Please be patient as the page loads
|
P52K_HUMAN
|
||||||
| SwissProt Accessions | O43422, Q8WTW1, Q9Y3Z4 | Gene names | PRKRIR, DAP4, P52RIPK, THAP0 | |||
|
Domain Architecture |

|
|||||
| Description | 52 kDa repressor of the inhibitor of the protein kinase (p58IPK- interacting protein) (58 kDa interferon-induced protein kinase- interacting protein) (P52rIPK) (Death-associated protein 4) (THAP domain-containing protein 0). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
P52K_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | O43422, Q8WTW1, Q9Y3Z4 | Gene names | PRKRIR, DAP4, P52RIPK, THAP0 | |||
|
Domain Architecture |

|
|||||
| Description | 52 kDa repressor of the inhibitor of the protein kinase (p58IPK- interacting protein) (58 kDa interferon-induced protein kinase- interacting protein) (P52rIPK) (Death-associated protein 4) (THAP domain-containing protein 0). | |||||
|
P52K_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.968505 (rank : 2) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9CUX1, Q80Y58 | Gene names | Prkrir, Thap0 | |||
|
Domain Architecture |

|
|||||
| Description | 52 kDa repressor of the inhibitor of the protein kinase (p58IPK- interacting protein) (58 kDa interferon-induced protein kinase- interacting protein) (P52rIPK) (THAP domain-containing protein 0). | |||||
|
ZMYM1_HUMAN
|
||||||
| θ value | 1.12514e-34 (rank : 3) | NC score | 0.493913 (rank : 3) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5SVZ6, Q7Z3Q4 | Gene names | ZMYM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 1. | |||||
|
THAP2_HUMAN
|
||||||
| θ value | 1.133e-10 (rank : 4) | NC score | 0.459322 (rank : 4) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H0W7 | Gene names | THAP2 | |||
|
Domain Architecture |
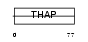
|
|||||
| Description | THAP domain-containing protein 2. | |||||
|
THAP2_MOUSE
|
||||||
| θ value | 5.62301e-10 (rank : 5) | NC score | 0.452429 (rank : 5) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9D305 | Gene names | Thap2 | |||
|
Domain Architecture |
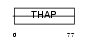
|
|||||
| Description | THAP domain-containing protein 2. | |||||
|
THAP7_MOUSE
|
||||||
| θ value | 1.09739e-05 (rank : 6) | NC score | 0.348550 (rank : 7) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8VCZ3 | Gene names | Thap7 | |||
|
Domain Architecture |
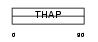
|
|||||
| Description | THAP domain-containing protein 7. | |||||
|
THAP7_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 7) | NC score | 0.347841 (rank : 8) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BT49 | Gene names | THAP7 | |||
|
Domain Architecture |
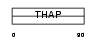
|
|||||
| Description | THAP domain-containing protein 7. | |||||
|
THAP4_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 8) | NC score | 0.294654 (rank : 10) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8WY91, Q53NU7, Q6GRN0, Q6IPJ3, Q9NW26, Q9Y325 | Gene names | THAP4 | |||
|
Domain Architecture |
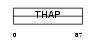
|
|||||
| Description | THAP domain-containing protein 4. | |||||
|
THAP6_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 9) | NC score | 0.354023 (rank : 6) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8TBB0 | Gene names | THAP6 | |||
|
Domain Architecture |
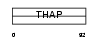
|
|||||
| Description | THAP domain-containing protein 6. | |||||
|
THA11_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 10) | NC score | 0.273054 (rank : 13) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96EK4, O94795 | Gene names | THAP11 | |||
|
Domain Architecture |
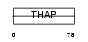
|
|||||
| Description | THAP domain-containing protein 11. | |||||
|
THAP8_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 11) | NC score | 0.322377 (rank : 9) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8NA92, Q96M21 | Gene names | THAP8 | |||
|
Domain Architecture |
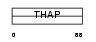
|
|||||
| Description | THAP domain-containing protein 8. | |||||
|
ZN452_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 12) | NC score | 0.062967 (rank : 24) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6R2W3, Q2NKL9, Q5SRJ3, Q8TCN2, Q96PW3 | Gene names | ZNF452, KIAA1925 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF452. | |||||
|
THA11_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 13) | NC score | 0.243461 (rank : 15) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JJD0 | Gene names | Thap11 | |||
|
Domain Architecture |
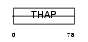
|
|||||
| Description | THAP domain-containing protein 11. | |||||
|
THAP4_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 14) | NC score | 0.268544 (rank : 14) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6P3Z3, Q3US28, Q6NZN6, Q6PEN6, Q91WR2, Q9CVE5, Q9CVG3 | Gene names | Thap4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THAP domain-containing protein 4. | |||||
|
THAP1_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 15) | NC score | 0.288628 (rank : 11) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NVV9, Q53FQ1, Q6IA99 | Gene names | THAP1 | |||
|
Domain Architecture |
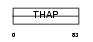
|
|||||
| Description | THAP domain-containing protein 1. | |||||
|
CALD1_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 16) | NC score | 0.041254 (rank : 45) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
NARGL_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 17) | NC score | 0.083330 (rank : 19) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6N069, Q5VSP9, Q6P2D5, Q8N5J3, Q8N870 | Gene names | NARG1L, NAT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NMDA receptor-regulated 1-like protein (NARG1-like protein). | |||||
|
THAP1_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 18) | NC score | 0.276383 (rank : 12) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CHW1 | Gene names | Thap1 | |||
|
Domain Architecture |
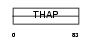
|
|||||
| Description | THAP domain-containing protein 1. | |||||
|
DHX29_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 19) | NC score | 0.039591 (rank : 52) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7Z478, O75549, Q63HN0, Q63HN3, Q8IWW2, Q8N3A1, Q9UMH2 | Gene names | DHX29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX29 (EC 3.6.1.-) (DEAH box protein 29) (Nucleic acid helicase DDXx). | |||||
|
RPGR1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 20) | NC score | 0.053195 (rank : 35) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
TXLNA_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 21) | NC score | 0.058676 (rank : 26) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 846 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6PAM1, Q6P1E5 | Gene names | Txlna, Txln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
TXLNA_HUMAN
|
||||||
| θ value | 0.125558 (rank : 22) | NC score | 0.057147 (rank : 31) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 982 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P40222, Q66K62, Q86T54, Q86T85, Q86T86, Q86Y86, Q86YW3, Q8N2Y3 | Gene names | TXLNA, TXLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
LRRF2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 23) | NC score | 0.048052 (rank : 37) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 791 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Y608, Q68CV3, Q9NXH5 | Gene names | LRRFIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 2 (LRR FLII- interacting protein 2). | |||||
|
PHF14_MOUSE
|
||||||
| θ value | 0.163984 (rank : 24) | NC score | 0.040063 (rank : 50) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9D4H9, Q810Z6, Q8C284, Q8CGF8, Q9D1Q8 | Gene names | Phf14 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 14. | |||||
|
VDP_MOUSE
|
||||||
| θ value | 0.21417 (rank : 25) | NC score | 0.042299 (rank : 43) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 469 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Z1Z0, Q3T9L9, Q3TH58, Q3U1C7, Q3UMW6, Q91WE7, Q99JZ5 | Gene names | Vdp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General vesicular transport factor p115 (Transcytosis-associated protein) (TAP) (Vesicle docking protein). | |||||
|
IL33_HUMAN
|
||||||
| θ value | 0.279714 (rank : 26) | NC score | 0.063254 (rank : 23) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95760, Q2YEJ5 | Gene names | IL33, C9orf26, IL1F11, NFHEV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-33 precursor (IL-33) (Interleukin-1 family member 11) (IL- 1F11) (Nuclear factor from high endothelial venules) (NF-HEV). | |||||
|
CD2L2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 27) | NC score | 0.012050 (rank : 131) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
RAD50_HUMAN
|
||||||
| θ value | 0.365318 (rank : 28) | NC score | 0.040661 (rank : 49) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
EPMIP_MOUSE
|
||||||
| θ value | 0.47712 (rank : 29) | NC score | 0.057294 (rank : 30) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VEH5, Q80TS4 | Gene names | Epm2aip1, Kiaa0766 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EPM2A-interacting protein 1 (Laforin-interacting protein). | |||||
|
RAD50_MOUSE
|
||||||
| θ value | 0.47712 (rank : 30) | NC score | 0.040920 (rank : 47) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
RENT2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 31) | NC score | 0.050392 (rank : 36) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
ATRX_HUMAN
|
||||||
| θ value | 0.62314 (rank : 32) | NC score | 0.032071 (rank : 66) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
CCAR1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 33) | NC score | 0.041543 (rank : 44) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8CH18, Q6AXC9, Q6PAR2, Q80XE4, Q8BJY0, Q8BVN2, Q8CGG1, Q9CSR5 | Gene names | Ccar1, Carp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle and apoptosis regulator protein 1 (Cell cycle and apoptosis regulatory protein 1) (CARP-1). | |||||
|
CAF1A_MOUSE
|
||||||
| θ value | 0.813845 (rank : 34) | NC score | 0.045619 (rank : 39) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9QWF0 | Gene names | Chaf1a, Caip150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
NARGL_MOUSE
|
||||||
| θ value | 0.813845 (rank : 35) | NC score | 0.073359 (rank : 20) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9DBB4, Q3U7V2 | Gene names | Narg1l, Nat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NMDA receptor-regulated 1-like protein (NARG1-like protein). | |||||
|
CAF1A_HUMAN
|
||||||
| θ value | 1.06291 (rank : 36) | NC score | 0.044475 (rank : 40) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q13111, Q7Z7K3, Q9UJY8 | Gene names | CHAF1A, CAF, CAF1P150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
CD2L1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 37) | NC score | 0.011199 (rank : 134) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
CD2L1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 38) | NC score | 0.011534 (rank : 133) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1463 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P24788, Q3UI03, Q61399, Q7TST4, Q8BP53 | Gene names | Cdc2l1 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1). | |||||
|
CROP_HUMAN
|
||||||
| θ value | 1.06291 (rank : 39) | NC score | 0.037318 (rank : 56) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O95232, Q6PHR9, Q9NUY0, Q9P2S7 | Gene names | CROP, CREAP1, O48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cisplatin resistance-associated overexpressed protein (cAMP regulatory element-associated protein 1) (CRE-associated protein 1) (CREAP-1) (Luc7A) (Okadaic acid-inducible phosphoprotein OA48-18). | |||||
|
DNL1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 40) | NC score | 0.028638 (rank : 76) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P18858 | Gene names | LIG1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 1 (EC 6.5.1.1) (DNA ligase I) (Polydeoxyribonucleotide synthase [ATP] 1). | |||||
|
PELP1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 41) | NC score | 0.037199 (rank : 57) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
WBP11_MOUSE
|
||||||
| θ value | 1.06291 (rank : 42) | NC score | 0.033924 (rank : 63) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
CROP_MOUSE
|
||||||
| θ value | 1.38821 (rank : 43) | NC score | 0.035652 (rank : 59) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5SUF2, Q3U9D5, Q8BUJ5, Q921Z3, Q9CRS7, Q9CTY4 | Gene names | Crop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cisplatin resistance-associated overexpressed protein. | |||||
|
DPOE3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 44) | NC score | 0.047649 (rank : 38) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NRF9, Q5W0U1, Q8N758, Q8NCE5, Q9NR32 | Gene names | POLE3, CHRAC17 | |||
|
Domain Architecture |
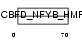
|
|||||
| Description | DNA polymerase epsilon subunit 3 (EC 2.7.7.7) (DNA polymerase II subunit 3) (DNA polymerase epsilon subunit p17) (Chromatin accessibility complex 17) (HuCHRAC17) (CHRAC-17) (Arsenic- transactivated protein) (AsTP). | |||||
|
FGD5_MOUSE
|
||||||
| θ value | 1.38821 (rank : 45) | NC score | 0.017376 (rank : 110) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80UZ0, Q8BHM5 | Gene names | Fgd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 5. | |||||
|
HOME1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 46) | NC score | 0.030012 (rank : 69) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 564 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q86YM7, O96003, Q86YM5 | Gene names | HOMER1, SYN47 | |||
|
Domain Architecture |
|
|||||
| Description | Homer protein homolog 1. | |||||
|
MRCKG_HUMAN
|
||||||
| θ value | 1.38821 (rank : 47) | NC score | 0.007707 (rank : 144) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1519 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6DT37, O00565 | Gene names | CDC42BPG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK gamma (EC 2.7.11.1) (CDC42- binding protein kinase gamma) (Myotonic dystrophy kinase-related CDC42-binding kinase gamma) (Myotonic dystrophy protein kinase-like alpha) (MRCK gamma) (DMPK-like gamma). | |||||
|
MYST4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 48) | NC score | 0.027751 (rank : 80) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
NARG1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 49) | NC score | 0.070861 (rank : 22) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BXJ9, Q8IWH4, Q8NEV2, Q9H8P6 | Gene names | NARG1, GA19, NATH, TBDN100 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NMDA receptor-regulated protein 1 (N-terminal acetyltransferase) (Tubedown-1 protein) (Tbdn100) (Gastric cancer antigen Ga19). | |||||
|
NARG1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 50) | NC score | 0.071020 (rank : 21) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80UM3, Q811Z9, Q9JID5 | Gene names | Narg1, Nat1, Tbdn-1, Tubedown | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NMDA receptor-regulated protein 1 (N-terminal acetyltransferase 1) (Protein Tubedown-1). | |||||
|
NFH_HUMAN
|
||||||
| θ value | 1.38821 (rank : 51) | NC score | 0.016234 (rank : 116) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
RIF1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 52) | NC score | 0.034938 (rank : 62) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6PR54, Q6T8D9, Q7TPD8, Q8BTU2, Q8C408, Q9CS77 | Gene names | Rif1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomere-associated protein RIF1 (Rap1-interacting factor 1 homolog) (mRif1). | |||||
|
TXLNB_HUMAN
|
||||||
| θ value | 1.38821 (rank : 53) | NC score | 0.053663 (rank : 34) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8N3L3, Q76L25, Q86T52, Q8N3S2 | Gene names | TXLNB, C6orf198, MDP77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77) (hMDP77). | |||||
|
ARI4A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 54) | NC score | 0.038345 (rank : 53) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P29374, Q15991, Q15992, Q15993 | Gene names | ARID4A, RBBP1, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 4A (ARID domain- containing protein 4A) (Retinoblastoma-binding protein 1) (RBBP-1). | |||||
|
CF170_HUMAN
|
||||||
| θ value | 1.81305 (rank : 55) | NC score | 0.057356 (rank : 29) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96NH3, Q6ZMY4, Q6ZUR7, Q8NB47 | Gene names | C6orf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf170. | |||||
|
DHX29_MOUSE
|
||||||
| θ value | 1.81305 (rank : 56) | NC score | 0.030649 (rank : 68) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PGC1, Q8BQJ4, Q8BT01, Q8C9B7, Q8C9H9 | Gene names | Dhx29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX29 (EC 3.6.1.-) (DEAH box protein 29). | |||||
|
DNMT1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 57) | NC score | 0.033374 (rank : 64) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P26358, Q9UHG5, Q9ULA2, Q9UMZ6 | Gene names | DNMT1, AIM, DNMT | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 1 (EC 2.1.1.37) (Dnmt1) (DNA methyltransferase HsaI) (DNA MTase HsaI) (MCMT) (M.HsaI). | |||||
|
ZEP1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 58) | NC score | 0.011700 (rank : 132) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1036 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P15822, Q14122 | Gene names | HIVEP1, ZNF40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 40 (Human immunodeficiency virus type I enhancer- binding protein 1) (HIV-EP1) (Major histocompatibility complex-binding protein 1) (MBP-1) (Positive regulatory domain II-binding factor 1) (PRDII-BF1). | |||||
|
AT1B4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 59) | NC score | 0.018473 (rank : 106) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UN42, Q9UN41 | Gene names | ATP1B4 | |||
|
Domain Architecture |
|
|||||
| Description | X/potassium-transporting ATPase subunit beta-m (X,K-ATPase beta-m subunit). | |||||
|
CJ006_MOUSE
|
||||||
| θ value | 2.36792 (rank : 60) | NC score | 0.028967 (rank : 75) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6P9P0, Q8C041, Q8C0J5 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf6 homolog. | |||||
|
DDX46_MOUSE
|
||||||
| θ value | 2.36792 (rank : 61) | NC score | 0.010171 (rank : 137) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q569Z5, Q6ZQ42, Q8R0R6 | Gene names | Ddx46, Kiaa0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46). | |||||
|
IF2P_HUMAN
|
||||||
| θ value | 2.36792 (rank : 62) | NC score | 0.029328 (rank : 72) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
ITSN2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 63) | NC score | 0.022852 (rank : 93) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1371 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Z0R6, Q9Z0R5 | Gene names | Itsn2, Ese2, Sh3d1B | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (EH and SH3 domains protein 2) (EH domain and SH3 domain regulator of endocytosis 2). | |||||
|
MYT1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 64) | NC score | 0.024283 (rank : 89) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q01538, O94922, Q9UPV2 | Gene names | MYT1, KIAA0835, KIAA1050, MTF1, MYTI, PLPB1 | |||
|
Domain Architecture |

|
|||||
| Description | Myelin transcription factor 1 (MyT1) (MyTI) (Proteolipid protein- binding protein) (PLPB1). | |||||
|
PININ_HUMAN
|
||||||
| θ value | 2.36792 (rank : 65) | NC score | 0.037799 (rank : 54) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9H307, O60899, Q53EM7, Q6P5X4, Q7KYL1, Q99738, Q9UHZ9, Q9UQR9 | Gene names | PNN, DRS, MEMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin (140 kDa nuclear and cell adhesion-related phosphoprotein) (Domain-rich serine protein) (DRS-protein) (DRSP) (Melanoma metastasis clone A protein) (Desmosome-associated protein) (SR-like protein) (Nuclear protein SDK3). | |||||
|
TAF3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 66) | NC score | 0.025737 (rank : 83) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
TCF8_HUMAN
|
||||||
| θ value | 2.36792 (rank : 67) | NC score | 0.005769 (rank : 148) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P37275, Q12924, Q13800 | Gene names | TCF8, AREB6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (NIL-2-A zinc finger protein) (Negative regulator of IL2). | |||||
|
TRI44_MOUSE
|
||||||
| θ value | 2.36792 (rank : 68) | NC score | 0.024964 (rank : 85) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9QXA7, Q80SV9, Q9JHR8 | Gene names | Trim44, Dipb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB) (Protein Mc7). | |||||
|
HOME1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 69) | NC score | 0.028142 (rank : 79) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 630 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Z2Y3, Q8K3E1, Q8K4M8, Q9Z0E9, Q9Z216 | Gene names | Homer1, Vesl1 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 1 (VASP/Ena-related gene up-regulated during seizure and LTP) (Vesl-1). | |||||
|
HS90B_MOUSE
|
||||||
| θ value | 3.0926 (rank : 70) | NC score | 0.024497 (rank : 88) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P11499, O89078 | Gene names | Hsp90ab1, Hsp84, Hsp84-1, Hspcb | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-beta (HSP 84) (Tumor-specific transplantation 84 kDa antigen) (TSTA). | |||||
|
NFM_MOUSE
|
||||||
| θ value | 3.0926 (rank : 71) | NC score | 0.016952 (rank : 112) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
PININ_MOUSE
|
||||||
| θ value | 3.0926 (rank : 72) | NC score | 0.039830 (rank : 51) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
SNIP1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 73) | NC score | 0.027282 (rank : 82) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TAD8, Q96SP9, Q9H9T7 | Gene names | SNIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Smad nuclear-interacting protein 1. | |||||
|
SRPK2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 74) | NC score | 0.010021 (rank : 139) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O54781, Q8VCD9 | Gene names | Srpk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SRPK2 (EC 2.7.11.1) (Serine/arginine- rich protein-specific kinase 2) (SR-protein-specific kinase 2) (SFRS protein kinase 2). | |||||
|
TOP1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 75) | NC score | 0.029865 (rank : 71) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P11387, O43256, Q12855, Q12856, Q15610, Q5TFY3, Q9UJN0 | Gene names | TOP1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 1 (EC 5.99.1.2) (DNA topoisomerase I). | |||||
|
CHM4B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | 0.029004 (rank : 74) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H444, Q53ZD6 | Gene names | CHMP4B, C20orf178, SHAX1 | |||
|
Domain Architecture |
|
|||||
| Description | Charged multivesicular body protein 4b (Chromatin-modifying protein 4b) (CHMP4b) (Vacuolar protein sorting 7-2) (SNF7-2) (hSnf7-2) (SNF7 homolog associated with Alix 1) (hVps32). | |||||
|
CHM4B_MOUSE
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.029005 (rank : 73) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D8B3, Q3TXM7, Q91VM7, Q922P1 | Gene names | Chmp4b | |||
|
Domain Architecture |

|
|||||
| Description | Charged multivesicular body protein 4b (Chromatin-modifying protein 4b) (CHMP4b). | |||||
|
CN145_HUMAN
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | 0.034956 (rank : 61) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1196 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q6ZU80, Q96ML4 | Gene names | C14orf145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145. | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.029973 (rank : 70) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
MIER1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.019941 (rank : 103) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N108, Q5T104, Q6AHY8, Q86TB4, Q8N156, Q8N161, Q8NC37, Q8NES4, Q8NES5, Q8NES6, Q8WWG2, Q9HCG2 | Gene names | MIER1, KIAA1610 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm induction early response protein 1 (Mi-er1) (hmi-er1) (Early response 1) (Er1). | |||||
|
MYST3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 81) | NC score | 0.023710 (rank : 91) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
NOC2L_MOUSE
|
||||||
| θ value | 4.03905 (rank : 82) | NC score | 0.023939 (rank : 90) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9WV70, Q8VE16 | Gene names | Noc2l | |||
|
Domain Architecture |
|
|||||
| Description | Nucleolar complex protein 2 homolog (Protein NOC2 homolog) (NOC2- like). | |||||
|
PELP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.032759 (rank : 65) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8IZL8, O15450, Q5EGN3, Q6NTE6, Q96FT1, Q9BU60 | Gene names | PELP1, HMX3, MNAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3). | |||||
|
PP2CG_MOUSE
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | 0.016317 (rank : 115) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61074 | Gene names | Ppm1g, Fin13, Ppm1c | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform gamma (EC 3.1.3.16) (PP2C-gamma) (Protein phosphatase magnesium-dependent 1 gamma) (Protein phosphatase 1C) (Fibroblast growth factor-inducible protein 13) (FIN13). | |||||
|
PTMS_HUMAN
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.043160 (rank : 41) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P20962 | Gene names | PTMS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
RBM25_HUMAN
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | 0.021278 (rank : 100) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P49756, Q9UEQ5, Q9UIE9 | Gene names | RBM25, RNPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable RNA-binding protein 25 (RNA-binding motif protein 25) (RNA- binding region-containing protein 7) (Protein S164). | |||||
|
TCF8_MOUSE
|
||||||
| θ value | 4.03905 (rank : 87) | NC score | 0.003890 (rank : 150) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1067 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q64318, Q62519 | Gene names | Tcf8, Zfhx1a, Zfx1a, Zfx1ha | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (Zinc finger homeobox protein 1a) (MEB1) (Delta EF1). | |||||
|
ZEP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 88) | NC score | 0.009758 (rank : 140) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||
|
AT2B1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.007141 (rank : 145) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P20020, Q12992, Q12993, Q13819, Q13820, Q13821, Q16504, Q93082 | Gene names | ATP2B1, PMCA1 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 1 (EC 3.6.3.8) (PMCA1) (Plasma membrane calcium pump isoform 1) (Plasma membrane calcium ATPase isoform 1). | |||||
|
CALR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.013485 (rank : 125) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P27797 | Gene names | CALR, CRTC | |||
|
Domain Architecture |

|
|||||
| Description | Calreticulin precursor (CRP55) (Calregulin) (HACBP) (ERp60) (grp60). | |||||
|
CCAR1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.031639 (rank : 67) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8IX12, Q32NE3, Q5VUP6, Q6PIZ0, Q6X935, Q9H8N4, Q9NVA7, Q9NVQ0, Q9NWM6 | Gene names | CCAR1, CARP1, DIS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle and apoptosis regulator protein 1 (Cell cycle and apoptosis regulatory protein 1) (CARP-1) (Death inducer with SAP domain). | |||||
|
CG301_HUMAN
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.028299 (rank : 78) | |||
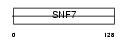
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P59074, Q8NFA9 | Gene names | ames=CGI-301 | |||
|
Domain Architecture |
|
|||||
| Description | Protein CGI-301. | |||||
|
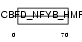
DPOE3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.035622 (rank : 60) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JKP7, Q8R198 | Gene names | Pole3 | |||
|
Domain Architecture |
|
|||||
| Description | DNA polymerase epsilon subunit 3 (EC 2.7.7.7) (DNA polymerase II subunit 3) (DNA polymerase epsilon subunit p17) (YB-like protein 1) (YBL1) (NF-YB-like protein). | |||||
|
FA43A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.017909 (rank : 109) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N2R8, Q8IXP4, Q8WZ07 | Gene names | FAM43A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM43A. | |||||
|
FAM9B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.027584 (rank : 81) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IZU0 | Gene names | FAM9B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM9B. | |||||
|
HS90A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 96) | NC score | 0.024942 (rank : 87) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P07900, Q2PP14, Q9BVQ5 | Gene names | HSP90AA1, HSP90A, HSPC1, HSPCA | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-alpha (HSP 86) (NY-REN-38 antigen). | |||||
|
MTX1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 97) | NC score | 0.019299 (rank : 104) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13505 | Gene names | MTX1, MTX, MTXN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metaxin-1. | |||||
|
MYPT1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 98) | NC score | 0.009542 (rank : 141) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O14974, Q86WU3, Q8NFR6, Q9BYH0 | Gene names | PPP1R12A, MBS, MYPT1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1) (Protein phosphatase myosin-binding subunit). | |||||
|
MYT1L_HUMAN
|
||||||
| θ value | 5.27518 (rank : 99) | NC score | 0.022063 (rank : 97) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UL68, Q6IQ17, Q9UPP6 | Gene names | MYT1L, KIAA1106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (MyT1-L). | |||||
|
NFL_MOUSE
|
||||||
| θ value | 5.27518 (rank : 100) | NC score | 0.011089 (rank : 136) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 695 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P08551, Q8K0Z0 | Gene names | Nefl, Nf68, Nfl | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet L protein (68 kDa neurofilament protein) (Neurofilament light polypeptide) (NF-L). | |||||
|
PHF20_MOUSE
|
||||||
| θ value | 5.27518 (rank : 101) | NC score | 0.043061 (rank : 42) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
SMRC1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 102) | NC score | 0.021624 (rank : 98) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P97496, Q7TS80, Q7TT29 | Gene names | Smarcc1, Baf155, Srg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155) (SWI3-related protein). | |||||
|
SRCH_HUMAN
|
||||||
| θ value | 5.27518 (rank : 103) | NC score | 0.022380 (rank : 95) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P23327, Q504Y6 | Gene names | HRC, HCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcoplasmic reticulum histidine-rich calcium-binding protein precursor. | |||||
|
TEKT1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 104) | NC score | 0.018261 (rank : 107) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q969V4 | Gene names | TEKT1 | |||
|
Domain Architecture |

|
|||||
| Description | Tektin-1. | |||||
|
TRI44_HUMAN
|
||||||
| θ value | 5.27518 (rank : 105) | NC score | 0.020224 (rank : 102) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96DX7, Q96QY2, Q9UGK0 | Gene names | TRIM44, DIPB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB). | |||||
|
ANR21_HUMAN
|
||||||
| θ value | 6.88961 (rank : 106) | NC score | 0.024946 (rank : 86) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q86YR6 | Gene names | ANKRD21, POTE | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 21 (Protein POTE) (Prostate, ovary, testis-expressed protein). | |||||
|
CLSPN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 107) | NC score | 0.023002 (rank : 92) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9HAW4, Q5VYG0, Q6P6H5, Q8IWI1 | Gene names | CLSPN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin (hClaspin) (Hu-Claspin). | |||||
|
EHMT2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 108) | NC score | 0.005932 (rank : 147) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Z148, Q6PE08, Q8K4R6, Q8K4R7, Q9Z149 | Gene names | Ehmt2, Bat8, G9a, Ng36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
HS90B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.021524 (rank : 99) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P08238, Q5T9W7, Q9NQW0 | Gene names | HSP90AB1, HSP90B, HSPC2, HSPCB | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-beta (HSP 84) (HSP 90). | |||||
|
MYH4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.015994 (rank : 118) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
MYH8_MOUSE
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.018251 (rank : 108) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
NUDC_MOUSE
|
||||||
| θ value | 6.88961 (rank : 112) | NC score | 0.016077 (rank : 117) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35685, Q3UJS7 | Gene names | Nudc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear migration protein nudC (Nuclear distribution protein C homolog) (Silica-induced gene 92 protein) (SIG-92). | |||||
|
PHF14_HUMAN
|
||||||
| θ value | 6.88961 (rank : 113) | NC score | 0.028373 (rank : 77) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O94880 | Gene names | PHF14, KIAA0783 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 14. | |||||
|
POT15_HUMAN
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.018852 (rank : 105) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q6S5H4, Q6NXN7, Q6S5H7 | Gene names | POTE15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prostate, ovary, testis-expressed protein on chromosome 15. | |||||
|
RPA1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.011095 (rank : 135) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95602, Q9UEH0, Q9UFT9 | Gene names | POLR1A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase I largest subunit (EC 2.7.7.6) (RNA polymerase I 194 kDa subunit) (RPA194). | |||||
|
SETD2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 116) | NC score | 0.014785 (rank : 122) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
TOP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 117) | NC score | 0.025453 (rank : 84) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q04750 | Gene names | Top1, Top-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 1 (EC 5.99.1.2) (DNA topoisomerase I). | |||||
|
TXLNG_HUMAN
|
||||||
| θ value | 6.88961 (rank : 118) | NC score | 0.040672 (rank : 48) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9NUQ3, Q9P0X1 | Gene names | TXLNG, CXorf15, LSR5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gamma-taxilin (Lipopolysaccharide-specific response protein 5). | |||||
|
TXLNG_MOUSE
|
||||||
| θ value | 6.88961 (rank : 119) | NC score | 0.041208 (rank : 46) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 552 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BHN1, Q8BP11 | Gene names | Txlng | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gamma-taxilin. | |||||
|
UBP34_HUMAN
|
||||||
| θ value | 6.88961 (rank : 120) | NC score | 0.015918 (rank : 119) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q70CQ2, O60316, O94834, Q3B777, Q6P6C9, Q7L8P6, Q8N3T9, Q8TBW2, Q9UGA1 | Gene names | USP34, KIAA0570, KIAA0729 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
UBP34_MOUSE
|
||||||
| θ value | 6.88961 (rank : 121) | NC score | 0.015411 (rank : 120) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6ZQ93, Q3UPN0, Q6P563, Q7TMJ6, Q8CCH0 | Gene names | Usp34, Kiaa0570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
ZN326_MOUSE
|
||||||
| θ value | 6.88961 (rank : 122) | NC score | 0.012883 (rank : 129) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88291, Q8BSJ5, Q8K1X9, Q9CYG9 | Gene names | Znf326, Zan75, Zfp326 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 326 (Zinc finger protein-associated with nuclear matrix of 75 kDa). | |||||
|
CSTN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | 0.005301 (rank : 149) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O94985 | Gene names | CLSTN1, CS1, KIAA0911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calsyntenin-1 precursor. | |||||
|
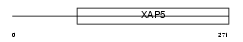
FA50A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.012504 (rank : 130) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14320, Q5HY37 | Gene names | FAM50A, DXS9928E, HXC26, XAP5 | |||
|
Domain Architecture |
|
|||||
| Description | Protein FAM50A (Protein XAP-5) (Protein HXC-26). | |||||
|
FAM9A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.022466 (rank : 94) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IZU1 | Gene names | FAM9A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM9A. | |||||
|
GNL3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 126) | NC score | 0.014909 (rank : 121) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CI11, Q811R9, Q811S8, Q8BK21 | Gene names | Gnl3, Ns | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein-like 3 (Nucleolar GTP-binding protein 3) (Nucleostemin). | |||||
|
GRIPE_HUMAN
|
||||||
| θ value | 8.99809 (rank : 127) | NC score | 0.010107 (rank : 138) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6GYQ0, O94960, Q6GYP9, Q6ZT23, Q86YF3, Q86YF5, Q8ND69 | Gene names | GARNL1, KIAA0884, TULIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTPase-activating Rap/Ran-GAP domain-like 1 (GAP-related-interacting partner to E12) (GRIPE) (Tuberin-like protein 1). | |||||
|
HTSF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.013894 (rank : 124) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43719, Q59G06, Q99730 | Gene names | HTATSF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 (Tat-SF1). | |||||
|
ITSN2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | 0.013482 (rank : 126) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1173 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NZM3, O95062, Q15812, Q9HAK4, Q9NXE6, Q9NYG0, Q9NZM2, Q9ULG4 | Gene names | ITSN2, KIAA1256, SH3D1B, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (SH3P18) (SH3P18-like WASP-associated protein). | |||||
|
KI21A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | 0.016398 (rank : 113) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1543 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q7Z4S6, Q6UKL9, Q7Z668, Q86WZ5, Q8IVZ8, Q9C0F5, Q9NXU4, Q9Y590 | Gene names | KIF21A, KIAA1708, KIF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A (Kinesin-like protein KIF2) (NY-REN-62 antigen). | |||||
|
LRC50_MOUSE
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.009359 (rank : 142) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D2H9, Q9CVS9 | Gene names | Lrrc50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 50. | |||||
|
LRRF2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | 0.036828 (rank : 58) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q91WK0, Q8BVD1, Q9CU89 | Gene names | Lrrfip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 2 (LRR FLII- interacting protein 2). | |||||
|
LYRIC_HUMAN
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | 0.014757 (rank : 123) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q86UE4, Q6PK07, Q8TCX3 | Gene names | MTDH, AEG1, LYRIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LYRIC (Lysine-rich CEACAM1 co-isolated protein) (3D3/lyric) (Metastasis adhesion protein) (Metadherin) (Astrocyte elevated gene-1 protein) (AEG-1). | |||||
|
NOL10_MOUSE
|
||||||
| θ value | 8.99809 (rank : 134) | NC score | 0.013299 (rank : 127) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5RJG1 | Gene names | Nol10, Gm67 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 10. | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 135) | NC score | 0.037347 (rank : 55) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
PLCB4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 136) | NC score | 0.020689 (rank : 101) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q15147, Q5JYS8, Q5JYT0, Q5JYT4, Q9BQW5, Q9BQW6, Q9BQW8, Q9UJQ2 | Gene names | PLCB4 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 4 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-4) (PLC-beta-4). | |||||
|
PLEC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 137) | NC score | 0.013100 (rank : 128) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
RPO3G_MOUSE
|
||||||
| θ value | 8.99809 (rank : 138) | NC score | 0.017371 (rank : 111) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6NXY9, Q8K0W5, Q9CV05 | Gene names | Polr3g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-directed RNA polymerase III subunit G (EC 2.7.7.6) (DNA-directed RNA polymerase III 32 kDa polypeptide) (RNA polymerase III C32 subunit). | |||||
|
S12A6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 139) | NC score | 0.006162 (rank : 146) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UHW9, Q8TDD4, Q9UFR2, Q9Y642, Q9Y665 | Gene names | SLC12A6, KCC3 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 6 (Electroneutral potassium-chloride cotransporter 3) (K-Cl cotransporter 3). | |||||
|
WBP11_HUMAN
|
||||||
| θ value | 8.99809 (rank : 140) | NC score | 0.022257 (rank : 96) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
WDHD1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 141) | NC score | 0.009093 (rank : 143) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P59328, Q6P408 | Gene names | Wdhd1, And1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat and HMG-box DNA-binding protein 1 (Acidic nucleoplasmic DNA- binding protein 1) (And-1). | |||||
|
XE7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 142) | NC score | 0.016370 (rank : 114) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 738 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q02040, Q02832 | Gene names | CXYorf3, DXYS155E, XE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-lymphocyte antigen precursor (B-lymphocyte surface antigen) (721P) (Protein XE7). | |||||
|
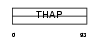
THA10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.061109 (rank : 25) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9P2Z0 | Gene names | THAP10 | |||
|
Domain Architecture |
|
|||||
| Description | THAP domain-containing protein 10. | |||||
|
THAP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.159395 (rank : 17) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8WTV1, Q569K1, Q5TH66, Q5TH67, Q8N8T6, Q9BSC7, Q9Y3H2, Q9Y3H3 | Gene names | THAP3 | |||
|
Domain Architecture |
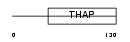
|
|||||
| Description | THAP domain-containing protein 3. | |||||
|
THAP3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.160837 (rank : 16) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BJ25, Q8BII6 | Gene names | Thap3 | |||
|
Domain Architecture |
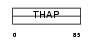
|
|||||
| Description | THAP domain-containing protein 3. | |||||
|
ZMYM3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.056916 (rank : 32) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14202, O15089 | Gene names | ZMYM3, DXS6673E, KIAA0385, ZNF261 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 3 (Zinc finger protein 261). | |||||
|
ZMYM3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.057538 (rank : 28) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JLM4, Q80U17 | Gene names | Zmym3, Kiaa0385, Zfp261, Znf261 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 3 (Zinc finger protein 261) (DXHXS6673E protein). | |||||
|
ZMYM5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.056620 (rank : 33) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9UJ78, Q5T6E2, Q96IY6, Q9NZY5, Q9UBW0, Q9UJ77 | Gene names | ZMYM5, ZNF198L1, ZNF237 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 5 (Zinc finger protein 237) (Zinc finger protein 198-like 1). | |||||
|
ZMYM6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.104717 (rank : 18) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | O95789, Q96IY0 | Gene names | ZMYM6, ZNF258 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 6 (Zinc finger protein 258). | |||||
|
ZN198_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.058251 (rank : 27) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UBW7, O43212, O43434, O60898, Q5W0Q4, Q63HP0, Q8NE39, Q9H538, Q9UEU2 | Gene names | ZNF198, FIM, RAMP, ZMYM2 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 198 (Zinc finger MYM-type protein 2) (Fused in myeloproliferative disorders protein) (Rearranged in atypical myeloproliferative disorder protein). | |||||
|
P52K_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | O43422, Q8WTW1, Q9Y3Z4 | Gene names | PRKRIR, DAP4, P52RIPK, THAP0 | |||
|
Domain Architecture |

|
|||||
| Description | 52 kDa repressor of the inhibitor of the protein kinase (p58IPK- interacting protein) (58 kDa interferon-induced protein kinase- interacting protein) (P52rIPK) (Death-associated protein 4) (THAP domain-containing protein 0). | |||||
|
P52K_MOUSE
|
||||||
| NC score | 0.968505 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9CUX1, Q80Y58 | Gene names | Prkrir, Thap0 | |||
|
Domain Architecture |

|
|||||
| Description | 52 kDa repressor of the inhibitor of the protein kinase (p58IPK- interacting protein) (58 kDa interferon-induced protein kinase- interacting protein) (P52rIPK) (THAP domain-containing protein 0). | |||||
|
ZMYM1_HUMAN
|
||||||
| NC score | 0.493913 (rank : 3) | θ value | 1.12514e-34 (rank : 3) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5SVZ6, Q7Z3Q4 | Gene names | ZMYM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 1. | |||||
|
THAP2_HUMAN
|
||||||
| NC score | 0.459322 (rank : 4) | θ value | 1.133e-10 (rank : 4) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H0W7 | Gene names | THAP2 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 2. | |||||
|
THAP2_MOUSE
|
||||||
| NC score | 0.452429 (rank : 5) | θ value | 5.62301e-10 (rank : 5) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9D305 | Gene names | Thap2 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 2. | |||||
|
THAP6_HUMAN
|
||||||
| NC score | 0.354023 (rank : 6) | θ value | 0.000602161 (rank : 9) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8TBB0 | Gene names | THAP6 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 6. | |||||
|
THAP7_MOUSE
|
||||||
| NC score | 0.348550 (rank : 7) | θ value | 1.09739e-05 (rank : 6) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8VCZ3 | Gene names | Thap7 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 7. | |||||
|
THAP7_HUMAN
|
||||||
| NC score | 0.347841 (rank : 8) | θ value | 1.43324e-05 (rank : 7) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BT49 | Gene names | THAP7 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 7. | |||||
|
THAP8_HUMAN
|
||||||
| NC score | 0.322377 (rank : 9) | θ value | 0.00134147 (rank : 11) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8NA92, Q96M21 | Gene names | THAP8 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 8. | |||||
|
THAP4_HUMAN
|
||||||
| NC score | 0.294654 (rank : 10) | θ value | 9.29e-05 (rank : 8) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8WY91, Q53NU7, Q6GRN0, Q6IPJ3, Q9NW26, Q9Y325 | Gene names | THAP4 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 4. | |||||
|
THAP1_HUMAN
|
||||||
| NC score | 0.288628 (rank : 11) | θ value | 0.00665767 (rank : 15) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NVV9, Q53FQ1, Q6IA99 | Gene names | THAP1 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 1. | |||||
|
THAP1_MOUSE
|
||||||
| NC score | 0.276383 (rank : 12) | θ value | 0.0252991 (rank : 18) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CHW1 | Gene names | Thap1 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 1. | |||||
|
THA11_HUMAN
|
||||||
| NC score | 0.273054 (rank : 13) | θ value | 0.00102713 (rank : 10) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96EK4, O94795 | Gene names | THAP11 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 11. | |||||
|
THAP4_MOUSE
|
||||||
| NC score | 0.268544 (rank : 14) | θ value | 0.00298849 (rank : 14) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6P3Z3, Q3US28, Q6NZN6, Q6PEN6, Q91WR2, Q9CVE5, Q9CVG3 | Gene names | Thap4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THAP domain-containing protein 4. | |||||
|
THA11_MOUSE
|
||||||
| NC score | 0.243461 (rank : 15) | θ value | 0.00298849 (rank : 13) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JJD0 | Gene names | Thap11 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 11. | |||||
|
THAP3_MOUSE
|
||||||
| NC score | 0.160837 (rank : 16) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BJ25, Q8BII6 | Gene names | Thap3 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 3. | |||||
|
THAP3_HUMAN
|
||||||
| NC score | 0.159395 (rank : 17) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8WTV1, Q569K1, Q5TH66, Q5TH67, Q8N8T6, Q9BSC7, Q9Y3H2, Q9Y3H3 | Gene names | THAP3 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 3. | |||||
|
ZMYM6_HUMAN
|
||||||
| NC score | 0.104717 (rank : 18) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | O95789, Q96IY0 | Gene names | ZMYM6, ZNF258 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 6 (Zinc finger protein 258). | |||||
|
NARGL_HUMAN
|
||||||
| NC score | 0.083330 (rank : 19) | θ value | 0.0252991 (rank : 17) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6N069, Q5VSP9, Q6P2D5, Q8N5J3, Q8N870 | Gene names | NARG1L, NAT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NMDA receptor-regulated 1-like protein (NARG1-like protein). | |||||
|
NARGL_MOUSE
|
||||||
| NC score | 0.073359 (rank : 20) | θ value | 0.813845 (rank : 35) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9DBB4, Q3U7V2 | Gene names | Narg1l, Nat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NMDA receptor-regulated 1-like protein (NARG1-like protein). | |||||
|
NARG1_MOUSE
|
||||||
| NC score | 0.071020 (rank : 21) | θ value | 1.38821 (rank : 50) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80UM3, Q811Z9, Q9JID5 | Gene names | Narg1, Nat1, Tbdn-1, Tubedown | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NMDA receptor-regulated protein 1 (N-terminal acetyltransferase 1) (Protein Tubedown-1). | |||||
|
NARG1_HUMAN
|
||||||
| NC score | 0.070861 (rank : 22) | θ value | 1.38821 (rank : 49) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BXJ9, Q8IWH4, Q8NEV2, Q9H8P6 | Gene names | NARG1, GA19, NATH, TBDN100 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NMDA receptor-regulated protein 1 (N-terminal acetyltransferase) (Tubedown-1 protein) (Tbdn100) (Gastric cancer antigen Ga19). | |||||
|
IL33_HUMAN
|
||||||
| NC score | 0.063254 (rank : 23) | θ value | 0.279714 (rank : 26) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95760, Q2YEJ5 | Gene names | IL33, C9orf26, IL1F11, NFHEV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-33 precursor (IL-33) (Interleukin-1 family member 11) (IL- 1F11) (Nuclear factor from high endothelial venules) (NF-HEV). | |||||
|
ZN452_HUMAN
|
||||||
| NC score | 0.062967 (rank : 24) | θ value | 0.00228821 (rank : 12) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6R2W3, Q2NKL9, Q5SRJ3, Q8TCN2, Q96PW3 | Gene names | ZNF452, KIAA1925 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF452. | |||||
|
THA10_HUMAN
|
||||||
| NC score | 0.061109 (rank : 25) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9P2Z0 | Gene names | THAP10 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 10. | |||||
|
TXLNA_MOUSE
|
||||||
| NC score | 0.058676 (rank : 26) | θ value | 0.0961366 (rank : 21) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 846 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6PAM1, Q6P1E5 | Gene names | Txlna, Txln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
ZN198_HUMAN
|
||||||
| NC score | 0.058251 (rank : 27) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UBW7, O43212, O43434, O60898, Q5W0Q4, Q63HP0, Q8NE39, Q9H538, Q9UEU2 | Gene names | ZNF198, FIM, RAMP, ZMYM2 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 198 (Zinc finger MYM-type protein 2) (Fused in myeloproliferative disorders protein) (Rearranged in atypical myeloproliferative disorder protein). | |||||
|
ZMYM3_MOUSE
|
||||||
| NC score | 0.057538 (rank : 28) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JLM4, Q80U17 | Gene names | Zmym3, Kiaa0385, Zfp261, Znf261 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 3 (Zinc finger protein 261) (DXHXS6673E protein). | |||||
|
CF170_HUMAN
|
||||||
| NC score | 0.057356 (rank : 29) | θ value | 1.81305 (rank : 55) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96NH3, Q6ZMY4, Q6ZUR7, Q8NB47 | Gene names | C6orf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf170. | |||||
|
EPMIP_MOUSE
|
||||||
| NC score | 0.057294 (rank : 30) | θ value | 0.47712 (rank : 29) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VEH5, Q80TS4 | Gene names | Epm2aip1, Kiaa0766 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EPM2A-interacting protein 1 (Laforin-interacting protein). | |||||
|
TXLNA_HUMAN
|
||||||
| NC score | 0.057147 (rank : 31) | θ value | 0.125558 (rank : 22) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 982 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P40222, Q66K62, Q86T54, Q86T85, Q86T86, Q86Y86, Q86YW3, Q8N2Y3 | Gene names | TXLNA, TXLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
ZMYM3_HUMAN
|
||||||
| NC score | 0.056916 (rank : 32) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14202, O15089 | Gene names | ZMYM3, DXS6673E, KIAA0385, ZNF261 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 3 (Zinc finger protein 261). | |||||
|
ZMYM5_HUMAN
|
||||||
| NC score | 0.056620 (rank : 33) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9UJ78, Q5T6E2, Q96IY6, Q9NZY5, Q9UBW0, Q9UJ77 | Gene names | ZMYM5, ZNF198L1, ZNF237 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 5 (Zinc finger protein 237) (Zinc finger protein 198-like 1). | |||||
|
TXLNB_HUMAN
|
||||||
| NC score | 0.053663 (rank : 34) | θ value | 1.38821 (rank : 53) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8N3L3, Q76L25, Q86T52, Q8N3S2 | Gene names | TXLNB, C6orf198, MDP77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77) (hMDP77). | |||||
|
RPGR1_MOUSE
|
||||||
| NC score | 0.053195 (rank : 35) | θ value | 0.0563607 (rank : 20) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
RENT2_HUMAN
|
||||||
| NC score | 0.050392 (rank : 36) | θ value | 0.47712 (rank : 31) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
LRRF2_HUMAN
|
||||||
| NC score | 0.048052 (rank : 37) | θ value | 0.163984 (rank : 23) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 791 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Y608, Q68CV3, Q9NXH5 | Gene names | LRRFIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 2 (LRR FLII- interacting protein 2). | |||||
|
DPOE3_HUMAN
|
||||||
| NC score | 0.047649 (rank : 38) | θ value | 1.38821 (rank : 44) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NRF9, Q5W0U1, Q8N758, Q8NCE5, Q9NR32 | Gene names | POLE3, CHRAC17 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase epsilon subunit 3 (EC 2.7.7.7) (DNA polymerase II subunit 3) (DNA polymerase epsilon subunit p17) (Chromatin accessibility complex 17) (HuCHRAC17) (CHRAC-17) (Arsenic- transactivated protein) (AsTP). | |||||
|
CAF1A_MOUSE
|
||||||
| NC score | 0.045619 (rank : 39) | θ value | 0.813845 (rank : 34) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9QWF0 | Gene names | Chaf1a, Caip150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
CAF1A_HUMAN
|
||||||
| NC score | 0.044475 (rank : 40) | θ value | 1.06291 (rank : 36) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q13111, Q7Z7K3, Q9UJY8 | Gene names | CHAF1A, CAF, CAF1P150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
PTMS_HUMAN
|
||||||
| NC score | 0.043160 (rank : 41) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P20962 | Gene names | PTMS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
PHF20_MOUSE
|
||||||
| NC score | 0.043061 (rank : 42) | θ value | 5.27518 (rank : 101) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
VDP_MOUSE
|
||||||
| NC score | 0.042299 (rank : 43) | θ value | 0.21417 (rank : 25) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 469 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Z1Z0, Q3T9L9, Q3TH58, Q3U1C7, Q3UMW6, Q91WE7, Q99JZ5 | Gene names | Vdp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General vesicular transport factor p115 (Transcytosis-associated protein) (TAP) (Vesicle docking protein). | |||||
|
CCAR1_MOUSE
|
||||||
| NC score | 0.041543 (rank : 44) | θ value | 0.62314 (rank : 33) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8CH18, Q6AXC9, Q6PAR2, Q80XE4, Q8BJY0, Q8BVN2, Q8CGG1, Q9CSR5 | Gene names | Ccar1, Carp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle and apoptosis regulator protein 1 (Cell cycle and apoptosis regulatory protein 1) (CARP-1). | |||||
|
CALD1_HUMAN
|
||||||
| NC score | 0.041254 (rank : 45) | θ value | 0.0193708 (rank : 16) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
TXLNG_MOUSE
|
||||||
| NC score | 0.041208 (rank : 46) | θ value | 6.88961 (rank : 119) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 552 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BHN1, Q8BP11 | Gene names | Txlng | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gamma-taxilin. | |||||
|
RAD50_MOUSE
|
||||||
| NC score | 0.040920 (rank : 47) | θ value | 0.47712 (rank : 30) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
TXLNG_HUMAN
|
||||||
| NC score | 0.040672 (rank : 48) | θ value | 6.88961 (rank : 118) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9NUQ3, Q9P0X1 | Gene names | TXLNG, CXorf15, LSR5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gamma-taxilin (Lipopolysaccharide-specific response protein 5). | |||||
|
RAD50_HUMAN
|
||||||
| NC score | 0.040661 (rank : 49) | θ value | 0.365318 (rank : 28) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
PHF14_MOUSE
|
||||||
| NC score | 0.040063 (rank : 50) | θ value | 0.163984 (rank : 24) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9D4H9, Q810Z6, Q8C284, Q8CGF8, Q9D1Q8 | Gene names | Phf14 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 14. | |||||
|
PININ_MOUSE
|
||||||
| NC score | 0.039830 (rank : 51) | θ value | 3.0926 (rank : 72) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
DHX29_HUMAN
|
||||||
| NC score | 0.039591 (rank : 52) | θ value | 0.0330416 (rank : 19) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7Z478, O75549, Q63HN0, Q63HN3, Q8IWW2, Q8N3A1, Q9UMH2 | Gene names | DHX29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX29 (EC 3.6.1.-) (DEAH box protein 29) (Nucleic acid helicase DDXx). | |||||
|
ARI4A_HUMAN
|
||||||
| NC score | 0.038345 (rank : 53) | θ value | 1.81305 (rank : 54) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P29374, Q15991, Q15992, Q15993 | Gene names | ARID4A, RBBP1, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 4A (ARID domain- containing protein 4A) (Retinoblastoma-binding protein 1) (RBBP-1). | |||||
|
PININ_HUMAN
|
||||||
| NC score | 0.037799 (rank : 54) | θ value | 2.36792 (rank : 65) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9H307, O60899, Q53EM7, Q6P5X4, Q7KYL1, Q99738, Q9UHZ9, Q9UQR9 | Gene names | PNN, DRS, MEMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin (140 kDa nuclear and cell adhesion-related phosphoprotein) (Domain-rich serine protein) (DRS-protein) (DRSP) (Melanoma metastasis clone A protein) (Desmosome-associated protein) (SR-like protein) (Nuclear protein SDK3). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.037347 (rank : 55) | θ value | 8.99809 (rank : 135) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
CROP_HUMAN
|
||||||
| NC score | 0.037318 (rank : 56) | θ value | 1.06291 (rank : 39) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O95232, Q6PHR9, Q9NUY0, Q9P2S7 | Gene names | CROP, CREAP1, O48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cisplatin resistance-associated overexpressed protein (cAMP regulatory element-associated protein 1) (CRE-associated protein 1) (CREAP-1) (Luc7A) (Okadaic acid-inducible phosphoprotein OA48-18). | |||||
|
PELP1_MOUSE
|
||||||
| NC score | 0.037199 (rank : 57) | θ value | 1.06291 (rank : 41) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
LRRF2_MOUSE
|
||||||
| NC score | 0.036828 (rank : 58) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q91WK0, Q8BVD1, Q9CU89 | Gene names | Lrrfip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 2 (LRR FLII- interacting protein 2). | |||||
|
CROP_MOUSE
|
||||||
| NC score | 0.035652 (rank : 59) | θ value | 1.38821 (rank : 43) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5SUF2, Q3U9D5, Q8BUJ5, Q921Z3, Q9CRS7, Q9CTY4 | Gene names | Crop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cisplatin resistance-associated overexpressed protein. | |||||
|
DPOE3_MOUSE
|
||||||
| NC score | 0.035622 (rank : 60) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JKP7, Q8R198 | Gene names | Pole3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase epsilon subunit 3 (EC 2.7.7.7) (DNA polymerase II subunit 3) (DNA polymerase epsilon subunit p17) (YB-like protein 1) (YBL1) (NF-YB-like protein). | |||||
|
CN145_HUMAN
|
||||||
| NC score | 0.034956 (rank : 61) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1196 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q6ZU80, Q96ML4 | Gene names | C14orf145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145. | |||||
|
RIF1_MOUSE
|
||||||
| NC score | 0.034938 (rank : 62) | θ value | 1.38821 (rank : 52) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6PR54, Q6T8D9, Q7TPD8, Q8BTU2, Q8C408, Q9CS77 | Gene names | Rif1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomere-associated protein RIF1 (Rap1-interacting factor 1 homolog) (mRif1). | |||||
|
WBP11_MOUSE
|
||||||
| NC score | 0.033924 (rank : 63) | θ value | 1.06291 (rank : 42) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
DNMT1_HUMAN
|
||||||
| NC score | 0.033374 (rank : 64) | θ value | 1.81305 (rank : 57) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P26358, Q9UHG5, Q9ULA2, Q9UMZ6 | Gene names | DNMT1, AIM, DNMT | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 1 (EC 2.1.1.37) (Dnmt1) (DNA methyltransferase HsaI) (DNA MTase HsaI) (MCMT) (M.HsaI). | |||||
|
PELP1_HUMAN
|
||||||
| NC score | 0.032759 (rank : 65) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8IZL8, O15450, Q5EGN3, Q6NTE6, Q96FT1, Q9BU60 | Gene names | PELP1, HMX3, MNAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3). | |||||
|
ATRX_HUMAN
|
||||||
| NC score | 0.032071 (rank : 66) | θ value | 0.62314 (rank : 32) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
CCAR1_HUMAN
|
||||||
| NC score | 0.031639 (rank : 67) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8IX12, Q32NE3, Q5VUP6, Q6PIZ0, Q6X935, Q9H8N4, Q9NVA7, Q9NVQ0, Q9NWM6 | Gene names | CCAR1, CARP1, DIS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle and apoptosis regulator protein 1 (Cell cycle and apoptosis regulatory protein 1) (CARP-1) (Death inducer with SAP domain). | |||||
|
DHX29_MOUSE
|
||||||
| NC score | 0.030649 (rank : 68) | θ value | 1.81305 (rank : 56) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PGC1, Q8BQJ4, Q8BT01, Q8C9B7, Q8C9H9 | Gene names | Dhx29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX29 (EC 3.6.1.-) (DEAH box protein 29). | |||||
|
HOME1_HUMAN
|
||||||
| NC score | 0.030012 (rank : 69) | θ value | 1.38821 (rank : 46) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 564 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q86YM7, O96003, Q86YM5 | Gene names | HOMER1, SYN47 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 1. | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.029973 (rank : 70) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
TOP1_HUMAN
|
||||||
| NC score | 0.029865 (rank : 71) | θ value | 3.0926 (rank : 75) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P11387, O43256, Q12855, Q12856, Q15610, Q5TFY3, Q9UJN0 | Gene names | TOP1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 1 (EC 5.99.1.2) (DNA topoisomerase I). | |||||
|
IF2P_HUMAN
|
||||||
| NC score | 0.029328 (rank : 72) | θ value | 2.36792 (rank : 62) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
CHM4B_MOUSE
|
||||||
| NC score | 0.029005 (rank : 73) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D8B3, Q3TXM7, Q91VM7, Q922P1 | Gene names | Chmp4b | |||
|
Domain Architecture |

|
|||||
| Description | Charged multivesicular body protein 4b (Chromatin-modifying protein 4b) (CHMP4b). | |||||
|
CHM4B_HUMAN
|
||||||
| NC score | 0.029004 (rank : 74) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H444, Q53ZD6 | Gene names | CHMP4B, C20orf178, SHAX1 | |||
|
Domain Architecture |

|
|||||
| Description | Charged multivesicular body protein 4b (Chromatin-modifying protein 4b) (CHMP4b) (Vacuolar protein sorting 7-2) (SNF7-2) (hSnf7-2) (SNF7 homolog associated with Alix 1) (hVps32). | |||||
|
CJ006_MOUSE
|
||||||
| NC score | 0.028967 (rank : 75) | θ value | 2.36792 (rank : 60) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6P9P0, Q8C041, Q8C0J5 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf6 homolog. | |||||
|
DNL1_HUMAN
|
||||||
| NC score | 0.028638 (rank : 76) | θ value | 1.06291 (rank : 40) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P18858 | Gene names | LIG1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 1 (EC 6.5.1.1) (DNA ligase I) (Polydeoxyribonucleotide synthase [ATP] 1). | |||||
|
PHF14_HUMAN
|
||||||
| NC score | 0.028373 (rank : 77) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O94880 | Gene names | PHF14, KIAA0783 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 14. | |||||
|
CG301_HUMAN
|
||||||
| NC score | 0.028299 (rank : 78) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P59074, Q8NFA9 | Gene names | ames=CGI-301 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CGI-301. | |||||
|
HOME1_MOUSE
|
||||||
| NC score | 0.028142 (rank : 79) | θ value | 3.0926 (rank : 69) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 630 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Z2Y3, Q8K3E1, Q8K4M8, Q9Z0E9, Q9Z216 | Gene names | Homer1, Vesl1 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 1 (VASP/Ena-related gene up-regulated during seizure and LTP) (Vesl-1). | |||||
|
MYST4_HUMAN
|
||||||
| NC score | 0.027751 (rank : 80) | θ value | 1.38821 (rank : 48) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
FAM9B_HUMAN
|
||||||
| NC score | 0.027584 (rank : 81) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IZU0 | Gene names | FAM9B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM9B. | |||||
|
SNIP1_HUMAN
|
||||||
| NC score | 0.027282 (rank : 82) | θ value | 3.0926 (rank : 73) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TAD8, Q96SP9, Q9H9T7 | Gene names | SNIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Smad nuclear-interacting protein 1. | |||||
|
TAF3_HUMAN
|
||||||
| NC score | 0.025737 (rank : 83) | θ value | 2.36792 (rank : 66) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
TOP1_MOUSE
|
||||||
| NC score | 0.025453 (rank : 84) | θ value | 6.88961 (rank : 117) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q04750 | Gene names | Top1, Top-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 1 (EC 5.99.1.2) (DNA topoisomerase I). | |||||
|
TRI44_MOUSE
|
||||||
| NC score | 0.024964 (rank : 85) | θ value | 2.36792 (rank : 68) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9QXA7, Q80SV9, Q9JHR8 | Gene names | Trim44, Dipb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB) (Protein Mc7). | |||||
|
ANR21_HUMAN
|
||||||
| NC score | 0.024946 (rank : 86) | θ value | 6.88961 (rank : 106) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q86YR6 | Gene names | ANKRD21, POTE | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 21 (Protein POTE) (Prostate, ovary, testis-expressed protein). | |||||
|
HS90A_HUMAN
|
||||||
| NC score | 0.024942 (rank : 87) | θ value | 5.27518 (rank : 96) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P07900, Q2PP14, Q9BVQ5 | Gene names | HSP90AA1, HSP90A, HSPC1, HSPCA | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-alpha (HSP 86) (NY-REN-38 antigen). | |||||
|
HS90B_MOUSE
|
||||||
| NC score | 0.024497 (rank : 88) | θ value | 3.0926 (rank : 70) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P11499, O89078 | Gene names | Hsp90ab1, Hsp84, Hsp84-1, Hspcb | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-beta (HSP 84) (Tumor-specific transplantation 84 kDa antigen) (TSTA). | |||||
|
MYT1_HUMAN
|
||||||
| NC score | 0.024283 (rank : 89) | θ value | 2.36792 (rank : 64) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q01538, O94922, Q9UPV2 | Gene names | MYT1, KIAA0835, KIAA1050, MTF1, MYTI, PLPB1 | |||
|
Domain Architecture |

|
|||||
| Description | Myelin transcription factor 1 (MyT1) (MyTI) (Proteolipid protein- binding protein) (PLPB1). | |||||
|
NOC2L_MOUSE
|
||||||
| NC score | 0.023939 (rank : 90) | θ value | 4.03905 (rank : 82) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9WV70, Q8VE16 | Gene names | Noc2l | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar complex protein 2 homolog (Protein NOC2 homolog) (NOC2- like). | |||||
|
MYST3_MOUSE
|
||||||
| NC score | 0.023710 (rank : 91) | θ value | 4.03905 (rank : 81) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
CLSPN_HUMAN
|
||||||
| NC score | 0.023002 (rank : 92) | θ value | 6.88961 (rank : 107) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9HAW4, Q5VYG0, Q6P6H5, Q8IWI1 | Gene names | CLSPN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin (hClaspin) (Hu-Claspin). | |||||
|
ITSN2_MOUSE
|
||||||
| NC score | 0.022852 (rank : 93) | θ value | 2.36792 (rank : 63) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1371 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Z0R6, Q9Z0R5 | Gene names | Itsn2, Ese2, Sh3d1B | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (EH and SH3 domains protein 2) (EH domain and SH3 domain regulator of endocytosis 2). | |||||
|
FAM9A_HUMAN
|
||||||
| NC score | 0.022466 (rank : 94) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IZU1 | Gene names | FAM9A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM9A. | |||||
|
SRCH_HUMAN
|
||||||
| NC score | 0.022380 (rank : 95) | θ value | 5.27518 (rank : 103) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P23327, Q504Y6 | Gene names | HRC, HCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcoplasmic reticulum histidine-rich calcium-binding protein precursor. | |||||
|
WBP11_HUMAN
|
||||||
| NC score | 0.022257 (rank : 96) | θ value | 8.99809 (rank : 140) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
MYT1L_HUMAN
|
||||||
| NC score | 0.022063 (rank : 97) | θ value | 5.27518 (rank : 99) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UL68, Q6IQ17, Q9UPP6 | Gene names | MYT1L, KIAA1106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (MyT1-L). | |||||
|
SMRC1_MOUSE
|
||||||
| NC score | 0.021624 (rank : 98) | θ value | 5.27518 (rank : 102) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P97496, Q7TS80, Q7TT29 | Gene names | Smarcc1, Baf155, Srg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155) (SWI3-related protein). | |||||
|
HS90B_HUMAN
|
||||||
| NC score | 0.021524 (rank : 99) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P08238, Q5T9W7, Q9NQW0 | Gene names | HSP90AB1, HSP90B, HSPC2, HSPCB | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-beta (HSP 84) (HSP 90). | |||||
|
RBM25_HUMAN
|
||||||
| NC score | 0.021278 (rank : 100) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P49756, Q9UEQ5, Q9UIE9 | Gene names | RBM25, RNPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable RNA-binding protein 25 (RNA-binding motif protein 25) (RNA- binding region-containing protein 7) (Protein S164). | |||||
|
PLCB4_HUMAN
|
||||||
| NC score | 0.020689 (rank : 101) | θ value | 8.99809 (rank : 136) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q15147, Q5JYS8, Q5JYT0, Q5JYT4, Q9BQW5, Q9BQW6, Q9BQW8, Q9UJQ2 | Gene names | PLCB4 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 4 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-4) (PLC-beta-4). | |||||
|
TRI44_HUMAN
|
||||||
| NC score | 0.020224 (rank : 102) | θ value | 5.27518 (rank : 105) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96DX7, Q96QY2, Q9UGK0 | Gene names | TRIM44, DIPB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB). | |||||
|
MIER1_HUMAN
|
||||||
| NC score | 0.019941 (rank : 103) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N108, Q5T104, Q6AHY8, Q86TB4, Q8N156, Q8N161, Q8NC37, Q8NES4, Q8NES5, Q8NES6, Q8WWG2, Q9HCG2 | Gene names | MIER1, KIAA1610 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm induction early response protein 1 (Mi-er1) (hmi-er1) (Early response 1) (Er1). | |||||
|
MTX1_HUMAN
|
||||||
| NC score | 0.019299 (rank : 104) | θ value | 5.27518 (rank : 97) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13505 | Gene names | MTX1, MTX, MTXN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metaxin-1. | |||||
|
POT15_HUMAN
|
||||||
| NC score | 0.018852 (rank : 105) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q6S5H4, Q6NXN7, Q6S5H7 | Gene names | POTE15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prostate, ovary, testis-expressed protein on chromosome 15. | |||||
|
AT1B4_HUMAN
|
||||||
| NC score | 0.018473 (rank : 106) | θ value | 2.36792 (rank : 59) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UN42, Q9UN41 | Gene names | ATP1B4 | |||
|
Domain Architecture |

|
|||||
| Description | X/potassium-transporting ATPase subunit beta-m (X,K-ATPase beta-m subunit). | |||||
|
TEKT1_HUMAN
|
||||||
| NC score | 0.018261 (rank : 107) | θ value | 5.27518 (rank : 104) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q969V4 | Gene names | TEKT1 | |||
|
Domain Architecture |

|
|||||
| Description | Tektin-1. | |||||
|
MYH8_MOUSE
|
||||||
| NC score | 0.018251 (rank : 108) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
FA43A_HUMAN
|
||||||
| NC score | 0.017909 (rank : 109) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N2R8, Q8IXP4, Q8WZ07 | Gene names | FAM43A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM43A. | |||||
|
FGD5_MOUSE
|
||||||
| NC score | 0.017376 (rank : 110) | θ value | 1.38821 (rank : 45) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80UZ0, Q8BHM5 | Gene names | Fgd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 5. | |||||
|
RPO3G_MOUSE
|
||||||
| NC score | 0.017371 (rank : 111) | θ value | 8.99809 (rank : 138) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6NXY9, Q8K0W5, Q9CV05 | Gene names | Polr3g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-directed RNA polymerase III subunit G (EC 2.7.7.6) (DNA-directed RNA polymerase III 32 kDa polypeptide) (RNA polymerase III C32 subunit). | |||||
|
NFM_MOUSE
|
||||||
| NC score | 0.016952 (rank : 112) | θ value | 3.0926 (rank : 71) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
KI21A_HUMAN
|
||||||
| NC score | 0.016398 (rank : 113) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1543 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q7Z4S6, Q6UKL9, Q7Z668, Q86WZ5, Q8IVZ8, Q9C0F5, Q9NXU4, Q9Y590 | Gene names | KIF21A, KIAA1708, KIF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A (Kinesin-like protein KIF2) (NY-REN-62 antigen). | |||||
|
XE7_HUMAN
|
||||||
| NC score | 0.016370 (rank : 114) | θ value | 8.99809 (rank : 142) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 738 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q02040, Q02832 | Gene names | CXYorf3, DXYS155E, XE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-lymphocyte antigen precursor (B-lymphocyte surface antigen) (721P) (Protein XE7). | |||||
|
PP2CG_MOUSE
|
||||||
| NC score | 0.016317 (rank : 115) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61074 | Gene names | Ppm1g, Fin13, Ppm1c | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform gamma (EC 3.1.3.16) (PP2C-gamma) (Protein phosphatase magnesium-dependent 1 gamma) (Protein phosphatase 1C) (Fibroblast growth factor-inducible protein 13) (FIN13). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.016234 (rank : 116) | θ value | 1.38821 (rank : 51) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
NUDC_MOUSE
|
||||||
| NC score | 0.016077 (rank : 117) | θ value | 6.88961 (rank : 112) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35685, Q3UJS7 | Gene names | Nudc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear migration protein nudC (Nuclear distribution protein C homolog) (Silica-induced gene 92 protein) (SIG-92). | |||||
|
MYH4_MOUSE
|
||||||
| NC score | 0.015994 (rank : 118) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
UBP34_HUMAN
|
||||||
| NC score | 0.015918 (rank : 119) | θ value | 6.88961 (rank : 120) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q70CQ2, O60316, O94834, Q3B777, Q6P6C9, Q7L8P6, Q8N3T9, Q8TBW2, Q9UGA1 | Gene names | USP34, KIAA0570, KIAA0729 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
UBP34_MOUSE
|
||||||
| NC score | 0.015411 (rank : 120) | θ value | 6.88961 (rank : 121) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6ZQ93, Q3UPN0, Q6P563, Q7TMJ6, Q8CCH0 | Gene names | Usp34, Kiaa0570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
GNL3_MOUSE
|
||||||
| NC score | 0.014909 (rank : 121) | θ value | 8.99809 (rank : 126) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CI11, Q811R9, Q811S8, Q8BK21 | Gene names | Gnl3, Ns | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein-like 3 (Nucleolar GTP-binding protein 3) (Nucleostemin). | |||||
|
SETD2_HUMAN
|
||||||
| NC score | 0.014785 (rank : 122) | θ value | 6.88961 (rank : 116) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
LYRIC_HUMAN
|
||||||
| NC score | 0.014757 (rank : 123) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q86UE4, Q6PK07, Q8TCX3 | Gene names | MTDH, AEG1, LYRIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LYRIC (Lysine-rich CEACAM1 co-isolated protein) (3D3/lyric) (Metastasis adhesion protein) (Metadherin) (Astrocyte elevated gene-1 protein) (AEG-1). | |||||
|
HTSF1_HUMAN
|
||||||
| NC score | 0.013894 (rank : 124) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43719, Q59G06, Q99730 | Gene names | HTATSF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 (Tat-SF1). | |||||
|
CALR_HUMAN
|
||||||
| NC score | 0.013485 (rank : 125) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P27797 | Gene names | CALR, CRTC | |||
|
Domain Architecture |

|
|||||
| Description | Calreticulin precursor (CRP55) (Calregulin) (HACBP) (ERp60) (grp60). | |||||
|
ITSN2_HUMAN
|
||||||
| NC score | 0.013482 (rank : 126) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1173 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NZM3, O95062, Q15812, Q9HAK4, Q9NXE6, Q9NYG0, Q9NZM2, Q9ULG4 | Gene names | ITSN2, KIAA1256, SH3D1B, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (SH3P18) (SH3P18-like WASP-associated protein). | |||||
|
NOL10_MOUSE
|
||||||
| NC score | 0.013299 (rank : 127) | θ value | 8.99809 (rank : 134) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5RJG1 | Gene names | Nol10, Gm67 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 10. | |||||
|
PLEC1_HUMAN
|
||||||
| NC score | 0.013100 (rank : 128) | θ value | 8.99809 (rank : 137) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
ZN326_MOUSE
|
||||||
| NC score | 0.012883 (rank : 129) | θ value | 6.88961 (rank : 122) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88291, Q8BSJ5, Q8K1X9, Q9CYG9 | Gene names | Znf326, Zan75, Zfp326 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 326 (Zinc finger protein-associated with nuclear matrix of 75 kDa). | |||||
|
FA50A_HUMAN
|
||||||
| NC score | 0.012504 (rank : 130) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14320, Q5HY37 | Gene names | FAM50A, DXS9928E, HXC26, XAP5 | |||
|
Domain Architecture |

|
|||||
| Description | Protein FAM50A (Protein XAP-5) (Protein HXC-26). | |||||
|
CD2L2_HUMAN
|
||||||
| NC score | 0.012050 (rank : 131) | θ value | 0.365318 (rank : 27) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
ZEP1_HUMAN
|
||||||
| NC score | 0.011700 (rank : 132) | θ value | 1.81305 (rank : 58) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1036 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P15822, Q14122 | Gene names | HIVEP1, ZNF40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 40 (Human immunodeficiency virus type I enhancer- binding protein 1) (HIV-EP1) (Major histocompatibility complex-binding protein 1) (MBP-1) (Positive regulatory domain II-binding factor 1) (PRDII-BF1). | |||||
|
CD2L1_MOUSE
|
||||||
| NC score | 0.011534 (rank : 133) | θ value | 1.06291 (rank : 38) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1463 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P24788, Q3UI03, Q61399, Q7TST4, Q8BP53 | Gene names | Cdc2l1 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1). | |||||
|
CD2L1_HUMAN
|
||||||
| NC score | 0.011199 (rank : 134) | θ value | 1.06291 (rank : 37) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
RPA1_HUMAN
|
||||||
| NC score | 0.011095 (rank : 135) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95602, Q9UEH0, Q9UFT9 | Gene names | POLR1A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase I largest subunit (EC 2.7.7.6) (RNA polymerase I 194 kDa subunit) (RPA194). | |||||
|
NFL_MOUSE
|
||||||
| NC score | 0.011089 (rank : 136) | θ value | 5.27518 (rank : 100) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 695 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P08551, Q8K0Z0 | Gene names | Nefl, Nf68, Nfl | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet L protein (68 kDa neurofilament protein) (Neurofilament light polypeptide) (NF-L). | |||||
|
DDX46_MOUSE
|
||||||
| NC score | 0.010171 (rank : 137) | θ value | 2.36792 (rank : 61) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q569Z5, Q6ZQ42, Q8R0R6 | Gene names | Ddx46, Kiaa0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46). | |||||
|
GRIPE_HUMAN
|
||||||
| NC score | 0.010107 (rank : 138) | θ value | 8.99809 (rank : 127) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6GYQ0, O94960, Q6GYP9, Q6ZT23, Q86YF3, Q86YF5, Q8ND69 | Gene names | GARNL1, KIAA0884, TULIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTPase-activating Rap/Ran-GAP domain-like 1 (GAP-related-interacting partner to E12) (GRIPE) (Tuberin-like protein 1). | |||||
|
SRPK2_MOUSE
|
||||||
| NC score | 0.010021 (rank : 139) | θ value | 3.0926 (rank : 74) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O54781, Q8VCD9 | Gene names | Srpk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SRPK2 (EC 2.7.11.1) (Serine/arginine- rich protein-specific kinase 2) (SR-protein-specific kinase 2) (SFRS protein kinase 2). | |||||
|
ZEP1_MOUSE
|
||||||
| NC score | 0.009758 (rank : 140) | θ value | 4.03905 (rank : 88) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||
|
MYPT1_HUMAN
|
||||||
| NC score | 0.009542 (rank : 141) | θ value | 5.27518 (rank : 98) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O14974, Q86WU3, Q8NFR6, Q9BYH0 | Gene names | PPP1R12A, MBS, MYPT1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1) (Protein phosphatase myosin-binding subunit). | |||||
|
LRC50_MOUSE
|
||||||
| NC score | 0.009359 (rank : 142) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D2H9, Q9CVS9 | Gene names | Lrrc50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 50. | |||||
|
WDHD1_MOUSE
|
||||||
| NC score | 0.009093 (rank : 143) | θ value | 8.99809 (rank : 141) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P59328, Q6P408 | Gene names | Wdhd1, And1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat and HMG-box DNA-binding protein 1 (Acidic nucleoplasmic DNA- binding protein 1) (And-1). | |||||
|
MRCKG_HUMAN
|
||||||
| NC score | 0.007707 (rank : 144) | θ value | 1.38821 (rank : 47) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1519 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6DT37, O00565 | Gene names | CDC42BPG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK gamma (EC 2.7.11.1) (CDC42- binding protein kinase gamma) (Myotonic dystrophy kinase-related CDC42-binding kinase gamma) (Myotonic dystrophy protein kinase-like alpha) (MRCK gamma) (DMPK-like gamma). | |||||
|
AT2B1_HUMAN
|
||||||
| NC score | 0.007141 (rank : 145) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P20020, Q12992, Q12993, Q13819, Q13820, Q13821, Q16504, Q93082 | Gene names | ATP2B1, PMCA1 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 1 (EC 3.6.3.8) (PMCA1) (Plasma membrane calcium pump isoform 1) (Plasma membrane calcium ATPase isoform 1). | |||||
|
S12A6_HUMAN
|
||||||
| NC score | 0.006162 (rank : 146) | θ value | 8.99809 (rank : 139) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UHW9, Q8TDD4, Q9UFR2, Q9Y642, Q9Y665 | Gene names | SLC12A6, KCC3 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 6 (Electroneutral potassium-chloride cotransporter 3) (K-Cl cotransporter 3). | |||||
|
EHMT2_MOUSE
|
||||||
| NC score | 0.005932 (rank : 147) | θ value | 6.88961 (rank : 108) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Z148, Q6PE08, Q8K4R6, Q8K4R7, Q9Z149 | Gene names | Ehmt2, Bat8, G9a, Ng36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
TCF8_HUMAN
|
||||||
| NC score | 0.005769 (rank : 148) | θ value | 2.36792 (rank : 67) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P37275, Q12924, Q13800 | Gene names | TCF8, AREB6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (NIL-2-A zinc finger protein) (Negative regulator of IL2). | |||||
|
CSTN1_HUMAN
|
||||||
| NC score | 0.005301 (rank : 149) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O94985 | Gene names | CLSTN1, CS1, KIAA0911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calsyntenin-1 precursor. | |||||
|
TCF8_MOUSE
|
||||||
| NC score | 0.003890 (rank : 150) | θ value | 4.03905 (rank : 87) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1067 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q64318, Q62519 | Gene names | Tcf8, Zfhx1a, Zfx1a, Zfx1ha | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (Zinc finger homeobox protein 1a) (MEB1) (Delta EF1). | |||||