Please be patient as the page loads
|
MIA2_MOUSE
|
||||||
| SwissProt Accessions | Q91ZV0 | Gene names | Mia2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma inhibitory activity protein 2 precursor. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MIA2_MOUSE
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 109 | |
| SwissProt Accessions | Q91ZV0 | Gene names | Mia2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma inhibitory activity protein 2 precursor. | |||||
|
MIA2_HUMAN
|
||||||
| θ value | 2.26584e-152 (rank : 2) | NC score | 0.937681 (rank : 2) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96PC5, Q9H6C1 | Gene names | MIA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma inhibitory activity protein 2 precursor. | |||||
|
OTOR_HUMAN
|
||||||
| θ value | 8.67504e-11 (rank : 3) | NC score | 0.584114 (rank : 3) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NRC9 | Gene names | OTOR, FDP, MIAL | |||
|
Domain Architecture |
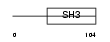
|
|||||
| Description | Otoraplin precursor (Fibrocyte-derived protein) (Melanoma inhibitory activity-like protein). | |||||
|
MIA_HUMAN
|
||||||
| θ value | 5.62301e-10 (rank : 4) | NC score | 0.564074 (rank : 4) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q16674 | Gene names | MIA | |||
|
Domain Architecture |
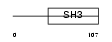
|
|||||
| Description | Melanoma-derived growth regulatory protein precursor (Melanoma inhibitory activity). | |||||
|
OTOR_MOUSE
|
||||||
| θ value | 4.76016e-09 (rank : 5) | NC score | 0.559086 (rank : 5) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JIE3 | Gene names | Otor, Mial | |||
|
Domain Architecture |
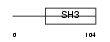
|
|||||
| Description | Otoraplin precursor (Melanoma inhibitory activity-like protein). | |||||
|
MIA_MOUSE
|
||||||
| θ value | 1.38499e-08 (rank : 6) | NC score | 0.551854 (rank : 6) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61865, O09086, P97495 | Gene names | Mia, Cdrap, Mia1 | |||
|
Domain Architecture |
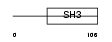
|
|||||
| Description | Melanoma-derived growth regulatory protein precursor (Melanoma inhibitory activity) (Cartilage-derived retinoic acid-sensitive protein) (CD-RAP). | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 7) | NC score | 0.090196 (rank : 9) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
APLP2_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 8) | NC score | 0.060077 (rank : 27) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q06481 | Gene names | APLP2, APPL2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 2 precursor (Amyloid protein homolog) (APPH) (CDEI box-binding protein) (CDEBP). | |||||
|
HTSF1_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 9) | NC score | 0.103643 (rank : 8) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BGC0, Q1WWK0, Q9CT41, Q9DAU3 | Gene names | Htatsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 homolog. | |||||
|
ERCC6_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 10) | NC score | 0.037913 (rank : 61) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q03468 | Gene names | ERCC6, CSB | |||
|
Domain Architecture |

|
|||||
| Description | DNA excision repair protein ERCC-6 (EC 3.6.1.-) (ATP-dependent helicase ERCC6) (Cockayne syndrome protein CSB). | |||||
|
MYT1_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 11) | NC score | 0.054075 (rank : 29) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q01538, O94922, Q9UPV2 | Gene names | MYT1, KIAA0835, KIAA1050, MTF1, MYTI, PLPB1 | |||
|
Domain Architecture |

|
|||||
| Description | Myelin transcription factor 1 (MyT1) (MyTI) (Proteolipid protein- binding protein) (PLPB1). | |||||
|
AHI1_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 12) | NC score | 0.078199 (rank : 13) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8K3E5, Q7TNV2, Q9CVY1 | Gene names | Ahi1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Jouberin (Abelson helper integration site 1 protein) (AHI-1). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 13) | NC score | 0.109448 (rank : 7) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
VAV2_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 14) | NC score | 0.065577 (rank : 19) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q60992 | Gene names | Vav2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-2. | |||||
|
AHI1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 15) | NC score | 0.082107 (rank : 12) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 537 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8N157, Q504T3, Q5TCP9, Q6P098, Q6PIT6, Q8NDX0, Q9H0H2 | Gene names | AHI1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Jouberin (Abelson helper integration site 1 protein homolog) (AHI-1). | |||||
|
VAV2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 16) | NC score | 0.064323 (rank : 20) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P52735 | Gene names | VAV2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-2. | |||||
|
PP2CG_HUMAN
|
||||||
| θ value | 0.163984 (rank : 17) | NC score | 0.052013 (rank : 33) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O15355 | Gene names | PPM1G, PPM1C | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform gamma (EC 3.1.3.16) (PP2C-gamma) (Protein phosphatase magnesium-dependent 1 gamma) (Protein phosphatase 1C). | |||||
|
PELP1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 18) | NC score | 0.074585 (rank : 14) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8IZL8, O15450, Q5EGN3, Q6NTE6, Q96FT1, Q9BU60 | Gene names | PELP1, HMX3, MNAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3). | |||||
|
CENPB_MOUSE
|
||||||
| θ value | 0.279714 (rank : 19) | NC score | 0.062157 (rank : 24) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P27790 | Gene names | Cenpb, Cenp-b | |||
|
Domain Architecture |

|
|||||
| Description | Major centromere autoantigen B (Centromere protein B) (CENP-B). | |||||
|
NFM_MOUSE
|
||||||
| θ value | 0.279714 (rank : 20) | NC score | 0.018879 (rank : 96) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
RPGR1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 21) | NC score | 0.049889 (rank : 37) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
TRI44_HUMAN
|
||||||
| θ value | 0.365318 (rank : 22) | NC score | 0.052295 (rank : 32) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96DX7, Q96QY2, Q9UGK0 | Gene names | TRIM44, DIPB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB). | |||||
|
PELP1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 23) | NC score | 0.071947 (rank : 15) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
ATBF1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 24) | NC score | 0.027282 (rank : 82) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
HTSF1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 25) | NC score | 0.082999 (rank : 11) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O43719, Q59G06, Q99730 | Gene names | HTATSF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 (Tat-SF1). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 0.62314 (rank : 26) | NC score | 0.034265 (rank : 65) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
TRDN_HUMAN
|
||||||
| θ value | 0.62314 (rank : 27) | NC score | 0.042596 (rank : 48) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
ITSN2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 28) | NC score | 0.039270 (rank : 55) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1371 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Z0R6, Q9Z0R5 | Gene names | Itsn2, Ese2, Sh3d1B | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (EH and SH3 domains protein 2) (EH domain and SH3 domain regulator of endocytosis 2). | |||||
|
CLSPN_HUMAN
|
||||||
| θ value | 1.06291 (rank : 29) | NC score | 0.044496 (rank : 46) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9HAW4, Q5VYG0, Q6P6H5, Q8IWI1 | Gene names | CLSPN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin (hClaspin) (Hu-Claspin). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 30) | NC score | 0.028093 (rank : 77) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
POT15_HUMAN
|
||||||
| θ value | 1.06291 (rank : 31) | NC score | 0.030558 (rank : 74) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6S5H4, Q6NXN7, Q6S5H7 | Gene names | POTE15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prostate, ovary, testis-expressed protein on chromosome 15. | |||||
|
SRGP1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 32) | NC score | 0.039998 (rank : 54) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7Z6B7, Q9H8A3, Q9P2P2 | Gene names | SRGAP1, ARHGAP13, KIAA1304 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 1 (srGAP1) (Rho-GTPase- activating protein 13). | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | 1.06291 (rank : 33) | NC score | 0.047245 (rank : 42) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
CCDC9_MOUSE
|
||||||
| θ value | 1.38821 (rank : 34) | NC score | 0.045047 (rank : 45) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8VC31 | Gene names | Ccdc9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 9. | |||||
|
ITSN2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 35) | NC score | 0.040826 (rank : 52) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1173 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NZM3, O95062, Q15812, Q9HAK4, Q9NXE6, Q9NYG0, Q9NZM2, Q9ULG4 | Gene names | ITSN2, KIAA1256, SH3D1B, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (SH3P18) (SH3P18-like WASP-associated protein). | |||||
|
SRCA_HUMAN
|
||||||
| θ value | 1.38821 (rank : 36) | NC score | 0.038657 (rank : 59) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
TCGAP_MOUSE
|
||||||
| θ value | 1.38821 (rank : 37) | NC score | 0.045244 (rank : 44) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q80YF9, Q6P7W6 | Gene names | Snx26, Tcgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
TRI44_MOUSE
|
||||||
| θ value | 1.38821 (rank : 38) | NC score | 0.047302 (rank : 41) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9QXA7, Q80SV9, Q9JHR8 | Gene names | Trim44, Dipb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB) (Protein Mc7). | |||||
|
AN32E_HUMAN
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.024822 (rank : 85) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BTT0, Q8N1S4, Q8WWW9 | Gene names | ANP32E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member E (LANP- like protein) (LANP-L). | |||||
|
ATBF1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.022781 (rank : 89) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
B3GL2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 41) | NC score | 0.030702 (rank : 73) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NCR0, Q59GR3, Q5TCI3, Q96AL7 | Gene names | B3GALNT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UDP-GalNAc:beta-1,3-N-acetylgalactosaminyltransferase 2 (EC 2.4.1.-) (Beta-1,3-N-acetylgalactosaminyltransferase II) (Beta-3-GalNAc-T2). | |||||
|
CNGB1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.021278 (rank : 93) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14028, Q14029 | Gene names | CNGB1, CNCG4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel 4 (CNG channel 4) (CNG-4) (CNG4) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
GLU2B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 43) | NC score | 0.032937 (rank : 67) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P14314, Q96BU9, Q96D06, Q9P0W9 | Gene names | PRKCSH, G19P1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glucosidase 2 subunit beta precursor (Glucosidase II subunit beta) (Protein kinase C substrate, 60.1 kDa protein, heavy chain) (PKCSH) (80K-H protein). | |||||
|
HCLS1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 44) | NC score | 0.053101 (rank : 30) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P14317 | Gene names | HCLS1, HS1 | |||
|
Domain Architecture |

|
|||||
| Description | Hematopoietic lineage cell-specific protein (Hematopoietic cell- specific LYN substrate 1) (LckBP1) (p75). | |||||
|
PININ_HUMAN
|
||||||
| θ value | 1.81305 (rank : 45) | NC score | 0.033068 (rank : 66) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9H307, O60899, Q53EM7, Q6P5X4, Q7KYL1, Q99738, Q9UHZ9, Q9UQR9 | Gene names | PNN, DRS, MEMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin (140 kDa nuclear and cell adhesion-related phosphoprotein) (Domain-rich serine protein) (DRS-protein) (DRSP) (Melanoma metastasis clone A protein) (Desmosome-associated protein) (SR-like protein) (Nuclear protein SDK3). | |||||
|
SET1A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 46) | NC score | 0.041554 (rank : 49) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
ANR21_HUMAN
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.028012 (rank : 79) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q86YR6 | Gene names | ANKRD21, POTE | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 21 (Protein POTE) (Prostate, ovary, testis-expressed protein). | |||||
|
AP3B2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 48) | NC score | 0.027493 (rank : 81) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q13367, O14808 | Gene names | AP3B2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (AP-3 complex beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain) (Neuron-specific vesicle coat protein beta-NAP). | |||||
|
BRD8_HUMAN
|
||||||
| θ value | 2.36792 (rank : 49) | NC score | 0.051455 (rank : 35) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9H0E9, O43178, Q15355, Q59GN0, Q969M9 | Gene names | BRD8, SMAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8 (p120) (Skeletal muscle abundant protein) (Thyroid hormone receptor coactivating protein 120kDa) (TrCP120). | |||||
|
NEUM_MOUSE
|
||||||
| θ value | 2.36792 (rank : 50) | NC score | 0.043418 (rank : 47) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P06837 | Gene names | Gap43, Basp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (Calmodulin-binding protein P-57). | |||||
|
PACN1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 51) | NC score | 0.047450 (rank : 40) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 320 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BY11, Q9P2G8 | Gene names | PACSIN1, KIAA1379 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C and casein kinase substrate in neurons protein 1. | |||||
|
PACN1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 52) | NC score | 0.047911 (rank : 39) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q61644 | Gene names | Pacsin1, Pacsin | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C and casein kinase substrate in neurons protein 1. | |||||
|
C2TA_MOUSE
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | 0.022950 (rank : 88) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P79621, O46787, O78036, O78109, Q31115, Q9TPP1 | Gene names | Ciita, C2ta, Mhc2ta | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
CD2L2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.004975 (rank : 111) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
DAXX_HUMAN
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.039003 (rank : 56) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UER7, O14747, O15141, O15208, Q9BWI3 | Gene names | DAXX, BING2, DAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Death domain-associated protein 6 (Daxx) (hDaxx) (Fas death domain- associated protein) (ETS1-associated protein 1) (EAP1). | |||||
|
DMP1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | 0.062566 (rank : 23) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
DYSF_MOUSE
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.027901 (rank : 80) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9ESD7, Q9QXC0 | Gene names | Dysf, Fer1l1 | |||
|
Domain Architecture |

|
|||||
| Description | Dysferlin (Dystrophy-associated fer-1-like protein) (Fer-1-like protein 1). | |||||
|
IMA3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.019118 (rank : 95) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35344 | Gene names | Kpna3 | |||
|
Domain Architecture |
|
|||||
| Description | Importin alpha-3 subunit (Karyopherin alpha-3 subunit) (Importin alpha Q2). | |||||
|
RGP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.030003 (rank : 75) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P46061, Q60801 | Gene names | Rangap1, Fug1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
CDN1C_MOUSE
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.038820 (rank : 57) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P49919 | Gene names | Cdkn1c, Kip2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
CTR9_MOUSE
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.038684 (rank : 58) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q62018, Q3UFF5, Q3UY40, Q66JX4, Q7TPS6, Q8BND9, Q8BRD1, Q8C9W7, Q8C9Y3, Q8CHI1 | Gene names | Ctr9, Kiaa0155, Sh2bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1) (Tetratricopeptide repeat-containing, SH2-binding phosphoprotein of 150 kDa) (TPR-containing, SH2-binding phosphoprotein of 150 kDa) (p150TSP). | |||||
|
FA48A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.026358 (rank : 83) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7TT00, Q8BG53, Q9JLS9 | Gene names | Fam48a, D3Ertd300e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM48A (p38-interacting protein) (p38IP). | |||||
|
KIFA3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.032884 (rank : 68) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P70188, P70189 | Gene names | Kifap3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-associated protein 3 (KAP3). | |||||
|
LAS1L_HUMAN
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.034496 (rank : 64) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y4W2, Q5JXQ0, Q8TEN5, Q9H9V5 | Gene names | LAS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LAS1-like protein. | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.004907 (rank : 112) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
SMRC2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 66) | NC score | 0.032724 (rank : 70) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6PDG5, Q6P6P3 | Gene names | Smarcc2, Baf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
TEX14_HUMAN
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.017121 (rank : 100) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1103 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8IWB6, Q7RTP3, Q8ND97, Q9BXT9 | Gene names | TEX14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed protein 14 (Testis-expressed sequence 14). | |||||
|
TTBK1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.009733 (rank : 106) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1058 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5TCY1, Q2L6C6, Q8N444, Q96JH2 | Gene names | TTBK1, BDTK, KIAA1855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 1 (EC 2.7.11.1) (Brain-derived tau kinase). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.017923 (rank : 97) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
BRD8_MOUSE
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.051192 (rank : 36) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
CCD96_HUMAN
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.023847 (rank : 87) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 595 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q2M329, Q8N2I7 | Gene names | CCDC96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 96. | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.021511 (rank : 92) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
FNBP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.036358 (rank : 63) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O75044 | Gene names | SRGAP2, FNBP2, KIAA0456 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 2 (srGAP2) (Formin-binding protein 2). | |||||
|
IMA3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.017490 (rank : 99) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00505, O00191, O43195, Q96AA7 | Gene names | KPNA3 | |||
|
Domain Architecture |
|
|||||
| Description | Importin alpha-3 subunit (Karyopherin alpha-3 subunit) (SRP1-gamma). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.084770 (rank : 10) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
MELPH_MOUSE
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.021663 (rank : 91) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91V27, Q99N53 | Gene names | Mlph, Ln, Slac2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanophilin (Exophilin-3) (Leaden protein) (Synaptotagmin-like protein 2a) (Slac2-a) (Slp homolog lacking C2 domains a). | |||||
|
NUCL_HUMAN
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.014320 (rank : 103) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P19338, Q53SK1, Q8NB06 | Gene names | NCL | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
PACN3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.046755 (rank : 43) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UKS6, Q9H331, Q9NWV9 | Gene names | PACSIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C and casein kinase substrate in neurons protein 3 (SH3 domain-containing protein 6511) (Endophilin I). | |||||
|
PACN3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.048792 (rank : 38) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q99JB8, Q9EQP9 | Gene names | Pacsin3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C and casein kinase II substrate protein 3. | |||||
|
RCOR1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.013267 (rank : 104) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CFE3, Q3V092, Q8BK28, Q8CHI2 | Gene names | Rcor1, D12Wsu95e, Kiaa0071 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 1 (Protein CoREST). | |||||
|
RPGR_MOUSE
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.040072 (rank : 53) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9R0X5, O88408, Q9CU92 | Gene names | Rpgr | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator (mRpgr). | |||||
|
SALL4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.003738 (rank : 116) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 799 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UJQ4 | Gene names | SALL4 | |||
|
Domain Architecture |

|
|||||
| Description | Sal-like protein 4 (Zinc finger protein SALL4). | |||||
|
ZN649_HUMAN
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | -0.000448 (rank : 118) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BS31, Q9H9N2 | Gene names | ZNF649 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 649. | |||||
|
CCD27_HUMAN
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.015919 (rank : 102) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 594 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q2M243, Q5TBV3, Q96M50 | Gene names | CCDC27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 27. | |||||
|
DMP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.063769 (rank : 21) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
DYSF_HUMAN
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.025895 (rank : 84) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75923, O75696, Q9UEN7 | Gene names | DYSF, FER1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Dysferlin (Dystrophy-associated fer-1-like protein) (Fer-1-like protein 1). | |||||
|
IASPP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.032850 (rank : 69) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
KI13B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.005974 (rank : 110) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NQT8, O75134, Q9BYJ6 | Gene names | KIF13B, GAKIN, KIAA0639 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF13B (Kinesin-like protein GAKIN). | |||||
|
MDM2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.017830 (rank : 98) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P23804, Q61040, Q64330 | Gene names | Mdm2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3 Mdm2 (EC 6.3.2.-) (p53-binding protein Mdm2) (Oncoprotein Mdm2) (Double minute 2 protein). | |||||
|
STAC3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.038136 (rank : 60) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96MF2, Q96HU5 | Gene names | STAC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3 and cysteine-rich domain-containing protein 3. | |||||
|
STAC3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.037817 (rank : 62) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BZ71 | Gene names | Stac3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3 and cysteine-rich domain-containing protein 3. | |||||
|
STOX1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.024590 (rank : 86) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZVD7, Q4F8Q6, Q5I946, Q5I947, Q5I948, Q5VX38, Q5VX39, Q6ZRY3, Q96LR3, Q96LS0 | Gene names | STOX1, C10orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Storkhead-box protein 1 (Winged helix domain-containing protein). | |||||
|
TCF8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.007198 (rank : 109) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P37275, Q12924, Q13800 | Gene names | TCF8, AREB6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (NIL-2-A zinc finger protein) (Negative regulator of IL2). | |||||
|
VCX1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.040970 (rank : 51) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H320, Q9P0H3 | Gene names | VCX, VCX1, VCX10R, VCXB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 1 (VCX-B1 protein) (Variably charged protein X-B1) (Variable charge protein on X with ten repeats) (VCX- 10r). | |||||
|
YTDC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.041037 (rank : 50) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
ZBTB4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.002616 (rank : 117) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 926 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9P1Z0, Q7Z697, Q86XJ4, Q8N4V8 | Gene names | ZBTB4, KIAA1538 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 4 (KAISO-like zinc finger protein 1) (KAISO-L1). | |||||
|
AATF_MOUSE
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.020461 (rank : 94) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JKX4, Q7TQN1, Q8C5Q2, Q99P89 | Gene names | Aatf, Che1, Trb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein AATF (Apoptosis-antagonizing transcription factor) (Rb-binding protein Che-1) (Traube protein). | |||||
|
ABLM1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.004705 (rank : 113) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K4G5, Q80U86, Q8BIR9, Q8K4G3, Q8K4G4 | Gene names | Ablim1, Ablim, Kiaa0059 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding LIM protein 1 (Actin-binding LIM protein family member 1) (abLIM-1). | |||||
|
CDN2D_MOUSE
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.008840 (rank : 108) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q60773, Q60794 | Gene names | Cdkn2d | |||
|
Domain Architecture |
|
|||||
| Description | Cyclin-dependent kinase 4 inhibitor D (p19-INK4d). | |||||
|
CENPF_HUMAN
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.009612 (rank : 107) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
CTR9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.032653 (rank : 71) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6PD62, Q15015 | Gene names | CTR9, KIAA0155, SH2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1). | |||||
|
DDX10_HUMAN
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.004511 (rank : 114) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13206, Q5BJD8 | Gene names | DDX10 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
GNAS3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.030803 (rank : 72) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z0F1, Q9QXW5, Q9Z0H2 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroendocrine secretory protein 55 (NESP55) [Contains: LHAL tetrapeptide; GPIPIRRH peptide]. | |||||
|
KIFA3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.028029 (rank : 78) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92845, Q8NHU7, Q9H416 | Gene names | KIFAP3, SMAP | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-associated protein 3 (Smg GDS-associated protein). | |||||
|
LSP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.029277 (rank : 76) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P19973, P97339, Q04950, Q62022, Q62023, Q62024, Q8CD28, Q99L65 | Gene names | Lsp1, Pp52, S37, Wp34 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte-specific protein 1 (Protein pp52) (52 kDa phosphoprotein) (Lymphocyte-specific antigen WP34) (S37 protein). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.021983 (rank : 90) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
SV2C_HUMAN
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.010265 (rank : 105) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q496J9, Q496K1, Q9UPU8 | Gene names | SV2C, KIAA1054 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptic vesicle glycoprotein 2C. | |||||
|
TE2IP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.016786 (rank : 101) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91VL8, Q543F4, Q9JJE8, Q9JJE9 | Gene names | Terf2ip, Rap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomeric repeat-binding factor 2-interacting protein 1 (TRF2- interacting telomeric protein Rap1). | |||||
|
VCAM1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.004501 (rank : 115) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 320 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P29533 | Gene names | Vcam1, Vcam-1 | |||
|
Domain Architecture |

|
|||||
| Description | Vascular cell adhesion protein 1 precursor (V-CAM 1). | |||||
|
AKA12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.060297 (rank : 26) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
ERIC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.068905 (rank : 17) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
LEO1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.063671 (rank : 22) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.066332 (rank : 18) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
NASP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.061570 (rank : 25) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
NSBP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.069727 (rank : 16) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
TCAL5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.056960 (rank : 28) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
VAV3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.052333 (rank : 31) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 398 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UKW4, O95230, Q9Y5X8 | Gene names | VAV3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-3. | |||||
|
VAV3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.051787 (rank : 34) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 404 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9R0C8, Q7TS85 | Gene names | Vav3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-3. | |||||
|
MIA2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 109 | |
| SwissProt Accessions | Q91ZV0 | Gene names | Mia2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma inhibitory activity protein 2 precursor. | |||||
|
MIA2_HUMAN
|
||||||
| NC score | 0.937681 (rank : 2) | θ value | 2.26584e-152 (rank : 2) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96PC5, Q9H6C1 | Gene names | MIA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma inhibitory activity protein 2 precursor. | |||||
|
OTOR_HUMAN
|
||||||
| NC score | 0.584114 (rank : 3) | θ value | 8.67504e-11 (rank : 3) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NRC9 | Gene names | OTOR, FDP, MIAL | |||
|
Domain Architecture |

|
|||||
| Description | Otoraplin precursor (Fibrocyte-derived protein) (Melanoma inhibitory activity-like protein). | |||||
|
MIA_HUMAN
|
||||||
| NC score | 0.564074 (rank : 4) | θ value | 5.62301e-10 (rank : 4) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q16674 | Gene names | MIA | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-derived growth regulatory protein precursor (Melanoma inhibitory activity). | |||||
|
OTOR_MOUSE
|
||||||
| NC score | 0.559086 (rank : 5) | θ value | 4.76016e-09 (rank : 5) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JIE3 | Gene names | Otor, Mial | |||
|
Domain Architecture |

|
|||||
| Description | Otoraplin precursor (Melanoma inhibitory activity-like protein). | |||||
|
MIA_MOUSE
|
||||||
| NC score | 0.551854 (rank : 6) | θ value | 1.38499e-08 (rank : 6) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61865, O09086, P97495 | Gene names | Mia, Cdrap, Mia1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-derived growth regulatory protein precursor (Melanoma inhibitory activity) (Cartilage-derived retinoic acid-sensitive protein) (CD-RAP). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.109448 (rank : 7) | θ value | 0.0431538 (rank : 13) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
HTSF1_MOUSE
|
||||||
| NC score | 0.103643 (rank : 8) | θ value | 0.0113563 (rank : 9) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BGC0, Q1WWK0, Q9CT41, Q9DAU3 | Gene names | Htatsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 homolog. | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.090196 (rank : 9) | θ value | 0.000270298 (rank : 7) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.084770 (rank : 10) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
HTSF1_HUMAN
|
||||||
| NC score | 0.082999 (rank : 11) | θ value | 0.62314 (rank : 25) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O43719, Q59G06, Q99730 | Gene names | HTATSF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 (Tat-SF1). | |||||
|
AHI1_HUMAN
|
||||||
| NC score | 0.082107 (rank : 12) | θ value | 0.0736092 (rank : 15) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 537 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8N157, Q504T3, Q5TCP9, Q6P098, Q6PIT6, Q8NDX0, Q9H0H2 | Gene names | AHI1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Jouberin (Abelson helper integration site 1 protein homolog) (AHI-1). | |||||
|
AHI1_MOUSE
|
||||||
| NC score | 0.078199 (rank : 13) | θ value | 0.0330416 (rank : 12) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8K3E5, Q7TNV2, Q9CVY1 | Gene names | Ahi1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Jouberin (Abelson helper integration site 1 protein) (AHI-1). | |||||
|
PELP1_HUMAN
|
||||||
| NC score | 0.074585 (rank : 14) | θ value | 0.21417 (rank : 18) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8IZL8, O15450, Q5EGN3, Q6NTE6, Q96FT1, Q9BU60 | Gene names | PELP1, HMX3, MNAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3). | |||||
|
PELP1_MOUSE
|
||||||
| NC score | 0.071947 (rank : 15) | θ value | 0.47712 (rank : 23) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
NSBP1_MOUSE
|
||||||
| NC score | 0.069727 (rank : 16) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
ERIC1_HUMAN
|
||||||
| NC score | 0.068905 (rank : 17) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.066332 (rank : 18) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
VAV2_MOUSE
|
||||||
| NC score | 0.065577 (rank : 19) | θ value | 0.0563607 (rank : 14) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q60992 | Gene names | Vav2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-2. | |||||
|
VAV2_HUMAN
|
||||||
| NC score | 0.064323 (rank : 20) | θ value | 0.0961366 (rank : 16) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P52735 | Gene names | VAV2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-2. | |||||
|
DMP1_MOUSE
|
||||||
| NC score | 0.063769 (rank : 21) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.063671 (rank : 22) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
DMP1_HUMAN
|
||||||
| NC score | 0.062566 (rank : 23) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
CENPB_MOUSE
|
||||||
| NC score | 0.062157 (rank : 24) | θ value | 0.279714 (rank : 19) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P27790 | Gene names | Cenpb, Cenp-b | |||
|
Domain Architecture |

|
|||||
| Description | Major centromere autoantigen B (Centromere protein B) (CENP-B). | |||||
|
NASP_HUMAN
|
||||||
| NC score | 0.061570 (rank : 25) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.060297 (rank : 26) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
APLP2_HUMAN
|
||||||
| NC score | 0.060077 (rank : 27) | θ value | 0.0113563 (rank : 8) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q06481 | Gene names | APLP2, APPL2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 2 precursor (Amyloid protein homolog) (APPH) (CDEI box-binding protein) (CDEBP). | |||||
|
TCAL5_HUMAN
|
||||||
| NC score | 0.056960 (rank : 28) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
MYT1_HUMAN
|
||||||
| NC score | 0.054075 (rank : 29) | θ value | 0.0252991 (rank : 11) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q01538, O94922, Q9UPV2 | Gene names | MYT1, KIAA0835, KIAA1050, MTF1, MYTI, PLPB1 | |||
|
Domain Architecture |

|
|||||
| Description | Myelin transcription factor 1 (MyT1) (MyTI) (Proteolipid protein- binding protein) (PLPB1). | |||||
|
HCLS1_HUMAN
|
||||||
| NC score | 0.053101 (rank : 30) | θ value | 1.81305 (rank : 44) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P14317 | Gene names | HCLS1, HS1 | |||
|
Domain Architecture |

|
|||||
| Description | Hematopoietic lineage cell-specific protein (Hematopoietic cell- specific LYN substrate 1) (LckBP1) (p75). | |||||
|
VAV3_HUMAN
|
||||||
| NC score | 0.052333 (rank : 31) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 398 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UKW4, O95230, Q9Y5X8 | Gene names | VAV3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-3. | |||||
|
TRI44_HUMAN
|
||||||
| NC score | 0.052295 (rank : 32) | θ value | 0.365318 (rank : 22) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96DX7, Q96QY2, Q9UGK0 | Gene names | TRIM44, DIPB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB). | |||||
|
PP2CG_HUMAN
|
||||||
| NC score | 0.052013 (rank : 33) | θ value | 0.163984 (rank : 17) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O15355 | Gene names | PPM1G, PPM1C | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform gamma (EC 3.1.3.16) (PP2C-gamma) (Protein phosphatase magnesium-dependent 1 gamma) (Protein phosphatase 1C). | |||||
|
VAV3_MOUSE
|
||||||
| NC score | 0.051787 (rank : 34) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 404 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9R0C8, Q7TS85 | Gene names | Vav3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-3. | |||||
|
BRD8_HUMAN
|
||||||
| NC score | 0.051455 (rank : 35) | θ value | 2.36792 (rank : 49) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9H0E9, O43178, Q15355, Q59GN0, Q969M9 | Gene names | BRD8, SMAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8 (p120) (Skeletal muscle abundant protein) (Thyroid hormone receptor coactivating protein 120kDa) (TrCP120). | |||||
|
BRD8_MOUSE
|
||||||
| NC score | 0.051192 (rank : 36) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
RPGR1_MOUSE
|
||||||
| NC score | 0.049889 (rank : 37) | θ value | 0.279714 (rank : 21) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
PACN3_MOUSE
|
||||||
| NC score | 0.048792 (rank : 38) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q99JB8, Q9EQP9 | Gene names | Pacsin3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C and casein kinase II substrate protein 3. | |||||
|
PACN1_MOUSE
|
||||||
| NC score | 0.047911 (rank : 39) | θ value | 2.36792 (rank : 52) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q61644 | Gene names | Pacsin1, Pacsin | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C and casein kinase substrate in neurons protein 1. | |||||
|
PACN1_HUMAN
|
||||||
| NC score | 0.047450 (rank : 40) | θ value | 2.36792 (rank : 51) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 320 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BY11, Q9P2G8 | Gene names | PACSIN1, KIAA1379 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C and casein kinase substrate in neurons protein 1. | |||||
|
TRI44_MOUSE
|
||||||
| NC score | 0.047302 (rank : 41) | θ value | 1.38821 (rank : 38) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9QXA7, Q80SV9, Q9JHR8 | Gene names | Trim44, Dipb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB) (Protein Mc7). | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.047245 (rank : 42) | θ value | 1.06291 (rank : 33) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
PACN3_HUMAN
|
||||||
| NC score | 0.046755 (rank : 43) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UKS6, Q9H331, Q9NWV9 | Gene names | PACSIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C and casein kinase substrate in neurons protein 3 (SH3 domain-containing protein 6511) (Endophilin I). | |||||
|
TCGAP_MOUSE
|
||||||
| NC score | 0.045244 (rank : 44) | θ value | 1.38821 (rank : 37) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q80YF9, Q6P7W6 | Gene names | Snx26, Tcgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
CCDC9_MOUSE
|
||||||
| NC score | 0.045047 (rank : 45) | θ value | 1.38821 (rank : 34) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8VC31 | Gene names | Ccdc9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 9. | |||||
|
CLSPN_HUMAN
|
||||||
| NC score | 0.044496 (rank : 46) | θ value | 1.06291 (rank : 29) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9HAW4, Q5VYG0, Q6P6H5, Q8IWI1 | Gene names | CLSPN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin (hClaspin) (Hu-Claspin). | |||||
|
NEUM_MOUSE
|
||||||
| NC score | 0.043418 (rank : 47) | θ value | 2.36792 (rank : 50) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P06837 | Gene names | Gap43, Basp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (Calmodulin-binding protein P-57). | |||||
|
TRDN_HUMAN
|
||||||
| NC score | 0.042596 (rank : 48) | θ value | 0.62314 (rank : 27) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.041554 (rank : 49) | θ value | 1.81305 (rank : 46) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
YTDC1_HUMAN
|
||||||
| NC score | 0.041037 (rank : 50) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
VCX1_HUMAN
|
||||||
| NC score | 0.040970 (rank : 51) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H320, Q9P0H3 | Gene names | VCX, VCX1, VCX10R, VCXB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 1 (VCX-B1 protein) (Variably charged protein X-B1) (Variable charge protein on X with ten repeats) (VCX- 10r). | |||||
|
ITSN2_HUMAN
|
||||||
| NC score | 0.040826 (rank : 52) | θ value | 1.38821 (rank : 35) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1173 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NZM3, O95062, Q15812, Q9HAK4, Q9NXE6, Q9NYG0, Q9NZM2, Q9ULG4 | Gene names | ITSN2, KIAA1256, SH3D1B, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (SH3P18) (SH3P18-like WASP-associated protein). | |||||
|
RPGR_MOUSE
|
||||||
| NC score | 0.040072 (rank : 53) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9R0X5, O88408, Q9CU92 | Gene names | Rpgr | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator (mRpgr). | |||||
|
SRGP1_HUMAN
|
||||||
| NC score | 0.039998 (rank : 54) | θ value | 1.06291 (rank : 32) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7Z6B7, Q9H8A3, Q9P2P2 | Gene names | SRGAP1, ARHGAP13, KIAA1304 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 1 (srGAP1) (Rho-GTPase- activating protein 13). | |||||
|
ITSN2_MOUSE
|
||||||
| NC score | 0.039270 (rank : 55) | θ value | 0.813845 (rank : 28) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1371 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Z0R6, Q9Z0R5 | Gene names | Itsn2, Ese2, Sh3d1B | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (EH and SH3 domains protein 2) (EH domain and SH3 domain regulator of endocytosis 2). | |||||
|
DAXX_HUMAN
|
||||||
| NC score | 0.039003 (rank : 56) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UER7, O14747, O15141, O15208, Q9BWI3 | Gene names | DAXX, BING2, DAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Death domain-associated protein 6 (Daxx) (hDaxx) (Fas death domain- associated protein) (ETS1-associated protein 1) (EAP1). | |||||
|
CDN1C_MOUSE
|
||||||
| NC score | 0.038820 (rank : 57) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P49919 | Gene names | Cdkn1c, Kip2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
CTR9_MOUSE
|
||||||
| NC score | 0.038684 (rank : 58) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q62018, Q3UFF5, Q3UY40, Q66JX4, Q7TPS6, Q8BND9, Q8BRD1, Q8C9W7, Q8C9Y3, Q8CHI1 | Gene names | Ctr9, Kiaa0155, Sh2bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1) (Tetratricopeptide repeat-containing, SH2-binding phosphoprotein of 150 kDa) (TPR-containing, SH2-binding phosphoprotein of 150 kDa) (p150TSP). | |||||
|
SRCA_HUMAN
|
||||||
| NC score | 0.038657 (rank : 59) | θ value | 1.38821 (rank : 36) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
STAC3_HUMAN
|
||||||
| NC score | 0.038136 (rank : 60) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96MF2, Q96HU5 | Gene names | STAC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3 and cysteine-rich domain-containing protein 3. | |||||
|
ERCC6_HUMAN
|
||||||
| NC score | 0.037913 (rank : 61) | θ value | 0.0252991 (rank : 10) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q03468 | Gene names | ERCC6, CSB | |||
|
Domain Architecture |

|
|||||
| Description | DNA excision repair protein ERCC-6 (EC 3.6.1.-) (ATP-dependent helicase ERCC6) (Cockayne syndrome protein CSB). | |||||
|
STAC3_MOUSE
|
||||||
| NC score | 0.037817 (rank : 62) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BZ71 | Gene names | Stac3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3 and cysteine-rich domain-containing protein 3. | |||||
|
FNBP2_HUMAN
|
||||||
| NC score | 0.036358 (rank : 63) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O75044 | Gene names | SRGAP2, FNBP2, KIAA0456 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 2 (srGAP2) (Formin-binding protein 2). | |||||
|
LAS1L_HUMAN
|
||||||
| NC score | 0.034496 (rank : 64) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y4W2, Q5JXQ0, Q8TEN5, Q9H9V5 | Gene names | LAS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LAS1-like protein. | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.034265 (rank : 65) | θ value | 0.62314 (rank : 26) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PININ_HUMAN
|
||||||
| NC score | 0.033068 (rank : 66) | θ value | 1.81305 (rank : 45) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9H307, O60899, Q53EM7, Q6P5X4, Q7KYL1, Q99738, Q9UHZ9, Q9UQR9 | Gene names | PNN, DRS, MEMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin (140 kDa nuclear and cell adhesion-related phosphoprotein) (Domain-rich serine protein) (DRS-protein) (DRSP) (Melanoma metastasis clone A protein) (Desmosome-associated protein) (SR-like protein) (Nuclear protein SDK3). | |||||
|
GLU2B_HUMAN
|
||||||
| NC score | 0.032937 (rank : 67) | θ value | 1.81305 (rank : 43) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P14314, Q96BU9, Q96D06, Q9P0W9 | Gene names | PRKCSH, G19P1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glucosidase 2 subunit beta precursor (Glucosidase II subunit beta) (Protein kinase C substrate, 60.1 kDa protein, heavy chain) (PKCSH) (80K-H protein). | |||||
|
KIFA3_MOUSE
|
||||||
| NC score | 0.032884 (rank : 68) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P70188, P70189 | Gene names | Kifap3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-associated protein 3 (KAP3). | |||||
|
IASPP_MOUSE
|
||||||
| NC score | 0.032850 (rank : 69) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
SMRC2_MOUSE
|
||||||
| NC score | 0.032724 (rank : 70) | θ value | 4.03905 (rank : 66) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6PDG5, Q6P6P3 | Gene names | Smarcc2, Baf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
CTR9_HUMAN
|
||||||
| NC score | 0.032653 (rank : 71) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6PD62, Q15015 | Gene names | CTR9, KIAA0155, SH2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1). | |||||
|
GNAS3_MOUSE
|
||||||
| NC score | 0.030803 (rank : 72) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z0F1, Q9QXW5, Q9Z0H2 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroendocrine secretory protein 55 (NESP55) [Contains: LHAL tetrapeptide; GPIPIRRH peptide]. | |||||
|
B3GL2_HUMAN
|
||||||
| NC score | 0.030702 (rank : 73) | θ value | 1.81305 (rank : 41) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NCR0, Q59GR3, Q5TCI3, Q96AL7 | Gene names | B3GALNT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UDP-GalNAc:beta-1,3-N-acetylgalactosaminyltransferase 2 (EC 2.4.1.-) (Beta-1,3-N-acetylgalactosaminyltransferase II) (Beta-3-GalNAc-T2). | |||||
|
POT15_HUMAN
|
||||||
| NC score | 0.030558 (rank : 74) | θ value | 1.06291 (rank : 31) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6S5H4, Q6NXN7, Q6S5H7 | Gene names | POTE15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prostate, ovary, testis-expressed protein on chromosome 15. | |||||
|
RGP1_MOUSE
|
||||||
| NC score | 0.030003 (rank : 75) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P46061, Q60801 | Gene names | Rangap1, Fug1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
LSP1_MOUSE
|
||||||
| NC score | 0.029277 (rank : 76) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P19973, P97339, Q04950, Q62022, Q62023, Q62024, Q8CD28, Q99L65 | Gene names | Lsp1, Pp52, S37, Wp34 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte-specific protein 1 (Protein pp52) (52 kDa phosphoprotein) (Lymphocyte-specific antigen WP34) (S37 protein). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.028093 (rank : 77) | θ value | 1.06291 (rank : 30) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
KIFA3_HUMAN
|
||||||
| NC score | 0.028029 (rank : 78) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92845, Q8NHU7, Q9H416 | Gene names | KIFAP3, SMAP | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-associated protein 3 (Smg GDS-associated protein). | |||||
|
ANR21_HUMAN
|
||||||
| NC score | 0.028012 (rank : 79) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q86YR6 | Gene names | ANKRD21, POTE | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 21 (Protein POTE) (Prostate, ovary, testis-expressed protein). | |||||
|
DYSF_MOUSE
|
||||||
| NC score | 0.027901 (rank : 80) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9ESD7, Q9QXC0 | Gene names | Dysf, Fer1l1 | |||
|
Domain Architecture |

|
|||||
| Description | Dysferlin (Dystrophy-associated fer-1-like protein) (Fer-1-like protein 1). | |||||
|
AP3B2_HUMAN
|
||||||
| NC score | 0.027493 (rank : 81) | θ value | 2.36792 (rank : 48) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q13367, O14808 | Gene names | AP3B2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (AP-3 complex beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain) (Neuron-specific vesicle coat protein beta-NAP). | |||||
|
ATBF1_MOUSE
|
||||||
| NC score | 0.027282 (rank : 82) | θ value | 0.62314 (rank : 24) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
FA48A_MOUSE
|
||||||
| NC score | 0.026358 (rank : 83) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7TT00, Q8BG53, Q9JLS9 | Gene names | Fam48a, D3Ertd300e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM48A (p38-interacting protein) (p38IP). | |||||
|
DYSF_HUMAN
|
||||||
| NC score | 0.025895 (rank : 84) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75923, O75696, Q9UEN7 | Gene names | DYSF, FER1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Dysferlin (Dystrophy-associated fer-1-like protein) (Fer-1-like protein 1). | |||||
|
AN32E_HUMAN
|
||||||
| NC score | 0.024822 (rank : 85) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BTT0, Q8N1S4, Q8WWW9 | Gene names | ANP32E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member E (LANP- like protein) (LANP-L). | |||||
|
STOX1_HUMAN
|
||||||
| NC score | 0.024590 (rank : 86) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZVD7, Q4F8Q6, Q5I946, Q5I947, Q5I948, Q5VX38, Q5VX39, Q6ZRY3, Q96LR3, Q96LS0 | Gene names | STOX1, C10orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Storkhead-box protein 1 (Winged helix domain-containing protein). | |||||
|
CCD96_HUMAN
|
||||||
| NC score | 0.023847 (rank : 87) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 595 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q2M329, Q8N2I7 | Gene names | CCDC96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 96. | |||||
|
C2TA_MOUSE
|
||||||
| NC score | 0.022950 (rank : 88) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P79621, O46787, O78036, O78109, Q31115, Q9TPP1 | Gene names | Ciita, C2ta, Mhc2ta | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
ATBF1_HUMAN
|
||||||
| NC score | 0.022781 (rank : 89) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.021983 (rank : 90) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
MELPH_MOUSE
|
||||||
| NC score | 0.021663 (rank : 91) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91V27, Q99N53 | Gene names | Mlph, Ln, Slac2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanophilin (Exophilin-3) (Leaden protein) (Synaptotagmin-like protein 2a) (Slac2-a) (Slp homolog lacking C2 domains a). | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.021511 (rank : 92) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
CNGB1_HUMAN
|
||||||
| NC score | 0.021278 (rank : 93) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14028, Q14029 | Gene names | CNGB1, CNCG4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel 4 (CNG channel 4) (CNG-4) (CNG4) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
AATF_MOUSE
|
||||||
| NC score | 0.020461 (rank : 94) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JKX4, Q7TQN1, Q8C5Q2, Q99P89 | Gene names | Aatf, Che1, Trb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein AATF (Apoptosis-antagonizing transcription factor) (Rb-binding protein Che-1) (Traube protein). | |||||
|
IMA3_MOUSE
|
||||||
| NC score | 0.019118 (rank : 95) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35344 | Gene names | Kpna3 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-3 subunit (Karyopherin alpha-3 subunit) (Importin alpha Q2). | |||||
|
NFM_MOUSE
|
||||||
| NC score | 0.018879 (rank : 96) | θ value | 0.279714 (rank : 20) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.017923 (rank : 97) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
MDM2_MOUSE
|
||||||
| NC score | 0.017830 (rank : 98) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P23804, Q61040, Q64330 | Gene names | Mdm2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3 Mdm2 (EC 6.3.2.-) (p53-binding protein Mdm2) (Oncoprotein Mdm2) (Double minute 2 protein). | |||||
|
IMA3_HUMAN
|
||||||
| NC score | 0.017490 (rank : 99) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00505, O00191, O43195, Q96AA7 | Gene names | KPNA3 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-3 subunit (Karyopherin alpha-3 subunit) (SRP1-gamma). | |||||
|
TEX14_HUMAN
|
||||||
| NC score | 0.017121 (rank : 100) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1103 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8IWB6, Q7RTP3, Q8ND97, Q9BXT9 | Gene names | TEX14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed protein 14 (Testis-expressed sequence 14). | |||||
|
TE2IP_MOUSE
|
||||||
| NC score | 0.016786 (rank : 101) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91VL8, Q543F4, Q9JJE8, Q9JJE9 | Gene names | Terf2ip, Rap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomeric repeat-binding factor 2-interacting protein 1 (TRF2- interacting telomeric protein Rap1). | |||||
|
CCD27_HUMAN
|
||||||
| NC score | 0.015919 (rank : 102) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 594 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q2M243, Q5TBV3, Q96M50 | Gene names | CCDC27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 27. | |||||
|
NUCL_HUMAN
|
||||||
| NC score | 0.014320 (rank : 103) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P19338, Q53SK1, Q8NB06 | Gene names | NCL | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
RCOR1_MOUSE
|
||||||
| NC score | 0.013267 (rank : 104) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CFE3, Q3V092, Q8BK28, Q8CHI2 | Gene names | Rcor1, D12Wsu95e, Kiaa0071 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 1 (Protein CoREST). | |||||
|
SV2C_HUMAN
|
||||||
| NC score | 0.010265 (rank : 105) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q496J9, Q496K1, Q9UPU8 | Gene names | SV2C, KIAA1054 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptic vesicle glycoprotein 2C. | |||||
|
TTBK1_HUMAN
|
||||||
| NC score | 0.009733 (rank : 106) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1058 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5TCY1, Q2L6C6, Q8N444, Q96JH2 | Gene names | TTBK1, BDTK, KIAA1855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 1 (EC 2.7.11.1) (Brain-derived tau kinase). | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.009612 (rank : 107) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
CDN2D_MOUSE
|
||||||
| NC score | 0.008840 (rank : 108) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q60773, Q60794 | Gene names | Cdkn2d | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase 4 inhibitor D (p19-INK4d). | |||||
|
TCF8_HUMAN
|
||||||
| NC score | 0.007198 (rank : 109) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P37275, Q12924, Q13800 | Gene names | TCF8, AREB6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (NIL-2-A zinc finger protein) (Negative regulator of IL2). | |||||
|
KI13B_HUMAN
|
||||||
| NC score | 0.005974 (rank : 110) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NQT8, O75134, Q9BYJ6 | Gene names | KIF13B, GAKIN, KIAA0639 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF13B (Kinesin-like protein GAKIN). | |||||
|
CD2L2_HUMAN
|
||||||
| NC score | 0.004975 (rank : 111) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
MAST4_HUMAN
|
||||||
| NC score | 0.004907 (rank : 112) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
ABLM1_MOUSE
|
||||||
| NC score | 0.004705 (rank : 113) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K4G5, Q80U86, Q8BIR9, Q8K4G3, Q8K4G4 | Gene names | Ablim1, Ablim, Kiaa0059 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding LIM protein 1 (Actin-binding LIM protein family member 1) (abLIM-1). | |||||
|
DDX10_HUMAN
|
||||||
| NC score | 0.004511 (rank : 114) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13206, Q5BJD8 | Gene names | DDX10 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
VCAM1_MOUSE
|
||||||
| NC score | 0.004501 (rank : 115) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 320 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P29533 | Gene names | Vcam1, Vcam-1 | |||
|
Domain Architecture |

|
|||||
| Description | Vascular cell adhesion protein 1 precursor (V-CAM 1). | |||||
|
SALL4_HUMAN
|
||||||
| NC score | 0.003738 (rank : 116) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 799 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UJQ4 | Gene names | SALL4 | |||
|
Domain Architecture |

|
|||||
| Description | Sal-like protein 4 (Zinc finger protein SALL4). | |||||
|
ZBTB4_HUMAN
|
||||||
| NC score | 0.002616 (rank : 117) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 926 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9P1Z0, Q7Z697, Q86XJ4, Q8N4V8 | Gene names | ZBTB4, KIAA1538 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 4 (KAISO-like zinc finger protein 1) (KAISO-L1). | |||||
|
ZN649_HUMAN
|
||||||
| NC score | -0.000448 (rank : 118) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BS31, Q9H9N2 | Gene names | ZNF649 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 649. | |||||