Please be patient as the page loads
|
RCOR1_MOUSE
|
||||||
| SwissProt Accessions | Q8CFE3, Q3V092, Q8BK28, Q8CHI2 | Gene names | Rcor1, D12Wsu95e, Kiaa0071 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 1 (Protein CoREST). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
RCOR1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.997288 (rank : 2) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9UKL0, Q15044, Q6P2I9, Q86VG5 | Gene names | RCOR1, KIAA0071, RCOR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 1 (Protein CoREST). | |||||
|
RCOR1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8CFE3, Q3V092, Q8BK28, Q8CHI2 | Gene names | Rcor1, D12Wsu95e, Kiaa0071 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 1 (Protein CoREST). | |||||
|
RCOR3_HUMAN
|
||||||
| θ value | 2.4321e-138 (rank : 3) | NC score | 0.967115 (rank : 3) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9P2K3, Q5VT47, Q7L9I5, Q8N5U3, Q9NV83 | Gene names | RCOR3, KIAA1343 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 3. | |||||
|
RCOR2_MOUSE
|
||||||
| θ value | 2.9938e-112 (rank : 4) | NC score | 0.964387 (rank : 5) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8C796, Q9JMK4 | Gene names | Rcor2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 2 (M-CoREST). | |||||
|
RCOR2_HUMAN
|
||||||
| θ value | 5.64607e-111 (rank : 5) | NC score | 0.965144 (rank : 4) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IZ40, Q96FP3 | Gene names | RCOR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 2. | |||||
|
RCOR3_MOUSE
|
||||||
| θ value | 3.20603e-106 (rank : 6) | NC score | 0.957749 (rank : 6) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6PGA0 | Gene names | Rcor3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 3. | |||||
|
NCOR2_HUMAN
|
||||||
| θ value | 6.20254e-17 (rank : 7) | NC score | 0.477644 (rank : 15) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
MTA3_MOUSE
|
||||||
| θ value | 8.95645e-16 (rank : 8) | NC score | 0.540044 (rank : 7) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q924K8, Q8VC18 | Gene names | Mta3 | |||
|
Domain Architecture |
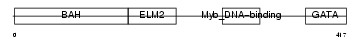
|
|||||
| Description | Metastasis-associated protein MTA3. | |||||
|
NCOR2_MOUSE
|
||||||
| θ value | 1.68911e-14 (rank : 9) | NC score | 0.466318 (rank : 17) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
NCOR1_MOUSE
|
||||||
| θ value | 2.20605e-14 (rank : 10) | NC score | 0.477893 (rank : 14) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
NCOR1_HUMAN
|
||||||
| θ value | 3.76295e-14 (rank : 11) | NC score | 0.472138 (rank : 16) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
MTA2_HUMAN
|
||||||
| θ value | 4.16044e-13 (rank : 12) | NC score | 0.530602 (rank : 8) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O94776, Q9UQB5 | Gene names | MTA2, MTA1L1, PID | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA2 (Metastasis-associated 1-like 1) (MTA1-L1 protein) (p53 target protein in deacetylase complex). | |||||
|
MTA2_MOUSE
|
||||||
| θ value | 7.09661e-13 (rank : 13) | NC score | 0.527173 (rank : 10) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9R190, Q3TVT8 | Gene names | Mta2, Mta1l1 | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA2 (Metastasis-associated 1-like 1). | |||||
|
MTA3_HUMAN
|
||||||
| θ value | 3.52202e-12 (rank : 14) | NC score | 0.528336 (rank : 9) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BTC8, Q9NSP2, Q9ULF4 | Gene names | MTA3, KIAA1266 | |||
|
Domain Architecture |
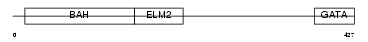
|
|||||
| Description | Metastasis-associated protein MTA3. | |||||
|
MTA1_HUMAN
|
||||||
| θ value | 6.64225e-11 (rank : 15) | NC score | 0.500051 (rank : 12) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13330, Q8NFI8, Q96GI8 | Gene names | MTA1 | |||
|
Domain Architecture |
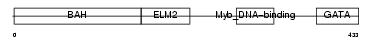
|
|||||
| Description | Metastasis-associated protein MTA1. | |||||
|
MTA1_MOUSE
|
||||||
| θ value | 2.52405e-10 (rank : 16) | NC score | 0.495422 (rank : 13) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8K4B0, Q80UI1, Q8K4D4, Q924K9 | Gene names | Mta1 | |||
|
Domain Architecture |
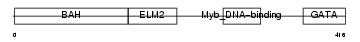
|
|||||
| Description | Metastasis-associated protein MTA1. | |||||
|
ZN541_HUMAN
|
||||||
| θ value | 1.06045e-08 (rank : 17) | NC score | 0.521614 (rank : 11) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H0D2, Q8NDK8 | Gene names | ZNF541 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 541. | |||||
|
TREF1_MOUSE
|
||||||
| θ value | 5.26297e-08 (rank : 18) | NC score | 0.376262 (rank : 21) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BXJ2, Q6PCQ4, Q80X25, Q810H8, Q8BY31 | Gene names | Trerf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132). | |||||
|
TREF1_HUMAN
|
||||||
| θ value | 2.61198e-07 (rank : 19) | NC score | 0.379015 (rank : 20) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96PN7, Q7Z6T2, Q7Z6T3, Q9NQ72, Q9NQ73, Q9NUN9 | Gene names | TRERF1, RAPA, TREP132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132) (Zinc finger protein rapa). | |||||
|
RERE_MOUSE
|
||||||
| θ value | 2.21117e-06 (rank : 20) | NC score | 0.339731 (rank : 22) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
RERE_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 21) | NC score | 0.336566 (rank : 23) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
MIER1_MOUSE
|
||||||
| θ value | 1.09739e-05 (rank : 22) | NC score | 0.465248 (rank : 18) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5UAK0, Q5UAK1, Q5UAK2, Q5UAK3, Q6ZPL6, Q9D402 | Gene names | Mier1, Kiaa1610 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm induction early response protein 1 (Mi-er1). | |||||
|
MIER1_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 23) | NC score | 0.461054 (rank : 19) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8N108, Q5T104, Q6AHY8, Q86TB4, Q8N156, Q8N161, Q8NC37, Q8NES4, Q8NES5, Q8NES6, Q8WWG2, Q9HCG2 | Gene names | MIER1, KIAA1610 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm induction early response protein 1 (Mi-er1) (hmi-er1) (Early response 1) (Er1). | |||||
|
MYSM1_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 24) | NC score | 0.238100 (rank : 25) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5VVJ2, Q7Z3G8, Q96PX3 | Gene names | MYSM1, KIAA1915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein MYSM1 (Myb-like, SWIRM and MPN domain-containing protein 1). | |||||
|
MYSM1_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 25) | NC score | 0.239399 (rank : 24) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q69Z66, Q3TPV7, Q8BRP5, Q8C4N1 | Gene names | Mysm1, Kiaa1915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein MYSM1 (Myb-like, SWIRM and MPN domain-containing protein 1). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 26) | NC score | 0.058498 (rank : 27) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
NIN_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 27) | NC score | 0.035100 (rank : 36) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
KTN1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 28) | NC score | 0.016658 (rank : 50) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1324 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q61595, Q8BG49, Q8BHF4, Q8BHM8, Q8C9Y5, Q8CG51, Q8CG52, Q8CG53, Q8CG54, Q8CG55, Q8CG56, Q8CG57, Q8CG58, Q8CG59, Q8CG60, Q8CG61, Q8CG62, Q8CG63 | Gene names | Ktn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin. | |||||
|
TRAIP_MOUSE
|
||||||
| θ value | 0.279714 (rank : 29) | NC score | 0.037377 (rank : 35) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8VIG6, O08854, Q922M8, Q9CPP4 | Gene names | Traip, Trip | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
MYH3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 30) | NC score | 0.015094 (rank : 52) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
CUTL2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 31) | NC score | 0.034324 (rank : 37) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O14529 | Gene names | CUTL2, KIAA0293 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
PNMA1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 32) | NC score | 0.029120 (rank : 38) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8ND90, O95144, Q8NG07 | Gene names | PNMA1, MA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paraneoplastic antigen Ma1 (Neuron- and testis-specific protein 1) (37 kDa neuronal protein). | |||||
|
RHG12_HUMAN
|
||||||
| θ value | 1.06291 (rank : 33) | NC score | 0.024094 (rank : 42) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IWW6, Q86UB3, Q8IWW7, Q8N3L1, Q9NT76 | Gene names | ARHGAP12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 1.38821 (rank : 34) | NC score | 0.024990 (rank : 41) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
PLCB1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 35) | NC score | 0.019431 (rank : 47) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NQ66, O60325, Q5TFF7, Q5TGC9, Q8IV93, Q9BQW2, Q9H4H2, Q9H8H5, Q9NQ65, Q9NQH9, Q9NTH4, Q9UJP6, Q9UM26 | Gene names | PLCB1, KIAA0581 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-1) (PLC-beta-1) (PLC-I) (PLC-154). | |||||
|
RHG12_MOUSE
|
||||||
| θ value | 1.38821 (rank : 36) | NC score | 0.022876 (rank : 44) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8C0D4, Q8BVP8 | Gene names | Arhgap12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
GSTK1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 37) | NC score | 0.037897 (rank : 34) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2Q3, Q9P1S4 | Gene names | GSTK1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutathione S-transferase kappa 1 (EC 2.5.1.18) (GST 13-13) (Glutathione S-transferase subunit 13) (GST class-kappa) (GSTK1-1) (hGSTK1). | |||||
|
MINT_HUMAN
|
||||||
| θ value | 2.36792 (rank : 38) | NC score | 0.020145 (rank : 45) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MYH7_HUMAN
|
||||||
| θ value | 2.36792 (rank : 39) | NC score | 0.012011 (rank : 55) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
PININ_HUMAN
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.023499 (rank : 43) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9H307, O60899, Q53EM7, Q6P5X4, Q7KYL1, Q99738, Q9UHZ9, Q9UQR9 | Gene names | PNN, DRS, MEMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin (140 kDa nuclear and cell adhesion-related phosphoprotein) (Domain-rich serine protein) (DRS-protein) (DRSP) (Melanoma metastasis clone A protein) (Desmosome-associated protein) (SR-like protein) (Nuclear protein SDK3). | |||||
|
SPTA1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.014303 (rank : 53) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P08032, P97502 | Gene names | Spta1, Spna1, Spta | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
CNGB3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 42) | NC score | 0.008137 (rank : 59) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NQW8, Q9NRE9 | Gene names | CNGB3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel beta 3 (CNG channel beta 3) (Cyclic nucleotide-gated channel beta 3) (Cone photoreceptor cGMP- gated channel subunit beta) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
TPR_HUMAN
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.019955 (rank : 46) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
BIRC6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 44) | NC score | 0.017437 (rank : 49) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NR09, Q9ULD1 | Gene names | BIRC6, KIAA1289 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 6 (Ubiquitin-conjugating BIR-domain enzyme apollon). | |||||
|
FILA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 45) | NC score | 0.018597 (rank : 48) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
MD1L1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 46) | NC score | 0.026180 (rank : 39) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y6D9, Q13312, Q75MI0, Q86UM4, Q9UNH0 | Gene names | MAD1L1, MAD1, TXBP181 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1) (Mitotic checkpoint MAD1 protein-homolog) (HsMAD1) (hMAD1) (Tax-binding protein 181). | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 5.27518 (rank : 47) | NC score | 0.010361 (rank : 57) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
CT075_HUMAN
|
||||||
| θ value | 5.27518 (rank : 48) | NC score | 0.005552 (rank : 61) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WUT4, Q5JWV6, Q9H419 | Gene names | C20orf75 | |||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf75 precursor. | |||||
|
MIA2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 49) | NC score | 0.013267 (rank : 54) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91ZV0 | Gene names | Mia2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma inhibitory activity protein 2 precursor. | |||||
|
TRAIP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 50) | NC score | 0.025976 (rank : 40) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BWF2, O00467 | Gene names | TRAIP, TRIP | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
FCER2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 51) | NC score | 0.005167 (rank : 62) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P20693, Q61556, Q61557 | Gene names | Fcer2, Fcer2a | |||
|
Domain Architecture |

|
|||||
| Description | Low affinity immunoglobulin epsilon Fc receptor (Lymphocyte IGE receptor) (Fc-epsilon-RII) (CD23 antigen). | |||||
|
JHD2C_MOUSE
|
||||||
| θ value | 6.88961 (rank : 52) | NC score | 0.011098 (rank : 56) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q69ZK6, Q6NV48, Q8BUF5, Q8C4I5, Q8C5Q9 | Gene names | Jmjd1c, Jhdm2c, Kiaa1380 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable JmjC domain-containing histone demethylation protein 2C (EC 1.14.11.-) (Jumonji domain-containing protein 1C). | |||||
|
SMC2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 53) | NC score | 0.015735 (rank : 51) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
LONM_HUMAN
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.010137 (rank : 58) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P36776, P36777, Q9UQ95 | Gene names | PRSS15 | |||
|
Domain Architecture |

|
|||||
| Description | Lon protease homolog, mitochondrial precursor (EC 3.4.21.-) (Lon protease-like protein) (LONP) (LONHs). | |||||
|
PARVA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.005895 (rank : 60) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EPC1, Q9JJ65 | Gene names | Parva, Actp | |||
|
Domain Architecture |
|
|||||
| Description | Alpha-parvin (Actopaxin). | |||||
|
ATN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.065577 (rank : 26) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
CJ108_HUMAN
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.055025 (rank : 31) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N8Z3 | Gene names | C10orf108 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative uncharacterized protein C10orf108. | |||||
|
DCP1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.058110 (rank : 28) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NPI6 | Gene names | DCP1A, SMIF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA decapping enzyme 1A (EC 3.-.-.-) (Transcription factor SMIF) (Smad4-interacting transcriptional co-activator). | |||||
|
FA47B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.055312 (rank : 30) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
GA2L2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.054354 (rank : 33) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5SSG4 | Gene names | Gas2l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.057679 (rank : 29) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
SON_HUMAN
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.054567 (rank : 32) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
RCOR1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8CFE3, Q3V092, Q8BK28, Q8CHI2 | Gene names | Rcor1, D12Wsu95e, Kiaa0071 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 1 (Protein CoREST). | |||||
|
RCOR1_HUMAN
|
||||||
| NC score | 0.997288 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9UKL0, Q15044, Q6P2I9, Q86VG5 | Gene names | RCOR1, KIAA0071, RCOR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 1 (Protein CoREST). | |||||
|
RCOR3_HUMAN
|
||||||
| NC score | 0.967115 (rank : 3) | θ value | 2.4321e-138 (rank : 3) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9P2K3, Q5VT47, Q7L9I5, Q8N5U3, Q9NV83 | Gene names | RCOR3, KIAA1343 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 3. | |||||
|
RCOR2_HUMAN
|
||||||
| NC score | 0.965144 (rank : 4) | θ value | 5.64607e-111 (rank : 5) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IZ40, Q96FP3 | Gene names | RCOR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 2. | |||||
|
RCOR2_MOUSE
|
||||||
| NC score | 0.964387 (rank : 5) | θ value | 2.9938e-112 (rank : 4) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8C796, Q9JMK4 | Gene names | Rcor2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 2 (M-CoREST). | |||||
|
RCOR3_MOUSE
|
||||||
| NC score | 0.957749 (rank : 6) | θ value | 3.20603e-106 (rank : 6) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6PGA0 | Gene names | Rcor3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 3. | |||||
|
MTA3_MOUSE
|
||||||
| NC score | 0.540044 (rank : 7) | θ value | 8.95645e-16 (rank : 8) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q924K8, Q8VC18 | Gene names | Mta3 | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA3. | |||||
|
MTA2_HUMAN
|
||||||
| NC score | 0.530602 (rank : 8) | θ value | 4.16044e-13 (rank : 12) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O94776, Q9UQB5 | Gene names | MTA2, MTA1L1, PID | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA2 (Metastasis-associated 1-like 1) (MTA1-L1 protein) (p53 target protein in deacetylase complex). | |||||
|
MTA3_HUMAN
|
||||||
| NC score | 0.528336 (rank : 9) | θ value | 3.52202e-12 (rank : 14) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BTC8, Q9NSP2, Q9ULF4 | Gene names | MTA3, KIAA1266 | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA3. | |||||
|
MTA2_MOUSE
|
||||||
| NC score | 0.527173 (rank : 10) | θ value | 7.09661e-13 (rank : 13) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9R190, Q3TVT8 | Gene names | Mta2, Mta1l1 | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA2 (Metastasis-associated 1-like 1). | |||||
|
ZN541_HUMAN
|
||||||
| NC score | 0.521614 (rank : 11) | θ value | 1.06045e-08 (rank : 17) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H0D2, Q8NDK8 | Gene names | ZNF541 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 541. | |||||
|
MTA1_HUMAN
|
||||||
| NC score | 0.500051 (rank : 12) | θ value | 6.64225e-11 (rank : 15) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13330, Q8NFI8, Q96GI8 | Gene names | MTA1 | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA1. | |||||
|
MTA1_MOUSE
|
||||||
| NC score | 0.495422 (rank : 13) | θ value | 2.52405e-10 (rank : 16) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8K4B0, Q80UI1, Q8K4D4, Q924K9 | Gene names | Mta1 | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA1. | |||||
|
NCOR1_MOUSE
|
||||||
| NC score | 0.477893 (rank : 14) | θ value | 2.20605e-14 (rank : 10) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
NCOR2_HUMAN
|
||||||
| NC score | 0.477644 (rank : 15) | θ value | 6.20254e-17 (rank : 7) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
NCOR1_HUMAN
|
||||||
| NC score | 0.472138 (rank : 16) | θ value | 3.76295e-14 (rank : 11) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
NCOR2_MOUSE
|
||||||
| NC score | 0.466318 (rank : 17) | θ value | 1.68911e-14 (rank : 9) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
MIER1_MOUSE
|
||||||
| NC score | 0.465248 (rank : 18) | θ value | 1.09739e-05 (rank : 22) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5UAK0, Q5UAK1, Q5UAK2, Q5UAK3, Q6ZPL6, Q9D402 | Gene names | Mier1, Kiaa1610 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm induction early response protein 1 (Mi-er1). | |||||
|
MIER1_HUMAN
|
||||||
| NC score | 0.461054 (rank : 19) | θ value | 3.19293e-05 (rank : 23) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8N108, Q5T104, Q6AHY8, Q86TB4, Q8N156, Q8N161, Q8NC37, Q8NES4, Q8NES5, Q8NES6, Q8WWG2, Q9HCG2 | Gene names | MIER1, KIAA1610 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm induction early response protein 1 (Mi-er1) (hmi-er1) (Early response 1) (Er1). | |||||
|
TREF1_HUMAN
|
||||||
| NC score | 0.379015 (rank : 20) | θ value | 2.61198e-07 (rank : 19) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96PN7, Q7Z6T2, Q7Z6T3, Q9NQ72, Q9NQ73, Q9NUN9 | Gene names | TRERF1, RAPA, TREP132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132) (Zinc finger protein rapa). | |||||
|
TREF1_MOUSE
|
||||||
| NC score | 0.376262 (rank : 21) | θ value | 5.26297e-08 (rank : 18) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BXJ2, Q6PCQ4, Q80X25, Q810H8, Q8BY31 | Gene names | Trerf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132). | |||||
|
RERE_MOUSE
|
||||||
| NC score | 0.339731 (rank : 22) | θ value | 2.21117e-06 (rank : 20) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
RERE_HUMAN
|
||||||
| NC score | 0.336566 (rank : 23) | θ value | 6.43352e-06 (rank : 21) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
MYSM1_MOUSE
|
||||||
| NC score | 0.239399 (rank : 24) | θ value | 0.00869519 (rank : 25) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q69Z66, Q3TPV7, Q8BRP5, Q8C4N1 | Gene names | Mysm1, Kiaa1915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein MYSM1 (Myb-like, SWIRM and MPN domain-containing protein 1). | |||||
|
MYSM1_HUMAN
|
||||||
| NC score | 0.238100 (rank : 25) | θ value | 0.00298849 (rank : 24) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5VVJ2, Q7Z3G8, Q96PX3 | Gene names | MYSM1, KIAA1915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein MYSM1 (Myb-like, SWIRM and MPN domain-containing protein 1). | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.065577 (rank : 26) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.058498 (rank : 27) | θ value | 0.0431538 (rank : 26) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
DCP1A_HUMAN
|
||||||
| NC score | 0.058110 (rank : 28) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NPI6 | Gene names | DCP1A, SMIF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA decapping enzyme 1A (EC 3.-.-.-) (Transcription factor SMIF) (Smad4-interacting transcriptional co-activator). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.057679 (rank : 29) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
FA47B_HUMAN
|
||||||
| NC score | 0.055312 (rank : 30) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
CJ108_HUMAN
|
||||||
| NC score | 0.055025 (rank : 31) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N8Z3 | Gene names | C10orf108 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative uncharacterized protein C10orf108. | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.054567 (rank : 32) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
GA2L2_MOUSE
|
||||||
| NC score | 0.054354 (rank : 33) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5SSG4 | Gene names | Gas2l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2). | |||||
|
GSTK1_HUMAN
|
||||||
| NC score | 0.037897 (rank : 34) | θ value | 2.36792 (rank : 37) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2Q3, Q9P1S4 | Gene names | GSTK1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutathione S-transferase kappa 1 (EC 2.5.1.18) (GST 13-13) (Glutathione S-transferase subunit 13) (GST class-kappa) (GSTK1-1) (hGSTK1). | |||||
|
TRAIP_MOUSE
|
||||||
| NC score | 0.037377 (rank : 35) | θ value | 0.279714 (rank : 29) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8VIG6, O08854, Q922M8, Q9CPP4 | Gene names | Traip, Trip | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
NIN_HUMAN
|
||||||
| NC score | 0.035100 (rank : 36) | θ value | 0.0431538 (rank : 27) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
CUTL2_HUMAN
|
||||||
| NC score | 0.034324 (rank : 37) | θ value | 0.47712 (rank : 31) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O14529 | Gene names | CUTL2, KIAA0293 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
PNMA1_HUMAN
|
||||||
| NC score | 0.029120 (rank : 38) | θ value | 1.06291 (rank : 32) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8ND90, O95144, Q8NG07 | Gene names | PNMA1, MA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paraneoplastic antigen Ma1 (Neuron- and testis-specific protein 1) (37 kDa neuronal protein). | |||||
|
MD1L1_HUMAN
|
||||||
| NC score | 0.026180 (rank : 39) | θ value | 4.03905 (rank : 46) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y6D9, Q13312, Q75MI0, Q86UM4, Q9UNH0 | Gene names | MAD1L1, MAD1, TXBP181 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1) (Mitotic checkpoint MAD1 protein-homolog) (HsMAD1) (hMAD1) (Tax-binding protein 181). | |||||
|
TRAIP_HUMAN
|
||||||
| NC score | 0.025976 (rank : 40) | θ value | 5.27518 (rank : 50) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BWF2, O00467 | Gene names | TRAIP, TRIP | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.024990 (rank : 41) | θ value | 1.38821 (rank : 34) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
RHG12_HUMAN
|
||||||
| NC score | 0.024094 (rank : 42) | θ value | 1.06291 (rank : 33) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IWW6, Q86UB3, Q8IWW7, Q8N3L1, Q9NT76 | Gene names | ARHGAP12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
PININ_HUMAN
|
||||||
| NC score | 0.023499 (rank : 43) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9H307, O60899, Q53EM7, Q6P5X4, Q7KYL1, Q99738, Q9UHZ9, Q9UQR9 | Gene names | PNN, DRS, MEMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin (140 kDa nuclear and cell adhesion-related phosphoprotein) (Domain-rich serine protein) (DRS-protein) (DRSP) (Melanoma metastasis clone A protein) (Desmosome-associated protein) (SR-like protein) (Nuclear protein SDK3). | |||||
|
RHG12_MOUSE
|
||||||
| NC score | 0.022876 (rank : 44) | θ value | 1.38821 (rank : 36) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8C0D4, Q8BVP8 | Gene names | Arhgap12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.020145 (rank : 45) | θ value | 2.36792 (rank : 38) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.019955 (rank : 46) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
PLCB1_HUMAN
|
||||||
| NC score | 0.019431 (rank : 47) | θ value | 1.38821 (rank : 35) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NQ66, O60325, Q5TFF7, Q5TGC9, Q8IV93, Q9BQW2, Q9H4H2, Q9H8H5, Q9NQ65, Q9NQH9, Q9NTH4, Q9UJP6, Q9UM26 | Gene names | PLCB1, KIAA0581 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-1) (PLC-beta-1) (PLC-I) (PLC-154). | |||||
|
FILA_HUMAN
|
||||||
| NC score | 0.018597 (rank : 48) | θ value | 4.03905 (rank : 45) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
BIRC6_HUMAN
|
||||||
| NC score | 0.017437 (rank : 49) | θ value | 4.03905 (rank : 44) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NR09, Q9ULD1 | Gene names | BIRC6, KIAA1289 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 6 (Ubiquitin-conjugating BIR-domain enzyme apollon). | |||||
|
KTN1_MOUSE
|
||||||
| NC score | 0.016658 (rank : 50) | θ value | 0.279714 (rank : 28) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1324 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q61595, Q8BG49, Q8BHF4, Q8BHM8, Q8C9Y5, Q8CG51, Q8CG52, Q8CG53, Q8CG54, Q8CG55, Q8CG56, Q8CG57, Q8CG58, Q8CG59, Q8CG60, Q8CG61, Q8CG62, Q8CG63 | Gene names | Ktn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin. | |||||
|
SMC2_HUMAN
|
||||||
| NC score | 0.015735 (rank : 51) | θ value | 6.88961 (rank : 53) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
MYH3_HUMAN
|
||||||
| NC score | 0.015094 (rank : 52) | θ value | 0.365318 (rank : 30) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
SPTA1_MOUSE
|
||||||
| NC score | 0.014303 (rank : 53) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P08032, P97502 | Gene names | Spta1, Spna1, Spta | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
MIA2_MOUSE
|
||||||
| NC score | 0.013267 (rank : 54) | θ value | 5.27518 (rank : 49) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91ZV0 | Gene names | Mia2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma inhibitory activity protein 2 precursor. | |||||
|
MYH7_HUMAN
|
||||||
| NC score | 0.012011 (rank : 55) | θ value | 2.36792 (rank : 39) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
JHD2C_MOUSE
|
||||||
| NC score | 0.011098 (rank : 56) | θ value | 6.88961 (rank : 52) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q69ZK6, Q6NV48, Q8BUF5, Q8C4I5, Q8C5Q9 | Gene names | Jmjd1c, Jhdm2c, Kiaa1380 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable JmjC domain-containing histone demethylation protein 2C (EC 1.14.11.-) (Jumonji domain-containing protein 1C). | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.010361 (rank : 57) | θ value | 5.27518 (rank : 47) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
LONM_HUMAN
|
||||||
| NC score | 0.010137 (rank : 58) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P36776, P36777, Q9UQ95 | Gene names | PRSS15 | |||
|
Domain Architecture |

|
|||||
| Description | Lon protease homolog, mitochondrial precursor (EC 3.4.21.-) (Lon protease-like protein) (LONP) (LONHs). | |||||
|
CNGB3_HUMAN
|
||||||
| NC score | 0.008137 (rank : 59) | θ value | 3.0926 (rank : 42) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NQW8, Q9NRE9 | Gene names | CNGB3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel beta 3 (CNG channel beta 3) (Cyclic nucleotide-gated channel beta 3) (Cone photoreceptor cGMP- gated channel subunit beta) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
PARVA_MOUSE
|
||||||
| NC score | 0.005895 (rank : 60) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EPC1, Q9JJ65 | Gene names | Parva, Actp | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-parvin (Actopaxin). | |||||
|
CT075_HUMAN
|
||||||
| NC score | 0.005552 (rank : 61) | θ value | 5.27518 (rank : 48) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WUT4, Q5JWV6, Q9H419 | Gene names | C20orf75 | |||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf75 precursor. | |||||
|
FCER2_MOUSE
|
||||||
| NC score | 0.005167 (rank : 62) | θ value | 6.88961 (rank : 51) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P20693, Q61556, Q61557 | Gene names | Fcer2, Fcer2a | |||
|
Domain Architecture |

|
|||||
| Description | Low affinity immunoglobulin epsilon Fc receptor (Lymphocyte IGE receptor) (Fc-epsilon-RII) (CD23 antigen). | |||||