Please be patient as the page loads
|
DYN1_MOUSE
|
||||||
| SwissProt Accessions | P39053, Q61358, Q61359, Q61360, Q8JZZ4, Q9CSY7, Q9QXX1 | Gene names | Dnm1, Dnm | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-1 (EC 3.6.5.5). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DYN1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.993902 (rank : 2) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 155 | |
| SwissProt Accessions | Q05193 | Gene names | DNM1, DNM | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-1 (EC 3.6.5.5). | |||||
|
DYN1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 168 | |
| SwissProt Accessions | P39053, Q61358, Q61359, Q61360, Q8JZZ4, Q9CSY7, Q9QXX1 | Gene names | Dnm1, Dnm | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-1 (EC 3.6.5.5). | |||||
|
DYN2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.981951 (rank : 3) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P50570, Q7Z5S3, Q9UPH4 | Gene names | DNM2, DYN2 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-2 (EC 3.6.5.5). | |||||
|
DYN2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.980376 (rank : 4) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P39054, Q9DBE1 | Gene names | Dnm2, Dyn2 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-2 (EC 3.6.5.5) (Dynamin UDNM). | |||||
|
DYN3_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.980064 (rank : 5) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9UQ16, O14982, O95555, Q5W129, Q9H0P3, Q9H548, Q9NQ68, Q9NQN6 | Gene names | DNM3, KIAA0820 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-3 (EC 3.6.5.5) (Dynamin, testicular) (T-dynamin). | |||||
|
DNM1L_HUMAN
|
||||||
| θ value | 1.20984e-129 (rank : 6) | NC score | 0.944844 (rank : 7) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O00429, O14541, O60709, Q7L6B3, Q8TBT7, Q9BWM1, Q9Y5J2 | Gene names | DNM1L, DLP1, DRP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynamin-1-like protein (EC 3.6.5.5) (Dynamin-like protein) (Dnm1p/Vps1p-like protein) (DVLP) (Dynamin family member proline-rich carboxyl-terminal domain less) (Dymple) (Dynamin-related protein 1) (Dynamin-like protein 4) (Dynamin-like protein IV) (HdynIV). | |||||
|
DNM1L_MOUSE
|
||||||
| θ value | 6.00439e-129 (rank : 7) | NC score | 0.945051 (rank : 6) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8K1M6, Q8BNQ5, Q8BQ64, Q8CGD0, Q8K1A1 | Gene names | Dnm1l, Drp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynamin-1-like protein (EC 3.6.5.5) (Dynamin-related protein 1) (Dynamin family member proline-rich carboxyl-terminal domain less) (Dymple). | |||||
|
MX1_HUMAN
|
||||||
| θ value | 4.84666e-62 (rank : 8) | NC score | 0.862047 (rank : 8) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P20591 | Gene names | MX1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced GTP-binding protein Mx1 (Interferon-regulated resistance GTP-binding protein MxA) (Interferon-induced protein p78) (IFI-78K). | |||||
|
MX2_MOUSE
|
||||||
| θ value | 3.14151e-61 (rank : 9) | NC score | 0.860214 (rank : 9) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9WVP9, Q922L4 | Gene names | Mx2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-induced GTP-binding protein Mx2. | |||||
|
MX1_MOUSE
|
||||||
| θ value | 1.90589e-58 (rank : 10) | NC score | 0.859960 (rank : 10) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P09922 | Gene names | Mx1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced GTP-binding protein Mx1 (Influenza resistance protein). | |||||
|
MX2_HUMAN
|
||||||
| θ value | 1.97229e-55 (rank : 11) | NC score | 0.854048 (rank : 11) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P20592 | Gene names | MX2 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced GTP-binding protein Mx2 (Interferon-regulated resistance GTP-binding protein MxB) (p78-related protein). | |||||
|
OPA1_HUMAN
|
||||||
| θ value | 2.59989e-23 (rank : 12) | NC score | 0.686077 (rank : 13) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O60313 | Gene names | OPA1, KIAA0567 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-like 120 kDa protein, mitochondrial precursor (Optic atrophy protein 1) [Contains: Dynamin-like 120 kDa protein, form S1]. | |||||
|
OPA1_MOUSE
|
||||||
| θ value | 2.59989e-23 (rank : 13) | NC score | 0.688954 (rank : 12) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P58281, Q3ULA5, Q8BKU7, Q8BLL3, Q8BM08, Q8R3J7 | Gene names | Opa1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-like 120 kDa protein, mitochondrial precursor (Optic atrophy protein 1 homolog) (Large GTP-binding protein) (LargeG) [Contains: Dynamin-like 120 kDa protein, form S1]. | |||||
|
CEL_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 14) | NC score | 0.075218 (rank : 33) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
CYH1_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 15) | NC score | 0.132221 (rank : 17) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q15438, Q9P123, Q9P124 | Gene names | PSCD1, D17S811E | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-1 (SEC7 homolog B2-1). | |||||
|
CYH1_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 16) | NC score | 0.131527 (rank : 18) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9QX11, O88817 | Gene names | Pscd1 | |||
|
Domain Architecture |
|
|||||
| Description | Cytohesin-1 (CLM1). | |||||
|
CYH3_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 17) | NC score | 0.129425 (rank : 19) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O43739 | Gene names | PSCD3, ARNO3, GRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-3 (ARF nucleotide-binding site opener 3) (ARNO3 protein) (General receptor of phosphoinositides 1) (Grp1). | |||||
|
CYH2_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 18) | NC score | 0.123307 (rank : 21) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q99418, Q92958 | Gene names | PSCD2, ARNO | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-2 (ARF nucleotide-binding site opener) (ARNO protein) (ARF exchange factor). | |||||
|
CYH2_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 19) | NC score | 0.123177 (rank : 22) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P63034, O89099, P97695, Q3UP39 | Gene names | Pscd2, Sec7b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytohesin-2 (CLM2) (SEC7 homolog B) (msec7-2). | |||||
|
CYH3_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 20) | NC score | 0.126472 (rank : 20) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O08967, Q8CI93 | Gene names | Pscd3, Grp1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-3 (ARF nucleotide-binding site opener 3) (ARNO3 protein) (General receptor of phosphoinositides 1) (Grp1) (CLM-3). | |||||
|
CNKR1_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 21) | NC score | 0.152735 (rank : 14) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q969H4, O95381 | Gene names | CNKSR1, CNK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 1 (Connector enhancer of KSR1) (hCNK1) (Connector enhancer of KSR-like) (CNK homolog protein 1). | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 22) | NC score | 0.074916 (rank : 34) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
HXB3_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 23) | NC score | 0.016735 (rank : 135) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P14651, O95615, P17484 | Gene names | HOXB3, HOX2G | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B3 (Hox-2G) (Hox-2.7). | |||||
|
CNKR2_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 24) | NC score | 0.133502 (rank : 15) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8WXI2, O94976, Q8WXI1 | Gene names | CNKSR2, CNK2, KIAA0902, KSR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 2 (Connector enhancer of KSR2) (CNK2). | |||||
|
CNKR2_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 25) | NC score | 0.133383 (rank : 16) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q80YA9, Q80TP2 | Gene names | Cnksr2, Kiaa0902 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 2 (Connector enhancer of KSR2) (CNK2). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 26) | NC score | 0.082812 (rank : 28) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
ROCK1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 27) | NC score | 0.017397 (rank : 129) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
SLK_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 28) | NC score | 0.011522 (rank : 153) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1798 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O54988, Q80U65, Q8CAU2, Q8CDW2, Q9WU41 | Gene names | Slk, Kiaa0204, Stk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (mSLK) (Serine/threonine-protein kinase 2) (STE20-related kinase SMAK) (Etk4). | |||||
|
AATK_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 29) | NC score | 0.016852 (rank : 133) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1243 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q6ZMQ8, O75136, Q6ZN31, Q86X28 | Gene names | AATK, KIAA0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase) (CDK5-binding protein) (p35-binding protein) (p35BP). | |||||
|
CE016_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 30) | NC score | 0.054477 (rank : 53) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NC24, Q6P4E7, Q6UXY2 | Gene names | C5orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C5orf16. | |||||
|
EPN2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 31) | NC score | 0.049006 (rank : 61) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O95208, O95207, Q9H7Z2, Q9UPT7 | Gene names | EPN2, KIAA1065 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-2 (EPS-15-interacting protein 2). | |||||
|
MRIP_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 32) | NC score | 0.077285 (rank : 31) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 569 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P97434, Q3TBR5, Q3TC85, Q3TRS0, Q3U1Y8, Q3U3E4, Q5SWZ3, Q5SWZ4, Q5SWZ7, Q6P559, Q80YC8 | Gene names | Mrip, Kiaa0864, Rhoip3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (p116Rip) (RIP3). | |||||
|
ROCK1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 33) | NC score | 0.017691 (rank : 127) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
TLE1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 34) | NC score | 0.036008 (rank : 80) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q62440 | Gene names | Tle1, Grg1 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 1 (Groucho-related protein 1) (Grg- 1). | |||||
|
TLE4_HUMAN
|
||||||
| θ value | 0.125558 (rank : 35) | NC score | 0.034229 (rank : 87) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q04727, Q5T1Y2, Q9BZ07, Q9BZ08, Q9BZ09, Q9NSL3, Q9ULF9 | Gene names | TLE4, KIAA1261 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 4. | |||||
|
TLE4_MOUSE
|
||||||
| θ value | 0.125558 (rank : 36) | NC score | 0.034216 (rank : 88) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q62441, Q9JKQ9 | Gene names | Tle4, Grg4 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 4 (Groucho-related protein 4) (Grg- 4). | |||||
|
AKAP2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 37) | NC score | 0.040829 (rank : 69) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Y2D5, Q9UG26 | Gene names | AKAP2, KIAA0920 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 2 (Protein kinase A-anchoring protein 2) (PRKA2) (AKAP-2). | |||||
|
IF4B_HUMAN
|
||||||
| θ value | 0.163984 (rank : 38) | NC score | 0.062236 (rank : 43) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P23588 | Gene names | EIF4B | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
JPH2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 39) | NC score | 0.038501 (rank : 72) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
SC24A_MOUSE
|
||||||
| θ value | 0.163984 (rank : 40) | NC score | 0.060355 (rank : 45) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q3U2P1, Q3TQ05, Q3TRG7, Q8BIS0 | Gene names | Sec24a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein transport protein Sec24A (SEC24-related protein A). | |||||
|
TARA_MOUSE
|
||||||
| θ value | 0.163984 (rank : 41) | NC score | 0.074331 (rank : 35) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 42) | NC score | 0.043014 (rank : 66) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
MRIP_HUMAN
|
||||||
| θ value | 0.21417 (rank : 43) | NC score | 0.077885 (rank : 30) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q6WCQ1, Q3KQZ5, Q5FB94, Q6DHW2, Q7Z5Y2, Q8N390, Q96G40 | Gene names | MRIP, KIAA0864, RHOIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (M-RIP) (RIP3) (p116Rip). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 0.21417 (rank : 44) | NC score | 0.076696 (rank : 32) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
SYNE2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 45) | NC score | 0.021272 (rank : 119) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
CK024_HUMAN
|
||||||
| θ value | 0.279714 (rank : 46) | NC score | 0.051447 (rank : 58) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
K1802_MOUSE
|
||||||
| θ value | 0.279714 (rank : 47) | NC score | 0.034629 (rank : 84) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 0.279714 (rank : 48) | NC score | 0.059518 (rank : 46) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
RHG25_HUMAN
|
||||||
| θ value | 0.279714 (rank : 49) | NC score | 0.072024 (rank : 39) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 550 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P42331, Q8IXQ2 | Gene names | ARHGAP25, KIAA0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
S23IP_MOUSE
|
||||||
| θ value | 0.279714 (rank : 50) | NC score | 0.045274 (rank : 64) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6NZC7, Q8BXY6 | Gene names | Sec23ip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SEC23-interacting protein. | |||||
|
SLK_HUMAN
|
||||||
| θ value | 0.279714 (rank : 51) | NC score | 0.010566 (rank : 157) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1775 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9H2G2, O00211, Q6P1Z4, Q86WU7, Q86WW1, Q92603, Q9NQL0, Q9NQL1 | Gene names | SLK, KIAA0204, STK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (hSLK) (Serine/threonine-protein kinase 2) (CTCL tumor antigen se20-9). | |||||
|
MYO15_HUMAN
|
||||||
| θ value | 0.365318 (rank : 52) | NC score | 0.022452 (rank : 115) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
PKHA5_HUMAN
|
||||||
| θ value | 0.365318 (rank : 53) | NC score | 0.093828 (rank : 26) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9HAU0, Q8N3K6, Q96DY9, Q9BVR4, Q9C0H7, Q9H924, Q9NVK8 | Gene names | PLEKHA5, KIAA1686, PEPP2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 5 (Phosphoinositol 3-phosphate-binding protein 2) (PEPP-2). | |||||
|
SC24C_HUMAN
|
||||||
| θ value | 0.365318 (rank : 54) | NC score | 0.056768 (rank : 51) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
TLE1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 55) | NC score | 0.034562 (rank : 85) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q04724, Q5T3G4, Q969V9 | Gene names | TLE1 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 1 (ESG1) (E(Sp1) homolog). | |||||
|
BCL9_MOUSE
|
||||||
| θ value | 0.47712 (rank : 56) | NC score | 0.062335 (rank : 42) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
C2TA_HUMAN
|
||||||
| θ value | 0.47712 (rank : 57) | NC score | 0.033282 (rank : 90) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P33076 | Gene names | CIITA, MHC2TA | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
CECR2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 58) | NC score | 0.035393 (rank : 82) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9BXF3, Q658Z4, Q96P58, Q9C0C3 | Gene names | CECR2, KIAA1740 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 2. | |||||
|
EPN3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 59) | NC score | 0.051942 (rank : 56) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q91W69, Q9CV55 | Gene names | Epn3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 0.47712 (rank : 60) | NC score | 0.053657 (rank : 54) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
RHG25_MOUSE
|
||||||
| θ value | 0.47712 (rank : 61) | NC score | 0.073912 (rank : 36) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BYW1, Q2VPR2, Q3TE04, Q3TVX0, Q3UVB7, Q6A0E0, Q8BX98 | Gene names | Arhgap25, Kiaa0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
RUNX1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 62) | NC score | 0.018117 (rank : 125) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q03347, O08598, Q62049, Q9ESB9, Q9ET65 | Gene names | Runx1, Aml1, Cbfa2, Pebp2ab | |||
|
Domain Architecture |
|
|||||
| Description | Runt-related transcription factor 1 (Core-binding factor, alpha 2 subunit) (CBF-alpha 2) (Acute myeloid leukemia 1 protein) (Oncogene AML-1) (Polyomavirus enhancer-binding protein 2 alpha B subunit) (PEBP2-alpha B) (PEA2-alpha B) (SL3-3 enhancer factor 1 alpha B subunit) (SL3/AKV core-binding factor alpha B subunit). | |||||
|
IF4B_MOUSE
|
||||||
| θ value | 0.62314 (rank : 63) | NC score | 0.057664 (rank : 48) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BGD9, Q3TD64 | Gene names | Eif4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
K1802_HUMAN
|
||||||
| θ value | 0.62314 (rank : 64) | NC score | 0.034374 (rank : 86) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
ZIMP7_HUMAN
|
||||||
| θ value | 0.62314 (rank : 65) | NC score | 0.047128 (rank : 63) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8NF64, O94790, Q659A8, Q6JKL5, Q8WTX8, Q96Q01, Q9BQH7 | Gene names | ZIMP7, KIAA1886 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
BCAR1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 66) | NC score | 0.024241 (rank : 111) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q61140, Q60869 | Gene names | Bcar1, Cas, Crkas | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 67) | NC score | 0.040516 (rank : 70) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
PELP1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 68) | NC score | 0.049155 (rank : 60) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
RP3A_MOUSE
|
||||||
| θ value | 0.813845 (rank : 69) | NC score | 0.018716 (rank : 123) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P47708 | Gene names | Rph3a | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
SPEG_HUMAN
|
||||||
| θ value | 0.813845 (rank : 70) | NC score | 0.012587 (rank : 150) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
CSTF2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 71) | NC score | 0.019109 (rank : 121) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P33240, Q5H951, Q6LA74, Q8N502 | Gene names | CSTF2 | |||
|
Domain Architecture |
|
|||||
| Description | Cleavage stimulation factor 64 kDa subunit (CSTF 64 kDa subunit) (CF-1 64 kDa subunit) (CstF-64). | |||||
|
DESM_MOUSE
|
||||||
| θ value | 1.06291 (rank : 72) | NC score | 0.010738 (rank : 155) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P31001 | Gene names | Des | |||
|
Domain Architecture |
|
|||||
| Description | Desmin. | |||||
|
PKHB1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 73) | NC score | 0.078157 (rank : 29) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UF11, Q9UBF5, Q9UI37, Q9UI44 | Gene names | PLEKHB1, KPL1, PHR1 | |||
|
Domain Architecture |
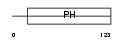
|
|||||
| Description | Pleckstrin homology domain-containing family B member 1 (Pleckstrin homology domain retinal protein 1) (PH domain-containing protein in retina 1) (PHRET1). | |||||
|
PKHB1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 74) | NC score | 0.072251 (rank : 38) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9QYE9, Q9QYB3, Q9QYB4, Q9QYD2, Q9QYD3 | Gene names | Plekhb1, Phr1 | |||
|
Domain Architecture |
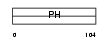
|
|||||
| Description | Pleckstrin homology domain-containing family B member 1 (Pleckstrin homology domain retinal protein 1) (PH domain-containing protein in retina 1) (PHRET1). | |||||
|
PSMF1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 75) | NC score | 0.036521 (rank : 79) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BHL8, Q8C0G9 | Gene names | Psmf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteasome inhibitor PI31 subunit. | |||||
|
RIF1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 76) | NC score | 0.025411 (rank : 105) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6PR54, Q6T8D9, Q7TPD8, Q8BTU2, Q8C408, Q9CS77 | Gene names | Rif1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomere-associated protein RIF1 (Rap1-interacting factor 1 homolog) (mRif1). | |||||
|
ZIM10_HUMAN
|
||||||
| θ value | 1.06291 (rank : 77) | NC score | 0.043707 (rank : 65) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
ANGL1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 78) | NC score | 0.009642 (rank : 160) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q640P2, Q3UQL1, Q9CZ81 | Gene names | Angptl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiopoietin-related protein 1 precursor (Angiopoietin-like 1). | |||||
|
ASTL_HUMAN
|
||||||
| θ value | 1.38821 (rank : 79) | NC score | 0.024444 (rank : 110) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6HA08 | Gene names | ASTL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Astacin-like metalloendopeptidase precursor (EC 3.4.-.-) (Oocyte astacin) (Ovastacin). | |||||
|
BAG4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 80) | NC score | 0.032841 (rank : 91) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O95429, O95818 | Gene names | BAG4, SODD | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 4 (BCL2-associated athanogene 4) (BAG-4) (Silencer of death domains). | |||||
|
BMX_HUMAN
|
||||||
| θ value | 1.38821 (rank : 81) | NC score | 0.006526 (rank : 170) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P51813, O60564, Q12871 | Gene names | BMX | |||
|
Domain Architecture |

|
|||||
| Description | Cytoplasmic tyrosine-protein kinase BMX (EC 2.7.10.2) (Bone marrow tyrosine kinase gene in chromosome X protein) (Epithelial and endothelial tyrosine kinase) (ETK) (NTK38). | |||||
|
CSKI1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 82) | NC score | 0.023067 (rank : 114) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
CYH4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 83) | NC score | 0.105342 (rank : 23) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q80YW0 | Gene names | Pscd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytohesin-4. | |||||
|
EGR4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 84) | NC score | 0.014178 (rank : 142) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q05215 | Gene names | EGR4 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 4 (EGR-4) (AT133). | |||||
|
HPS4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 85) | NC score | 0.031274 (rank : 94) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NQG7, Q5H8V6, Q96LX6, Q9BY93, Q9UH37, Q9UH38 | Gene names | HPS4, KIAA1667 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hermansky-Pudlak syndrome 4 protein (Light-ear protein homolog). | |||||
|
MUCDL_HUMAN
|
||||||
| θ value | 1.38821 (rank : 86) | NC score | 0.032083 (rank : 93) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
NCOA3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 87) | NC score | 0.017082 (rank : 131) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O09000, Q9CRD5 | Gene names | Ncoa3, Aib1, Pcip, Rac3, Tram1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein homolog) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP- interacting protein) (p/CIP) (pCIP). | |||||
|
NFAC1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 88) | NC score | 0.027449 (rank : 102) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O95644, Q12865, Q15793 | Gene names | NFATC1, NFAT2, NFATC | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 1 (NFAT transcription complex cytosolic component) (NF-ATc1) (NF-ATc). | |||||
|
ZN385_HUMAN
|
||||||
| θ value | 1.38821 (rank : 89) | NC score | 0.025201 (rank : 108) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96PM9, Q5VH53, Q9H7R6, Q9UFU3 | Gene names | ZNF385, HZF, RZF | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 385 (Hematopoietic zinc finger protein) (Retinal zinc finger protein). | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 1.81305 (rank : 90) | NC score | 0.012731 (rank : 149) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
EPN3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 91) | NC score | 0.037429 (rank : 75) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9H201, Q9BVN6, Q9NWK2 | Gene names | EPN3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
FYCO1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 92) | NC score | 0.023404 (rank : 113) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1149 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9BQS8, Q3MJE6, Q86T41, Q86TB1, Q8TEF9, Q96IV5, Q9H8P9 | Gene names | FYCO1, ZFYVE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1 (Zinc finger FYVE domain-containing protein 7). | |||||
|
K2C6B_MOUSE
|
||||||
| θ value | 1.81305 (rank : 93) | NC score | 0.014654 (rank : 141) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Z331 | Gene names | Krt6b, K6-beta, Krt2-6b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin, type II cytoskeletal 6B (Cytokeratin-6B) (CK 6B) (K6b keratin) (Keratin-6 beta) (mK6-beta). | |||||
|
MEGF9_MOUSE
|
||||||
| θ value | 1.81305 (rank : 94) | NC score | 0.018651 (rank : 124) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
NRX2A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 95) | NC score | 0.025088 (rank : 109) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
NRX2B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 96) | NC score | 0.037884 (rank : 73) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
PKHA6_HUMAN
|
||||||
| θ value | 1.81305 (rank : 97) | NC score | 0.098819 (rank : 25) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
SCEL_HUMAN
|
||||||
| θ value | 1.81305 (rank : 98) | NC score | 0.026183 (rank : 104) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95171 | Gene names | SCEL | |||
|
Domain Architecture |

|
|||||
| Description | Sciellin. | |||||
|
SMRD2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 99) | NC score | 0.017354 (rank : 130) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99JR8 | Gene names | Smarcd2, Baf60b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily D member 2 (60 kDa BRG-1/Brm-associated factor subunit B) (BRG1-associated factor 60B). | |||||
|
ALDR_HUMAN
|
||||||
| θ value | 2.36792 (rank : 100) | NC score | 0.010599 (rank : 156) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P15121, Q9BS21, Q9UCI9 | Gene names | AKR1B1, ALDR1 | |||
|
Domain Architecture |
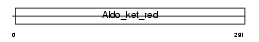
|
|||||
| Description | Aldose reductase (EC 1.1.1.21) (AR) (Aldehyde reductase). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 2.36792 (rank : 101) | NC score | 0.041033 (rank : 67) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CO9A1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 102) | NC score | 0.013934 (rank : 144) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q05722, Q61269, Q61270, Q61433, Q61940 | Gene names | Col9a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
ELOA2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 103) | NC score | 0.017056 (rank : 132) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8IYF1, Q9P2V9 | Gene names | TCEB3B, TCEB3L | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II transcription factor SIII subunit A2 (Elongin-A2) (EloA2) (Transcription elongation factor B polypeptide 3B). | |||||
|
GLYG2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 104) | NC score | 0.024127 (rank : 112) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15488, O15485, O15486, O15487, O15489, O15490 | Gene names | GYG2 | |||
|
Domain Architecture |

|
|||||
| Description | Glycogenin-2 (EC 2.4.1.186) (GN-2) (GN2). | |||||
|
SIM2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 105) | NC score | 0.008335 (rank : 164) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14190, O60766, Q15470, Q15471, Q15472, Q15473, Q16532 | Gene names | SIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2. | |||||
|
COG8_MOUSE
|
||||||
| θ value | 3.0926 (rank : 106) | NC score | 0.025358 (rank : 107) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JJA2 | Gene names | Cog8 | |||
|
Domain Architecture |

|
|||||
| Description | Conserved oligomeric Golgi complex component 8. | |||||
|
CP250_MOUSE
|
||||||
| θ value | 3.0926 (rank : 107) | NC score | 0.013102 (rank : 146) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
CYH4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 108) | NC score | 0.100229 (rank : 24) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UIA0, Q5R3F9, Q9UGT6 | Gene names | PSCD4, CYT4 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-4. | |||||
|
GRAP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 109) | NC score | 0.015913 (rank : 137) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1234 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8VD04, O35693, Q3T9C3, Q69ZP9 | Gene names | Gripap1, DXImx47e, Kiaa1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1) (HCMV-interacting protein). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 110) | NC score | 0.036578 (rank : 78) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 101 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
RBCC1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 111) | NC score | 0.018953 (rank : 122) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1048 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9ESK9, Q3TXX2, Q61384, Q8BRY9, Q9JK14 | Gene names | Rb1cc1, Cc1, Kiaa0203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1 (Coiled-coil-forming protein 1) (LaXp180 protein). | |||||
|
SPEG_MOUSE
|
||||||
| θ value | 3.0926 (rank : 112) | NC score | 0.012343 (rank : 151) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
SYNP2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 113) | NC score | 0.022271 (rank : 116) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UMS6, Q9UK89 | Gene names | SYNPO2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin-2 (Myopodin) (Genethonin 2). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 3.0926 (rank : 114) | NC score | 0.008651 (rank : 163) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
UGGG2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 115) | NC score | 0.013973 (rank : 143) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NYU1, Q9UFC4 | Gene names | UGCGL2, UGT2 | |||
|
Domain Architecture |

|
|||||
| Description | UDP-glucose:glycoprotein glucosyltransferase 2 precursor (EC 2.4.1.-) (UDP-glucose ceramide glucosyltransferase-like 1) (UDP-- Glc:glycoprotein glucosyltransferase 2) (HUGT2). | |||||
|
CAC1B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 116) | NC score | 0.010895 (rank : 154) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CCD37_MOUSE
|
||||||
| θ value | 4.03905 (rank : 117) | NC score | 0.021416 (rank : 118) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80VN0, Q6AXD6 | Gene names | Ccdc37 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 37. | |||||
|
CIC_HUMAN
|
||||||
| θ value | 4.03905 (rank : 118) | NC score | 0.033868 (rank : 89) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
DESM_HUMAN
|
||||||
| θ value | 4.03905 (rank : 119) | NC score | 0.008718 (rank : 162) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P17661, Q15787, Q8IZR1, Q8IZR6, Q8NES2, Q8NEU6, Q8TAC4, Q8TCX2, Q8TD99, Q9UHN5, Q9UJ80 | Gene names | DES | |||
|
Domain Architecture |
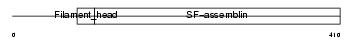
|
|||||
| Description | Desmin. | |||||
|
GLI2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 120) | NC score | 0.000277 (rank : 178) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 971 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P10070, O60252, O60253, O60254, O60255, Q15590, Q15591 | Gene names | GLI2, THP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI2 (Tax helper protein). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 121) | NC score | 0.013644 (rank : 145) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
MAML2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 122) | NC score | 0.028200 (rank : 98) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8IZL2, Q6AI23, Q6Y3A3, Q8IUL3, Q96JK6 | Gene names | MAML2, KIAA1819 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 2 (Mam-2). | |||||
|
MAST1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 123) | NC score | 0.005917 (rank : 173) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1146 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y2H9, O00114, Q8N6X0 | Gene names | MAST1, KIAA0973 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 124) | NC score | 0.032127 (rank : 92) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
PKHA6_MOUSE
|
||||||
| θ value | 4.03905 (rank : 125) | NC score | 0.092861 (rank : 27) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
TROAP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 126) | NC score | 0.030767 (rank : 95) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q12815 | Gene names | TROAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trophinin-associated protein (Tastin) (Trophinin-assisting protein). | |||||
|
ZIMP7_MOUSE
|
||||||
| θ value | 4.03905 (rank : 127) | NC score | 0.040209 (rank : 71) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8CIE2, Q6P558, Q6P9Q5, Q8BJ00 | Gene names | Zimp7, D11Bwg0280e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
AMOL1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 128) | NC score | 0.014719 (rank : 140) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9D4H4, Q571F1 | Gene names | Amotl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin-like protein 1. | |||||
|
AP3D1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 129) | NC score | 0.009937 (rank : 158) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O54774 | Gene names | Ap3d1, Ap3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin) (mBLVR1). | |||||
|
BMX_MOUSE
|
||||||
| θ value | 5.27518 (rank : 130) | NC score | 0.005124 (rank : 174) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P97504 | Gene names | Bmx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic tyrosine-protein kinase BMX (EC 2.7.10.2) (Bone marrow tyrosine kinase gene in chromosome X protein homolog). | |||||
|
CING_MOUSE
|
||||||
| θ value | 5.27518 (rank : 131) | NC score | 0.012184 (rank : 152) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
CRTC1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 132) | NC score | 0.037099 (rank : 77) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q6UUV9, O75114, Q6Y3A3, Q7LDZ2, Q8IUL3, Q8IZ34, Q8IZL1, Q8N6W3, Q96AI8, Q9H801 | Gene names | CRTC1, KIAA0616, MECT1, TORC1, WAMTP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Transducer of regulated cAMP response element-binding protein 1) (Transducer of CREB protein 1) (Mucoepidermoid carcinoma translocated protein 1). | |||||
|
ICK_MOUSE
|
||||||
| θ value | 5.27518 (rank : 133) | NC score | 0.003429 (rank : 175) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 860 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JKV2, Q8K138 | Gene names | Ick | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase ICK (EC 2.7.11.22) (Intestinal cell kinase) (mICK) (MAK-related kinase) (MRK). | |||||
|
IL9R_HUMAN
|
||||||
| θ value | 5.27518 (rank : 134) | NC score | 0.015943 (rank : 136) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q01113, Q14634, Q8WWU1, Q96TF0 | Gene names | IL9R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-9 receptor precursor (IL-9R) (CD129 antigen). | |||||
|
KI67_HUMAN
|
||||||
| θ value | 5.27518 (rank : 135) | NC score | 0.021193 (rank : 120) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
LSD1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 136) | NC score | 0.012950 (rank : 148) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O60341, Q5TH94, Q5TH95, Q86VT7, Q8IXK4, Q8NDP6, Q8TAZ3, Q96AW4 | Gene names | AOF2, KIAA0601, LSD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
MFN1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 137) | NC score | 0.052020 (rank : 55) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IWA4, O15323, O60639, Q9BZB5, Q9NWQ2 | Gene names | MFN1 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane GTPase MFN1 (EC 3.6.5.-) (Mitofusin-1) (Fzo homolog). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 5.27518 (rank : 138) | NC score | 0.035298 (rank : 83) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
PAK4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 139) | NC score | 0.006770 (rank : 169) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1098 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8BTW9, Q6ZPX0, Q80Z97, Q9CS71 | Gene names | Pak4, Kiaa1142 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PAK 4 (EC 2.7.11.1) (p21-activated kinase 4) (PAK-4). | |||||
|
POMT2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 140) | NC score | 0.012972 (rank : 147) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BGQ4, Q8R4Z0 | Gene names | Pomt2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein O-mannosyl-transferase 2 (EC 2.4.1.109) (Dolichyl-phosphate- mannose--protein mannosyltransferase 2). | |||||
|
SON_HUMAN
|
||||||
| θ value | 5.27518 (rank : 141) | NC score | 0.047245 (rank : 62) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 142) | NC score | 0.037878 (rank : 74) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
ZIM10_MOUSE
|
||||||
| θ value | 5.27518 (rank : 143) | NC score | 0.035593 (rank : 81) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6P1E1, Q6PDK9, Q6PF85, Q8BW47 | Gene names | Rai17, Zimp10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
ZN516_MOUSE
|
||||||
| θ value | 5.27518 (rank : 144) | NC score | 0.006257 (rank : 171) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q7TSH3 | Gene names | Znf516, Kiaa0222, Zfp516 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 516. | |||||
|
AP3D1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 145) | NC score | 0.008830 (rank : 161) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O14617, O00202, O75262, Q59HF5, Q96G11, Q9H3C6 | Gene names | AP3D1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin). | |||||
|
BIM_HUMAN
|
||||||
| θ value | 6.88961 (rank : 146) | NC score | 0.017524 (rank : 128) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43521, O43522 | Gene names | BCL2L11, BIM | |||
|
Domain Architecture |

|
|||||
| Description | Bcl-2-like protein 11 (Bcl2-interacting mediator of cell death). | |||||
|
KCNQ4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 147) | NC score | 0.007154 (rank : 168) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P56696, O96025 | Gene names | KCNQ4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 4 (Voltage-gated potassium channel subunit Kv7.4) (Potassium channel subunit alpha KvLQT4) (KQT-like 4). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 148) | NC score | 0.026247 (rank : 103) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
MTSS1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 149) | NC score | 0.029065 (rank : 97) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8R1S4, Q8BMM3, Q99LB3 | Gene names | Mtss1, Mim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 6.88961 (rank : 150) | NC score | 0.028061 (rank : 99) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
RBCC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 151) | NC score | 0.017797 (rank : 126) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1023 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8TDY2, Q8WVU9, Q92601 | Gene names | RB1CC1, KIAA0203, RBICC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1. | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 152) | NC score | 0.037275 (rank : 76) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
SYNG_MOUSE
|
||||||
| θ value | 6.88961 (rank : 153) | NC score | 0.015540 (rank : 138) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5SV85, Q5SV84, Q6PHT6 | Gene names | Ap1gbp1, Syng | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP1 subunit gamma-binding protein 1 (Gamma-synergin). | |||||
|
TBCD1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 154) | NC score | 0.009890 (rank : 159) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q86TI0, Q96K82, Q9UPP4 | Gene names | TBC1D1, KIAA1108 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 1. | |||||
|
ZN598_HUMAN
|
||||||
| θ value | 6.88961 (rank : 155) | NC score | 0.027817 (rank : 100) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q86UK7, Q8IW49, Q8N3D9, Q96FG3, Q9H7J3 | Gene names | ZNF598 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 598. | |||||
|
AT2A3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 156) | NC score | 0.002821 (rank : 177) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q64518, O70625, Q64517 | Gene names | Atp2a3 | |||
|
Domain Architecture |

|
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 3 (EC 3.6.3.8) (Calcium pump 3) (SERCA3) (SR Ca(2+)-ATPase 3). | |||||
|
COKA1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 157) | NC score | 0.006201 (rank : 172) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9P218, Q4VXQ4, Q8WUT2, Q96CY9, Q9BQU6, Q9BQU7 | Gene names | COL20A1, KIAA1510 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XX) chain precursor. | |||||
|
CRSP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 158) | NC score | 0.027672 (rank : 101) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O60244, Q9UNB3 | Gene names | CRSP2, ARC150, DRIP150, EXLM1, TRAP170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 2 (Cofactor required for Sp1 transcriptional activation subunit 2) (Transcriptional coactivator CRSP150) (Vitamin D3 receptor-interacting protein complex 150 kDa component) (DRIP150) (Thyroid hormone receptor-associated protein complex 170 kDa component) (Trap170) (Activator-recruited cofactor 150 kDa component) (ARC150). | |||||
|
CU086_HUMAN
|
||||||
| θ value | 8.99809 (rank : 159) | NC score | 0.040874 (rank : 68) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P59089 | Gene names | C21orf86 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C21orf86. | |||||
|
FOG1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 160) | NC score | 0.007317 (rank : 166) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1237 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O35615 | Gene names | Zfpm1, Fog, Fog1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
HXD3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 161) | NC score | 0.007280 (rank : 167) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P31249, Q99955, Q9BSC5 | Gene names | HOXD3, HOX4A | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D3 (Hox-4A). | |||||
|
ICK_HUMAN
|
||||||
| θ value | 8.99809 (rank : 162) | NC score | 0.002943 (rank : 176) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UPZ9, O75985, Q5THL2, Q8IYH8, Q9BX17, Q9NYX3 | Gene names | ICK, KIAA0936 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase ICK (EC 2.7.11.22) (Intestinal cell kinase) (hICK) (MAK-related kinase) (MRK) (Laryngeal cancer kinase 2) (LCK2). | |||||
|
JIP3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 163) | NC score | 0.016810 (rank : 134) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
MILK1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 164) | NC score | 0.022188 (rank : 117) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 8.99809 (rank : 165) | NC score | 0.030423 (rank : 96) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PHC2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 166) | NC score | 0.015086 (rank : 139) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8IXK0, Q5T0C1, Q6NUJ6, Q6ZQR1, Q8N306, Q8TAG8, Q96BL4, Q9Y4Y7 | Gene names | PHC2, EDR2, PH2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 2 (hPH2) (Early development regulatory protein 2). | |||||
|
SAPS1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 167) | NC score | 0.025394 (rank : 106) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
SOX17_MOUSE
|
||||||
| θ value | 8.99809 (rank : 168) | NC score | 0.007537 (rank : 165) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q61473, Q61472, Q62248 | Gene names | Sox17, Sox-17 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-17. | |||||
|
CN032_HUMAN
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.055152 (rank : 52) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
CN032_MOUSE
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.051813 (rank : 57) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
PKHA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.050779 (rank : 59) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9HB21, Q9BVK0 | Gene names | PLEKHA1, TAPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
PKHA4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.072674 (rank : 37) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9H4M7, Q8N4M8, Q8N658 | Gene names | PLEKHA4, PEPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
PKHA4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.064017 (rank : 41) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8VC98, Q8R3N3 | Gene names | Plekha4, Pepp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
PLEK_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.058147 (rank : 47) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P08567, Q53SU8, Q6FGM8, Q8WV81 | Gene names | PLEK, P47 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin (Platelet p47 protein). | |||||
|
PRB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.057400 (rank : 49) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.056809 (rank : 50) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PRP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.061765 (rank : 44) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.069109 (rank : 40) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
DYN1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 168 | |
| SwissProt Accessions | P39053, Q61358, Q61359, Q61360, Q8JZZ4, Q9CSY7, Q9QXX1 | Gene names | Dnm1, Dnm | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-1 (EC 3.6.5.5). | |||||
|
DYN1_HUMAN
|
||||||
| NC score | 0.993902 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 155 | |
| SwissProt Accessions | Q05193 | Gene names | DNM1, DNM | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-1 (EC 3.6.5.5). | |||||
|
DYN2_HUMAN
|
||||||
| NC score | 0.981951 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P50570, Q7Z5S3, Q9UPH4 | Gene names | DNM2, DYN2 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-2 (EC 3.6.5.5). | |||||
|
DYN2_MOUSE
|
||||||
| NC score | 0.980376 (rank : 4) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P39054, Q9DBE1 | Gene names | Dnm2, Dyn2 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-2 (EC 3.6.5.5) (Dynamin UDNM). | |||||
|
DYN3_HUMAN
|
||||||
| NC score | 0.980064 (rank : 5) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9UQ16, O14982, O95555, Q5W129, Q9H0P3, Q9H548, Q9NQ68, Q9NQN6 | Gene names | DNM3, KIAA0820 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-3 (EC 3.6.5.5) (Dynamin, testicular) (T-dynamin). | |||||
|
DNM1L_MOUSE
|
||||||
| NC score | 0.945051 (rank : 6) | θ value | 6.00439e-129 (rank : 7) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8K1M6, Q8BNQ5, Q8BQ64, Q8CGD0, Q8K1A1 | Gene names | Dnm1l, Drp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynamin-1-like protein (EC 3.6.5.5) (Dynamin-related protein 1) (Dynamin family member proline-rich carboxyl-terminal domain less) (Dymple). | |||||
|
DNM1L_HUMAN
|
||||||
| NC score | 0.944844 (rank : 7) | θ value | 1.20984e-129 (rank : 6) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O00429, O14541, O60709, Q7L6B3, Q8TBT7, Q9BWM1, Q9Y5J2 | Gene names | DNM1L, DLP1, DRP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynamin-1-like protein (EC 3.6.5.5) (Dynamin-like protein) (Dnm1p/Vps1p-like protein) (DVLP) (Dynamin family member proline-rich carboxyl-terminal domain less) (Dymple) (Dynamin-related protein 1) (Dynamin-like protein 4) (Dynamin-like protein IV) (HdynIV). | |||||
|
MX1_HUMAN
|
||||||
| NC score | 0.862047 (rank : 8) | θ value | 4.84666e-62 (rank : 8) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P20591 | Gene names | MX1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced GTP-binding protein Mx1 (Interferon-regulated resistance GTP-binding protein MxA) (Interferon-induced protein p78) (IFI-78K). | |||||
|
MX2_MOUSE
|
||||||
| NC score | 0.860214 (rank : 9) | θ value | 3.14151e-61 (rank : 9) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9WVP9, Q922L4 | Gene names | Mx2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-induced GTP-binding protein Mx2. | |||||
|
MX1_MOUSE
|
||||||
| NC score | 0.859960 (rank : 10) | θ value | 1.90589e-58 (rank : 10) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P09922 | Gene names | Mx1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced GTP-binding protein Mx1 (Influenza resistance protein). | |||||
|
MX2_HUMAN
|
||||||
| NC score | 0.854048 (rank : 11) | θ value | 1.97229e-55 (rank : 11) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P20592 | Gene names | MX2 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced GTP-binding protein Mx2 (Interferon-regulated resistance GTP-binding protein MxB) (p78-related protein). | |||||
|
OPA1_MOUSE
|
||||||
| NC score | 0.688954 (rank : 12) | θ value | 2.59989e-23 (rank : 13) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P58281, Q3ULA5, Q8BKU7, Q8BLL3, Q8BM08, Q8R3J7 | Gene names | Opa1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-like 120 kDa protein, mitochondrial precursor (Optic atrophy protein 1 homolog) (Large GTP-binding protein) (LargeG) [Contains: Dynamin-like 120 kDa protein, form S1]. | |||||
|
OPA1_HUMAN
|
||||||
| NC score | 0.686077 (rank : 13) | θ value | 2.59989e-23 (rank : 12) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O60313 | Gene names | OPA1, KIAA0567 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-like 120 kDa protein, mitochondrial precursor (Optic atrophy protein 1) [Contains: Dynamin-like 120 kDa protein, form S1]. | |||||
|
CNKR1_HUMAN
|
||||||
| NC score | 0.152735 (rank : 14) | θ value | 0.0330416 (rank : 21) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q969H4, O95381 | Gene names | CNKSR1, CNK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 1 (Connector enhancer of KSR1) (hCNK1) (Connector enhancer of KSR-like) (CNK homolog protein 1). | |||||
|
CNKR2_HUMAN
|
||||||
| NC score | 0.133502 (rank : 15) | θ value | 0.0736092 (rank : 24) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8WXI2, O94976, Q8WXI1 | Gene names | CNKSR2, CNK2, KIAA0902, KSR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 2 (Connector enhancer of KSR2) (CNK2). | |||||
|
CNKR2_MOUSE
|
||||||
| NC score | 0.133383 (rank : 16) | θ value | 0.0736092 (rank : 25) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q80YA9, Q80TP2 | Gene names | Cnksr2, Kiaa0902 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 2 (Connector enhancer of KSR2) (CNK2). | |||||
|
CYH1_HUMAN
|
||||||
| NC score | 0.132221 (rank : 17) | θ value | 0.00134147 (rank : 15) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q15438, Q9P123, Q9P124 | Gene names | PSCD1, D17S811E | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-1 (SEC7 homolog B2-1). | |||||
|
CYH1_MOUSE
|
||||||
| NC score | 0.131527 (rank : 18) | θ value | 0.00134147 (rank : 16) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9QX11, O88817 | Gene names | Pscd1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-1 (CLM1). | |||||
|
CYH3_HUMAN
|
||||||
| NC score | 0.129425 (rank : 19) | θ value | 0.0148317 (rank : 17) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O43739 | Gene names | PSCD3, ARNO3, GRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-3 (ARF nucleotide-binding site opener 3) (ARNO3 protein) (General receptor of phosphoinositides 1) (Grp1). | |||||
|
CYH3_MOUSE
|
||||||
| NC score | 0.126472 (rank : 20) | θ value | 0.0252991 (rank : 20) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O08967, Q8CI93 | Gene names | Pscd3, Grp1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-3 (ARF nucleotide-binding site opener 3) (ARNO3 protein) (General receptor of phosphoinositides 1) (Grp1) (CLM-3). | |||||
|
CYH2_HUMAN
|
||||||
| NC score | 0.123307 (rank : 21) | θ value | 0.0252991 (rank : 18) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q99418, Q92958 | Gene names | PSCD2, ARNO | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-2 (ARF nucleotide-binding site opener) (ARNO protein) (ARF exchange factor). | |||||
|
CYH2_MOUSE
|
||||||
| NC score | 0.123177 (rank : 22) | θ value | 0.0252991 (rank : 19) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P63034, O89099, P97695, Q3UP39 | Gene names | Pscd2, Sec7b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytohesin-2 (CLM2) (SEC7 homolog B) (msec7-2). | |||||
|
CYH4_MOUSE
|
||||||
| NC score | 0.105342 (rank : 23) | θ value | 1.38821 (rank : 83) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q80YW0 | Gene names | Pscd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytohesin-4. | |||||
|
CYH4_HUMAN
|
||||||
| NC score | 0.100229 (rank : 24) | θ value | 3.0926 (rank : 108) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UIA0, Q5R3F9, Q9UGT6 | Gene names | PSCD4, CYT4 | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-4. | |||||
|
PKHA6_HUMAN
|
||||||
| NC score | 0.098819 (rank : 25) | θ value | 1.81305 (rank : 97) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
PKHA5_HUMAN
|
||||||
| NC score | 0.093828 (rank : 26) | θ value | 0.365318 (rank : 53) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9HAU0, Q8N3K6, Q96DY9, Q9BVR4, Q9C0H7, Q9H924, Q9NVK8 | Gene names | PLEKHA5, KIAA1686, PEPP2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 5 (Phosphoinositol 3-phosphate-binding protein 2) (PEPP-2). | |||||
|
PKHA6_MOUSE
|
||||||
| NC score | 0.092861 (rank : 27) | θ value | 4.03905 (rank : 125) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.082812 (rank : 28) | θ value | 0.0736092 (rank : 26) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
PKHB1_HUMAN
|
||||||
| NC score | 0.078157 (rank : 29) | θ value | 1.06291 (rank : 73) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UF11, Q9UBF5, Q9UI37, Q9UI44 | Gene names | PLEKHB1, KPL1, PHR1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family B member 1 (Pleckstrin homology domain retinal protein 1) (PH domain-containing protein in retina 1) (PHRET1). | |||||
|
MRIP_HUMAN
|
||||||
| NC score | 0.077885 (rank : 30) | θ value | 0.21417 (rank : 43) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q6WCQ1, Q3KQZ5, Q5FB94, Q6DHW2, Q7Z5Y2, Q8N390, Q96G40 | Gene names | MRIP, KIAA0864, RHOIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (M-RIP) (RIP3) (p116Rip). | |||||
|
MRIP_MOUSE
|
||||||
| NC score | 0.077285 (rank : 31) | θ value | 0.0961366 (rank : 32) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 569 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P97434, Q3TBR5, Q3TC85, Q3TRS0, Q3U1Y8, Q3U3E4, Q5SWZ3, Q5SWZ4, Q5SWZ7, Q6P559, Q80YC8 | Gene names | Mrip, Kiaa0864, Rhoip3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (p116Rip) (RIP3). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.076696 (rank : 32) | θ value | 0.21417 (rank : 44) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.075218 (rank : 33) | θ value | 4.92598e-06 (rank : 14) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.074916 (rank : 34) | θ value | 0.0431538 (rank : 22) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.074331 (rank : 35) | θ value | 0.163984 (rank : 41) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
RHG25_MOUSE
|
||||||
| NC score | 0.073912 (rank : 36) | θ value | 0.47712 (rank : 61) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BYW1, Q2VPR2, Q3TE04, Q3TVX0, Q3UVB7, Q6A0E0, Q8BX98 | Gene names | Arhgap25, Kiaa0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
PKHA4_HUMAN
|
||||||
| NC score | 0.072674 (rank : 37) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9H4M7, Q8N4M8, Q8N658 | Gene names | PLEKHA4, PEPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
PKHB1_MOUSE
|
||||||
| NC score | 0.072251 (rank : 38) | θ value | 1.06291 (rank : 74) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9QYE9, Q9QYB3, Q9QYB4, Q9QYD2, Q9QYD3 | Gene names | Plekhb1, Phr1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family B member 1 (Pleckstrin homology domain retinal protein 1) (PH domain-containing protein in retina 1) (PHRET1). | |||||
|
RHG25_HUMAN
|
||||||
| NC score | 0.072024 (rank : 39) | θ value | 0.279714 (rank : 49) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 550 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P42331, Q8IXQ2 | Gene names | ARHGAP25, KIAA0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.069109 (rank : 40) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PKHA4_MOUSE
|
||||||
| NC score | 0.064017 (rank : 41) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8VC98, Q8R3N3 | Gene names | Plekha4, Pepp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
BCL9_MOUSE
|
||||||
| NC score | 0.062335 (rank : 42) | θ value | 0.47712 (rank : 56) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
IF4B_HUMAN
|
||||||
| NC score | 0.062236 (rank : 43) | θ value | 0.163984 (rank : 38) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P23588 | Gene names | EIF4B | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.061765 (rank : 44) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
SC24A_MOUSE
|
||||||
| NC score | 0.060355 (rank : 45) | θ value | 0.163984 (rank : 40) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q3U2P1, Q3TQ05, Q3TRG7, Q8BIS0 | Gene names | Sec24a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein transport protein Sec24A (SEC24-related protein A). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.059518 (rank : 46) | θ value | 0.279714 (rank : 48) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
PLEK_HUMAN
|
||||||
| NC score | 0.058147 (rank : 47) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P08567, Q53SU8, Q6FGM8, Q8WV81 | Gene names | PLEK, P47 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin (Platelet p47 protein). | |||||
|
IF4B_MOUSE
|
||||||
| NC score | 0.057664 (rank : 48) | θ value | 0.62314 (rank : 63) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BGD9, Q3TD64 | Gene names | Eif4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
PRB3_HUMAN
|
||||||
| NC score | 0.057400 (rank : 49) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.056809 (rank : 50) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
SC24C_HUMAN
|
||||||
| NC score | 0.056768 (rank : 51) | θ value | 0.365318 (rank : 54) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
CN032_HUMAN
|
||||||
| NC score | 0.055152 (rank : 52) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
CE016_HUMAN
|
||||||
| NC score | 0.054477 (rank : 53) | θ value | 0.0961366 (rank : 30) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NC24, Q6P4E7, Q6UXY2 | Gene names | C5orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C5orf16. | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.053657 (rank : 54) | θ value | 0.47712 (rank : 60) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
MFN1_HUMAN
|
||||||
| NC score | 0.052020 (rank : 55) | θ value | 5.27518 (rank : 137) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IWA4, O15323, O60639, Q9BZB5, Q9NWQ2 | Gene names | MFN1 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane GTPase MFN1 (EC 3.6.5.-) (Mitofusin-1) (Fzo homolog). | |||||
|
EPN3_MOUSE
|
||||||
| NC score | 0.051942 (rank : 56) | θ value | 0.47712 (rank : 59) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q91W69, Q9CV55 | Gene names | Epn3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
CN032_MOUSE
|
||||||
| NC score | 0.051813 (rank : 57) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
CK024_HUMAN
|
||||||
| NC score | 0.051447 (rank : 58) | θ value | 0.279714 (rank : 46) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
PKHA1_HUMAN
|
||||||
| NC score | 0.050779 (rank : 59) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9HB21, Q9BVK0 | Gene names | PLEKHA1, TAPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
PELP1_MOUSE
|
||||||
| NC score | 0.049155 (rank : 60) | θ value | 0.813845 (rank : 68) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
EPN2_HUMAN
|
||||||
| NC score | 0.049006 (rank : 61) | θ value | 0.0961366 (rank : 31) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O95208, O95207, Q9H7Z2, Q9UPT7 | Gene names | EPN2, KIAA1065 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-2 (EPS-15-interacting protein 2). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.047245 (rank : 62) | θ value | 5.27518 (rank : 141) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
ZIMP7_HUMAN
|
||||||
| NC score | 0.047128 (rank : 63) | θ value | 0.62314 (rank : 65) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8NF64, O94790, Q659A8, Q6JKL5, Q8WTX8, Q96Q01, Q9BQH7 | Gene names | ZIMP7, KIAA1886 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
S23IP_MOUSE
|
||||||
| NC score | 0.045274 (rank : 64) | θ value | 0.279714 (rank : 50) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6NZC7, Q8BXY6 | Gene names | Sec23ip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SEC23-interacting protein. | |||||
|
ZIM10_HUMAN
|
||||||
| NC score | 0.043707 (rank : 65) | θ value | 1.06291 (rank : 77) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.043014 (rank : 66) | θ value | 0.21417 (rank : 42) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.041033 (rank : 67) | θ value | 2.36792 (rank : 101) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CU086_HUMAN
|
||||||
| NC score | 0.040874 (rank : 68) | θ value | 8.99809 (rank : 159) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P59089 | Gene names | C21orf86 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C21orf86. | |||||
|
AKAP2_HUMAN
|
||||||
| NC score | 0.040829 (rank : 69) | θ value | 0.163984 (rank : 37) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Y2D5, Q9UG26 | Gene names | AKAP2, KIAA0920 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 2 (Protein kinase A-anchoring protein 2) (PRKA2) (AKAP-2). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.040516 (rank : 70) | θ value | 0.813845 (rank : 67) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
ZIMP7_MOUSE
|
||||||
| NC score | 0.040209 (rank : 71) | θ value | 4.03905 (rank : 127) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8CIE2, Q6P558, Q6P9Q5, Q8BJ00 | Gene names | Zimp7, D11Bwg0280e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
JPH2_HUMAN
|
||||||
| NC score | 0.038501 (rank : 72) | θ value | 0.163984 (rank : 39) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
NRX2B_HUMAN
|
||||||
| NC score | 0.037884 (rank : 73) | θ value | 1.81305 (rank : 96) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.037878 (rank : 74) | θ value | 5.27518 (rank : 142) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
EPN3_HUMAN
|
||||||
| NC score | 0.037429 (rank : 75) | θ value | 1.81305 (rank : 91) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9H201, Q9BVN6, Q9NWK2 | Gene names | EPN3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.037275 (rank : 76) | θ value | 6.88961 (rank : 152) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
CRTC1_HUMAN
|
||||||
| NC score | 0.037099 (rank : 77) | θ value | 5.27518 (rank : 132) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q6UUV9, O75114, Q6Y3A3, Q7LDZ2, Q8IUL3, Q8IZ34, Q8IZL1, Q8N6W3, Q96AI8, Q9H801 | Gene names | CRTC1, KIAA0616, MECT1, TORC1, WAMTP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Transducer of regulated cAMP response element-binding protein 1) (Transducer of CREB protein 1) (Mucoepidermoid carcinoma translocated protein 1). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.036578 (rank : 78) | θ value | 3.0926 (rank : 110) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 101 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
PSMF1_MOUSE
|
||||||
| NC score | 0.036521 (rank : 79) | θ value | 1.06291 (rank : 75) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BHL8, Q8C0G9 | Gene names | Psmf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteasome inhibitor PI31 subunit. | |||||
|
TLE1_MOUSE
|
||||||
| NC score | 0.036008 (rank : 80) | θ value | 0.0961366 (rank : 34) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q62440 | Gene names | Tle1, Grg1 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 1 (Groucho-related protein 1) (Grg- 1). | |||||
|
ZIM10_MOUSE
|
||||||
| NC score | 0.035593 (rank : 81) | θ value | 5.27518 (rank : 143) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6P1E1, Q6PDK9, Q6PF85, Q8BW47 | Gene names | Rai17, Zimp10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
CECR2_HUMAN
|
||||||
| NC score | 0.035393 (rank : 82) | θ value | 0.47712 (rank : 58) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9BXF3, Q658Z4, Q96P58, Q9C0C3 | Gene names | CECR2, KIAA1740 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 2. | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.035298 (rank : 83) | θ value | 5.27518 (rank : 138) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.034629 (rank : 84) | θ value | 0.279714 (rank : 47) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
TLE1_HUMAN
|
||||||
| NC score | 0.034562 (rank : 85) | θ value | 0.365318 (rank : 55) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q04724, Q5T3G4, Q969V9 | Gene names | TLE1 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 1 (ESG1) (E(Sp1) homolog). | |||||
|
K1802_HUMAN
|
||||||
| NC score | 0.034374 (rank : 86) | θ value | 0.62314 (rank : 64) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
TLE4_HUMAN
|
||||||
| NC score | 0.034229 (rank : 87) | θ value | 0.125558 (rank : 35) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q04727, Q5T1Y2, Q9BZ07, Q9BZ08, Q9BZ09, Q9NSL3, Q9ULF9 | Gene names | TLE4, KIAA1261 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 4. | |||||
|
TLE4_MOUSE
|
||||||
| NC score | 0.034216 (rank : 88) | θ value | 0.125558 (rank : 36) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q62441, Q9JKQ9 | Gene names | Tle4, Grg4 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 4 (Groucho-related protein 4) (Grg- 4). | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.033868 (rank : 89) | θ value | 4.03905 (rank : 118) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
C2TA_HUMAN
|
||||||
| NC score | 0.033282 (rank : 90) | θ value | 0.47712 (rank : 57) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P33076 | Gene names | CIITA, MHC2TA | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
BAG4_HUMAN
|
||||||
| NC score | 0.032841 (rank : 91) | θ value | 1.38821 (rank : 80) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O95429, O95818 | Gene names | BAG4, SODD | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 4 (BCL2-associated athanogene 4) (BAG-4) (Silencer of death domains). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.032127 (rank : 92) | θ value | 4.03905 (rank : 124) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
MUCDL_HUMAN
|
||||||
| NC score | 0.032083 (rank : 93) | θ value | 1.38821 (rank : 86) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
HPS4_HUMAN
|
||||||
| NC score | 0.031274 (rank : 94) | θ value | 1.38821 (rank : 85) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NQG7, Q5H8V6, Q96LX6, Q9BY93, Q9UH37, Q9UH38 | Gene names | HPS4, KIAA1667 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hermansky-Pudlak syndrome 4 protein (Light-ear protein homolog). | |||||
|
TROAP_HUMAN
|
||||||
| NC score | 0.030767 (rank : 95) | θ value | 4.03905 (rank : 126) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q12815 | Gene names | TROAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trophinin-associated protein (Tastin) (Trophinin-assisting protein). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.030423 (rank : 96) | θ value | 8.99809 (rank : 165) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
MTSS1_MOUSE
|
||||||
| NC score | 0.029065 (rank : 97) | θ value | 6.88961 (rank : 149) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8R1S4, Q8BMM3, Q99LB3 | Gene names | Mtss1, Mim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein). | |||||
|
MAML2_HUMAN
|
||||||
| NC score | 0.028200 (rank : 98) | θ value | 4.03905 (rank : 122) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8IZL2, Q6AI23, Q6Y3A3, Q8IUL3, Q96JK6 | Gene names | MAML2, KIAA1819 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 2 (Mam-2). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.028061 (rank : 99) | θ value | 6.88961 (rank : 150) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
ZN598_HUMAN
|
||||||
| NC score | 0.027817 (rank : 100) | θ value | 6.88961 (rank : 155) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q86UK7, Q8IW49, Q8N3D9, Q96FG3, Q9H7J3 | Gene names | ZNF598 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 598. | |||||
|
CRSP2_HUMAN
|
||||||
| NC score | 0.027672 (rank : 101) | θ value | 8.99809 (rank : 158) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O60244, Q9UNB3 | Gene names | CRSP2, ARC150, DRIP150, EXLM1, TRAP170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 2 (Cofactor required for Sp1 transcriptional activation subunit 2) (Transcriptional coactivator CRSP150) (Vitamin D3 receptor-interacting protein complex 150 kDa component) (DRIP150) (Thyroid hormone receptor-associated protein complex 170 kDa component) (Trap170) (Activator-recruited cofactor 150 kDa component) (ARC150). | |||||
|
NFAC1_HUMAN
|
||||||
| NC score | 0.027449 (rank : 102) | θ value | 1.38821 (rank : 88) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O95644, Q12865, Q15793 | Gene names | NFATC1, NFAT2, NFATC | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 1 (NFAT transcription complex cytosolic component) (NF-ATc1) (NF-ATc). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.026247 (rank : 103) | θ value | 6.88961 (rank : 148) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
SCEL_HUMAN
|
||||||
| NC score | 0.026183 (rank : 104) | θ value | 1.81305 (rank : 98) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95171 | Gene names | SCEL | |||
|
Domain Architecture |

|
|||||
| Description | Sciellin. | |||||
|
RIF1_MOUSE
|
||||||
| NC score | 0.025411 (rank : 105) | θ value | 1.06291 (rank : 76) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6PR54, Q6T8D9, Q7TPD8, Q8BTU2, Q8C408, Q9CS77 | Gene names | Rif1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomere-associated protein RIF1 (Rap1-interacting factor 1 homolog) (mRif1). | |||||
|
SAPS1_HUMAN
|
||||||
| NC score | 0.025394 (rank : 106) | θ value | 8.99809 (rank : 167) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
COG8_MOUSE
|
||||||
| NC score | 0.025358 (rank : 107) | θ value | 3.0926 (rank : 106) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JJA2 | Gene names | Cog8 | |||
|
Domain Architecture |

|
|||||
| Description | Conserved oligomeric Golgi complex component 8. | |||||
|
ZN385_HUMAN
|
||||||
| NC score | 0.025201 (rank : 108) | θ value | 1.38821 (rank : 89) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96PM9, Q5VH53, Q9H7R6, Q9UFU3 | Gene names | ZNF385, HZF, RZF | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 385 (Hematopoietic zinc finger protein) (Retinal zinc finger protein). | |||||
|
NRX2A_HUMAN
|
||||||
| NC score | 0.025088 (rank : 109) | θ value | 1.81305 (rank : 95) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
ASTL_HUMAN
|
||||||
| NC score | 0.024444 (rank : 110) | θ value | 1.38821 (rank : 79) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6HA08 | Gene names | ASTL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Astacin-like metalloendopeptidase precursor (EC 3.4.-.-) (Oocyte astacin) (Ovastacin). | |||||
|
BCAR1_MOUSE
|
||||||
| NC score | 0.024241 (rank : 111) | θ value | 0.813845 (rank : 66) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q61140, Q60869 | Gene names | Bcar1, Cas, Crkas | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
GLYG2_HUMAN
|
||||||
| NC score | 0.024127 (rank : 112) | θ value | 2.36792 (rank : 104) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15488, O15485, O15486, O15487, O15489, O15490 | Gene names | GYG2 | |||
|
Domain Architecture |

|
|||||
| Description | Glycogenin-2 (EC 2.4.1.186) (GN-2) (GN2). | |||||
|
FYCO1_HUMAN
|
||||||
| NC score | 0.023404 (rank : 113) | θ value | 1.81305 (rank : 92) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1149 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9BQS8, Q3MJE6, Q86T41, Q86TB1, Q8TEF9, Q96IV5, Q9H8P9 | Gene names | FYCO1, ZFYVE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1 (Zinc finger FYVE domain-containing protein 7). | |||||
|
CSKI1_HUMAN
|
||||||
| NC score | 0.023067 (rank : 114) | θ value | 1.38821 (rank : 82) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
MYO15_HUMAN
|
||||||
| NC score | 0.022452 (rank : 115) | θ value | 0.365318 (rank : 52) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
SYNP2_HUMAN
|
||||||
| NC score | 0.022271 (rank : 116) | θ value | 3.0926 (rank : 113) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UMS6, Q9UK89 | Gene names | SYNPO2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin-2 (Myopodin) (Genethonin 2). | |||||
|
MILK1_MOUSE
|
||||||
| NC score | 0.022188 (rank : 117) | θ value | 8.99809 (rank : 164) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
CCD37_MOUSE
|
||||||
| NC score | 0.021416 (rank : 118) | θ value | 4.03905 (rank : 117) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80VN0, Q6AXD6 | Gene names | Ccdc37 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 37. | |||||
|
SYNE2_HUMAN
|
||||||
| NC score | 0.021272 (rank : 119) | θ value | 0.21417 (rank : 45) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
KI67_HUMAN
|
||||||
| NC score | 0.021193 (rank : 120) | θ value | 5.27518 (rank : 135) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
CSTF2_HUMAN
|
||||||
| NC score | 0.019109 (rank : 121) | θ value | 1.06291 (rank : 71) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P33240, Q5H951, Q6LA74, Q8N502 | Gene names | CSTF2 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage stimulation factor 64 kDa subunit (CSTF 64 kDa subunit) (CF-1 64 kDa subunit) (CstF-64). | |||||
|
RBCC1_MOUSE
|
||||||
| NC score | 0.018953 (rank : 122) | θ value | 3.0926 (rank : 111) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1048 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9ESK9, Q3TXX2, Q61384, Q8BRY9, Q9JK14 | Gene names | Rb1cc1, Cc1, Kiaa0203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1 (Coiled-coil-forming protein 1) (LaXp180 protein). | |||||
|
RP3A_MOUSE
|
||||||
| NC score | 0.018716 (rank : 123) | θ value | 0.813845 (rank : 69) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P47708 | Gene names | Rph3a | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
MEGF9_MOUSE
|
||||||
| NC score | 0.018651 (rank : 124) | θ value | 1.81305 (rank : 94) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
RUNX1_MOUSE
|
||||||
| NC score | 0.018117 (rank : 125) | θ value | 0.47712 (rank : 62) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q03347, O08598, Q62049, Q9ESB9, Q9ET65 | Gene names | Runx1, Aml1, Cbfa2, Pebp2ab | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 1 (Core-binding factor, alpha 2 subunit) (CBF-alpha 2) (Acute myeloid leukemia 1 protein) (Oncogene AML-1) (Polyomavirus enhancer-binding protein 2 alpha B subunit) (PEBP2-alpha B) (PEA2-alpha B) (SL3-3 enhancer factor 1 alpha B subunit) (SL3/AKV core-binding factor alpha B subunit). | |||||
|
RBCC1_HUMAN
|
||||||
| NC score | 0.017797 (rank : 126) | θ value | 6.88961 (rank : 151) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1023 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8TDY2, Q8WVU9, Q92601 | Gene names | RB1CC1, KIAA0203, RBICC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1. | |||||
|
ROCK1_HUMAN
|
||||||
| NC score | 0.017691 (rank : 127) | θ value | 0.0961366 (rank : 33) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
BIM_HUMAN
|
||||||
| NC score | 0.017524 (rank : 128) | θ value | 6.88961 (rank : 146) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43521, O43522 | Gene names | BCL2L11, BIM | |||
|
Domain Architecture |

|
|||||
| Description | Bcl-2-like protein 11 (Bcl2-interacting mediator of cell death). | |||||
|
ROCK1_MOUSE
|
||||||
| NC score | 0.017397 (rank : 129) | θ value | 0.0736092 (rank : 27) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
SMRD2_MOUSE
|
||||||
| NC score | 0.017354 (rank : 130) | θ value | 1.81305 (rank : 99) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99JR8 | Gene names | Smarcd2, Baf60b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily D member 2 (60 kDa BRG-1/Brm-associated factor subunit B) (BRG1-associated factor 60B). | |||||
|
NCOA3_MOUSE
|
||||||
| NC score | 0.017082 (rank : 131) | θ value | 1.38821 (rank : 87) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O09000, Q9CRD5 | Gene names | Ncoa3, Aib1, Pcip, Rac3, Tram1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein homolog) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP- interacting protein) (p/CIP) (pCIP). | |||||
|
ELOA2_HUMAN
|
||||||
| NC score | 0.017056 (rank : 132) | θ value | 2.36792 (rank : 103) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8IYF1, Q9P2V9 | Gene names | TCEB3B, TCEB3L | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II transcription factor SIII subunit A2 (Elongin-A2) (EloA2) (Transcription elongation factor B polypeptide 3B). | |||||
|
AATK_HUMAN
|
||||||
| NC score | 0.016852 (rank : 133) | θ value | 0.0961366 (rank : 29) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1243 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q6ZMQ8, O75136, Q6ZN31, Q86X28 | Gene names | AATK, KIAA0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase) (CDK5-binding protein) (p35-binding protein) (p35BP). | |||||
|
JIP3_HUMAN
|
||||||
| NC score | 0.016810 (rank : 134) | θ value | 8.99809 (rank : 163) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
HXB3_HUMAN
|
||||||
| NC score | 0.016735 (rank : 135) | θ value | 0.0431538 (rank : 23) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P14651, O95615, P17484 | Gene names | HOXB3, HOX2G | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B3 (Hox-2G) (Hox-2.7). | |||||
|
IL9R_HUMAN
|
||||||
| NC score | 0.015943 (rank : 136) | θ value | 5.27518 (rank : 134) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q01113, Q14634, Q8WWU1, Q96TF0 | Gene names | IL9R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-9 receptor precursor (IL-9R) (CD129 antigen). | |||||
|
GRAP1_MOUSE
|
||||||
| NC score | 0.015913 (rank : 137) | θ value | 3.0926 (rank : 109) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1234 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8VD04, O35693, Q3T9C3, Q69ZP9 | Gene names | Gripap1, DXImx47e, Kiaa1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1) (HCMV-interacting protein). | |||||
|
SYNG_MOUSE
|
||||||
| NC score | 0.015540 (rank : 138) | θ value | 6.88961 (rank : 153) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5SV85, Q5SV84, Q6PHT6 | Gene names | Ap1gbp1, Syng | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP1 subunit gamma-binding protein 1 (Gamma-synergin). | |||||
|
PHC2_HUMAN
|
||||||
| NC score | 0.015086 (rank : 139) | θ value | 8.99809 (rank : 166) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8IXK0, Q5T0C1, Q6NUJ6, Q6ZQR1, Q8N306, Q8TAG8, Q96BL4, Q9Y4Y7 | Gene names | PHC2, EDR2, PH2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 2 (hPH2) (Early development regulatory protein 2). | |||||
|
AMOL1_MOUSE
|
||||||
| NC score | 0.014719 (rank : 140) | θ value | 5.27518 (rank : 128) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9D4H4, Q571F1 | Gene names | Amotl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin-like protein 1. | |||||
|
K2C6B_MOUSE
|
||||||
| NC score | 0.014654 (rank : 141) | θ value | 1.81305 (rank : 93) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Z331 | Gene names | Krt6b, K6-beta, Krt2-6b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin, type II cytoskeletal 6B (Cytokeratin-6B) (CK 6B) (K6b keratin) (Keratin-6 beta) (mK6-beta). | |||||
|
EGR4_HUMAN
|
||||||
| NC score | 0.014178 (rank : 142) | θ value | 1.38821 (rank : 84) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q05215 | Gene names | EGR4 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 4 (EGR-4) (AT133). | |||||
|
UGGG2_HUMAN
|
||||||
| NC score | 0.013973 (rank : 143) | θ value | 3.0926 (rank : 115) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NYU1, Q9UFC4 | Gene names | UGCGL2, UGT2 | |||
|
Domain Architecture |

|
|||||
| Description | UDP-glucose:glycoprotein glucosyltransferase 2 precursor (EC 2.4.1.-) (UDP-glucose ceramide glucosyltransferase-like 1) (UDP-- Glc:glycoprotein glucosyltransferase 2) (HUGT2). | |||||
|
CO9A1_MOUSE
|
||||||
| NC score | 0.013934 (rank : 144) | θ value | 2.36792 (rank : 102) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q05722, Q61269, Q61270, Q61433, Q61940 | Gene names | Col9a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.013644 (rank : 145) | θ value | 4.03905 (rank : 121) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
CP250_MOUSE
|
||||||
| NC score | 0.013102 (rank : 146) | θ value | 3.0926 (rank : 107) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
POMT2_MOUSE
|
||||||
| NC score | 0.012972 (rank : 147) | θ value | 5.27518 (rank : 140) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BGQ4, Q8R4Z0 | Gene names | Pomt2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein O-mannosyl-transferase 2 (EC 2.4.1.109) (Dolichyl-phosphate- mannose--protein mannosyltransferase 2). | |||||
|
LSD1_HUMAN
|
||||||
| NC score | 0.012950 (rank : 148) | θ value | 5.27518 (rank : 136) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O60341, Q5TH94, Q5TH95, Q86VT7, Q8IXK4, Q8NDP6, Q8TAZ3, Q96AW4 | Gene names | AOF2, KIAA0601, LSD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.012731 (rank : 149) | θ value | 1.81305 (rank : 90) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
SPEG_HUMAN
|
||||||
| NC score | 0.012587 (rank : 150) | θ value | 0.813845 (rank : 70) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
SPEG_MOUSE
|
||||||
| NC score | 0.012343 (rank : 151) | θ value | 3.0926 (rank : 112) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
CING_MOUSE
|
||||||
| NC score | 0.012184 (rank : 152) | θ value | 5.27518 (rank : 131) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
SLK_MOUSE
|
||||||
| NC score | 0.011522 (rank : 153) | θ value | 0.0736092 (rank : 28) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1798 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O54988, Q80U65, Q8CAU2, Q8CDW2, Q9WU41 | Gene names | Slk, Kiaa0204, Stk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (mSLK) (Serine/threonine-protein kinase 2) (STE20-related kinase SMAK) (Etk4). | |||||
|
CAC1B_HUMAN
|
||||||
| NC score | 0.010895 (rank : 154) | θ value | 4.03905 (rank : 116) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
DESM_MOUSE
|
||||||
| NC score | 0.010738 (rank : 155) | θ value | 1.06291 (rank : 72) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P31001 | Gene names | Des | |||
|
Domain Architecture |

|
|||||
| Description | Desmin. | |||||
|
ALDR_HUMAN
|
||||||
| NC score | 0.010599 (rank : 156) | θ value | 2.36792 (rank : 100) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P15121, Q9BS21, Q9UCI9 | Gene names | AKR1B1, ALDR1 | |||
|
Domain Architecture |
|
|||||
| Description | Aldose reductase (EC 1.1.1.21) (AR) (Aldehyde reductase). | |||||
|
SLK_HUMAN
|
||||||
| NC score | 0.010566 (rank : 157) | θ value | 0.279714 (rank : 51) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1775 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9H2G2, O00211, Q6P1Z4, Q86WU7, Q86WW1, Q92603, Q9NQL0, Q9NQL1 | Gene names | SLK, KIAA0204, STK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (hSLK) (Serine/threonine-protein kinase 2) (CTCL tumor antigen se20-9). | |||||
|
AP3D1_MOUSE
|
||||||
| NC score | 0.009937 (rank : 158) | θ value | 5.27518 (rank : 129) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O54774 | Gene names | Ap3d1, Ap3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin) (mBLVR1). | |||||
|
TBCD1_HUMAN
|
||||||
| NC score | 0.009890 (rank : 159) | θ value | 6.88961 (rank : 154) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q86TI0, Q96K82, Q9UPP4 | Gene names | TBC1D1, KIAA1108 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 1. | |||||
|
ANGL1_MOUSE
|
||||||
| NC score | 0.009642 (rank : 160) | θ value | 1.38821 (rank : 78) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q640P2, Q3UQL1, Q9CZ81 | Gene names | Angptl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiopoietin-related protein 1 precursor (Angiopoietin-like 1). | |||||
|
AP3D1_HUMAN
|
||||||
| NC score | 0.008830 (rank : 161) | θ value | 6.88961 (rank : 145) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O14617, O00202, O75262, Q59HF5, Q96G11, Q9H3C6 | Gene names | AP3D1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin). | |||||
|
DESM_HUMAN
|
||||||
| NC score | 0.008718 (rank : 162) | θ value | 4.03905 (rank : 119) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P17661, Q15787, Q8IZR1, Q8IZR6, Q8NES2, Q8NEU6, Q8TAC4, Q8TCX2, Q8TD99, Q9UHN5, Q9UJ80 | Gene names | DES | |||
|
Domain Architecture |

|
|||||
| Description | Desmin. | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.008651 (rank : 163) | θ value | 3.0926 (rank : 114) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
SIM2_HUMAN
|
||||||
| NC score | 0.008335 (rank : 164) | θ value | 2.36792 (rank : 105) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14190, O60766, Q15470, Q15471, Q15472, Q15473, Q16532 | Gene names | SIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2. | |||||
|
SOX17_MOUSE
|
||||||
| NC score | 0.007537 (rank : 165) | θ value | 8.99809 (rank : 168) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q61473, Q61472, Q62248 | Gene names | Sox17, Sox-17 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-17. | |||||
|
FOG1_MOUSE
|
||||||
| NC score | 0.007317 (rank : 166) | θ value | 8.99809 (rank : 160) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1237 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O35615 | Gene names | Zfpm1, Fog, Fog1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
HXD3_HUMAN
|
||||||
| NC score | 0.007280 (rank : 167) | θ value | 8.99809 (rank : 161) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P31249, Q99955, Q9BSC5 | Gene names | HOXD3, HOX4A | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D3 (Hox-4A). | |||||
|
KCNQ4_HUMAN
|
||||||
| NC score | 0.007154 (rank : 168) | θ value | 6.88961 (rank : 147) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P56696, O96025 | Gene names | KCNQ4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 4 (Voltage-gated potassium channel subunit Kv7.4) (Potassium channel subunit alpha KvLQT4) (KQT-like 4). | |||||
|
PAK4_MOUSE
|
||||||
| NC score | 0.006770 (rank : 169) | θ value | 5.27518 (rank : 139) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1098 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8BTW9, Q6ZPX0, Q80Z97, Q9CS71 | Gene names | Pak4, Kiaa1142 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PAK 4 (EC 2.7.11.1) (p21-activated kinase 4) (PAK-4). | |||||
|
BMX_HUMAN
|
||||||
| NC score | 0.006526 (rank : 170) | θ value | 1.38821 (rank : 81) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P51813, O60564, Q12871 | Gene names | BMX | |||
|
Domain Architecture |

|
|||||
| Description | Cytoplasmic tyrosine-protein kinase BMX (EC 2.7.10.2) (Bone marrow tyrosine kinase gene in chromosome X protein) (Epithelial and endothelial tyrosine kinase) (ETK) (NTK38). | |||||
|
ZN516_MOUSE
|
||||||
| NC score | 0.006257 (rank : 171) | θ value | 5.27518 (rank : 144) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q7TSH3 | Gene names | Znf516, Kiaa0222, Zfp516 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 516. | |||||
|
COKA1_HUMAN
|
||||||
| NC score | 0.006201 (rank : 172) | θ value | 8.99809 (rank : 157) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9P218, Q4VXQ4, Q8WUT2, Q96CY9, Q9BQU6, Q9BQU7 | Gene names | COL20A1, KIAA1510 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XX) chain precursor. | |||||
|
MAST1_HUMAN
|
||||||
| NC score | 0.005917 (rank : 173) | θ value | 4.03905 (rank : 123) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1146 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y2H9, O00114, Q8N6X0 | Gene names | MAST1, KIAA0973 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
BMX_MOUSE
|
||||||
| NC score | 0.005124 (rank : 174) | θ value | 5.27518 (rank : 130) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P97504 | Gene names | Bmx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic tyrosine-protein kinase BMX (EC 2.7.10.2) (Bone marrow tyrosine kinase gene in chromosome X protein homolog). | |||||
|
ICK_MOUSE
|
||||||
| NC score | 0.003429 (rank : 175) | θ value | 5.27518 (rank : 133) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 860 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JKV2, Q8K138 | Gene names | Ick | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase ICK (EC 2.7.11.22) (Intestinal cell kinase) (mICK) (MAK-related kinase) (MRK). | |||||
|
ICK_HUMAN
|
||||||
| NC score | 0.002943 (rank : 176) | θ value | 8.99809 (rank : 162) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UPZ9, O75985, Q5THL2, Q8IYH8, Q9BX17, Q9NYX3 | Gene names | ICK, KIAA0936 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase ICK (EC 2.7.11.22) (Intestinal cell kinase) (hICK) (MAK-related kinase) (MRK) (Laryngeal cancer kinase 2) (LCK2). | |||||
|
AT2A3_MOUSE
|
||||||
| NC score | 0.002821 (rank : 177) | θ value | 8.99809 (rank : 156) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q64518, O70625, Q64517 | Gene names | Atp2a3 | |||
|
Domain Architecture |

|
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 3 (EC 3.6.3.8) (Calcium pump 3) (SERCA3) (SR Ca(2+)-ATPase 3). | |||||
|
GLI2_HUMAN
|
||||||
| NC score | 0.000277 (rank : 178) | θ value | 4.03905 (rank : 120) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 971 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P10070, O60252, O60253, O60254, O60255, Q15590, Q15591 | Gene names | GLI2, THP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI2 (Tax helper protein). | |||||