Please be patient as the page loads
|
MIA2_HUMAN
|
||||||
| SwissProt Accessions | Q96PC5, Q9H6C1 | Gene names | MIA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma inhibitory activity protein 2 precursor. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MIA2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96PC5, Q9H6C1 | Gene names | MIA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma inhibitory activity protein 2 precursor. | |||||
|
MIA2_MOUSE
|
||||||
| θ value | 2.26584e-152 (rank : 2) | NC score | 0.937681 (rank : 2) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q91ZV0 | Gene names | Mia2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma inhibitory activity protein 2 precursor. | |||||
|
OTOR_HUMAN
|
||||||
| θ value | 2.0648e-12 (rank : 3) | NC score | 0.621471 (rank : 3) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NRC9 | Gene names | OTOR, FDP, MIAL | |||
|
Domain Architecture |
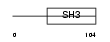
|
|||||
| Description | Otoraplin precursor (Fibrocyte-derived protein) (Melanoma inhibitory activity-like protein). | |||||
|
MIA_HUMAN
|
||||||
| θ value | 1.133e-10 (rank : 4) | NC score | 0.584551 (rank : 5) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q16674 | Gene names | MIA | |||
|
Domain Architecture |
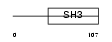
|
|||||
| Description | Melanoma-derived growth regulatory protein precursor (Melanoma inhibitory activity). | |||||
|
MIA_MOUSE
|
||||||
| θ value | 1.9326e-10 (rank : 5) | NC score | 0.574578 (rank : 6) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61865, O09086, P97495 | Gene names | Mia, Cdrap, Mia1 | |||
|
Domain Architecture |
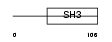
|
|||||
| Description | Melanoma-derived growth regulatory protein precursor (Melanoma inhibitory activity) (Cartilage-derived retinoic acid-sensitive protein) (CD-RAP). | |||||
|
OTOR_MOUSE
|
||||||
| θ value | 7.34386e-10 (rank : 6) | NC score | 0.591270 (rank : 4) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JIE3 | Gene names | Otor, Mial | |||
|
Domain Architecture |
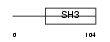
|
|||||
| Description | Otoraplin precursor (Melanoma inhibitory activity-like protein). | |||||
|
MYPT2_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 7) | NC score | 0.028525 (rank : 16) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 707 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60237, Q8N179, Q9HCB7, Q9HCB8 | Gene names | PPP1R12B, MYPT2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12B (Myosin phosphatase- targeting subunit 2) (Myosin phosphatase target subunit 2). | |||||
|
NPAS3_MOUSE
|
||||||
| θ value | 0.21417 (rank : 8) | NC score | 0.046967 (rank : 12) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QZQ0, Q9EQP4 | Gene names | Npas3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
TP53B_HUMAN
|
||||||
| θ value | 0.21417 (rank : 9) | NC score | 0.055574 (rank : 8) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q12888, Q2M1Z7, Q4LE46, Q5FWZ3, Q7Z3U4 | Gene names | TP53BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 10) | NC score | 0.049210 (rank : 11) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
NPAS3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 11) | NC score | 0.044942 (rank : 13) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IXF0, Q86US6, Q86US7, Q8IXF2, Q9BY81, Q9H323, Q9Y4L8 | Gene names | NPAS3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
SPT16_MOUSE
|
||||||
| θ value | 1.06291 (rank : 12) | NC score | 0.056034 (rank : 7) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q920B9, Q3TZ48, Q3UFH0, Q921H4 | Gene names | Supt16h, Fact140, Factp140 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FACT complex subunit SPT16 (Facilitates chromatin transcription complex subunit SPT16) (FACT 140 kDa subunit) (FACTp140) (Chromatin- specific transcription elongation factor 140 kDa subunit). | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 13) | NC score | 0.053125 (rank : 9) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
PIAS3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 14) | NC score | 0.020425 (rank : 21) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y6X2, Q9UFI3 | Gene names | PIAS3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein inhibitor of activated STAT protein 3. | |||||
|
RPGR_MOUSE
|
||||||
| θ value | 3.0926 (rank : 15) | NC score | 0.028714 (rank : 15) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9R0X5, O88408, Q9CU92 | Gene names | Rpgr | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator (mRpgr). | |||||
|
SPT16_HUMAN
|
||||||
| θ value | 3.0926 (rank : 16) | NC score | 0.050287 (rank : 10) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y5B9, Q6GMT8, Q6P2F1, Q6PJM1, Q9NRX0 | Gene names | SUPT16H, FACT140, FACTP140 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FACT complex subunit SPT16 (Facilitates chromatin transcription complex subunit SPT16) (hSPT16) (FACT 140 kDa subunit) (FACTp140) (Chromatin-specific transcription elongation factor 140 kDa subunit). | |||||
|
CF055_HUMAN
|
||||||
| θ value | 6.88961 (rank : 17) | NC score | 0.021765 (rank : 20) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NP79, Q9H0R2, Q9H3K9, Q9P0Q0 | Gene names | C6orf55 | |||
|
Domain Architecture |
|
|||||
| Description | Protein C6orf55 (Dopamine-responsive protein DRG-1). | |||||
|
CFDP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 18) | NC score | 0.024397 (rank : 17) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UEE9, O00393, O00404, Q9UEF0, Q9UEF1, Q9UEF8 | Gene names | CFDP1, BCNT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Craniofacial development protein 1 (Bucentaur). | |||||
|
CM007_MOUSE
|
||||||
| θ value | 6.88961 (rank : 19) | NC score | 0.014221 (rank : 22) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K2Y0, Q9CYU1, Q9D1Y0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C13orf7 homolog. | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 20) | NC score | 0.022346 (rank : 19) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
NFM_MOUSE
|
||||||
| θ value | 6.88961 (rank : 21) | NC score | 0.011167 (rank : 23) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
TRS85_HUMAN
|
||||||
| θ value | 6.88961 (rank : 22) | NC score | 0.024290 (rank : 18) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y2L5, Q9H0L2 | Gene names | KIAA1012 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein TRS85 homolog. | |||||
|
CI093_HUMAN
|
||||||
| θ value | 8.99809 (rank : 23) | NC score | 0.005393 (rank : 25) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6TFL3, Q5SU58, Q6P1W1, Q6ZR13, Q8N1Z4, Q8N8L3, Q8TBR2 | Gene names | C9orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf93. | |||||
|
DMP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 24) | NC score | 0.030806 (rank : 14) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
MYST4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 25) | NC score | 0.010296 (rank : 24) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
RIPK1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 26) | NC score | 0.001452 (rank : 26) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 891 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13546, Q13180 | Gene names | RIPK1, RIP | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 1 (EC 2.7.11.1) (Serine/threonine-protein kinase RIP) (Cell death protein RIP) (Receptor-interacting protein). | |||||
|
YES_HUMAN
|
||||||
| θ value | 8.99809 (rank : 27) | NC score | 0.001253 (rank : 28) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P07947 | Gene names | YES1, YES | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase Yes (EC 2.7.10.2) (p61-Yes) (c- Yes). | |||||
|
YES_MOUSE
|
||||||
| θ value | 8.99809 (rank : 28) | NC score | 0.001261 (rank : 27) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 1052 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q04736 | Gene names | Yes1, Yes | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase Yes (EC 2.7.10.2) (p61-Yes) (c- Yes). | |||||
|
MIA2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96PC5, Q9H6C1 | Gene names | MIA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma inhibitory activity protein 2 precursor. | |||||
|
MIA2_MOUSE
|
||||||
| NC score | 0.937681 (rank : 2) | θ value | 2.26584e-152 (rank : 2) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q91ZV0 | Gene names | Mia2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma inhibitory activity protein 2 precursor. | |||||
|
OTOR_HUMAN
|
||||||
| NC score | 0.621471 (rank : 3) | θ value | 2.0648e-12 (rank : 3) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NRC9 | Gene names | OTOR, FDP, MIAL | |||
|
Domain Architecture |

|
|||||
| Description | Otoraplin precursor (Fibrocyte-derived protein) (Melanoma inhibitory activity-like protein). | |||||
|
OTOR_MOUSE
|
||||||
| NC score | 0.591270 (rank : 4) | θ value | 7.34386e-10 (rank : 6) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JIE3 | Gene names | Otor, Mial | |||
|
Domain Architecture |

|
|||||
| Description | Otoraplin precursor (Melanoma inhibitory activity-like protein). | |||||
|
MIA_HUMAN
|
||||||
| NC score | 0.584551 (rank : 5) | θ value | 1.133e-10 (rank : 4) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q16674 | Gene names | MIA | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-derived growth regulatory protein precursor (Melanoma inhibitory activity). | |||||
|
MIA_MOUSE
|
||||||
| NC score | 0.574578 (rank : 6) | θ value | 1.9326e-10 (rank : 5) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61865, O09086, P97495 | Gene names | Mia, Cdrap, Mia1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-derived growth regulatory protein precursor (Melanoma inhibitory activity) (Cartilage-derived retinoic acid-sensitive protein) (CD-RAP). | |||||
|
SPT16_MOUSE
|
||||||
| NC score | 0.056034 (rank : 7) | θ value | 1.06291 (rank : 12) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q920B9, Q3TZ48, Q3UFH0, Q921H4 | Gene names | Supt16h, Fact140, Factp140 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FACT complex subunit SPT16 (Facilitates chromatin transcription complex subunit SPT16) (FACT 140 kDa subunit) (FACTp140) (Chromatin- specific transcription elongation factor 140 kDa subunit). | |||||
|
TP53B_HUMAN
|
||||||
| NC score | 0.055574 (rank : 8) | θ value | 0.21417 (rank : 9) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q12888, Q2M1Z7, Q4LE46, Q5FWZ3, Q7Z3U4 | Gene names | TP53BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.053125 (rank : 9) | θ value | 1.38821 (rank : 13) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
SPT16_HUMAN
|
||||||
| NC score | 0.050287 (rank : 10) | θ value | 3.0926 (rank : 16) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y5B9, Q6GMT8, Q6P2F1, Q6PJM1, Q9NRX0 | Gene names | SUPT16H, FACT140, FACTP140 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FACT complex subunit SPT16 (Facilitates chromatin transcription complex subunit SPT16) (hSPT16) (FACT 140 kDa subunit) (FACTp140) (Chromatin-specific transcription elongation factor 140 kDa subunit). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.049210 (rank : 11) | θ value | 0.365318 (rank : 10) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
NPAS3_MOUSE
|
||||||
| NC score | 0.046967 (rank : 12) | θ value | 0.21417 (rank : 8) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QZQ0, Q9EQP4 | Gene names | Npas3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
NPAS3_HUMAN
|
||||||
| NC score | 0.044942 (rank : 13) | θ value | 0.47712 (rank : 11) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IXF0, Q86US6, Q86US7, Q8IXF2, Q9BY81, Q9H323, Q9Y4L8 | Gene names | NPAS3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
DMP1_MOUSE
|
||||||
| NC score | 0.030806 (rank : 14) | θ value | 8.99809 (rank : 24) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
RPGR_MOUSE
|
||||||
| NC score | 0.028714 (rank : 15) | θ value | 3.0926 (rank : 15) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9R0X5, O88408, Q9CU92 | Gene names | Rpgr | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator (mRpgr). | |||||
|
MYPT2_HUMAN
|
||||||
| NC score | 0.028525 (rank : 16) | θ value | 0.0431538 (rank : 7) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 707 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60237, Q8N179, Q9HCB7, Q9HCB8 | Gene names | PPP1R12B, MYPT2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12B (Myosin phosphatase- targeting subunit 2) (Myosin phosphatase target subunit 2). | |||||
|
CFDP1_HUMAN
|
||||||
| NC score | 0.024397 (rank : 17) | θ value | 6.88961 (rank : 18) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UEE9, O00393, O00404, Q9UEF0, Q9UEF1, Q9UEF8 | Gene names | CFDP1, BCNT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Craniofacial development protein 1 (Bucentaur). | |||||
|
TRS85_HUMAN
|
||||||
| NC score | 0.024290 (rank : 18) | θ value | 6.88961 (rank : 22) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y2L5, Q9H0L2 | Gene names | KIAA1012 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein TRS85 homolog. | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.022346 (rank : 19) | θ value | 6.88961 (rank : 20) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
CF055_HUMAN
|
||||||
| NC score | 0.021765 (rank : 20) | θ value | 6.88961 (rank : 17) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NP79, Q9H0R2, Q9H3K9, Q9P0Q0 | Gene names | C6orf55 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C6orf55 (Dopamine-responsive protein DRG-1). | |||||
|
PIAS3_HUMAN
|
||||||
| NC score | 0.020425 (rank : 21) | θ value | 1.81305 (rank : 14) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y6X2, Q9UFI3 | Gene names | PIAS3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein inhibitor of activated STAT protein 3. | |||||
|
CM007_MOUSE
|
||||||
| NC score | 0.014221 (rank : 22) | θ value | 6.88961 (rank : 19) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K2Y0, Q9CYU1, Q9D1Y0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C13orf7 homolog. | |||||
|
NFM_MOUSE
|
||||||
| NC score | 0.011167 (rank : 23) | θ value | 6.88961 (rank : 21) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
MYST4_MOUSE
|
||||||
| NC score | 0.010296 (rank : 24) | θ value | 8.99809 (rank : 25) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
CI093_HUMAN
|
||||||
| NC score | 0.005393 (rank : 25) | θ value | 8.99809 (rank : 23) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6TFL3, Q5SU58, Q6P1W1, Q6ZR13, Q8N1Z4, Q8N8L3, Q8TBR2 | Gene names | C9orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf93. | |||||
|
RIPK1_HUMAN
|
||||||
| NC score | 0.001452 (rank : 26) | θ value | 8.99809 (rank : 26) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 891 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13546, Q13180 | Gene names | RIPK1, RIP | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 1 (EC 2.7.11.1) (Serine/threonine-protein kinase RIP) (Cell death protein RIP) (Receptor-interacting protein). | |||||
|
YES_MOUSE
|
||||||
| NC score | 0.001261 (rank : 27) | θ value | 8.99809 (rank : 28) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 1052 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q04736 | Gene names | Yes1, Yes | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase Yes (EC 2.7.10.2) (p61-Yes) (c- Yes). | |||||
|
YES_HUMAN
|
||||||
| NC score | 0.001253 (rank : 28) | θ value | 8.99809 (rank : 27) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P07947 | Gene names | YES1, YES | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase Yes (EC 2.7.10.2) (p61-Yes) (c- Yes). | |||||