Please be patient as the page loads
|
PIAS3_HUMAN
|
||||||
| SwissProt Accessions | Q9Y6X2, Q9UFI3 | Gene names | PIAS3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein inhibitor of activated STAT protein 3. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PIAS1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.987636 (rank : 3) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O75925, Q99751, Q9UN02 | Gene names | PIAS1, DDXBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein inhibitor of activated STAT protein 1 (Gu-binding protein) (GBP) (RNA helicase II-binding protein) (DEAD/H box-binding protein 1). | |||||
|
PIAS3_HUMAN
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9Y6X2, Q9UFI3 | Gene names | PIAS3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein inhibitor of activated STAT protein 3. | |||||
|
PIAS3_MOUSE
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.997174 (rank : 2) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O54714, Q80WF8, Q8R598 | Gene names | Pias3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein inhibitor of activated STAT protein 3. | |||||
|
PIAS1_MOUSE
|
||||||
| θ value | 2.24492e-184 (rank : 4) | NC score | 0.987588 (rank : 4) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O88907, Q8C6H5 | Gene names | Pias1, Ddxbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein inhibitor of activated STAT protein 1 (DEAD/H box-binding protein 1). | |||||
|
PIAS2_HUMAN
|
||||||
| θ value | 2.75699e-166 (rank : 5) | NC score | 0.986861 (rank : 6) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O75928, O75927, Q96BT5, Q96KE3 | Gene names | PIAS2, PIASX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein inhibitor of activated STAT2 (Protein inhibitor of activated STAT x) (Msx-interacting zinc finger protein) (Miz1) (DAB2-interacting protein) (DIP) (Androgen receptor-interacting protein 3) (ARIP3) (PIAS-NY protein). | |||||
|
PIAS2_MOUSE
|
||||||
| θ value | 4.70271e-166 (rank : 6) | NC score | 0.986957 (rank : 5) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8C5D8, O54987, Q8C384, Q8CDQ8, Q8K208, Q99JX5, Q9D5W7, Q9QZ63 | Gene names | Pias2, Miz1, Piasx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein inhibitor of activated STAT2 (Protein inhibitor of activated STAT x) (Msx-interacting zinc finger protein) (DAB2-interacting protein) (DIP) (Androgen receptor-interacting protein 3) (ARIP3). | |||||
|
PIAS4_HUMAN
|
||||||
| θ value | 6.228e-118 (rank : 7) | NC score | 0.974818 (rank : 7) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N2W9, O75926, Q96G19, Q9UN16 | Gene names | PIAS4, PIASG | |||
|
Domain Architecture |

|
|||||
| Description | Protein inhibitor of activated STAT protein 4 (Protein inhibitor of activated STAT protein gamma) (PIAS-gamma) (PIASy). | |||||
|
PIAS4_MOUSE
|
||||||
| θ value | 4.4633e-116 (rank : 8) | NC score | 0.974741 (rank : 8) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JM05, Q8R165 | Gene names | Pias4, Piasg | |||
|
Domain Architecture |

|
|||||
| Description | Protein inhibitor of activated STAT protein 4 (Protein inhibitor of activated STAT protein gamma) (PIAS-gamma) (PIASy). | |||||
|
ZIM10_MOUSE
|
||||||
| θ value | 8.10077e-17 (rank : 9) | NC score | 0.523340 (rank : 10) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6P1E1, Q6PDK9, Q6PF85, Q8BW47 | Gene names | Rai17, Zimp10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
ZIM10_HUMAN
|
||||||
| θ value | 1.80466e-16 (rank : 10) | NC score | 0.484040 (rank : 12) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
ZIMP7_HUMAN
|
||||||
| θ value | 2.60593e-15 (rank : 11) | NC score | 0.563638 (rank : 9) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8NF64, O94790, Q659A8, Q6JKL5, Q8WTX8, Q96Q01, Q9BQH7 | Gene names | ZIMP7, KIAA1886 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
ZIMP7_MOUSE
|
||||||
| θ value | 3.40345e-15 (rank : 12) | NC score | 0.506461 (rank : 11) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8CIE2, Q6P558, Q6P9Q5, Q8BJ00 | Gene names | Zimp7, D11Bwg0280e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
FBX7_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 13) | NC score | 0.086015 (rank : 13) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y3I1, Q5TGC4, Q96HM6, Q9UF21, Q9UKT2 | Gene names | FBXO7, FBX7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 7. | |||||
|
BC11B_HUMAN
|
||||||
| θ value | 0.163984 (rank : 14) | NC score | 0.005659 (rank : 50) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 761 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9C0K0, Q9H162 | Gene names | BCL11B, CTIP2, RIT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma/leukemia 11B (B-cell CLL/lymphoma 11B) (Radiation- induced tumor suppressor gene 1 protein) (hRit1) (COUP-TF-interacting protein 2). | |||||
|
EPN2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 15) | NC score | 0.037590 (rank : 20) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CHU3, O70447, Q8BZ85 | Gene names | Epn2 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-2 (EPS-15-interacting protein 2) (Intersectin-EH-binding protein 2) (Ibp2). | |||||
|
HEY1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 16) | NC score | 0.037910 (rank : 19) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WV93 | Gene names | Hey1, Hesr1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1). | |||||
|
ZN364_MOUSE
|
||||||
| θ value | 0.813845 (rank : 17) | NC score | 0.054297 (rank : 14) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D0C1, Q8R5A1, Q9D885 | Gene names | Znf364, Zfp364 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 364 (Rabring 7). | |||||
|
TENS1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 18) | NC score | 0.039111 (rank : 18) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9HBL0 | Gene names | TNS1, TNS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-1. | |||||
|
ZN364_HUMAN
|
||||||
| θ value | 1.06291 (rank : 19) | NC score | 0.050887 (rank : 16) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y4L5, Q5T2V9, Q7Z2J2 | Gene names | ZNF364, RNF115 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 364 (Rabring 7) (RING finger protein 115). | |||||
|
CUED1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 20) | NC score | 0.041957 (rank : 17) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R3V6 | Gene names | Cuedc1 | |||
|
Domain Architecture |
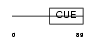
|
|||||
| Description | CUE domain-containing protein 1. | |||||
|
MIA2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 21) | NC score | 0.020425 (rank : 33) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96PC5, Q9H6C1 | Gene names | MIA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma inhibitory activity protein 2 precursor. | |||||
|
ZN690_HUMAN
|
||||||
| θ value | 1.81305 (rank : 22) | NC score | -0.000206 (rank : 57) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IWY8, Q32M75, Q32M76, Q8NA40 | Gene names | ZNF690 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 690. | |||||
|
ANKS6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 23) | NC score | 0.004824 (rank : 51) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 411 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q68DC2, Q5VSL0, Q5VSL2, Q5VSL3, Q5VSL4, Q68DB8, Q6P2R2, Q8N9L6, Q96D62 | Gene names | ANKS6, ANKRD14, SAMD6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 6 (Sterile alpha motif domain-containing protein 6) (Ankyrin repeat domain-containing protein 14). | |||||
|
ATX1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 24) | NC score | 0.031651 (rank : 24) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P54253, Q9UJG2, Q9Y4J1 | Gene names | ATXN1, ATX1, SCA1 | |||
|
Domain Architecture |

|
|||||
| Description | Ataxin-1 (Spinocerebellar ataxia type 1 protein). | |||||
|
COKA1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 25) | NC score | 0.015162 (rank : 43) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9P218, Q4VXQ4, Q8WUT2, Q96CY9, Q9BQU6, Q9BQU7 | Gene names | COL20A1, KIAA1510 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XX) chain precursor. | |||||
|
EPN2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 26) | NC score | 0.034187 (rank : 21) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O95208, O95207, Q9H7Z2, Q9UPT7 | Gene names | EPN2, KIAA1065 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-2 (EPS-15-interacting protein 2). | |||||
|
ATX1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 27) | NC score | 0.032214 (rank : 22) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P54254 | Gene names | Atxn1, Sca1 | |||
|
Domain Architecture |

|
|||||
| Description | Ataxin-1 (Spinocerebellar ataxia type 1 protein homolog). | |||||
|
CBP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 28) | NC score | 0.029627 (rank : 25) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CU058_HUMAN
|
||||||
| θ value | 3.0926 (rank : 29) | NC score | 0.054185 (rank : 15) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P58505 | Gene names | C21orf58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C21orf58. | |||||
|
HEY1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 30) | NC score | 0.027677 (rank : 27) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y5J3, Q5TZS3, Q9NYP4 | Gene names | HEY1, CHF2, HESR1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1) (Cardiovascular helix-loop-helix factor 2) (HES-related repressor protein 2) (HERP2). | |||||
|
I11RB_MOUSE
|
||||||
| θ value | 3.0926 (rank : 31) | NC score | 0.020883 (rank : 32) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P70225, O09074, Q78DW8 | Gene names | Il11ra2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-11 receptor alpha-2 chain precursor (IL-11R-alpha-2) (Interleukin-11 receptor beta chain) (IL-11R-beta). | |||||
|
BRCA1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 32) | NC score | 0.014991 (rank : 44) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48754, Q60957, Q60983 | Gene names | Brca1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 1 susceptibility protein homolog. | |||||
|
EPN1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 33) | NC score | 0.029288 (rank : 26) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80VP1, O70446 | Gene names | Epn1 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-1 (EPS-15-interacting protein 1) (Intersectin-EH-binding protein 1) (Ibp1). | |||||
|
GAW2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 34) | NC score | 0.023929 (rank : 30) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NRZ4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-W_3q26.32 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein (Gag P15); Capsid protein (Core shell protein) (Gag P30)]. | |||||
|
I11RA_MOUSE
|
||||||
| θ value | 4.03905 (rank : 35) | NC score | 0.019568 (rank : 34) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q64385, Q6NSQ0 | Gene names | Il11ra1, Etl2, Il11ra | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-11 receptor alpha-1 chain precursor (IL-11R-alpha-1) (IL- 11RA1) (IL-11R-alpha) (IL-11RA) (Enhancer trap locus homolog 2) (Etl- 2) (NR-1). | |||||
|
LGMN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 36) | NC score | 0.018188 (rank : 37) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99538, O00123 | Gene names | LGMN, PRSC1 | |||
|
Domain Architecture |

|
|||||
| Description | Legumain precursor (EC 3.4.22.34) (Asparaginyl endopeptidase) (Protease, cysteine 1). | |||||
|
MYCD_HUMAN
|
||||||
| θ value | 4.03905 (rank : 37) | NC score | 0.009890 (rank : 49) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IZQ8, Q8N7Q1 | Gene names | MYOCD, MYCD | |||
|
Domain Architecture |

|
|||||
| Description | Myocardin. | |||||
|
NRX2A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 38) | NC score | 0.012384 (rank : 46) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
NRX2B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 39) | NC score | 0.018395 (rank : 36) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
ZSWM5_MOUSE
|
||||||
| θ value | 4.03905 (rank : 40) | NC score | 0.017613 (rank : 39) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80TC6 | Gene names | Zswim5, Kiaa1511 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger SWIM domain-containing protein 5. | |||||
|
CBP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 41) | NC score | 0.023848 (rank : 31) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
EPN1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 42) | NC score | 0.032109 (rank : 23) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y6I3, Q86ST3, Q9HA18 | Gene names | EPN1 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-1 (EPS-15-interacting protein 1) (EH domain-binding mitotic phosphoprotein). | |||||
|
IRAK1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 43) | NC score | 0.000472 (rank : 56) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q62406, Q6Y3Z5, Q6Y3Z6 | Gene names | Irak1, Il1rak | |||
|
Domain Architecture |
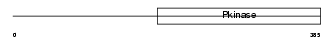
|
|||||
| Description | Interleukin-1 receptor-associated kinase 1 (EC 2.7.11.1) (IRAK-1) (IRAK) (Pelle-like protein kinase) (mPLK). | |||||
|
MARK4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 44) | NC score | -0.002064 (rank : 58) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96L34, Q8NG37, Q96JG7, Q96SQ2, Q9BYD8 | Gene names | MARK4, KIAA1860, MARKL1 | |||
|
Domain Architecture |

|
|||||
| Description | MAP/microtubule affinity-regulating kinase 4 (EC 2.7.11.1) (MAP/microtubule affinity-regulating kinase-like 1). | |||||
|
POGZ_HUMAN
|
||||||
| θ value | 5.27518 (rank : 45) | NC score | 0.002181 (rank : 52) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7Z3K3, O75049, Q8TDZ7, Q9Y4X7 | Gene names | POGZ, KIAA0461 | |||
|
Domain Architecture |

|
|||||
| Description | Pogo transposable element with ZNF domain. | |||||
|
THOC2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 46) | NC score | 0.010568 (rank : 48) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NI27, Q9H8I6 | Gene names | THOC2, CXorf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THO complex subunit 2 (Tho2). | |||||
|
TIG2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 47) | NC score | 0.027039 (rank : 28) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DD06, Q8CHU8 | Gene names | Rarres2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid receptor responder protein 2 precursor. | |||||
|
UST_MOUSE
|
||||||
| θ value | 5.27518 (rank : 48) | NC score | 0.018826 (rank : 35) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BUB6 | Gene names | Ust | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uronyl 2-sulfotransferase (EC 2.8.2.-). | |||||
|
ZSWM5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 49) | NC score | 0.014182 (rank : 45) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P217, Q5SXQ9 | Gene names | ZSWIM5, KIAA1511 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger SWIM domain-containing protein 5. | |||||
|
IL3B2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 50) | NC score | 0.017156 (rank : 40) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P26954 | Gene names | Csf2rb2, Ai2ca, Il3r, Il3rb2 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-3 receptor class 2 beta chain precursor (Interleukin-3 receptor class II beta chain) (Colony-stimulating factor 2 receptor, beta 2 chain). | |||||
|
SON_HUMAN
|
||||||
| θ value | 6.88961 (rank : 51) | NC score | 0.017821 (rank : 38) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
CUED1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 52) | NC score | 0.025425 (rank : 29) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NWM3, Q9NWD0 | Gene names | CUEDC1 | |||
|
Domain Architecture |
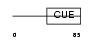
|
|||||
| Description | CUE domain-containing protein 1. | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | 0.016845 (rank : 42) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
IL3RB_MOUSE
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.017064 (rank : 41) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P26955 | Gene names | Csf2rb, Aic2b, Csf2rb1, Il3rb1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytokine receptor common beta chain precursor (GM-CSF/IL-3/IL-5 receptor common beta-chain) (CD131 antigen). | |||||
|
KLF5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.000564 (rank : 55) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z0Z7, Q9JMI2 | Gene names | Klf5, Bteb2, Iklf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 5 (Intestinal-enriched krueppel-like factor) (Transcription factor BTEB2) (Basic transcription element-binding protein 2) (BTE-binding protein 2). | |||||
|
SC11A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.002115 (rank : 53) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UI33, Q68K15, Q8NDX3, Q9UHE0, Q9UHM0 | Gene names | SCN11A, PN5, SCN12A, SNS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (Peripheral nerve sodium channel 5) (hNaN). | |||||
|
VGLL1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.010643 (rank : 47) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99NC0 | Gene names | Vgll1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription cofactor vestigial-like protein 1 (Vgl-1) (Vestigial- related factor). | |||||
|
ZN687_HUMAN
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.002004 (rank : 54) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8N1G0, Q68DQ8, Q9H937, Q9P2A7 | Gene names | ZNF687, KIAA1441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
PIAS3_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9Y6X2, Q9UFI3 | Gene names | PIAS3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein inhibitor of activated STAT protein 3. | |||||
|
PIAS3_MOUSE
|
||||||
| NC score | 0.997174 (rank : 2) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O54714, Q80WF8, Q8R598 | Gene names | Pias3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein inhibitor of activated STAT protein 3. | |||||
|
PIAS1_HUMAN
|
||||||
| NC score | 0.987636 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O75925, Q99751, Q9UN02 | Gene names | PIAS1, DDXBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein inhibitor of activated STAT protein 1 (Gu-binding protein) (GBP) (RNA helicase II-binding protein) (DEAD/H box-binding protein 1). | |||||
|
PIAS1_MOUSE
|
||||||
| NC score | 0.987588 (rank : 4) | θ value | 2.24492e-184 (rank : 4) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O88907, Q8C6H5 | Gene names | Pias1, Ddxbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein inhibitor of activated STAT protein 1 (DEAD/H box-binding protein 1). | |||||
|
PIAS2_MOUSE
|
||||||
| NC score | 0.986957 (rank : 5) | θ value | 4.70271e-166 (rank : 6) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8C5D8, O54987, Q8C384, Q8CDQ8, Q8K208, Q99JX5, Q9D5W7, Q9QZ63 | Gene names | Pias2, Miz1, Piasx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein inhibitor of activated STAT2 (Protein inhibitor of activated STAT x) (Msx-interacting zinc finger protein) (DAB2-interacting protein) (DIP) (Androgen receptor-interacting protein 3) (ARIP3). | |||||
|
PIAS2_HUMAN
|
||||||
| NC score | 0.986861 (rank : 6) | θ value | 2.75699e-166 (rank : 5) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O75928, O75927, Q96BT5, Q96KE3 | Gene names | PIAS2, PIASX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein inhibitor of activated STAT2 (Protein inhibitor of activated STAT x) (Msx-interacting zinc finger protein) (Miz1) (DAB2-interacting protein) (DIP) (Androgen receptor-interacting protein 3) (ARIP3) (PIAS-NY protein). | |||||
|
PIAS4_HUMAN
|
||||||
| NC score | 0.974818 (rank : 7) | θ value | 6.228e-118 (rank : 7) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N2W9, O75926, Q96G19, Q9UN16 | Gene names | PIAS4, PIASG | |||
|
Domain Architecture |

|
|||||
| Description | Protein inhibitor of activated STAT protein 4 (Protein inhibitor of activated STAT protein gamma) (PIAS-gamma) (PIASy). | |||||
|
PIAS4_MOUSE
|
||||||
| NC score | 0.974741 (rank : 8) | θ value | 4.4633e-116 (rank : 8) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JM05, Q8R165 | Gene names | Pias4, Piasg | |||
|
Domain Architecture |

|
|||||
| Description | Protein inhibitor of activated STAT protein 4 (Protein inhibitor of activated STAT protein gamma) (PIAS-gamma) (PIASy). | |||||
|
ZIMP7_HUMAN
|
||||||
| NC score | 0.563638 (rank : 9) | θ value | 2.60593e-15 (rank : 11) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8NF64, O94790, Q659A8, Q6JKL5, Q8WTX8, Q96Q01, Q9BQH7 | Gene names | ZIMP7, KIAA1886 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
ZIM10_MOUSE
|
||||||
| NC score | 0.523340 (rank : 10) | θ value | 8.10077e-17 (rank : 9) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6P1E1, Q6PDK9, Q6PF85, Q8BW47 | Gene names | Rai17, Zimp10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
ZIMP7_MOUSE
|
||||||
| NC score | 0.506461 (rank : 11) | θ value | 3.40345e-15 (rank : 12) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8CIE2, Q6P558, Q6P9Q5, Q8BJ00 | Gene names | Zimp7, D11Bwg0280e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
ZIM10_HUMAN
|
||||||
| NC score | 0.484040 (rank : 12) | θ value | 1.80466e-16 (rank : 10) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
FBX7_HUMAN
|
||||||
| NC score | 0.086015 (rank : 13) | θ value | 0.0563607 (rank : 13) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y3I1, Q5TGC4, Q96HM6, Q9UF21, Q9UKT2 | Gene names | FBXO7, FBX7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 7. | |||||
|
ZN364_MOUSE
|
||||||
| NC score | 0.054297 (rank : 14) | θ value | 0.813845 (rank : 17) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D0C1, Q8R5A1, Q9D885 | Gene names | Znf364, Zfp364 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 364 (Rabring 7). | |||||
|
CU058_HUMAN
|
||||||
| NC score | 0.054185 (rank : 15) | θ value | 3.0926 (rank : 29) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P58505 | Gene names | C21orf58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C21orf58. | |||||
|
ZN364_HUMAN
|
||||||
| NC score | 0.050887 (rank : 16) | θ value | 1.06291 (rank : 19) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y4L5, Q5T2V9, Q7Z2J2 | Gene names | ZNF364, RNF115 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 364 (Rabring 7) (RING finger protein 115). | |||||
|
CUED1_MOUSE
|
||||||
| NC score | 0.041957 (rank : 17) | θ value | 1.38821 (rank : 20) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R3V6 | Gene names | Cuedc1 | |||
|
Domain Architecture |

|
|||||
| Description | CUE domain-containing protein 1. | |||||
|
TENS1_HUMAN
|
||||||
| NC score | 0.039111 (rank : 18) | θ value | 1.06291 (rank : 18) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9HBL0 | Gene names | TNS1, TNS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-1. | |||||
|
HEY1_MOUSE
|
||||||
| NC score | 0.037910 (rank : 19) | θ value | 0.62314 (rank : 16) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WV93 | Gene names | Hey1, Hesr1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1). | |||||
|
EPN2_MOUSE
|
||||||
| NC score | 0.037590 (rank : 20) | θ value | 0.62314 (rank : 15) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CHU3, O70447, Q8BZ85 | Gene names | Epn2 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-2 (EPS-15-interacting protein 2) (Intersectin-EH-binding protein 2) (Ibp2). | |||||
|
EPN2_HUMAN
|
||||||
| NC score | 0.034187 (rank : 21) | θ value | 2.36792 (rank : 26) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O95208, O95207, Q9H7Z2, Q9UPT7 | Gene names | EPN2, KIAA1065 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-2 (EPS-15-interacting protein 2). | |||||
|
ATX1_MOUSE
|
||||||
| NC score | 0.032214 (rank : 22) | θ value | 3.0926 (rank : 27) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P54254 | Gene names | Atxn1, Sca1 | |||
|
Domain Architecture |

|
|||||
| Description | Ataxin-1 (Spinocerebellar ataxia type 1 protein homolog). | |||||
|
EPN1_HUMAN
|
||||||
| NC score | 0.032109 (rank : 23) | θ value | 5.27518 (rank : 42) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y6I3, Q86ST3, Q9HA18 | Gene names | EPN1 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-1 (EPS-15-interacting protein 1) (EH domain-binding mitotic phosphoprotein). | |||||
|
ATX1_HUMAN
|
||||||
| NC score | 0.031651 (rank : 24) | θ value | 2.36792 (rank : 24) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P54253, Q9UJG2, Q9Y4J1 | Gene names | ATXN1, ATX1, SCA1 | |||
|
Domain Architecture |

|
|||||
| Description | Ataxin-1 (Spinocerebellar ataxia type 1 protein). | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.029627 (rank : 25) | θ value | 3.0926 (rank : 28) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
EPN1_MOUSE
|
||||||
| NC score | 0.029288 (rank : 26) | θ value | 4.03905 (rank : 33) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80VP1, O70446 | Gene names | Epn1 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-1 (EPS-15-interacting protein 1) (Intersectin-EH-binding protein 1) (Ibp1). | |||||
|
HEY1_HUMAN
|
||||||
| NC score | 0.027677 (rank : 27) | θ value | 3.0926 (rank : 30) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y5J3, Q5TZS3, Q9NYP4 | Gene names | HEY1, CHF2, HESR1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1) (Cardiovascular helix-loop-helix factor 2) (HES-related repressor protein 2) (HERP2). | |||||
|
TIG2_MOUSE
|
||||||
| NC score | 0.027039 (rank : 28) | θ value | 5.27518 (rank : 47) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DD06, Q8CHU8 | Gene names | Rarres2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid receptor responder protein 2 precursor. | |||||
|
CUED1_HUMAN
|
||||||
| NC score | 0.025425 (rank : 29) | θ value | 8.99809 (rank : 52) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NWM3, Q9NWD0 | Gene names | CUEDC1 | |||
|
Domain Architecture |

|
|||||
| Description | CUE domain-containing protein 1. | |||||
|
GAW2_HUMAN
|
||||||
| NC score | 0.023929 (rank : 30) | θ value | 4.03905 (rank : 34) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NRZ4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-W_3q26.32 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein (Gag P15); Capsid protein (Core shell protein) (Gag P30)]. | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.023848 (rank : 31) | θ value | 5.27518 (rank : 41) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
I11RB_MOUSE
|
||||||
| NC score | 0.020883 (rank : 32) | θ value | 3.0926 (rank : 31) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P70225, O09074, Q78DW8 | Gene names | Il11ra2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-11 receptor alpha-2 chain precursor (IL-11R-alpha-2) (Interleukin-11 receptor beta chain) (IL-11R-beta). | |||||
|
MIA2_HUMAN
|
||||||
| NC score | 0.020425 (rank : 33) | θ value | 1.81305 (rank : 21) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96PC5, Q9H6C1 | Gene names | MIA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma inhibitory activity protein 2 precursor. | |||||
|
I11RA_MOUSE
|
||||||
| NC score | 0.019568 (rank : 34) | θ value | 4.03905 (rank : 35) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q64385, Q6NSQ0 | Gene names | Il11ra1, Etl2, Il11ra | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-11 receptor alpha-1 chain precursor (IL-11R-alpha-1) (IL- 11RA1) (IL-11R-alpha) (IL-11RA) (Enhancer trap locus homolog 2) (Etl- 2) (NR-1). | |||||
|
UST_MOUSE
|
||||||
| NC score | 0.018826 (rank : 35) | θ value | 5.27518 (rank : 48) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BUB6 | Gene names | Ust | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uronyl 2-sulfotransferase (EC 2.8.2.-). | |||||
|
NRX2B_HUMAN
|
||||||
| NC score | 0.018395 (rank : 36) | θ value | 4.03905 (rank : 39) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
LGMN_HUMAN
|
||||||
| NC score | 0.018188 (rank : 37) | θ value | 4.03905 (rank : 36) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99538, O00123 | Gene names | LGMN, PRSC1 | |||
|
Domain Architecture |

|
|||||
| Description | Legumain precursor (EC 3.4.22.34) (Asparaginyl endopeptidase) (Protease, cysteine 1). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.017821 (rank : 38) | θ value | 6.88961 (rank : 51) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
ZSWM5_MOUSE
|
||||||
| NC score | 0.017613 (rank : 39) | θ value | 4.03905 (rank : 40) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80TC6 | Gene names | Zswim5, Kiaa1511 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger SWIM domain-containing protein 5. | |||||
|
IL3B2_MOUSE
|
||||||
| NC score | 0.017156 (rank : 40) | θ value | 6.88961 (rank : 50) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P26954 | Gene names | Csf2rb2, Ai2ca, Il3r, Il3rb2 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-3 receptor class 2 beta chain precursor (Interleukin-3 receptor class II beta chain) (Colony-stimulating factor 2 receptor, beta 2 chain). | |||||
|
IL3RB_MOUSE
|
||||||
| NC score | 0.017064 (rank : 41) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P26955 | Gene names | Csf2rb, Aic2b, Csf2rb1, Il3rb1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytokine receptor common beta chain precursor (GM-CSF/IL-3/IL-5 receptor common beta-chain) (CD131 antigen). | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.016845 (rank : 42) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
COKA1_HUMAN
|
||||||
| NC score | 0.015162 (rank : 43) | θ value | 2.36792 (rank : 25) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9P218, Q4VXQ4, Q8WUT2, Q96CY9, Q9BQU6, Q9BQU7 | Gene names | COL20A1, KIAA1510 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XX) chain precursor. | |||||
|
BRCA1_MOUSE
|
||||||
| NC score | 0.014991 (rank : 44) | θ value | 4.03905 (rank : 32) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48754, Q60957, Q60983 | Gene names | Brca1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 1 susceptibility protein homolog. | |||||
|
ZSWM5_HUMAN
|
||||||
| NC score | 0.014182 (rank : 45) | θ value | 5.27518 (rank : 49) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P217, Q5SXQ9 | Gene names | ZSWIM5, KIAA1511 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger SWIM domain-containing protein 5. | |||||
|
NRX2A_HUMAN
|
||||||
| NC score | 0.012384 (rank : 46) | θ value | 4.03905 (rank : 38) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
VGLL1_MOUSE
|
||||||
| NC score | 0.010643 (rank : 47) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99NC0 | Gene names | Vgll1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription cofactor vestigial-like protein 1 (Vgl-1) (Vestigial- related factor). | |||||
|
THOC2_HUMAN
|
||||||
| NC score | 0.010568 (rank : 48) | θ value | 5.27518 (rank : 46) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NI27, Q9H8I6 | Gene names | THOC2, CXorf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THO complex subunit 2 (Tho2). | |||||
|
MYCD_HUMAN
|
||||||
| NC score | 0.009890 (rank : 49) | θ value | 4.03905 (rank : 37) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IZQ8, Q8N7Q1 | Gene names | MYOCD, MYCD | |||
|
Domain Architecture |

|
|||||
| Description | Myocardin. | |||||
|
BC11B_HUMAN
|
||||||
| NC score | 0.005659 (rank : 50) | θ value | 0.163984 (rank : 14) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 761 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9C0K0, Q9H162 | Gene names | BCL11B, CTIP2, RIT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma/leukemia 11B (B-cell CLL/lymphoma 11B) (Radiation- induced tumor suppressor gene 1 protein) (hRit1) (COUP-TF-interacting protein 2). | |||||
|
ANKS6_HUMAN
|
||||||
| NC score | 0.004824 (rank : 51) | θ value | 2.36792 (rank : 23) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 411 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q68DC2, Q5VSL0, Q5VSL2, Q5VSL3, Q5VSL4, Q68DB8, Q6P2R2, Q8N9L6, Q96D62 | Gene names | ANKS6, ANKRD14, SAMD6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 6 (Sterile alpha motif domain-containing protein 6) (Ankyrin repeat domain-containing protein 14). | |||||
|
POGZ_HUMAN
|
||||||
| NC score | 0.002181 (rank : 52) | θ value | 5.27518 (rank : 45) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7Z3K3, O75049, Q8TDZ7, Q9Y4X7 | Gene names | POGZ, KIAA0461 | |||
|
Domain Architecture |

|
|||||
| Description | Pogo transposable element with ZNF domain. | |||||
|
SC11A_HUMAN
|
||||||
| NC score | 0.002115 (rank : 53) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UI33, Q68K15, Q8NDX3, Q9UHE0, Q9UHM0 | Gene names | SCN11A, PN5, SCN12A, SNS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (Peripheral nerve sodium channel 5) (hNaN). | |||||
|
ZN687_HUMAN
|
||||||
| NC score | 0.002004 (rank : 54) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8N1G0, Q68DQ8, Q9H937, Q9P2A7 | Gene names | ZNF687, KIAA1441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
KLF5_MOUSE
|
||||||
| NC score | 0.000564 (rank : 55) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z0Z7, Q9JMI2 | Gene names | Klf5, Bteb2, Iklf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 5 (Intestinal-enriched krueppel-like factor) (Transcription factor BTEB2) (Basic transcription element-binding protein 2) (BTE-binding protein 2). | |||||
|
IRAK1_MOUSE
|
||||||
| NC score | 0.000472 (rank : 56) | θ value | 5.27518 (rank : 43) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q62406, Q6Y3Z5, Q6Y3Z6 | Gene names | Irak1, Il1rak | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-1 receptor-associated kinase 1 (EC 2.7.11.1) (IRAK-1) (IRAK) (Pelle-like protein kinase) (mPLK). | |||||
|
ZN690_HUMAN
|
||||||
| NC score | -0.000206 (rank : 57) | θ value | 1.81305 (rank : 22) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IWY8, Q32M75, Q32M76, Q8NA40 | Gene names | ZNF690 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 690. | |||||
|
MARK4_HUMAN
|
||||||
| NC score | -0.002064 (rank : 58) | θ value | 5.27518 (rank : 44) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96L34, Q8NG37, Q96JG7, Q96SQ2, Q9BYD8 | Gene names | MARK4, KIAA1860, MARKL1 | |||
|
Domain Architecture |

|
|||||
| Description | MAP/microtubule affinity-regulating kinase 4 (EC 2.7.11.1) (MAP/microtubule affinity-regulating kinase-like 1). | |||||