Please be patient as the page loads
|
EPN2_HUMAN
|
||||||
| SwissProt Accessions | O95208, O95207, Q9H7Z2, Q9UPT7 | Gene names | EPN2, KIAA1065 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-2 (EPS-15-interacting protein 2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
EPN2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 156 | |
| SwissProt Accessions | O95208, O95207, Q9H7Z2, Q9UPT7 | Gene names | EPN2, KIAA1065 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-2 (EPS-15-interacting protein 2). | |||||
|
EPN2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.960883 (rank : 2) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8CHU3, O70447, Q8BZ85 | Gene names | Epn2 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-2 (EPS-15-interacting protein 2) (Intersectin-EH-binding protein 2) (Ibp2). | |||||
|
EPN3_HUMAN
|
||||||
| θ value | 2.63489e-92 (rank : 3) | NC score | 0.896010 (rank : 3) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9H201, Q9BVN6, Q9NWK2 | Gene names | EPN3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
EPN3_MOUSE
|
||||||
| θ value | 1.22395e-89 (rank : 4) | NC score | 0.879599 (rank : 6) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q91W69, Q9CV55 | Gene names | Epn3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
EPN1_HUMAN
|
||||||
| θ value | 1.8332e-77 (rank : 5) | NC score | 0.883752 (rank : 5) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9Y6I3, Q86ST3, Q9HA18 | Gene names | EPN1 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-1 (EPS-15-interacting protein 1) (EH domain-binding mitotic phosphoprotein). | |||||
|
EPN1_MOUSE
|
||||||
| θ value | 2.73936e-73 (rank : 6) | NC score | 0.884352 (rank : 4) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q80VP1, O70446 | Gene names | Epn1 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-1 (EPS-15-interacting protein 1) (Intersectin-EH-binding protein 1) (Ibp1). | |||||
|
EPN4_HUMAN
|
||||||
| θ value | 8.89434e-40 (rank : 7) | NC score | 0.824570 (rank : 7) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14677, Q8NAF1, Q96E05 | Gene names | CLINT1, ENTH, EPN4, EPNR, KIAA0171 | |||
|
Domain Architecture |
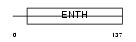
|
|||||
| Description | Clathrin interactor 1 (Epsin-4) (Epsin-related protein) (EpsinR) (Enthoprotin) (Clathrin-interacting protein localized in the trans- Golgi region) (Clint). | |||||
|
EPN4_MOUSE
|
||||||
| θ value | 8.89434e-40 (rank : 8) | NC score | 0.822377 (rank : 8) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99KN9, Q8CFH4 | Gene names | Clint1, Enth, Epn4, Epnr, Kiaa0171 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin interactor 1 (Epsin-4) (Epsin-related protein) (EpsinR) (Enthoprotin). | |||||
|
NU214_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 9) | NC score | 0.127246 (rank : 10) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
DSCAM_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 10) | NC score | 0.023770 (rank : 112) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60469, O60468 | Gene names | DSCAM | |||
|
Domain Architecture |

|
|||||
| Description | Down syndrome cell adhesion molecule precursor (CHD2). | |||||
|
CN032_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 11) | NC score | 0.141636 (rank : 9) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
K1802_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 12) | NC score | 0.084902 (rank : 21) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 13) | NC score | 0.104781 (rank : 13) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
MUC5B_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 14) | NC score | 0.058622 (rank : 53) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 15) | NC score | 0.105399 (rank : 12) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
PO121_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 16) | NC score | 0.083434 (rank : 23) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
BORG5_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 17) | NC score | 0.068055 (rank : 38) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q00587, Q96GN1 | Gene names | CDC42EP1, BORG5, CEP1, MSE55 | |||
|
Domain Architecture |
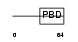
|
|||||
| Description | Cdc42 effector protein 1 (Binder of Rho GTPases 5) (Serum protein MSE55). | |||||
|
HRBL_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 18) | NC score | 0.102212 (rank : 14) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O95081, O75429, Q96AB9, Q96GL4 | Gene names | HRBL, RABR | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
NRX2A_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 19) | NC score | 0.048933 (rank : 71) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
NRX2B_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 20) | NC score | 0.069720 (rank : 35) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
CSKI2_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 21) | NC score | 0.031264 (rank : 97) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8WXE0, Q7LG69, Q9ULT1 | Gene names | CASKIN2, KIAA1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
DYN1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 22) | NC score | 0.049610 (rank : 69) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q05193 | Gene names | DNM1, DNM | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-1 (EC 3.6.5.5). | |||||
|
DYN1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 23) | NC score | 0.049006 (rank : 70) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P39053, Q61358, Q61359, Q61360, Q8JZZ4, Q9CSY7, Q9QXX1 | Gene names | Dnm1, Dnm | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-1 (EC 3.6.5.5). | |||||
|
SALL3_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 24) | NC score | 0.016915 (rank : 134) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 1066 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9BXA9, Q9UGH1 | Gene names | SALL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 3 (Zinc finger protein SALL3) (hSALL3). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 25) | NC score | 0.057839 (rank : 56) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
CJ078_MOUSE
|
||||||
| θ value | 0.125558 (rank : 26) | NC score | 0.052782 (rank : 66) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
NUP62_MOUSE
|
||||||
| θ value | 0.125558 (rank : 27) | NC score | 0.069373 (rank : 37) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q63850, Q99JN7 | Gene names | Nup62 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore glycoprotein p62 (62 kDa nucleoporin). | |||||
|
TNR6A_HUMAN
|
||||||
| θ value | 0.125558 (rank : 28) | NC score | 0.066366 (rank : 42) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8NDV7, O15408, Q658L5, Q6NVB5, Q8NEZ0, Q8TBT8, Q8TCR0, Q9NV59, Q9P268 | Gene names | TNRC6A, CAGH26, KIAA1460, TNRC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trinucleotide repeat-containing gene 6A protein (CAG repeat protein 26) (Glycine-tryptophan protein of 182 kDa) (GW182 autoantigen) (Protein GW1) (EMSY interactor protein). | |||||
|
MUC2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 29) | NC score | 0.071942 (rank : 30) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 0.163984 (rank : 30) | NC score | 0.097018 (rank : 15) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 110 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
DAAM2_MOUSE
|
||||||
| θ value | 0.21417 (rank : 31) | NC score | 0.022818 (rank : 115) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 371 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q80U19, Q810J5 | Gene names | Daam2, Kiaa0381 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 2. | |||||
|
KCNE4_HUMAN
|
||||||
| θ value | 0.21417 (rank : 32) | NC score | 0.073468 (rank : 28) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WWG9, Q96CC4 | Gene names | KCNE4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily E member 4 (Minimum potassium ion channel-related peptide 3) (Potassium channel subunit beta MiRP3) (MinK-related peptide 3). | |||||
|
KCNE4_MOUSE
|
||||||
| θ value | 0.279714 (rank : 33) | NC score | 0.071471 (rank : 31) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WTW3 | Gene names | Kcne4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily E member 4 (Minimum potassium ion channel-related peptide 3) (Potassium channel subunit beta MiRP3) (MinK-related peptide 3). | |||||
|
TAF4_HUMAN
|
||||||
| θ value | 0.279714 (rank : 34) | NC score | 0.086339 (rank : 20) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
CIC_MOUSE
|
||||||
| θ value | 0.365318 (rank : 35) | NC score | 0.069736 (rank : 34) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CO002_HUMAN
|
||||||
| θ value | 0.365318 (rank : 36) | NC score | 0.083761 (rank : 22) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9NZP6 | Gene names | C15orf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C15orf2. | |||||
|
ENAH_MOUSE
|
||||||
| θ value | 0.365318 (rank : 37) | NC score | 0.064054 (rank : 45) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
NSD1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 38) | NC score | 0.035927 (rank : 87) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
ROBO4_MOUSE
|
||||||
| θ value | 0.47712 (rank : 39) | NC score | 0.030076 (rank : 101) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8C310, Q9DBW1 | Gene names | Robo4 | |||
|
Domain Architecture |
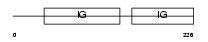
|
|||||
| Description | Roundabout homolog 4 precursor. | |||||
|
FOXJ3_MOUSE
|
||||||
| θ value | 0.62314 (rank : 40) | NC score | 0.014081 (rank : 145) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BUR3, Q3UTD8 | Gene names | Foxj3, Kiaa1041 | |||
|
Domain Architecture |
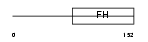
|
|||||
| Description | Forkhead box protein J3. | |||||
|
IRAK1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 41) | NC score | 0.007089 (rank : 166) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P51617, Q7Z5V4, Q96RL2 | Gene names | IRAK1, IRAK | |||
|
Domain Architecture |
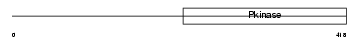
|
|||||
| Description | Interleukin-1 receptor-associated kinase 1 (EC 2.7.11.1) (IRAK-1). | |||||
|
NCOR2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 42) | NC score | 0.046635 (rank : 74) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
NU153_HUMAN
|
||||||
| θ value | 0.62314 (rank : 43) | NC score | 0.051571 (rank : 68) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P49790 | Gene names | NUP153 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup153 (Nucleoporin Nup153) (153 kDa nucleoporin). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 0.62314 (rank : 44) | NC score | 0.052632 (rank : 67) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
SNIP_MOUSE
|
||||||
| θ value | 0.62314 (rank : 45) | NC score | 0.040962 (rank : 81) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9QWI6, O70298 | Gene names | P140, Kiaa1684 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | p130Cas-associated protein (p140Cap) (SNAP-25-interacting protein) (SNIP). | |||||
|
SRA1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 46) | NC score | 0.044076 (rank : 78) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9HD15, Q6NVU9, Q8IXM1, Q9HD13, Q9HD14 | Gene names | SRA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Steroid receptor RNA activator 1 (Steroid receptor RNA activator protein) (SRAP). | |||||
|
SYNG_HUMAN
|
||||||
| θ value | 0.62314 (rank : 47) | NC score | 0.042601 (rank : 79) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UMZ2, Q6ZT17 | Gene names | AP1GBP1, SYNG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP1 subunit gamma-binding protein 1 (Gamma-synergin). | |||||
|
AP180_MOUSE
|
||||||
| θ value | 0.813845 (rank : 48) | NC score | 0.094918 (rank : 17) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q61548, Q61547 | Gene names | Snap91 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin coat assembly protein AP180 (Clathrin coat-associated protein AP180) (91 kDa synaptosomal-associated protein) (Phosphoprotein F1- 20). | |||||
|
ARMX2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 49) | NC score | 0.044329 (rank : 77) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6A058, Q9CXI9 | Gene names | Armcx2, Kiaa0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2. | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 50) | NC score | 0.061732 (rank : 48) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
MYPT2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 51) | NC score | 0.016016 (rank : 137) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 707 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O60237, Q8N179, Q9HCB7, Q9HCB8 | Gene names | PPP1R12B, MYPT2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12B (Myosin phosphatase- targeting subunit 2) (Myosin phosphatase target subunit 2). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 0.813845 (rank : 52) | NC score | 0.074234 (rank : 27) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
PO2F1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 53) | NC score | 0.013622 (rank : 147) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P14859, Q5TBT7, Q6PK46, Q8NEU9, Q9BPV1 | Gene names | POU2F1, OCT1, OTF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 2, transcription factor 1 (Octamer-binding transcription factor 1) (Oct-1) (OTF-1) (NF-A1). | |||||
|
AFF3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 54) | NC score | 0.027004 (rank : 107) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P51826 | Gene names | AFF3, LAF4 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 3 (Protein LAF-4) (Lymphoid nuclear protein related to AF4). | |||||
|
ATS12_MOUSE
|
||||||
| θ value | 1.06291 (rank : 55) | NC score | 0.007383 (rank : 165) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q811B3, Q8BK92, Q8BKY1 | Gene names | Adamts12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAMTS-12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 12) (ADAM-TS 12) (ADAM-TS12). | |||||
|
EMID2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 56) | NC score | 0.023268 (rank : 114) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96A83 | Gene names | EMID2, COL26A1, EMU2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XXVI) chain precursor (EMI domain-containing protein 2) (Protein Emu2) (Emilin and multimerin domain-containing protein 2). | |||||
|
IF4G1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 57) | NC score | 0.041506 (rank : 80) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q04637, O43177, O95066, Q5HYG0, Q6ZN21, Q8N102 | Gene names | EIF4G1, EIF4G, EIF4GI | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1) (p220). | |||||
|
MUCDL_HUMAN
|
||||||
| θ value | 1.06291 (rank : 58) | NC score | 0.048452 (rank : 72) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
NPAS4_MOUSE
|
||||||
| θ value | 1.06291 (rank : 59) | NC score | 0.037543 (rank : 85) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8BGD7, Q3V3U3 | Gene names | Npas4, Nxf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF) (Limbic-enhanced PAS protein) (LE-PAS). | |||||
|
PDLI7_HUMAN
|
||||||
| θ value | 1.06291 (rank : 60) | NC score | 0.029526 (rank : 102) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 547 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9NR12, Q14250, Q5XG82, Q6NVZ5, Q96C91, Q9BXB8, Q9BXB9 | Gene names | PDLIM7, ENIGMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
CLCN6_MOUSE
|
||||||
| θ value | 1.38821 (rank : 61) | NC score | 0.020825 (rank : 124) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35454 | Gene names | Clcn6, Clc6 | |||
|
Domain Architecture |

|
|||||
| Description | Chloride channel protein 6 (ClC-6). | |||||
|
LDB3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 62) | NC score | 0.019729 (rank : 128) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9JKS4, Q6A038, Q811P2, Q811P3, Q811P4, Q811P5, Q9D130, Q9JKS3, Q9R0Z1, Q9WVH1, Q9WVH2 | Gene names | Ldb3, Kiaa0613 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher) (Protein oracle). | |||||
|
MILK1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 63) | NC score | 0.027582 (rank : 106) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8N3F8, Q5TI16, Q7RTP5, Q8N3N8, Q9BVL9, Q9BY92, Q9UH43, Q9UH44, Q9UH45 | Gene names | MIRAB13, KIAA1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
NFAC1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 64) | NC score | 0.021563 (rank : 121) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O95644, Q12865, Q15793 | Gene names | NFATC1, NFAT2, NFATC | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 1 (NFAT transcription complex cytosolic component) (NF-ATc1) (NF-ATc). | |||||
|
NGN2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 65) | NC score | 0.014777 (rank : 140) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70447, P70237 | Gene names | Neurog2, Ath4a, Atoh4, Ngn2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 2 (Atonal protein homolog 4) (Helix-loop-helix protein mATH-4A) (MATH4A). | |||||
|
RAI2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 66) | NC score | 0.027647 (rank : 105) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QVY8 | Gene names | Rai2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 2 (3f8). | |||||
|
SMP1L_MOUSE
|
||||||
| θ value | 1.38821 (rank : 67) | NC score | 0.025964 (rank : 110) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7TN29, Q3U798 | Gene names | Smap1l, Smap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1-like (Stromal membrane- associated protein 2). | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | 1.38821 (rank : 68) | NC score | 0.059587 (rank : 51) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
TCGAP_MOUSE
|
||||||
| θ value | 1.38821 (rank : 69) | NC score | 0.021857 (rank : 120) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80YF9, Q6P7W6 | Gene names | Snx26, Tcgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 70) | NC score | 0.026824 (rank : 109) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
CN032_MOUSE
|
||||||
| θ value | 1.81305 (rank : 71) | NC score | 0.107782 (rank : 11) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
FARP1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 72) | NC score | 0.008697 (rank : 158) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y4F1, Q6IQ29 | Gene names | FARP1, CDEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 1 (Chondrocyte- derived ezrin-like protein). | |||||
|
GPC2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 73) | NC score | 0.013782 (rank : 146) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N158 | Gene names | GPC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glypican-2 precursor. | |||||
|
MYH3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 74) | NC score | 0.002594 (rank : 171) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
PIAS3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 75) | NC score | 0.032912 (rank : 90) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O54714, Q80WF8, Q8R598 | Gene names | Pias3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein inhibitor of activated STAT protein 3. | |||||
|
SHAN1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 76) | NC score | 0.038169 (rank : 84) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
AMOT_HUMAN
|
||||||
| θ value | 2.36792 (rank : 77) | NC score | 0.020996 (rank : 123) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q4VCS5, Q504X5, Q9HD27, Q9UPT1 | Gene names | AMOT, KIAA1071 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin. | |||||
|
ATBF1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 78) | NC score | 0.016099 (rank : 135) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
GNRP_HUMAN
|
||||||
| θ value | 2.36792 (rank : 79) | NC score | 0.015255 (rank : 139) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13972, Q16027 | Gene names | RASGRF1, CDC25 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (Ras-specific guanine nucleotide-releasing factor). | |||||
|
MARCS_HUMAN
|
||||||
| θ value | 2.36792 (rank : 80) | NC score | 0.031886 (rank : 92) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
MEF2A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 81) | NC score | 0.014461 (rank : 141) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q02078, O43814, Q14223, Q14224, Q96D14 | Gene names | MEF2A, MEF2 | |||
|
Domain Architecture |
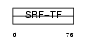
|
|||||
| Description | Myocyte-specific enhancer factor 2A (Serum response factor-like protein 1). | |||||
|
NUP62_HUMAN
|
||||||
| θ value | 2.36792 (rank : 82) | NC score | 0.071004 (rank : 32) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P37198, Q503A4, Q96C43, Q9NSL1 | Gene names | NUP62 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore glycoprotein p62 (62 kDa nucleoporin). | |||||
|
PHC3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 83) | NC score | 0.021947 (rank : 119) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8CHP6, Q3TLV5 | Gene names | Phc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 3. | |||||
|
PHF14_MOUSE
|
||||||
| θ value | 2.36792 (rank : 84) | NC score | 0.018288 (rank : 131) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9D4H9, Q810Z6, Q8C284, Q8CGF8, Q9D1Q8 | Gene names | Phf14 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 14. | |||||
|
PIAS3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 85) | NC score | 0.034187 (rank : 88) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y6X2, Q9UFI3 | Gene names | PIAS3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein inhibitor of activated STAT protein 3. | |||||
|
RNF12_MOUSE
|
||||||
| θ value | 2.36792 (rank : 86) | NC score | 0.014313 (rank : 142) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WTV7 | Gene names | Rnf12, Rlim | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 12 (LIM domain-interacting RING finger protein) (RING finger LIM domain-binding protein) (R-LIM). | |||||
|
SYNG_MOUSE
|
||||||
| θ value | 2.36792 (rank : 87) | NC score | 0.045896 (rank : 75) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q5SV85, Q5SV84, Q6PHT6 | Gene names | Ap1gbp1, Syng | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP1 subunit gamma-binding protein 1 (Gamma-synergin). | |||||
|
TR10C_HUMAN
|
||||||
| θ value | 2.36792 (rank : 88) | NC score | 0.028114 (rank : 104) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O14798, O14755, Q6UXM5 | Gene names | TNFRSF10C, DCR1, LIT, TRAILR3, TRID | |||
|
Domain Architecture |
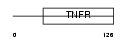
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 10C precursor (Decoy receptor 1) (DcR1) (Decoy TRAIL receptor without death domain) (TNF- related apoptosis-inducing ligand receptor 3) (TRAIL receptor 3) (TRAIL-R3) (Trail receptor without an intracellular domain) (Lymphocyte inhibitor of TRAIL) (Antagonist decoy receptor for TRAIL/Apo-2L) (CD263 antigen). | |||||
|
BCOR_HUMAN
|
||||||
| θ value | 3.0926 (rank : 89) | NC score | 0.031572 (rank : 95) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6W2J9, Q6P4B6, Q7Z2K7, Q8TEB4, Q96DB3, Q9H232, Q9H233, Q9HCJ7, Q9NXF2 | Gene names | BCOR, KIAA1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
BCOR_MOUSE
|
||||||
| θ value | 3.0926 (rank : 90) | NC score | 0.028360 (rank : 103) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8CGN4, Q6PDK5, Q6ZPM3, Q8BKF5, Q8CGN1, Q8CGN2, Q8CGN3 | Gene names | Bcor, Kiaa1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
CAC1I_HUMAN
|
||||||
| θ value | 3.0926 (rank : 91) | NC score | 0.009808 (rank : 154) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9P0X4, O95504, Q5JZ88, Q7Z6S9, Q8NFX6, Q9NZC8, Q9UH15, Q9UH30, Q9ULU9, Q9UNE6 | Gene names | CACNA1I, KIAA1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1I (Voltage- gated calcium channel subunit alpha Cav3.3) (Ca(v)3.3). | |||||
|
DEN1A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 92) | NC score | 0.032431 (rank : 91) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8K382 | Gene names | Dennd1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 1A. | |||||
|
K0460_MOUSE
|
||||||
| θ value | 3.0926 (rank : 93) | NC score | 0.031388 (rank : 96) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6NXI6, Q3U3L8, Q6ZQA7 | Gene names | Kiaa0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
MEFV_MOUSE
|
||||||
| θ value | 3.0926 (rank : 94) | NC score | 0.012918 (rank : 151) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JJ26 | Gene names | Mefv | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
PRP1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 95) | NC score | 0.030681 (rank : 99) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
SFTPD_HUMAN
|
||||||
| θ value | 3.0926 (rank : 96) | NC score | 0.007810 (rank : 161) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P35247, Q86YK9, Q9UCJ2, Q9UCJ3 | Gene names | SFTPD, PSPD, SFTP4 | |||
|
Domain Architecture |

|
|||||
| Description | Pulmonary surfactant-associated protein D precursor (SP-D) (PSP-D). | |||||
|
ZHX1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 97) | NC score | 0.016043 (rank : 136) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P70121, Q8BQ68, Q8C6T4, Q8CJG3 | Gene names | Zhx1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc fingers and homeoboxes protein 1. | |||||
|
AP180_HUMAN
|
||||||
| θ value | 4.03905 (rank : 98) | NC score | 0.072503 (rank : 29) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O60641, Q5JX13, Q68DL9, Q6P9D3, Q9NTY7 | Gene names | SNAP91, KIAA0656 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin coat assembly protein AP180 (Clathrin coat-associated protein AP180) (91 kDa synaptosomal-associated protein). | |||||
|
ATF2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 99) | NC score | 0.031805 (rank : 94) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P15336, Q13000 | Gene names | ATF2, CREB2, CREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (HB16). | |||||
|
ATF2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 100) | NC score | 0.031878 (rank : 93) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P16951, Q64089, Q64090, Q64091 | Gene names | Atf2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (MXBP protein). | |||||
|
ATX2L_MOUSE
|
||||||
| θ value | 4.03905 (rank : 101) | NC score | 0.026863 (rank : 108) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q7TQH0, Q80XN9, Q8K059 | Gene names | Atxn2l, A2lp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein. | |||||
|
CU056_MOUSE
|
||||||
| θ value | 4.03905 (rank : 102) | NC score | 0.019988 (rank : 125) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D9W0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative protein C21orf56 homolog. | |||||
|
CUTL1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 103) | NC score | 0.012957 (rank : 150) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
EGR2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 104) | NC score | 0.000923 (rank : 173) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 728 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P11161, Q8IV26, Q9UNA6 | Gene names | EGR2, KROX20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early growth response protein 2 (EGR-2) (Krox-20 protein) (AT591). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 105) | NC score | 0.014220 (rank : 144) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
PLCB1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 106) | NC score | 0.009280 (rank : 156) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NQ66, O60325, Q5TFF7, Q5TGC9, Q8IV93, Q9BQW2, Q9H4H2, Q9H8H5, Q9NQ65, Q9NQH9, Q9NTH4, Q9UJP6, Q9UM26 | Gene names | PLCB1, KIAA0581 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-1) (PLC-beta-1) (PLC-I) (PLC-154). | |||||
|
SMP1L_HUMAN
|
||||||
| θ value | 4.03905 (rank : 107) | NC score | 0.022503 (rank : 117) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8WU79, Q5QPL2, Q96C93, Q9NST2, Q9UJL8 | Gene names | SMAP1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1-like. | |||||
|
SNIP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 108) | NC score | 0.033379 (rank : 89) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9C0H9, Q8N4W8 | Gene names | P140, KIAA1684 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | p130Cas-associated protein (p140Cap) (SNAP-25-interacting protein) (SNIP). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 109) | NC score | 0.056006 (rank : 59) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
TERF2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 110) | NC score | 0.019903 (rank : 127) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O35144 | Gene names | Terf2, Trf2 | |||
|
Domain Architecture |

|
|||||
| Description | Telomeric repeat-binding factor 2 (TTAGGG repeat-binding factor 2) (Telomeric DNA-binding protein). | |||||
|
TP53B_MOUSE
|
||||||
| θ value | 4.03905 (rank : 111) | NC score | 0.045230 (rank : 76) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
ZN750_HUMAN
|
||||||
| θ value | 4.03905 (rank : 112) | NC score | 0.025276 (rank : 111) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q32MQ0, Q9H899 | Gene names | ZNF750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
CLCN6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 113) | NC score | 0.017143 (rank : 133) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P51797, O60818, O60819, O60820, O60821, P78520, P78521, Q5SNW2, Q5SNW3, Q5SNX1, Q5SNX2, Q5SNX3, Q99427, Q99428, Q99429 | Gene names | CLCN6, KIAA0046 | |||
|
Domain Architecture |

|
|||||
| Description | Chloride channel protein 6 (ClC-6). | |||||
|
FOG1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 114) | NC score | 0.006770 (rank : 167) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 981 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8IX07 | Gene names | ZFPM1, FOG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
HUWE1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 115) | NC score | 0.010126 (rank : 153) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
MILK1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 116) | NC score | 0.039158 (rank : 83) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
NOTC1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 117) | NC score | 0.004922 (rank : 169) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q01705, Q06007, Q61905, Q99JC2, Q9QW58, Q9R0X7 | Gene names | Notch1, Motch | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (Motch A) (mT14) (p300) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
PTN23_MOUSE
|
||||||
| θ value | 5.27518 (rank : 118) | NC score | 0.015757 (rank : 138) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
SIG10_HUMAN
|
||||||
| θ value | 5.27518 (rank : 119) | NC score | 0.009719 (rank : 155) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96LC7, Q6UXI8, Q96G54, Q96LC8 | Gene names | SIGLEC10, SLG2 | |||
|
Domain Architecture |

|
|||||
| Description | Sialic acid-binding Ig-like lectin 10 precursor (Siglec-10) (Siglec- like protein 2). | |||||
|
SON_HUMAN
|
||||||
| θ value | 5.27518 (rank : 120) | NC score | 0.063229 (rank : 46) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
CUTL1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 121) | NC score | 0.018944 (rank : 130) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
DDEF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 122) | NC score | 0.014231 (rank : 143) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9QWY8, O08612, Q80TG8, Q80UV6, Q99LV8, Q9Z2B6 | Gene names | Ddef1, Asap1, Kiaa1249, Shag1 | |||
|
Domain Architecture |

|
|||||
| Description | 130 kDa phosphatidylinositol 4,5-biphosphate-dependent ARF1 GTPase- activating protein (PIP2-dependent ARF1 GAP) (ADP-ribosylation factor- directed GTPase-activating protein 1) (ARF GTPase-activating protein 1) (Development and differentiation-enhancing factor 1) (Differentiation-enhancing factor 1) (DEF-1). | |||||
|
DLG5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 123) | NC score | 0.008649 (rank : 159) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8TDM6, Q5T1H7, Q5T1H8, Q6DKG3, Q86WC0, Q8TDM7, Q9UE73, Q9Y4E3 | Gene names | DLG5, KIAA0583, PDLG | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 5 (Placenta and prostate DLG) (Discs large protein P-dlg). | |||||
|
EMID2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 124) | NC score | 0.023493 (rank : 113) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q91VF6, Q8K4P3 | Gene names | Emid2, Col26a, Col26a1, Emu2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XXVI) chain precursor (EMI domain-containing protein 2) (Protein Emu2) (Emilin and multimerin domain-containing protein 2). | |||||
|
FOXC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 125) | NC score | 0.007706 (rank : 162) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q12948, Q86UP7, Q9BYM1, Q9NUE5, Q9UDD0, Q9UP06 | Gene names | FOXC1, FKHL7, FREAC3 | |||
|
Domain Architecture |
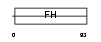
|
|||||
| Description | Forkhead box protein C1 (Forkhead-related protein FKHL7) (Forkhead- related transcription factor 3) (FREAC-3). | |||||
|
IF4G1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 126) | NC score | 0.036086 (rank : 86) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
ING4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 127) | NC score | 0.013103 (rank : 149) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UNL4, Q96E15, Q9H3J0 | Gene names | ING4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 4 (p29ING4). | |||||
|
ING4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 128) | NC score | 0.013498 (rank : 148) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C0D7, Q8C1S7, Q8K3Q5, Q8K3Q6, Q8K3Q7, Q9D7F9 | Gene names | Ing4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 4 (p29ING4). | |||||
|
KLF12_HUMAN
|
||||||
| θ value | 6.88961 (rank : 129) | NC score | -0.000381 (rank : 175) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 717 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y4X4, Q5VZM7, Q9UHZ0 | Gene names | KLF12, AP2REP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 12 (Transcriptional repressor AP-2rep). | |||||
|
MAML1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 130) | NC score | 0.030489 (rank : 100) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q92585, Q9NZ12 | Gene names | MAML1, KIAA0200 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 1 (Mam-1). | |||||
|
MPP5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 131) | NC score | 0.004558 (rank : 170) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N3R9, Q7Z631, Q86T98, Q8N7I5, Q9H9Q0 | Gene names | MPP5 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 5. | |||||
|
NLGN2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 132) | NC score | 0.007682 (rank : 163) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NFZ4, Q9P2I1 | Gene names | NLGN2, KIAA1366 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-2 precursor. | |||||
|
NLGN2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 133) | NC score | 0.007655 (rank : 164) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q69ZK9, Q5F288 | Gene names | Nlgn2, Kiaa1366 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroligin-2 precursor. | |||||
|
NRK_MOUSE
|
||||||
| θ value | 6.88961 (rank : 134) | NC score | -0.002402 (rank : 178) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 899 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9R0G8, Q6NV55, Q8C9S9, Q9R0S4 | Gene names | Nrk, Nesk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nik-related protein kinase (EC 2.7.11.1) (Nck-interacting kinase-like embryo specific kinase) (NIK-like embryo-specific kinase) (NESK). | |||||
|
PCQAP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 135) | NC score | 0.019468 (rank : 129) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q924H2 | Gene names | Pcqap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (mPcqap). | |||||
|
SELPL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 136) | NC score | 0.019980 (rank : 126) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14242, Q12775 | Gene names | SELPLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand) (CD162 antigen). | |||||
|
TCRG1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 137) | NC score | 0.021486 (rank : 122) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
ZBT38_HUMAN
|
||||||
| θ value | 6.88961 (rank : 138) | NC score | -0.000009 (rank : 174) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 885 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8NAP3 | Gene names | ZBTB38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and BTB domain-containing protein 38. | |||||
|
BCL9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 139) | NC score | 0.047567 (rank : 73) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
CAC1H_HUMAN
|
||||||
| θ value | 8.99809 (rank : 140) | NC score | 0.012203 (rank : 152) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
CIC_HUMAN
|
||||||
| θ value | 8.99809 (rank : 141) | NC score | 0.069613 (rank : 36) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CNOT3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 142) | NC score | 0.030942 (rank : 98) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O75175, Q9NZN7, Q9UF76 | Gene names | CNOT3, KIAA0691, NOT3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 143) | NC score | 0.055998 (rank : 60) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
ENAH_HUMAN
|
||||||
| θ value | 8.99809 (rank : 144) | NC score | 0.022733 (rank : 116) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8N8S7, Q502W5, Q5T5M7, Q5VTQ9, Q5VTR0, Q9NVF3, Q9UFB8 | Gene names | ENAH, MENA | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog. | |||||
|
EP300_HUMAN
|
||||||
| θ value | 8.99809 (rank : 145) | NC score | 0.040140 (rank : 82) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
FOXM1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 146) | NC score | 0.008602 (rank : 160) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O08696 | Gene names | Foxm1, Fkh16 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein M1 (Forkhead homolog 16) (Winged-helix transcription factor Trident). | |||||
|
GPR39_HUMAN
|
||||||
| θ value | 8.99809 (rank : 147) | NC score | -0.002229 (rank : 177) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 880 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43194, Q53R01 | Gene names | GPR39 | |||
|
Domain Architecture |
|
|||||
| Description | Probable G-protein coupled receptor 39. | |||||
|
K1802_MOUSE
|
||||||
| θ value | 8.99809 (rank : 148) | NC score | 0.067899 (rank : 39) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
MARK2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 149) | NC score | -0.001872 (rank : 176) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 1018 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q05512, Q6PDR4, Q8BR95 | Gene names | Mark2, Emk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase MARK2 (EC 2.7.11.1) (MAP/microtubule affinity-regulating kinase 2) (ELKL Motif Kinase) (EMK1). | |||||
|
MISS_MOUSE
|
||||||
| θ value | 8.99809 (rank : 150) | NC score | 0.093451 (rank : 18) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
POGZ_HUMAN
|
||||||
| θ value | 8.99809 (rank : 151) | NC score | 0.002386 (rank : 172) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q7Z3K3, O75049, Q8TDZ7, Q9Y4X7 | Gene names | POGZ, KIAA0461 | |||
|
Domain Architecture |

|
|||||
| Description | Pogo transposable element with ZNF domain. | |||||
|
PRG4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 152) | NC score | 0.062940 (rank : 47) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRP5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 153) | NC score | 0.022347 (rank : 118) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P04281 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide IB-1. | |||||
|
SHAN2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 154) | NC score | 0.017430 (rank : 132) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q80Z38, Q3UTK4, Q5DU07 | Gene names | Shank2, Kiaa1022 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 2 (Shank2) (Cortactin- binding protein 1) (CortBP1). | |||||
|
SNX1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 155) | NC score | 0.005617 (rank : 168) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WV80, Q9EQZ9 | Gene names | Snx1 | |||
|
Domain Architecture |
|
|||||
| Description | Sorting nexin-1. | |||||
|
TIF1G_MOUSE
|
||||||
| θ value | 8.99809 (rank : 156) | NC score | 0.008743 (rank : 157) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q99PP7, Q6SI71, Q6ZPX5 | Gene names | Trim33, Kiaa1113 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33). | |||||
|
BCL9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.056615 (rank : 58) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
BSN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.066406 (rank : 41) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CEL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.060641 (rank : 49) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
CS016_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.076251 (rank : 25) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8ND99 | Gene names | C19orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf16. | |||||
|
HRBL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.054445 (rank : 63) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q80WC7, Q8BKS5, Q99J67 | Gene names | Hrbl, Rabr | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.060168 (rank : 50) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.058952 (rank : 52) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.058352 (rank : 55) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
MLL3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.066286 (rank : 43) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.095452 (rank : 16) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.083390 (rank : 24) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.069746 (rank : 33) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
PI5PA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.058547 (rank : 54) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
PO121_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.065951 (rank : 44) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.086886 (rank : 19) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
PS1C2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.076095 (rank : 26) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UIG4 | Gene names | PSORS1C2, C6orf17, SPR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Psoriasis susceptibility 1 candidate gene 2 protein precursor (SPR1 protein). | |||||
|
PS1C2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.066703 (rank : 40) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80ZC9 | Gene names | Psors1c2, Spr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Psoriasis susceptibility 1 candidate gene 2 protein homolog precursor (SPR1 protein). | |||||
|
RPB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.053641 (rank : 64) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P24928 | Gene names | POLR2A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
RPB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.053640 (rank : 65) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P08775 | Gene names | Polr2a, Rpii215, Rpo2-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
SON_MOUSE
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.056638 (rank : 57) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
SSBP4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.054473 (rank : 62) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
TCF19_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.055032 (rank : 61) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y242, Q13176, Q15967, Q5SQ89, Q5STD6, Q5STF5, Q9BUM2, Q9UBH7 | Gene names | TCF19, SC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 19 (Transcription factor SC1). | |||||
|
EPN2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 156 | |
| SwissProt Accessions | O95208, O95207, Q9H7Z2, Q9UPT7 | Gene names | EPN2, KIAA1065 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-2 (EPS-15-interacting protein 2). | |||||
|
EPN2_MOUSE
|
||||||
| NC score | 0.960883 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8CHU3, O70447, Q8BZ85 | Gene names | Epn2 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-2 (EPS-15-interacting protein 2) (Intersectin-EH-binding protein 2) (Ibp2). | |||||
|
EPN3_HUMAN
|
||||||
| NC score | 0.896010 (rank : 3) | θ value | 2.63489e-92 (rank : 3) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9H201, Q9BVN6, Q9NWK2 | Gene names | EPN3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
EPN1_MOUSE
|
||||||
| NC score | 0.884352 (rank : 4) | θ value | 2.73936e-73 (rank : 6) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q80VP1, O70446 | Gene names | Epn1 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-1 (EPS-15-interacting protein 1) (Intersectin-EH-binding protein 1) (Ibp1). | |||||
|
EPN1_HUMAN
|
||||||
| NC score | 0.883752 (rank : 5) | θ value | 1.8332e-77 (rank : 5) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9Y6I3, Q86ST3, Q9HA18 | Gene names | EPN1 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-1 (EPS-15-interacting protein 1) (EH domain-binding mitotic phosphoprotein). | |||||
|
EPN3_MOUSE
|
||||||
| NC score | 0.879599 (rank : 6) | θ value | 1.22395e-89 (rank : 4) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q91W69, Q9CV55 | Gene names | Epn3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
EPN4_HUMAN
|
||||||
| NC score | 0.824570 (rank : 7) | θ value | 8.89434e-40 (rank : 7) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14677, Q8NAF1, Q96E05 | Gene names | CLINT1, ENTH, EPN4, EPNR, KIAA0171 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin interactor 1 (Epsin-4) (Epsin-related protein) (EpsinR) (Enthoprotin) (Clathrin-interacting protein localized in the trans- Golgi region) (Clint). | |||||
|
EPN4_MOUSE
|
||||||
| NC score | 0.822377 (rank : 8) | θ value | 8.89434e-40 (rank : 8) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99KN9, Q8CFH4 | Gene names | Clint1, Enth, Epn4, Epnr, Kiaa0171 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin interactor 1 (Epsin-4) (Epsin-related protein) (EpsinR) (Enthoprotin). | |||||
|
CN032_HUMAN
|
||||||
| NC score | 0.141636 (rank : 9) | θ value | 0.0148317 (rank : 11) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
NU214_HUMAN
|
||||||
| NC score | 0.127246 (rank : 10) | θ value | 0.00134147 (rank : 9) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
CN032_MOUSE
|
||||||
| NC score | 0.107782 (rank : 11) | θ value | 1.81305 (rank : 71) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.105399 (rank : 12) | θ value | 0.0252991 (rank : 15) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.104781 (rank : 13) | θ value | 0.0193708 (rank : 13) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
HRBL_HUMAN
|
||||||
| NC score | 0.102212 (rank : 14) | θ value | 0.0431538 (rank : 18) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O95081, O75429, Q96AB9, Q96GL4 | Gene names | HRBL, RABR | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.097018 (rank : 15) | θ value | 0.163984 (rank : 30) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 110 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.095452 (rank : 16) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
AP180_MOUSE
|
||||||
| NC score | 0.094918 (rank : 17) | θ value | 0.813845 (rank : 48) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q61548, Q61547 | Gene names | Snap91 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin coat assembly protein AP180 (Clathrin coat-associated protein AP180) (91 kDa synaptosomal-associated protein) (Phosphoprotein F1- 20). | |||||
|
MISS_MOUSE
|
||||||
| NC score | 0.093451 (rank : 18) | θ value | 8.99809 (rank : 150) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.086886 (rank : 19) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.086339 (rank : 20) | θ value | 0.279714 (rank : 34) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
K1802_HUMAN
|
||||||
| NC score | 0.084902 (rank : 21) | θ value | 0.0148317 (rank : 12) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
CO002_HUMAN
|
||||||
| NC score | 0.083761 (rank : 22) | θ value | 0.365318 (rank : 36) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9NZP6 | Gene names | C15orf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C15orf2. | |||||
|
PO121_MOUSE
|
||||||
| NC score | 0.083434 (rank : 23) | θ value | 0.0330416 (rank : 16) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.083390 (rank : 24) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
CS016_HUMAN
|
||||||
| NC score | 0.076251 (rank : 25) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8ND99 | Gene names | C19orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf16. | |||||
|
PS1C2_HUMAN
|
||||||
| NC score | 0.076095 (rank : 26) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UIG4 | Gene names | PSORS1C2, C6orf17, SPR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Psoriasis susceptibility 1 candidate gene 2 protein precursor (SPR1 protein). | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.074234 (rank : 27) | θ value | 0.813845 (rank : 52) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
KCNE4_HUMAN
|
||||||
| NC score | 0.073468 (rank : 28) | θ value | 0.21417 (rank : 32) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WWG9, Q96CC4 | Gene names | KCNE4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily E member 4 (Minimum potassium ion channel-related peptide 3) (Potassium channel subunit beta MiRP3) (MinK-related peptide 3). | |||||
|
AP180_HUMAN
|
||||||
| NC score | 0.072503 (rank : 29) | θ value | 4.03905 (rank : 98) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O60641, Q5JX13, Q68DL9, Q6P9D3, Q9NTY7 | Gene names | SNAP91, KIAA0656 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin coat assembly protein AP180 (Clathrin coat-associated protein AP180) (91 kDa synaptosomal-associated protein). | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.071942 (rank : 30) | θ value | 0.163984 (rank : 29) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
KCNE4_MOUSE
|
||||||
| NC score | 0.071471 (rank : 31) | θ value | 0.279714 (rank : 33) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WTW3 | Gene names | Kcne4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily E member 4 (Minimum potassium ion channel-related peptide 3) (Potassium channel subunit beta MiRP3) (MinK-related peptide 3). | |||||
|
NUP62_HUMAN
|
||||||
| NC score | 0.071004 (rank : 32) | θ value | 2.36792 (rank : 82) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P37198, Q503A4, Q96C43, Q9NSL1 | Gene names | NUP62 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore glycoprotein p62 (62 kDa nucleoporin). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.069746 (rank : 33) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
CIC_MOUSE
|
||||||
| NC score | 0.069736 (rank : 34) | θ value | 0.365318 (rank : 35) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
NRX2B_HUMAN
|
||||||
| NC score | 0.069720 (rank : 35) | θ value | 0.0563607 (rank : 20) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.069613 (rank : 36) | θ value | 8.99809 (rank : 141) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
NUP62_MOUSE
|
||||||
| NC score | 0.069373 (rank : 37) | θ value | 0.125558 (rank : 27) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q63850, Q99JN7 | Gene names | Nup62 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore glycoprotein p62 (62 kDa nucleoporin). | |||||
|
BORG5_HUMAN
|
||||||
| NC score | 0.068055 (rank : 38) | θ value | 0.0431538 (rank : 17) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q00587, Q96GN1 | Gene names | CDC42EP1, BORG5, CEP1, MSE55 | |||
|
Domain Architecture |

|
|||||
| Description | Cdc42 effector protein 1 (Binder of Rho GTPases 5) (Serum protein MSE55). | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.067899 (rank : 39) | θ value | 8.99809 (rank : 148) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
PS1C2_MOUSE
|
||||||
| NC score | 0.066703 (rank : 40) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80ZC9 | Gene names | Psors1c2, Spr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Psoriasis susceptibility 1 candidate gene 2 protein homolog precursor (SPR1 protein). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.066406 (rank : 41) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
TNR6A_HUMAN
|
||||||
| NC score | 0.066366 (rank : 42) | θ value | 0.125558 (rank : 28) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8NDV7, O15408, Q658L5, Q6NVB5, Q8NEZ0, Q8TBT8, Q8TCR0, Q9NV59, Q9P268 | Gene names | TNRC6A, CAGH26, KIAA1460, TNRC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trinucleotide repeat-containing gene 6A protein (CAG repeat protein 26) (Glycine-tryptophan protein of 182 kDa) (GW182 autoantigen) (Protein GW1) (EMSY interactor protein). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.066286 (rank : 43) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
PO121_HUMAN
|
||||||
| NC score | 0.065951 (rank : 44) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
ENAH_MOUSE
|
||||||
| NC score | 0.064054 (rank : 45) | θ value | 0.365318 (rank : 37) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.063229 (rank : 46) | θ value | 5.27518 (rank : 120) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.062940 (rank : 47) | θ value | 8.99809 (rank : 152) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.061732 (rank : 48) | θ value | 0.813845 (rank : 50) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.060641 (rank : 49) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.060168 (rank : 50) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.059587 (rank : 51) | θ value | 1.38821 (rank : 68) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.058952 (rank : 52) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MUC5B_HUMAN
|
||||||
| NC score | 0.058622 (rank : 53) | θ value | 0.0193708 (rank : 14) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
PI5PA_HUMAN
|
||||||
| NC score | 0.058547 (rank : 54) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.058352 (rank : 55) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.057839 (rank : 56) | θ value | 0.0961366 (rank : 25) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.056638 (rank : 57) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.056615 (rank : 58) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.056006 (rank : 59) | θ value | 4.03905 (rank : 109) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.055998 (rank : 60) | θ value | 8.99809 (rank : 143) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
TCF19_HUMAN
|
||||||
| NC score | 0.055032 (rank : 61) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y242, Q13176, Q15967, Q5SQ89, Q5STD6, Q5STF5, Q9BUM2, Q9UBH7 | Gene names | TCF19, SC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 19 (Transcription factor SC1). | |||||
|
SSBP4_HUMAN
|
||||||
| NC score | 0.054473 (rank : 62) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
HRBL_MOUSE
|
||||||
| NC score | 0.054445 (rank : 63) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q80WC7, Q8BKS5, Q99J67 | Gene names | Hrbl, Rabr | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
RPB1_HUMAN
|
||||||
| NC score | 0.053641 (rank : 64) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P24928 | Gene names | POLR2A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
RPB1_MOUSE
|
||||||
| NC score | 0.053640 (rank : 65) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P08775 | Gene names | Polr2a, Rpii215, Rpo2-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
CJ078_MOUSE
|
||||||
| NC score | 0.052782 (rank : 66) | θ value | 0.125558 (rank : 26) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.052632 (rank : 67) | θ value | 0.62314 (rank : 44) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
NU153_HUMAN
|
||||||
| NC score | 0.051571 (rank : 68) | θ value | 0.62314 (rank : 43) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P49790 | Gene names | NUP153 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup153 (Nucleoporin Nup153) (153 kDa nucleoporin). | |||||
|
DYN1_HUMAN
|
||||||
| NC score | 0.049610 (rank : 69) | θ value | 0.0961366 (rank : 22) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q05193 | Gene names | DNM1, DNM | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-1 (EC 3.6.5.5). | |||||
|
DYN1_MOUSE
|
||||||
| NC score | 0.049006 (rank : 70) | θ value | 0.0961366 (rank : 23) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P39053, Q61358, Q61359, Q61360, Q8JZZ4, Q9CSY7, Q9QXX1 | Gene names | Dnm1, Dnm | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-1 (EC 3.6.5.5). | |||||
|
NRX2A_HUMAN
|
||||||
| NC score | 0.048933 (rank : 71) | θ value | 0.0563607 (rank : 19) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
MUCDL_HUMAN
|
||||||
| NC score | 0.048452 (rank : 72) | θ value | 1.06291 (rank : 58) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
BCL9_MOUSE
|
||||||
| NC score | 0.047567 (rank : 73) | θ value | 8.99809 (rank : 139) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
NCOR2_HUMAN
|
||||||
| NC score | 0.046635 (rank : 74) | θ value | 0.62314 (rank : 42) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
SYNG_MOUSE
|
||||||
| NC score | 0.045896 (rank : 75) | θ value | 2.36792 (rank : 87) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q5SV85, Q5SV84, Q6PHT6 | Gene names | Ap1gbp1, Syng | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP1 subunit gamma-binding protein 1 (Gamma-synergin). | |||||
|
TP53B_MOUSE
|
||||||
| NC score | 0.045230 (rank : 76) | θ value | 4.03905 (rank : 111) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
ARMX2_MOUSE
|
||||||
| NC score | 0.044329 (rank : 77) | θ value | 0.813845 (rank : 49) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6A058, Q9CXI9 | Gene names | Armcx2, Kiaa0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2. | |||||
|
SRA1_HUMAN
|
||||||
| NC score | 0.044076 (rank : 78) | θ value | 0.62314 (rank : 46) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9HD15, Q6NVU9, Q8IXM1, Q9HD13, Q9HD14 | Gene names | SRA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Steroid receptor RNA activator 1 (Steroid receptor RNA activator protein) (SRAP). | |||||
|
SYNG_HUMAN
|
||||||
| NC score | 0.042601 (rank : 79) | θ value | 0.62314 (rank : 47) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UMZ2, Q6ZT17 | Gene names | AP1GBP1, SYNG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP1 subunit gamma-binding protein 1 (Gamma-synergin). | |||||
|
IF4G1_HUMAN
|
||||||
| NC score | 0.041506 (rank : 80) | θ value | 1.06291 (rank : 57) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q04637, O43177, O95066, Q5HYG0, Q6ZN21, Q8N102 | Gene names | EIF4G1, EIF4G, EIF4GI | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1) (p220). | |||||
|
SNIP_MOUSE
|
||||||
| NC score | 0.040962 (rank : 81) | θ value | 0.62314 (rank : 45) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9QWI6, O70298 | Gene names | P140, Kiaa1684 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | p130Cas-associated protein (p140Cap) (SNAP-25-interacting protein) (SNIP). | |||||
|
EP300_HUMAN
|
||||||
| NC score | 0.040140 (rank : 82) | θ value | 8.99809 (rank : 145) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
MILK1_MOUSE
|
||||||
| NC score | 0.039158 (rank : 83) | θ value | 5.27518 (rank : 116) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
SHAN1_HUMAN
|
||||||
| NC score | 0.038169 (rank : 84) | θ value | 1.81305 (rank : 76) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
NPAS4_MOUSE
|
||||||
| NC score | 0.037543 (rank : 85) | θ value | 1.06291 (rank : 59) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8BGD7, Q3V3U3 | Gene names | Npas4, Nxf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF) (Limbic-enhanced PAS protein) (LE-PAS). | |||||
|
IF4G1_MOUSE
|
||||||
| NC score | 0.036086 (rank : 86) | θ value | 6.88961 (rank : 126) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
NSD1_HUMAN
|
||||||
| NC score | 0.035927 (rank : 87) | θ value | 0.365318 (rank : 38) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
PIAS3_HUMAN
|
||||||
| NC score | 0.034187 (rank : 88) | θ value | 2.36792 (rank : 85) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y6X2, Q9UFI3 | Gene names | PIAS3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein inhibitor of activated STAT protein 3. | |||||
|
SNIP_HUMAN
|
||||||
| NC score | 0.033379 (rank : 89) | θ value | 4.03905 (rank : 108) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9C0H9, Q8N4W8 | Gene names | P140, KIAA1684 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | p130Cas-associated protein (p140Cap) (SNAP-25-interacting protein) (SNIP). | |||||
|
PIAS3_MOUSE
|
||||||
| NC score | 0.032912 (rank : 90) | θ value | 1.81305 (rank : 75) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O54714, Q80WF8, Q8R598 | Gene names | Pias3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein inhibitor of activated STAT protein 3. | |||||
|
DEN1A_MOUSE
|
||||||
| NC score | 0.032431 (rank : 91) | θ value | 3.0926 (rank : 92) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8K382 | Gene names | Dennd1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 1A. | |||||
|
MARCS_HUMAN
|
||||||
| NC score | 0.031886 (rank : 92) | θ value | 2.36792 (rank : 80) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
ATF2_MOUSE
|
||||||
| NC score | 0.031878 (rank : 93) | θ value | 4.03905 (rank : 100) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P16951, Q64089, Q64090, Q64091 | Gene names | Atf2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (MXBP protein). | |||||
|
ATF2_HUMAN
|
||||||
| NC score | 0.031805 (rank : 94) | θ value | 4.03905 (rank : 99) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P15336, Q13000 | Gene names | ATF2, CREB2, CREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (HB16). | |||||
|
BCOR_HUMAN
|
||||||
| NC score | 0.031572 (rank : 95) | θ value | 3.0926 (rank : 89) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6W2J9, Q6P4B6, Q7Z2K7, Q8TEB4, Q96DB3, Q9H232, Q9H233, Q9HCJ7, Q9NXF2 | Gene names | BCOR, KIAA1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
K0460_MOUSE
|
||||||
| NC score | 0.031388 (rank : 96) | θ value | 3.0926 (rank : 93) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6NXI6, Q3U3L8, Q6ZQA7 | Gene names | Kiaa0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
CSKI2_HUMAN
|
||||||
| NC score | 0.031264 (rank : 97) | θ value | 0.0736092 (rank : 21) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8WXE0, Q7LG69, Q9ULT1 | Gene names | CASKIN2, KIAA1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
CNOT3_HUMAN
|
||||||
| NC score | 0.030942 (rank : 98) | θ value | 8.99809 (rank : 142) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O75175, Q9NZN7, Q9UF76 | Gene names | CNOT3, KIAA0691, NOT3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.030681 (rank : 99) | θ value | 3.0926 (rank : 95) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
MAML1_HUMAN
|
||||||
| NC score | 0.030489 (rank : 100) | θ value | 6.88961 (rank : 130) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q92585, Q9NZ12 | Gene names | MAML1, KIAA0200 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 1 (Mam-1). | |||||
|
ROBO4_MOUSE
|
||||||
| NC score | 0.030076 (rank : 101) | θ value | 0.47712 (rank : 39) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8C310, Q9DBW1 | Gene names | Robo4 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 4 precursor. | |||||
|
PDLI7_HUMAN
|
||||||
| NC score | 0.029526 (rank : 102) | θ value | 1.06291 (rank : 60) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 547 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9NR12, Q14250, Q5XG82, Q6NVZ5, Q96C91, Q9BXB8, Q9BXB9 | Gene names | PDLIM7, ENIGMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
BCOR_MOUSE
|
||||||
| NC score | 0.028360 (rank : 103) | θ value | 3.0926 (rank : 90) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8CGN4, Q6PDK5, Q6ZPM3, Q8BKF5, Q8CGN1, Q8CGN2, Q8CGN3 | Gene names | Bcor, Kiaa1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
TR10C_HUMAN
|
||||||
| NC score | 0.028114 (rank : 104) | θ value | 2.36792 (rank : 88) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O14798, O14755, Q6UXM5 | Gene names | TNFRSF10C, DCR1, LIT, TRAILR3, TRID | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 10C precursor (Decoy receptor 1) (DcR1) (Decoy TRAIL receptor without death domain) (TNF- related apoptosis-inducing ligand receptor 3) (TRAIL receptor 3) (TRAIL-R3) (Trail receptor without an intracellular domain) (Lymphocyte inhibitor of TRAIL) (Antagonist decoy receptor for TRAIL/Apo-2L) (CD263 antigen). | |||||
|
RAI2_MOUSE
|
||||||
| NC score | 0.027647 (rank : 105) | θ value | 1.38821 (rank : 66) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QVY8 | Gene names | Rai2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 2 (3f8). | |||||
|
MILK1_HUMAN
|
||||||
| NC score | 0.027582 (rank : 106) | θ value | 1.38821 (rank : 63) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8N3F8, Q5TI16, Q7RTP5, Q8N3N8, Q9BVL9, Q9BY92, Q9UH43, Q9UH44, Q9UH45 | Gene names | MIRAB13, KIAA1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
AFF3_HUMAN
|
||||||
| NC score | 0.027004 (rank : 107) | θ value | 1.06291 (rank : 54) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P51826 | Gene names | AFF3, LAF4 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 3 (Protein LAF-4) (Lymphoid nuclear protein related to AF4). | |||||
|
ATX2L_MOUSE
|
||||||
| NC score | 0.026863 (rank : 108) | θ value | 4.03905 (rank : 101) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q7TQH0, Q80XN9, Q8K059 | Gene names | Atxn2l, A2lp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein. | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.026824 (rank : 109) | θ value | 1.81305 (rank : 70) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
SMP1L_MOUSE
|
||||||
| NC score | 0.025964 (rank : 110) | θ value | 1.38821 (rank : 67) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7TN29, Q3U798 | Gene names | Smap1l, Smap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1-like (Stromal membrane- associated protein 2). | |||||
|
ZN750_HUMAN
|
||||||
| NC score | 0.025276 (rank : 111) | θ value | 4.03905 (rank : 112) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q32MQ0, Q9H899 | Gene names | ZNF750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
DSCAM_HUMAN
|
||||||
| NC score | 0.023770 (rank : 112) | θ value | 0.00390308 (rank : 10) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60469, O60468 | Gene names | DSCAM | |||
|
Domain Architecture |

|
|||||
| Description | Down syndrome cell adhesion molecule precursor (CHD2). | |||||
|
EMID2_MOUSE
|
||||||
| NC score | 0.023493 (rank : 113) | θ value | 6.88961 (rank : 124) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q91VF6, Q8K4P3 | Gene names | Emid2, Col26a, Col26a1, Emu2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XXVI) chain precursor (EMI domain-containing protein 2) (Protein Emu2) (Emilin and multimerin domain-containing protein 2). | |||||
|
EMID2_HUMAN
|
||||||
| NC score | 0.023268 (rank : 114) | θ value | 1.06291 (rank : 56) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96A83 | Gene names | EMID2, COL26A1, EMU2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XXVI) chain precursor (EMI domain-containing protein 2) (Protein Emu2) (Emilin and multimerin domain-containing protein 2). | |||||
|
DAAM2_MOUSE
|
||||||
| NC score | 0.022818 (rank : 115) | θ value | 0.21417 (rank : 31) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 371 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q80U19, Q810J5 | Gene names | Daam2, Kiaa0381 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 2. | |||||
|
ENAH_HUMAN
|
||||||
| NC score | 0.022733 (rank : 116) | θ value | 8.99809 (rank : 144) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8N8S7, Q502W5, Q5T5M7, Q5VTQ9, Q5VTR0, Q9NVF3, Q9UFB8 | Gene names | ENAH, MENA | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog. | |||||
|
SMP1L_HUMAN
|
||||||
| NC score | 0.022503 (rank : 117) | θ value | 4.03905 (rank : 107) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8WU79, Q5QPL2, Q96C93, Q9NST2, Q9UJL8 | Gene names | SMAP1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1-like. | |||||
|
PRP5_HUMAN
|
||||||
| NC score | 0.022347 (rank : 118) | θ value | 8.99809 (rank : 153) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P04281 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide IB-1. | |||||
|
PHC3_MOUSE
|
||||||
| NC score | 0.021947 (rank : 119) | θ value | 2.36792 (rank : 83) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8CHP6, Q3TLV5 | Gene names | Phc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 3. | |||||
|
TCGAP_MOUSE
|
||||||
| NC score | 0.021857 (rank : 120) | θ value | 1.38821 (rank : 69) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80YF9, Q6P7W6 | Gene names | Snx26, Tcgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
NFAC1_HUMAN
|
||||||
| NC score | 0.021563 (rank : 121) | θ value | 1.38821 (rank : 64) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O95644, Q12865, Q15793 | Gene names | NFATC1, NFAT2, NFATC | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 1 (NFAT transcription complex cytosolic component) (NF-ATc1) (NF-ATc). | |||||
|
TCRG1_MOUSE
|
||||||
| NC score | 0.021486 (rank : 122) | θ value | 6.88961 (rank : 137) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
AMOT_HUMAN
|
||||||
| NC score | 0.020996 (rank : 123) | θ value | 2.36792 (rank : 77) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q4VCS5, Q504X5, Q9HD27, Q9UPT1 | Gene names | AMOT, KIAA1071 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin. | |||||
|
CLCN6_MOUSE
|
||||||
| NC score | 0.020825 (rank : 124) | θ value | 1.38821 (rank : 61) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35454 | Gene names | Clcn6, Clc6 | |||
|
Domain Architecture |

|
|||||
| Description | Chloride channel protein 6 (ClC-6). | |||||
|
CU056_MOUSE
|
||||||
| NC score | 0.019988 (rank : 125) | θ value | 4.03905 (rank : 102) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D9W0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative protein C21orf56 homolog. | |||||
|
SELPL_HUMAN
|
||||||
| NC score | 0.019980 (rank : 126) | θ value | 6.88961 (rank : 136) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14242, Q12775 | Gene names | SELPLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand) (CD162 antigen). | |||||
|
TERF2_MOUSE
|
||||||
| NC score | 0.019903 (rank : 127) | θ value | 4.03905 (rank : 110) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O35144 | Gene names | Terf2, Trf2 | |||
|
Domain Architecture |

|
|||||
| Description | Telomeric repeat-binding factor 2 (TTAGGG repeat-binding factor 2) (Telomeric DNA-binding protein). | |||||
|
LDB3_MOUSE
|
||||||
| NC score | 0.019729 (rank : 128) | θ value | 1.38821 (rank : 62) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9JKS4, Q6A038, Q811P2, Q811P3, Q811P4, Q811P5, Q9D130, Q9JKS3, Q9R0Z1, Q9WVH1, Q9WVH2 | Gene names | Ldb3, Kiaa0613 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher) (Protein oracle). | |||||
|
PCQAP_MOUSE
|
||||||
| NC score | 0.019468 (rank : 129) | θ value | 6.88961 (rank : 135) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q924H2 | Gene names | Pcqap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (mPcqap). | |||||
|
CUTL1_MOUSE
|
||||||
| NC score | 0.018944 (rank : 130) | θ value | 6.88961 (rank : 121) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
PHF14_MOUSE
|
||||||
| NC score | 0.018288 (rank : 131) | θ value | 2.36792 (rank : 84) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9D4H9, Q810Z6, Q8C284, Q8CGF8, Q9D1Q8 | Gene names | Phf14 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 14. | |||||
|
SHAN2_MOUSE
|
||||||
| NC score | 0.017430 (rank : 132) | θ value | 8.99809 (rank : 154) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q80Z38, Q3UTK4, Q5DU07 | Gene names | Shank2, Kiaa1022 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 2 (Shank2) (Cortactin- binding protein 1) (CortBP1). | |||||
|
CLCN6_HUMAN
|
||||||
| NC score | 0.017143 (rank : 133) | θ value | 5.27518 (rank : 113) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P51797, O60818, O60819, O60820, O60821, P78520, P78521, Q5SNW2, Q5SNW3, Q5SNX1, Q5SNX2, Q5SNX3, Q99427, Q99428, Q99429 | Gene names | CLCN6, KIAA0046 | |||
|
Domain Architecture |

|
|||||
| Description | Chloride channel protein 6 (ClC-6). | |||||
|
SALL3_HUMAN
|
||||||
| NC score | 0.016915 (rank : 134) | θ value | 0.0961366 (rank : 24) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 1066 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9BXA9, Q9UGH1 | Gene names | SALL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 3 (Zinc finger protein SALL3) (hSALL3). | |||||
|
ATBF1_MOUSE
|
||||||
| NC score | 0.016099 (rank : 135) | θ value | 2.36792 (rank : 78) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
ZHX1_MOUSE
|
||||||
| NC score | 0.016043 (rank : 136) | θ value | 3.0926 (rank : 97) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P70121, Q8BQ68, Q8C6T4, Q8CJG3 | Gene names | Zhx1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc fingers and homeoboxes protein 1. | |||||
|
MYPT2_HUMAN
|
||||||
| NC score | 0.016016 (rank : 137) | θ value | 0.813845 (rank : 51) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 707 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O60237, Q8N179, Q9HCB7, Q9HCB8 | Gene names | PPP1R12B, MYPT2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12B (Myosin phosphatase- targeting subunit 2) (Myosin phosphatase target subunit 2). | |||||
|
PTN23_MOUSE
|
||||||
| NC score | 0.015757 (rank : 138) | θ value | 5.27518 (rank : 118) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
GNRP_HUMAN
|
||||||
| NC score | 0.015255 (rank : 139) | θ value | 2.36792 (rank : 79) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13972, Q16027 | Gene names | RASGRF1, CDC25 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-releasing protein (GNRP) (Ras-specific nucleotide exchange factor CDC25) (Ras-specific guanine nucleotide-releasing factor). | |||||
|
NGN2_MOUSE
|
||||||
| NC score | 0.014777 (rank : 140) | θ value | 1.38821 (rank : 65) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70447, P70237 | Gene names | Neurog2, Ath4a, Atoh4, Ngn2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 2 (Atonal protein homolog 4) (Helix-loop-helix protein mATH-4A) (MATH4A). | |||||
|
MEF2A_HUMAN
|
||||||
| NC score | 0.014461 (rank : 141) | θ value | 2.36792 (rank : 81) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q02078, O43814, Q14223, Q14224, Q96D14 | Gene names | MEF2A, MEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2A (Serum response factor-like protein 1). | |||||
|
RNF12_MOUSE
|
||||||
| NC score | 0.014313 (rank : 142) | θ value | 2.36792 (rank : 86) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WTV7 | Gene names | Rnf12, Rlim | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 12 (LIM domain-interacting RING finger protein) (RING finger LIM domain-binding protein) (R-LIM). | |||||
|
DDEF1_MOUSE
|
||||||
| NC score | 0.014231 (rank : 143) | θ value | 6.88961 (rank : 122) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9QWY8, O08612, Q80TG8, Q80UV6, Q99LV8, Q9Z2B6 | Gene names | Ddef1, Asap1, Kiaa1249, Shag1 | |||
|
Domain Architecture |

|
|||||
| Description | 130 kDa phosphatidylinositol 4,5-biphosphate-dependent ARF1 GTPase- activating protein (PIP2-dependent ARF1 GAP) (ADP-ribosylation factor- directed GTPase-activating protein 1) (ARF GTPase-activating protein 1) (Development and differentiation-enhancing factor 1) (Differentiation-enhancing factor 1) (DEF-1). | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.014220 (rank : 144) | θ value | 4.03905 (rank : 105) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
FOXJ3_MOUSE
|
||||||
| NC score | 0.014081 (rank : 145) | θ value | 0.62314 (rank : 40) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BUR3, Q3UTD8 | Gene names | Foxj3, Kiaa1041 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein J3. | |||||
|
GPC2_HUMAN
|
||||||
| NC score | 0.013782 (rank : 146) | θ value | 1.81305 (rank : 73) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N158 | Gene names | GPC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glypican-2 precursor. | |||||
|
PO2F1_HUMAN
|
||||||
| NC score | 0.013622 (rank : 147) | θ value | 0.813845 (rank : 53) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P14859, Q5TBT7, Q6PK46, Q8NEU9, Q9BPV1 | Gene names | POU2F1, OCT1, OTF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 2, transcription factor 1 (Octamer-binding transcription factor 1) (Oct-1) (OTF-1) (NF-A1). | |||||
|
ING4_MOUSE
|
||||||
| NC score | 0.013498 (rank : 148) | θ value | 6.88961 (rank : 128) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C0D7, Q8C1S7, Q8K3Q5, Q8K3Q6, Q8K3Q7, Q9D7F9 | Gene names | Ing4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 4 (p29ING4). | |||||
|
ING4_HUMAN
|
||||||
| NC score | 0.013103 (rank : 149) | θ value | 6.88961 (rank : 127) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UNL4, Q96E15, Q9H3J0 | Gene names | ING4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 4 (p29ING4). | |||||
|
CUTL1_HUMAN
|
||||||
| NC score | 0.012957 (rank : 150) | θ value | 4.03905 (rank : 103) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
MEFV_MOUSE
|
||||||
| NC score | 0.012918 (rank : 151) | θ value | 3.0926 (rank : 94) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JJ26 | Gene names | Mefv | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
CAC1H_HUMAN
|
||||||
| NC score | 0.012203 (rank : 152) | θ value | 8.99809 (rank : 140) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
HUWE1_MOUSE
|
||||||
| NC score | 0.010126 (rank : 153) | θ value | 5.27518 (rank : 115) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
CAC1I_HUMAN
|
||||||
| NC score | 0.009808 (rank : 154) | θ value | 3.0926 (rank : 91) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9P0X4, O95504, Q5JZ88, Q7Z6S9, Q8NFX6, Q9NZC8, Q9UH15, Q9UH30, Q9ULU9, Q9UNE6 | Gene names | CACNA1I, KIAA1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1I (Voltage- gated calcium channel subunit alpha Cav3.3) (Ca(v)3.3). | |||||
|
SIG10_HUMAN
|
||||||
| NC score | 0.009719 (rank : 155) | θ value | 5.27518 (rank : 119) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96LC7, Q6UXI8, Q96G54, Q96LC8 | Gene names | SIGLEC10, SLG2 | |||
|
Domain Architecture |

|
|||||
| Description | Sialic acid-binding Ig-like lectin 10 precursor (Siglec-10) (Siglec- like protein 2). | |||||
|
PLCB1_HUMAN
|
||||||
| NC score | 0.009280 (rank : 156) | θ value | 4.03905 (rank : 106) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NQ66, O60325, Q5TFF7, Q5TGC9, Q8IV93, Q9BQW2, Q9H4H2, Q9H8H5, Q9NQ65, Q9NQH9, Q9NTH4, Q9UJP6, Q9UM26 | Gene names | PLCB1, KIAA0581 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-1) (PLC-beta-1) (PLC-I) (PLC-154). | |||||
|
TIF1G_MOUSE
|
||||||
| NC score | 0.008743 (rank : 157) | θ value | 8.99809 (rank : 156) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q99PP7, Q6SI71, Q6ZPX5 | Gene names | Trim33, Kiaa1113 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33). | |||||
|
FARP1_HUMAN
|
||||||
| NC score | 0.008697 (rank : 158) | θ value | 1.81305 (rank : 72) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y4F1, Q6IQ29 | Gene names | FARP1, CDEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 1 (Chondrocyte- derived ezrin-like protein). | |||||
|
DLG5_HUMAN
|
||||||
| NC score | 0.008649 (rank : 159) | θ value | 6.88961 (rank : 123) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8TDM6, Q5T1H7, Q5T1H8, Q6DKG3, Q86WC0, Q8TDM7, Q9UE73, Q9Y4E3 | Gene names | DLG5, KIAA0583, PDLG | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 5 (Placenta and prostate DLG) (Discs large protein P-dlg). | |||||
|
FOXM1_MOUSE
|
||||||
| NC score | 0.008602 (rank : 160) | θ value | 8.99809 (rank : 146) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O08696 | Gene names | Foxm1, Fkh16 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein M1 (Forkhead homolog 16) (Winged-helix transcription factor Trident). | |||||
|
SFTPD_HUMAN
|
||||||
| NC score | 0.007810 (rank : 161) | θ value | 3.0926 (rank : 96) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P35247, Q86YK9, Q9UCJ2, Q9UCJ3 | Gene names | SFTPD, PSPD, SFTP4 | |||
|
Domain Architecture |

|
|||||
| Description | Pulmonary surfactant-associated protein D precursor (SP-D) (PSP-D). | |||||
|
FOXC1_HUMAN
|
||||||
| NC score | 0.007706 (rank : 162) | θ value | 6.88961 (rank : 125) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q12948, Q86UP7, Q9BYM1, Q9NUE5, Q9UDD0, Q9UP06 | Gene names | FOXC1, FKHL7, FREAC3 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein C1 (Forkhead-related protein FKHL7) (Forkhead- related transcription factor 3) (FREAC-3). | |||||
|
NLGN2_HUMAN
|
||||||
| NC score | 0.007682 (rank : 163) | θ value | 6.88961 (rank : 132) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NFZ4, Q9P2I1 | Gene names | NLGN2, KIAA1366 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-2 precursor. | |||||
|
NLGN2_MOUSE
|
||||||
| NC score | 0.007655 (rank : 164) | θ value | 6.88961 (rank : 133) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q69ZK9, Q5F288 | Gene names | Nlgn2, Kiaa1366 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroligin-2 precursor. | |||||
|
ATS12_MOUSE
|
||||||
| NC score | 0.007383 (rank : 165) | θ value | 1.06291 (rank : 55) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q811B3, Q8BK92, Q8BKY1 | Gene names | Adamts12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAMTS-12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 12) (ADAM-TS 12) (ADAM-TS12). | |||||
|
IRAK1_HUMAN
|
||||||
| NC score | 0.007089 (rank : 166) | θ value | 0.62314 (rank : 41) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P51617, Q7Z5V4, Q96RL2 | Gene names | IRAK1, IRAK | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-1 receptor-associated kinase 1 (EC 2.7.11.1) (IRAK-1). | |||||
|
FOG1_HUMAN
|
||||||
| NC score | 0.006770 (rank : 167) | θ value | 5.27518 (rank : 114) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 981 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8IX07 | Gene names | ZFPM1, FOG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
SNX1_MOUSE
|
||||||
| NC score | 0.005617 (rank : 168) | θ value | 8.99809 (rank : 155) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WV80, Q9EQZ9 | Gene names | Snx1 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-1. | |||||
|
NOTC1_MOUSE
|
||||||
| NC score | 0.004922 (rank : 169) | θ value | 5.27518 (rank : 117) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q01705, Q06007, Q61905, Q99JC2, Q9QW58, Q9R0X7 | Gene names | Notch1, Motch | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (Motch A) (mT14) (p300) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
MPP5_HUMAN
|
||||||
| NC score | 0.004558 (rank : 170) | θ value | 6.88961 (rank : 131) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N3R9, Q7Z631, Q86T98, Q8N7I5, Q9H9Q0 | Gene names | MPP5 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 5. | |||||
|
MYH3_HUMAN
|
||||||
| NC score | 0.002594 (rank : 171) | θ value | 1.81305 (rank : 74) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
POGZ_HUMAN
|
||||||
| NC score | 0.002386 (rank : 172) | θ value | 8.99809 (rank : 151) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q7Z3K3, O75049, Q8TDZ7, Q9Y4X7 | Gene names | POGZ, KIAA0461 | |||
|
Domain Architecture |

|
|||||
| Description | Pogo transposable element with ZNF domain. | |||||
|
EGR2_HUMAN
|
||||||
| NC score | 0.000923 (rank : 173) | θ value | 4.03905 (rank : 104) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 728 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P11161, Q8IV26, Q9UNA6 | Gene names | EGR2, KROX20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early growth response protein 2 (EGR-2) (Krox-20 protein) (AT591). | |||||
|
ZBT38_HUMAN
|
||||||
| NC score | -0.000009 (rank : 174) | θ value | 6.88961 (rank : 138) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 885 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8NAP3 | Gene names | ZBTB38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and BTB domain-containing protein 38. | |||||
|
KLF12_HUMAN
|
||||||
| NC score | -0.000381 (rank : 175) | θ value | 6.88961 (rank : 129) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 717 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y4X4, Q5VZM7, Q9UHZ0 | Gene names | KLF12, AP2REP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 12 (Transcriptional repressor AP-2rep). | |||||
|
MARK2_MOUSE
|
||||||
| NC score | -0.001872 (rank : 176) | θ value | 8.99809 (rank : 149) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 1018 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q05512, Q6PDR4, Q8BR95 | Gene names | Mark2, Emk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase MARK2 (EC 2.7.11.1) (MAP/microtubule affinity-regulating kinase 2) (ELKL Motif Kinase) (EMK1). | |||||
|
GPR39_HUMAN
|
||||||
| NC score | -0.002229 (rank : 177) | θ value | 8.99809 (rank : 147) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 880 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43194, Q53R01 | Gene names | GPR39 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 39. | |||||
|
NRK_MOUSE
|
||||||
| NC score | -0.002402 (rank : 178) | θ value | 6.88961 (rank : 134) | |||
| Query Neighborhood Hits | 156 | Target Neighborhood Hits | 899 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9R0G8, Q6NV55, Q8C9S9, Q9R0S4 | Gene names | Nrk, Nesk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nik-related protein kinase (EC 2.7.11.1) (Nck-interacting kinase-like embryo specific kinase) (NIK-like embryo-specific kinase) (NESK). | |||||