Please be patient as the page loads
|
ZN198_HUMAN
|
||||||
| SwissProt Accessions | Q9UBW7, O43212, O43434, O60898, Q5W0Q4, Q63HP0, Q8NE39, Q9H538, Q9UEU2 | Gene names | ZNF198, FIM, RAMP, ZMYM2 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 198 (Zinc finger MYM-type protein 2) (Fused in myeloproliferative disorders protein) (Rearranged in atypical myeloproliferative disorder protein). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ZMYM3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.915078 (rank : 3) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q14202, O15089 | Gene names | ZMYM3, DXS6673E, KIAA0385, ZNF261 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 3 (Zinc finger protein 261). | |||||
|
ZMYM3_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.921565 (rank : 2) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JLM4, Q80U17 | Gene names | Zmym3, Kiaa0385, Zfp261, Znf261 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 3 (Zinc finger protein 261) (DXHXS6673E protein). | |||||
|
ZN198_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 128 | |
| SwissProt Accessions | Q9UBW7, O43212, O43434, O60898, Q5W0Q4, Q63HP0, Q8NE39, Q9H538, Q9UEU2 | Gene names | ZNF198, FIM, RAMP, ZMYM2 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 198 (Zinc finger MYM-type protein 2) (Fused in myeloproliferative disorders protein) (Rearranged in atypical myeloproliferative disorder protein). | |||||
|
ZMYM6_HUMAN
|
||||||
| θ value | 7.63087e-108 (rank : 4) | NC score | 0.885907 (rank : 4) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O95789, Q96IY0 | Gene names | ZMYM6, ZNF258 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 6 (Zinc finger protein 258). | |||||
|
ZMYM5_HUMAN
|
||||||
| θ value | 2.39424e-77 (rank : 5) | NC score | 0.841695 (rank : 5) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UJ78, Q5T6E2, Q96IY6, Q9NZY5, Q9UBW0, Q9UJ77 | Gene names | ZMYM5, ZNF198L1, ZNF237 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 5 (Zinc finger protein 237) (Zinc finger protein 198-like 1). | |||||
|
ZMYM1_HUMAN
|
||||||
| θ value | 2.76489e-41 (rank : 6) | NC score | 0.725486 (rank : 6) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5SVZ6, Q7Z3Q4 | Gene names | ZMYM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 1. | |||||
|
K1958_HUMAN
|
||||||
| θ value | 2.61198e-07 (rank : 7) | NC score | 0.269889 (rank : 8) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N8K9, Q5T252, Q8TF43, Q96N02 | Gene names | KIAA1958 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1958. | |||||
|
K1958_MOUSE
|
||||||
| θ value | 2.61198e-07 (rank : 8) | NC score | 0.273730 (rank : 7) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8C4P0, Q8BN62, Q8K1X8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1958 homolog. | |||||
|
ZN11B_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 9) | NC score | 0.039586 (rank : 18) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 802 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q06732, Q86XY8, Q8NDW3 | Gene names | ZNF11B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 11B. | |||||
|
DSPP_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 10) | NC score | 0.045279 (rank : 13) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
ZN236_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 11) | NC score | 0.034731 (rank : 27) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UL36, Q9UL37 | Gene names | ZNF236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 236. | |||||
|
ZBT17_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 12) | NC score | 0.037042 (rank : 20) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q60821, Q60699 | Gene names | Zbtb17, Zfp100, Znf151 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger and BTB domain-containing protein 17 (Zinc finger protein 151) (Zinc finger protein 100) (Zfp-100) (Polyomavirus late initiator promoter-binding protein) (LP-1) (Zinc finger protein Z13). | |||||
|
ZN408_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 13) | NC score | 0.033330 (rank : 32) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9H9D4 | Gene names | ZNF408, PFM14, PRDM17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 408 (PR-domain zinc finger protein 17). | |||||
|
ZNF12_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 14) | NC score | 0.033155 (rank : 33) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P17014, Q9ULZ6 | Gene names | ZNF12, GIOT3, KOX3, ZNF325 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 12 (Zinc finger protein KOX3) (Zinc finger protein 325) (Gonadotropin-inducible transcription repressor 3) (GIOT-3). | |||||
|
SETX_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 15) | NC score | 0.044754 (rank : 14) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7Z333, O75120, Q3KQX4, Q68DW5, Q6AZD7, Q7Z3J6, Q8WX33, Q9H9D1, Q9NVP9 | Gene names | SETX, ALS4, KIAA0625, SCAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable helicase senataxin (EC 3.6.1.-) (SEN1 homolog). | |||||
|
ZN248_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 16) | NC score | 0.032982 (rank : 34) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8NDW4, Q9UMP3 | Gene names | ZNF248 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 248. | |||||
|
ZN502_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 17) | NC score | 0.033483 (rank : 31) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 762 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8TBZ5 | Gene names | ZNF502 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 502. | |||||
|
ZN334_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 18) | NC score | 0.037463 (rank : 19) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 778 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9HCZ1, Q5T6U2, Q9NVW4 | Gene names | ZNF334 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 334. | |||||
|
DMP1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 19) | NC score | 0.050237 (rank : 11) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
FRAS1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 20) | NC score | 0.028277 (rank : 65) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q80T14, Q80TC7, Q811H8, Q8BPZ4 | Gene names | Fras1, Kiaa1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 21) | NC score | 0.027529 (rank : 66) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
KLK11_HUMAN
|
||||||
| θ value | 0.21417 (rank : 22) | NC score | 0.007910 (rank : 117) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UBX7, O75837, Q9NS65 | Gene names | KLK11, PRSS20, TLSP | |||
|
Domain Architecture |
|
|||||
| Description | Kallikrein-11 precursor (EC 3.4.21.-) (Hippostasin) (Trypsin-like protease). | |||||
|
MATN2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 23) | NC score | 0.018244 (rank : 81) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O00339, Q6UWA5, Q7Z5X1, Q8NDE6, Q96FT5, Q9NSZ1 | Gene names | MATN2 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-2 precursor. | |||||
|
PRDM5_HUMAN
|
||||||
| θ value | 0.279714 (rank : 24) | NC score | 0.036922 (rank : 21) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9NQX1 | Gene names | PRDM5, PFM2 | |||
|
Domain Architecture |
|
|||||
| Description | PR domain zinc finger protein 5 (PR domain-containing protein 5). | |||||
|
KR107_HUMAN
|
||||||
| θ value | 0.62314 (rank : 25) | NC score | 0.022023 (rank : 74) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P60409, Q70LJ2 | Gene names | KRTAP10-7, KAP10.7, KAP18-7, KRTAP10.7, KRTAP18-7, KRTAP18.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-7 (Keratin-associated protein 10.7) (High sulfur keratin-associated protein 10.7) (Keratin-associated protein 18-7) (Keratin-associated protein 18.7). | |||||
|
LAP2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 26) | NC score | 0.017834 (rank : 84) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80TH2, Q8BQ14, Q8CE41, Q8K171, Q99JU3, Q9JI47 | Gene names | Erbb2ip, Erbin, Kiaa1225, Lap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP2 (Erbb2-interacting protein) (Erbin) (Densin-180-like protein). | |||||
|
UB7I1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 27) | NC score | 0.044115 (rank : 15) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NWF9, Q6Y691, Q75ML7, Q7Z2H7, Q7Z7C1, Q8NHW7, Q9NYT1 | Gene names | UBCE7IP1, ZIN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin ligase TRIAD3 (EC 6.3.2.-) (Ubiquitin-conjugating enzyme 7-interacting protein 1) (Zinc finger protein inhibiting NF-kappa-B) (Triad domain-containing protein 3). | |||||
|
UB7I1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 28) | NC score | 0.043177 (rank : 17) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P58283, Q3U493, Q3UE56, Q3UGM3, Q68FN0, Q6P1H8, Q6PWY5, Q8BN27, Q8C1U3 | Gene names | Ubce7ip1, Triad3, Uip83, Zin | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin ligase TRIAD3 (EC 6.3.2.-) (Ubiquitin-conjugating enzyme 7-interacting protein 1) (UbcM4-interacting protein 83) (Triad domain- containing protein 3). | |||||
|
KR101_HUMAN
|
||||||
| θ value | 0.813845 (rank : 29) | NC score | 0.025369 (rank : 69) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P60331 | Gene names | KRTAP10-1, KAP10.1, KAP18-1, KRTAP10.1, KRTAP18-1, KRTAP18.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-1 (Keratin-associated protein 10.1) (High sulfur keratin-associated protein 10.1) (Keratin-associated protein 18-1) (Keratin-associated protein 18.1). | |||||
|
NU153_HUMAN
|
||||||
| θ value | 0.813845 (rank : 30) | NC score | 0.034121 (rank : 29) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P49790 | Gene names | NUP153 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup153 (Nucleoporin Nup153) (153 kDa nucleoporin). | |||||
|
DEK_MOUSE
|
||||||
| θ value | 1.06291 (rank : 31) | NC score | 0.034493 (rank : 28) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7TNV0, Q80VC5, Q8BZV6 | Gene names | Dek | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
GON4L_HUMAN
|
||||||
| θ value | 1.06291 (rank : 32) | NC score | 0.035175 (rank : 25) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
GON4L_MOUSE
|
||||||
| θ value | 1.06291 (rank : 33) | NC score | 0.033486 (rank : 30) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9DB00, Q80TB4, Q91YI9 | Gene names | Gon4l, Gon4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
LAP2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 34) | NC score | 0.017943 (rank : 83) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96RT1, Q86W38, Q9NR18, Q9NW48, Q9ULJ5 | Gene names | ERBB2IP, ERBIN, KIAA1225, LAP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP2 (Erbb2-interacting protein) (Erbin) (Densin-180-like protein). | |||||
|
TCF17_MOUSE
|
||||||
| θ value | 1.06291 (rank : 35) | NC score | 0.030667 (rank : 45) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q61751 | Gene names | Znf354a, Kid1, Tcf17, Zfp354a | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 354A (Transcription factor 17) (Renal transcription factor Kid-1) (Kidney, ischemia, and developmentally- regulated protein 1). | |||||
|
ZBT17_HUMAN
|
||||||
| θ value | 1.06291 (rank : 36) | NC score | 0.035291 (rank : 22) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 925 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q13105, Q15932, Q5JYB2, Q9NUC9 | Gene names | ZBTB17, MIZ1, ZNF151 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 17 (Zinc finger protein 151) (Myc-interacting zinc finger protein) (Miz-1 protein). | |||||
|
ARHG7_HUMAN
|
||||||
| θ value | 1.38821 (rank : 37) | NC score | 0.013701 (rank : 97) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14155, Q6P9G3, Q6PII2, Q86W63, Q8N3M1 | Gene names | ARHGEF7, COOL1, KIAA0142, P85SPR, PAK3BP, PIXB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 7 (PAK-interacting exchange factor beta) (Beta-Pix) (COOL-1) (p85). | |||||
|
ZFP59_MOUSE
|
||||||
| θ value | 1.38821 (rank : 38) | NC score | 0.030949 (rank : 42) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P16373, Q61849 | Gene names | Zfp59, Mfg2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 59 (Zfp-59) (Zinc finger protein Mfg-2). | |||||
|
ZN567_HUMAN
|
||||||
| θ value | 1.38821 (rank : 39) | NC score | 0.031300 (rank : 37) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 769 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8N184, Q6N044 | Gene names | ZNF567 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 567. | |||||
|
ANR12_HUMAN
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.009554 (rank : 111) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6UB98, O94951, Q658K1, Q6QMF7, Q9H231, Q9H784 | Gene names | ANKRD12, ANCO2, KIAA0874 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 12 (Ankyrin repeat-containing cofactor 2) (GAC-1 protein). | |||||
|
KR18_HUMAN
|
||||||
| θ value | 1.81305 (rank : 41) | NC score | 0.030743 (rank : 44) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 763 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9HCG1, Q96JC5, Q9H7N6 | Gene names | KIAA1611 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein Kr18 (HKr18). | |||||
|
TIE2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.001928 (rank : 128) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1204 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q02858 | Gene names | Tek, Hyk, Tie-2, Tie2 | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-1 receptor precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor TIE-2) (mTIE2) (Tyrosine-protein kinase receptor TEK) (p140 TEK) (Tunica interna endothelial cell kinase) (HYK) (STK1) (CD202b antigen). | |||||
|
ZN337_HUMAN
|
||||||
| θ value | 1.81305 (rank : 43) | NC score | 0.030823 (rank : 43) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 781 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Y3M9, Q9Y3Y5 | Gene names | ZNF337 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 337. | |||||
|
ZN557_HUMAN
|
||||||
| θ value | 1.81305 (rank : 44) | NC score | 0.029919 (rank : 53) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 752 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8N988, Q6PEJ3, Q9BTZ1 | Gene names | ZNF557 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 557. | |||||
|
EYA1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 45) | NC score | 0.012890 (rank : 102) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99502 | Gene names | EYA1 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 1 (EC 3.1.3.48). | |||||
|
HTSF1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 46) | NC score | 0.021343 (rank : 77) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O43719, Q59G06, Q99730 | Gene names | HTATSF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 (Tat-SF1). | |||||
|
PRDM5_MOUSE
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.035115 (rank : 26) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9CXE0 | Gene names | Prdm5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PR domain zinc finger protein 5 (PR domain-containing protein 5). | |||||
|
XPC_MOUSE
|
||||||
| θ value | 2.36792 (rank : 48) | NC score | 0.022391 (rank : 72) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P51612, P54732, Q3TKI2, Q920M1, Q9DBW7 | Gene names | Xpc | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-C cells homolog (Xeroderma pigmentosum group C-complementing protein homolog) (p125). | |||||
|
ZIM3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 49) | NC score | 0.030317 (rank : 47) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q96PE6 | Gene names | ZIM3 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger imprinted 3. | |||||
|
AIOL_MOUSE
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.029209 (rank : 59) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O08900 | Gene names | Ikzf3, Zfpn1a3, Znfn1a3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein Aiolos (IKAROS family zinc finger protein 3). | |||||
|
DAXX_HUMAN
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.027177 (rank : 68) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UER7, O14747, O15141, O15208, Q9BWI3 | Gene names | DAXX, BING2, DAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Death domain-associated protein 6 (Daxx) (hDaxx) (Fas death domain- associated protein) (ETS1-associated protein 1) (EAP1). | |||||
|
ELOA1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.022140 (rank : 73) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14241, Q8IXH1 | Gene names | TCEB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor B polypeptide 3 (RNA polymerase II transcription factor SIII subunit A1) (SIII p110) (Elongin-A) (EloA) (Elongin 110 kDa subunit). | |||||
|
INHBA_MOUSE
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | 0.008821 (rank : 112) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q04998 | Gene names | Inhba | |||
|
Domain Architecture |
|
|||||
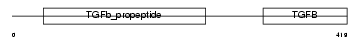
| Description | Inhibin beta A chain precursor (Activin beta-A chain). | |||||
|
K0423_HUMAN
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.018128 (rank : 82) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y4F4, Q68D66, Q6PG27 | Gene names | KIAA0423 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0423. | |||||
|
MEP1B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.012982 (rank : 101) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q16820, Q670J1 | Gene names | MEP1B | |||
|
Domain Architecture |

|
|||||
| Description | Meprin A subunit beta precursor (EC 3.4.24.18) (Endopeptidase-2) (N- benzoyl-L-tyrosyl-P-amino-benzoic acid hydrolase subunit beta) (PABA peptide hydrolase) (PPH beta). | |||||
|
PHC1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | 0.021434 (rank : 76) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P78364, Q8WVM3, Q9BU63 | Gene names | PHC1, EDR1, PH1 | |||
|
Domain Architecture |

|
|||||
| Description | Polyhomeotic-like protein 1 (hPH1) (Early development regulatory protein 1). | |||||
|
PHC1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.021931 (rank : 75) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q64028, P70359, Q64307, Q8BZ80 | Gene names | Phc1, Edr, Edr1, Rae28 | |||
|
Domain Architecture |

|
|||||
| Description | Polyhomeotic-like protein 1 (mPH1) (Early development regulatory protein 1) (RAE-28). | |||||
|
ZN155_HUMAN
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.031010 (rank : 40) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q12901, Q9UIE1, Q9UK14 | Gene names | ZNF155 | |||
|
Domain Architecture |
|
|||||
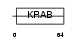
| Description | Zinc finger protein 155. | |||||
|
ZN33A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.035189 (rank : 24) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q06730, Q5VZ86 | Gene names | ZNF33A, KIAA0065, KOX31, ZNF33 | |||
|
Domain Architecture |
|
|||||
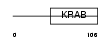
| Description | Zinc finger protein 33A (Zinc finger protein KOX31) (Zinc finger and ZAK-associated protein with KRAB domain) (ZZaPK). | |||||
|
BRCA1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.015983 (rank : 92) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P48754, Q60957, Q60983 | Gene names | Brca1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 1 susceptibility protein homolog. | |||||
|
GZF1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.031129 (rank : 39) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 854 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9H116, Q96N08, Q9BQK9, Q9H117, Q9H6W6 | Gene names | GZF1, ZBTB23, ZNF336 | |||
|
Domain Architecture |
|
|||||
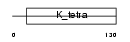
| Description | GDNF-inducible zinc finger protein 1 (Zinc finger protein 336) (Zinc finger and BTB domain-containing protein 23). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.047836 (rank : 12) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
LRRC1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.012343 (rank : 103) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BTT6, Q9HAC0, Q9NVF1 | Gene names | LRRC1, LANO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 1 (LAP and no PDZ protein) (LANO adapter protein). | |||||
|
MYST3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.010306 (rank : 107) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
PRD15_HUMAN
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.035205 (rank : 23) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 805 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P57071, Q8N0X3, Q8NEX0, Q9NQV3 | Gene names | PRDM15, C21orf83, ZNF298 | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 15 (PR domain-containing protein 15) (Zinc finger protein 298). | |||||
|
PSMD1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 66) | NC score | 0.017189 (rank : 85) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99460, Q53TI2, Q6GMU5, Q6P2P4, Q6PJM7, Q6PKG9, Q86VU1, Q8IV79 | Gene names | PSMD1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 1 (26S proteasome regulatory subunit RPN2) (26S proteasome regulatory subunit S1) (26S proteasome subunit p112). | |||||
|
TOPB1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.015587 (rank : 93) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6ZQF0, Q6P6P0, Q80Y33, Q8BUI0, Q8BUK1, Q8R348, Q91VX3, Q922X8 | Gene names | Topbp1, Kiaa0259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA topoisomerase II-binding protein 1 (DNA topoisomerase IIbeta- binding protein 1) (TopBP1). | |||||
|
TRI32_HUMAN
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.015233 (rank : 95) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13049, Q9NQP8 | Gene names | TRIM32, HT2A | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 32 (EC 6.3.2.-) (Zinc finger protein HT2A) (72 kDa Tat-interacting protein). | |||||
|
TRI32_MOUSE
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.015302 (rank : 94) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CH72, Q8K055 | Gene names | Trim32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 32 (EC 6.3.2.-). | |||||
|
XPA_MOUSE
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.028521 (rank : 63) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q64267 | Gene names | Xpa, Xpac | |||
|
Domain Architecture |
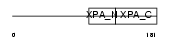
|
|||||
| Description | DNA-repair protein complementing XP-A cells homolog (Xeroderma pigmentosum group A-complementing protein homolog). | |||||
|
ZN221_HUMAN
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.031506 (rank : 36) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UK13, Q9P1U8 | Gene names | ZNF221 | |||
|
Domain Architecture |
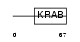
|
|||||
| Description | Zinc finger protein 221. | |||||
|
ZN343_HUMAN
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.030137 (rank : 49) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q6P1L6, Q5JXU8, Q5JXU9, Q8N8F2, Q96EX8, Q96NA5, Q9BQB0 | Gene names | ZNF343 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 343. | |||||
|
ZN347_HUMAN
|
||||||
| θ value | 4.03905 (rank : 73) | NC score | 0.030275 (rank : 48) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 756 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96SE7, Q8TCN1 | Gene names | ZNF347, ZNF1111 | |||
|
Domain Architecture |
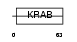
|
|||||
| Description | Zinc finger protein 347 (Zinc finger protein 1111). | |||||
|
PHF7_MOUSE
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.013378 (rank : 98) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DAG9, Q6PG81 | Gene names | Phf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 7 (Testis development protein NYD-SP6). | |||||
|
PTPRN_MOUSE
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.004591 (rank : 122) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60673, Q62129 | Gene names | Ptprn, Ptp35 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase-like N precursor (R-PTP-N) (PTP IA-2). | |||||
|
RAPH1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.010310 (rank : 106) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
RBBP6_MOUSE
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.016867 (rank : 86) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
TCF17_HUMAN
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.029932 (rank : 52) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 797 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O60765, Q9UNJ8 | Gene names | ZNF354A, EZNF, HKL1, TCF17 | |||
|
Domain Architecture |
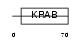
|
|||||
| Description | Zinc finger protein 354A (Transcription factor 17) (Zinc finger protein eZNF). | |||||
|
ZCH10_MOUSE
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.027247 (rank : 67) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CX48 | Gene names | Zcchc10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
ZN449_HUMAN
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.028540 (rank : 62) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 758 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q6P9G9, Q5JRZ8, Q6NZX2, Q8N3Q1, Q8TED7 | Gene names | ZNF449, ZSCAN19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 449 (Zinc finger and SCAN domain-containing protein 19). | |||||
|
ZN605_HUMAN
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.030103 (rank : 50) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q86T29, Q86T91 | Gene names | ZNF605 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 605. | |||||
|
ZN718_HUMAN
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.029064 (rank : 60) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 761 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q3SXZ3, Q3SXZ4, Q3SXZ5 | Gene names | ZNF718 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 718. | |||||
|
ZNF43_HUMAN
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.031836 (rank : 35) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P17038, P28160, Q96DG1 | Gene names | ZNF43, KOX27, ZNF39 | |||
|
Domain Architecture |
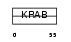
|
|||||
| Description | Zinc finger protein 43 (Zinc protein HTF6) (Zinc finger protein KOX27). | |||||
|
ARHG7_MOUSE
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.010098 (rank : 109) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ES28, O08757, Q9ES27 | Gene names | Arhgef7, Pak3bp | |||
|
Domain Architecture |
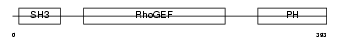
|
|||||
| Description | Rho guanine nucleotide exchange factor 7 (PAK-interacting exchange factor beta) (Beta-Pix) (p85SPR). | |||||
|
ATS1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.003140 (rank : 127) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UHI8, Q9NSJ8, Q9P2K0, Q9UH83, Q9UP80 | Gene names | ADAMTS1, KIAA1346, METH1 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-1 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 1) (ADAM-TS 1) (ADAM-TS1) (METH-1). | |||||
|
BORIS_HUMAN
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.031174 (rank : 38) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 743 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8NI51, Q5JUG4, Q9BZ30, Q9NQJ3 | Gene names | CTCFL, BORIS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional repressor CTCFL (CCCTC-binding factor) (Brother of the regulator of imprinted sites) (Zinc finger protein CTCF-T) (CTCF paralog). | |||||
|
DLX1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.003823 (rank : 125) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P56177, Q53ZU4, Q8IYB2 | Gene names | DLX1 | |||
|
Domain Architecture |
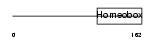
|
|||||
| Description | Homeobox protein DLX-1. | |||||
|
DLX1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.003818 (rank : 126) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q64317 | Gene names | Dlx1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein DLX-1. | |||||
|
DSPP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.028385 (rank : 64) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
ELOA1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.018375 (rank : 80) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CB77, Q80VB2, Q9R0Q5 | Gene names | Tceb3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor B polypeptide 3 (RNA polymerase II transcription factor SIII subunit A) (SIII p110) (Elongin-A) (EloA) (Elongin 110 kDa subunit). | |||||
|
KRA93_HUMAN
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.016771 (rank : 87) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BYQ3 | Gene names | KRTAP9-3, KAP9.3, KRTAP9.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-3 (Keratin-associated protein 9.3) (Ultrahigh sulfur keratin-associated protein 9.3). | |||||
|
NEBL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.008581 (rank : 114) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O76041, Q9UIC4 | Gene names | NEBL | |||
|
Domain Architecture |

|
|||||
| Description | Nebulette (Actin-binding Z-disk protein). | |||||
|
PRDM4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.029960 (rank : 51) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9UKN5, Q9UFA6 | Gene names | PRDM4, PFM1 | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 4 (PR domain-containing protein 4). | |||||
|
PRDM4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.029915 (rank : 54) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q80V63 | Gene names | Prdm4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PR domain zinc finger protein 4 (PR domain-containing protein 4). | |||||
|
SDCG1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.014874 (rank : 96) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60524, Q8WW70, Q9NWG1 | Gene names | SDCCAG1 | |||
|
Domain Architecture |
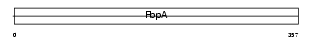
|
|||||
| Description | Serologically defined colon cancer antigen 1 (Antigen NY-CO-1). | |||||
|
SMG1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.008038 (rank : 116) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BKX6, Q5DU42, Q6ZQC0, Q8BLU4, Q8BWJ5, Q8BXD3 | Gene names | Smg1, Kiaa0421 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1). | |||||
|
SYCP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.016752 (rank : 88) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BX26, O75763, Q5JX11, Q9NTX8, Q9UG27 | Gene names | SYCP2, SCP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptonemal complex protein 2 (SCP-2) (Synaptonemal complex lateral element protein) (hsSCP2). | |||||
|
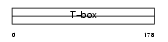
TACD1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.016066 (rank : 91) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P16422, P18180, Q96C47 | Gene names | TACSTD1, GA733-2, M1S2, TROP1 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor-associated calcium signal transducer 1 precursor (Major gastrointestinal tumor-associated protein GA733-2) (Epithelial cell surface antigen) (Epithelial glycoprotein) (EGP) (Adenocarcinoma- associated antigen) (KSA) (KS 1/4 antigen) (Cell surface glycoprotein Trop-1) (CD326 antigen). | |||||
|
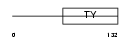
TBX5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.004779 (rank : 121) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70326, Q9R003 | Gene names | Tbx5 | |||
|
Domain Architecture |
|
|||||
| Description | T-box transcription factor TBX5 (T-box protein 5). | |||||
|
TR13B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.018444 (rank : 79) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ET35, Q9DBZ3 | Gene names | Tnfrsf13b, Taci | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor necrosis factor receptor superfamily member 13B (Transmembrane activator and CAML interactor). | |||||
|
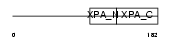
XPA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.025309 (rank : 71) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P23025, Q6LCW7 | Gene names | XPA, XPAC | |||
|
Domain Architecture |
|
|||||
| Description | DNA-repair protein complementing XP-A cells (Xeroderma pigmentosum group A-complementing protein). | |||||
|
ZBT24_HUMAN
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | 0.028597 (rank : 61) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O43167, Q5TED5, Q8N455 | Gene names | ZBTB24, KIAA0441, ZNF450 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and BTB domain-containing protein 24 (Zinc finger protein 450). | |||||
|
ZN669_HUMAN
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | 0.029597 (rank : 56) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 746 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q96BR6, Q5VT39, Q9H9Q6 | Gene names | ZNF669 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 669. | |||||
|
ZNF44_HUMAN
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | 0.029588 (rank : 57) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 757 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P15621, P17018, Q9ULZ7 | Gene names | ZNF44, KOX7 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 44 (Zinc finger protein KOX7) (Gonadotropin- inducible transcription repressor 2) (GIOT-2). | |||||
|
ZNF92_HUMAN
|
||||||
| θ value | 6.88961 (rank : 105) | NC score | 0.029763 (rank : 55) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q03936, Q8N492, Q8NB35 | Gene names | ZNF92 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 92 (Zinc finger protein HTF12). | |||||
|
AP3B2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.008757 (rank : 113) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13367, O14808 | Gene names | AP3B2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (AP-3 complex beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain) (Neuron-specific vesicle coat protein beta-NAP). | |||||
|
CHD2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.004475 (rank : 123) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O14647 | Gene names | CHD2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 2 (EC 3.6.1.-) (ATP- dependent helicase CHD2) (CHD-2). | |||||
|
DX26B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.010399 (rank : 105) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BND4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DDX26B. | |||||
|
FBN1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.006769 (rank : 119) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 563 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61554, Q60826 | Gene names | Fbn1, Fbn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-1 precursor. | |||||
|
FHL5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.025350 (rank : 70) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 523 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q5TD97, Q5TD98, Q8WW21, Q9NQU2 | Gene names | FHL5, ACT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Four and a half LIM domains protein 5 (FHL-5) (Activator of cAMP- responsive element modulator in testis) (Activator of CREM in testis). | |||||
|
FRAS1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.019294 (rank : 78) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q86XX4, Q86UZ4, Q8N3U9, Q8NAU7, Q96JW7, Q9H6N9, Q9P228 | Gene names | FRAS1, KIAA1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.044040 (rank : 16) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
KRA98_HUMAN
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.016195 (rank : 90) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BYQ0 | Gene names | KRTAP9-8, KAP9.8, KRTAP9.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-8 (Keratin-associated protein 9.8) (Ultrahigh sulfur keratin-associated protein 9.8). | |||||
|
LRRC1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.011016 (rank : 104) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80VQ1, Q3UI50, Q8BKR1, Q8BUS9 | Gene names | Lrrc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 1. | |||||
|
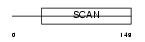
MZF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.029217 (rank : 58) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P28698, Q9NRY0, Q9UBW2 | Gene names | MZF1, MZF, ZNF42, ZSCAN6 | |||
|
Domain Architecture |
|
|||||
| Description | Myeloid zinc finger 1 (MZF-1) (Zinc finger protein 42) (Zinc finger and SCAN domain-containing protein 6). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.013075 (rank : 100) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
NELL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.008490 (rank : 115) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q92832, Q9Y472 | Gene names | NELL1, NRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein NELL1 precursor (NEL-like protein 1) (Nel-related protein 1). | |||||
|
NTRK3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | -0.001151 (rank : 130) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1115 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q16288, Q12827, Q16289 | Gene names | NTRK3, TRKC | |||
|
Domain Architecture |

|
|||||
| Description | NT-3 growth factor receptor precursor (EC 2.7.10.1) (Neurotrophic tyrosine kinase receptor type 3) (TrkC tyrosine kinase) (GP145-TrkC) (Trk-C). | |||||
|
OTOF_MOUSE
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.006016 (rank : 120) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ESF1, Q9ESF2 | Gene names | Otof, Fer1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Otoferlin (Fer-1-like protein 2). | |||||
|
RBM19_MOUSE
|
||||||
| θ value | 8.99809 (rank : 120) | NC score | 0.004085 (rank : 124) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R3C6, Q8BHR0, Q9CW63 | Gene names | Rbm19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable RNA-binding protein 19 (RNA-binding motif protein 19). | |||||
|
SMBT1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 121) | NC score | 0.009897 (rank : 110) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JMD1, Q6NZD3, Q8CFS1 | Gene names | Sfmbt1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scm-like with four MBT domains protein 1. | |||||
|
SSPO_MOUSE
|
||||||
| θ value | 8.99809 (rank : 122) | NC score | 0.006994 (rank : 118) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
TIE2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | 0.000534 (rank : 129) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1192 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q02763 | Gene names | TEK, TIE2 | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-1 receptor precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor TIE-2) (hTIE2) (Tyrosine-protein kinase receptor TEK) (p140 TEK) (Tunica interna endothelial cell kinase) (CD202b antigen). | |||||
|
TMM24_MOUSE
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.010210 (rank : 108) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80X80 | Gene names | Tmem24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 24. | |||||
|
UB7I4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.013208 (rank : 99) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P50876 | Gene names | RNF144, KIAA0161, UBCE7IP4 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-conjugating enzyme 7-interacting protein 4 (UbcM4- interacting protein 4) (RING finger protein 144). | |||||
|
ZBT38_HUMAN
|
||||||
| θ value | 8.99809 (rank : 126) | NC score | 0.030586 (rank : 46) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 885 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8NAP3 | Gene names | ZBTB38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and BTB domain-containing protein 38. | |||||
|
ZN624_HUMAN
|
||||||
| θ value | 8.99809 (rank : 127) | NC score | 0.030950 (rank : 41) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9P2J8 | Gene names | ZNF624, KIAA1349 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 624. | |||||
|
ZNFX1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.016210 (rank : 89) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8R151, Q3U016, Q3UD71, Q3UEZ0 | Gene names | ZNFX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NFX1-type zinc finger-containing protein 1. | |||||
|
P52K_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.058251 (rank : 10) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43422, Q8WTW1, Q9Y3Z4 | Gene names | PRKRIR, DAP4, P52RIPK, THAP0 | |||
|
Domain Architecture |

|
|||||
| Description | 52 kDa repressor of the inhibitor of the protein kinase (p58IPK- interacting protein) (58 kDa interferon-induced protein kinase- interacting protein) (P52rIPK) (Death-associated protein 4) (THAP domain-containing protein 0). | |||||
|
P52K_MOUSE
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.059316 (rank : 9) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9CUX1, Q80Y58 | Gene names | Prkrir, Thap0 | |||
|
Domain Architecture |

|
|||||
| Description | 52 kDa repressor of the inhibitor of the protein kinase (p58IPK- interacting protein) (58 kDa interferon-induced protein kinase- interacting protein) (P52rIPK) (THAP domain-containing protein 0). | |||||
|
ZN198_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 128 | |
| SwissProt Accessions | Q9UBW7, O43212, O43434, O60898, Q5W0Q4, Q63HP0, Q8NE39, Q9H538, Q9UEU2 | Gene names | ZNF198, FIM, RAMP, ZMYM2 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 198 (Zinc finger MYM-type protein 2) (Fused in myeloproliferative disorders protein) (Rearranged in atypical myeloproliferative disorder protein). | |||||
|
ZMYM3_MOUSE
|
||||||
| NC score | 0.921565 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JLM4, Q80U17 | Gene names | Zmym3, Kiaa0385, Zfp261, Znf261 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 3 (Zinc finger protein 261) (DXHXS6673E protein). | |||||
|
ZMYM3_HUMAN
|
||||||
| NC score | 0.915078 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q14202, O15089 | Gene names | ZMYM3, DXS6673E, KIAA0385, ZNF261 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 3 (Zinc finger protein 261). | |||||
|
ZMYM6_HUMAN
|
||||||
| NC score | 0.885907 (rank : 4) | θ value | 7.63087e-108 (rank : 4) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O95789, Q96IY0 | Gene names | ZMYM6, ZNF258 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 6 (Zinc finger protein 258). | |||||
|
ZMYM5_HUMAN
|
||||||
| NC score | 0.841695 (rank : 5) | θ value | 2.39424e-77 (rank : 5) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UJ78, Q5T6E2, Q96IY6, Q9NZY5, Q9UBW0, Q9UJ77 | Gene names | ZMYM5, ZNF198L1, ZNF237 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 5 (Zinc finger protein 237) (Zinc finger protein 198-like 1). | |||||
|
ZMYM1_HUMAN
|
||||||
| NC score | 0.725486 (rank : 6) | θ value | 2.76489e-41 (rank : 6) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5SVZ6, Q7Z3Q4 | Gene names | ZMYM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 1. | |||||
|
K1958_MOUSE
|
||||||
| NC score | 0.273730 (rank : 7) | θ value | 2.61198e-07 (rank : 8) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8C4P0, Q8BN62, Q8K1X8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1958 homolog. | |||||
|
K1958_HUMAN
|
||||||
| NC score | 0.269889 (rank : 8) | θ value | 2.61198e-07 (rank : 7) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N8K9, Q5T252, Q8TF43, Q96N02 | Gene names | KIAA1958 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1958. | |||||
|
P52K_MOUSE
|
||||||
| NC score | 0.059316 (rank : 9) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9CUX1, Q80Y58 | Gene names | Prkrir, Thap0 | |||
|
Domain Architecture |

|
|||||
| Description | 52 kDa repressor of the inhibitor of the protein kinase (p58IPK- interacting protein) (58 kDa interferon-induced protein kinase- interacting protein) (P52rIPK) (THAP domain-containing protein 0). | |||||
|
P52K_HUMAN
|
||||||
| NC score | 0.058251 (rank : 10) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43422, Q8WTW1, Q9Y3Z4 | Gene names | PRKRIR, DAP4, P52RIPK, THAP0 | |||
|
Domain Architecture |

|
|||||
| Description | 52 kDa repressor of the inhibitor of the protein kinase (p58IPK- interacting protein) (58 kDa interferon-induced protein kinase- interacting protein) (P52rIPK) (Death-associated protein 4) (THAP domain-containing protein 0). | |||||
|
DMP1_HUMAN
|
||||||
| NC score | 0.050237 (rank : 11) | θ value | 0.163984 (rank : 19) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.047836 (rank : 12) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.045279 (rank : 13) | θ value | 0.0252991 (rank : 10) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
SETX_HUMAN
|
||||||
| NC score | 0.044754 (rank : 14) | θ value | 0.0736092 (rank : 15) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7Z333, O75120, Q3KQX4, Q68DW5, Q6AZD7, Q7Z3J6, Q8WX33, Q9H9D1, Q9NVP9 | Gene names | SETX, ALS4, KIAA0625, SCAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable helicase senataxin (EC 3.6.1.-) (SEN1 homolog). | |||||
|
UB7I1_HUMAN
|
||||||
| NC score | 0.044115 (rank : 15) | θ value | 0.62314 (rank : 27) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NWF9, Q6Y691, Q75ML7, Q7Z2H7, Q7Z7C1, Q8NHW7, Q9NYT1 | Gene names | UBCE7IP1, ZIN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin ligase TRIAD3 (EC 6.3.2.-) (Ubiquitin-conjugating enzyme 7-interacting protein 1) (Zinc finger protein inhibiting NF-kappa-B) (Triad domain-containing protein 3). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.044040 (rank : 16) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
UB7I1_MOUSE
|
||||||
| NC score | 0.043177 (rank : 17) | θ value | 0.62314 (rank : 28) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P58283, Q3U493, Q3UE56, Q3UGM3, Q68FN0, Q6P1H8, Q6PWY5, Q8BN27, Q8C1U3 | Gene names | Ubce7ip1, Triad3, Uip83, Zin | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin ligase TRIAD3 (EC 6.3.2.-) (Ubiquitin-conjugating enzyme 7-interacting protein 1) (UbcM4-interacting protein 83) (Triad domain- containing protein 3). | |||||
|
ZN11B_HUMAN
|
||||||
| NC score | 0.039586 (rank : 18) | θ value | 0.0113563 (rank : 9) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 802 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q06732, Q86XY8, Q8NDW3 | Gene names | ZNF11B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 11B. | |||||
|
ZN334_HUMAN
|
||||||
| NC score | 0.037463 (rank : 19) | θ value | 0.0961366 (rank : 18) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 778 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9HCZ1, Q5T6U2, Q9NVW4 | Gene names | ZNF334 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 334. | |||||
|
ZBT17_MOUSE
|
||||||
| NC score | 0.037042 (rank : 20) | θ value | 0.0563607 (rank : 12) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q60821, Q60699 | Gene names | Zbtb17, Zfp100, Znf151 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 17 (Zinc finger protein 151) (Zinc finger protein 100) (Zfp-100) (Polyomavirus late initiator promoter-binding protein) (LP-1) (Zinc finger protein Z13). | |||||
|
PRDM5_HUMAN
|
||||||
| NC score | 0.036922 (rank : 21) | θ value | 0.279714 (rank : 24) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9NQX1 | Gene names | PRDM5, PFM2 | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 5 (PR domain-containing protein 5). | |||||
|
ZBT17_HUMAN
|
||||||
| NC score | 0.035291 (rank : 22) | θ value | 1.06291 (rank : 36) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 925 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q13105, Q15932, Q5JYB2, Q9NUC9 | Gene names | ZBTB17, MIZ1, ZNF151 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 17 (Zinc finger protein 151) (Myc-interacting zinc finger protein) (Miz-1 protein). | |||||
|
PRD15_HUMAN
|
||||||
| NC score | 0.035205 (rank : 23) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 805 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P57071, Q8N0X3, Q8NEX0, Q9NQV3 | Gene names | PRDM15, C21orf83, ZNF298 | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 15 (PR domain-containing protein 15) (Zinc finger protein 298). | |||||
|
ZN33A_HUMAN
|
||||||
| NC score | 0.035189 (rank : 24) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q06730, Q5VZ86 | Gene names | ZNF33A, KIAA0065, KOX31, ZNF33 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 33A (Zinc finger protein KOX31) (Zinc finger and ZAK-associated protein with KRAB domain) (ZZaPK). | |||||
|
GON4L_HUMAN
|
||||||
| NC score | 0.035175 (rank : 25) | θ value | 1.06291 (rank : 32) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
PRDM5_MOUSE
|
||||||
| NC score | 0.035115 (rank : 26) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9CXE0 | Gene names | Prdm5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PR domain zinc finger protein 5 (PR domain-containing protein 5). | |||||
|
ZN236_HUMAN
|
||||||
| NC score | 0.034731 (rank : 27) | θ value | 0.0252991 (rank : 11) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UL36, Q9UL37 | Gene names | ZNF236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 236. | |||||
|
DEK_MOUSE
|
||||||
| NC score | 0.034493 (rank : 28) | θ value | 1.06291 (rank : 31) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7TNV0, Q80VC5, Q8BZV6 | Gene names | Dek | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
NU153_HUMAN
|
||||||
| NC score | 0.034121 (rank : 29) | θ value | 0.813845 (rank : 30) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P49790 | Gene names | NUP153 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup153 (Nucleoporin Nup153) (153 kDa nucleoporin). | |||||
|
GON4L_MOUSE
|
||||||
| NC score | 0.033486 (rank : 30) | θ value | 1.06291 (rank : 33) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9DB00, Q80TB4, Q91YI9 | Gene names | Gon4l, Gon4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
ZN502_HUMAN
|
||||||
| NC score | 0.033483 (rank : 31) | θ value | 0.0736092 (rank : 17) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 762 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8TBZ5 | Gene names | ZNF502 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 502. | |||||
|
ZN408_HUMAN
|
||||||
| NC score | 0.033330 (rank : 32) | θ value | 0.0563607 (rank : 13) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9H9D4 | Gene names | ZNF408, PFM14, PRDM17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 408 (PR-domain zinc finger protein 17). | |||||
|
ZNF12_HUMAN
|
||||||
| NC score | 0.033155 (rank : 33) | θ value | 0.0563607 (rank : 14) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P17014, Q9ULZ6 | Gene names | ZNF12, GIOT3, KOX3, ZNF325 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 12 (Zinc finger protein KOX3) (Zinc finger protein 325) (Gonadotropin-inducible transcription repressor 3) (GIOT-3). | |||||
|
ZN248_HUMAN
|
||||||
| NC score | 0.032982 (rank : 34) | θ value | 0.0736092 (rank : 16) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8NDW4, Q9UMP3 | Gene names | ZNF248 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 248. | |||||
|
ZNF43_HUMAN
|
||||||
| NC score | 0.031836 (rank : 35) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P17038, P28160, Q96DG1 | Gene names | ZNF43, KOX27, ZNF39 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 43 (Zinc protein HTF6) (Zinc finger protein KOX27). | |||||
|
ZN221_HUMAN
|
||||||
| NC score | 0.031506 (rank : 36) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UK13, Q9P1U8 | Gene names | ZNF221 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 221. | |||||
|
ZN567_HUMAN
|
||||||
| NC score | 0.031300 (rank : 37) | θ value | 1.38821 (rank : 39) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 769 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8N184, Q6N044 | Gene names | ZNF567 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 567. | |||||
|
BORIS_HUMAN
|
||||||
| NC score | 0.031174 (rank : 38) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 743 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8NI51, Q5JUG4, Q9BZ30, Q9NQJ3 | Gene names | CTCFL, BORIS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional repressor CTCFL (CCCTC-binding factor) (Brother of the regulator of imprinted sites) (Zinc finger protein CTCF-T) (CTCF paralog). | |||||
|
GZF1_HUMAN
|
||||||
| NC score | 0.031129 (rank : 39) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 854 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9H116, Q96N08, Q9BQK9, Q9H117, Q9H6W6 | Gene names | GZF1, ZBTB23, ZNF336 | |||
|
Domain Architecture |

|
|||||
| Description | GDNF-inducible zinc finger protein 1 (Zinc finger protein 336) (Zinc finger and BTB domain-containing protein 23). | |||||
|
ZN155_HUMAN
|
||||||
| NC score | 0.031010 (rank : 40) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q12901, Q9UIE1, Q9UK14 | Gene names | ZNF155 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 155. | |||||
|
ZN624_HUMAN
|
||||||
| NC score | 0.030950 (rank : 41) | θ value | 8.99809 (rank : 127) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9P2J8 | Gene names | ZNF624, KIAA1349 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 624. | |||||
|
ZFP59_MOUSE
|
||||||
| NC score | 0.030949 (rank : 42) | θ value | 1.38821 (rank : 38) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P16373, Q61849 | Gene names | Zfp59, Mfg2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 59 (Zfp-59) (Zinc finger protein Mfg-2). | |||||
|
ZN337_HUMAN
|
||||||
| NC score | 0.030823 (rank : 43) | θ value | 1.81305 (rank : 43) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 781 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Y3M9, Q9Y3Y5 | Gene names | ZNF337 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 337. | |||||
|
KR18_HUMAN
|
||||||
| NC score | 0.030743 (rank : 44) | θ value | 1.81305 (rank : 41) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 763 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9HCG1, Q96JC5, Q9H7N6 | Gene names | KIAA1611 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein Kr18 (HKr18). | |||||
|
TCF17_MOUSE
|
||||||
| NC score | 0.030667 (rank : 45) | θ value | 1.06291 (rank : 35) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q61751 | Gene names | Znf354a, Kid1, Tcf17, Zfp354a | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 354A (Transcription factor 17) (Renal transcription factor Kid-1) (Kidney, ischemia, and developmentally- regulated protein 1). | |||||
|
ZBT38_HUMAN
|
||||||
| NC score | 0.030586 (rank : 46) | θ value | 8.99809 (rank : 126) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 885 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8NAP3 | Gene names | ZBTB38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and BTB domain-containing protein 38. | |||||
|
ZIM3_HUMAN
|
||||||
| NC score | 0.030317 (rank : 47) | θ value | 2.36792 (rank : 49) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q96PE6 | Gene names | ZIM3 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger imprinted 3. | |||||
|
ZN347_HUMAN
|
||||||
| NC score | 0.030275 (rank : 48) | θ value | 4.03905 (rank : 73) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 756 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96SE7, Q8TCN1 | Gene names | ZNF347, ZNF1111 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 347 (Zinc finger protein 1111). | |||||
|
ZN343_HUMAN
|
||||||
| NC score | 0.030137 (rank : 49) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q6P1L6, Q5JXU8, Q5JXU9, Q8N8F2, Q96EX8, Q96NA5, Q9BQB0 | Gene names | ZNF343 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 343. | |||||
|
ZN605_HUMAN
|
||||||
| NC score | 0.030103 (rank : 50) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q86T29, Q86T91 | Gene names | ZNF605 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 605. | |||||
|
PRDM4_HUMAN
|
||||||
| NC score | 0.029960 (rank : 51) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9UKN5, Q9UFA6 | Gene names | PRDM4, PFM1 | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 4 (PR domain-containing protein 4). | |||||
|
TCF17_HUMAN
|
||||||
| NC score | 0.029932 (rank : 52) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 797 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O60765, Q9UNJ8 | Gene names | ZNF354A, EZNF, HKL1, TCF17 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 354A (Transcription factor 17) (Zinc finger protein eZNF). | |||||
|
ZN557_HUMAN
|
||||||
| NC score | 0.029919 (rank : 53) | θ value | 1.81305 (rank : 44) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 752 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8N988, Q6PEJ3, Q9BTZ1 | Gene names | ZNF557 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 557. | |||||
|
PRDM4_MOUSE
|
||||||
| NC score | 0.029915 (rank : 54) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q80V63 | Gene names | Prdm4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PR domain zinc finger protein 4 (PR domain-containing protein 4). | |||||
|
ZNF92_HUMAN
|
||||||
| NC score | 0.029763 (rank : 55) | θ value | 6.88961 (rank : 105) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q03936, Q8N492, Q8NB35 | Gene names | ZNF92 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 92 (Zinc finger protein HTF12). | |||||
|
ZN669_HUMAN
|
||||||
| NC score | 0.029597 (rank : 56) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 746 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q96BR6, Q5VT39, Q9H9Q6 | Gene names | ZNF669 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 669. | |||||
|
ZNF44_HUMAN
|
||||||
| NC score | 0.029588 (rank : 57) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 757 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P15621, P17018, Q9ULZ7 | Gene names | ZNF44, KOX7 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 44 (Zinc finger protein KOX7) (Gonadotropin- inducible transcription repressor 2) (GIOT-2). | |||||
|
MZF1_HUMAN
|
||||||
| NC score | 0.029217 (rank : 58) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P28698, Q9NRY0, Q9UBW2 | Gene names | MZF1, MZF, ZNF42, ZSCAN6 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid zinc finger 1 (MZF-1) (Zinc finger protein 42) (Zinc finger and SCAN domain-containing protein 6). | |||||
|
AIOL_MOUSE
|
||||||
| NC score | 0.029209 (rank : 59) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O08900 | Gene names | Ikzf3, Zfpn1a3, Znfn1a3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein Aiolos (IKAROS family zinc finger protein 3). | |||||
|
ZN718_HUMAN
|
||||||
| NC score | 0.029064 (rank : 60) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 761 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q3SXZ3, Q3SXZ4, Q3SXZ5 | Gene names | ZNF718 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 718. | |||||
|
ZBT24_HUMAN
|
||||||
| NC score | 0.028597 (rank : 61) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O43167, Q5TED5, Q8N455 | Gene names | ZBTB24, KIAA0441, ZNF450 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and BTB domain-containing protein 24 (Zinc finger protein 450). | |||||
|
ZN449_HUMAN
|
||||||
| NC score | 0.028540 (rank : 62) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 758 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q6P9G9, Q5JRZ8, Q6NZX2, Q8N3Q1, Q8TED7 | Gene names | ZNF449, ZSCAN19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 449 (Zinc finger and SCAN domain-containing protein 19). | |||||
|
XPA_MOUSE
|
||||||
| NC score | 0.028521 (rank : 63) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q64267 | Gene names | Xpa, Xpac | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-A cells homolog (Xeroderma pigmentosum group A-complementing protein homolog). | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.028385 (rank : 64) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
FRAS1_MOUSE
|
||||||
| NC score | 0.028277 (rank : 65) | θ value | 0.163984 (rank : 20) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q80T14, Q80TC7, Q811H8, Q8BPZ4 | Gene names | Fras1, Kiaa1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.027529 (rank : 66) | θ value | 0.163984 (rank : 21) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
ZCH10_MOUSE
|
||||||
| NC score | 0.027247 (rank : 67) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CX48 | Gene names | Zcchc10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
DAXX_HUMAN
|
||||||
| NC score | 0.027177 (rank : 68) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UER7, O14747, O15141, O15208, Q9BWI3 | Gene names | DAXX, BING2, DAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Death domain-associated protein 6 (Daxx) (hDaxx) (Fas death domain- associated protein) (ETS1-associated protein 1) (EAP1). | |||||
|
KR101_HUMAN
|
||||||
| NC score | 0.025369 (rank : 69) | θ value | 0.813845 (rank : 29) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P60331 | Gene names | KRTAP10-1, KAP10.1, KAP18-1, KRTAP10.1, KRTAP18-1, KRTAP18.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-1 (Keratin-associated protein 10.1) (High sulfur keratin-associated protein 10.1) (Keratin-associated protein 18-1) (Keratin-associated protein 18.1). | |||||
|
FHL5_HUMAN
|
||||||
| NC score | 0.025350 (rank : 70) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 523 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q5TD97, Q5TD98, Q8WW21, Q9NQU2 | Gene names | FHL5, ACT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Four and a half LIM domains protein 5 (FHL-5) (Activator of cAMP- responsive element modulator in testis) (Activator of CREM in testis). | |||||
|
XPA_HUMAN
|
||||||
| NC score | 0.025309 (rank : 71) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P23025, Q6LCW7 | Gene names | XPA, XPAC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-A cells (Xeroderma pigmentosum group A-complementing protein). | |||||
|
XPC_MOUSE
|
||||||
| NC score | 0.022391 (rank : 72) | θ value | 2.36792 (rank : 48) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P51612, P54732, Q3TKI2, Q920M1, Q9DBW7 | Gene names | Xpc | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-C cells homolog (Xeroderma pigmentosum group C-complementing protein homolog) (p125). | |||||
|
ELOA1_HUMAN
|
||||||
| NC score | 0.022140 (rank : 73) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14241, Q8IXH1 | Gene names | TCEB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor B polypeptide 3 (RNA polymerase II transcription factor SIII subunit A1) (SIII p110) (Elongin-A) (EloA) (Elongin 110 kDa subunit). | |||||
|
KR107_HUMAN
|
||||||
| NC score | 0.022023 (rank : 74) | θ value | 0.62314 (rank : 25) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P60409, Q70LJ2 | Gene names | KRTAP10-7, KAP10.7, KAP18-7, KRTAP10.7, KRTAP18-7, KRTAP18.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-7 (Keratin-associated protein 10.7) (High sulfur keratin-associated protein 10.7) (Keratin-associated protein 18-7) (Keratin-associated protein 18.7). | |||||
|
PHC1_MOUSE
|
||||||
| NC score | 0.021931 (rank : 75) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q64028, P70359, Q64307, Q8BZ80 | Gene names | Phc1, Edr, Edr1, Rae28 | |||
|
Domain Architecture |

|
|||||
| Description | Polyhomeotic-like protein 1 (mPH1) (Early development regulatory protein 1) (RAE-28). | |||||
|
PHC1_HUMAN
|
||||||
| NC score | 0.021434 (rank : 76) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P78364, Q8WVM3, Q9BU63 | Gene names | PHC1, EDR1, PH1 | |||
|
Domain Architecture |

|
|||||
| Description | Polyhomeotic-like protein 1 (hPH1) (Early development regulatory protein 1). | |||||
|
HTSF1_HUMAN
|
||||||
| NC score | 0.021343 (rank : 77) | θ value | 2.36792 (rank : 46) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O43719, Q59G06, Q99730 | Gene names | HTATSF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 (Tat-SF1). | |||||
|
FRAS1_HUMAN
|
||||||
| NC score | 0.019294 (rank : 78) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q86XX4, Q86UZ4, Q8N3U9, Q8NAU7, Q96JW7, Q9H6N9, Q9P228 | Gene names | FRAS1, KIAA1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
TR13B_MOUSE
|
||||||
| NC score | 0.018444 (rank : 79) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ET35, Q9DBZ3 | Gene names | Tnfrsf13b, Taci | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor necrosis factor receptor superfamily member 13B (Transmembrane activator and CAML interactor). | |||||
|
ELOA1_MOUSE
|
||||||
| NC score | 0.018375 (rank : 80) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CB77, Q80VB2, Q9R0Q5 | Gene names | Tceb3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor B polypeptide 3 (RNA polymerase II transcription factor SIII subunit A) (SIII p110) (Elongin-A) (EloA) (Elongin 110 kDa subunit). | |||||
|
MATN2_HUMAN
|
||||||
| NC score | 0.018244 (rank : 81) | θ value | 0.21417 (rank : 23) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O00339, Q6UWA5, Q7Z5X1, Q8NDE6, Q96FT5, Q9NSZ1 | Gene names | MATN2 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-2 precursor. | |||||
|
K0423_HUMAN
|
||||||
| NC score | 0.018128 (rank : 82) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y4F4, Q68D66, Q6PG27 | Gene names | KIAA0423 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0423. | |||||
|
LAP2_HUMAN
|
||||||
| NC score | 0.017943 (rank : 83) | θ value | 1.06291 (rank : 34) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96RT1, Q86W38, Q9NR18, Q9NW48, Q9ULJ5 | Gene names | ERBB2IP, ERBIN, KIAA1225, LAP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP2 (Erbb2-interacting protein) (Erbin) (Densin-180-like protein). | |||||
|
LAP2_MOUSE
|
||||||
| NC score | 0.017834 (rank : 84) | θ value | 0.62314 (rank : 26) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80TH2, Q8BQ14, Q8CE41, Q8K171, Q99JU3, Q9JI47 | Gene names | Erbb2ip, Erbin, Kiaa1225, Lap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP2 (Erbb2-interacting protein) (Erbin) (Densin-180-like protein). | |||||
|
PSMD1_HUMAN
|
||||||
| NC score | 0.017189 (rank : 85) | θ value | 4.03905 (rank : 66) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99460, Q53TI2, Q6GMU5, Q6P2P4, Q6PJM7, Q6PKG9, Q86VU1, Q8IV79 | Gene names | PSMD1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 1 (26S proteasome regulatory subunit RPN2) (26S proteasome regulatory subunit S1) (26S proteasome subunit p112). | |||||
|
RBBP6_MOUSE
|
||||||
| NC score | 0.016867 (rank : 86) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
KRA93_HUMAN
|
||||||
| NC score | 0.016771 (rank : 87) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BYQ3 | Gene names | KRTAP9-3, KAP9.3, KRTAP9.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-3 (Keratin-associated protein 9.3) (Ultrahigh sulfur keratin-associated protein 9.3). | |||||
|
SYCP2_HUMAN
|
||||||
| NC score | 0.016752 (rank : 88) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BX26, O75763, Q5JX11, Q9NTX8, Q9UG27 | Gene names | SYCP2, SCP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptonemal complex protein 2 (SCP-2) (Synaptonemal complex lateral element protein) (hsSCP2). | |||||
|
ZNFX1_MOUSE
|
||||||
| NC score | 0.016210 (rank : 89) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8R151, Q3U016, Q3UD71, Q3UEZ0 | Gene names | ZNFX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NFX1-type zinc finger-containing protein 1. | |||||
|
KRA98_HUMAN
|
||||||
| NC score | 0.016195 (rank : 90) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BYQ0 | Gene names | KRTAP9-8, KAP9.8, KRTAP9.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-8 (Keratin-associated protein 9.8) (Ultrahigh sulfur keratin-associated protein 9.8). | |||||
|
TACD1_HUMAN
|
||||||
| NC score | 0.016066 (rank : 91) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P16422, P18180, Q96C47 | Gene names | TACSTD1, GA733-2, M1S2, TROP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor-associated calcium signal transducer 1 precursor (Major gastrointestinal tumor-associated protein GA733-2) (Epithelial cell surface antigen) (Epithelial glycoprotein) (EGP) (Adenocarcinoma- associated antigen) (KSA) (KS 1/4 antigen) (Cell surface glycoprotein Trop-1) (CD326 antigen). | |||||
|
BRCA1_MOUSE
|
||||||
| NC score | 0.015983 (rank : 92) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P48754, Q60957, Q60983 | Gene names | Brca1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 1 susceptibility protein homolog. | |||||
|
TOPB1_MOUSE
|
||||||
| NC score | 0.015587 (rank : 93) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6ZQF0, Q6P6P0, Q80Y33, Q8BUI0, Q8BUK1, Q8R348, Q91VX3, Q922X8 | Gene names | Topbp1, Kiaa0259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA topoisomerase II-binding protein 1 (DNA topoisomerase IIbeta- binding protein 1) (TopBP1). | |||||
|
TRI32_MOUSE
|
||||||
| NC score | 0.015302 (rank : 94) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CH72, Q8K055 | Gene names | Trim32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 32 (EC 6.3.2.-). | |||||
|
TRI32_HUMAN
|
||||||
| NC score | 0.015233 (rank : 95) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13049, Q9NQP8 | Gene names | TRIM32, HT2A | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 32 (EC 6.3.2.-) (Zinc finger protein HT2A) (72 kDa Tat-interacting protein). | |||||
|
SDCG1_HUMAN
|
||||||
| NC score | 0.014874 (rank : 96) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60524, Q8WW70, Q9NWG1 | Gene names | SDCCAG1 | |||
|
Domain Architecture |

|
|||||
| Description | Serologically defined colon cancer antigen 1 (Antigen NY-CO-1). | |||||
|
ARHG7_HUMAN
|
||||||
| NC score | 0.013701 (rank : 97) | θ value | 1.38821 (rank : 37) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14155, Q6P9G3, Q6PII2, Q86W63, Q8N3M1 | Gene names | ARHGEF7, COOL1, KIAA0142, P85SPR, PAK3BP, PIXB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 7 (PAK-interacting exchange factor beta) (Beta-Pix) (COOL-1) (p85). | |||||
|
PHF7_MOUSE
|
||||||
| NC score | 0.013378 (rank : 98) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DAG9, Q6PG81 | Gene names | Phf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 7 (Testis development protein NYD-SP6). | |||||
|
UB7I4_HUMAN
|
||||||
| NC score | 0.013208 (rank : 99) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P50876 | Gene names | RNF144, KIAA0161, UBCE7IP4 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-conjugating enzyme 7-interacting protein 4 (UbcM4- interacting protein 4) (RING finger protein 144). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.013075 (rank : 100) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
MEP1B_HUMAN
|
||||||
| NC score | 0.012982 (rank : 101) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q16820, Q670J1 | Gene names | MEP1B | |||
|
Domain Architecture |

|
|||||
| Description | Meprin A subunit beta precursor (EC 3.4.24.18) (Endopeptidase-2) (N- benzoyl-L-tyrosyl-P-amino-benzoic acid hydrolase subunit beta) (PABA peptide hydrolase) (PPH beta). | |||||
|
EYA1_HUMAN
|
||||||
| NC score | 0.012890 (rank : 102) | θ value | 2.36792 (rank : 45) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99502 | Gene names | EYA1 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 1 (EC 3.1.3.48). | |||||
|
LRRC1_HUMAN
|
||||||
| NC score | 0.012343 (rank : 103) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BTT6, Q9HAC0, Q9NVF1 | Gene names | LRRC1, LANO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 1 (LAP and no PDZ protein) (LANO adapter protein). | |||||
|
LRRC1_MOUSE
|
||||||
| NC score | 0.011016 (rank : 104) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80VQ1, Q3UI50, Q8BKR1, Q8BUS9 | Gene names | Lrrc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 1. | |||||
|
DX26B_MOUSE
|
||||||
| NC score | 0.010399 (rank : 105) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BND4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DDX26B. | |||||
|
RAPH1_HUMAN
|
||||||
| NC score | 0.010310 (rank : 106) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.010306 (rank : 107) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
TMM24_MOUSE
|
||||||
| NC score | 0.010210 (rank : 108) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80X80 | Gene names | Tmem24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 24. | |||||
|
ARHG7_MOUSE
|
||||||
| NC score | 0.010098 (rank : 109) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ES28, O08757, Q9ES27 | Gene names | Arhgef7, Pak3bp | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 7 (PAK-interacting exchange factor beta) (Beta-Pix) (p85SPR). | |||||
|
SMBT1_MOUSE
|
||||||
| NC score | 0.009897 (rank : 110) | θ value | 8.99809 (rank : 121) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JMD1, Q6NZD3, Q8CFS1 | Gene names | Sfmbt1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scm-like with four MBT domains protein 1. | |||||
|
ANR12_HUMAN
|
||||||
| NC score | 0.009554 (rank : 111) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6UB98, O94951, Q658K1, Q6QMF7, Q9H231, Q9H784 | Gene names | ANKRD12, ANCO2, KIAA0874 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 12 (Ankyrin repeat-containing cofactor 2) (GAC-1 protein). | |||||
|
INHBA_MOUSE
|
||||||
| NC score | 0.008821 (rank : 112) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q04998 | Gene names | Inhba | |||
|
Domain Architecture |

|
|||||
| Description | Inhibin beta A chain precursor (Activin beta-A chain). | |||||
|
AP3B2_HUMAN
|
||||||
| NC score | 0.008757 (rank : 113) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13367, O14808 | Gene names | AP3B2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (AP-3 complex beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain) (Neuron-specific vesicle coat protein beta-NAP). | |||||
|
NEBL_HUMAN
|
||||||
| NC score | 0.008581 (rank : 114) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O76041, Q9UIC4 | Gene names | NEBL | |||
|
Domain Architecture |

|
|||||
| Description | Nebulette (Actin-binding Z-disk protein). | |||||
|
NELL1_HUMAN
|
||||||
| NC score | 0.008490 (rank : 115) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q92832, Q9Y472 | Gene names | NELL1, NRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein NELL1 precursor (NEL-like protein 1) (Nel-related protein 1). | |||||
|
SMG1_MOUSE
|
||||||
| NC score | 0.008038 (rank : 116) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BKX6, Q5DU42, Q6ZQC0, Q8BLU4, Q8BWJ5, Q8BXD3 | Gene names | Smg1, Kiaa0421 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1). | |||||
|
KLK11_HUMAN
|
||||||
| NC score | 0.007910 (rank : 117) | θ value | 0.21417 (rank : 22) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UBX7, O75837, Q9NS65 | Gene names | KLK11, PRSS20, TLSP | |||
|
Domain Architecture |

|
|||||
| Description | Kallikrein-11 precursor (EC 3.4.21.-) (Hippostasin) (Trypsin-like protease). | |||||
|
SSPO_MOUSE
|
||||||
| NC score | 0.006994 (rank : 118) | θ value | 8.99809 (rank : 122) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
FBN1_MOUSE
|
||||||
| NC score | 0.006769 (rank : 119) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 563 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61554, Q60826 | Gene names | Fbn1, Fbn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-1 precursor. | |||||
|
OTOF_MOUSE
|
||||||
| NC score | 0.006016 (rank : 120) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ESF1, Q9ESF2 | Gene names | Otof, Fer1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Otoferlin (Fer-1-like protein 2). | |||||
|
TBX5_MOUSE
|
||||||
| NC score | 0.004779 (rank : 121) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70326, Q9R003 | Gene names | Tbx5 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX5 (T-box protein 5). | |||||
|
PTPRN_MOUSE
|
||||||
| NC score | 0.004591 (rank : 122) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60673, Q62129 | Gene names | Ptprn, Ptp35 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase-like N precursor (R-PTP-N) (PTP IA-2). | |||||
|
CHD2_HUMAN
|
||||||
| NC score | 0.004475 (rank : 123) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O14647 | Gene names | CHD2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 2 (EC 3.6.1.-) (ATP- dependent helicase CHD2) (CHD-2). | |||||
|
RBM19_MOUSE
|
||||||
| NC score | 0.004085 (rank : 124) | θ value | 8.99809 (rank : 120) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R3C6, Q8BHR0, Q9CW63 | Gene names | Rbm19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable RNA-binding protein 19 (RNA-binding motif protein 19). | |||||
|
DLX1_HUMAN
|
||||||
| NC score | 0.003823 (rank : 125) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P56177, Q53ZU4, Q8IYB2 | Gene names | DLX1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein DLX-1. | |||||
|
DLX1_MOUSE
|
||||||
| NC score | 0.003818 (rank : 126) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q64317 | Gene names | Dlx1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein DLX-1. | |||||
|
ATS1_HUMAN
|
||||||
| NC score | 0.003140 (rank : 127) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UHI8, Q9NSJ8, Q9P2K0, Q9UH83, Q9UP80 | Gene names | ADAMTS1, KIAA1346, METH1 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-1 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 1) (ADAM-TS 1) (ADAM-TS1) (METH-1). | |||||
|
TIE2_MOUSE
|
||||||
| NC score | 0.001928 (rank : 128) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1204 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q02858 | Gene names | Tek, Hyk, Tie-2, Tie2 | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-1 receptor precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor TIE-2) (mTIE2) (Tyrosine-protein kinase receptor TEK) (p140 TEK) (Tunica interna endothelial cell kinase) (HYK) (STK1) (CD202b antigen). | |||||
|
TIE2_HUMAN
|
||||||
| NC score | 0.000534 (rank : 129) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1192 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q02763 | Gene names | TEK, TIE2 | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-1 receptor precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor TIE-2) (hTIE2) (Tyrosine-protein kinase receptor TEK) (p140 TEK) (Tunica interna endothelial cell kinase) (CD202b antigen). | |||||
|
NTRK3_HUMAN
|
||||||
| NC score | -0.001151 (rank : 130) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1115 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q16288, Q12827, Q16289 | Gene names | NTRK3, TRKC | |||
|
Domain Architecture |

|
|||||
| Description | NT-3 growth factor receptor precursor (EC 2.7.10.1) (Neurotrophic tyrosine kinase receptor type 3) (TrkC tyrosine kinase) (GP145-TrkC) (Trk-C). | |||||