0.00102713 (rank : 13) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8BUJ9, Q8BWM9 | Gene names | Lrp12 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 12 precursor. | |||||
|
LRP12_HUMAN
|
||||||
| NC score | 0.289346 (rank : 18) | θ value | 0.00102713 (rank : 12) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Y561 | Gene names | LRP12, ST7 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 12 precursor (Suppressor of tumorigenicity protein 7). | |||||
|
VLDLR_HUMAN
|
||||||
| NC score | 0.288996 (rank : 19) | θ value | 0.00228821 (rank : 19) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P98155, Q5VVF6 | Gene names | VLDLR | |||
|
Domain Architecture |

|
|||||
| Description | Very low-density lipoprotein receptor precursor (VLDL receptor) (VLDL- R). | |||||
|
LDLR_MOUSE
|
||||||
| NC score | 0.287315 (rank : 20) | θ value | 0.00509761 (rank : 23) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P35951 | Gene names | Ldlr | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor precursor (LDL receptor). | |||||
|
VLDLR_MOUSE
|
||||||
| NC score | 0.286376 (rank : 21) | θ value | 0.00390308 (rank : 22) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P98156, Q64022 | Gene names | Vldlr | |||
|
Domain Architecture |

|
|||||
| Description | Very low-density lipoprotein receptor precursor (VLDL receptor) (VLDL- R). | |||||
|
LRP2_HUMAN
|
||||||
| NC score | 0.284353 (rank : 22) | θ value | 0.00020696 (rank : 7) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P98164, O00711, Q16215 | Gene names | LRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 2 precursor (Megalin) (Glycoprotein 330) (gp330). | |||||
|
LRP8_MOUSE
|
||||||
| NC score | 0.282565 (rank : 23) | θ value | 0.00228821 (rank : 18) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q924X6, Q8CAK9, Q8CDF5, Q921B6 | Gene names | Lrp8, Apoer2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 8 precursor (Apolipoprotein E receptor 2). | |||||
|
LRP8_HUMAN
|
||||||
| NC score | 0.282025 (rank : 24) | θ value | 0.0193708 (rank : 29) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q14114, O14968, Q86V27, Q99876, Q9BR78 | Gene names | LRP8, APOER2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 8 precursor (Apolipoprotein E receptor 2). | |||||
|
LDLR_HUMAN
|
||||||
| NC score | 0.279598 (rank : 25) | θ value | 0.0113563 (rank : 27) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P01130, Q53ZD9 | Gene names | LDLR | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor precursor (LDL receptor). | |||||
|
SORL_MOUSE
|
||||||
| NC score | 0.271863 (rank : 26) | θ value | 0.0193708 (rank : 30) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O88307, O54711, O70581 | Gene names | Sorl1 | |||
|
Domain Architecture |

|
|||||
| Description | Sortilin-related receptor precursor (Sorting protein-related receptor containing LDLR class A repeats) (mSorLA) (SorLA-1) (Low-density lipoprotein receptor relative with 11 ligand-binding repeats) (LDLR relative with 11 ligand-binding repeats) (LR11) (Gp250). | |||||
|
SORL_HUMAN
|
||||||
| NC score | 0.270490 (rank : 27) | θ value | 0.0431538 (rank : 34) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q92673, Q92856 | Gene names | SORL1 | |||
|
Domain Architecture |

|
|||||
| Description | Sortilin-related receptor precursor (Sorting protein-related receptor containing LDLR class A repeats) (SorLA) (SorLA-1) (Low-density lipoprotein receptor relative with 11 ligand-binding repeats) (LDLR relative with 11 ligand-binding repeats) (LR11). | |||||
|
LRP1B_MOUSE
|
||||||
| NC score | 0.262981 (rank : 28) | θ value | 0.00175202 (rank : 17) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 606 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9JI18, Q8BZD3, Q8BZM7 | Gene names | Lrp1b, Lrpdit | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1B precursor (Low- density lipoprotein receptor-related protein-deleted in tumor) (LRP- DIT). | |||||
|
LRP1_HUMAN
|
||||||
| NC score | 0.262814 (rank : 29) | θ value | 0.000786445 (rank : 9) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q07954, Q2PP12, Q8IVG8 | Gene names | LRP1, A2MR | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1 precursor (LRP) (Alpha-2-macroglobulin receptor) (A2MR) (Apolipoprotein E receptor) (APOER) (CD91 antigen). | |||||
|
LRP1B_HUMAN
|
||||||
| NC score | 0.262403 (rank : 30) | θ value | 0.00175202 (rank : 16) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 602 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9NZR2, Q8WY29, Q8WY30, Q8WY31 | Gene names | LRP1B, LRPDIT | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1B precursor (Low- density lipoprotein receptor-related protein-deleted in tumor) (LRP- DIT). | |||||
|
CO6_HUMAN
|
||||||
| NC score | 0.245451 (rank : 31) | θ value | 0.000121331 (rank : 6) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P13671 | Gene names | C6 | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C6 precursor. | |||||
|
CO8B_HUMAN
|
||||||
| NC score | 0.234655 (rank : 32) | θ value | 0.0431538 (rank : 32) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P07358 | Gene names | C8B | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C8 beta chain precursor (Complement component 8 subunit beta). | |||||
|
CO8B_MOUSE
|
||||||
| NC score | 0.227941 (rank : 33) | θ value | 0.0431538 (rank : 33) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8BH35, Q4VAH1, Q8CHJ5, Q8CHJ6, Q8VC14 | Gene names | C8b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement component C8 beta chain precursor (Complement component 8 subunit beta). | |||||
|
CO8A_MOUSE
|
||||||
| NC score | 0.226259 (rank : 34) | θ value | 0.62314 (rank : 41) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8K182, Q8CHJ9, Q8CHK1 | Gene names | C8a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement component C8 alpha chain precursor (Complement component 8 subunit alpha). | |||||
|
SSPO_MOUSE
|
||||||
| NC score | 0.219731 (rank : 35) | θ value | 9.29e-05 (rank : 5) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
CO8A_HUMAN
|
||||||
| NC score | 0.217326 (rank : 36) | θ value | 1.06291 (rank : 46) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P07357, Q13668, Q9H130 | Gene names | C8A | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C8 alpha chain precursor (Complement component 8 subunit alpha). | |||||
|
CO7_HUMAN
|
||||||
| NC score | 0.196091 (rank : 37) | θ value | 0.21417 (rank : 37) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P10643, Q6P3T5, Q92489 | Gene names | C7 | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C7 precursor. | |||||
|
CORIN_HUMAN
|
||||||
| NC score | 0.174376 (rank : 38) | θ value | 0.00134147 (rank : 14) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9Y5Q5, Q9UHY2 | Gene names | CORIN, CRN, TMPRSS10 | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuteric peptide-converting enzyme (EC 3.4.21.-) (pro-ANP- converting enzyme) (Corin) (Heart-specific serine proteinase ATC2) (Transmembrane protease, serine 10). | |||||
|
CORIN_MOUSE
|
||||||
| NC score | 0.174344 (rank : 39) | θ value | 0.00102713 (rank : 11) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9Z319 | Gene names | Corin, Crn, Lrp4 | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuteric peptide-converting enzyme (EC 3.4.21.-) (pro-ANP- converting enzyme) (Corin) (Low density lipoprotein receptor-related protein 4). | |||||
|
LRP11_MOUSE
|
||||||
| NC score | 0.168885 (rank : 40) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8CB67, Q8C7Y7 | Gene names | Lrp11 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 11 precursor. | |||||
|
ST14_MOUSE
|
||||||
| NC score | 0.168330 (rank : 41) | θ value | 0.00390308 (rank : 21) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P56677 | Gene names | St14, Prss14 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of tumorigenicity protein 14 (EC 3.4.21.-) (Serine protease 14) (Epithin). | |||||
|
ST14_HUMAN
|
||||||
| NC score | 0.167090 (rank : 42) | θ value | 0.0113563 (rank : 28) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9Y5Y6, Q9BS01, Q9H3S0, Q9HB36, Q9HCA3 | Gene names | ST14, PRSS14, SNC19, TADG15 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of tumorigenicity protein 14 (EC 3.4.21.-) (Serine protease 14) (Matriptase) (Membrane-type serine protease 1) (MT-SP1) (Prostamin) (Serine protease TADG-15) (Tumor-associated differentially-expressed gene 15 protein). | |||||
|
IDD_HUMAN
|
||||||
| NC score | 0.162637 (rank : 43) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P98153 | Gene names | DGCR2, IDD, KIAA0163 | |||
|
Domain Architecture |

|
|||||
| Description | Integral membrane protein DGCR2/IDD precursor. | |||||
|
LRP11_HUMAN
|
||||||
| NC score | 0.151255 (rank : 44) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q86VZ4, Q5VYC0, Q96SN6 | Gene names | LRP11 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 11 precursor. | |||||
|
IDD_MOUSE
|
||||||
| NC score | 0.147889 (rank : 45) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P98154, Q61844 | Gene names | Dgcr2, Dgsc, Idd, Sez-12, Sez12 | |||
|
Domain Architecture |

|
|||||
| Description | Integral membrane protein DGCR2/IDD precursor (Seizure-related membrane-bound adhesion protein). | |||||
|
SPIT1_HUMAN
|
||||||
| NC score | 0.146332 (rank : 46) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O43278, Q7Z7D2 | Gene names | SPINT1, HAI1 | |||
|
Domain Architecture |

|
|||||
| Description | Kunitz-type protease inhibitor 1 precursor (Hepatocyte growth factor activator inhibitor type 1) (HAI-1). | |||||
|
SPIT1_MOUSE
|
||||||
| NC score | 0.145940 (rank : 47) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9R097 | Gene names | Spint1, Hai1 | |||
|
Domain Architecture |

|
|||||
| Description | Kunitz-type protease inhibitor 1 precursor (Hepatocyte growth factor activator inhibitor type 1) (HAI-1). | |||||
|
EGF_HUMAN
|
||||||
| NC score | 0.140414 (rank : 48) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P01133 | Gene names | EGF | |||
|
Domain Architecture |

|
|||||
| Description | Pro-epidermal growth factor precursor (EGF) [Contains: Epidermal growth factor (Urogastrone)]. | |||||
|
EGF_MOUSE
|
||||||
| NC score | 0.136178 (rank : 49) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P01132, Q6P9J2 | Gene names | Egf | |||
|
Domain Architecture |

|
|||||
| Description | Pro-epidermal growth factor precursor (EGF) [Contains: Epidermal growth factor]. | |||||
|
TMPS7_MOUSE
|
||||||
| NC score | 0.131370 (rank : 50) | θ value | 1.81305 (rank : 52) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8BIK6 | Gene names | Tmprss7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 7 precursor (EC 3.4.21.-). | |||||
|
MFRP_HUMAN
|
||||||
| NC score | 0.127706 (rank : 51) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9BY79, Q335M3, Q96DQ9 | Gene names | MFRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane frizzled-related protein (Membrane-type frizzled-related protein). | |||||
|
TMPS7_HUMAN
|
||||||
| NC score | 0.125001 (rank : 52) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q7RTY8 | Gene names | TMPRSS7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 7 precursor (EC 3.4.21.-). | |||||
|
CFAI_HUMAN
|
||||||
| NC score | 0.122698 (rank : 53) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P05156, O60442 | Gene names | CFI, IF | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor I precursor (EC 3.4.21.45) (C3B/C4B inactivator) [Contains: Complement factor I heavy chain; Complement factor I light chain]. | |||||
|
TMPS6_HUMAN
|
||||||
| NC score | 0.121621 (rank : 54) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8IU80, Q5TI06, Q6UXD8, Q8IUE2, Q8IXV8 | Gene names | TMPRSS6 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 6 (EC 3.4.21.-) (Matriptase-2). | |||||
|
PGBM_MOUSE
|
||||||
| NC score | 0.119887 (rank : 55) | θ value | 1.81305 (rank : 50) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q05793 | Gene names | Hspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
CFAI_MOUSE
|
||||||
| NC score | 0.118964 (rank : 56) | θ value | 1.38821 (rank : 48) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q61129, Q9WU07 | Gene names | Cfi, If | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor I precursor (EC 3.4.21.45) (C3B/C4B inactivator) [Contains: Complement factor I heavy chain; Complement factor I light chain]. | |||||
|
TMPS6_MOUSE
|
||||||
| NC score | 0.118582 (rank : 57) | θ value | 1.81305 (rank : 51) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9DBI0, Q6PF94 | Gene names | Tmprss6 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 6 (EC 3.4.21.-) (Matriptase-2). | |||||
|
MFRP_MOUSE
|
||||||
| NC score | 0.116974 (rank : 58) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8K480, Q8BPP4 | Gene names | Mfrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane frizzled-related protein (Membrane-type frizzled-related protein). | |||||
|
NID1_MOUSE
|
||||||
| NC score | 0.114722 (rank : 59) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P10493, Q8BQI3, Q8C3U8, Q8C9P6 | Gene names | Nid1, Ent | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-1 precursor (Entactin). | |||||
|
PGBM_HUMAN
|
||||||
| NC score | 0.114647 (rank : 60) | θ value | 1.06291 (rank : 47) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P98160, Q16287, Q9H3V5 | Gene names | HSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
NID1_HUMAN
|
||||||
| NC score | 0.113492 (rank : 61) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P14543, Q14942, Q59FL2, Q5TAF2, Q5TAF3, Q86XD7 | Gene names | NID1, NID | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-1 precursor (Entactin). | |||||
|
GLU2B_HUMAN
|
||||||
| NC score | 0.110362 (rank : 62) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P14314, Q96BU9, Q96D06, Q9P0W9 | Gene names | PRKCSH, G19P1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glucosidase 2 subunit beta precursor (Glucosidase II subunit beta) (Protein kinase C substrate, 60.1 kDa protein, heavy chain) (PKCSH) (80K-H protein). | |||||
|
NID2_MOUSE
|
||||||
| NC score | 0.107593 (rank : 63) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O88322 | Gene names | Nid2 | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-2 precursor (NID-2) (Entactin-2). | |||||
|
NID2_HUMAN
|
||||||
| NC score | 0.105986 (rank : 64) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q14112, O43710 | Gene names | NID2 | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-2 precursor (NID-2) (Osteonidogen). | |||||
|
GLU2B_MOUSE
|
||||||
| NC score | 0.104247 (rank : 65) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O08795, Q921X2 | Gene names | Prkcsh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glucosidase 2 subunit beta precursor (Glucosidase II subunit beta) (Protein kinase C substrate, 60.1 kDa protein, heavy chain) (PKCSH) (80K-H protein). | |||||
|
ENTK_MOUSE
|
||||||
| NC score | 0.093224 (rank : 66) | θ value | 0.62314 (rank : 42) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P97435 | Gene names | Prss7, Entk | |||
|
Domain Architecture |

|
|||||
| Description | Enteropeptidase (EC 3.4.21.9) (Enterokinase) [Contains: Enteropeptidase non-catalytic heavy chain; Enteropeptidase catalytic light chain]. | |||||
|
TMPS9_HUMAN
|
||||||
| NC score | 0.085393 (rank : 67) | θ value | 0.62314 (rank : 44) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q7Z410, Q6ZND6, Q7Z411 | Gene names | TMPRSS9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 9 (EC 3.4.21.-) (Polyserase-1) (Polyserase-I) (Polyserine protease 1) [Contains: Serase-1; Serase-2; Serase-3]. | |||||
|
ENTK_HUMAN
|
||||||
| NC score | 0.084135 (rank : 68) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P98073 | Gene names | PRSS7, ENTK | |||
|
Domain Architecture |

|
|||||
| Description | Enteropeptidase precursor (EC 3.4.21.9) (Enterokinase) [Contains: Enteropeptidase non-catalytic heavy chain; Enteropeptidase catalytic light chain]. | |||||
|
TMPS9_MOUSE
|
||||||
| NC score | 0.083327 (rank : 69) | θ value | 0.62314 (rank : 45) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P69525 | Gene names | Tmprss9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 9 (EC 3.4.21.-) (Polyserase-1) (Polyserine protease 1) (Polyserase-I) [Contains: Serase-1; Serase-2; Serase-3]. | |||||
|
GRN_HUMAN
|
||||||
| NC score | 0.082583 (rank : 70) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P28799, P23781, P23782, P23783, P23784, Q9BWE7, Q9UCH0 | Gene names | GRN | |||
|
Domain Architecture |

|
|||||
| Description | Granulins precursor (Proepithelin) (PEPI) [Contains: Acrogranin; Paragranulin; Granulin-1 (Granulin G); Granulin-2 (Granulin F); Granulin-3 (Granulin B); Granulin-4 (Granulin A); Granulin-5 (Granulin C); Granulin-6 (Granulin D); Granulin-7 (Granulin E)]. | |||||
|
SCUB3_HUMAN
|
||||||
| NC score | 0.074447 (rank : 71) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8IX30, Q5CZB3, Q86UZ9, Q8NAU9 | Gene names | SCUBE3, CEGF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal peptide, CUB and EGF-like domain-containing protein 3 precursor. | |||||
|
SCUB3_MOUSE
|
||||||
| NC score | 0.074125 (rank : 72) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q66PY1, Q68FG9 | Gene names | Scube3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal peptide, CUB and EGF-like domain-containing protein 3 precursor. | |||||
|
GAB1_MOUSE
|
||||||
| NC score | 0.070299 (rank : 73) | θ value | 0.62314 (rank : 43) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9QYY0, Q91VW7 | Gene names | Gab1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
ZAN_MOUSE
|
||||||
| NC score | 0.069270 (rank : 74) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
NETO1_HUMAN
|
||||||
| NC score | 0.067940 (rank : 75) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8TDF5, Q86W85, Q8ND78, Q8TDF4 | Gene names | NETO1, BTCL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 1 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 1). | |||||
|
NETO1_MOUSE
|
||||||
| NC score | 0.067649 (rank : 76) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8R4I7, Q80X39, Q8C4S3, Q8CCM2 | Gene names | Neto1, Btcl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 1 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 1). | |||||
|
CD248_MOUSE
|
||||||
| NC score | 0.067599 (rank : 77) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q91V98, Q3UMV6, Q91ZV1 | Gene names | Cd248, Tem1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
CD248_HUMAN
|
||||||
| NC score | 0.066665 (rank : 78) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9HCU0, Q3SX55, Q96KB6 | Gene names | CD248, CD164L1, TEM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
GAS6_MOUSE
|
||||||
| NC score | 0.064924 (rank : 79) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q61592, Q99K57 | Gene names | Gas6 | |||
|
Domain Architecture |

|
|||||
| Description | Growth-arrest-specific protein 6 precursor (GAS-6). | |||||
|
NETO2_HUMAN
|
||||||
| NC score | 0.064712 (rank : 80) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8NC67, Q7Z381, Q8ND51, Q96SP4, Q9NVY8 | Gene names | NETO2, BTCL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 2 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 2). | |||||
|
FBLN1_MOUSE
|
||||||
| NC score | 0.064690 (rank : 81) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q08879, Q08878, Q8C3B1, Q91ZC9, Q922K8 | Gene names | Fbln1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-1 precursor (Basement-membrane protein 90) (BM-90). | |||||
|
NETO2_MOUSE
|
||||||
| NC score | 0.064484 (rank : 82) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8BNJ6, Q5VM49, Q8C4Q8 | Gene names | Neto2, Btcl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 2 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 2). | |||||
|
GAS6_HUMAN
|
||||||
| NC score | 0.063975 (rank : 83) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q14393, Q7Z7N3 | Gene names | GAS6 | |||
|
Domain Architecture |

|
|||||
| Description | Growth-arrest-specific protein 6 precursor (GAS-6). | |||||
|
LTBP2_HUMAN
|
||||||
| NC score | 0.063624 (rank : 84) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 552 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q14767, Q99907, Q9NS51 | Gene names | LTBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
TENA_HUMAN
|
||||||
| NC score | 0.063506 (rank : 85) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 631 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P24821, Q14583, Q15567 | Gene names | TNC, HXB | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin precursor (TN) (Tenascin-C) (TN-C) (Hexabrachion) (Cytotactin) (Neuronectin) (GMEM) (JI) (Myotendinous antigen) (Glioma- associated-extracellular matrix antigen) (GP 150-225). | |||||
|
TENA_MOUSE
|
||||||
| NC score | 0.063476 (rank : 86) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q80YX1, Q64706, Q80YX2 | Gene names | Tnc, Hxb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tenascin precursor (TN) (Tenascin-C) (TN-C) (Hexabrachion). | |||||
|
TMPS2_MOUSE
|
||||||
| NC score | 0.063399 (rank : 87) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9JIQ8, Q9JKC4, Q9QY82 | Gene names | Tmprss2 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 2 (EC 3.4.21.-) (Epitheliasin) (Plasmic transmembrane protein X) [Contains: Transmembrane protease, serine 2 non-catalytic chain; Transmembrane protease, serine 2 catalytic chain]. | |||||
|
LTB1L_HUMAN
|
||||||
| NC score | 0.063125 (rank : 88) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q14766 | Gene names | LTBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1L precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
FBLN2_MOUSE
|
||||||
| NC score | 0.062957 (rank : 89) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P37889, Q9WUI2 | Gene names | Fbln2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-2 precursor. | |||||
|
TRBM_MOUSE
|
||||||
| NC score | 0.062590 (rank : 90) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P15306 | Gene names | Thbd | |||
|
Domain Architecture |

|
|||||
| Description | Thrombomodulin precursor (TM) (Fetomodulin) (CD141 antigen). | |||||
|
PROZ_HUMAN
|
||||||
| NC score | 0.061986 (rank : 91) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P22891, Q15213 | Gene names | PROZ | |||
|
Domain Architecture |

|
|||||
| Description | Vitamin K-dependent protein Z precursor. | |||||
|
PROS_HUMAN
|
||||||
| NC score | 0.061963 (rank : 92) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P07225, Q15518, Q7Z715 | Gene names | PROS1, PROS | |||
|
Domain Architecture |

|
|||||
| Description | Vitamin K-dependent protein S precursor. | |||||
|
FBLN1_HUMAN
|
||||||
| NC score | 0.061364 (rank : 93) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P23142, P23143, P23144, P37888, Q5TIC4, Q8TBH8, Q9HBQ5, Q9UC21, Q9UGR4, Q9UH41 | Gene names | FBLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-1 precursor. | |||||
|
LTB1S_HUMAN
|
||||||
| NC score | 0.061311 (rank : 94) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P22064, Q8TD95 | Gene names | LTBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1S precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
GNAS3_MOUSE
|
||||||
| NC score | 0.061009 (rank : 95) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z0F1, Q9QXW5, Q9Z0H2 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroendocrine secretory protein 55 (NESP55) [Contains: LHAL tetrapeptide; GPIPIRRH peptide]. | |||||
|
TRBM_HUMAN
|
||||||
| NC score | 0.060963 (rank : 96) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P07204, Q9UC32 | Gene names | THBD, THRM | |||
|
Domain Architecture |

|
|||||
| Description | Thrombomodulin precursor (TM) (Fetomodulin) (CD141 antigen). | |||||
|
C1QR1_HUMAN
|
||||||
| NC score | 0.060293 (rank : 97) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NPY3, O00274 | Gene names | CD93, C1QR1, MXRA4 | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C1q receptor precursor (Complement component 1 q subcomponent receptor 1) (C1qR) (C1qRp) (C1qR(p)) (C1q/MBL/SPA receptor) (CD93 antigen) (CDw93). | |||||
|
PROS_MOUSE
|
||||||
| NC score | 0.059993 (rank : 98) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q08761, P43483 | Gene names | Pros1, Pros | |||
|
Domain Architecture |

|
|||||
| Description | Vitamin K-dependent protein S precursor. | |||||
|
FBLN2_HUMAN
|
||||||
| NC score | 0.059840 (rank : 99) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P98095 | Gene names | FBLN2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-2 precursor. | |||||
|
FBN1_MOUSE
|
||||||
| NC score | 0.059800 (rank : 100) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 563 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61554, Q60826 | Gene names | Fbn1, Fbn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-1 precursor. | |||||
|
FBN1_HUMAN
|
||||||
| NC score | 0.059655 (rank : 101) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P35555, Q15972, Q75N87 | Gene names | FBN1, FBN | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-1 precursor. | |||||
|
TMPS3_HUMAN
|
||||||
| NC score | 0.059637 (rank : 102) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P57727, Q6ZMC3 | Gene names | TMPRSS3, ECHOS1, TADG12 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 3 (EC 3.4.21.-) (Serine protease TADG- 12) (Tumor-associated differentially-expressed gene 12 protein). | |||||
|
GRN_MOUSE
|
||||||
| NC score | 0.059449 (rank : 103) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P28798 | Gene names | Grn | |||
|
Domain Architecture |

|
|||||
| Description | Granulins precursor (Proepithelin) (PEPI) (PC cell-derived growth factor) (PCDGF) [Contains: Acrogranin; Granulin-1; Granulin-2; Granulin-3; Granulin-4; Granulin-5; Granulin-6; Granulin-7]. | |||||
|
FBN3_HUMAN
|
||||||
| NC score | 0.059205 (rank : 104) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q75N90, Q75N91, Q75N92, Q75N93, Q86SJ5, Q96JP8 | Gene names | FBN3, KIAA1776 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibrillin-3 precursor. | |||||
|
LTB1L_MOUSE
|
||||||
| NC score | 0.058945 (rank : 105) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 514 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8CG19, O88349, Q8BNW7, Q8C7F5, Q8CIR0 | Gene names | Ltbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1L precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
FBN2_HUMAN
|
||||||
| NC score | 0.058474 (rank : 106) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P35556 | Gene names | FBN2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-2 precursor. | |||||
|
TLL1_MOUSE
|
||||||
| NC score | 0.058048 (rank : 107) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q62381, Q3UTT9, Q8BNP5 | Gene names | Tll1, Tll | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 1 precursor (EC 3.4.24.-) (mTll). | |||||
|
FBLN5_HUMAN
|
||||||
| NC score | 0.057928 (rank : 108) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UBX5, O75966, Q6UWA3 | Gene names | FBLN5, DANCE | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-5 precursor (FIBL-5) (Developmental arteries and neural crest EGF-like protein) (Dance) (Urine p50 protein) (UP50). | |||||
|
FBN2_MOUSE
|
||||||
| NC score | 0.057924 (rank : 109) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61555, Q63957 | Gene names | Fbn2, Fbn-2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-2 precursor. | |||||
|
TLL1_HUMAN
|
||||||
| NC score | 0.057882 (rank : 110) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O43897, Q96AN3, Q9NQS4 | Gene names | TLL1, TLL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 1 precursor (EC 3.4.24.-). | |||||
|
LTB1S_MOUSE
|
||||||
| NC score | 0.057857 (rank : 111) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8CG18, Q8BNW7, Q8C7F5, Q8CIR0 | Gene names | Ltbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1S precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
PDZD8_HUMAN
|
||||||
| NC score | 0.057634 (rank : 112) | θ value | 2.36792 (rank : 53) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NEN9, Q86WE0, Q86WE5, Q9UFF1 | Gene names | PDZD8, PDZK8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 8 (Sarcoma antigen NY-SAR-84/NY-SAR- 104). | |||||
|
FBLN5_MOUSE
|
||||||
| NC score | 0.057392 (rank : 113) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9WVH9, Q541Z7 | Gene names | Fbln5, Dance | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-5 precursor (FIBL-5) (Developmental arteries and neural crest EGF-like protein) (Dance). | |||||
|
C1QR1_MOUSE
|
||||||
| NC score | 0.056638 (rank : 114) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O89103, Q3U2X0 | Gene names | Cd93, Aa4, C1qr1, C1qrp, Ly68 | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C1q receptor precursor (Complement component 1 q subcomponent receptor 1) (C1qRp) (C1qR(p)) (C1q/MBL/SPA receptor) (CD93 antigen) (Cell surface antigen AA4) (Lymphocyte antigen 68). | |||||
|
TMPS2_HUMAN
|
||||||
| NC score | 0.056634 (rank : 115) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O15393, Q9BXX1 | Gene names | TMPRSS2, PRSS10 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 2 precursor (EC 3.4.21.-) [Contains: Transmembrane protease, serine 2 non-catalytic chain; Transmembrane protease, serine 2 catalytic chain]. | |||||
|
MATN2_HUMAN
|
||||||
| NC score | 0.056512 (rank : 116) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O00339, Q6UWA5, Q7Z5X1, Q8NDE6, Q96FT5, Q9NSZ1 | Gene names | MATN2 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-2 precursor. | |||||
|
BMP1_MOUSE
|
||||||
| NC score | 0.055971 (rank : 117) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P98063 | Gene names | Bmp1 | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein 1 precursor (EC 3.4.24.19) (BMP-1) (Procollagen C-proteinase) (PCP) (Mammalian tolloid protein) (mTld). | |||||
|
PROZ_MOUSE
|
||||||
| NC score | 0.055853 (rank : 118) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9CQW3 | Gene names | Proz | |||
|
Domain Architecture |

|
|||||
| Description | Vitamin K-dependent protein Z precursor. | |||||
|
CUBN_MOUSE
|
||||||
| NC score | 0.055788 (rank : 119) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9JLB4 | Gene names | Cubn, Ifcr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cubilin precursor (Intrinsic factor-cobalamin receptor). | |||||
|
TLL2_MOUSE
|
||||||
| NC score | 0.055782 (rank : 120) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9WVM6 | Gene names | Tll2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 2 precursor (EC 3.4.24.-). | |||||
|
BMP1_HUMAN
|
||||||
| NC score | 0.055692 (rank : 121) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P13497, Q13292, Q13872, Q14874, Q99421, Q99422, Q99423, Q9UL38 | Gene names | BMP1, PCOLC | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein 1 precursor (EC 3.4.24.19) (BMP-1) (Procollagen C-proteinase) (PCP) (Mammalian tolloid protein) (mTld). | |||||
|
MEGF6_HUMAN
|
||||||
| NC score | 0.055650 (rank : 122) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O75095, Q5VV39 | Gene names | MEGF6, EGFL3, KIAA0815 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple epidermal growth factor-like domains 6 precursor (EGF-like domain-containing protein 3) (Multiple EGF-like domain protein 3). | |||||
|
PERF_MOUSE
|
||||||
| NC score | 0.055599 (rank : 123) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P10820 | Gene names | Prf1, Pfp | |||
|
Domain Architecture |

|
|||||
| Description | Perforin-1 precursor (P1) (Lymphocyte pore-forming protein) (Cytolysin). | |||||
|
RXFP2_HUMAN
|
||||||
| NC score | 0.055524 (rank : 124) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8WXD0 | Gene names | RXFP2, GPR106, GREAT, LGR8 | |||
|
Domain Architecture |

|
|||||
| Description | Relaxin receptor 2 (Relaxin family peptide receptor 2) (Leucine-rich repeat-containing G-protein coupled receptor 8) (G-protein coupled receptor affecting testicular descent) (G-protein coupled receptor 106). | |||||
|
TMPS3_MOUSE
|
||||||
| NC score | 0.054814 (rank : 125) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8K1T0, Q8VDE0 | Gene names | Tmprss3 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 3 (EC 3.4.21.-). | |||||
|
JAG2_MOUSE
|
||||||
| NC score | 0.054720 (rank : 126) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9QYE5, O55139, O70219 | Gene names | Jag2 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-2 precursor (Jagged2). | |||||
|
TLL2_HUMAN
|
||||||
| NC score | 0.054709 (rank : 127) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y6L7, Q6PJN5, Q9UQ00 | Gene names | TLL2, KIAA0932 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 2 precursor (EC 3.4.24.-). | |||||
|
JAG2_HUMAN
|
||||||
| NC score | 0.054500 (rank : 128) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y219, Q9UE17, Q9UE99, Q9UNK8, Q9Y6P9, Q9Y6Q0 | Gene names | JAG2 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-2 precursor (Jagged2) (HJ2). | |||||
|
FBLN4_MOUSE
|
||||||
| NC score | 0.054198 (rank : 129) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9WVJ9 | Gene names | Efemp2, Fbln4, Mbp1 | |||
|
Domain Architecture |

|
|||||
| Description | EGF-containing fibulin-like extracellular matrix protein 2 precursor (Fibulin-4) (FIBL-4) (Mutant p53-binding protein 1). | |||||
|
MASP1_HUMAN
|
||||||
| NC score | 0.054133 (rank : 130) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P48740, O95570, Q9UF09 | Gene names | MASP1, CRARF, CRARF1, PRSS5 | |||
|
Domain Architecture |

|
|||||
| Description | Complement-activating component of Ra-reactive factor precursor (EC 3.4.21.-) (Ra-reactive factor serine protease p100) (RaRF) (Mannan-binding lectin serine protease 1) (Mannose-binding protein- associated serine protease) (MASP-1) [Contains: Complement-activating component of Ra-reactive factor heavy chain; Complement-activating component of Ra-reactive factor light chain]. | |||||
|
FBLN4_HUMAN
|
||||||
| NC score | 0.054122 (rank : 131) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O95967, O75967 | Gene names | EFEMP2, FBLN4 | |||
|
Domain Architecture |

|
|||||
| Description | EGF-containing fibulin-like extracellular matrix protein 2 precursor (Fibulin-4) (FIBL-4) (UPH1 protein). | |||||
|
LTBP4_MOUSE
|
||||||
| NC score | 0.053976 (rank : 132) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8K4G1, Q8K4G0 | Gene names | Ltbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 4 precursor (LTBP-4). | |||||
|
FBLN3_HUMAN
|
||||||
| NC score | 0.053880 (rank : 133) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q12805 | Gene names | EFEMP1, FBLN3, FBNL | |||
|
Domain Architecture |

|
|||||
| Description | EGF-containing fibulin-like extracellular matrix protein 1 precursor (Fibulin-3) (FIBL-3) (Fibrillin-like protein) (Extracellular protein S1-5). | |||||
|
MASP2_HUMAN
|
||||||
| NC score | 0.053303 (rank : 134) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O00187, O75754, Q5TEQ5, Q5TER0, Q96QG4, Q9BZH0, Q9H498, Q9H499, Q9UBP3, Q9ULC7, Q9UMV3, Q9Y270 | Gene names | MASP2 | |||
|
Domain Architecture |

|
|||||
| Description | Mannan-binding lectin serine protease 2 precursor (EC 3.4.21.104) (Mannose-binding protein-associated serine protease 2) (MASP-2) (MBL- associated serine protease 2) [Contains: Mannan-binding lectin serine protease 2 A chain; Mannan-binding lectin serine protease 2 B chain]. | |||||
|
CREL1_MOUSE
|
||||||
| NC score | 0.053218 (rank : 135) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q91XD7, Q8BGJ8 | Gene names | Creld1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich with EGF-like domain protein 1 precursor. | |||||
|
LCE3B_HUMAN
|
||||||
| NC score | 0.053163 (rank : 136) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5TA77 | Gene names | LCE3B, LEP14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 3B (Late envelope protein 14). | |||||
|
MATN4_HUMAN
|
||||||
| NC score | 0.053147 (rank : 137) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O95460, Q5QPU2, Q5QPU3, Q5QPU4, Q8N2M5, Q8N2M7, Q9H1F8, Q9H1F9 | Gene names | MATN4 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-4 precursor. | |||||
|
DLL1_MOUSE
|
||||||
| NC score | 0.053143 (rank : 138) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61483 | Gene names | Dll1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 1 precursor (Drosophila Delta homolog 1) (Delta1). | |||||
|
PROC_MOUSE
|
||||||
| NC score | 0.053024 (rank : 139) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P33587, O35498, Q91WN8, Q99PC6 | Gene names | Proc | |||
|
Domain Architecture |

|
|||||
| Description | Vitamin K-dependent protein C precursor (EC 3.4.21.69) (Autoprothrombin IIA) (Anticoagulant protein C) (Blood coagulation factor XIV) [Contains: Vitamin K-dependent protein C light chain; Vitamin K-dependent protein C heavy chain; Activation peptide]. | |||||
|
DLL1_HUMAN
|
||||||
| NC score | 0.052916 (rank : 140) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O00548, Q9NU41, Q9UJV2 | Gene names | DLL1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 1 precursor (Drosophila Delta homolog 1) (Delta1) (H-Delta-1). | |||||
|
CREL1_HUMAN
|
||||||
| NC score | 0.052835 (rank : 141) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96HD1, Q6I9X5, Q8NFT4, Q9Y409 | Gene names | CRELD1, CIRRIN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich with EGF-like domain protein 1 precursor. | |||||
|
MATN4_MOUSE
|
||||||
| NC score | 0.052815 (rank : 142) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O89029, O89030, Q9QWS3 | Gene names | Matn4 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-4 precursor (MAT-4). | |||||
|
LTBP3_MOUSE
|
||||||
| NC score | 0.052581 (rank : 143) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61810, Q8BNQ6 | Gene names | Ltbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 3 precursor (LTBP-3). | |||||
|
C1RA_MOUSE
|
||||||
| NC score | 0.052393 (rank : 144) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8CG16, Q99KI6, Q9ET60 | Gene names | C1ra, C1r | |||
|
Domain Architecture |

|
|||||
| Description | Complement C1r-A subcomponent precursor (EC 3.4.21.41) (Complement component 1, r-A subcomponent) [Contains: Complement C1r-A subcomponent heavy chain; Complement C1r-A subcomponent light chain]. | |||||
|
MASP1_MOUSE
|
||||||
| NC score | 0.052316 (rank : 145) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P98064 | Gene names | Masp1, Crarf | |||
|
Domain Architecture |

|
|||||
| Description | Complement-activating component of Ra-reactive factor precursor (EC 3.4.21.-) (Ra-reactive factor serine protease p100) (RaRF) (Mannan-binding lectin serine protease 1) [Contains: Complement- activating component of Ra-reactive factor heavy chain; Complement- activating component of Ra-reactive factor light chain]. | |||||
|
FA9_HUMAN
|
||||||
| NC score | 0.052286 (rank : 146) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P00740 | Gene names | F9 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor IX precursor (EC 3.4.21.22) (Christmas factor) (Plasma thromboplastin component) (PTC) [Contains: Coagulation factor IXa light chain; Coagulation factor IXa heavy chain]. | |||||
|
LTBP2_MOUSE
|
||||||
| NC score | 0.052091 (rank : 147) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O08999, Q8C6W9 | Gene names | Ltbp2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
NELL2_HUMAN
|
||||||
| NC score | 0.052032 (rank : 148) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 449 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99435 | Gene names | NELL2, NRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein NELL2 precursor (NEL-like protein 2) (Nel-related protein 2). | |||||
|
LTBP3_HUMAN
|
||||||
| NC score | 0.051967 (rank : 149) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9NS15, O15107, Q96HB9, Q9H7K2, Q9UFN4 | Gene names | LTBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 3 precursor (LTBP-3). | |||||
|
C1RB_MOUSE
|
||||||
| NC score | 0.051664 (rank : 150) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8CFG9 | Gene names | C1rb, C1r | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement C1r-B subcomponent precursor (EC 3.4.21.41) (Complement component 1, r-B subcomponent) [Contains: Complement C1r-B subcomponent heavy chain; Complement C1r-B subcomponent light chain]. | |||||
|
JAG1_MOUSE
|
||||||
| NC score | 0.051646 (rank : 151) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9QXX0 | Gene names | Jag1 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-1 precursor (Jagged1) (CD339 antigen). | |||||
|
C1R_HUMAN
|
||||||
| NC score | 0.051555 (rank : 152) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P00736, Q8J012 | Gene names | C1R | |||
|
Domain Architecture |

|
|||||
| Description | Complement C1r subcomponent precursor (EC 3.4.21.41) (Complement component 1, r subcomponent) [Contains: Complement C1r subcomponent heavy chain; Complement C1r subcomponent light chain]. | |||||
|
NOTC1_MOUSE
|
||||||
| NC score | 0.051458 (rank : 153) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q01705, Q06007, Q61905, Q99JC2, Q9QW58, Q9R0X7 | Gene names | Notch1, Motch | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (Motch A) (mT14) (p300) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
FBLN3_MOUSE
|
||||||
| NC score | 0.051388 (rank : 154) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BPB5, Q6Y3N6 | Gene names | Efemp1, Fbln3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EGF-containing fibulin-like extracellular matrix protein 1 precursor (Fibulin-3) (FIBL-3). | |||||
|
FA10_MOUSE
|
||||||
| NC score | 0.051310 (rank : 155) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O88947, O54740, Q99L32 | Gene names | F10 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor X precursor (EC 3.4.21.6) (Stuart factor) [Contains: Factor X light chain; Factor X heavy chain; Activated factor Xa heavy chain]. | |||||
|
NELL2_MOUSE
|
||||||
| NC score | 0.051148 (rank : 156) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q61220 | Gene names | Nell2, Mel91 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein NELL2 precursor (NEL-like protein 2) (MEL91 protein). | |||||
|
MASP2_MOUSE
|
||||||
| NC score | 0.051097 (rank : 157) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q91WP0, Q9QXA4, Q9QXD2, Q9QXD5, Q9Z338 | Gene names | Masp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mannan-binding lectin serine protease 2 precursor (EC 3.4.21.104) (Mannose-binding protein-associated serine protease 2) (MASP-2) (MBL- associated serine protease 2) [Contains: Mannan-binding lectin serine protease 2 A chain; Mannan-binding lectin serine protease 2 B chain]. | |||||
|
EGFL7_HUMAN
|
||||||
| NC score | 0.051005 (rank : 158) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UHF1, Q5M7Y5, Q5VUD5, Q96EG0 | Gene names | EGFL7, MEGF7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EGF-like domain-containing protein 7 precursor (Multiple EGF-like domain protein 7) (Multiple epidermal growth factor-like domain protein 7) (Vascular endothelial statin) (VE-statin) (NOTCH4-like protein) (ZNEU1). | |||||
|
MATN2_MOUSE
|
||||||
| NC score | 0.050739 (rank : 159) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O08746 | Gene names | Matn2 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-2 precursor. | |||||
|
JAG1_HUMAN
|
||||||
| NC score | 0.050711 (rank : 160) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 514 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P78504, O14902, O15122, Q15816 | Gene names | JAG1, JAGL1 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-1 precursor (Jagged1) (hJ1) (CD339 antigen). | |||||
|
KR107_HUMAN
|
||||||
| NC score | 0.050635 (rank : 161) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P60409, Q70LJ2 | Gene names | KRTAP10-7, KAP10.7, KAP18-7, KRTAP10.7, KRTAP18-7, KRTAP18.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-7 (Keratin-associated protein 10.7) (High sulfur keratin-associated protein 10.7) (Keratin-associated protein 18-7) (Keratin-associated protein 18.7). | |||||
|
CRUM1_HUMAN
|
||||||
| NC score | 0.050373 (rank : 162) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P82279, Q5K3A6, Q5TC28, Q5VUT1, Q6N027, Q8WWY0 | Gene names | CRB1 | |||
|
Domain Architecture |

|
|||||
| Description | Crumbs homolog 1 precursor. | |||||
|
NELL1_HUMAN
|
||||||
| NC score | 0.050089 (rank : 163) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q92832, Q9Y472 | Gene names | NELL1, NRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein NELL1 precursor (NEL-like protein 1) (Nel-related protein 1). | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.036312 (rank : 164) | θ value | 0.47712 (rank : 39) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.035836 (rank : 165) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.035067 (rank : 166) | θ value | 0.47712 (rank : 40) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
ATS9_HUMAN
|
||||||
| NC score | 0.025839 (rank : 167) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9P2N4, Q9NR29 | Gene names | ADAMTS9, KIAA1312 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-9 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 9) (ADAM-TS 9) (ADAM-TS9). | |||||
|
5NT1B_MOUSE
|
||||||
| NC score | 0.021115 (rank : 168) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91YE9, Q91Y48 | Gene names | Nt5c1b, Airp | |||
|
Domain Architecture |

|
|||||
| Description | Cytosolic 5'-nucleotidase 1B (EC 3.1.3.5) (Cytosolic 5'-nucleotidase IB) (cN1B) (cN-IB) (Autoimmune infertility-related protein). | |||||
|
CELR2_HUMAN
|
||||||
| NC score | 0.018162 (rank : 169) | θ value | 8.99809 (rank : 67) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9HCU4, Q92566 | Gene names | CELSR2, CDHF10, EGFL2, KIAA0279, MEGF3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 2 precursor (Epidermal growth factor-like 2) (Multiple epidermal growth factor-like domains 3) (Flamingo 1). | |||||
|
MCP_MOUSE
|
||||||
| NC score | 0.017796 (rank : 170) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88174, Q9R0R9 | Gene names | Cd46, Mcp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane cofactor protein precursor (CD46 antigen). | |||||
|
LPIN3_HUMAN
|
||||||
| NC score | 0.014259 (rank : 171) | θ value | 8.99809 (rank : 68) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BQK8, Q5TDB9, Q9NPY8, Q9UJE5 | Gene names | LPIN3, LIPN3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipin-3 (Lipin 3-like). | |||||
|
CELR2_MOUSE
|
||||||
| NC score | 0.013648 (rank : 172) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9R0M0, Q99K26, Q9Z2R4 | Gene names | Celsr2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 2 precursor (Flamingo 1) (mFmi1). | |||||
|
SMAD4_MOUSE
|
||||||
| NC score | 0.009630 (rank : 173) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P97471, Q9CW56 | Gene names | Smad4, Dpc4, Madh4 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 4 (SMAD 4) (Mothers against DPP homolog 4) (Deletion target in pancreatic carcinoma 4 homolog) (Smad4). | |||||
|
CASR_MOUSE
|
||||||
| NC score | 0.009621 (rank : 174) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QY96, O08968, O88519, Q9QY95, Q9QZU8, Q9R1D6, Q9R1Y2 | Gene names | Casr, Gprc2a | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular calcium-sensing receptor precursor (CaSR) (Parathyroid Cell calcium-sensing receptor). | |||||
|
GPR63_MOUSE
|
||||||
| NC score | 0.001264 (rank : 175) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9EQQ3 | Gene names | Gpr63 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 63 (PSP24-beta) (PSP24-2). | |||||