Please be patient as the page loads
|
MIER1_HUMAN
|
||||||
| SwissProt Accessions | Q8N108, Q5T104, Q6AHY8, Q86TB4, Q8N156, Q8N161, Q8NC37, Q8NES4, Q8NES5, Q8NES6, Q8WWG2, Q9HCG2 | Gene names | MIER1, KIAA1610 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm induction early response protein 1 (Mi-er1) (hmi-er1) (Early response 1) (Er1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MIER1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q8N108, Q5T104, Q6AHY8, Q86TB4, Q8N156, Q8N161, Q8NC37, Q8NES4, Q8NES5, Q8NES6, Q8WWG2, Q9HCG2 | Gene names | MIER1, KIAA1610 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm induction early response protein 1 (Mi-er1) (hmi-er1) (Early response 1) (Er1). | |||||
|
MIER1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.991988 (rank : 2) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q5UAK0, Q5UAK1, Q5UAK2, Q5UAK3, Q6ZPL6, Q9D402 | Gene names | Mier1, Kiaa1610 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm induction early response protein 1 (Mi-er1). | |||||
|
MTA2_MOUSE
|
||||||
| θ value | 3.89403e-11 (rank : 3) | NC score | 0.524813 (rank : 3) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9R190, Q3TVT8 | Gene names | Mta2, Mta1l1 | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA2 (Metastasis-associated 1-like 1). | |||||
|
MTA2_HUMAN
|
||||||
| θ value | 1.133e-10 (rank : 4) | NC score | 0.522788 (rank : 4) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O94776, Q9UQB5 | Gene names | MTA2, MTA1L1, PID | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA2 (Metastasis-associated 1-like 1) (MTA1-L1 protein) (p53 target protein in deacetylase complex). | |||||
|
MTA3_HUMAN
|
||||||
| θ value | 1.63604e-09 (rank : 5) | NC score | 0.516237 (rank : 5) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BTC8, Q9NSP2, Q9ULF4 | Gene names | MTA3, KIAA1266 | |||
|
Domain Architecture |
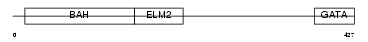
|
|||||
| Description | Metastasis-associated protein MTA3. | |||||
|
RERE_MOUSE
|
||||||
| θ value | 1.38499e-08 (rank : 6) | NC score | 0.384491 (rank : 16) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
RERE_HUMAN
|
||||||
| θ value | 3.08544e-08 (rank : 7) | NC score | 0.383494 (rank : 17) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
MTA3_MOUSE
|
||||||
| θ value | 8.97725e-08 (rank : 8) | NC score | 0.503304 (rank : 6) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q924K8, Q8VC18 | Gene names | Mta3 | |||
|
Domain Architecture |
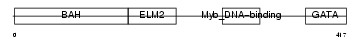
|
|||||
| Description | Metastasis-associated protein MTA3. | |||||
|
MTA1_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 9) | NC score | 0.494241 (rank : 7) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13330, Q8NFI8, Q96GI8 | Gene names | MTA1 | |||
|
Domain Architecture |
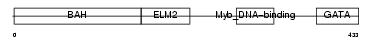
|
|||||
| Description | Metastasis-associated protein MTA1. | |||||
|
MTA1_MOUSE
|
||||||
| θ value | 1.99992e-07 (rank : 10) | NC score | 0.493141 (rank : 8) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8K4B0, Q80UI1, Q8K4D4, Q924K9 | Gene names | Mta1 | |||
|
Domain Architecture |
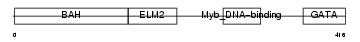
|
|||||
| Description | Metastasis-associated protein MTA1. | |||||
|
RCOR3_MOUSE
|
||||||
| θ value | 3.77169e-06 (rank : 11) | NC score | 0.451061 (rank : 11) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6PGA0 | Gene names | Rcor3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 3. | |||||
|
ZN541_HUMAN
|
||||||
| θ value | 3.77169e-06 (rank : 12) | NC score | 0.426187 (rank : 13) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9H0D2, Q8NDK8 | Gene names | ZNF541 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 541. | |||||
|
RCOR3_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 13) | NC score | 0.449597 (rank : 12) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9P2K3, Q5VT47, Q7L9I5, Q8N5U3, Q9NV83 | Gene names | RCOR3, KIAA1343 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 3. | |||||
|
NCOR2_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 14) | NC score | 0.297545 (rank : 20) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
RCOR1_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 15) | NC score | 0.461054 (rank : 9) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8CFE3, Q3V092, Q8BK28, Q8CHI2 | Gene names | Rcor1, D12Wsu95e, Kiaa0071 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 1 (Protein CoREST). | |||||
|
TREF1_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 16) | NC score | 0.308707 (rank : 18) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96PN7, Q7Z6T2, Q7Z6T3, Q9NQ72, Q9NQ73, Q9NUN9 | Gene names | TRERF1, RAPA, TREP132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132) (Zinc finger protein rapa). | |||||
|
RCOR1_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 17) | NC score | 0.458124 (rank : 10) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UKL0, Q15044, Q6P2I9, Q86VG5 | Gene names | RCOR1, KIAA0071, RCOR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 1 (Protein CoREST). | |||||
|
NCOR1_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 18) | NC score | 0.294956 (rank : 22) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
NCOR1_MOUSE
|
||||||
| θ value | 7.1131e-05 (rank : 19) | NC score | 0.297016 (rank : 21) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
TREF1_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 20) | NC score | 0.299552 (rank : 19) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BXJ2, Q6PCQ4, Q80X25, Q810H8, Q8BY31 | Gene names | Trerf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132). | |||||
|
NCOR2_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 21) | NC score | 0.279786 (rank : 23) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 22) | NC score | 0.115976 (rank : 26) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
RCOR2_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 23) | NC score | 0.426064 (rank : 14) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8IZ40, Q96FP3 | Gene names | RCOR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 2. | |||||
|
RCOR2_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 24) | NC score | 0.425462 (rank : 15) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8C796, Q9JMK4 | Gene names | Rcor2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 2 (M-CoREST). | |||||
|
SEC63_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 25) | NC score | 0.135127 (rank : 24) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
SEC63_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 26) | NC score | 0.128916 (rank : 25) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
TCAL5_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 27) | NC score | 0.089034 (rank : 31) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
IFIT1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 28) | NC score | 0.060140 (rank : 46) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P09914, Q96QM5 | Gene names | IFIT1, G10P1, IFI56 | |||
|
Domain Architecture |
|
|||||
| Description | Interferon-induced protein with tetratricopeptide repeats 1 (IFIT-1) (Interferon-induced 56 kDa protein) (IFI-56K). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 29) | NC score | 0.107332 (rank : 27) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
RAG2_MOUSE
|
||||||
| θ value | 0.125558 (rank : 30) | NC score | 0.106407 (rank : 28) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P21784 | Gene names | Rag2, Rag-2 | |||
|
Domain Architecture |

|
|||||
| Description | V(D)J recombination-activating protein 2 (RAG-2). | |||||
|
EP400_MOUSE
|
||||||
| θ value | 0.163984 (rank : 31) | NC score | 0.040567 (rank : 60) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
GNL3_MOUSE
|
||||||
| θ value | 0.365318 (rank : 32) | NC score | 0.061462 (rank : 45) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CI11, Q811R9, Q811S8, Q8BK21 | Gene names | Gnl3, Ns | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein-like 3 (Nucleolar GTP-binding protein 3) (Nucleostemin). | |||||
|
SMRC2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 33) | NC score | 0.073959 (rank : 39) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6PDG5, Q6P6P3 | Gene names | Smarcc2, Baf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
SNUT1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 34) | NC score | 0.092746 (rank : 30) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z315, Q8K155, Q9R1I9, Q9R270 | Gene names | Sart1, Haf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 1 (Squamous cell carcinoma antigen recognized by T-cells 1) (SART-1) (mSART-1) (Hypoxia- associated factor). | |||||
|
SMRC2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 35) | NC score | 0.080745 (rank : 34) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8TAQ2, Q92923, Q96E12, Q96GY4 | Gene names | SMARCC2, BAF170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
CHP2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 36) | NC score | 0.023472 (rank : 73) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43745 | Gene names | CHP2, HCA520 | |||
|
Domain Architecture |
|
|||||
| Description | Calcineurin B homologous protein 2 (Hepatocellular carcinoma- associated antigen 520). | |||||
|
DMP1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 37) | NC score | 0.067225 (rank : 41) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
TXLNB_MOUSE
|
||||||
| θ value | 0.813845 (rank : 38) | NC score | 0.021600 (rank : 75) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 640 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VBT1, Q3UVB8, Q8BUK2 | Gene names | Txlnb, Mdp77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77). | |||||
|
NSBP1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 39) | NC score | 0.059019 (rank : 48) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
RAG2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 40) | NC score | 0.084813 (rank : 33) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P55895, Q8TBL4 | Gene names | RAG2 | |||
|
Domain Architecture |

|
|||||
| Description | V(D)J recombination-activating protein 2 (RAG-2). | |||||
|
SNUT1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 41) | NC score | 0.085522 (rank : 32) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43290, Q53GB5 | Gene names | SART1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 1 (U4/U6.U5 tri-snRNP-associated 110 kDa protein) (Squamous cell carcinoma antigen recognized by T cells 1) (SART-1) (hSART-1) (hSnu66). | |||||
|
AKA12_HUMAN
|
||||||
| θ value | 1.38821 (rank : 42) | NC score | 0.064727 (rank : 42) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
BRD8_HUMAN
|
||||||
| θ value | 1.38821 (rank : 43) | NC score | 0.036738 (rank : 63) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H0E9, O43178, Q15355, Q59GN0, Q969M9 | Gene names | BRD8, SMAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8 (p120) (Skeletal muscle abundant protein) (Thyroid hormone receptor coactivating protein 120kDa) (TrCP120). | |||||
|
EDEM3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 44) | NC score | 0.044571 (rank : 59) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BZQ6, Q5TEZ0, Q9HCW1, Q9UFV7 | Gene names | EDEM3, C1orf22 | |||
|
Domain Architecture |

|
|||||
| Description | ER degradation-enhancing alpha-mannosidase-like 3. | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 45) | NC score | 0.055722 (rank : 51) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
UBP8_HUMAN
|
||||||
| θ value | 1.38821 (rank : 46) | NC score | 0.027581 (rank : 69) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P40818, Q7Z3U2, Q86VA0, Q8IWI7 | Gene names | USP8, KIAA0055, UBPY | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (hUBPy). | |||||
|
E41L3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 47) | NC score | 0.012532 (rank : 83) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y2J2, O95713, Q9BRP5 | Gene names | EPB41L3, DAL1, KIAA0987 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 3 (4.1B) (Differentially expressed in adenocarcinoma of the lung protein 1) (DAL-1). | |||||
|
JOSD3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.063495 (rank : 44) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H5J8, Q6I9Y6 | Gene names | JOSD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein JOSD3. | |||||
|
PPRB_MOUSE
|
||||||
| θ value | 1.81305 (rank : 49) | NC score | 0.037189 (rank : 62) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q925J9, O88323, Q3UHV0, Q6AXD5, Q8BW37, Q8BX19, Q8VDQ7, Q925K0 | Gene names | Pparbp, Trip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2). | |||||
|
SMRC1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 50) | NC score | 0.073837 (rank : 40) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P97496, Q7TS80, Q7TT29 | Gene names | Smarcc1, Baf155, Srg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155) (SWI3-related protein). | |||||
|
UBP33_HUMAN
|
||||||
| θ value | 2.36792 (rank : 51) | NC score | 0.028545 (rank : 68) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8TEY7, Q8TEY6, Q96AV6, Q9H9F0, Q9UPQ5 | Gene names | USP33, KIAA1097, VDU1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 33 (EC 3.1.2.15) (Ubiquitin thioesterase 33) (Ubiquitin-specific-processing protease 33) (Deubiquitinating enzyme 33) (VHL-interacting deubiquitinating enzyme 1). | |||||
|
ARX_HUMAN
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.012604 (rank : 81) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96QS3 | Gene names | ARX | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein ARX (Aristaless-related homeobox). | |||||
|
BRD8_MOUSE
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | 0.040385 (rank : 61) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
CENPJ_HUMAN
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.034564 (rank : 65) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9HC77, Q5T6R5, Q96KS5, Q9C067 | Gene names | CENPJ, CPAP, LAP, LIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein J (CENP-J) (Centrosomal P4.1-associated protein) (LAG-3-associated protein) (LYST-interacting protein 1). | |||||
|
KCNB2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.008213 (rank : 85) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92953, Q9BXD3 | Gene names | KCNB2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 2 (Voltage-gated potassium channel subunit Kv2.2). | |||||
|
ARX_MOUSE
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.012561 (rank : 82) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 398 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O35085, Q9QYT4 | Gene names | Arx | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein ARX (Aristaless-related homeobox). | |||||
|
P52K_HUMAN
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.019941 (rank : 77) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43422, Q8WTW1, Q9Y3Z4 | Gene names | PRKRIR, DAP4, P52RIPK, THAP0 | |||
|
Domain Architecture |

|
|||||
| Description | 52 kDa repressor of the inhibitor of the protein kinase (p58IPK- interacting protein) (58 kDa interferon-induced protein kinase- interacting protein) (P52rIPK) (Death-associated protein 4) (THAP domain-containing protein 0). | |||||
|
PDPN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.036504 (rank : 64) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86YL7, O60836, O95128, Q7L375, Q8NBQ8, Q8NBR3 | Gene names | PDPN, GP36 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Podoplanin precursor (Glycoprotein 36) (Gp36) (PA2.26 antigen) (T1A) (T1-alpha) (Aggrus). | |||||
|
PHF14_MOUSE
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | 0.025805 (rank : 71) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9D4H9, Q810Z6, Q8C284, Q8CGF8, Q9D1Q8 | Gene names | Phf14 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 14. | |||||
|
AFF2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.015877 (rank : 80) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P51816, O43786, O60215, P78407, Q13521, Q14323, Q9UNA5 | Gene names | AFF2, FMR2, OX19 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 2 (Fragile X mental retardation 2 protein) (Protein FMR-2) (FMR2P) (Protein Ox19) (Fragile X E mental retardation syndrome protein). | |||||
|
DSPP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.059170 (rank : 47) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
KI67_HUMAN
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.054061 (rank : 54) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
TB182_HUMAN
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.020954 (rank : 76) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9C0C2 | Gene names | TNKS1BP1, KIAA1741, TAB182 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 182 kDa tankyrase 1-binding protein. | |||||
|
TCAL3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.051605 (rank : 57) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q969E4 | Gene names | TCEAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 3 (TCEA-like protein 3) (Transcription elongation factor S-II protein-like 3). | |||||
|
BPTF_HUMAN
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.027448 (rank : 70) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
CC47_MOUSE
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.022470 (rank : 74) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9D024, Q3TSB5, Q3V1S7, Q8C5D9, Q920S6 | Gene names | Ccdc47, Asp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 47 precursor (Adipocyte-specific protein 4). | |||||
|
FA53B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.016274 (rank : 79) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BGR5, Q3UGW1, Q8BI13 | Gene names | Fam53b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM53B. | |||||
|
MN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.030792 (rank : 66) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q10571 | Gene names | MN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable tumor suppressor protein MN1. | |||||
|
RBBP5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.017521 (rank : 78) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15291 | Gene names | RBBP5, RBQ3 | |||
|
Domain Architecture |
|
|||||
| Description | Retinoblastoma-binding protein 5 (RBBP-5) (Retinoblastoma-binding protein RBQ-3). | |||||
|
TCAL6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.054390 (rank : 52) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6IPX3, Q5H9J8 | Gene names | TCEAL6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 6 (TCEA-like protein 6) (Transcription elongation factor S-II protein-like 6). | |||||
|
TEX14_HUMAN
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.009478 (rank : 84) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1103 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8IWB6, Q7RTP3, Q8ND97, Q9BXT9 | Gene names | TEX14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed protein 14 (Testis-expressed sequence 14). | |||||
|
C8AP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.029637 (rank : 67) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 548 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UKL3, Q6PH76, Q7LCQ7, Q86YD9, Q9NUQ4, Q9NZV9, Q9P2N1, Q9Y563 | Gene names | CASP8AP2, FLASH, KIAA1315, RIP25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
DGKD_HUMAN
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.005875 (rank : 87) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q16760, Q14158, Q6PK55, Q8NG53 | Gene names | DGKD, KIAA0145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase delta (EC 2.7.1.107) (Diglyceride kinase delta) (DGK-delta) (DAG kinase delta) (130 kDa diacylglycerol kinase). | |||||
|
MTMR7_MOUSE
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.007244 (rank : 86) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z2C9, Q8C4J6 | Gene names | Mtmr7 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 7 (EC 3.1.3.-). | |||||
|
PHF14_HUMAN
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.023745 (rank : 72) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O94880 | Gene names | PHF14, KIAA0783 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 14. | |||||
|
ATN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.094944 (rank : 29) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
DMP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.052451 (rank : 55) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.050140 (rank : 58) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.051950 (rank : 56) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
MYSM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.080253 (rank : 35) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5VVJ2, Q7Z3G8, Q96PX3 | Gene names | MYSM1, KIAA1915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein MYSM1 (Myb-like, SWIRM and MPN domain-containing protein 1). | |||||
|
MYSM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.079218 (rank : 37) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q69Z66, Q3TPV7, Q8BRP5, Q8C4N1 | Gene names | Mysm1, Kiaa1915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein MYSM1 (Myb-like, SWIRM and MPN domain-containing protein 1). | |||||
|
NSBP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.056372 (rank : 50) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
PTMA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.079759 (rank : 36) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
PTMA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.074137 (rank : 38) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
PTMS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.056568 (rank : 49) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P20962 | Gene names | PTMS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
PTMS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.064492 (rank : 43) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9D0J8 | Gene names | Ptms | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
SMRC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.054185 (rank : 53) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q92922, Q6P172, Q8IWH2 | Gene names | SMARCC1, BAF155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155). | |||||
|
MIER1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q8N108, Q5T104, Q6AHY8, Q86TB4, Q8N156, Q8N161, Q8NC37, Q8NES4, Q8NES5, Q8NES6, Q8WWG2, Q9HCG2 | Gene names | MIER1, KIAA1610 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm induction early response protein 1 (Mi-er1) (hmi-er1) (Early response 1) (Er1). | |||||
|
MIER1_MOUSE
|
||||||
| NC score | 0.991988 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q5UAK0, Q5UAK1, Q5UAK2, Q5UAK3, Q6ZPL6, Q9D402 | Gene names | Mier1, Kiaa1610 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm induction early response protein 1 (Mi-er1). | |||||
|
MTA2_MOUSE
|
||||||
| NC score | 0.524813 (rank : 3) | θ value | 3.89403e-11 (rank : 3) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9R190, Q3TVT8 | Gene names | Mta2, Mta1l1 | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA2 (Metastasis-associated 1-like 1). | |||||
|
MTA2_HUMAN
|
||||||
| NC score | 0.522788 (rank : 4) | θ value | 1.133e-10 (rank : 4) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O94776, Q9UQB5 | Gene names | MTA2, MTA1L1, PID | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA2 (Metastasis-associated 1-like 1) (MTA1-L1 protein) (p53 target protein in deacetylase complex). | |||||
|
MTA3_HUMAN
|
||||||
| NC score | 0.516237 (rank : 5) | θ value | 1.63604e-09 (rank : 5) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BTC8, Q9NSP2, Q9ULF4 | Gene names | MTA3, KIAA1266 | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA3. | |||||
|
MTA3_MOUSE
|
||||||
| NC score | 0.503304 (rank : 6) | θ value | 8.97725e-08 (rank : 8) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q924K8, Q8VC18 | Gene names | Mta3 | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA3. | |||||
|
MTA1_HUMAN
|
||||||
| NC score | 0.494241 (rank : 7) | θ value | 1.53129e-07 (rank : 9) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13330, Q8NFI8, Q96GI8 | Gene names | MTA1 | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA1. | |||||
|
MTA1_MOUSE
|
||||||
| NC score | 0.493141 (rank : 8) | θ value | 1.99992e-07 (rank : 10) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8K4B0, Q80UI1, Q8K4D4, Q924K9 | Gene names | Mta1 | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA1. | |||||
|
RCOR1_MOUSE
|
||||||
| NC score | 0.461054 (rank : 9) | θ value | 3.19293e-05 (rank : 15) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8CFE3, Q3V092, Q8BK28, Q8CHI2 | Gene names | Rcor1, D12Wsu95e, Kiaa0071 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 1 (Protein CoREST). | |||||
|
RCOR1_HUMAN
|
||||||
| NC score | 0.458124 (rank : 10) | θ value | 4.1701e-05 (rank : 17) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UKL0, Q15044, Q6P2I9, Q86VG5 | Gene names | RCOR1, KIAA0071, RCOR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 1 (Protein CoREST). | |||||
|
RCOR3_MOUSE
|
||||||
| NC score | 0.451061 (rank : 11) | θ value | 3.77169e-06 (rank : 11) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6PGA0 | Gene names | Rcor3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 3. | |||||
|
RCOR3_HUMAN
|
||||||
| NC score | 0.449597 (rank : 12) | θ value | 4.92598e-06 (rank : 13) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9P2K3, Q5VT47, Q7L9I5, Q8N5U3, Q9NV83 | Gene names | RCOR3, KIAA1343 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 3. | |||||
|
ZN541_HUMAN
|
||||||
| NC score | 0.426187 (rank : 13) | θ value | 3.77169e-06 (rank : 12) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9H0D2, Q8NDK8 | Gene names | ZNF541 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 541. | |||||
|
RCOR2_HUMAN
|
||||||
| NC score | 0.426064 (rank : 14) | θ value | 0.000786445 (rank : 23) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8IZ40, Q96FP3 | Gene names | RCOR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 2. | |||||
|
RCOR2_MOUSE
|
||||||
| NC score | 0.425462 (rank : 15) | θ value | 0.000786445 (rank : 24) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8C796, Q9JMK4 | Gene names | Rcor2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 2 (M-CoREST). | |||||
|
RERE_MOUSE
|
||||||
| NC score | 0.384491 (rank : 16) | θ value | 1.38499e-08 (rank : 6) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
RERE_HUMAN
|
||||||
| NC score | 0.383494 (rank : 17) | θ value | 3.08544e-08 (rank : 7) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
TREF1_HUMAN
|
||||||
| NC score | 0.308707 (rank : 18) | θ value | 3.19293e-05 (rank : 16) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96PN7, Q7Z6T2, Q7Z6T3, Q9NQ72, Q9NQ73, Q9NUN9 | Gene names | TRERF1, RAPA, TREP132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132) (Zinc finger protein rapa). | |||||
|
TREF1_MOUSE
|
||||||
| NC score | 0.299552 (rank : 19) | θ value | 9.29e-05 (rank : 20) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BXJ2, Q6PCQ4, Q80X25, Q810H8, Q8BY31 | Gene names | Trerf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132). | |||||
|
NCOR2_HUMAN
|
||||||
| NC score | 0.297545 (rank : 20) | θ value | 1.87187e-05 (rank : 14) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
NCOR1_MOUSE
|
||||||
| NC score | 0.297016 (rank : 21) | θ value | 7.1131e-05 (rank : 19) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
NCOR1_HUMAN
|
||||||
| NC score | 0.294956 (rank : 22) | θ value | 7.1131e-05 (rank : 18) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
NCOR2_MOUSE
|
||||||
| NC score | 0.279786 (rank : 23) | θ value | 0.000270298 (rank : 21) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
SEC63_HUMAN
|
||||||
| NC score | 0.135127 (rank : 24) | θ value | 0.00390308 (rank : 25) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
SEC63_MOUSE
|
||||||
| NC score | 0.128916 (rank : 25) | θ value | 0.00869519 (rank : 26) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.115976 (rank : 26) | θ value | 0.000602161 (rank : 22) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.107332 (rank : 27) | θ value | 0.0431538 (rank : 29) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
RAG2_MOUSE
|
||||||
| NC score | 0.106407 (rank : 28) | θ value | 0.125558 (rank : 30) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P21784 | Gene names | Rag2, Rag-2 | |||
|
Domain Architecture |

|
|||||
| Description | V(D)J recombination-activating protein 2 (RAG-2). | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.094944 (rank : 29) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
SNUT1_MOUSE
|
||||||
| NC score | 0.092746 (rank : 30) | θ value | 0.47712 (rank : 34) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z315, Q8K155, Q9R1I9, Q9R270 | Gene names | Sart1, Haf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 1 (Squamous cell carcinoma antigen recognized by T-cells 1) (SART-1) (mSART-1) (Hypoxia- associated factor). | |||||
|
TCAL5_HUMAN
|
||||||
| NC score | 0.089034 (rank : 31) | θ value | 0.0193708 (rank : 27) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
SNUT1_HUMAN
|
||||||
| NC score | 0.085522 (rank : 32) | θ value | 1.06291 (rank : 41) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43290, Q53GB5 | Gene names | SART1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 1 (U4/U6.U5 tri-snRNP-associated 110 kDa protein) (Squamous cell carcinoma antigen recognized by T cells 1) (SART-1) (hSART-1) (hSnu66). | |||||
|
RAG2_HUMAN
|
||||||
| NC score | 0.084813 (rank : 33) | θ value | 1.06291 (rank : 40) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P55895, Q8TBL4 | Gene names | RAG2 | |||
|
Domain Architecture |

|
|||||
| Description | V(D)J recombination-activating protein 2 (RAG-2). | |||||
|
SMRC2_HUMAN
|
||||||
| NC score | 0.080745 (rank : 34) | θ value | 0.62314 (rank : 35) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8TAQ2, Q92923, Q96E12, Q96GY4 | Gene names | SMARCC2, BAF170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
MYSM1_HUMAN
|
||||||
| NC score | 0.080253 (rank : 35) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5VVJ2, Q7Z3G8, Q96PX3 | Gene names | MYSM1, KIAA1915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein MYSM1 (Myb-like, SWIRM and MPN domain-containing protein 1). | |||||
|
PTMA_HUMAN
|
||||||
| NC score | 0.079759 (rank : 36) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
MYSM1_MOUSE
|
||||||
| NC score | 0.079218 (rank : 37) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q69Z66, Q3TPV7, Q8BRP5, Q8C4N1 | Gene names | Mysm1, Kiaa1915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein MYSM1 (Myb-like, SWIRM and MPN domain-containing protein 1). | |||||
|
PTMA_MOUSE
|
||||||
| NC score | 0.074137 (rank : 38) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
SMRC2_MOUSE
|
||||||
| NC score | 0.073959 (rank : 39) | θ value | 0.47712 (rank : 33) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6PDG5, Q6P6P3 | Gene names | Smarcc2, Baf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
SMRC1_MOUSE
|
||||||
| NC score | 0.073837 (rank : 40) | θ value | 1.81305 (rank : 50) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P97496, Q7TS80, Q7TT29 | Gene names | Smarcc1, Baf155, Srg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155) (SWI3-related protein). | |||||
|
DMP1_HUMAN
|
||||||
| NC score | 0.067225 (rank : 41) | θ value | 0.813845 (rank : 37) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.064727 (rank : 42) | θ value | 1.38821 (rank : 42) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
PTMS_MOUSE
|
||||||
| NC score | 0.064492 (rank : 43) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9D0J8 | Gene names | Ptms | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
JOSD3_HUMAN
|
||||||
| NC score | 0.063495 (rank : 44) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H5J8, Q6I9Y6 | Gene names | JOSD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein JOSD3. | |||||
|
GNL3_MOUSE
|
||||||
| NC score | 0.061462 (rank : 45) | θ value | 0.365318 (rank : 32) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CI11, Q811R9, Q811S8, Q8BK21 | Gene names | Gnl3, Ns | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein-like 3 (Nucleolar GTP-binding protein 3) (Nucleostemin). | |||||
|
IFIT1_HUMAN
|
||||||
| NC score | 0.060140 (rank : 46) | θ value | 0.0431538 (rank : 28) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P09914, Q96QM5 | Gene names | IFIT1, G10P1, IFI56 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced protein with tetratricopeptide repeats 1 (IFIT-1) (Interferon-induced 56 kDa protein) (IFI-56K). | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.059170 (rank : 47) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
NSBP1_MOUSE
|
||||||
| NC score | 0.059019 (rank : 48) | θ value | 1.06291 (rank : 39) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
PTMS_HUMAN
|
||||||
| NC score | 0.056568 (rank : 49) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P20962 | Gene names | PTMS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
NSBP1_HUMAN
|
||||||
| NC score | 0.056372 (rank : 50) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.055722 (rank : 51) | θ value | 1.38821 (rank : 45) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
TCAL6_HUMAN
|
||||||
| NC score | 0.054390 (rank : 52) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6IPX3, Q5H9J8 | Gene names | TCEAL6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 6 (TCEA-like protein 6) (Transcription elongation factor S-II protein-like 6). | |||||
|
SMRC1_HUMAN
|
||||||
| NC score | 0.054185 (rank : 53) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q92922, Q6P172, Q8IWH2 | Gene names | SMARCC1, BAF155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155). | |||||
|
KI67_HUMAN
|
||||||
| NC score | 0.054061 (rank : 54) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
DMP1_MOUSE
|
||||||
| NC score | 0.052451 (rank : 55) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.051950 (rank : 56) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
TCAL3_HUMAN
|
||||||
| NC score | 0.051605 (rank : 57) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q969E4 | Gene names | TCEAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 3 (TCEA-like protein 3) (Transcription elongation factor S-II protein-like 3). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.050140 (rank : 58) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
EDEM3_HUMAN
|
||||||
| NC score | 0.044571 (rank : 59) | θ value | 1.38821 (rank : 44) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BZQ6, Q5TEZ0, Q9HCW1, Q9UFV7 | Gene names | EDEM3, C1orf22 | |||
|
Domain Architecture |

|
|||||
| Description | ER degradation-enhancing alpha-mannosidase-like 3. | |||||
|
EP400_MOUSE
|
||||||
| NC score | 0.040567 (rank : 60) | θ value | 0.163984 (rank : 31) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
BRD8_MOUSE
|
||||||
| NC score | 0.040385 (rank : 61) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
PPRB_MOUSE
|
||||||
| NC score | 0.037189 (rank : 62) | θ value | 1.81305 (rank : 49) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q925J9, O88323, Q3UHV0, Q6AXD5, Q8BW37, Q8BX19, Q8VDQ7, Q925K0 | Gene names | Pparbp, Trip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2). | |||||
|
BRD8_HUMAN
|
||||||
| NC score | 0.036738 (rank : 63) | θ value | 1.38821 (rank : 43) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H0E9, O43178, Q15355, Q59GN0, Q969M9 | Gene names | BRD8, SMAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8 (p120) (Skeletal muscle abundant protein) (Thyroid hormone receptor coactivating protein 120kDa) (TrCP120). | |||||
|
PDPN_HUMAN
|
||||||
| NC score | 0.036504 (rank : 64) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86YL7, O60836, O95128, Q7L375, Q8NBQ8, Q8NBR3 | Gene names | PDPN, GP36 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Podoplanin precursor (Glycoprotein 36) (Gp36) (PA2.26 antigen) (T1A) (T1-alpha) (Aggrus). | |||||
|
CENPJ_HUMAN
|
||||||
| NC score | 0.034564 (rank : 65) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9HC77, Q5T6R5, Q96KS5, Q9C067 | Gene names | CENPJ, CPAP, LAP, LIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein J (CENP-J) (Centrosomal P4.1-associated protein) (LAG-3-associated protein) (LYST-interacting protein 1). | |||||
|
MN1_HUMAN
|
||||||
| NC score | 0.030792 (rank : 66) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q10571 | Gene names | MN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable tumor suppressor protein MN1. | |||||
|
C8AP2_HUMAN
|
||||||
| NC score | 0.029637 (rank : 67) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 548 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UKL3, Q6PH76, Q7LCQ7, Q86YD9, Q9NUQ4, Q9NZV9, Q9P2N1, Q9Y563 | Gene names | CASP8AP2, FLASH, KIAA1315, RIP25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
UBP33_HUMAN
|
||||||
| NC score | 0.028545 (rank : 68) | θ value | 2.36792 (rank : 51) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8TEY7, Q8TEY6, Q96AV6, Q9H9F0, Q9UPQ5 | Gene names | USP33, KIAA1097, VDU1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 33 (EC 3.1.2.15) (Ubiquitin thioesterase 33) (Ubiquitin-specific-processing protease 33) (Deubiquitinating enzyme 33) (VHL-interacting deubiquitinating enzyme 1). | |||||
|
UBP8_HUMAN
|
||||||
| NC score | 0.027581 (rank : 69) | θ value | 1.38821 (rank : 46) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P40818, Q7Z3U2, Q86VA0, Q8IWI7 | Gene names | USP8, KIAA0055, UBPY | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (hUBPy). | |||||
|
BPTF_HUMAN
|
||||||
| NC score | 0.027448 (rank : 70) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
PHF14_MOUSE
|
||||||
| NC score | 0.025805 (rank : 71) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9D4H9, Q810Z6, Q8C284, Q8CGF8, Q9D1Q8 | Gene names | Phf14 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 14. | |||||
|
PHF14_HUMAN
|
||||||
| NC score | 0.023745 (rank : 72) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O94880 | Gene names | PHF14, KIAA0783 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 14. | |||||
|
CHP2_HUMAN
|
||||||
| NC score | 0.023472 (rank : 73) | θ value | 0.813845 (rank : 36) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43745 | Gene names | CHP2, HCA520 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin B homologous protein 2 (Hepatocellular carcinoma- associated antigen 520). | |||||
|
CC47_MOUSE
|
||||||
| NC score | 0.022470 (rank : 74) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9D024, Q3TSB5, Q3V1S7, Q8C5D9, Q920S6 | Gene names | Ccdc47, Asp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 47 precursor (Adipocyte-specific protein 4). | |||||
|
TXLNB_MOUSE
|
||||||
| NC score | 0.021600 (rank : 75) | θ value | 0.813845 (rank : 38) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 640 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VBT1, Q3UVB8, Q8BUK2 | Gene names | Txlnb, Mdp77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77). | |||||
|
TB182_HUMAN
|
||||||
| NC score | 0.020954 (rank : 76) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9C0C2 | Gene names | TNKS1BP1, KIAA1741, TAB182 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 182 kDa tankyrase 1-binding protein. | |||||
|
P52K_HUMAN
|
||||||
| NC score | 0.019941 (rank : 77) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43422, Q8WTW1, Q9Y3Z4 | Gene names | PRKRIR, DAP4, P52RIPK, THAP0 | |||
|
Domain Architecture |

|
|||||
| Description | 52 kDa repressor of the inhibitor of the protein kinase (p58IPK- interacting protein) (58 kDa interferon-induced protein kinase- interacting protein) (P52rIPK) (Death-associated protein 4) (THAP domain-containing protein 0). | |||||
|
RBBP5_HUMAN
|
||||||
| NC score | 0.017521 (rank : 78) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15291 | Gene names | RBBP5, RBQ3 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoblastoma-binding protein 5 (RBBP-5) (Retinoblastoma-binding protein RBQ-3). | |||||
|
FA53B_MOUSE
|
||||||
| NC score | 0.016274 (rank : 79) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BGR5, Q3UGW1, Q8BI13 | Gene names | Fam53b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM53B. | |||||
|
AFF2_HUMAN
|
||||||
| NC score | 0.015877 (rank : 80) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P51816, O43786, O60215, P78407, Q13521, Q14323, Q9UNA5 | Gene names | AFF2, FMR2, OX19 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 2 (Fragile X mental retardation 2 protein) (Protein FMR-2) (FMR2P) (Protein Ox19) (Fragile X E mental retardation syndrome protein). | |||||
|
ARX_HUMAN
|
||||||
| NC score | 0.012604 (rank : 81) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96QS3 | Gene names | ARX | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein ARX (Aristaless-related homeobox). | |||||
|
ARX_MOUSE
|
||||||
| NC score | 0.012561 (rank : 82) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 398 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O35085, Q9QYT4 | Gene names | Arx | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein ARX (Aristaless-related homeobox). | |||||
|
E41L3_HUMAN
|
||||||
| NC score | 0.012532 (rank : 83) | θ value | 1.81305 (rank : 47) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y2J2, O95713, Q9BRP5 | Gene names | EPB41L3, DAL1, KIAA0987 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 3 (4.1B) (Differentially expressed in adenocarcinoma of the lung protein 1) (DAL-1). | |||||
|
TEX14_HUMAN
|
||||||
| NC score | 0.009478 (rank : 84) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1103 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8IWB6, Q7RTP3, Q8ND97, Q9BXT9 | Gene names | TEX14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed protein 14 (Testis-expressed sequence 14). | |||||
|
KCNB2_HUMAN
|
||||||
| NC score | 0.008213 (rank : 85) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92953, Q9BXD3 | Gene names | KCNB2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 2 (Voltage-gated potassium channel subunit Kv2.2). | |||||
|
MTMR7_MOUSE
|
||||||
| NC score | 0.007244 (rank : 86) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z2C9, Q8C4J6 | Gene names | Mtmr7 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 7 (EC 3.1.3.-). | |||||
|
DGKD_HUMAN
|
||||||
| NC score | 0.005875 (rank : 87) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q16760, Q14158, Q6PK55, Q8NG53 | Gene names | DGKD, KIAA0145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase delta (EC 2.7.1.107) (Diglyceride kinase delta) (DGK-delta) (DAG kinase delta) (130 kDa diacylglycerol kinase). | |||||