Please be patient as the page loads
|
ZMYM3_MOUSE
|
||||||
| SwissProt Accessions | Q9JLM4, Q80U17 | Gene names | Zmym3, Kiaa0385, Zfp261, Znf261 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 3 (Zinc finger protein 261) (DXHXS6673E protein). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ZMYM3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.972312 (rank : 2) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | Q14202, O15089 | Gene names | ZMYM3, DXS6673E, KIAA0385, ZNF261 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 3 (Zinc finger protein 261). | |||||
|
ZMYM3_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 177 | |
| SwissProt Accessions | Q9JLM4, Q80U17 | Gene names | Zmym3, Kiaa0385, Zfp261, Znf261 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 3 (Zinc finger protein 261) (DXHXS6673E protein). | |||||
|
ZN198_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.921565 (rank : 3) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UBW7, O43212, O43434, O60898, Q5W0Q4, Q63HP0, Q8NE39, Q9H538, Q9UEU2 | Gene names | ZNF198, FIM, RAMP, ZMYM2 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 198 (Zinc finger MYM-type protein 2) (Fused in myeloproliferative disorders protein) (Rearranged in atypical myeloproliferative disorder protein). | |||||
|
ZMYM6_HUMAN
|
||||||
| θ value | 3.45727e-76 (rank : 4) | NC score | 0.853362 (rank : 4) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O95789, Q96IY0 | Gene names | ZMYM6, ZNF258 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 6 (Zinc finger protein 258). | |||||
|
ZMYM1_HUMAN
|
||||||
| θ value | 2.11701e-41 (rank : 5) | NC score | 0.700171 (rank : 5) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5SVZ6, Q7Z3Q4 | Gene names | ZMYM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 1. | |||||
|
ZMYM5_HUMAN
|
||||||
| θ value | 8.10077e-17 (rank : 6) | NC score | 0.698302 (rank : 6) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UJ78, Q5T6E2, Q96IY6, Q9NZY5, Q9UBW0, Q9UJ77 | Gene names | ZMYM5, ZNF198L1, ZNF237 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 5 (Zinc finger protein 237) (Zinc finger protein 198-like 1). | |||||
|
K1958_HUMAN
|
||||||
| θ value | 1.29631e-06 (rank : 7) | NC score | 0.267247 (rank : 8) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N8K9, Q5T252, Q8TF43, Q96N02 | Gene names | KIAA1958 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1958. | |||||
|
K1958_MOUSE
|
||||||
| θ value | 1.29631e-06 (rank : 8) | NC score | 0.270623 (rank : 7) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8C4P0, Q8BN62, Q8K1X8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1958 homolog. | |||||
|
ZN271_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 9) | NC score | 0.053443 (rank : 23) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 769 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q14591, Q96T29, Q9BSX2, Q9UN33, Q9Y5B7 | Gene names | ZNF271, ZNFEB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 271 (Zinc finger protein HZF7) (Zinc finger protein ZNFphex133) (Epstein-Barr virus-induced zinc finger protein) (ZNF-EB) (CT-ZFP48) (Zinc finger protein dp) (ZNF-dp). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 10) | NC score | 0.045032 (rank : 63) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
ZN11B_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 11) | NC score | 0.055658 (rank : 19) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 802 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q06732, Q86XY8, Q8NDW3 | Gene names | ZNF11B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 11B. | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 12) | NC score | 0.092888 (rank : 9) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 13) | NC score | 0.051302 (rank : 29) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
ZN334_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 14) | NC score | 0.052703 (rank : 24) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 778 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9HCZ1, Q5T6U2, Q9NVW4 | Gene names | ZNF334 | |||
|
Domain Architecture |
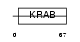
|
|||||
| Description | Zinc finger protein 334. | |||||
|
GAK_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 15) | NC score | 0.017862 (rank : 138) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99KY4, Q6P1I8, Q6P9S5, Q8BM74, Q8K0Q4 | Gene names | Gak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||
|
ARHG1_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 16) | NC score | 0.035093 (rank : 92) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61210, O89074, Q80YE8, Q80YE9, Q91VL3 | Gene names | Arhgef1, Lbcl2, Lsc | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 1 (Lymphoid blast crisis-like 2) (Lbc's second cousin). | |||||
|
BCL9_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 17) | NC score | 0.074580 (rank : 10) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
ZN518_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 18) | NC score | 0.057495 (rank : 18) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q6AHZ1, O15044, Q32MP4 | Gene names | ZNF518, KIAA0335 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 518. | |||||
|
ZNF28_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 19) | NC score | 0.050166 (rank : 35) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P17035, Q5HYM9, Q6ZML9, Q6ZN56 | Gene names | ZNF28, KOX24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 28 (Zinc finger protein KOX24). | |||||
|
CSPG2_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 20) | NC score | 0.018138 (rank : 136) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q62059, Q62058, Q9CUU0 | Gene names | Cspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M). | |||||
|
FYV1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 21) | NC score | 0.064188 (rank : 12) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y2I7, Q8NB67 | Gene names | PIP5K3, KIAA0981, PIKFYVE | |||
|
Domain Architecture |

|
|||||
| Description | FYVE finger-containing phosphoinositide kinase (EC 2.7.1.68) (1- phosphatidylinositol-4-phosphate 5-kinase) (Phosphatidylinositol-3- phosphate 5-kinase type III) (PIP5K) (PtdIns(4)P-5-kinase) (PIKfyve) (p235). | |||||
|
FYV1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 22) | NC score | 0.064615 (rank : 11) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z1T6 | Gene names | Pip5k3, Pikfyve | |||
|
Domain Architecture |

|
|||||
| Description | FYVE finger-containing phosphoinositide kinase (EC 2.7.1.68) (1- phosphatidylinositol-4-phosphate 5-kinase) (PIP5K) (PtdIns(4)P-5- kinase) (PIKfyve) (p235). | |||||
|
PRD15_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 23) | NC score | 0.052237 (rank : 26) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 805 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P57071, Q8N0X3, Q8NEX0, Q9NQV3 | Gene names | PRDM15, C21orf83, ZNF298 | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 15 (PR domain-containing protein 15) (Zinc finger protein 298). | |||||
|
ZN319_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 24) | NC score | 0.051019 (rank : 32) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 762 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9ERR8 | Gene names | Znf319, Zfp319 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 319. | |||||
|
PDLI7_HUMAN
|
||||||
| θ value | 0.125558 (rank : 25) | NC score | 0.029754 (rank : 104) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 547 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NR12, Q14250, Q5XG82, Q6NVZ5, Q96C91, Q9BXB8, Q9BXB9 | Gene names | PDLIM7, ENIGMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
PRDM5_HUMAN
|
||||||
| θ value | 0.125558 (rank : 26) | NC score | 0.052476 (rank : 25) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9NQX1 | Gene names | PRDM5, PFM2 | |||
|
Domain Architecture |
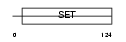
|
|||||
| Description | PR domain zinc finger protein 5 (PR domain-containing protein 5). | |||||
|
FHL5_MOUSE
|
||||||
| θ value | 0.163984 (rank : 27) | NC score | 0.047787 (rank : 44) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9WTX7 | Gene names | Fhl5, Act | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Four and a half LIM domains protein 5 (FHL-5) (Activator of cAMP- responsive element modulator in testis) (Activator of CREM in testis). | |||||
|
ZNF14_HUMAN
|
||||||
| θ value | 0.163984 (rank : 28) | NC score | 0.048808 (rank : 40) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P17017, Q9ULZ5 | Gene names | ZNF14, GIOT4, KOX6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 14 (Zinc finger protein KOX6) (Gonadotropin- inducible transcription repressor 4) (GIOT-4). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 29) | NC score | 0.060761 (rank : 14) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
TXLNA_HUMAN
|
||||||
| θ value | 0.21417 (rank : 30) | NC score | 0.026768 (rank : 112) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 982 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P40222, Q66K62, Q86T54, Q86T85, Q86T86, Q86Y86, Q86YW3, Q8N2Y3 | Gene names | TXLNA, TXLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
ZHX1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 31) | NC score | 0.041266 (rank : 85) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UKY1, Q8IWD8 | Gene names | ZHX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc fingers and homeoboxes protein 1. | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 32) | NC score | 0.061122 (rank : 13) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
MCM3A_HUMAN
|
||||||
| θ value | 0.279714 (rank : 33) | NC score | 0.052109 (rank : 27) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60318, Q9UMT4 | Gene names | MCM3AP, GANP, KIAA0572, MAP80 | |||
|
Domain Architecture |

|
|||||
| Description | 80 kDa MCM3-associated protein (GANP protein). | |||||
|
MEF2C_MOUSE
|
||||||
| θ value | 0.279714 (rank : 34) | NC score | 0.035909 (rank : 90) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CFN5, Q8R0H1, Q9D7L0, Q9QW20 | Gene names | Mef2c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myocyte-specific enhancer factor 2C. | |||||
|
OGFR_MOUSE
|
||||||
| θ value | 0.279714 (rank : 35) | NC score | 0.051195 (rank : 30) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q99PG2, Q99L33 | Gene names | Ogfr | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor). | |||||
|
ZFP11_MOUSE
|
||||||
| θ value | 0.279714 (rank : 36) | NC score | 0.049442 (rank : 38) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 763 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P10751, Q505E4, Q6PDR5, Q8BSI2, Q8C3R5 | Gene names | Zfp11, Krox-6.1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 11 (Zfp-11) (Krox-6.1A protein). | |||||
|
ZN319_HUMAN
|
||||||
| θ value | 0.279714 (rank : 37) | NC score | 0.049755 (rank : 36) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9P2F9 | Gene names | ZNF319, KIAA1388 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 319. | |||||
|
ASPP1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 38) | NC score | 0.032808 (rank : 99) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
PRDM5_MOUSE
|
||||||
| θ value | 0.365318 (rank : 39) | NC score | 0.050353 (rank : 34) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9CXE0 | Gene names | Prdm5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PR domain zinc finger protein 5 (PR domain-containing protein 5). | |||||
|
ZN324_HUMAN
|
||||||
| θ value | 0.365318 (rank : 40) | NC score | 0.046664 (rank : 56) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O75467 | Gene names | ZNF324 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 324 (Zinc finger protein ZF5128). | |||||
|
CHL1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 41) | NC score | 0.010073 (rank : 161) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70232, Q8BS24, Q8C6W0, Q8C823, Q8VBY7 | Gene names | Chl1, Call | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural cell adhesion molecule L1-like protein precursor (Cell adhesion molecule with homology to L1CAM) (Close homolog of L1) (Chl1-like protein). | |||||
|
HXB3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 42) | NC score | 0.014411 (rank : 142) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P14651, O95615, P17484 | Gene names | HOXB3, HOX2G | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B3 (Hox-2G) (Hox-2.7). | |||||
|
ZBT17_HUMAN
|
||||||
| θ value | 0.47712 (rank : 43) | NC score | 0.050406 (rank : 33) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 925 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q13105, Q15932, Q5JYB2, Q9NUC9 | Gene names | ZBTB17, MIZ1, ZNF151 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 17 (Zinc finger protein 151) (Myc-interacting zinc finger protein) (Miz-1 protein). | |||||
|
ZBT17_MOUSE
|
||||||
| θ value | 0.47712 (rank : 44) | NC score | 0.051138 (rank : 31) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q60821, Q60699 | Gene names | Zbtb17, Zfp100, Znf151 | |||
|
Domain Architecture |
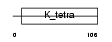
|
|||||
| Description | Zinc finger and BTB domain-containing protein 17 (Zinc finger protein 151) (Zinc finger protein 100) (Zfp-100) (Polyomavirus late initiator promoter-binding protein) (LP-1) (Zinc finger protein Z13). | |||||
|
ZN226_HUMAN
|
||||||
| θ value | 0.47712 (rank : 45) | NC score | 0.046853 (rank : 51) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9NYT6, Q96TE6, Q9NS44 | Gene names | ZNF226 | |||
|
Domain Architecture |
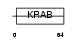
|
|||||
| Description | Zinc finger protein 226. | |||||
|
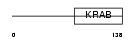
ZN268_HUMAN
|
||||||
| θ value | 0.47712 (rank : 46) | NC score | 0.048937 (rank : 39) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q14587, Q96RH4, Q9BZJ9 | Gene names | ZNF268 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 268 (Zinc finger protein HZF3). | |||||
|
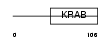
ZN33A_HUMAN
|
||||||
| θ value | 0.47712 (rank : 47) | NC score | 0.051330 (rank : 28) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q06730, Q5VZ86 | Gene names | ZNF33A, KIAA0065, KOX31, ZNF33 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 33A (Zinc finger protein KOX31) (Zinc finger and ZAK-associated protein with KRAB domain) (ZZaPK). | |||||
|
ZN560_HUMAN
|
||||||
| θ value | 0.47712 (rank : 48) | NC score | 0.048761 (rank : 41) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q96MR9, Q495S9, Q495T1 | Gene names | ZNF560 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 560. | |||||
|
ZNF35_HUMAN
|
||||||
| θ value | 0.47712 (rank : 49) | NC score | 0.047101 (rank : 50) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 761 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P13682, Q96D01 | Gene names | ZNF35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 35 (Zinc finger protein HF.10). | |||||
|
CO5A1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 50) | NC score | 0.032132 (rank : 100) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P20908, Q15094, Q5SUX4 | Gene names | COL5A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(V) chain precursor. | |||||
|
HXB3_MOUSE
|
||||||
| θ value | 0.62314 (rank : 51) | NC score | 0.013598 (rank : 147) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P09026, P10285, Q4PJ06, Q61680 | Gene names | Hoxb3, Hox-2.7, Hoxb-3 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B3 (Hox-2.7) (Homeobox protein MH-23). | |||||
|
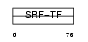
MEF2C_HUMAN
|
||||||
| θ value | 0.62314 (rank : 52) | NC score | 0.034160 (rank : 96) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q06413 | Gene names | MEF2C | |||
|
Domain Architecture |
|
|||||
| Description | Myocyte-specific enhancer factor 2C. | |||||
|
PTN22_HUMAN
|
||||||
| θ value | 0.62314 (rank : 53) | NC score | 0.011247 (rank : 156) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y2R2, O95063, O95064 | Gene names | PTPN22, PTPN8 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 22 (EC 3.1.3.48) (Hematopoietic cell protein-tyrosine phosphatase 70Z-PEP) (Lymphoid phosphatase) (LyP). | |||||
|
RAI2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 54) | NC score | 0.046804 (rank : 53) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y5P3, Q8N6X7 | Gene names | RAI2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 2. | |||||
|
ZN256_HUMAN
|
||||||
| θ value | 0.62314 (rank : 55) | NC score | 0.047186 (rank : 49) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9Y2P7, Q9BV71 | Gene names | ZNF256, BMZF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 256 (Bone marrow zinc finger 3) (BMZF-3). | |||||
|
GA2L2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 56) | NC score | 0.025456 (rank : 114) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8NHY3, Q8NHY4 | Gene names | GAS2L2, GAR17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2) (GAS2-related protein on chromosome 17) (GAR17 protein). | |||||
|
ZFP62_MOUSE
|
||||||
| θ value | 0.813845 (rank : 57) | NC score | 0.049487 (rank : 37) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8C827, Q3U3H0, Q62510, Q8BT00, Q8C2S6, Q8R5D1 | Gene names | Zfp62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 62 (Zfp-62) (ZT3). | |||||
|
ZN206_HUMAN
|
||||||
| θ value | 0.813845 (rank : 58) | NC score | 0.047331 (rank : 48) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q96SZ4 | Gene names | ZNF206, ZSCAN10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 206 (Zinc finger and SCAN domain-containing protein 10). | |||||
|
ZN674_HUMAN
|
||||||
| θ value | 0.813845 (rank : 59) | NC score | 0.044923 (rank : 64) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q2M3X9 | Gene names | ZNF674 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 674. | |||||
|
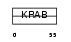
ZNF43_HUMAN
|
||||||
| θ value | 0.813845 (rank : 60) | NC score | 0.048020 (rank : 43) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P17038, P28160, Q96DG1 | Gene names | ZNF43, KOX27, ZNF39 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 43 (Zinc protein HTF6) (Zinc finger protein KOX27). | |||||
|
MTSS1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 61) | NC score | 0.055165 (rank : 20) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O43312, Q8TCA2, Q96RX2 | Gene names | MTSS1, KIAA0429, MIM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein) (Metastasis suppressor YGL-1). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 1.06291 (rank : 62) | NC score | 0.035438 (rank : 91) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
ZN267_HUMAN
|
||||||
| θ value | 1.06291 (rank : 63) | NC score | 0.046713 (rank : 55) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q14586, Q8NE41, Q9NRJ0 | Gene names | ZNF267 | |||
|
Domain Architecture |
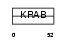
|
|||||
| Description | Zinc finger protein 267 (Zinc finger protein HZF2). | |||||
|
CNTRB_MOUSE
|
||||||
| θ value | 1.38821 (rank : 64) | NC score | 0.013672 (rank : 146) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8CB62, Q5NCF3 | Gene names | Cntrob, Lip8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
FHL2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 65) | NC score | 0.054475 (rank : 21) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 533 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q14192, Q13229, Q13644, Q9P294 | Gene names | FHL2, DRAL, SLIM3 | |||
|
Domain Architecture |
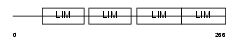
|
|||||
| Description | Four and a half LIM domains protein 2 (FHL-2) (Skeletal muscle LIM- protein 3) (SLIM 3) (LIM-domain protein DRAL). | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 66) | NC score | 0.008021 (rank : 168) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
MINT_HUMAN
|
||||||
| θ value | 1.38821 (rank : 67) | NC score | 0.034886 (rank : 94) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
PI5PA_MOUSE
|
||||||
| θ value | 1.38821 (rank : 68) | NC score | 0.031642 (rank : 101) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P59644 | Gene names | Pib5pa | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
ZN221_HUMAN
|
||||||
| θ value | 1.38821 (rank : 69) | NC score | 0.046283 (rank : 57) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9UK13, Q9P1U8 | Gene names | ZNF221 | |||
|
Domain Architecture |
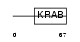
|
|||||
| Description | Zinc finger protein 221. | |||||
|
ZN433_HUMAN
|
||||||
| θ value | 1.38821 (rank : 70) | NC score | 0.047591 (rank : 46) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 782 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8N7K0 | Gene names | ZNF433 | |||
|
Domain Architecture |
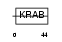
|
|||||
| Description | Zinc finger protein 433. | |||||
|
ZN610_HUMAN
|
||||||
| θ value | 1.38821 (rank : 71) | NC score | 0.042805 (rank : 81) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8N9Z0, Q86YH8, Q8NDS9 | Gene names | ZNF610 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 610. | |||||
|
GLU2B_MOUSE
|
||||||
| θ value | 1.81305 (rank : 72) | NC score | 0.041528 (rank : 84) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O08795, Q921X2 | Gene names | Prkcsh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glucosidase 2 subunit beta precursor (Glucosidase II subunit beta) (Protein kinase C substrate, 60.1 kDa protein, heavy chain) (PKCSH) (80K-H protein). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 73) | NC score | 0.036365 (rank : 89) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MKRN3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 74) | NC score | 0.027162 (rank : 110) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q60764, Q7TQE4 | Gene names | Mkrn3, Zfp127, Znf127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127). | |||||
|
PSL1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 75) | NC score | 0.034956 (rank : 93) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
TACC2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 76) | NC score | 0.021341 (rank : 123) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JJG0 | Gene names | Tacc2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2. | |||||
|
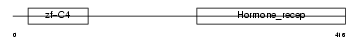
VDR_HUMAN
|
||||||
| θ value | 1.81305 (rank : 77) | NC score | 0.007954 (rank : 169) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P11473 | Gene names | VDR, NR1I1 | |||
|
Domain Architecture |
|
|||||
| Description | Vitamin D3 receptor (VDR) (1,25-dihydroxyvitamin D3 receptor). | |||||
|
DOCK4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 78) | NC score | 0.019488 (rank : 131) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P59764 | Gene names | Dock4, Kiaa0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
DSCL1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 79) | NC score | 0.007876 (rank : 170) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8TD84, Q76MU9, Q8IZY3, Q8IZY4, Q8WXU7, Q9ULT7 | Gene names | DSCAML1, DSCAM2, KIAA1132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Down syndrome cell adhesion molecule-like protein 1 precursor (Down syndrome cell adhesion molecule 2). | |||||
|
HIC2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 80) | NC score | 0.043553 (rank : 73) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q96JB3, Q96KR3, Q9NSM9, Q9UPX9 | Gene names | HIC2, HRG22, KIAA1020, ZBTB30 | |||
|
Domain Architecture |

|
|||||
| Description | Hypermethylated in cancer 2 protein (Hic-2) (Hic-3) (HIC1-related gene on chromosome 22) (Zinc finger and BTB domain-containing protein 30). | |||||
|
HS105_HUMAN
|
||||||
| θ value | 2.36792 (rank : 81) | NC score | 0.010439 (rank : 160) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92598, O95739, Q5TBM6, Q5TBM8, Q9UPC4 | Gene names | HSPH1, HSP105, HSP110, KIAA0201 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein 105 kDa (Heat shock 110 kDa protein) (Antigen NY- CO-25). | |||||
|
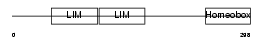
LHX9_MOUSE
|
||||||
| θ value | 2.36792 (rank : 82) | NC score | 0.017912 (rank : 137) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9WUH2, Q9QYQ5, Q9QYQ6, Q9QZ00, Q9WU44 | Gene names | Lhx9 | |||
|
Domain Architecture |
|
|||||
| Description | LIM/homeobox protein Lhx9. | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 83) | NC score | 0.034545 (rank : 95) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MTSS1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 84) | NC score | 0.046149 (rank : 59) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8R1S4, Q8BMM3, Q99LB3 | Gene names | Mtss1, Mim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein). | |||||
|
MUC2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 85) | NC score | 0.029676 (rank : 105) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
SL9A5_HUMAN
|
||||||
| θ value | 2.36792 (rank : 86) | NC score | 0.024506 (rank : 117) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14940, Q9Y626 | Gene names | SLC9A5, NHE5 | |||
|
Domain Architecture |
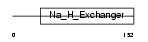
|
|||||
| Description | Sodium/hydrogen exchanger 5 (Na(+)/H(+) exchanger 5) (NHE-5) (Solute carrier family 9 member 5). | |||||
|
SOX13_MOUSE
|
||||||
| θ value | 2.36792 (rank : 87) | NC score | 0.012438 (rank : 149) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q04891, Q922L3 | Gene names | Sox13, Sox-13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein. | |||||
|
SPTN2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 88) | NC score | 0.010804 (rank : 159) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O15020, O14872, O14873 | Gene names | SPTBN2, KIAA0302 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 2 (Spectrin, non-erythroid beta chain 2) (Beta-III spectrin). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 89) | NC score | 0.038384 (rank : 87) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
TNR25_HUMAN
|
||||||
| θ value | 2.36792 (rank : 90) | NC score | 0.033941 (rank : 97) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q93038, O00275, O00276, O00277, O00278, O00279, O00280, O14865, O14866, P78507, P78515, Q92983, Q93036, Q93037, Q99722, Q99830, Q99831, Q9BY86, Q9UME0, Q9UME1, Q9UME5 | Gene names | TNFRSF25, APO3, DDR3, DR3, TNFRSF12, WSL, WSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 25 precursor (WSL-1 protein) (Apoptosis-mediating receptor DR3) (Apoptosis-mediating receptor TRAMP) (Death domain receptor 3) (WSL protein) (Apoptosis- inducing receptor AIR) (Apo-3) (Lymphocyte-associated receptor of death) (LARD). | |||||
|
ZHX1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 91) | NC score | 0.027838 (rank : 108) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P70121, Q8BQ68, Q8C6T4, Q8CJG3 | Gene names | Zhx1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc fingers and homeoboxes protein 1. | |||||
|
ZN509_MOUSE
|
||||||
| θ value | 2.36792 (rank : 92) | NC score | 0.042482 (rank : 83) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 990 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q8BXX2, Q8CID4, Q8K2Z6 | Gene names | Znf509, Zfp509 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 509. | |||||
|
ACHA4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 93) | NC score | 0.006826 (rank : 172) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P43681, Q4JGR7 | Gene names | CHRNA4, NACRA4 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal acetylcholine receptor protein subunit alpha-4 precursor. | |||||
|
CCD96_MOUSE
|
||||||
| θ value | 3.0926 (rank : 94) | NC score | 0.024619 (rank : 116) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9CR92 | Gene names | Ccdc96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 96. | |||||
|
HIC2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 95) | NC score | 0.043276 (rank : 75) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9JLZ6, Q3U030, Q3ULP4, Q5K036, Q8BSZ9, Q8C3T5 | Gene names | Hic2, Kiaa1020 | |||
|
Domain Architecture |

|
|||||
| Description | Hypermethylated in cancer 2 protein (Hic-2). | |||||
|
JHD1A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 96) | NC score | 0.020892 (rank : 127) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y2K7, Q49A21, Q4G0M3, Q69YY8, Q9BVH5, Q9H7H5, Q9UK66 | Gene names | FBXL11, FBL7, JHDM1A, KIAA1004 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 1A (EC 1.14.11.-) (F-box/LRR-repeat protein 11) (F-box and leucine-rich repeat protein 11) (F-box protein FBL7) (F-box protein Lilina). | |||||
|
JHD1A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 97) | NC score | 0.020905 (rank : 126) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P59997, Q3U1M5, Q3UR56, Q3V3Q1, Q69ZT4 | Gene names | Fbxl11, Jhdm1a, Kiaa1004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 1A (EC 1.14.11.-) (F-box/LRR-repeat protein 11) (F-box and leucine-rich repeat protein 11). | |||||
|
MIZF_MOUSE
|
||||||
| θ value | 3.0926 (rank : 98) | NC score | 0.045555 (rank : 62) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8K1K9, Q8BWY0 | Gene names | Mizf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MBD2-interacting zinc finger protein (Methyl-CpG-binding protein 2- interacting zinc finger protein). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 99) | NC score | 0.023506 (rank : 120) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
PALB2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 100) | NC score | 0.024504 (rank : 118) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q3U0P1, Q6NZG9, Q7TMQ4, Q8CEA9 | Gene names | Palb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Partner and localizer of BRCA2. | |||||
|
RN125_MOUSE
|
||||||
| θ value | 3.0926 (rank : 101) | NC score | 0.028384 (rank : 107) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D9R0, Q52KL4, Q8C7F2 | Gene names | Rnf125 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 125 (EC 6.3.2.-). | |||||
|
SNPC4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 102) | NC score | 0.025591 (rank : 113) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5SXM2, Q9Y6P7 | Gene names | SNAPC4, SNAP190 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor subunit alpha) (PSE-binding factor subunit alpha) (PTF subunit alpha). | |||||
|
SYGP1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 103) | NC score | 0.010965 (rank : 157) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96PV0, Q8TCS2, Q9UGE2 | Gene names | SYNGAP1, KIAA1938 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein SynGAP (Synaptic Ras-GTPase-activating protein 1) (Synaptic Ras-GAP 1) (Neuronal RasGAP). | |||||
|
TNR21_MOUSE
|
||||||
| θ value | 3.0926 (rank : 104) | NC score | 0.015454 (rank : 141) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9EPU5, Q91W77, Q91XH9 | Gene names | Tnfrsf21, Dr6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 21 precursor (TNFR- related death receptor 6) (Death receptor 6). | |||||
|
ZN473_HUMAN
|
||||||
| θ value | 3.0926 (rank : 105) | NC score | 0.047584 (rank : 47) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8WTR7, Q9ULS9, Q9Y4Q7 | Gene names | ZNF473, KIAA1141, ZFP100 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 473 (Zinc finger protein 100 homolog) (Zfp-100). | |||||
|
ZN669_HUMAN
|
||||||
| θ value | 3.0926 (rank : 106) | NC score | 0.043584 (rank : 71) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 746 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q96BR6, Q5VT39, Q9H9Q6 | Gene names | ZNF669 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 669. | |||||
|
BCAS1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 107) | NC score | 0.044561 (rank : 65) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80YN3, Q9CVA1 | Gene names | Bcas1, Nabc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast carcinoma amplified sequence 1 homolog (Novel amplified in breast cancer 1 homolog). | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 4.03905 (rank : 108) | NC score | 0.003274 (rank : 180) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
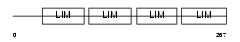
FHL1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 109) | NC score | 0.043513 (rank : 74) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 462 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P97447, O55181, Q8K318 | Gene names | Fhl1 | |||
|
Domain Architecture |
|
|||||
| Description | Four and a half LIM domains protein 1 (FHL-1) (Skeletal muscle LIM- protein 1) (SLIM 1) (SLIM) (KyoT) (RBP-associated molecule 14-1) (RAM14-1). | |||||
|
INCE_HUMAN
|
||||||
| θ value | 4.03905 (rank : 110) | NC score | 0.012104 (rank : 150) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
K1802_MOUSE
|
||||||
| θ value | 4.03905 (rank : 111) | NC score | 0.058006 (rank : 15) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
MATN2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 112) | NC score | 0.011571 (rank : 154) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O00339, Q6UWA5, Q7Z5X1, Q8NDE6, Q96FT5, Q9NSZ1 | Gene names | MATN2 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-2 precursor. | |||||
|
PRGR_MOUSE
|
||||||
| θ value | 4.03905 (rank : 113) | NC score | 0.008348 (rank : 166) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q00175 | Gene names | Pgr, Nr3c3, Pr | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone receptor (PR). | |||||
|
RPC11_MOUSE
|
||||||
| θ value | 4.03905 (rank : 114) | NC score | 0.021270 (rank : 125) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CQZ7, Q9D1U1 | Gene names | Polr3k | |||
|
Domain Architecture |
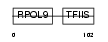
|
|||||
| Description | DNA-directed RNA polymerases III 12.5 kDa polypeptide (EC 2.7.7.6) (RNA polymerase III subunit K) (RNA polymerase III C11 subunit). | |||||
|
ZFP62_HUMAN
|
||||||
| θ value | 4.03905 (rank : 115) | NC score | 0.048215 (rank : 42) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8NB50 | Gene names | ZFP62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 62 homolog (Zfp-62). | |||||
|
ZN443_HUMAN
|
||||||
| θ value | 4.03905 (rank : 116) | NC score | 0.046780 (rank : 54) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9Y2A4 | Gene names | ZNF443 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 443 (Krueppel-type zinc finger protein ZK1). | |||||
|
ZN502_HUMAN
|
||||||
| θ value | 4.03905 (rank : 117) | NC score | 0.047646 (rank : 45) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 762 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8TBZ5 | Gene names | ZNF502 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 502. | |||||
|
ANK3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 118) | NC score | 0.006153 (rank : 174) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
ASPP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 119) | NC score | 0.016208 (rank : 139) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q13625, Q12892, Q86X75, Q96KQ3 | Gene names | TP53BP2, ASPP2, BBP | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 2 (Tumor suppressor p53-binding protein 2) (p53-binding protein 2) (p53BP2) (53BP2) (Bcl2-binding protein) (Bbp) (NY-REN-51 antigen). | |||||
|
CEAM5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 120) | NC score | 0.007800 (rank : 171) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P06731 | Gene names | CEACAM5, CEA | |||
|
Domain Architecture |

|
|||||
| Description | Carcinoembryonic antigen-related cell adhesion molecule 5 precursor (Carcinoembryonic antigen) (CEA) (Meconium antigen 100) (CD66e antigen). | |||||
|
CO002_HUMAN
|
||||||
| θ value | 5.27518 (rank : 121) | NC score | 0.029110 (rank : 106) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NZP6 | Gene names | C15orf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C15orf2. | |||||
|
CO4A1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 122) | NC score | 0.013696 (rank : 145) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P02462 | Gene names | COL4A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IV) chain precursor. | |||||
|
DAXX_MOUSE
|
||||||
| θ value | 5.27518 (rank : 123) | NC score | 0.019041 (rank : 133) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35613, Q9QWT8, Q9QWV3 | Gene names | Daxx | |||
|
Domain Architecture |

|
|||||
| Description | Death domain-associated protein 6 (Daxx). | |||||
|
GZF1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 124) | NC score | 0.044455 (rank : 67) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 854 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9H116, Q96N08, Q9BQK9, Q9H117, Q9H6W6 | Gene names | GZF1, ZBTB23, ZNF336 | |||
|
Domain Architecture |
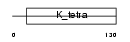
|
|||||
| Description | GDNF-inducible zinc finger protein 1 (Zinc finger protein 336) (Zinc finger and BTB domain-containing protein 23). | |||||
|
IRF5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 125) | NC score | 0.008439 (rank : 165) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P56477 | Gene names | Irf5 | |||
|
Domain Architecture |
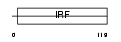
|
|||||
| Description | Interferon regulatory factor 5 (IRF-5). | |||||
|
ITA7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 126) | NC score | 0.005795 (rank : 175) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13683, O43197, Q9NY89, Q9UET0, Q9UEV2 | Gene names | ITGA7 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-7 precursor [Contains: Integrin alpha-7 heavy chain; Integrin alpha-7 light chain]. | |||||
|
K1543_MOUSE
|
||||||
| θ value | 5.27518 (rank : 127) | NC score | 0.027771 (rank : 109) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q80VC9, Q5DTW9, Q8BUZ0 | Gene names | Kiaa1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 5.27518 (rank : 128) | NC score | 0.039419 (rank : 86) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 129) | NC score | 0.037124 (rank : 88) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
TUR8_MOUSE
|
||||||
| θ value | 5.27518 (rank : 130) | NC score | 0.032813 (rank : 98) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P19473 | Gene names | Trap1a, P1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor rejection antigen P815A. | |||||
|
TXND2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 131) | NC score | 0.045914 (rank : 61) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
ZFY21_HUMAN
|
||||||
| θ value | 5.27518 (rank : 132) | NC score | 0.030289 (rank : 103) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BQ24, Q86T05, Q96LT1 | Gene names | ZFYVE21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 21. | |||||
|
ZN264_HUMAN
|
||||||
| θ value | 5.27518 (rank : 133) | NC score | 0.043875 (rank : 70) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O43296, Q9P1V0 | Gene names | ZNF264, KIAA0412 | |||
|
Domain Architecture |
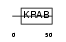
|
|||||
| Description | Zinc finger protein 264. | |||||
|
ADDA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 134) | NC score | 0.018515 (rank : 135) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9QYC0, Q9JLE3 | Gene names | Add1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-adducin (Erythrocyte adducin subunit alpha). | |||||
|
CSKI1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 135) | NC score | 0.018966 (rank : 134) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
DZIP3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 136) | NC score | 0.008248 (rank : 167) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TPV2, Q80TU4, Q8BYK7 | Gene names | Dzip3, Kiaa0675 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein DZIP3 (EC 6.3.2.-) (DAZ-interacting protein 3 homolog). | |||||
|
HPS4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 137) | NC score | 0.014387 (rank : 143) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99KG7 | Gene names | Hps4, Le | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hermansky-Pudlak syndrome 4 protein homolog (Light-ear protein) (Le protein). | |||||
|
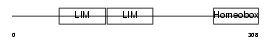
LHX9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 138) | NC score | 0.013999 (rank : 144) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NQ69, Q5VUE3, Q9BYU6, Q9NQ70 | Gene names | LHX9 | |||
|
Domain Architecture |
|
|||||
| Description | LIM/homeobox protein Lhx9. | |||||
|
LRFN3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 139) | NC score | 0.004575 (rank : 178) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BTN0, Q6UY10 | Gene names | LRFN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat and fibronectin type-III domain-containing protein 3 precursor. | |||||
|
MEGF8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 140) | NC score | 0.011959 (rank : 151) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7Z7M0, O75097 | Gene names | MEGF8, EGFL4, KIAA0817 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 8 (EGF-like domain- containing protein 4) (Multiple EGF-like domain protein 4). | |||||
|
MEGF8_MOUSE
|
||||||
| θ value | 6.88961 (rank : 141) | NC score | 0.011840 (rank : 152) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 520 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P60882, Q80TR3, Q80V41, Q8BMN9, Q8JZW7, Q8K0J3 | Gene names | Megf8, Egfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 8 (EGF-like domain- containing protein 4) (Multiple EGF-like domain protein 4). | |||||
|
NID2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 142) | NC score | 0.005165 (rank : 176) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14112, O43710 | Gene names | NID2 | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-2 precursor (NID-2) (Osteonidogen). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 143) | NC score | 0.019862 (rank : 128) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
PRDM4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 144) | NC score | 0.042900 (rank : 80) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9UKN5, Q9UFA6 | Gene names | PRDM4, PFM1 | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 4 (PR domain-containing protein 4). | |||||
|
PRDM4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 145) | NC score | 0.042937 (rank : 78) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q80V63 | Gene names | Prdm4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PR domain zinc finger protein 4 (PR domain-containing protein 4). | |||||
|
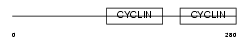
TF2B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 146) | NC score | 0.019498 (rank : 130) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q00403, Q5JS30 | Gene names | GTF2B, TF2B, TFIIB | |||
|
Domain Architecture |
|
|||||
| Description | Transcription initiation factor IIB (General transcription factor TFIIB) (S300-II). | |||||
|
TF2B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 147) | NC score | 0.019499 (rank : 129) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P62915, P29053 | Gene names | Gtf2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor IIB (General transcription factor TFIIB) (RNA polymerase II alpha initiation factor). | |||||
|
TIF1G_MOUSE
|
||||||
| θ value | 6.88961 (rank : 148) | NC score | 0.009418 (rank : 163) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99PP7, Q6SI71, Q6ZPX5 | Gene names | Trim33, Kiaa1113 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33). | |||||
|
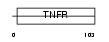
TNR22_MOUSE
|
||||||
| θ value | 6.88961 (rank : 149) | NC score | 0.025153 (rank : 115) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ER62, Q8VHB9, Q9CZA4 | Gene names | Tnfrsf22, Dctrailr2, Tnfrh2, Tnfrsf1al2 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 22 (Tumor necrosis factor receptor p60 homolog 2) (TNF receptor family member SOBa) (Decoy TRAIL receptor 2) (TNF receptor homolog 2). | |||||
|
ZBT38_HUMAN
|
||||||
| θ value | 6.88961 (rank : 150) | NC score | 0.043565 (rank : 72) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 885 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8NAP3 | Gene names | ZBTB38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and BTB domain-containing protein 38. | |||||
|
ZN561_HUMAN
|
||||||
| θ value | 6.88961 (rank : 151) | NC score | 0.042908 (rank : 79) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 734 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8N587, Q6PJS0 | Gene names | ZNF561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 561. | |||||
|
ZN595_HUMAN
|
||||||
| θ value | 6.88961 (rank : 152) | NC score | 0.044467 (rank : 66) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 798 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8IYB9 | Gene names | ZNF595 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 595. | |||||
|
ZN628_HUMAN
|
||||||
| θ value | 6.88961 (rank : 153) | NC score | 0.046818 (rank : 52) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 814 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q5EBL2, Q86X34 | Gene names | ZNF628 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 628. | |||||
|
ZN628_MOUSE
|
||||||
| θ value | 6.88961 (rank : 154) | NC score | 0.046052 (rank : 60) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 815 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8CJ78, Q3U2L5, Q6P5B3 | Gene names | Znf628, Zec, Zfp628 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 628 (Zinc finger protein expressed in embryonal cells and certain adult organs). | |||||
|
ZN649_HUMAN
|
||||||
| θ value | 6.88961 (rank : 155) | NC score | 0.042490 (rank : 82) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9BS31, Q9H9N2 | Gene names | ZNF649 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 649. | |||||
|
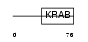
ZNF20_HUMAN
|
||||||
| θ value | 6.88961 (rank : 156) | NC score | 0.043896 (rank : 69) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P17024, Q9UG41 | Gene names | ZNF20, KOX13 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 20 (Zinc finger protein KOX13). | |||||
|
ZNF41_HUMAN
|
||||||
| θ value | 6.88961 (rank : 157) | NC score | 0.046188 (rank : 58) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 769 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P51814, Q96LE8, Q9UMC4, Q9UMV5, Q9UMV6, Q9UMV7, Q9UMV8, Q9UMV9, Q9UMW0, Q9UMW1 | Gene names | ZNF41 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 41. | |||||
|
ZNF85_HUMAN
|
||||||
| θ value | 6.88961 (rank : 158) | NC score | 0.044032 (rank : 68) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q03923 | Gene names | ZNF85 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 85 (Zinc finger protein HPF4) (HTF1). | |||||
|
AP180_HUMAN
|
||||||
| θ value | 8.99809 (rank : 159) | NC score | 0.022484 (rank : 121) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O60641, Q5JX13, Q68DL9, Q6P9D3, Q9NTY7 | Gene names | SNAP91, KIAA0656 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin coat assembly protein AP180 (Clathrin coat-associated protein AP180) (91 kDa synaptosomal-associated protein). | |||||
|
ARHGB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 160) | NC score | 0.021416 (rank : 122) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O15085 | Gene names | ARHGEF11, KIAA0380 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 11 (PDZ-RhoGEF). | |||||
|
BMPER_HUMAN
|
||||||
| θ value | 8.99809 (rank : 161) | NC score | 0.010806 (rank : 158) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N8U9 | Gene names | BMPER | |||
|
Domain Architecture |

|
|||||
| Description | BMP-binding endothelial regulator protein precursor (Bone morphogenetic protein-binding endothelial cell precursor-derived regulator) (Protein crossveinless-2) (hCV2). | |||||
|
CD1E_HUMAN
|
||||||
| θ value | 8.99809 (rank : 162) | NC score | 0.003354 (rank : 179) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P15812, Q9UMM1, Q9Y5M3 | Gene names | CD1E | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface glycoprotein CD1e precursor (CD1e antigen) (R2G1). | |||||
|
CE016_HUMAN
|
||||||
| θ value | 8.99809 (rank : 163) | NC score | 0.011605 (rank : 153) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NC24, Q6P4E7, Q6UXY2 | Gene names | C5orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C5orf16. | |||||
|
CTR9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 164) | NC score | 0.016123 (rank : 140) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q62018, Q3UFF5, Q3UY40, Q66JX4, Q7TPS6, Q8BND9, Q8BRD1, Q8C9W7, Q8C9Y3, Q8CHI1 | Gene names | Ctr9, Kiaa0155, Sh2bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1) (Tetratricopeptide repeat-containing, SH2-binding phosphoprotein of 150 kDa) (TPR-containing, SH2-binding phosphoprotein of 150 kDa) (p150TSP). | |||||
|
LYST_MOUSE
|
||||||
| θ value | 8.99809 (rank : 165) | NC score | 0.004582 (rank : 177) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97412, Q62403, Q8VBS6 | Gene names | Lyst, Bg, Chs1 | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal-trafficking regulator (Beige protein) (CHS1 homolog). | |||||
|
NFAC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 166) | NC score | 0.012781 (rank : 148) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O95644, Q12865, Q15793 | Gene names | NFATC1, NFAT2, NFATC | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 1 (NFAT transcription complex cytosolic component) (NF-ATc1) (NF-ATc). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 167) | NC score | 0.019088 (rank : 132) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
NU214_HUMAN
|
||||||
| θ value | 8.99809 (rank : 168) | NC score | 0.026965 (rank : 111) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
OSBL5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 169) | NC score | 0.006463 (rank : 173) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H0X9, Q9BZB0, Q9P1Z4 | Gene names | OSBPL5, KIAA1534, ORP5 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 5 (OSBP-related protein 5) (ORP-5). | |||||
|
RAI1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 170) | NC score | 0.021339 (rank : 124) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
SIPA1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 171) | NC score | 0.009031 (rank : 164) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P46062, P70204 | Gene names | Sipa1, Spa-1, Spa1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal-induced proliferation-associated protein 1 (Sipa-1) (GTPase- activating protein Spa-1). | |||||
|
SPN90_MOUSE
|
||||||
| θ value | 8.99809 (rank : 172) | NC score | 0.030597 (rank : 102) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9ESJ4, Q68G72 | Gene names | Nckipsd, Spin90, Wasbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3 adapter protein SPIN90 (NCK-interacting protein with SH3 domain) (SH3 protein interacting with Nck, 90 kDa) (VacA-interacting protein, 54 kDa) (VIP54) (N-WASP-interacting protein, 90kDa) (Wiskott-Aldrich syndrome protein-binding protein WISH) (N-WASP-binding protein). | |||||
|
TMUB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 173) | NC score | 0.011290 (rank : 155) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BVT8 | Gene names | TMUB1, C7orf21 | |||
|
Domain Architecture |
|
|||||
| Description | Transmembrane and ubiquitin-like domain-containing protein 1 (Ubiquitin-like protein SB144). | |||||
|
ZFY28_HUMAN
|
||||||
| θ value | 8.99809 (rank : 174) | NC score | 0.023891 (rank : 119) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9HCC9 | Gene names | ZFYVE28, KIAA1643 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 28. | |||||
|
ZN382_HUMAN
|
||||||
| θ value | 8.99809 (rank : 175) | NC score | 0.043248 (rank : 76) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q96SR6 | Gene names | ZNF382 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 382 (KRAB/Zinc finger suppressor protein 1) (KS1). | |||||
|
ZN436_MOUSE
|
||||||
| θ value | 8.99809 (rank : 176) | NC score | 0.042989 (rank : 77) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 756 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8BPP0, Q69ZC6, Q8BQC2 | Gene names | Znf436, Kiaa1710, Zfp46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 436 (Zinc finger protein 46). | |||||
|
ZPBP2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 177) | NC score | 0.009999 (rank : 162) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6X786, Q6X785, Q9DA91 | Gene names | Zpbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zona pellucida-binding protein 2 precursor. | |||||
|
K1802_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.053584 (rank : 22) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
P52K_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.057538 (rank : 17) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43422, Q8WTW1, Q9Y3Z4 | Gene names | PRKRIR, DAP4, P52RIPK, THAP0 | |||
|
Domain Architecture |

|
|||||
| Description | 52 kDa repressor of the inhibitor of the protein kinase (p58IPK- interacting protein) (58 kDa interferon-induced protein kinase- interacting protein) (P52rIPK) (Death-associated protein 4) (THAP domain-containing protein 0). | |||||
|
P52K_MOUSE
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.057641 (rank : 16) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9CUX1, Q80Y58 | Gene names | Prkrir, Thap0 | |||
|
Domain Architecture |

|
|||||
| Description | 52 kDa repressor of the inhibitor of the protein kinase (p58IPK- interacting protein) (58 kDa interferon-induced protein kinase- interacting protein) (P52rIPK) (THAP domain-containing protein 0). | |||||
|
ZMYM3_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 177 | |
| SwissProt Accessions | Q9JLM4, Q80U17 | Gene names | Zmym3, Kiaa0385, Zfp261, Znf261 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 3 (Zinc finger protein 261) (DXHXS6673E protein). | |||||
|
ZMYM3_HUMAN
|
||||||
| NC score | 0.972312 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | Q14202, O15089 | Gene names | ZMYM3, DXS6673E, KIAA0385, ZNF261 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 3 (Zinc finger protein 261). | |||||
|
ZN198_HUMAN
|
||||||
| NC score | 0.921565 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UBW7, O43212, O43434, O60898, Q5W0Q4, Q63HP0, Q8NE39, Q9H538, Q9UEU2 | Gene names | ZNF198, FIM, RAMP, ZMYM2 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 198 (Zinc finger MYM-type protein 2) (Fused in myeloproliferative disorders protein) (Rearranged in atypical myeloproliferative disorder protein). | |||||
|
ZMYM6_HUMAN
|
||||||
| NC score | 0.853362 (rank : 4) | θ value | 3.45727e-76 (rank : 4) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O95789, Q96IY0 | Gene names | ZMYM6, ZNF258 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 6 (Zinc finger protein 258). | |||||
|
ZMYM1_HUMAN
|
||||||
| NC score | 0.700171 (rank : 5) | θ value | 2.11701e-41 (rank : 5) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5SVZ6, Q7Z3Q4 | Gene names | ZMYM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 1. | |||||
|
ZMYM5_HUMAN
|
||||||
| NC score | 0.698302 (rank : 6) | θ value | 8.10077e-17 (rank : 6) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UJ78, Q5T6E2, Q96IY6, Q9NZY5, Q9UBW0, Q9UJ77 | Gene names | ZMYM5, ZNF198L1, ZNF237 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 5 (Zinc finger protein 237) (Zinc finger protein 198-like 1). | |||||
|
K1958_MOUSE
|
||||||
| NC score | 0.270623 (rank : 7) | θ value | 1.29631e-06 (rank : 8) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8C4P0, Q8BN62, Q8K1X8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1958 homolog. | |||||
|
K1958_HUMAN
|
||||||
| NC score | 0.267247 (rank : 8) | θ value | 1.29631e-06 (rank : 7) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N8K9, Q5T252, Q8TF43, Q96N02 | Gene names | KIAA1958 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1958. | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.092888 (rank : 9) | θ value | 0.00175202 (rank : 12) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
BCL9_MOUSE
|
||||||
| NC score | 0.074580 (rank : 10) | θ value | 0.0431538 (rank : 17) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
FYV1_MOUSE
|
||||||
| NC score | 0.064615 (rank : 11) | θ value | 0.0961366 (rank : 22) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z1T6 | Gene names | Pip5k3, Pikfyve | |||
|
Domain Architecture |

|
|||||
| Description | FYVE finger-containing phosphoinositide kinase (EC 2.7.1.68) (1- phosphatidylinositol-4-phosphate 5-kinase) (PIP5K) (PtdIns(4)P-5- kinase) (PIKfyve) (p235). | |||||
|
FYV1_HUMAN
|
||||||
| NC score | 0.064188 (rank : 12) | θ value | 0.0961366 (rank : 21) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y2I7, Q8NB67 | Gene names | PIP5K3, KIAA0981, PIKFYVE | |||
|
Domain Architecture |

|
|||||
| Description | FYVE finger-containing phosphoinositide kinase (EC 2.7.1.68) (1- phosphatidylinositol-4-phosphate 5-kinase) (Phosphatidylinositol-3- phosphate 5-kinase type III) (PIP5K) (PtdIns(4)P-5-kinase) (PIKfyve) (p235). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.061122 (rank : 13) | θ value | 0.279714 (rank : 32) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.060761 (rank : 14) | θ value | 0.21417 (rank : 29) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.058006 (rank : 15) | θ value | 4.03905 (rank : 111) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
P52K_MOUSE
|
||||||
| NC score | 0.057641 (rank : 16) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9CUX1, Q80Y58 | Gene names | Prkrir, Thap0 | |||
|
Domain Architecture |

|
|||||
| Description | 52 kDa repressor of the inhibitor of the protein kinase (p58IPK- interacting protein) (58 kDa interferon-induced protein kinase- interacting protein) (P52rIPK) (THAP domain-containing protein 0). | |||||
|
P52K_HUMAN
|
||||||
| NC score | 0.057538 (rank : 17) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43422, Q8WTW1, Q9Y3Z4 | Gene names | PRKRIR, DAP4, P52RIPK, THAP0 | |||
|
Domain Architecture |

|
|||||
| Description | 52 kDa repressor of the inhibitor of the protein kinase (p58IPK- interacting protein) (58 kDa interferon-induced protein kinase- interacting protein) (P52rIPK) (Death-associated protein 4) (THAP domain-containing protein 0). | |||||
|
ZN518_HUMAN
|
||||||
| NC score | 0.057495 (rank : 18) | θ value | 0.0736092 (rank : 18) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q6AHZ1, O15044, Q32MP4 | Gene names | ZNF518, KIAA0335 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 518. | |||||
|
ZN11B_HUMAN
|
||||||
| NC score | 0.055658 (rank : 19) | θ value | 0.00102713 (rank : 11) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 802 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q06732, Q86XY8, Q8NDW3 | Gene names | ZNF11B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 11B. | |||||
|
MTSS1_HUMAN
|
||||||
| NC score | 0.055165 (rank : 20) | θ value | 1.06291 (rank : 61) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O43312, Q8TCA2, Q96RX2 | Gene names | MTSS1, KIAA0429, MIM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein) (Metastasis suppressor YGL-1). | |||||
|
FHL2_HUMAN
|
||||||
| NC score | 0.054475 (rank : 21) | θ value | 1.38821 (rank : 65) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 533 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q14192, Q13229, Q13644, Q9P294 | Gene names | FHL2, DRAL, SLIM3 | |||
|
Domain Architecture |

|
|||||
| Description | Four and a half LIM domains protein 2 (FHL-2) (Skeletal muscle LIM- protein 3) (SLIM 3) (LIM-domain protein DRAL). | |||||
|
K1802_HUMAN
|
||||||
| NC score | 0.053584 (rank : 22) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
ZN271_HUMAN
|
||||||
| NC score | 0.053443 (rank : 23) | θ value | 0.000786445 (rank : 9) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 769 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q14591, Q96T29, Q9BSX2, Q9UN33, Q9Y5B7 | Gene names | ZNF271, ZNFEB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 271 (Zinc finger protein HZF7) (Zinc finger protein ZNFphex133) (Epstein-Barr virus-induced zinc finger protein) (ZNF-EB) (CT-ZFP48) (Zinc finger protein dp) (ZNF-dp). | |||||
|
ZN334_HUMAN
|
||||||
| NC score | 0.052703 (rank : 24) | θ value | 0.00390308 (rank : 14) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 778 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9HCZ1, Q5T6U2, Q9NVW4 | Gene names | ZNF334 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 334. | |||||
|
PRDM5_HUMAN
|
||||||
| NC score | 0.052476 (rank : 25) | θ value | 0.125558 (rank : 26) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9NQX1 | Gene names | PRDM5, PFM2 | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 5 (PR domain-containing protein 5). | |||||
|
PRD15_HUMAN
|
||||||
| NC score | 0.052237 (rank : 26) | θ value | 0.0961366 (rank : 23) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 805 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P57071, Q8N0X3, Q8NEX0, Q9NQV3 | Gene names | PRDM15, C21orf83, ZNF298 | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 15 (PR domain-containing protein 15) (Zinc finger protein 298). | |||||
|
MCM3A_HUMAN
|
||||||
| NC score | 0.052109 (rank : 27) | θ value | 0.279714 (rank : 33) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60318, Q9UMT4 | Gene names | MCM3AP, GANP, KIAA0572, MAP80 | |||
|
Domain Architecture |

|
|||||
| Description | 80 kDa MCM3-associated protein (GANP protein). | |||||
|
ZN33A_HUMAN
|
||||||
| NC score | 0.051330 (rank : 28) | θ value | 0.47712 (rank : 47) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q06730, Q5VZ86 | Gene names | ZNF33A, KIAA0065, KOX31, ZNF33 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 33A (Zinc finger protein KOX31) (Zinc finger and ZAK-associated protein with KRAB domain) (ZZaPK). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.051302 (rank : 29) | θ value | 0.00175202 (rank : 13) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
OGFR_MOUSE
|
||||||
| NC score | 0.051195 (rank : 30) | θ value | 0.279714 (rank : 35) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q99PG2, Q99L33 | Gene names | Ogfr | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor). | |||||
|
ZBT17_MOUSE
|
||||||
| NC score | 0.051138 (rank : 31) | θ value | 0.47712 (rank : 44) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q60821, Q60699 | Gene names | Zbtb17, Zfp100, Znf151 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 17 (Zinc finger protein 151) (Zinc finger protein 100) (Zfp-100) (Polyomavirus late initiator promoter-binding protein) (LP-1) (Zinc finger protein Z13). | |||||
|
ZN319_MOUSE
|
||||||
| NC score | 0.051019 (rank : 32) | θ value | 0.0961366 (rank : 24) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 762 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9ERR8 | Gene names | Znf319, Zfp319 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 319. | |||||
|
ZBT17_HUMAN
|
||||||
| NC score | 0.050406 (rank : 33) | θ value | 0.47712 (rank : 43) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 925 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q13105, Q15932, Q5JYB2, Q9NUC9 | Gene names | ZBTB17, MIZ1, ZNF151 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 17 (Zinc finger protein 151) (Myc-interacting zinc finger protein) (Miz-1 protein). | |||||
|
PRDM5_MOUSE
|
||||||
| NC score | 0.050353 (rank : 34) | θ value | 0.365318 (rank : 39) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9CXE0 | Gene names | Prdm5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PR domain zinc finger protein 5 (PR domain-containing protein 5). | |||||
|
ZNF28_HUMAN
|
||||||
| NC score | 0.050166 (rank : 35) | θ value | 0.0736092 (rank : 19) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P17035, Q5HYM9, Q6ZML9, Q6ZN56 | Gene names | ZNF28, KOX24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 28 (Zinc finger protein KOX24). | |||||
|
ZN319_HUMAN
|
||||||
| NC score | 0.049755 (rank : 36) | θ value | 0.279714 (rank : 37) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9P2F9 | Gene names | ZNF319, KIAA1388 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 319. | |||||
|
ZFP62_MOUSE
|
||||||
| NC score | 0.049487 (rank : 37) | θ value | 0.813845 (rank : 57) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8C827, Q3U3H0, Q62510, Q8BT00, Q8C2S6, Q8R5D1 | Gene names | Zfp62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 62 (Zfp-62) (ZT3). | |||||
|
ZFP11_MOUSE
|
||||||
| NC score | 0.049442 (rank : 38) | θ value | 0.279714 (rank : 36) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 763 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P10751, Q505E4, Q6PDR5, Q8BSI2, Q8C3R5 | Gene names | Zfp11, Krox-6.1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 11 (Zfp-11) (Krox-6.1A protein). | |||||
|
ZN268_HUMAN
|
||||||
| NC score | 0.048937 (rank : 39) | θ value | 0.47712 (rank : 46) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q14587, Q96RH4, Q9BZJ9 | Gene names | ZNF268 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 268 (Zinc finger protein HZF3). | |||||
|
ZNF14_HUMAN
|
||||||
| NC score | 0.048808 (rank : 40) | θ value | 0.163984 (rank : 28) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P17017, Q9ULZ5 | Gene names | ZNF14, GIOT4, KOX6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 14 (Zinc finger protein KOX6) (Gonadotropin- inducible transcription repressor 4) (GIOT-4). | |||||
|
ZN560_HUMAN
|
||||||
| NC score | 0.048761 (rank : 41) | θ value | 0.47712 (rank : 48) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q96MR9, Q495S9, Q495T1 | Gene names | ZNF560 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 560. | |||||
|
ZFP62_HUMAN
|
||||||
| NC score | 0.048215 (rank : 42) | θ value | 4.03905 (rank : 115) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8NB50 | Gene names | ZFP62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 62 homolog (Zfp-62). | |||||
|
ZNF43_HUMAN
|
||||||
| NC score | 0.048020 (rank : 43) | θ value | 0.813845 (rank : 60) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P17038, P28160, Q96DG1 | Gene names | ZNF43, KOX27, ZNF39 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 43 (Zinc protein HTF6) (Zinc finger protein KOX27). | |||||
|
FHL5_MOUSE
|
||||||
| NC score | 0.047787 (rank : 44) | θ value | 0.163984 (rank : 27) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9WTX7 | Gene names | Fhl5, Act | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Four and a half LIM domains protein 5 (FHL-5) (Activator of cAMP- responsive element modulator in testis) (Activator of CREM in testis). | |||||
|
ZN502_HUMAN
|
||||||
| NC score | 0.047646 (rank : 45) | θ value | 4.03905 (rank : 117) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 762 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8TBZ5 | Gene names | ZNF502 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 502. | |||||
|
ZN433_HUMAN
|
||||||
| NC score | 0.047591 (rank : 46) | θ value | 1.38821 (rank : 70) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 782 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8N7K0 | Gene names | ZNF433 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 433. | |||||
|
ZN473_HUMAN
|
||||||
| NC score | 0.047584 (rank : 47) | θ value | 3.0926 (rank : 105) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8WTR7, Q9ULS9, Q9Y4Q7 | Gene names | ZNF473, KIAA1141, ZFP100 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 473 (Zinc finger protein 100 homolog) (Zfp-100). | |||||
|
ZN206_HUMAN
|
||||||
| NC score | 0.047331 (rank : 48) | θ value | 0.813845 (rank : 58) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q96SZ4 | Gene names | ZNF206, ZSCAN10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 206 (Zinc finger and SCAN domain-containing protein 10). | |||||
|
ZN256_HUMAN
|
||||||
| NC score | 0.047186 (rank : 49) | θ value | 0.62314 (rank : 55) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9Y2P7, Q9BV71 | Gene names | ZNF256, BMZF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 256 (Bone marrow zinc finger 3) (BMZF-3). | |||||
|
ZNF35_HUMAN
|
||||||
| NC score | 0.047101 (rank : 50) | θ value | 0.47712 (rank : 49) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 761 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P13682, Q96D01 | Gene names | ZNF35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 35 (Zinc finger protein HF.10). | |||||
|
ZN226_HUMAN
|
||||||
| NC score | 0.046853 (rank : 51) | θ value | 0.47712 (rank : 45) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9NYT6, Q96TE6, Q9NS44 | Gene names | ZNF226 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 226. | |||||
|
ZN628_HUMAN
|
||||||
| NC score | 0.046818 (rank : 52) | θ value | 6.88961 (rank : 153) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 814 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q5EBL2, Q86X34 | Gene names | ZNF628 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 628. | |||||
|
RAI2_HUMAN
|
||||||
| NC score | 0.046804 (rank : 53) | θ value | 0.62314 (rank : 54) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y5P3, Q8N6X7 | Gene names | RAI2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 2. | |||||
|
ZN443_HUMAN
|
||||||
| NC score | 0.046780 (rank : 54) | θ value | 4.03905 (rank : 116) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9Y2A4 | Gene names | ZNF443 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 443 (Krueppel-type zinc finger protein ZK1). | |||||
|
ZN267_HUMAN
|
||||||
| NC score | 0.046713 (rank : 55) | θ value | 1.06291 (rank : 63) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q14586, Q8NE41, Q9NRJ0 | Gene names | ZNF267 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 267 (Zinc finger protein HZF2). | |||||
|
ZN324_HUMAN
|
||||||
| NC score | 0.046664 (rank : 56) | θ value | 0.365318 (rank : 40) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O75467 | Gene names | ZNF324 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 324 (Zinc finger protein ZF5128). | |||||
|
ZN221_HUMAN
|
||||||
| NC score | 0.046283 (rank : 57) | θ value | 1.38821 (rank : 69) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9UK13, Q9P1U8 | Gene names | ZNF221 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 221. | |||||
|
ZNF41_HUMAN
|
||||||
| NC score | 0.046188 (rank : 58) | θ value | 6.88961 (rank : 157) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 769 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P51814, Q96LE8, Q9UMC4, Q9UMV5, Q9UMV6, Q9UMV7, Q9UMV8, Q9UMV9, Q9UMW0, Q9UMW1 | Gene names | ZNF41 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 41. | |||||
|
MTSS1_MOUSE
|
||||||
| NC score | 0.046149 (rank : 59) | θ value | 2.36792 (rank : 84) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8R1S4, Q8BMM3, Q99LB3 | Gene names | Mtss1, Mim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein). | |||||
|
ZN628_MOUSE
|
||||||
| NC score | 0.046052 (rank : 60) | θ value | 6.88961 (rank : 154) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 815 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8CJ78, Q3U2L5, Q6P5B3 | Gene names | Znf628, Zec, Zfp628 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 628 (Zinc finger protein expressed in embryonal cells and certain adult organs). | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.045914 (rank : 61) | θ value | 5.27518 (rank : 131) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
MIZF_MOUSE
|
||||||
| NC score | 0.045555 (rank : 62) | θ value | 3.0926 (rank : 98) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8K1K9, Q8BWY0 | Gene names | Mizf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MBD2-interacting zinc finger protein (Methyl-CpG-binding protein 2- interacting zinc finger protein). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.045032 (rank : 63) | θ value | 0.00102713 (rank : 10) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
ZN674_HUMAN
|
||||||
| NC score | 0.044923 (rank : 64) | θ value | 0.813845 (rank : 59) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q2M3X9 | Gene names | ZNF674 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 674. | |||||
|
BCAS1_MOUSE
|
||||||
| NC score | 0.044561 (rank : 65) | θ value | 4.03905 (rank : 107) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80YN3, Q9CVA1 | Gene names | Bcas1, Nabc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast carcinoma amplified sequence 1 homolog (Novel amplified in breast cancer 1 homolog). | |||||
|
ZN595_HUMAN
|
||||||
| NC score | 0.044467 (rank : 66) | θ value | 6.88961 (rank : 152) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 798 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8IYB9 | Gene names | ZNF595 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 595. | |||||
|
GZF1_HUMAN
|
||||||
| NC score | 0.044455 (rank : 67) | θ value | 5.27518 (rank : 124) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 854 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9H116, Q96N08, Q9BQK9, Q9H117, Q9H6W6 | Gene names | GZF1, ZBTB23, ZNF336 | |||
|
Domain Architecture |

|
|||||
| Description | GDNF-inducible zinc finger protein 1 (Zinc finger protein 336) (Zinc finger and BTB domain-containing protein 23). | |||||
|
ZNF85_HUMAN
|
||||||
| NC score | 0.044032 (rank : 68) | θ value | 6.88961 (rank : 158) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q03923 | Gene names | ZNF85 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 85 (Zinc finger protein HPF4) (HTF1). | |||||
|
ZNF20_HUMAN
|
||||||
| NC score | 0.043896 (rank : 69) | θ value | 6.88961 (rank : 156) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P17024, Q9UG41 | Gene names | ZNF20, KOX13 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 20 (Zinc finger protein KOX13). | |||||
|
ZN264_HUMAN
|
||||||
| NC score | 0.043875 (rank : 70) | θ value | 5.27518 (rank : 133) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O43296, Q9P1V0 | Gene names | ZNF264, KIAA0412 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 264. | |||||
|
ZN669_HUMAN
|
||||||
| NC score | 0.043584 (rank : 71) | θ value | 3.0926 (rank : 106) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 746 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q96BR6, Q5VT39, Q9H9Q6 | Gene names | ZNF669 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 669. | |||||
|
ZBT38_HUMAN
|
||||||
| NC score | 0.043565 (rank : 72) | θ value | 6.88961 (rank : 150) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 885 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8NAP3 | Gene names | ZBTB38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and BTB domain-containing protein 38. | |||||
|
HIC2_HUMAN
|
||||||
| NC score | 0.043553 (rank : 73) | θ value | 2.36792 (rank : 80) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q96JB3, Q96KR3, Q9NSM9, Q9UPX9 | Gene names | HIC2, HRG22, KIAA1020, ZBTB30 | |||
|
Domain Architecture |

|
|||||
| Description | Hypermethylated in cancer 2 protein (Hic-2) (Hic-3) (HIC1-related gene on chromosome 22) (Zinc finger and BTB domain-containing protein 30). | |||||
|
FHL1_MOUSE
|
||||||
| NC score | 0.043513 (rank : 74) | θ value | 4.03905 (rank : 109) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 462 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P97447, O55181, Q8K318 | Gene names | Fhl1 | |||
|
Domain Architecture |

|
|||||
| Description | Four and a half LIM domains protein 1 (FHL-1) (Skeletal muscle LIM- protein 1) (SLIM 1) (SLIM) (KyoT) (RBP-associated molecule 14-1) (RAM14-1). | |||||
|
HIC2_MOUSE
|
||||||
| NC score | 0.043276 (rank : 75) | θ value | 3.0926 (rank : 95) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9JLZ6, Q3U030, Q3ULP4, Q5K036, Q8BSZ9, Q8C3T5 | Gene names | Hic2, Kiaa1020 | |||
|
Domain Architecture |

|
|||||
| Description | Hypermethylated in cancer 2 protein (Hic-2). | |||||
|
ZN382_HUMAN
|
||||||
| NC score | 0.043248 (rank : 76) | θ value | 8.99809 (rank : 175) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q96SR6 | Gene names | ZNF382 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 382 (KRAB/Zinc finger suppressor protein 1) (KS1). | |||||
|
ZN436_MOUSE
|
||||||
| NC score | 0.042989 (rank : 77) | θ value | 8.99809 (rank : 176) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 756 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8BPP0, Q69ZC6, Q8BQC2 | Gene names | Znf436, Kiaa1710, Zfp46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 436 (Zinc finger protein 46). | |||||
|
PRDM4_MOUSE
|
||||||
| NC score | 0.042937 (rank : 78) | θ value | 6.88961 (rank : 145) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q80V63 | Gene names | Prdm4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PR domain zinc finger protein 4 (PR domain-containing protein 4). | |||||
|
ZN561_HUMAN
|
||||||
| NC score | 0.042908 (rank : 79) | θ value | 6.88961 (rank : 151) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 734 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8N587, Q6PJS0 | Gene names | ZNF561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 561. | |||||
|
PRDM4_HUMAN
|
||||||
| NC score | 0.042900 (rank : 80) | θ value | 6.88961 (rank : 144) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9UKN5, Q9UFA6 | Gene names | PRDM4, PFM1 | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 4 (PR domain-containing protein 4). | |||||
|
ZN610_HUMAN
|
||||||
| NC score | 0.042805 (rank : 81) | θ value | 1.38821 (rank : 71) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8N9Z0, Q86YH8, Q8NDS9 | Gene names | ZNF610 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 610. | |||||
|
ZN649_HUMAN
|
||||||
| NC score | 0.042490 (rank : 82) | θ value | 6.88961 (rank : 155) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9BS31, Q9H9N2 | Gene names | ZNF649 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 649. | |||||
|
ZN509_MOUSE
|
||||||
| NC score | 0.042482 (rank : 83) | θ value | 2.36792 (rank : 92) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 990 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q8BXX2, Q8CID4, Q8K2Z6 | Gene names | Znf509, Zfp509 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 509. | |||||
|
GLU2B_MOUSE
|
||||||
| NC score | 0.041528 (rank : 84) | θ value | 1.81305 (rank : 72) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O08795, Q921X2 | Gene names | Prkcsh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glucosidase 2 subunit beta precursor (Glucosidase II subunit beta) (Protein kinase C substrate, 60.1 kDa protein, heavy chain) (PKCSH) (80K-H protein). | |||||
|
ZHX1_HUMAN
|
||||||
| NC score | 0.041266 (rank : 85) | θ value | 0.21417 (rank : 31) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UKY1, Q8IWD8 | Gene names | ZHX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc fingers and homeoboxes protein 1. | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.039419 (rank : 86) | θ value | 5.27518 (rank : 128) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.038384 (rank : 87) | θ value | 2.36792 (rank : 89) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.037124 (rank : 88) | θ value | 5.27518 (rank : 129) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.036365 (rank : 89) | θ value | 1.81305 (rank : 73) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MEF2C_MOUSE
|
||||||
| NC score | 0.035909 (rank : 90) | θ value | 0.279714 (rank : 34) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CFN5, Q8R0H1, Q9D7L0, Q9QW20 | Gene names | Mef2c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myocyte-specific enhancer factor 2C. | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.035438 (rank : 91) | θ value | 1.06291 (rank : 62) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
ARHG1_MOUSE
|
||||||
| NC score | 0.035093 (rank : 92) | θ value | 0.0330416 (rank : 16) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61210, O89074, Q80YE8, Q80YE9, Q91VL3 | Gene names | Arhgef1, Lbcl2, Lsc | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 1 (Lymphoid blast crisis-like 2) (Lbc's second cousin). | |||||
|
PSL1_HUMAN
|
||||||
| NC score | 0.034956 (rank : 93) | θ value | 1.81305 (rank : 75) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.034886 (rank : 94) | θ value | 1.38821 (rank : 67) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.034545 (rank : 95) | θ value | 2.36792 (rank : 83) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MEF2C_HUMAN
|
||||||
| NC score | 0.034160 (rank : 96) | θ value | 0.62314 (rank : 52) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q06413 | Gene names | MEF2C | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2C. | |||||
|
TNR25_HUMAN
|
||||||
| NC score | 0.033941 (rank : 97) | θ value | 2.36792 (rank : 90) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q93038, O00275, O00276, O00277, O00278, O00279, O00280, O14865, O14866, P78507, P78515, Q92983, Q93036, Q93037, Q99722, Q99830, Q99831, Q9BY86, Q9UME0, Q9UME1, Q9UME5 | Gene names | TNFRSF25, APO3, DDR3, DR3, TNFRSF12, WSL, WSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 25 precursor (WSL-1 protein) (Apoptosis-mediating receptor DR3) (Apoptosis-mediating receptor TRAMP) (Death domain receptor 3) (WSL protein) (Apoptosis- inducing receptor AIR) (Apo-3) (Lymphocyte-associated receptor of death) (LARD). | |||||
|
TUR8_MOUSE
|
||||||
| NC score | 0.032813 (rank : 98) | θ value | 5.27518 (rank : 130) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P19473 | Gene names | Trap1a, P1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor rejection antigen P815A. | |||||
|
ASPP1_MOUSE
|
||||||
| NC score | 0.032808 (rank : 99) | θ value | 0.365318 (rank : 38) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
CO5A1_HUMAN
|
||||||
| NC score | 0.032132 (rank : 100) | θ value | 0.62314 (rank : 50) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P20908, Q15094, Q5SUX4 | Gene names | COL5A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(V) chain precursor. | |||||
|
PI5PA_MOUSE
|
||||||
| NC score | 0.031642 (rank : 101) | θ value | 1.38821 (rank : 68) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P59644 | Gene names | Pib5pa | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
SPN90_MOUSE
|
||||||
| NC score | 0.030597 (rank : 102) | θ value | 8.99809 (rank : 172) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9ESJ4, Q68G72 | Gene names | Nckipsd, Spin90, Wasbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3 adapter protein SPIN90 (NCK-interacting protein with SH3 domain) (SH3 protein interacting with Nck, 90 kDa) (VacA-interacting protein, 54 kDa) (VIP54) (N-WASP-interacting protein, 90kDa) (Wiskott-Aldrich syndrome protein-binding protein WISH) (N-WASP-binding protein). | |||||
|
ZFY21_HUMAN
|
||||||
| NC score | 0.030289 (rank : 103) | θ value | 5.27518 (rank : 132) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BQ24, Q86T05, Q96LT1 | Gene names | ZFYVE21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 21. | |||||
|
PDLI7_HUMAN
|
||||||
| NC score | 0.029754 (rank : 104) | θ value | 0.125558 (rank : 25) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 547 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NR12, Q14250, Q5XG82, Q6NVZ5, Q96C91, Q9BXB8, Q9BXB9 | Gene names | PDLIM7, ENIGMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.029676 (rank : 105) | θ value | 2.36792 (rank : 85) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
CO002_HUMAN
|
||||||
| NC score | 0.029110 (rank : 106) | θ value | 5.27518 (rank : 121) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NZP6 | Gene names | C15orf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C15orf2. | |||||
|
RN125_MOUSE
|
||||||
| NC score | 0.028384 (rank : 107) | θ value | 3.0926 (rank : 101) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D9R0, Q52KL4, Q8C7F2 | Gene names | Rnf125 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 125 (EC 6.3.2.-). | |||||
|
ZHX1_MOUSE
|
||||||
| NC score | 0.027838 (rank : 108) | θ value | 2.36792 (rank : 91) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P70121, Q8BQ68, Q8C6T4, Q8CJG3 | Gene names | Zhx1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc fingers and homeoboxes protein 1. | |||||
|
K1543_MOUSE
|
||||||
| NC score | 0.027771 (rank : 109) | θ value | 5.27518 (rank : 127) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q80VC9, Q5DTW9, Q8BUZ0 | Gene names | Kiaa1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
MKRN3_MOUSE
|
||||||
| NC score | 0.027162 (rank : 110) | θ value | 1.81305 (rank : 74) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q60764, Q7TQE4 | Gene names | Mkrn3, Zfp127, Znf127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127). | |||||
|
NU214_HUMAN
|
||||||
| NC score | 0.026965 (rank : 111) | θ value | 8.99809 (rank : 168) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
TXLNA_HUMAN
|
||||||
| NC score | 0.026768 (rank : 112) | θ value | 0.21417 (rank : 30) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 982 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P40222, Q66K62, Q86T54, Q86T85, Q86T86, Q86Y86, Q86YW3, Q8N2Y3 | Gene names | TXLNA, TXLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
SNPC4_HUMAN
|
||||||
| NC score | 0.025591 (rank : 113) | θ value | 3.0926 (rank : 102) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5SXM2, Q9Y6P7 | Gene names | SNAPC4, SNAP190 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor subunit alpha) (PSE-binding factor subunit alpha) (PTF subunit alpha). | |||||
|
GA2L2_HUMAN
|
||||||
| NC score | 0.025456 (rank : 114) | θ value | 0.813845 (rank : 56) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8NHY3, Q8NHY4 | Gene names | GAS2L2, GAR17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2) (GAS2-related protein on chromosome 17) (GAR17 protein). | |||||
|
TNR22_MOUSE
|
||||||
| NC score | 0.025153 (rank : 115) | θ value | 6.88961 (rank : 149) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ER62, Q8VHB9, Q9CZA4 | Gene names | Tnfrsf22, Dctrailr2, Tnfrh2, Tnfrsf1al2 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 22 (Tumor necrosis factor receptor p60 homolog 2) (TNF receptor family member SOBa) (Decoy TRAIL receptor 2) (TNF receptor homolog 2). | |||||
|
CCD96_MOUSE
|
||||||
| NC score | 0.024619 (rank : 116) | θ value | 3.0926 (rank : 94) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9CR92 | Gene names | Ccdc96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 96. | |||||
|
SL9A5_HUMAN
|
||||||
| NC score | 0.024506 (rank : 117) | θ value | 2.36792 (rank : 86) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14940, Q9Y626 | Gene names | SLC9A5, NHE5 | |||
|
Domain Architecture |
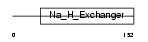
|
|||||
| Description | Sodium/hydrogen exchanger 5 (Na(+)/H(+) exchanger 5) (NHE-5) (Solute carrier family 9 member 5). | |||||
|
PALB2_MOUSE
|
||||||
| NC score | 0.024504 (rank : 118) | θ value | 3.0926 (rank : 100) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q3U0P1, Q6NZG9, Q7TMQ4, Q8CEA9 | Gene names | Palb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Partner and localizer of BRCA2. | |||||
|
ZFY28_HUMAN
|
||||||
| NC score | 0.023891 (rank : 119) | θ value | 8.99809 (rank : 174) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9HCC9 | Gene names | ZFYVE28, KIAA1643 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 28. | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.023506 (rank : 120) | θ value | 3.0926 (rank : 99) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
AP180_HUMAN
|
||||||
| NC score | 0.022484 (rank : 121) | θ value | 8.99809 (rank : 159) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O60641, Q5JX13, Q68DL9, Q6P9D3, Q9NTY7 | Gene names | SNAP91, KIAA0656 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin coat assembly protein AP180 (Clathrin coat-associated protein AP180) (91 kDa synaptosomal-associated protein). | |||||
|
ARHGB_HUMAN
|
||||||
| NC score | 0.021416 (rank : 122) | θ value | 8.99809 (rank : 160) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O15085 | Gene names | ARHGEF11, KIAA0380 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 11 (PDZ-RhoGEF). | |||||
|
TACC2_MOUSE
|
||||||
| NC score | 0.021341 (rank : 123) | θ value | 1.81305 (rank : 76) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JJG0 | Gene names | Tacc2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2. | |||||
|
RAI1_HUMAN
|
||||||
| NC score | 0.021339 (rank : 124) | θ value | 8.99809 (rank : 170) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
RPC11_MOUSE
|
||||||
| NC score | 0.021270 (rank : 125) | θ value | 4.03905 (rank : 114) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CQZ7, Q9D1U1 | Gene names | Polr3k | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerases III 12.5 kDa polypeptide (EC 2.7.7.6) (RNA polymerase III subunit K) (RNA polymerase III C11 subunit). | |||||
|
JHD1A_MOUSE
|
||||||
| NC score | 0.020905 (rank : 126) | θ value | 3.0926 (rank : 97) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P59997, Q3U1M5, Q3UR56, Q3V3Q1, Q69ZT4 | Gene names | Fbxl11, Jhdm1a, Kiaa1004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 1A (EC 1.14.11.-) (F-box/LRR-repeat protein 11) (F-box and leucine-rich repeat protein 11). | |||||
|
JHD1A_HUMAN
|
||||||
| NC score | 0.020892 (rank : 127) | θ value | 3.0926 (rank : 96) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y2K7, Q49A21, Q4G0M3, Q69YY8, Q9BVH5, Q9H7H5, Q9UK66 | Gene names | FBXL11, FBL7, JHDM1A, KIAA1004 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 1A (EC 1.14.11.-) (F-box/LRR-repeat protein 11) (F-box and leucine-rich repeat protein 11) (F-box protein FBL7) (F-box protein Lilina). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.019862 (rank : 128) | θ value | 6.88961 (rank : 143) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
TF2B_MOUSE
|
||||||
| NC score | 0.019499 (rank : 129) | θ value | 6.88961 (rank : 147) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P62915, P29053 | Gene names | Gtf2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor IIB (General transcription factor TFIIB) (RNA polymerase II alpha initiation factor). | |||||
|
TF2B_HUMAN
|
||||||
| NC score | 0.019498 (rank : 130) | θ value | 6.88961 (rank : 146) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q00403, Q5JS30 | Gene names | GTF2B, TF2B, TFIIB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIB (General transcription factor TFIIB) (S300-II). | |||||
|
DOCK4_MOUSE
|
||||||
| NC score | 0.019488 (rank : 131) | θ value | 2.36792 (rank : 78) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P59764 | Gene names | Dock4, Kiaa0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.019088 (rank : 132) | θ value | 8.99809 (rank : 167) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
DAXX_MOUSE
|
||||||
| NC score | 0.019041 (rank : 133) | θ value | 5.27518 (rank : 123) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35613, Q9QWT8, Q9QWV3 | Gene names | Daxx | |||
|
Domain Architecture |

|
|||||
| Description | Death domain-associated protein 6 (Daxx). | |||||
|
CSKI1_MOUSE
|
||||||
| NC score | 0.018966 (rank : 134) | θ value | 6.88961 (rank : 135) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
ADDA_MOUSE
|
||||||
| NC score | 0.018515 (rank : 135) | θ value | 6.88961 (rank : 134) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9QYC0, Q9JLE3 | Gene names | Add1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-adducin (Erythrocyte adducin subunit alpha). | |||||
|
CSPG2_MOUSE
|
||||||
| NC score | 0.018138 (rank : 136) | θ value | 0.0961366 (rank : 20) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q62059, Q62058, Q9CUU0 | Gene names | Cspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M). | |||||
|
LHX9_MOUSE
|
||||||
| NC score | 0.017912 (rank : 137) | θ value | 2.36792 (rank : 82) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9WUH2, Q9QYQ5, Q9QYQ6, Q9QZ00, Q9WU44 | Gene names | Lhx9 | |||
|
Domain Architecture |

|
|||||
| Description | LIM/homeobox protein Lhx9. | |||||
|
GAK_MOUSE
|
||||||
| NC score | 0.017862 (rank : 138) | θ value | 0.0252991 (rank : 15) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99KY4, Q6P1I8, Q6P9S5, Q8BM74, Q8K0Q4 | Gene names | Gak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||
|
ASPP2_HUMAN
|
||||||
| NC score | 0.016208 (rank : 139) | θ value | 5.27518 (rank : 119) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q13625, Q12892, Q86X75, Q96KQ3 | Gene names | TP53BP2, ASPP2, BBP | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 2 (Tumor suppressor p53-binding protein 2) (p53-binding protein 2) (p53BP2) (53BP2) (Bcl2-binding protein) (Bbp) (NY-REN-51 antigen). | |||||
|
CTR9_MOUSE
|
||||||
| NC score | 0.016123 (rank : 140) | θ value | 8.99809 (rank : 164) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q62018, Q3UFF5, Q3UY40, Q66JX4, Q7TPS6, Q8BND9, Q8BRD1, Q8C9W7, Q8C9Y3, Q8CHI1 | Gene names | Ctr9, Kiaa0155, Sh2bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1) (Tetratricopeptide repeat-containing, SH2-binding phosphoprotein of 150 kDa) (TPR-containing, SH2-binding phosphoprotein of 150 kDa) (p150TSP). | |||||
|
TNR21_MOUSE
|
||||||
| NC score | 0.015454 (rank : 141) | θ value | 3.0926 (rank : 104) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9EPU5, Q91W77, Q91XH9 | Gene names | Tnfrsf21, Dr6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 21 precursor (TNFR- related death receptor 6) (Death receptor 6). | |||||
|
HXB3_HUMAN
|
||||||
| NC score | 0.014411 (rank : 142) | θ value | 0.47712 (rank : 42) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P14651, O95615, P17484 | Gene names | HOXB3, HOX2G | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B3 (Hox-2G) (Hox-2.7). | |||||
|
HPS4_MOUSE
|
||||||
| NC score | 0.014387 (rank : 143) | θ value | 6.88961 (rank : 137) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99KG7 | Gene names | Hps4, Le | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hermansky-Pudlak syndrome 4 protein homolog (Light-ear protein) (Le protein). | |||||
|
LHX9_HUMAN
|
||||||
| NC score | 0.013999 (rank : 144) | θ value | 6.88961 (rank : 138) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NQ69, Q5VUE3, Q9BYU6, Q9NQ70 | Gene names | LHX9 | |||
|
Domain Architecture |

|
|||||
| Description | LIM/homeobox protein Lhx9. | |||||
|
CO4A1_HUMAN
|
||||||
| NC score | 0.013696 (rank : 145) | θ value | 5.27518 (rank : 122) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P02462 | Gene names | COL4A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IV) chain precursor. | |||||
|
CNTRB_MOUSE
|
||||||
| NC score | 0.013672 (rank : 146) | θ value | 1.38821 (rank : 64) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8CB62, Q5NCF3 | Gene names | Cntrob, Lip8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
HXB3_MOUSE
|
||||||
| NC score | 0.013598 (rank : 147) | θ value | 0.62314 (rank : 51) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P09026, P10285, Q4PJ06, Q61680 | Gene names | Hoxb3, Hox-2.7, Hoxb-3 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B3 (Hox-2.7) (Homeobox protein MH-23). | |||||
|
NFAC1_HUMAN
|
||||||
| NC score | 0.012781 (rank : 148) | θ value | 8.99809 (rank : 166) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O95644, Q12865, Q15793 | Gene names | NFATC1, NFAT2, NFATC | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 1 (NFAT transcription complex cytosolic component) (NF-ATc1) (NF-ATc). | |||||
|
SOX13_MOUSE
|
||||||
| NC score | 0.012438 (rank : 149) | θ value | 2.36792 (rank : 87) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q04891, Q922L3 | Gene names | Sox13, Sox-13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein. | |||||
|
INCE_HUMAN
|
||||||
| NC score | 0.012104 (rank : 150) | θ value | 4.03905 (rank : 110) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
MEGF8_HUMAN
|
||||||
| NC score | 0.011959 (rank : 151) | θ value | 6.88961 (rank : 140) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7Z7M0, O75097 | Gene names | MEGF8, EGFL4, KIAA0817 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 8 (EGF-like domain- containing protein 4) (Multiple EGF-like domain protein 4). | |||||
|
MEGF8_MOUSE
|
||||||
| NC score | 0.011840 (rank : 152) | θ value | 6.88961 (rank : 141) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 520 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P60882, Q80TR3, Q80V41, Q8BMN9, Q8JZW7, Q8K0J3 | Gene names | Megf8, Egfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 8 (EGF-like domain- containing protein 4) (Multiple EGF-like domain protein 4). | |||||
|
CE016_HUMAN
|
||||||
| NC score | 0.011605 (rank : 153) | θ value | 8.99809 (rank : 163) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NC24, Q6P4E7, Q6UXY2 | Gene names | C5orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C5orf16. | |||||
|
MATN2_HUMAN
|
||||||
| NC score | 0.011571 (rank : 154) | θ value | 4.03905 (rank : 112) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O00339, Q6UWA5, Q7Z5X1, Q8NDE6, Q96FT5, Q9NSZ1 | Gene names | MATN2 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-2 precursor. | |||||
|
TMUB1_HUMAN
|
||||||
| NC score | 0.011290 (rank : 155) | θ value | 8.99809 (rank : 173) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BVT8 | Gene names | TMUB1, C7orf21 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane and ubiquitin-like domain-containing protein 1 (Ubiquitin-like protein SB144). | |||||
|
PTN22_HUMAN
|
||||||
| NC score | 0.011247 (rank : 156) | θ value | 0.62314 (rank : 53) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y2R2, O95063, O95064 | Gene names | PTPN22, PTPN8 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 22 (EC 3.1.3.48) (Hematopoietic cell protein-tyrosine phosphatase 70Z-PEP) (Lymphoid phosphatase) (LyP). | |||||
|
SYGP1_HUMAN
|
||||||
| NC score | 0.010965 (rank : 157) | θ value | 3.0926 (rank : 103) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96PV0, Q8TCS2, Q9UGE2 | Gene names | SYNGAP1, KIAA1938 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein SynGAP (Synaptic Ras-GTPase-activating protein 1) (Synaptic Ras-GAP 1) (Neuronal RasGAP). | |||||
|
BMPER_HUMAN
|
||||||
| NC score | 0.010806 (rank : 158) | θ value | 8.99809 (rank : 161) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N8U9 | Gene names | BMPER | |||
|
Domain Architecture |

|
|||||
| Description | BMP-binding endothelial regulator protein precursor (Bone morphogenetic protein-binding endothelial cell precursor-derived regulator) (Protein crossveinless-2) (hCV2). | |||||
|
SPTN2_HUMAN
|
||||||
| NC score | 0.010804 (rank : 159) | θ value | 2.36792 (rank : 88) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O15020, O14872, O14873 | Gene names | SPTBN2, KIAA0302 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 2 (Spectrin, non-erythroid beta chain 2) (Beta-III spectrin). | |||||
|
HS105_HUMAN
|
||||||
| NC score | 0.010439 (rank : 160) | θ value | 2.36792 (rank : 81) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92598, O95739, Q5TBM6, Q5TBM8, Q9UPC4 | Gene names | HSPH1, HSP105, HSP110, KIAA0201 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein 105 kDa (Heat shock 110 kDa protein) (Antigen NY- CO-25). | |||||
|
CHL1_MOUSE
|
||||||
| NC score | 0.010073 (rank : 161) | θ value | 0.47712 (rank : 41) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70232, Q8BS24, Q8C6W0, Q8C823, Q8VBY7 | Gene names | Chl1, Call | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural cell adhesion molecule L1-like protein precursor (Cell adhesion molecule with homology to L1CAM) (Close homolog of L1) (Chl1-like protein). | |||||
|
ZPBP2_MOUSE
|
||||||
| NC score | 0.009999 (rank : 162) | θ value | 8.99809 (rank : 177) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6X786, Q6X785, Q9DA91 | Gene names | Zpbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zona pellucida-binding protein 2 precursor. | |||||
|
TIF1G_MOUSE
|
||||||
| NC score | 0.009418 (rank : 163) | θ value | 6.88961 (rank : 148) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99PP7, Q6SI71, Q6ZPX5 | Gene names | Trim33, Kiaa1113 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33). | |||||
|
SIPA1_MOUSE
|
||||||
| NC score | 0.009031 (rank : 164) | θ value | 8.99809 (rank : 171) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P46062, P70204 | Gene names | Sipa1, Spa-1, Spa1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal-induced proliferation-associated protein 1 (Sipa-1) (GTPase- activating protein Spa-1). | |||||
|
IRF5_MOUSE
|
||||||
| NC score | 0.008439 (rank : 165) | θ value | 5.27518 (rank : 125) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P56477 | Gene names | Irf5 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 5 (IRF-5). | |||||
|
PRGR_MOUSE
|
||||||
| NC score | 0.008348 (rank : 166) | θ value | 4.03905 (rank : 113) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q00175 | Gene names | Pgr, Nr3c3, Pr | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone receptor (PR). | |||||
|
DZIP3_MOUSE
|
||||||
| NC score | 0.008248 (rank : 167) | θ value | 6.88961 (rank : 136) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TPV2, Q80TU4, Q8BYK7 | Gene names | Dzip3, Kiaa0675 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein DZIP3 (EC 6.3.2.-) (DAZ-interacting protein 3 homolog). | |||||
|
MAST4_HUMAN
|
||||||
| NC score | 0.008021 (rank : 168) | θ value | 1.38821 (rank : 66) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
VDR_HUMAN
|
||||||
| NC score | 0.007954 (rank : 169) | θ value | 1.81305 (rank : 77) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P11473 | Gene names | VDR, NR1I1 | |||
|
Domain Architecture |

|
|||||
| Description | Vitamin D3 receptor (VDR) (1,25-dihydroxyvitamin D3 receptor). | |||||
|
DSCL1_HUMAN
|
||||||
| NC score | 0.007876 (rank : 170) | θ value | 2.36792 (rank : 79) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8TD84, Q76MU9, Q8IZY3, Q8IZY4, Q8WXU7, Q9ULT7 | Gene names | DSCAML1, DSCAM2, KIAA1132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Down syndrome cell adhesion molecule-like protein 1 precursor (Down syndrome cell adhesion molecule 2). | |||||
|
CEAM5_HUMAN
|
||||||
| NC score | 0.007800 (rank : 171) | θ value | 5.27518 (rank : 120) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P06731 | Gene names | CEACAM5, CEA | |||
|
Domain Architecture |

|
|||||
| Description | Carcinoembryonic antigen-related cell adhesion molecule 5 precursor (Carcinoembryonic antigen) (CEA) (Meconium antigen 100) (CD66e antigen). | |||||
|
ACHA4_HUMAN
|
||||||
| NC score | 0.006826 (rank : 172) | θ value | 3.0926 (rank : 93) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P43681, Q4JGR7 | Gene names | CHRNA4, NACRA4 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal acetylcholine receptor protein subunit alpha-4 precursor. | |||||
|
OSBL5_HUMAN
|
||||||
| NC score | 0.006463 (rank : 173) | θ value | 8.99809 (rank : 169) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H0X9, Q9BZB0, Q9P1Z4 | Gene names | OSBPL5, KIAA1534, ORP5 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 5 (OSBP-related protein 5) (ORP-5). | |||||
|
ANK3_HUMAN
|
||||||
| NC score | 0.006153 (rank : 174) | θ value | 5.27518 (rank : 118) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
ITA7_HUMAN
|
||||||
| NC score | 0.005795 (rank : 175) | θ value | 5.27518 (rank : 126) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13683, O43197, Q9NY89, Q9UET0, Q9UEV2 | Gene names | ITGA7 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-7 precursor [Contains: Integrin alpha-7 heavy chain; Integrin alpha-7 light chain]. | |||||
|
NID2_HUMAN
|
||||||
| NC score | 0.005165 (rank : 176) | θ value | 6.88961 (rank : 142) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14112, O43710 | Gene names | NID2 | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-2 precursor (NID-2) (Osteonidogen). | |||||
|
LYST_MOUSE
|
||||||
| NC score | 0.004582 (rank : 177) | θ value | 8.99809 (rank : 165) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97412, Q62403, Q8VBS6 | Gene names | Lyst, Bg, Chs1 | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal-trafficking regulator (Beige protein) (CHS1 homolog). | |||||
|
LRFN3_HUMAN
|
||||||
| NC score | 0.004575 (rank : 178) | θ value | 6.88961 (rank : 139) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BTN0, Q6UY10 | Gene names | LRFN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat and fibronectin type-III domain-containing protein 3 precursor. | |||||
|
CD1E_HUMAN
|
||||||
| NC score | 0.003354 (rank : 179) | θ value | 8.99809 (rank : 162) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P15812, Q9UMM1, Q9Y5M3 | Gene names | CD1E | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface glycoprotein CD1e precursor (CD1e antigen) (R2G1). | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.003274 (rank : 180) | θ value | 4.03905 (rank : 108) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||