Please be patient as the page loads
|
GNL3_MOUSE
|
||||||
| SwissProt Accessions | Q8CI11, Q811R9, Q811S8, Q8BK21 | Gene names | Gnl3, Ns | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein-like 3 (Nucleolar GTP-binding protein 3) (Nucleostemin). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
GNL3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.959770 (rank : 2) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9BVP2, Q96SV6, Q96SV7, Q9UJY0 | Gene names | GNL3, E2IG3, NS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein-like 3 (Nucleolar GTP-binding protein 3) (Nucleostemin) (E2-induced gene 3-protein) (Novel nucleolar protein 47) (NNP47). | |||||
|
GNL3_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | Q8CI11, Q811R9, Q811S8, Q8BK21 | Gene names | Gnl3, Ns | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein-like 3 (Nucleolar GTP-binding protein 3) (Nucleostemin). | |||||
|
NOG2_MOUSE
|
||||||
| θ value | 1.16164e-39 (rank : 3) | NC score | 0.770836 (rank : 3) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99LH1, Q8BIF8 | Gene names | Gnl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar GTP-binding protein 2. | |||||
|
NOG2_HUMAN
|
||||||
| θ value | 1.51715e-39 (rank : 4) | NC score | 0.759727 (rank : 4) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13823, Q9BWN7 | Gene names | GNL2, NGP1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar GTP-binding protein 2 (Autoantigen NGP-1). | |||||
|
GNL1_HUMAN
|
||||||
| θ value | 3.64472e-09 (rank : 5) | NC score | 0.495140 (rank : 5) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P36915 | Gene names | GNL1, HSR1 | |||
|
Domain Architecture |
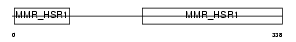
|
|||||
| Description | Guanine nucleotide-binding protein-like 1 (GTP-binding protein HSR1). | |||||
|
GNL1_MOUSE
|
||||||
| θ value | 3.64472e-09 (rank : 6) | NC score | 0.490405 (rank : 6) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P36916 | Gene names | Gnl1, Gna-rs1, Mmr1 | |||
|
Domain Architecture |
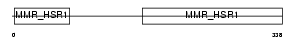
|
|||||
| Description | Guanine nucleotide-binding protein-like 1 (GTP-binding protein MMR1). | |||||
|
OPTN_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 7) | NC score | 0.069770 (rank : 20) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1120 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8K3K8 | Gene names | Optn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin. | |||||
|
TAOK1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 8) | NC score | 0.038711 (rank : 78) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1734 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q7L7X3, Q96L75, Q9H2K7, Q9H7S5, Q9P2I6 | Gene names | TAOK1, KIAA1361, MAP3K16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO1 (EC 2.7.11.1) (Thousand and one amino acid protein 1) (STE20-like kinase PSK2) (Kinase from chicken homolog B) (hKFC-B). | |||||
|
TAOK1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 9) | NC score | 0.038933 (rank : 75) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1720 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q5F2E8, Q69ZL2, Q8JZX2, Q91VG7 | Gene names | Taok1, Kiaa1361 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO1 (EC 2.7.11.1) (Thousand and one amino acid protein 1). | |||||
|
K0555_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 10) | NC score | 0.071726 (rank : 19) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 880 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q96AA8, O60302 | Gene names | KIAA0555 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0555. | |||||
|
ROCK2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 11) | NC score | 0.035593 (rank : 81) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 2073 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O75116, Q9UQN5 | Gene names | ROCK2, KIAA0619 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2) (Rho kinase 2). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 0.125558 (rank : 12) | NC score | 0.065305 (rank : 24) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
MIRO1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 13) | NC score | 0.050794 (rank : 56) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IXI2, Q6NUR3, Q6P9F8, Q6PJG1, Q6YMW8, Q86UB0, Q8IW28, Q8IXJ7, Q9H067, Q9H9N8, Q9NUZ2 | Gene names | RHOT1, ARHT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial Rho GTPase 1 (EC 3.6.5.-) (MIRO-1) (hMiro-1) (Ras homolog gene family member T1) (Rac-GTP-binding protein-like protein). | |||||
|
MIRO1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 14) | NC score | 0.050071 (rank : 61) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BG51, Q3TJB7, Q3UGU4, Q5SYC3, Q5SYC6, Q8BLW3, Q8BMH1, Q922N0, Q9D2R1, Q9JKB9 | Gene names | Rhot1, Arht1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial Rho GTPase 1 (EC 3.6.5.-) (MIRO-1) (Ras homolog gene family member T1). | |||||
|
SYCP1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 15) | NC score | 0.061351 (rank : 28) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
ROCK2_MOUSE
|
||||||
| θ value | 0.21417 (rank : 16) | NC score | 0.034831 (rank : 82) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 2072 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P70336, Q8BR64, Q8CBR0 | Gene names | Rock2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2). | |||||
|
TCRG1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 17) | NC score | 0.089580 (rank : 9) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
TCRG1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 18) | NC score | 0.088396 (rank : 10) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
CCAR1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 19) | NC score | 0.076097 (rank : 15) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8IX12, Q32NE3, Q5VUP6, Q6PIZ0, Q6X935, Q9H8N4, Q9NVA7, Q9NVQ0, Q9NWM6 | Gene names | CCAR1, CARP1, DIS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle and apoptosis regulator protein 1 (Cell cycle and apoptosis regulatory protein 1) (CARP-1) (Death inducer with SAP domain). | |||||
|
INVO_MOUSE
|
||||||
| θ value | 0.279714 (rank : 20) | NC score | 0.069588 (rank : 21) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P48997 | Gene names | Ivl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Involucrin. | |||||
|
CDKL2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 21) | NC score | 0.013320 (rank : 109) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 854 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9QUK0, Q3UL60, Q3V3X7, Q9QYI1, Q9QYI2 | Gene names | Cdkl2, Kkiamre, Kkm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-dependent kinase-like 2 (EC 2.7.11.22) (Serine/threonine- protein kinase KKIAMRE). | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 0.365318 (rank : 22) | NC score | 0.062623 (rank : 26) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
MIER1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 23) | NC score | 0.061462 (rank : 27) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N108, Q5T104, Q6AHY8, Q86TB4, Q8N156, Q8N161, Q8NC37, Q8NES4, Q8NES5, Q8NES6, Q8WWG2, Q9HCG2 | Gene names | MIER1, KIAA1610 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm induction early response protein 1 (Mi-er1) (hmi-er1) (Early response 1) (Er1). | |||||
|
CA065_HUMAN
|
||||||
| θ value | 0.47712 (rank : 24) | NC score | 0.073673 (rank : 16) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 469 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8N715, Q8N746, Q8NA93 | Gene names | C1orf65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf65. | |||||
|
ZN318_MOUSE
|
||||||
| θ value | 0.62314 (rank : 25) | NC score | 0.058496 (rank : 32) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q99PP2, Q3TZL5, Q9JJ01 | Gene names | Znf318, Tzf, Zfp318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Testicular zinc finger protein). | |||||
|
MIER1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 26) | NC score | 0.057134 (rank : 37) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5UAK0, Q5UAK1, Q5UAK2, Q5UAK3, Q6ZPL6, Q9D402 | Gene names | Mier1, Kiaa1610 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm induction early response protein 1 (Mi-er1). | |||||
|
CCD96_MOUSE
|
||||||
| θ value | 1.06291 (rank : 27) | NC score | 0.072858 (rank : 17) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9CR92 | Gene names | Ccdc96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 96. | |||||
|
GOGA5_MOUSE
|
||||||
| θ value | 1.06291 (rank : 28) | NC score | 0.053528 (rank : 44) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1434 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9QYE6, O88317, Q3TGE7, Q3U6S5, Q3UUF9 | Gene names | Golga5, Retii | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (Sumiko protein) (Ret-II protein). | |||||
|
OPTN_HUMAN
|
||||||
| θ value | 1.06291 (rank : 29) | NC score | 0.067071 (rank : 23) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 827 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q96CV9, Q5T672, Q5T673, Q5T674, Q5T675, Q7LDL9, Q8N562, Q9UET9, Q9UEV4, Q9Y218 | Gene names | OPTN, FIP2, GLC1E, NRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin (Optic neuropathy-inducing protein) (E3-14.7K-interacting protein) (FIP-2) (Huntingtin-interacting protein HYPL) (NEMO-related protein) (Transcription factor IIIA-interacting protein) (TFIIIA- IntP). | |||||
|
SM1L2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 30) | NC score | 0.060135 (rank : 29) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1019 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8NDV3, Q5TIC3, Q9Y3G5 | Gene names | SMC1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 2 protein (SMC1beta protein). | |||||
|
CT2NL_MOUSE
|
||||||
| θ value | 1.38821 (rank : 31) | NC score | 0.067903 (rank : 22) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 902 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q99LJ0, Q6ZPR0, Q8BSV1 | Gene names | Cttnbp2nl, Kiaa1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
PQBP1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 32) | NC score | 0.045535 (rank : 70) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91VJ5, Q80WW2, Q9ER43, Q9QYY2 | Gene names | Pqbp1, Npw38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyglutamine-binding protein 1 (Polyglutamine tract-binding protein 1) (PQBP-1) (38 kDa nuclear protein containing a WW domain) (Npw38). | |||||
|
TAOK2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 33) | NC score | 0.028510 (rank : 87) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1373 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6ZQ29, Q7TSS8 | Gene names | Taok2, Kiaa0881 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO2 (EC 2.7.11.1) (Thousand and one amino acid protein 2). | |||||
|
CCAR1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 34) | NC score | 0.071985 (rank : 18) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8CH18, Q6AXC9, Q6PAR2, Q80XE4, Q8BJY0, Q8BVN2, Q8CGG1, Q9CSR5 | Gene names | Ccar1, Carp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle and apoptosis regulator protein 1 (Cell cycle and apoptosis regulatory protein 1) (CARP-1). | |||||
|
CT2NL_HUMAN
|
||||||
| θ value | 1.81305 (rank : 35) | NC score | 0.063436 (rank : 25) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9P2B4, Q96B40 | Gene names | CTTNBP2NL, KIAA1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
MYH11_HUMAN
|
||||||
| θ value | 1.81305 (rank : 36) | NC score | 0.051843 (rank : 50) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
NFM_MOUSE
|
||||||
| θ value | 1.81305 (rank : 37) | NC score | 0.030424 (rank : 84) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
CE152_HUMAN
|
||||||
| θ value | 2.36792 (rank : 38) | NC score | 0.057931 (rank : 33) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O94986, Q6NTA0 | Gene names | CEP152, KIAA0912 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 152 kDa (Cep152 protein). | |||||
|
CJ006_MOUSE
|
||||||
| θ value | 2.36792 (rank : 39) | NC score | 0.029040 (rank : 86) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6P9P0, Q8C041, Q8C0J5 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf6 homolog. | |||||
|
GOGA3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.054566 (rank : 41) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
PTMA_HUMAN
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.053687 (rank : 42) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
SDPR_MOUSE
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.059565 (rank : 30) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q63918, Q3V1P6, Q78EC3, Q8CBT4 | Gene names | Sdpr, Sdr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein). | |||||
|
SH3K1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.027652 (rank : 90) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8R550, Q8CAL8, Q8CEF6, Q8R545, Q8R546, Q8R547, Q8R548, Q8R549, Q8R551, Q9CTQ9, Q9DC14, Q9JKC3 | Gene names | Sh3kbp1, Ruk, Seta | |||
|
Domain Architecture |

|
|||||
| Description | SH3-domain kinase-binding protein 1 (SH3-containing, expressed in tumorigenic astrocytes) (Regulator of ubiquitous kinase) (Ruk). | |||||
|
SMRC2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.023449 (rank : 98) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6PDG5, Q6P6P3 | Gene names | Smarcc2, Baf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
MY18A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.049843 (rank : 64) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
MYO5A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.049217 (rank : 65) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1117 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q99104 | Gene names | Myo5a, Dilute | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5A (Myosin Va) (Dilute myosin heavy chain, non-muscle). | |||||
|
PTD4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.057379 (rank : 36) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NTK5, Q8NCI8, Q96CU1, Q96SV2, Q9P1D3, Q9UNY9, Q9Y6G4 | Gene names | ames=PTD004, PRO2455 | |||
|
Domain Architecture |

|
|||||
| Description | Putative GTP-binding protein PTD004. | |||||
|
PTD4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.057407 (rank : 35) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CZ30, Q9CWE2, Q9CX91, Q9CYC9, Q9CZ56 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Putative GTP-binding protein PTD004 homolog. | |||||
|
RIMB1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.044237 (rank : 71) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
TMC2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.019509 (rank : 103) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8R4P4 | Gene names | Tmc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane cochlear-expressed protein 2. | |||||
|
AN32B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.018427 (rank : 105) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92688, O00655, P78458, P78459 | Gene names | ANP32B, APRIL, PHAPI2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member B (PHAPI2 protein) (Silver-stainable protein SSP29) (Acidic protein rich in leucines). | |||||
|
AN32B_MOUSE
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.018901 (rank : 104) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9EST5 | Gene names | Anp32b, Pal31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member B (Proliferation-related acidic leucine-rich protein PAL31). | |||||
|
CKAP2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.024831 (rank : 95) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q3V1H1, Q66LN2, Q8BSF0 | Gene names | Ckap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 2. | |||||
|
MYH10_HUMAN
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.048842 (rank : 66) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH10_MOUSE
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.048418 (rank : 67) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH11_MOUSE
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.051004 (rank : 53) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MYH14_HUMAN
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.048108 (rank : 68) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 995 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q7Z406, Q5CZ75, Q6XYE4, Q76B62, Q8WV23, Q96I22, Q9BT27, Q9BW35, Q9H882 | Gene names | MYH14, KIAA2034 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
MYH9_HUMAN
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.051628 (rank : 52) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MYH9_MOUSE
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | 0.051688 (rank : 51) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
RBBP6_MOUSE
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.037223 (rank : 80) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
TRI44_HUMAN
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.030861 (rank : 83) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96DX7, Q96QY2, Q9UGK0 | Gene names | TRIM44, DIPB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB). | |||||
|
TRI44_MOUSE
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.024844 (rank : 94) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9QXA7, Q80SV9, Q9JHR8 | Gene names | Trim44, Dipb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB) (Protein Mc7). | |||||
|
UBF1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.024704 (rank : 96) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
41_HUMAN
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.008413 (rank : 115) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P11171, P11176, Q14245, Q5TB35, Q5VXN8, Q8IXV9, Q9Y578, Q9Y579 | Gene names | EPB41, E41P | |||
|
Domain Architecture |

|
|||||
| Description | Protein 4.1 (Band 4.1) (P4.1) (EPB4.1) (4.1R). | |||||
|
GPC3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.012445 (rank : 110) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P51654 | Gene names | GPC3, OCI5 | |||
|
Domain Architecture |

|
|||||
| Description | Glypican-3 precursor (Intestinal protein OCI-5) (GTR2-2) (MXR7). | |||||
|
IF37_MOUSE
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.028378 (rank : 88) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O70194 | Gene names | Eif3s7 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 7 (eIF-3 zeta) (eIF3 p66) (eIF3d). | |||||
|
ITSN1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.040043 (rank : 74) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
KCC2G_HUMAN
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.009757 (rank : 112) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 841 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13555, O15378, Q13556, Q5SQZ4 | Gene names | CAMK2G | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium/calmodulin-dependent protein kinase type II gamma chain (EC 2.7.11.17) (CaM-kinase II gamma chain) (CaM kinase II gamma subunit) (CaMK-II subunit gamma). | |||||
|
KCC2G_MOUSE
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.009735 (rank : 113) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 838 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q923T9, Q3U3H3, Q8VED3 | Gene names | Camk2g | |||
|
Domain Architecture |
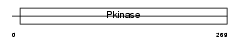
|
|||||
| Description | Calcium/calmodulin-dependent protein kinase type II gamma chain (EC 2.7.11.17) (CaM-kinase II gamma chain) (CaM kinase II gamma subunit) (CaMK-II subunit gamma). | |||||
|
LIPA2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.049897 (rank : 63) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1282 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8BSS9, Q8BN73 | Gene names | Ppfia2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
LNP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.020968 (rank : 102) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7TQ95, Q69ZC5, Q6PAQ1, Q6PEN8 | Gene names | Lnp, Kiaa1715, Uln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein lunapark (Protein ulnaless). | |||||
|
LRC45_MOUSE
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.050724 (rank : 57) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8CIM1, Q3U5Z2 | Gene names | Lrrc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
NOG1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.076806 (rank : 14) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9BZE4, O95446, Q9NVJ8 | Gene names | GTPBP4, CRFG, NOG1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar GTP-binding protein 1 (Chronic renal failure gene protein) (GTP-binding protein NGB). | |||||
|
NOG1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.080636 (rank : 11) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99ME9, Q99K16, Q99P78, Q9CT02 | Gene names | Gtpbp4, Crfg, Nog1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar GTP-binding protein 1 (Chronic renal failure gene protein) (GTP-binding protein NGB). | |||||
|
RBNS5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.038799 (rank : 77) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80Y56, Q8K0L6, Q9CTW0 | Gene names | Zfyve20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rabenosyn-5 (FYVE finger-containing Rab5 effector protein rabenosyn-5) (Zinc finger FYVE domain-containing protein 20). | |||||
|
STK10_MOUSE
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.030360 (rank : 85) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
BRD3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.021479 (rank : 101) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8K2F0, Q8C665, Q8CAX7, Q9JI25 | Gene names | Brd3, Fsrg2 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 3 (Bromodomain-containing FSH-like protein FSRG2). | |||||
|
DMP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.022841 (rank : 99) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
DRG1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.078754 (rank : 12) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y295, Q6FGP8, Q8WW69, Q9UGF2 | Gene names | DRG1 | |||
|
Domain Architecture |
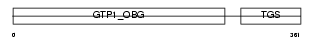
|
|||||
| Description | Developmentally-regulated GTP-binding protein 1 (DRG 1). | |||||
|
DRG1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.078719 (rank : 13) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P32233 | Gene names | Drg1, Drg, Nedd-3, Nedd3 | |||
|
Domain Architecture |
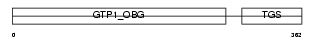
|
|||||
| Description | Developmentally-regulated GTP-binding protein 1 (DRG 1) (Protein NEDD3) (Neural precursor cell expressed developmentally down-regulated protein 3). | |||||
|
GSCR2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.025265 (rank : 92) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NZM5, Q9BTC6, Q9HAX6, Q9NPP1, Q9NPR4, Q9UFI2 | Gene names | GLTSCR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 2 protein (p60). | |||||
|
MYT1L_MOUSE
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.025295 (rank : 91) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P97500, O08996, Q8C643, Q8C7L4, Q8CHB4 | Gene names | Myt1l, Kiaa1106, Nzf1, Png1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (Postmitotic neural gene 1 protein) (Zinc finger protein Png-1) (Neural zinc finger factor 1) (NZF-1). | |||||
|
MYT1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.024040 (rank : 97) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8CFC2, O08995, Q8CFH1 | Gene names | Myt1, Kiaa0835, Nzf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1 (MyT1) (Neural zinc finger factor 2) (NZF-2). | |||||
|
PTMA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.037892 (rank : 79) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
AP3B1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.009197 (rank : 114) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Z1T1, Q91YR4 | Gene names | Ap3b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
BAZ1B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.028220 (rank : 89) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
CENPB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.013349 (rank : 108) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P07199 | Gene names | CENPB | |||
|
Domain Architecture |

|
|||||
| Description | Major centromere autoantigen B (Centromere protein B) (CENP-B). | |||||
|
ERAL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.096547 (rank : 7) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O75616, O75617, Q8WUY4, Q96LE2, Q96TC0 | Gene names | ERAL1, HERA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTP-binding protein era homolog (hERA) (ERA-W) (Conserved ERA-like GTPase) (CEGA). | |||||
|
ERAL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.091915 (rank : 8) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9CZU4, Q6NV78, Q925U1 | Gene names | Eral1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTP-binding protein era homolog (ERA-W) (Conserved ERA-like GTPase) (CEGA). | |||||
|
IF37_HUMAN
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.025158 (rank : 93) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15371, Q5M9Q6 | Gene names | EIF3S7 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 7 (eIF-3 zeta) (eIF3 p66) (eIF3d). | |||||
|
ITSN1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.038905 (rank : 76) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.040285 (rank : 73) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
MYO5C_HUMAN
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.043554 (rank : 72) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1206 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9NQX4 | Gene names | MYO5C | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5C (Myosin Vc). | |||||
|
NIN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.047757 (rank : 69) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
P52K_HUMAN
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.014909 (rank : 107) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43422, Q8WTW1, Q9Y3Z4 | Gene names | PRKRIR, DAP4, P52RIPK, THAP0 | |||
|
Domain Architecture |

|
|||||
| Description | 52 kDa repressor of the inhibitor of the protein kinase (p58IPK- interacting protein) (58 kDa interferon-induced protein kinase- interacting protein) (P52rIPK) (Death-associated protein 4) (THAP domain-containing protein 0). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.016932 (rank : 106) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.022090 (rank : 100) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
RPA1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.011641 (rank : 111) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35134, Q7TSA9 | Gene names | Polr1a, Rpa1, Rpo1-4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase I largest subunit (EC 2.7.7.6) (RNA polymerase I 194 kDa subunit) (RPA194). | |||||
|
SM1L2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.050862 (rank : 55) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q920F6 | Gene names | Smc1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 1-like 2 protein (SMC1beta protein). | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.050397 (rank : 59) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
CCD11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.050501 (rank : 58) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q96M91 | Gene names | CCDC11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 11. | |||||
|
CK5P2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.052729 (rank : 46) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
INCE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.055663 (rank : 38) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
INCE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.053195 (rank : 45) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9WU62, Q7TN28, Q8BGN4, Q8CGI4 | Gene names | Incenp | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
INVO_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.050919 (rank : 54) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P07476 | Gene names | IVL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Involucrin. | |||||
|
KTN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.051871 (rank : 49) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1324 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q61595, Q8BG49, Q8BHF4, Q8BHM8, Q8C9Y5, Q8CG51, Q8CG52, Q8CG53, Q8CG54, Q8CG55, Q8CG56, Q8CG57, Q8CG58, Q8CG59, Q8CG60, Q8CG61, Q8CG62, Q8CG63 | Gene names | Ktn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin. | |||||
|
NOP14_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.058673 (rank : 31) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P78316, Q7LGI5, Q7Z6K0, Q8TBR6 | Gene names | NOP14, C4orf9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable nucleolar complex protein 14. | |||||
|
RBCC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.050049 (rank : 62) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1048 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9ESK9, Q3TXX2, Q61384, Q8BRY9, Q9JK14 | Gene names | Rb1cc1, Cc1, Kiaa0203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1 (Coiled-coil-forming protein 1) (LaXp180 protein). | |||||
|
RBM25_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.052180 (rank : 48) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P49756, Q9UEQ5, Q9UIE9 | Gene names | RBM25, RNPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable RNA-binding protein 25 (RNA-binding motif protein 25) (RNA- binding region-containing protein 7) (Protein S164). | |||||
|
SMC1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.055281 (rank : 40) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
SMC1A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.057685 (rank : 34) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
SYCP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.052281 (rank : 47) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q15431, O14963 | Gene names | SYCP1, SCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
TPR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.050285 (rank : 60) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
TRHY_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.055470 (rank : 39) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
XE7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.053594 (rank : 43) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 738 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q02040, Q02832 | Gene names | CXYorf3, DXYS155E, XE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-lymphocyte antigen precursor (B-lymphocyte surface antigen) (721P) (Protein XE7). | |||||
|
GNL3_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | Q8CI11, Q811R9, Q811S8, Q8BK21 | Gene names | Gnl3, Ns | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein-like 3 (Nucleolar GTP-binding protein 3) (Nucleostemin). | |||||
|
GNL3_HUMAN
|
||||||
| NC score | 0.959770 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9BVP2, Q96SV6, Q96SV7, Q9UJY0 | Gene names | GNL3, E2IG3, NS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein-like 3 (Nucleolar GTP-binding protein 3) (Nucleostemin) (E2-induced gene 3-protein) (Novel nucleolar protein 47) (NNP47). | |||||
|
NOG2_MOUSE
|
||||||
| NC score | 0.770836 (rank : 3) | θ value | 1.16164e-39 (rank : 3) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99LH1, Q8BIF8 | Gene names | Gnl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar GTP-binding protein 2. | |||||
|
NOG2_HUMAN
|
||||||
| NC score | 0.759727 (rank : 4) | θ value | 1.51715e-39 (rank : 4) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13823, Q9BWN7 | Gene names | GNL2, NGP1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar GTP-binding protein 2 (Autoantigen NGP-1). | |||||
|
GNL1_HUMAN
|
||||||
| NC score | 0.495140 (rank : 5) | θ value | 3.64472e-09 (rank : 5) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P36915 | Gene names | GNL1, HSR1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-binding protein-like 1 (GTP-binding protein HSR1). | |||||
|
GNL1_MOUSE
|
||||||
| NC score | 0.490405 (rank : 6) | θ value | 3.64472e-09 (rank : 6) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P36916 | Gene names | Gnl1, Gna-rs1, Mmr1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-binding protein-like 1 (GTP-binding protein MMR1). | |||||
|
ERAL_HUMAN
|
||||||
| NC score | 0.096547 (rank : 7) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O75616, O75617, Q8WUY4, Q96LE2, Q96TC0 | Gene names | ERAL1, HERA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTP-binding protein era homolog (hERA) (ERA-W) (Conserved ERA-like GTPase) (CEGA). | |||||
|
ERAL_MOUSE
|
||||||
| NC score | 0.091915 (rank : 8) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9CZU4, Q6NV78, Q925U1 | Gene names | Eral1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTP-binding protein era homolog (ERA-W) (Conserved ERA-like GTPase) (CEGA). | |||||
|
TCRG1_HUMAN
|
||||||
| NC score | 0.089580 (rank : 9) | θ value | 0.21417 (rank : 17) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
TCRG1_MOUSE
|
||||||
| NC score | 0.088396 (rank : 10) | θ value | 0.21417 (rank : 18) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
NOG1_MOUSE
|
||||||
| NC score | 0.080636 (rank : 11) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99ME9, Q99K16, Q99P78, Q9CT02 | Gene names | Gtpbp4, Crfg, Nog1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar GTP-binding protein 1 (Chronic renal failure gene protein) (GTP-binding protein NGB). | |||||
|
DRG1_HUMAN
|
||||||
| NC score | 0.078754 (rank : 12) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y295, Q6FGP8, Q8WW69, Q9UGF2 | Gene names | DRG1 | |||
|
Domain Architecture |

|
|||||
| Description | Developmentally-regulated GTP-binding protein 1 (DRG 1). | |||||
|
DRG1_MOUSE
|
||||||
| NC score | 0.078719 (rank : 13) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P32233 | Gene names | Drg1, Drg, Nedd-3, Nedd3 | |||
|
Domain Architecture |

|
|||||
| Description | Developmentally-regulated GTP-binding protein 1 (DRG 1) (Protein NEDD3) (Neural precursor cell expressed developmentally down-regulated protein 3). | |||||
|
NOG1_HUMAN
|
||||||
| NC score | 0.076806 (rank : 14) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9BZE4, O95446, Q9NVJ8 | Gene names | GTPBP4, CRFG, NOG1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar GTP-binding protein 1 (Chronic renal failure gene protein) (GTP-binding protein NGB). | |||||
|
CCAR1_HUMAN
|
||||||
| NC score | 0.076097 (rank : 15) | θ value | 0.279714 (rank : 19) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8IX12, Q32NE3, Q5VUP6, Q6PIZ0, Q6X935, Q9H8N4, Q9NVA7, Q9NVQ0, Q9NWM6 | Gene names | CCAR1, CARP1, DIS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle and apoptosis regulator protein 1 (Cell cycle and apoptosis regulatory protein 1) (CARP-1) (Death inducer with SAP domain). | |||||
|
CA065_HUMAN
|
||||||
| NC score | 0.073673 (rank : 16) | θ value | 0.47712 (rank : 24) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 469 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8N715, Q8N746, Q8NA93 | Gene names | C1orf65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf65. | |||||
|
CCD96_MOUSE
|
||||||
| NC score | 0.072858 (rank : 17) | θ value | 1.06291 (rank : 27) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9CR92 | Gene names | Ccdc96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 96. | |||||
|
CCAR1_MOUSE
|
||||||
| NC score | 0.071985 (rank : 18) | θ value | 1.81305 (rank : 34) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8CH18, Q6AXC9, Q6PAR2, Q80XE4, Q8BJY0, Q8BVN2, Q8CGG1, Q9CSR5 | Gene names | Ccar1, Carp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle and apoptosis regulator protein 1 (Cell cycle and apoptosis regulatory protein 1) (CARP-1). | |||||
|
K0555_HUMAN
|
||||||
| NC score | 0.071726 (rank : 19) | θ value | 0.0736092 (rank : 10) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 880 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q96AA8, O60302 | Gene names | KIAA0555 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0555. | |||||
|
OPTN_MOUSE
|
||||||
| NC score | 0.069770 (rank : 20) | θ value | 0.0330416 (rank : 7) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1120 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8K3K8 | Gene names | Optn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin. | |||||
|
INVO_MOUSE
|
||||||
| NC score | 0.069588 (rank : 21) | θ value | 0.279714 (rank : 20) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P48997 | Gene names | Ivl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Involucrin. | |||||
|
CT2NL_MOUSE
|
||||||
| NC score | 0.067903 (rank : 22) | θ value | 1.38821 (rank : 31) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 902 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q99LJ0, Q6ZPR0, Q8BSV1 | Gene names | Cttnbp2nl, Kiaa1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
OPTN_HUMAN
|
||||||
| NC score | 0.067071 (rank : 23) | θ value | 1.06291 (rank : 29) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 827 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q96CV9, Q5T672, Q5T673, Q5T674, Q5T675, Q7LDL9, Q8N562, Q9UET9, Q9UEV4, Q9Y218 | Gene names | OPTN, FIP2, GLC1E, NRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin (Optic neuropathy-inducing protein) (E3-14.7K-interacting protein) (FIP-2) (Huntingtin-interacting protein HYPL) (NEMO-related protein) (Transcription factor IIIA-interacting protein) (TFIIIA- IntP). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.065305 (rank : 24) | θ value | 0.125558 (rank : 12) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
CT2NL_HUMAN
|
||||||
| NC score | 0.063436 (rank : 25) | θ value | 1.81305 (rank : 35) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9P2B4, Q96B40 | Gene names | CTTNBP2NL, KIAA1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.062623 (rank : 26) | θ value | 0.365318 (rank : 22) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
MIER1_HUMAN
|
||||||
| NC score | 0.061462 (rank : 27) | θ value | 0.365318 (rank : 23) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N108, Q5T104, Q6AHY8, Q86TB4, Q8N156, Q8N161, Q8NC37, Q8NES4, Q8NES5, Q8NES6, Q8WWG2, Q9HCG2 | Gene names | MIER1, KIAA1610 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm induction early response protein 1 (Mi-er1) (hmi-er1) (Early response 1) (Er1). | |||||
|
SYCP1_MOUSE
|
||||||
| NC score | 0.061351 (rank : 28) | θ value | 0.125558 (rank : 15) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
SM1L2_HUMAN
|
||||||
| NC score | 0.060135 (rank : 29) | θ value | 1.06291 (rank : 30) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1019 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8NDV3, Q5TIC3, Q9Y3G5 | Gene names | SMC1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 2 protein (SMC1beta protein). | |||||
|
SDPR_MOUSE
|
||||||
| NC score | 0.059565 (rank : 30) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q63918, Q3V1P6, Q78EC3, Q8CBT4 | Gene names | Sdpr, Sdr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein). | |||||
|
NOP14_HUMAN
|
||||||
| NC score | 0.058673 (rank : 31) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P78316, Q7LGI5, Q7Z6K0, Q8TBR6 | Gene names | NOP14, C4orf9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable nucleolar complex protein 14. | |||||
|
ZN318_MOUSE
|
||||||
| NC score | 0.058496 (rank : 32) | θ value | 0.62314 (rank : 25) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q99PP2, Q3TZL5, Q9JJ01 | Gene names | Znf318, Tzf, Zfp318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Testicular zinc finger protein). | |||||
|
CE152_HUMAN
|
||||||
| NC score | 0.057931 (rank : 33) | θ value | 2.36792 (rank : 38) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O94986, Q6NTA0 | Gene names | CEP152, KIAA0912 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 152 kDa (Cep152 protein). | |||||
|
SMC1A_MOUSE
|
||||||
| NC score | 0.057685 (rank : 34) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
PTD4_MOUSE
|
||||||
| NC score | 0.057407 (rank : 35) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CZ30, Q9CWE2, Q9CX91, Q9CYC9, Q9CZ56 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Putative GTP-binding protein PTD004 homolog. | |||||
|
PTD4_HUMAN
|
||||||
| NC score | 0.057379 (rank : 36) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NTK5, Q8NCI8, Q96CU1, Q96SV2, Q9P1D3, Q9UNY9, Q9Y6G4 | Gene names | ames=PTD004, PRO2455 | |||
|
Domain Architecture |

|
|||||
| Description | Putative GTP-binding protein PTD004. | |||||
|
MIER1_MOUSE
|
||||||
| NC score | 0.057134 (rank : 37) | θ value | 0.813845 (rank : 26) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5UAK0, Q5UAK1, Q5UAK2, Q5UAK3, Q6ZPL6, Q9D402 | Gene names | Mier1, Kiaa1610 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm induction early response protein 1 (Mi-er1). | |||||
|
INCE_HUMAN
|
||||||
| NC score | 0.055663 (rank : 38) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
TRHY_HUMAN
|
||||||
| NC score | 0.055470 (rank : 39) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
SMC1A_HUMAN
|
||||||
| NC score | 0.055281 (rank : 40) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
GOGA3_MOUSE
|
||||||
| NC score | 0.054566 (rank : 41) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
PTMA_HUMAN
|
||||||
| NC score | 0.053687 (rank : 42) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
XE7_HUMAN
|
||||||
| NC score | 0.053594 (rank : 43) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 738 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q02040, Q02832 | Gene names | CXYorf3, DXYS155E, XE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-lymphocyte antigen precursor (B-lymphocyte surface antigen) (721P) (Protein XE7). | |||||
|
GOGA5_MOUSE
|
||||||
| NC score | 0.053528 (rank : 44) | θ value | 1.06291 (rank : 28) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1434 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9QYE6, O88317, Q3TGE7, Q3U6S5, Q3UUF9 | Gene names | Golga5, Retii | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (Sumiko protein) (Ret-II protein). | |||||
|
INCE_MOUSE
|
||||||
| NC score | 0.053195 (rank : 45) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9WU62, Q7TN28, Q8BGN4, Q8CGI4 | Gene names | Incenp | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
CK5P2_HUMAN
|
||||||
| NC score | 0.052729 (rank : 46) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
SYCP1_HUMAN
|
||||||
| NC score | 0.052281 (rank : 47) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q15431, O14963 | Gene names | SYCP1, SCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
RBM25_HUMAN
|
||||||
| NC score | 0.052180 (rank : 48) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P49756, Q9UEQ5, Q9UIE9 | Gene names | RBM25, RNPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable RNA-binding protein 25 (RNA-binding motif protein 25) (RNA- binding region-containing protein 7) (Protein S164). | |||||
|
KTN1_MOUSE
|
||||||
| NC score | 0.051871 (rank : 49) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1324 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q61595, Q8BG49, Q8BHF4, Q8BHM8, Q8C9Y5, Q8CG51, Q8CG52, Q8CG53, Q8CG54, Q8CG55, Q8CG56, Q8CG57, Q8CG58, Q8CG59, Q8CG60, Q8CG61, Q8CG62, Q8CG63 | Gene names | Ktn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin. | |||||
|
MYH11_HUMAN
|
||||||
| NC score | 0.051843 (rank : 50) | θ value | 1.81305 (rank : 36) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MYH9_MOUSE
|
||||||
| NC score | 0.051688 (rank : 51) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MYH9_HUMAN
|
||||||
| NC score | 0.051628 (rank : 52) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MYH11_MOUSE
|
||||||
| NC score | 0.051004 (rank : 53) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
INVO_HUMAN
|
||||||
| NC score | 0.050919 (rank : 54) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P07476 | Gene names | IVL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Involucrin. | |||||
|
SM1L2_MOUSE
|
||||||
| NC score | 0.050862 (rank : 55) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q920F6 | Gene names | Smc1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 1-like 2 protein (SMC1beta protein). | |||||
|
MIRO1_HUMAN
|
||||||
| NC score | 0.050794 (rank : 56) | θ value | 0.125558 (rank : 13) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IXI2, Q6NUR3, Q6P9F8, Q6PJG1, Q6YMW8, Q86UB0, Q8IW28, Q8IXJ7, Q9H067, Q9H9N8, Q9NUZ2 | Gene names | RHOT1, ARHT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial Rho GTPase 1 (EC 3.6.5.-) (MIRO-1) (hMiro-1) (Ras homolog gene family member T1) (Rac-GTP-binding protein-like protein). | |||||
|
LRC45_MOUSE
|
||||||
| NC score | 0.050724 (rank : 57) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8CIM1, Q3U5Z2 | Gene names | Lrrc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
CCD11_HUMAN
|
||||||
| NC score | 0.050501 (rank : 58) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q96M91 | Gene names | CCDC11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 11. | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | 0.050397 (rank : 59) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.050285 (rank : 60) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
MIRO1_MOUSE
|
||||||
| NC score | 0.050071 (rank : 61) | θ value | 0.125558 (rank : 14) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BG51, Q3TJB7, Q3UGU4, Q5SYC3, Q5SYC6, Q8BLW3, Q8BMH1, Q922N0, Q9D2R1, Q9JKB9 | Gene names | Rhot1, Arht1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial Rho GTPase 1 (EC 3.6.5.-) (MIRO-1) (Ras homolog gene family member T1). | |||||
|
RBCC1_MOUSE
|
||||||
| NC score | 0.050049 (rank : 62) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1048 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9ESK9, Q3TXX2, Q61384, Q8BRY9, Q9JK14 | Gene names | Rb1cc1, Cc1, Kiaa0203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1 (Coiled-coil-forming protein 1) (LaXp180 protein). | |||||
|
LIPA2_MOUSE
|
||||||
| NC score | 0.049897 (rank : 63) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1282 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8BSS9, Q8BN73 | Gene names | Ppfia2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
MY18A_HUMAN
|
||||||
| NC score | 0.049843 (rank : 64) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
MYO5A_MOUSE
|
||||||
| NC score | 0.049217 (rank : 65) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1117 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q99104 | Gene names | Myo5a, Dilute | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5A (Myosin Va) (Dilute myosin heavy chain, non-muscle). | |||||
|
MYH10_HUMAN
|
||||||
| NC score | 0.048842 (rank : 66) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH10_MOUSE
|
||||||
| NC score | 0.048418 (rank : 67) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH14_HUMAN
|
||||||
| NC score | 0.048108 (rank : 68) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 995 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q7Z406, Q5CZ75, Q6XYE4, Q76B62, Q8WV23, Q96I22, Q9BT27, Q9BW35, Q9H882 | Gene names | MYH14, KIAA2034 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
NIN_MOUSE
|
||||||
| NC score | 0.047757 (rank : 69) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
PQBP1_MOUSE
|
||||||
| NC score | 0.045535 (rank : 70) | θ value | 1.38821 (rank : 32) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91VJ5, Q80WW2, Q9ER43, Q9QYY2 | Gene names | Pqbp1, Npw38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyglutamine-binding protein 1 (Polyglutamine tract-binding protein 1) (PQBP-1) (38 kDa nuclear protein containing a WW domain) (Npw38). | |||||
|
RIMB1_MOUSE
|
||||||
| NC score | 0.044237 (rank : 71) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
MYO5C_HUMAN
|
||||||
| NC score | 0.043554 (rank : 72) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1206 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9NQX4 | Gene names | MYO5C | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5C (Myosin Vc). | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.040285 (rank : 73) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
ITSN1_HUMAN
|
||||||
| NC score | 0.040043 (rank : 74) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
TAOK1_MOUSE
|
||||||
| NC score | 0.038933 (rank : 75) | θ value | 0.0563607 (rank : 9) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1720 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q5F2E8, Q69ZL2, Q8JZX2, Q91VG7 | Gene names | Taok1, Kiaa1361 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO1 (EC 2.7.11.1) (Thousand and one amino acid protein 1). | |||||
|
ITSN1_MOUSE
|
||||||
| NC score | 0.038905 (rank : 76) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
RBNS5_MOUSE
|
||||||
| NC score | 0.038799 (rank : 77) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80Y56, Q8K0L6, Q9CTW0 | Gene names | Zfyve20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rabenosyn-5 (FYVE finger-containing Rab5 effector protein rabenosyn-5) (Zinc finger FYVE domain-containing protein 20). | |||||
|
TAOK1_HUMAN
|
||||||
| NC score | 0.038711 (rank : 78) | θ value | 0.0563607 (rank : 8) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1734 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q7L7X3, Q96L75, Q9H2K7, Q9H7S5, Q9P2I6 | Gene names | TAOK1, KIAA1361, MAP3K16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO1 (EC 2.7.11.1) (Thousand and one amino acid protein 1) (STE20-like kinase PSK2) (Kinase from chicken homolog B) (hKFC-B). | |||||
|
PTMA_MOUSE
|
||||||
| NC score | 0.037892 (rank : 79) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
RBBP6_MOUSE
|
||||||
| NC score | 0.037223 (rank : 80) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
ROCK2_HUMAN
|
||||||
| NC score | 0.035593 (rank : 81) | θ value | 0.0961366 (rank : 11) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 2073 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O75116, Q9UQN5 | Gene names | ROCK2, KIAA0619 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2) (Rho kinase 2). | |||||
|
ROCK2_MOUSE
|
||||||
| NC score | 0.034831 (rank : 82) | θ value | 0.21417 (rank : 16) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 2072 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P70336, Q8BR64, Q8CBR0 | Gene names | Rock2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2). | |||||
|
TRI44_HUMAN
|
||||||
| NC score | 0.030861 (rank : 83) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96DX7, Q96QY2, Q9UGK0 | Gene names | TRIM44, DIPB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB). | |||||
|
NFM_MOUSE
|
||||||
| NC score | 0.030424 (rank : 84) | θ value | 1.81305 (rank : 37) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
STK10_MOUSE
|
||||||
| NC score | 0.030360 (rank : 85) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
CJ006_MOUSE
|
||||||
| NC score | 0.029040 (rank : 86) | θ value | 2.36792 (rank : 39) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6P9P0, Q8C041, Q8C0J5 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf6 homolog. | |||||
|
TAOK2_MOUSE
|
||||||
| NC score | 0.028510 (rank : 87) | θ value | 1.38821 (rank : 33) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1373 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6ZQ29, Q7TSS8 | Gene names | Taok2, Kiaa0881 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO2 (EC 2.7.11.1) (Thousand and one amino acid protein 2). | |||||
|
IF37_MOUSE
|
||||||
| NC score | 0.028378 (rank : 88) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O70194 | Gene names | Eif3s7 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 7 (eIF-3 zeta) (eIF3 p66) (eIF3d). | |||||
|
BAZ1B_HUMAN
|
||||||
| NC score | 0.028220 (rank : 89) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
SH3K1_MOUSE
|
||||||
| NC score | 0.027652 (rank : 90) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8R550, Q8CAL8, Q8CEF6, Q8R545, Q8R546, Q8R547, Q8R548, Q8R549, Q8R551, Q9CTQ9, Q9DC14, Q9JKC3 | Gene names | Sh3kbp1, Ruk, Seta | |||
|
Domain Architecture |

|
|||||
| Description | SH3-domain kinase-binding protein 1 (SH3-containing, expressed in tumorigenic astrocytes) (Regulator of ubiquitous kinase) (Ruk). | |||||
|
MYT1L_MOUSE
|
||||||
| NC score | 0.025295 (rank : 91) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P97500, O08996, Q8C643, Q8C7L4, Q8CHB4 | Gene names | Myt1l, Kiaa1106, Nzf1, Png1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (Postmitotic neural gene 1 protein) (Zinc finger protein Png-1) (Neural zinc finger factor 1) (NZF-1). | |||||
|
GSCR2_HUMAN
|
||||||
| NC score | 0.025265 (rank : 92) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NZM5, Q9BTC6, Q9HAX6, Q9NPP1, Q9NPR4, Q9UFI2 | Gene names | GLTSCR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 2 protein (p60). | |||||
|
IF37_HUMAN
|
||||||
| NC score | 0.025158 (rank : 93) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15371, Q5M9Q6 | Gene names | EIF3S7 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 7 (eIF-3 zeta) (eIF3 p66) (eIF3d). | |||||
|
TRI44_MOUSE
|
||||||
| NC score | 0.024844 (rank : 94) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9QXA7, Q80SV9, Q9JHR8 | Gene names | Trim44, Dipb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB) (Protein Mc7). | |||||
|
CKAP2_MOUSE
|
||||||
| NC score | 0.024831 (rank : 95) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q3V1H1, Q66LN2, Q8BSF0 | Gene names | Ckap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 2. | |||||
|
UBF1_HUMAN
|
||||||
| NC score | 0.024704 (rank : 96) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
MYT1_MOUSE
|
||||||
| NC score | 0.024040 (rank : 97) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8CFC2, O08995, Q8CFH1 | Gene names | Myt1, Kiaa0835, Nzf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1 (MyT1) (Neural zinc finger factor 2) (NZF-2). | |||||
|
SMRC2_MOUSE
|
||||||
| NC score | 0.023449 (rank : 98) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6PDG5, Q6P6P3 | Gene names | Smarcc2, Baf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
DMP1_HUMAN
|
||||||
| NC score | 0.022841 (rank : 99) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.022090 (rank : 100) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
BRD3_MOUSE
|
||||||
| NC score | 0.021479 (rank : 101) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8K2F0, Q8C665, Q8CAX7, Q9JI25 | Gene names | Brd3, Fsrg2 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 3 (Bromodomain-containing FSH-like protein FSRG2). | |||||
|
LNP_MOUSE
|
||||||
| NC score | 0.020968 (rank : 102) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7TQ95, Q69ZC5, Q6PAQ1, Q6PEN8 | Gene names | Lnp, Kiaa1715, Uln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein lunapark (Protein ulnaless). | |||||
|
TMC2_MOUSE
|
||||||
| NC score | 0.019509 (rank : 103) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8R4P4 | Gene names | Tmc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane cochlear-expressed protein 2. | |||||
|
AN32B_MOUSE
|
||||||
| NC score | 0.018901 (rank : 104) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9EST5 | Gene names | Anp32b, Pal31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member B (Proliferation-related acidic leucine-rich protein PAL31). | |||||
|
AN32B_HUMAN
|
||||||
| NC score | 0.018427 (rank : 105) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92688, O00655, P78458, P78459 | Gene names | ANP32B, APRIL, PHAPI2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member B (PHAPI2 protein) (Silver-stainable protein SSP29) (Acidic protein rich in leucines). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.016932 (rank : 106) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
P52K_HUMAN
|
||||||
| NC score | 0.014909 (rank : 107) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43422, Q8WTW1, Q9Y3Z4 | Gene names | PRKRIR, DAP4, P52RIPK, THAP0 | |||
|
Domain Architecture |

|
|||||
| Description | 52 kDa repressor of the inhibitor of the protein kinase (p58IPK- interacting protein) (58 kDa interferon-induced protein kinase- interacting protein) (P52rIPK) (Death-associated protein 4) (THAP domain-containing protein 0). | |||||
|
CENPB_HUMAN
|
||||||
| NC score | 0.013349 (rank : 108) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P07199 | Gene names | CENPB | |||
|
Domain Architecture |

|
|||||
| Description | Major centromere autoantigen B (Centromere protein B) (CENP-B). | |||||
|
CDKL2_MOUSE
|
||||||
| NC score | 0.013320 (rank : 109) | θ value | 0.365318 (rank : 21) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 854 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9QUK0, Q3UL60, Q3V3X7, Q9QYI1, Q9QYI2 | Gene names | Cdkl2, Kkiamre, Kkm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-dependent kinase-like 2 (EC 2.7.11.22) (Serine/threonine- protein kinase KKIAMRE). | |||||
|
GPC3_HUMAN
|
||||||
| NC score | 0.012445 (rank : 110) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P51654 | Gene names | GPC3, OCI5 | |||
|
Domain Architecture |

|
|||||
| Description | Glypican-3 precursor (Intestinal protein OCI-5) (GTR2-2) (MXR7). | |||||
|
RPA1_MOUSE
|
||||||
| NC score | 0.011641 (rank : 111) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35134, Q7TSA9 | Gene names | Polr1a, Rpa1, Rpo1-4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase I largest subunit (EC 2.7.7.6) (RNA polymerase I 194 kDa subunit) (RPA194). | |||||
|
KCC2G_HUMAN
|
||||||
| NC score | 0.009757 (rank : 112) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 841 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13555, O15378, Q13556, Q5SQZ4 | Gene names | CAMK2G | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium/calmodulin-dependent protein kinase type II gamma chain (EC 2.7.11.17) (CaM-kinase II gamma chain) (CaM kinase II gamma subunit) (CaMK-II subunit gamma). | |||||
|
KCC2G_MOUSE
|
||||||
| NC score | 0.009735 (rank : 113) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 838 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q923T9, Q3U3H3, Q8VED3 | Gene names | Camk2g | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent protein kinase type II gamma chain (EC 2.7.11.17) (CaM-kinase II gamma chain) (CaM kinase II gamma subunit) (CaMK-II subunit gamma). | |||||
|
AP3B1_MOUSE
|
||||||
| NC score | 0.009197 (rank : 114) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Z1T1, Q91YR4 | Gene names | Ap3b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
41_HUMAN
|
||||||
| NC score | 0.008413 (rank : 115) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P11171, P11176, Q14245, Q5TB35, Q5VXN8, Q8IXV9, Q9Y578, Q9Y579 | Gene names | EPB41, E41P | |||
|
Domain Architecture |

|
|||||
| Description | Protein 4.1 (Band 4.1) (P4.1) (EPB4.1) (4.1R). | |||||