Please be patient as the page loads
|
COG8_MOUSE
|
||||||
| SwissProt Accessions | Q9JJA2 | Gene names | Cog8 | |||
|
Domain Architecture |

|
|||||
| Description | Conserved oligomeric Golgi complex component 8. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
COG8_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.955665 (rank : 2) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96MW5, Q8WVV6, Q9H6F8 | Gene names | COG8 | |||
|
Domain Architecture |
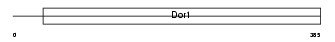
|
|||||
| Description | Conserved oligomeric Golgi complex component 8. | |||||
|
COG8_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 147 | |
| SwissProt Accessions | Q9JJA2 | Gene names | Cog8 | |||
|
Domain Architecture |

|
|||||
| Description | Conserved oligomeric Golgi complex component 8. | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 3) | NC score | 0.089700 (rank : 5) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
H6ST3_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 4) | NC score | 0.073731 (rank : 12) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QYK4 | Gene names | Hs6st3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan-sulfate 6-O-sulfotransferase 3 (EC 2.8.2.-) (HS6ST-3) (mHS6ST- 3). | |||||
|
ZN598_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 5) | NC score | 0.074001 (rank : 11) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q86UK7, Q8IW49, Q8N3D9, Q96FG3, Q9H7J3 | Gene names | ZNF598 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 598. | |||||
|
NEIL2_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 6) | NC score | 0.075781 (rank : 9) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q969S2, Q7Z3Q7, Q8N842, Q8NG52 | Gene names | NEIL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 2 (EC 3.2.2.-) (EC 4.2.99.18) (Nei-like 2) (DNA glycosylase/AP lyase Neil2) (DNA-(apurinic or apyrimidinic site) lyase Neil2) (NEH2). | |||||
|
DIAP3_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 7) | NC score | 0.062370 (rank : 23) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Z207 | Gene names | Diaph3, Diap3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 3 (Diaphanous-related formin-3) (DRF3) (mDIA2) (p134mDIA2). | |||||
|
GGN_MOUSE
|
||||||
| θ value | 0.125558 (rank : 8) | NC score | 0.073255 (rank : 13) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q80WJ1, Q5EBP4, Q80WI9, Q80WJ0 | Gene names | Ggn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
GLI3_MOUSE
|
||||||
| θ value | 0.125558 (rank : 9) | NC score | 0.017140 (rank : 131) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61602 | Gene names | Gli3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI3. | |||||
|
CPSF6_HUMAN
|
||||||
| θ value | 0.163984 (rank : 10) | NC score | 0.084469 (rank : 7) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q16630, Q53ES1, Q9BSJ7, Q9BW18 | Gene names | CPSF6, CFIM68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6 (Cleavage and polyadenylation specificity factor 68 kDa subunit) (CPSF 68 kDa subunit) (Pre-mRNA cleavage factor Im 68 kDa subunit) (Protein HPBRII- 4/7). | |||||
|
CPSF6_MOUSE
|
||||||
| θ value | 0.163984 (rank : 11) | NC score | 0.084675 (rank : 6) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6NVF9, Q8BX86, Q8BXI8 | Gene names | Cpsf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6. | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.21417 (rank : 12) | NC score | 0.102463 (rank : 4) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 106 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
ABI3_MOUSE
|
||||||
| θ value | 0.279714 (rank : 13) | NC score | 0.045827 (rank : 64) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BYZ1, Q6PE63, Q9D7S4 | Gene names | Abi3, Nesh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABI gene family member 3 (New molecule including SH3) (Nesh). | |||||
|
CASP_HUMAN
|
||||||
| θ value | 0.365318 (rank : 14) | NC score | 0.035572 (rank : 83) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q13948, Q53GU9, Q8TBS3 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CUTL1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 15) | NC score | 0.039023 (rank : 74) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
DLGP3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 16) | NC score | 0.052634 (rank : 45) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95886, Q5TDD5, Q9H3X7 | Gene names | DLGAP3, DAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large-associated protein 3 (DAP-3) (SAP90/PSD-95-associated protein 3) (SAPAP3) (PSD-95/SAP90-binding protein 3). | |||||
|
FMNL_HUMAN
|
||||||
| θ value | 0.365318 (rank : 17) | NC score | 0.072586 (rank : 14) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O95466, Q6DKG5, Q6IBP3, Q86UH1, Q8N671, Q8TDH1, Q96H10 | Gene names | FMNL1, C17orf1, C17orf1B, FMNL | |||
|
Domain Architecture |

|
|||||
| Description | Formin-like protein 1 (Leukocyte formin) (CLL-associated antigen KW- 13). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 0.365318 (rank : 18) | NC score | 0.074134 (rank : 10) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
DLGP3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 19) | NC score | 0.051882 (rank : 48) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PFD5, Q6PDX0, Q6XBF2 | Gene names | Dlgap3, Dap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large-associated protein 3 (DAP-3) (SAP90/PSD-95-associated protein 3) (SAPAP3) (PSD-95/SAP90-binding protein 3). | |||||
|
EFS_MOUSE
|
||||||
| θ value | 0.47712 (rank : 20) | NC score | 0.061963 (rank : 24) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q64355 | Gene names | Efs, Sin | |||
|
Domain Architecture |

|
|||||
| Description | Embryonal Fyn-associated substrate (SRC-interacting protein) (Signal- integrating protein). | |||||
|
GTSE1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 21) | NC score | 0.058658 (rank : 30) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8R080, O89015, Q9CSG9 | Gene names | Gtse1, B99 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G2 and S phase expressed protein 1 (Gtse-1) (B99 protein). | |||||
|
PRB3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 22) | NC score | 0.054606 (rank : 41) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
PRP5_HUMAN
|
||||||
| θ value | 0.47712 (rank : 23) | NC score | 0.109044 (rank : 3) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P04281 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide IB-1. | |||||
|
SET1A_HUMAN
|
||||||
| θ value | 0.47712 (rank : 24) | NC score | 0.048526 (rank : 59) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
WASF1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 25) | NC score | 0.064904 (rank : 18) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q92558 | Gene names | WASF1, KIAA0269, SCAR1, WAVE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 1 (WASP-family protein member 1) (Protein WAVE-1) (Verprolin homology domain-containing protein 1). | |||||
|
WASF1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 26) | NC score | 0.064742 (rank : 20) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8R5H6, Q91W51, Q9ERQ9 | Gene names | Wasf1, Wave1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 1 (WASP-family protein member 1) (Protein WAVE-1). | |||||
|
CASP_MOUSE
|
||||||
| θ value | 0.62314 (rank : 27) | NC score | 0.035070 (rank : 84) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1152 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P70403, Q7TQJ4, Q91WN6 | Gene names | Cutl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
PP1RA_MOUSE
|
||||||
| θ value | 0.62314 (rank : 28) | NC score | 0.059905 (rank : 27) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q80W00, Q811B6, Q8C6T7, Q8K2U8 | Gene names | Ppp1r10, Cat53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 1 regulatory subunit 10 (MHC class I region proline-rich protein CAT53). | |||||
|
SFPQ_MOUSE
|
||||||
| θ value | 0.62314 (rank : 29) | NC score | 0.042561 (rank : 69) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
DGKK_HUMAN
|
||||||
| θ value | 0.813845 (rank : 30) | NC score | 0.037067 (rank : 80) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5KSL6 | Gene names | DGKK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase kappa (EC 2.7.1.107) (Diglyceride kinase kappa) (DGK-kappa) (DAG kinase kappa) (142 kDa diacylglycerol kinase). | |||||
|
PELP1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 31) | NC score | 0.057502 (rank : 31) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
PRP1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 32) | NC score | 0.052557 (rank : 46) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
RHG05_MOUSE
|
||||||
| θ value | 0.813845 (rank : 33) | NC score | 0.040718 (rank : 72) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P97393 | Gene names | Arhgap5, Rhogap5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
FOG1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 34) | NC score | 0.019538 (rank : 121) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 981 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8IX07 | Gene names | ZFPM1, FOG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
GLI3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 35) | NC score | 0.016638 (rank : 133) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P10071, O75219, Q9UDT5, Q9UJ39 | Gene names | GLI3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI3. | |||||
|
GNDS_MOUSE
|
||||||
| θ value | 1.06291 (rank : 36) | NC score | 0.034051 (rank : 86) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q03385 | Gene names | Ralgds, Rgds | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator (RalGEF) (RalGDS). | |||||
|
MOT8_HUMAN
|
||||||
| θ value | 1.06291 (rank : 37) | NC score | 0.038125 (rank : 78) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P36021, Q7Z797 | Gene names | SLC16A2, MCT8, XPCT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Monocarboxylate transporter 8 (MCT 8) (MCT 7) (Solute carrier family 16 member 2) (X-linked PEST-containing transporter). | |||||
|
NFH_HUMAN
|
||||||
| θ value | 1.06291 (rank : 38) | NC score | 0.021906 (rank : 112) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
ATBF1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 39) | NC score | 0.020638 (rank : 118) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
EFS_HUMAN
|
||||||
| θ value | 1.38821 (rank : 40) | NC score | 0.055606 (rank : 37) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O43281, O43282 | Gene names | EFS | |||
|
Domain Architecture |

|
|||||
| Description | Embryonal Fyn-associated substrate (HEFS). | |||||
|
EP400_HUMAN
|
||||||
| θ value | 1.38821 (rank : 41) | NC score | 0.033705 (rank : 87) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
FMNL_MOUSE
|
||||||
| θ value | 1.38821 (rank : 42) | NC score | 0.067452 (rank : 15) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9JL26, Q6KAN4, Q9Z2V7 | Gene names | Fmnl1, Frl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-like protein 1 (Formin-related protein). | |||||
|
MMP15_MOUSE
|
||||||
| θ value | 1.38821 (rank : 43) | NC score | 0.013929 (rank : 136) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O54732 | Gene names | Mmp15 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-15 precursor (EC 3.4.24.-) (MMP-15) (Membrane-type matrix metalloproteinase 2) (MT-MMP 2) (MTMMP2) (Membrane-type-2 matrix metalloproteinase) (MT2-MMP) (MT2MMP). | |||||
|
PI5PA_MOUSE
|
||||||
| θ value | 1.38821 (rank : 44) | NC score | 0.051445 (rank : 51) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P59644 | Gene names | Pib5pa | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
RYR1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 45) | NC score | 0.026960 (rank : 98) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P21817, Q16314, Q16368, Q9NPK1, Q9P1U4 | Gene names | RYR1, RYDR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 1 (Skeletal muscle-type ryanodine receptor) (RyR1) (RYR-1) (Skeletal muscle calcium release channel). | |||||
|
SHAN1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 46) | NC score | 0.023280 (rank : 107) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
SYNJ2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 47) | NC score | 0.040927 (rank : 70) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9D2G5, O35404, O88399, O88400, O88401, O88402, O88403, O88404 | Gene names | Synj2, Kiaa0348 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-2 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 2). | |||||
|
TRI66_MOUSE
|
||||||
| θ value | 1.38821 (rank : 48) | NC score | 0.031598 (rank : 93) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q924W6 | Gene names | Trim66 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
CBP_HUMAN
|
||||||
| θ value | 1.81305 (rank : 49) | NC score | 0.045602 (rank : 65) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
H6ST3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 50) | NC score | 0.055285 (rank : 40) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IZP7, Q5W0L0, Q68CW6 | Gene names | HS6ST3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan-sulfate 6-O-sulfotransferase 3 (EC 2.8.2.-) (HS6ST-3). | |||||
|
HCN2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 51) | NC score | 0.021005 (rank : 117) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
PRPC_HUMAN
|
||||||
| θ value | 1.81305 (rank : 52) | NC score | 0.063823 (rank : 21) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P02810, Q4VBP2, Q53XA2, Q6P2F6 | Gene names | PRH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Salivary acidic proline-rich phosphoprotein 1/2 precursor (PRP-1/PRP- 2) (Parotid proline-rich protein 1/2) (Pr1/Pr2) (Protein C) (Parotid acidic protein) (Pa) (Parotid isoelectric focusing variant protein) (PIF-S) (Parotid double-band protein) (Db-s) [Contains: Salivary acidic proline-rich phosphoprotein 1/2; Salivary acidic proline-rich phosphoprotein 3/4 (PRP-3/PRP-4) (Protein A) (PIF-F) (Db-F); Peptide P-C]. | |||||
|
DIAP1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 53) | NC score | 0.050505 (rank : 56) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 839 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O08808 | Gene names | Diaph1, Diap1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1) (mDIA1) (p140mDIA). | |||||
|
EP400_MOUSE
|
||||||
| θ value | 2.36792 (rank : 54) | NC score | 0.034697 (rank : 85) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
M3K14_HUMAN
|
||||||
| θ value | 2.36792 (rank : 55) | NC score | 0.007459 (rank : 148) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 887 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99558, Q8IYN1 | Gene names | MAP3K14, NIK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 14 (EC 2.7.11.25) (NF- kappa beta-inducing kinase) (Serine/threonine-protein kinase NIK) (HsNIK). | |||||
|
NGB_HUMAN
|
||||||
| θ value | 2.36792 (rank : 56) | NC score | 0.060900 (rank : 26) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NPG2 | Gene names | NGB | |||
|
Domain Architecture |

|
|||||
| Description | Neuroglobin. | |||||
|
NOTC1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 57) | NC score | 0.011642 (rank : 141) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P46531 | Gene names | NOTCH1, TAN1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (hN1) (Translocation-associated notch protein TAN-1) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
PP1RA_HUMAN
|
||||||
| θ value | 2.36792 (rank : 58) | NC score | 0.057203 (rank : 33) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96QC0, O00405 | Gene names | PPP1R10, CAT53, FB19, PNUTS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 1 regulatory subunit 10 (Phosphatase 1 nuclear targeting subunit) (MHC class I region proline- rich protein CAT53) (FB19 protein) (PP1-binding protein of 114 kDa) (p99). | |||||
|
PRRT3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 59) | NC score | 0.036160 (rank : 82) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6PE13 | Gene names | Prrt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
SFPQ_HUMAN
|
||||||
| θ value | 2.36792 (rank : 60) | NC score | 0.038973 (rank : 76) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 2.36792 (rank : 61) | NC score | 0.007800 (rank : 147) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
WNK2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 62) | NC score | 0.012571 (rank : 138) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
AKA12_HUMAN
|
||||||
| θ value | 3.0926 (rank : 63) | NC score | 0.036969 (rank : 81) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
ANKS1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.020510 (rank : 119) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92625, Q5JYI9, Q5SYR2, Q86WQ7 | Gene names | ANKS1A, ANKS1, KIAA0229 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 1A (Odin). | |||||
|
ARHG1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.021510 (rank : 113) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q61210, O89074, Q80YE8, Q80YE9, Q91VL3 | Gene names | Arhgef1, Lbcl2, Lsc | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 1 (Lymphoid blast crisis-like 2) (Lbc's second cousin). | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.063241 (rank : 22) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
DYN1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 67) | NC score | 0.026268 (rank : 101) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q05193 | Gene names | DNM1, DNM | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-1 (EC 3.6.5.5). | |||||
|
DYN1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 68) | NC score | 0.025358 (rank : 104) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P39053, Q61358, Q61359, Q61360, Q8JZZ4, Q9CSY7, Q9QXX1 | Gene names | Dnm1, Dnm | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-1 (EC 3.6.5.5). | |||||
|
GNDS_HUMAN
|
||||||
| θ value | 3.0926 (rank : 69) | NC score | 0.033561 (rank : 89) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q12967, Q9HAX7, Q9HAY1, Q9HCT1 | Gene names | RALGDS, RGF | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator (RalGEF) (RalGDS). | |||||
|
HIC1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 70) | NC score | 0.005338 (rank : 156) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 863 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9R1Y5, Q9R1Y6, Q9R2B0 | Gene names | Hic1 | |||
|
Domain Architecture |
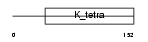
|
|||||
| Description | Hypermethylated in cancer 1 protein (Hic-1). | |||||
|
JPH2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 71) | NC score | 0.053846 (rank : 43) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
K1802_MOUSE
|
||||||
| θ value | 3.0926 (rank : 72) | NC score | 0.055433 (rank : 38) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
MA1C1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 73) | NC score | 0.018238 (rank : 126) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NR34, Q9Y545 | Gene names | MAN1C1, MAN1A3, MAN1C | |||
|
Domain Architecture |

|
|||||
| Description | Mannosyl-oligosaccharide 1,2-alpha-mannosidase IC (EC 3.2.1.113) (Processing alpha-1,2-mannosidase IC) (Alpha-1,2-mannosidase IC) (Mannosidase alpha class 1C member 1) (HMIC). | |||||
|
MAZ_HUMAN
|
||||||
| θ value | 3.0926 (rank : 74) | NC score | 0.005361 (rank : 155) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 757 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P56270, Q15703, Q99443 | Gene names | MAZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myc-associated zinc finger protein (MAZI) (Purine-binding transcription factor) (Pur-1) (ZF87) (ZIF87). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 75) | NC score | 0.037466 (rank : 79) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MUTED_HUMAN
|
||||||
| θ value | 3.0926 (rank : 76) | NC score | 0.044657 (rank : 66) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TDH9, Q5THS1, Q68D56, Q8N5F9, Q9NU16 | Gene names | MUTED | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Muted protein homolog. | |||||
|
PABP2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 77) | NC score | 0.023042 (rank : 110) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CCS6, O35935 | Gene names | Pabpn1, Pab2, Pabp2 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 2 (Poly(A)-binding protein 2) (Poly(A)- binding protein II) (PABII) (Polyadenylate-binding nuclear protein 1) (Nuclear poly(A)-binding protein 1). | |||||
|
SATB1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 78) | NC score | 0.023955 (rank : 106) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q60611 | Gene names | Satb1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SATB1 (Special AT-rich sequence-binding protein 1). | |||||
|
WASF2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 79) | NC score | 0.056109 (rank : 35) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y6W5, O60794, Q9UDY7 | Gene names | WASF2, WAVE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2) (Verprolin homology domain-containing protein 2). | |||||
|
ZYX_HUMAN
|
||||||
| θ value | 3.0926 (rank : 80) | NC score | 0.019446 (rank : 122) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q15942 | Gene names | ZYX | |||
|
Domain Architecture |

|
|||||
| Description | Zyxin (Zyxin-2). | |||||
|
ADDA_MOUSE
|
||||||
| θ value | 4.03905 (rank : 81) | NC score | 0.021123 (rank : 116) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9QYC0, Q9JLE3 | Gene names | Add1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-adducin (Erythrocyte adducin subunit alpha). | |||||
|
ANX11_HUMAN
|
||||||
| θ value | 4.03905 (rank : 82) | NC score | 0.017296 (rank : 128) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P50995 | Gene names | ANXA11, ANX11 | |||
|
Domain Architecture |
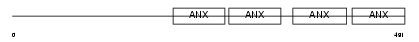
|
|||||
| Description | Annexin A11 (Annexin XI) (Calcyclin-associated annexin 50) (CAP-50) (56 kDa autoantigen). | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.052337 (rank : 47) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
CI079_HUMAN
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | 0.048245 (rank : 60) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6ZUB1, Q5SQC9, Q8NA41, Q8ND27 | Gene names | C9orf79 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf79. | |||||
|
CK002_HUMAN
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.054123 (rank : 42) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UID3, Q6PJV5, Q7L8A6, Q8WZ35, Q96DF4, Q96GR3 | Gene names | C11orf2, ANG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C11orf2 (Another new gene 2 protein). | |||||
|
DIAP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | 0.050876 (rank : 53) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O60610, Q9UC76 | Gene names | DIAPH1, DIAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1). | |||||
|
EMID2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 87) | NC score | 0.026675 (rank : 99) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96A83 | Gene names | EMID2, COL26A1, EMU2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XXVI) chain precursor (EMI domain-containing protein 2) (Protein Emu2) (Emilin and multimerin domain-containing protein 2). | |||||
|
ESX1L_HUMAN
|
||||||
| θ value | 4.03905 (rank : 88) | NC score | 0.013067 (rank : 137) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8N693, Q7Z6K7 | Gene names | ESX1L, ESX1R | |||
|
Domain Architecture |

|
|||||
| Description | Extraembryonic, spermatogenesis, homeobox 1-like protein. | |||||
|
FBLI1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 89) | NC score | 0.015447 (rank : 134) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q71FD7, Q99J35 | Gene names | Fblim1, Cal | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-binding LIM protein 1 (CSX-associated LIM). | |||||
|
MK07_HUMAN
|
||||||
| θ value | 4.03905 (rank : 90) | NC score | 0.005516 (rank : 153) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1087 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13164, Q16634 | Gene names | MAPK7, ERK4, ERK5, PRKM7 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase 7 (EC 2.7.11.24) (Extracellular signal-regulated kinase 5) (ERK-5) (ERK4) (BMK1 kinase). | |||||
|
PHLA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 91) | NC score | 0.030134 (rank : 95) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WV24, Q15184, Q2TAN2, Q9NZ17 | Gene names | PHLDA1, PHRIP, TDAG51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family A member 1 (T-cell death- associated gene 51 protein) (Apoptosis-associated nuclear protein) (Proline- and histidine-rich protein) (Proline- and glutamine-rich protein) (PQ-rich protein). | |||||
|
PRG4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 92) | NC score | 0.026336 (rank : 100) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRR12_HUMAN
|
||||||
| θ value | 4.03905 (rank : 93) | NC score | 0.044615 (rank : 67) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
SGIP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 94) | NC score | 0.032204 (rank : 92) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8VD37, Q3UFU3, Q3UGA0, Q8BXX4, Q8C034, Q9CXT2 | Gene names | Sgip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3-containing GRB2-like protein 3-interacting protein 1 (Endophilin- 3-interacting protein). | |||||
|
SNPC2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 95) | NC score | 0.033294 (rank : 90) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q91XA5, Q3UK17, Q8CBS9 | Gene names | Snapc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 2 (SNAPc subunit 2) (snRNA- activating protein complex 45 kDa subunit) (SNAPc 45 kDa subunit) (Small nuclear RNA-activating complex polypeptide 2). | |||||
|
SRCA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 96) | NC score | 0.023034 (rank : 111) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
SREC2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 97) | NC score | 0.012065 (rank : 140) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96GP6, Q8IXF3, Q9BW74 | Gene names | SCARF2, SREC2, SREPCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II) (SRECRP-1). | |||||
|
ZN385_HUMAN
|
||||||
| θ value | 4.03905 (rank : 98) | NC score | 0.059681 (rank : 28) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q96PM9, Q5VH53, Q9H7R6, Q9UFU3 | Gene names | ZNF385, HZF, RZF | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 385 (Hematopoietic zinc finger protein) (Retinal zinc finger protein). | |||||
|
ANR25_HUMAN
|
||||||
| θ value | 5.27518 (rank : 99) | NC score | 0.021316 (rank : 115) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q63ZY3, Q3KQZ3, Q6GUF5, Q9H8S4, Q9NUP0, Q9P210 | Gene names | ANKRD25, KIAA1518, SIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 25 (SRC-1-interacting protein) (SRC1-interacting protein). | |||||
|
ANX11_MOUSE
|
||||||
| θ value | 5.27518 (rank : 100) | NC score | 0.017157 (rank : 130) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P97384 | Gene names | Anxa11, Anx11 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A11 (Annexin XI) (Calcyclin-associated annexin 50) (CAP-50). | |||||
|
BAT3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 101) | NC score | 0.050584 (rank : 55) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P46379, O95874, Q5SQ35, Q5SQ36, Q5SQ37, Q5SRP8, Q96SA6, Q9BCN4 | Gene names | BAT3, G3 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3) (Protein G3). | |||||
|
CABL2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 102) | NC score | 0.023251 (rank : 108) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BTV7, Q5JWL0, Q9BYK0 | Gene names | CABLES2, C20orf150 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 and ABL1 enzyme substrate 2 (Interactor with CDK3 2) (Ik3-2). | |||||
|
COBA2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 103) | NC score | 0.019096 (rank : 123) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 501 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q64739, Q61432, Q9Z1W0 | Gene names | Col11a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(XI) chain precursor. | |||||
|
CTCF_MOUSE
|
||||||
| θ value | 5.27518 (rank : 104) | NC score | 0.003752 (rank : 157) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 796 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61164 | Gene names | Ctcf | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional repressor CTCF (CCCTC-binding factor) (CTCFL paralog) (11-zinc finger protein). | |||||
|
EMIL2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 105) | NC score | 0.017214 (rank : 129) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8K482 | Gene names | Emilin2 | |||
|
Domain Architecture |

|
|||||
| Description | EMILIN-2 precursor (Elastin microfibril interface-located protein 2) (Elastin microfibril interfacer 2) (Basilin). | |||||
|
FMN2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 106) | NC score | 0.046329 (rank : 62) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NZ56, Q59GF6, Q5VU37, Q9NZ55 | Gene names | FMN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-2. | |||||
|
JIP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.028197 (rank : 96) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q13387, Q96G62, Q99771, Q9NZ59, Q9UKQ4 | Gene names | MAPK8IP2, IB2, JIP2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 2 (JNK-interacting protein 2) (JIP-2) (JNK MAP kinase scaffold protein 2) (Islet-brain-2) (IB-2) (Mitogen-activated protein kinase 8-interacting protein 2). | |||||
|
KRHB1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 108) | NC score | 0.010422 (rank : 144) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14533, Q14846, Q16274, Q8WU52, Q9BR74 | Gene names | KRTHB1, MLN137 | |||
|
Domain Architecture |
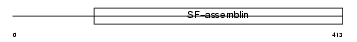
|
|||||
| Description | Keratin, type II cuticular Hb1 (Hair keratin, type II Hb1) (ghHKb1) (ghHb1) (MLN 137). | |||||
|
KRHB6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 109) | NC score | 0.010567 (rank : 143) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O43790, P78387 | Gene names | KRTHB6 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type II cuticular Hb6 (Hair keratin, type II Hb6) (ghHb6). | |||||
|
MOT8_MOUSE
|
||||||
| θ value | 5.27518 (rank : 110) | NC score | 0.032492 (rank : 91) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O70324, Q8K3S9 | Gene names | Slc16a2, Mct8, Xpct | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Monocarboxylate transporter 8 (MCT 8) (Solute carrier family 16 member 2) (X-linked PEST-containing transporter). | |||||
|
MRIP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 111) | NC score | 0.019915 (rank : 120) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6WCQ1, Q3KQZ5, Q5FB94, Q6DHW2, Q7Z5Y2, Q8N390, Q96G40 | Gene names | MRIP, KIAA0864, RHOIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (M-RIP) (RIP3) (p116Rip). | |||||
|
MTSS1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 112) | NC score | 0.057307 (rank : 32) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8R1S4, Q8BMM3, Q99LB3 | Gene names | Mtss1, Mim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein). | |||||
|
PI5PA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 113) | NC score | 0.065986 (rank : 16) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | 5.27518 (rank : 114) | NC score | 0.040206 (rank : 73) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
SSA27_MOUSE
|
||||||
| θ value | 5.27518 (rank : 115) | NC score | 0.027261 (rank : 97) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P56873 | Gene names | Sssca1, C184l | |||
|
Domain Architecture |
|
|||||
| Description | Sjoegren syndrome/scleroderma autoantigen 1 homolog (Autoantigen p27 homolog) (Protein C184L). | |||||
|
SYNJ1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 116) | NC score | 0.046010 (rank : 63) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O43426, O43425, O94984 | Gene names | SYNJ1, KIAA0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
TAF4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 117) | NC score | 0.038908 (rank : 77) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
ZN202_HUMAN
|
||||||
| θ value | 5.27518 (rank : 118) | NC score | 0.002129 (rank : 159) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 756 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O95125, Q4JG21, Q9H1B9, Q9NSM4 | Gene names | ZNF202 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 202. | |||||
|
ABI3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 119) | NC score | 0.030343 (rank : 94) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9P2A4, Q9H0P6 | Gene names | ABI3, NESH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABI gene family member 3 (New molecule including SH3) (Nesh). | |||||
|
BRD4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 120) | NC score | 0.025938 (rank : 102) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
FHOD1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 121) | NC score | 0.043205 (rank : 68) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y613, Q59F76, Q6Y1F2, Q76MS8, Q8N521 | Gene names | FHOD1, FHOS | |||
|
Domain Architecture |

|
|||||
| Description | FH1/FH2 domain-containing protein (Formin homolog overexpressed in spleen) (FHOS) (Formin homology 2 domain-containing protein 1). | |||||
|
FZD8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 122) | NC score | 0.006155 (rank : 152) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H461 | Gene names | FZD8 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-8 precursor (Fz-8) (hFz8). | |||||
|
GABP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 123) | NC score | 0.009633 (rank : 145) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q06547, Q06545, Q12940, Q12941, Q12942, Q8IYD0 | Gene names | GABPB2, E4TF1B, GABPB, GABPB1 | |||
|
Domain Architecture |
|
|||||
| Description | GA-binding protein beta chain (GABP subunit beta-2) (GABP-2) (GABP subunit beta-1) (GABPB-1) (Transcription factor E4TF1-53) (Transcription factor E4TF1-47) (Nuclear respiratory factor 2). | |||||
|
M3K12_HUMAN
|
||||||
| θ value | 6.88961 (rank : 124) | NC score | 0.001908 (rank : 160) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q12852, Q86VQ5, Q8WY25 | Gene names | MAP3K12, ZPK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 12 (EC 2.7.11.25) (Mixed lineage kinase) (Leucine-zipper protein kinase) (ZPK) (Dual leucine zipper bearing kinase) (DLK) (MAPK-upstream kinase) (MUK). | |||||
|
RP3A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 125) | NC score | 0.018435 (rank : 125) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y2J0, Q96AE0 | Gene names | RPH3A, KIAA0985 | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
SF3A2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 126) | NC score | 0.040798 (rank : 71) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SYNJ1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 127) | NC score | 0.047592 (rank : 61) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
WASF3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 128) | NC score | 0.048917 (rank : 58) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UPY6, O94974 | Gene names | WASF3, KIAA0900, SCAR3, WAVE3 | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein family member 3 (WASP-family protein member 3) (Protein WAVE-3) (Verprolin homology domain-containing protein 3). | |||||
|
WNK4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 129) | NC score | 0.007263 (rank : 149) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1043 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96J92, Q8N8X3, Q8N8Z2, Q96DT8, Q9BYS5 | Gene names | WNK4, PRKWNK4 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK4 (EC 2.7.11.1) (Protein kinase with no lysine 4) (Protein kinase, lysine-deficient 4). | |||||
|
ANKS1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | 0.017313 (rank : 127) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P59672, Q6ZQG0 | Gene names | Anks1a, Anks1, Kiaa0229 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 1A. | |||||
|
BRD4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.025404 (rank : 103) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
CO2A1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | 0.016671 (rank : 132) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 483 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P28481 | Gene names | Col2a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(II) chain precursor [Contains: Chondrocalcin]. | |||||
|
GLU2B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | 0.015057 (rank : 135) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P14314, Q96BU9, Q96D06, Q9P0W9 | Gene names | PRKCSH, G19P1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glucosidase 2 subunit beta precursor (Glucosidase II subunit beta) (Protein kinase C substrate, 60.1 kDa protein, heavy chain) (PKCSH) (80K-H protein). | |||||
|
GON4L_MOUSE
|
||||||
| θ value | 8.99809 (rank : 134) | NC score | 0.023046 (rank : 109) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9DB00, Q80TB4, Q91YI9 | Gene names | Gon4l, Gon4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
KIF3A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 135) | NC score | 0.005472 (rank : 154) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y496, Q86XE9, Q9Y6V4 | Gene names | KIF3A, KIF3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3A (Microtubule plus end-directed kinesin motor 3A). | |||||
|
KRHB2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 136) | NC score | 0.009341 (rank : 146) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NSB4 | Gene names | KRTHB2 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type II cuticular Hb2 (Hair keratin, type II Hb2). | |||||
|
PGCB_MOUSE
|
||||||
| θ value | 8.99809 (rank : 137) | NC score | 0.006379 (rank : 151) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61361 | Gene names | Bcan | |||
|
Domain Architecture |

|
|||||
| Description | Brevican core protein precursor. | |||||
|
QSCN6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 138) | NC score | 0.012074 (rank : 139) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O00391, Q59G29, Q5T2X0, Q8TDL6, Q8WVP4 | Gene names | QSCN6, QSOX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sulfhydryl oxidase 1 precursor (EC 1.8.3.2) (Quiescin Q6) (hQSOX). | |||||
|
RHG05_HUMAN
|
||||||
| θ value | 8.99809 (rank : 139) | NC score | 0.033660 (rank : 88) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13017 | Gene names | ARHGAP5, RHOGAP5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
RHG18_HUMAN
|
||||||
| θ value | 8.99809 (rank : 140) | NC score | 0.011591 (rank : 142) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N392, Q58EZ3, Q6P679, Q6PJD7, Q96S64 | Gene names | ARHGAP18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho-GTPase-activating protein 18 (MacGAP). | |||||
|
SF3A2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 141) | NC score | 0.038998 (rank : 75) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SFR15_HUMAN
|
||||||
| θ value | 8.99809 (rank : 142) | NC score | 0.024882 (rank : 105) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O95104, Q6P1M5, Q8N3I8, Q9UFM1, Q9ULP8 | Gene names | SFRS15, KIAA1172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 15 (CTD-binding SR-like protein RA4). | |||||
|
TAOK2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 143) | NC score | 0.002757 (rank : 158) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1172 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UL54, O94957, Q6UW73, Q7LC09, Q9NSW2 | Gene names | TAOK2, KIAA0881, MAP3K17, PSK, PSK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO2 (EC 2.7.11.1) (Thousand and one amino acid protein 2) (Prostate-derived STE20-like kinase 1) (PSK-1) (Kinase from chicken homolog C) (hKFC-C). | |||||
|
TRIB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 144) | NC score | 0.001755 (rank : 161) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 598 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96RU8, O15180, Q9H2Y8 | Gene names | TRIB1, C8FW, GIG2, TRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tribbles homolog 1 (TRB-1) (SKIP1) (G-protein-coupled receptor-induced protein 2) (GIG-2). | |||||
|
UBQL3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 145) | NC score | 0.018599 (rank : 124) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H347, Q9NRE0 | Gene names | UBQLN3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-3. | |||||
|
WASP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 146) | NC score | 0.021345 (rank : 114) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P42768, Q9BU11, Q9UNJ9 | Gene names | WAS, IMD2 | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein (WASp). | |||||
|
ZDHC9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 147) | NC score | 0.006760 (rank : 150) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y397, Q59EK4, Q5JSW5, Q8WWS7, Q9BPY4, Q9NSP0, Q9NVL0, Q9NVR6 | Gene names | ZDHHC9, ZNF379 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC9 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 9) (DHHC-9) (DHHC9) (Zinc finger protein 379). | |||||
|
BCL9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.050960 (rank : 52) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
BSN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.065042 (rank : 17) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CD2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.050483 (rank : 57) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P06729, Q96TE5 | Gene names | CD2 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface antigen CD2 precursor (T-cell surface antigen T11/Leu- 5) (LFA-2) (LFA-3 receptor) (Erythrocyte receptor) (Rosette receptor). | |||||
|
CEL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.052749 (rank : 44) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.050721 (rank : 54) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.064768 (rank : 19) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.055673 (rank : 36) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.055345 (rank : 39) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
OGFR_MOUSE
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.051606 (rank : 50) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q99PG2, Q99L33 | Gene names | Ogfr | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.079007 (rank : 8) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
RGS3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.057156 (rank : 34) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
SON_HUMAN
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.061460 (rank : 25) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
SPR1B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.058879 (rank : 29) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q62267 | Gene names | Sprr1b | |||
|
Domain Architecture |

|
|||||
| Description | Cornifin B (Small proline-rich protein 1B) (SPR1B) (SPR1 B). | |||||
|
SSBP4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.051816 (rank : 49) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
COG8_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 147 | |
| SwissProt Accessions | Q9JJA2 | Gene names | Cog8 | |||
|
Domain Architecture |

|
|||||
| Description | Conserved oligomeric Golgi complex component 8. | |||||
|
COG8_HUMAN
|
||||||
| NC score | 0.955665 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96MW5, Q8WVV6, Q9H6F8 | Gene names | COG8 | |||
|
Domain Architecture |

|
|||||
| Description | Conserved oligomeric Golgi complex component 8. | |||||
|
PRP5_HUMAN
|
||||||
| NC score | 0.109044 (rank : 3) | θ value | 0.47712 (rank : 23) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P04281 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide IB-1. | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.102463 (rank : 4) | θ value | 0.21417 (rank : 12) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 106 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.089700 (rank : 5) | θ value | 0.00228821 (rank : 3) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
CPSF6_MOUSE
|
||||||
| NC score | 0.084675 (rank : 6) | θ value | 0.163984 (rank : 11) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6NVF9, Q8BX86, Q8BXI8 | Gene names | Cpsf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6. | |||||
|
CPSF6_HUMAN
|
||||||
| NC score | 0.084469 (rank : 7) | θ value | 0.163984 (rank : 10) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q16630, Q53ES1, Q9BSJ7, Q9BW18 | Gene names | CPSF6, CFIM68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6 (Cleavage and polyadenylation specificity factor 68 kDa subunit) (CPSF 68 kDa subunit) (Pre-mRNA cleavage factor Im 68 kDa subunit) (Protein HPBRII- 4/7). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.079007 (rank : 8) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
NEIL2_HUMAN
|
||||||
| NC score | 0.075781 (rank : 9) | θ value | 0.0736092 (rank : 6) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q969S2, Q7Z3Q7, Q8N842, Q8NG52 | Gene names | NEIL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 2 (EC 3.2.2.-) (EC 4.2.99.18) (Nei-like 2) (DNA glycosylase/AP lyase Neil2) (DNA-(apurinic or apyrimidinic site) lyase Neil2) (NEH2). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.074134 (rank : 10) | θ value | 0.365318 (rank : 18) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
ZN598_HUMAN
|
||||||
| NC score | 0.074001 (rank : 11) | θ value | 0.0431538 (rank : 5) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q86UK7, Q8IW49, Q8N3D9, Q96FG3, Q9H7J3 | Gene names | ZNF598 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 598. | |||||
|
H6ST3_MOUSE
|
||||||
| NC score | 0.073731 (rank : 12) | θ value | 0.0148317 (rank : 4) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QYK4 | Gene names | Hs6st3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan-sulfate 6-O-sulfotransferase 3 (EC 2.8.2.-) (HS6ST-3) (mHS6ST- 3). | |||||
|
GGN_MOUSE
|
||||||
| NC score | 0.073255 (rank : 13) | θ value | 0.125558 (rank : 8) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q80WJ1, Q5EBP4, Q80WI9, Q80WJ0 | Gene names | Ggn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
FMNL_HUMAN
|
||||||
| NC score | 0.072586 (rank : 14) | θ value | 0.365318 (rank : 17) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O95466, Q6DKG5, Q6IBP3, Q86UH1, Q8N671, Q8TDH1, Q96H10 | Gene names | FMNL1, C17orf1, C17orf1B, FMNL | |||
|
Domain Architecture |

|
|||||
| Description | Formin-like protein 1 (Leukocyte formin) (CLL-associated antigen KW- 13). | |||||
|
FMNL_MOUSE
|
||||||
| NC score | 0.067452 (rank : 15) | θ value | 1.38821 (rank : 42) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9JL26, Q6KAN4, Q9Z2V7 | Gene names | Fmnl1, Frl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-like protein 1 (Formin-related protein). | |||||
|
PI5PA_HUMAN
|
||||||
| NC score | 0.065986 (rank : 16) | θ value | 5.27518 (rank : 113) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.065042 (rank : 17) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
WASF1_HUMAN
|
||||||
| NC score | 0.064904 (rank : 18) | θ value | 0.47712 (rank : 25) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q92558 | Gene names | WASF1, KIAA0269, SCAR1, WAVE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 1 (WASP-family protein member 1) (Protein WAVE-1) (Verprolin homology domain-containing protein 1). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.064768 (rank : 19) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
WASF1_MOUSE
|
||||||
| NC score | 0.064742 (rank : 20) | θ value | 0.47712 (rank : 26) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8R5H6, Q91W51, Q9ERQ9 | Gene names | Wasf1, Wave1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 1 (WASP-family protein member 1) (Protein WAVE-1). | |||||
|
PRPC_HUMAN
|
||||||
| NC score | 0.063823 (rank : 21) | θ value | 1.81305 (rank : 52) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P02810, Q4VBP2, Q53XA2, Q6P2F6 | Gene names | PRH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Salivary acidic proline-rich phosphoprotein 1/2 precursor (PRP-1/PRP- 2) (Parotid proline-rich protein 1/2) (Pr1/Pr2) (Protein C) (Parotid acidic protein) (Pa) (Parotid isoelectric focusing variant protein) (PIF-S) (Parotid double-band protein) (Db-s) [Contains: Salivary acidic proline-rich phosphoprotein 1/2; Salivary acidic proline-rich phosphoprotein 3/4 (PRP-3/PRP-4) (Protein A) (PIF-F) (Db-F); Peptide P-C]. | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.063241 (rank : 22) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
DIAP3_MOUSE
|
||||||
| NC score | 0.062370 (rank : 23) | θ value | 0.0961366 (rank : 7) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Z207 | Gene names | Diaph3, Diap3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 3 (Diaphanous-related formin-3) (DRF3) (mDIA2) (p134mDIA2). | |||||
|
EFS_MOUSE
|
||||||
| NC score | 0.061963 (rank : 24) | θ value | 0.47712 (rank : 20) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q64355 | Gene names | Efs, Sin | |||
|
Domain Architecture |

|
|||||
| Description | Embryonal Fyn-associated substrate (SRC-interacting protein) (Signal- integrating protein). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.061460 (rank : 25) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
NGB_HUMAN
|
||||||
| NC score | 0.060900 (rank : 26) | θ value | 2.36792 (rank : 56) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NPG2 | Gene names | NGB | |||
|
Domain Architecture |

|
|||||
| Description | Neuroglobin. | |||||
|
PP1RA_MOUSE
|
||||||
| NC score | 0.059905 (rank : 27) | θ value | 0.62314 (rank : 28) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q80W00, Q811B6, Q8C6T7, Q8K2U8 | Gene names | Ppp1r10, Cat53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 1 regulatory subunit 10 (MHC class I region proline-rich protein CAT53). | |||||
|
ZN385_HUMAN
|
||||||
| NC score | 0.059681 (rank : 28) | θ value | 4.03905 (rank : 98) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q96PM9, Q5VH53, Q9H7R6, Q9UFU3 | Gene names | ZNF385, HZF, RZF | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 385 (Hematopoietic zinc finger protein) (Retinal zinc finger protein). | |||||
|
SPR1B_MOUSE
|
||||||
| NC score | 0.058879 (rank : 29) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q62267 | Gene names | Sprr1b | |||
|
Domain Architecture |

|
|||||
| Description | Cornifin B (Small proline-rich protein 1B) (SPR1B) (SPR1 B). | |||||
|
GTSE1_MOUSE
|
||||||
| NC score | 0.058658 (rank : 30) | θ value | 0.47712 (rank : 21) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8R080, O89015, Q9CSG9 | Gene names | Gtse1, B99 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G2 and S phase expressed protein 1 (Gtse-1) (B99 protein). | |||||
|
PELP1_MOUSE
|
||||||
| NC score | 0.057502 (rank : 31) | θ value | 0.813845 (rank : 31) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
MTSS1_MOUSE
|
||||||
| NC score | 0.057307 (rank : 32) | θ value | 5.27518 (rank : 112) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8R1S4, Q8BMM3, Q99LB3 | Gene names | Mtss1, Mim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein). | |||||
|
PP1RA_HUMAN
|
||||||
| NC score | 0.057203 (rank : 33) | θ value | 2.36792 (rank : 58) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96QC0, O00405 | Gene names | PPP1R10, CAT53, FB19, PNUTS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 1 regulatory subunit 10 (Phosphatase 1 nuclear targeting subunit) (MHC class I region proline- rich protein CAT53) (FB19 protein) (PP1-binding protein of 114 kDa) (p99). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.057156 (rank : 34) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
WASF2_HUMAN
|
||||||
| NC score | 0.056109 (rank : 35) | θ value | 3.0926 (rank : 79) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y6W5, O60794, Q9UDY7 | Gene names | WASF2, WAVE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2) (Verprolin homology domain-containing protein 2). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.055673 (rank : 36) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
EFS_HUMAN
|
||||||
| NC score | 0.055606 (rank : 37) | θ value | 1.38821 (rank : 40) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O43281, O43282 | Gene names | EFS | |||
|
Domain Architecture |

|
|||||
| Description | Embryonal Fyn-associated substrate (HEFS). | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.055433 (rank : 38) | θ value | 3.0926 (rank : 72) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.055345 (rank : 39) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
H6ST3_HUMAN
|
||||||
| NC score | 0.055285 (rank : 40) | θ value | 1.81305 (rank : 50) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IZP7, Q5W0L0, Q68CW6 | Gene names | HS6ST3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan-sulfate 6-O-sulfotransferase 3 (EC 2.8.2.-) (HS6ST-3). | |||||
|
PRB3_HUMAN
|
||||||
| NC score | 0.054606 (rank : 41) | θ value | 0.47712 (rank : 22) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
CK002_HUMAN
|
||||||
| NC score | 0.054123 (rank : 42) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UID3, Q6PJV5, Q7L8A6, Q8WZ35, Q96DF4, Q96GR3 | Gene names | C11orf2, ANG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C11orf2 (Another new gene 2 protein). | |||||
|
JPH2_HUMAN
|
||||||
| NC score | 0.053846 (rank : 43) | θ value | 3.0926 (rank : 71) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.052749 (rank : 44) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
DLGP3_HUMAN
|
||||||
| NC score | 0.052634 (rank : 45) | θ value | 0.365318 (rank : 16) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95886, Q5TDD5, Q9H3X7 | Gene names | DLGAP3, DAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large-associated protein 3 (DAP-3) (SAP90/PSD-95-associated protein 3) (SAPAP3) (PSD-95/SAP90-binding protein 3). | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.052557 (rank : 46) | θ value | 0.813845 (rank : 32) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.052337 (rank : 47) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
DLGP3_MOUSE
|
||||||
| NC score | 0.051882 (rank : 48) | θ value | 0.47712 (rank : 19) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PFD5, Q6PDX0, Q6XBF2 | Gene names | Dlgap3, Dap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large-associated protein 3 (DAP-3) (SAP90/PSD-95-associated protein 3) (SAPAP3) (PSD-95/SAP90-binding protein 3). | |||||
|
SSBP4_HUMAN
|
||||||
| NC score | 0.051816 (rank : 49) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
OGFR_MOUSE
|
||||||
| NC score | 0.051606 (rank : 50) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q99PG2, Q99L33 | Gene names | Ogfr | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor). | |||||
|
PI5PA_MOUSE
|
||||||
| NC score | 0.051445 (rank : 51) | θ value | 1.38821 (rank : 44) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P59644 | Gene names | Pib5pa | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.050960 (rank : 52) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
DIAP1_HUMAN
|
||||||
| NC score | 0.050876 (rank : 53) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O60610, Q9UC76 | Gene names | DIAPH1, DIAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.050721 (rank : 54) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
BAT3_HUMAN
|
||||||
| NC score | 0.050584 (rank : 55) | θ value | 5.27518 (rank : 101) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P46379, O95874, Q5SQ35, Q5SQ36, Q5SQ37, Q5SRP8, Q96SA6, Q9BCN4 | Gene names | BAT3, G3 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3) (Protein G3). | |||||
|
DIAP1_MOUSE
|
||||||
| NC score | 0.050505 (rank : 56) | θ value | 2.36792 (rank : 53) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 839 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O08808 | Gene names | Diaph1, Diap1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1) (mDIA1) (p140mDIA). | |||||
|
CD2_HUMAN
|
||||||
| NC score | 0.050483 (rank : 57) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P06729, Q96TE5 | Gene names | CD2 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface antigen CD2 precursor (T-cell surface antigen T11/Leu- 5) (LFA-2) (LFA-3 receptor) (Erythrocyte receptor) (Rosette receptor). | |||||
|
WASF3_HUMAN
|
||||||
| NC score | 0.048917 (rank : 58) | θ value | 6.88961 (rank : 128) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UPY6, O94974 | Gene names | WASF3, KIAA0900, SCAR3, WAVE3 | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein family member 3 (WASP-family protein member 3) (Protein WAVE-3) (Verprolin homology domain-containing protein 3). | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.048526 (rank : 59) | θ value | 0.47712 (rank : 24) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
CI079_HUMAN
|
||||||
| NC score | 0.048245 (rank : 60) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6ZUB1, Q5SQC9, Q8NA41, Q8ND27 | Gene names | C9orf79 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf79. | |||||
|
SYNJ1_MOUSE
|
||||||
| NC score | 0.047592 (rank : 61) | θ value | 6.88961 (rank : 127) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
FMN2_HUMAN
|
||||||
| NC score | 0.046329 (rank : 62) | θ value | 5.27518 (rank : 106) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NZ56, Q59GF6, Q5VU37, Q9NZ55 | Gene names | FMN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-2. | |||||
|
SYNJ1_HUMAN
|
||||||
| NC score | 0.046010 (rank : 63) | θ value | 5.27518 (rank : 116) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O43426, O43425, O94984 | Gene names | SYNJ1, KIAA0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
ABI3_MOUSE
|
||||||
| NC score | 0.045827 (rank : 64) | θ value | 0.279714 (rank : 13) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BYZ1, Q6PE63, Q9D7S4 | Gene names | Abi3, Nesh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABI gene family member 3 (New molecule including SH3) (Nesh). | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.045602 (rank : 65) | θ value | 1.81305 (rank : 49) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
MUTED_HUMAN
|
||||||
| NC score | 0.044657 (rank : 66) | θ value | 3.0926 (rank : 76) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TDH9, Q5THS1, Q68D56, Q8N5F9, Q9NU16 | Gene names | MUTED | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Muted protein homolog. | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.044615 (rank : 67) | θ value | 4.03905 (rank : 93) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
FHOD1_HUMAN
|
||||||
| NC score | 0.043205 (rank : 68) | θ value | 6.88961 (rank : 121) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y613, Q59F76, Q6Y1F2, Q76MS8, Q8N521 | Gene names | FHOD1, FHOS | |||
|
Domain Architecture |

|
|||||
| Description | FH1/FH2 domain-containing protein (Formin homolog overexpressed in spleen) (FHOS) (Formin homology 2 domain-containing protein 1). | |||||
|
SFPQ_MOUSE
|
||||||
| NC score | 0.042561 (rank : 69) | θ value | 0.62314 (rank : 29) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
SYNJ2_MOUSE
|
||||||
| NC score | 0.040927 (rank : 70) | θ value | 1.38821 (rank : 47) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9D2G5, O35404, O88399, O88400, O88401, O88402, O88403, O88404 | Gene names | Synj2, Kiaa0348 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-2 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 2). | |||||
|
SF3A2_MOUSE
|
||||||
| NC score | 0.040798 (rank : 71) | θ value | 6.88961 (rank : 126) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
RHG05_MOUSE
|
||||||
| NC score | 0.040718 (rank : 72) | θ value | 0.813845 (rank : 33) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P97393 | Gene names | Arhgap5, Rhogap5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.040206 (rank : 73) | θ value | 5.27518 (rank : 114) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
CUTL1_HUMAN
|
||||||
| NC score | 0.039023 (rank : 74) | θ value | 0.365318 (rank : 15) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
SF3A2_HUMAN
|
||||||
| NC score | 0.038998 (rank : 75) | θ value | 8.99809 (rank : 141) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SFPQ_HUMAN
|
||||||
| NC score | 0.038973 (rank : 76) | θ value | 2.36792 (rank : 60) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.038908 (rank : 77) | θ value | 5.27518 (rank : 117) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
MOT8_HUMAN
|
||||||
| NC score | 0.038125 (rank : 78) | θ value | 1.06291 (rank : 37) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P36021, Q7Z797 | Gene names | SLC16A2, MCT8, XPCT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Monocarboxylate transporter 8 (MCT 8) (MCT 7) (Solute carrier family 16 member 2) (X-linked PEST-containing transporter). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.037466 (rank : 79) | θ value | 3.0926 (rank : 75) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
DGKK_HUMAN
|
||||||
| NC score | 0.037067 (rank : 80) | θ value | 0.813845 (rank : 30) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5KSL6 | Gene names | DGKK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase kappa (EC 2.7.1.107) (Diglyceride kinase kappa) (DGK-kappa) (DAG kinase kappa) (142 kDa diacylglycerol kinase). | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.036969 (rank : 81) | θ value | 3.0926 (rank : 63) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
PRRT3_MOUSE
|
||||||
| NC score | 0.036160 (rank : 82) | θ value | 2.36792 (rank : 59) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6PE13 | Gene names | Prrt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
CASP_HUMAN
|
||||||
| NC score | 0.035572 (rank : 83) | θ value | 0.365318 (rank : 14) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q13948, Q53GU9, Q8TBS3 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CASP_MOUSE
|
||||||
| NC score | 0.035070 (rank : 84) | θ value | 0.62314 (rank : 27) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1152 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P70403, Q7TQJ4, Q91WN6 | Gene names | Cutl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
EP400_MOUSE
|
||||||
| NC score | 0.034697 (rank : 85) | θ value | 2.36792 (rank : 54) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
GNDS_MOUSE
|
||||||
| NC score | 0.034051 (rank : 86) | θ value | 1.06291 (rank : 36) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q03385 | Gene names | Ralgds, Rgds | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator (RalGEF) (RalGDS). | |||||
|
EP400_HUMAN
|
||||||
| NC score | 0.033705 (rank : 87) | θ value | 1.38821 (rank : 41) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
RHG05_HUMAN
|
||||||
| NC score | 0.033660 (rank : 88) | θ value | 8.99809 (rank : 139) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13017 | Gene names | ARHGAP5, RHOGAP5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
GNDS_HUMAN
|
||||||
| NC score | 0.033561 (rank : 89) | θ value | 3.0926 (rank : 69) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q12967, Q9HAX7, Q9HAY1, Q9HCT1 | Gene names | RALGDS, RGF | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator (RalGEF) (RalGDS). | |||||
|
SNPC2_MOUSE
|
||||||
| NC score | 0.033294 (rank : 90) | θ value | 4.03905 (rank : 95) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q91XA5, Q3UK17, Q8CBS9 | Gene names | Snapc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 2 (SNAPc subunit 2) (snRNA- activating protein complex 45 kDa subunit) (SNAPc 45 kDa subunit) (Small nuclear RNA-activating complex polypeptide 2). | |||||
|
MOT8_MOUSE
|
||||||
| NC score | 0.032492 (rank : 91) | θ value | 5.27518 (rank : 110) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O70324, Q8K3S9 | Gene names | Slc16a2, Mct8, Xpct | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Monocarboxylate transporter 8 (MCT 8) (Solute carrier family 16 member 2) (X-linked PEST-containing transporter). | |||||
|
SGIP1_MOUSE
|
||||||
| NC score | 0.032204 (rank : 92) | θ value | 4.03905 (rank : 94) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8VD37, Q3UFU3, Q3UGA0, Q8BXX4, Q8C034, Q9CXT2 | Gene names | Sgip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3-containing GRB2-like protein 3-interacting protein 1 (Endophilin- 3-interacting protein). | |||||
|
TRI66_MOUSE
|
||||||
| NC score | 0.031598 (rank : 93) | θ value | 1.38821 (rank : 48) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q924W6 | Gene names | Trim66 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
ABI3_HUMAN
|
||||||
| NC score | 0.030343 (rank : 94) | θ value | 6.88961 (rank : 119) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9P2A4, Q9H0P6 | Gene names | ABI3, NESH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABI gene family member 3 (New molecule including SH3) (Nesh). | |||||
|
PHLA1_HUMAN
|
||||||
| NC score | 0.030134 (rank : 95) | θ value | 4.03905 (rank : 91) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WV24, Q15184, Q2TAN2, Q9NZ17 | Gene names | PHLDA1, PHRIP, TDAG51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family A member 1 (T-cell death- associated gene 51 protein) (Apoptosis-associated nuclear protein) (Proline- and histidine-rich protein) (Proline- and glutamine-rich protein) (PQ-rich protein). | |||||
|
JIP2_HUMAN
|
||||||
| NC score | 0.028197 (rank : 96) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q13387, Q96G62, Q99771, Q9NZ59, Q9UKQ4 | Gene names | MAPK8IP2, IB2, JIP2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 2 (JNK-interacting protein 2) (JIP-2) (JNK MAP kinase scaffold protein 2) (Islet-brain-2) (IB-2) (Mitogen-activated protein kinase 8-interacting protein 2). | |||||
|
SSA27_MOUSE
|
||||||
| NC score | 0.027261 (rank : 97) | θ value | 5.27518 (rank : 115) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P56873 | Gene names | Sssca1, C184l | |||
|
Domain Architecture |
|
|||||
| Description | Sjoegren syndrome/scleroderma autoantigen 1 homolog (Autoantigen p27 homolog) (Protein C184L). | |||||
|
RYR1_HUMAN
|
||||||
| NC score | 0.026960 (rank : 98) | θ value | 1.38821 (rank : 45) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P21817, Q16314, Q16368, Q9NPK1, Q9P1U4 | Gene names | RYR1, RYDR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 1 (Skeletal muscle-type ryanodine receptor) (RyR1) (RYR-1) (Skeletal muscle calcium release channel). | |||||
|
EMID2_HUMAN
|
||||||
| NC score | 0.026675 (rank : 99) | θ value | 4.03905 (rank : 87) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96A83 | Gene names | EMID2, COL26A1, EMU2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XXVI) chain precursor (EMI domain-containing protein 2) (Protein Emu2) (Emilin and multimerin domain-containing protein 2). | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.026336 (rank : 100) | θ value | 4.03905 (rank : 92) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
DYN1_HUMAN
|
||||||
| NC score | 0.026268 (rank : 101) | θ value | 3.0926 (rank : 67) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q05193 | Gene names | DNM1, DNM | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-1 (EC 3.6.5.5). | |||||
|
BRD4_MOUSE
|
||||||
| NC score | 0.025938 (rank : 102) | θ value | 6.88961 (rank : 120) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
BRD4_HUMAN
|
||||||
| NC score | 0.025404 (rank : 103) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
DYN1_MOUSE
|
||||||
| NC score | 0.025358 (rank : 104) | θ value | 3.0926 (rank : 68) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P39053, Q61358, Q61359, Q61360, Q8JZZ4, Q9CSY7, Q9QXX1 | Gene names | Dnm1, Dnm | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-1 (EC 3.6.5.5). | |||||
|
SFR15_HUMAN
|
||||||
| NC score | 0.024882 (rank : 105) | θ value | 8.99809 (rank : 142) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O95104, Q6P1M5, Q8N3I8, Q9UFM1, Q9ULP8 | Gene names | SFRS15, KIAA1172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 15 (CTD-binding SR-like protein RA4). | |||||
|
SATB1_MOUSE
|
||||||
| NC score | 0.023955 (rank : 106) | θ value | 3.0926 (rank : 78) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q60611 | Gene names | Satb1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SATB1 (Special AT-rich sequence-binding protein 1). | |||||
|
SHAN1_HUMAN
|
||||||
| NC score | 0.023280 (rank : 107) | θ value | 1.38821 (rank : 46) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
CABL2_HUMAN
|
||||||
| NC score | 0.023251 (rank : 108) | θ value | 5.27518 (rank : 102) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BTV7, Q5JWL0, Q9BYK0 | Gene names | CABLES2, C20orf150 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 and ABL1 enzyme substrate 2 (Interactor with CDK3 2) (Ik3-2). | |||||
|
GON4L_MOUSE
|
||||||
| NC score | 0.023046 (rank : 109) | θ value | 8.99809 (rank : 134) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9DB00, Q80TB4, Q91YI9 | Gene names | Gon4l, Gon4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
PABP2_MOUSE
|
||||||
| NC score | 0.023042 (rank : 110) | θ value | 3.0926 (rank : 77) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CCS6, O35935 | Gene names | Pabpn1, Pab2, Pabp2 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 2 (Poly(A)-binding protein 2) (Poly(A)- binding protein II) (PABII) (Polyadenylate-binding nuclear protein 1) (Nuclear poly(A)-binding protein 1). | |||||
|
SRCA_HUMAN
|
||||||
| NC score | 0.023034 (rank : 111) | θ value | 4.03905 (rank : 96) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.021906 (rank : 112) | θ value | 1.06291 (rank : 38) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
ARHG1_MOUSE
|
||||||
| NC score | 0.021510 (rank : 113) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q61210, O89074, Q80YE8, Q80YE9, Q91VL3 | Gene names | Arhgef1, Lbcl2, Lsc | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 1 (Lymphoid blast crisis-like 2) (Lbc's second cousin). | |||||
|
WASP_HUMAN
|
||||||
| NC score | 0.021345 (rank : 114) | θ value | 8.99809 (rank : 146) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P42768, Q9BU11, Q9UNJ9 | Gene names | WAS, IMD2 | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein (WASp). | |||||
|
ANR25_HUMAN
|
||||||
| NC score | 0.021316 (rank : 115) | θ value | 5.27518 (rank : 99) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q63ZY3, Q3KQZ3, Q6GUF5, Q9H8S4, Q9NUP0, Q9P210 | Gene names | ANKRD25, KIAA1518, SIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 25 (SRC-1-interacting protein) (SRC1-interacting protein). | |||||
|
ADDA_MOUSE
|
||||||
| NC score | 0.021123 (rank : 116) | θ value | 4.03905 (rank : 81) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9QYC0, Q9JLE3 | Gene names | Add1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-adducin (Erythrocyte adducin subunit alpha). | |||||
|
HCN2_HUMAN
|
||||||
| NC score | 0.021005 (rank : 117) | θ value | 1.81305 (rank : 51) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
ATBF1_MOUSE
|
||||||
| NC score | 0.020638 (rank : 118) | θ value | 1.38821 (rank : 39) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
ANKS1_HUMAN
|
||||||
| NC score | 0.020510 (rank : 119) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92625, Q5JYI9, Q5SYR2, Q86WQ7 | Gene names | ANKS1A, ANKS1, KIAA0229 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 1A (Odin). | |||||
|
MRIP_HUMAN
|
||||||
| NC score | 0.019915 (rank : 120) | θ value | 5.27518 (rank : 111) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6WCQ1, Q3KQZ5, Q5FB94, Q6DHW2, Q7Z5Y2, Q8N390, Q96G40 | Gene names | MRIP, KIAA0864, RHOIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (M-RIP) (RIP3) (p116Rip). | |||||
|
FOG1_HUMAN
|
||||||
| NC score | 0.019538 (rank : 121) | θ value | 1.06291 (rank : 34) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 981 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8IX07 | Gene names | ZFPM1, FOG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
ZYX_HUMAN
|
||||||
| NC score | 0.019446 (rank : 122) | θ value | 3.0926 (rank : 80) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q15942 | Gene names | ZYX | |||
|
Domain Architecture |

|
|||||
| Description | Zyxin (Zyxin-2). | |||||
|
COBA2_MOUSE
|
||||||
| NC score | 0.019096 (rank : 123) | θ value | 5.27518 (rank : 103) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 501 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q64739, Q61432, Q9Z1W0 | Gene names | Col11a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(XI) chain precursor. | |||||
|
UBQL3_HUMAN
|
||||||
| NC score | 0.018599 (rank : 124) | θ value | 8.99809 (rank : 145) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H347, Q9NRE0 | Gene names | UBQLN3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-3. | |||||
|
RP3A_HUMAN
|
||||||
| NC score | 0.018435 (rank : 125) | θ value | 6.88961 (rank : 125) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y2J0, Q96AE0 | Gene names | RPH3A, KIAA0985 | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
MA1C1_HUMAN
|
||||||
| NC score | 0.018238 (rank : 126) | θ value | 3.0926 (rank : 73) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NR34, Q9Y545 | Gene names | MAN1C1, MAN1A3, MAN1C | |||
|
Domain Architecture |

|
|||||
| Description | Mannosyl-oligosaccharide 1,2-alpha-mannosidase IC (EC 3.2.1.113) (Processing alpha-1,2-mannosidase IC) (Alpha-1,2-mannosidase IC) (Mannosidase alpha class 1C member 1) (HMIC). | |||||
|
ANKS1_MOUSE
|
||||||
| NC score | 0.017313 (rank : 127) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P59672, Q6ZQG0 | Gene names | Anks1a, Anks1, Kiaa0229 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 1A. | |||||
|
ANX11_HUMAN
|
||||||
| NC score | 0.017296 (rank : 128) | θ value | 4.03905 (rank : 82) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P50995 | Gene names | ANXA11, ANX11 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A11 (Annexin XI) (Calcyclin-associated annexin 50) (CAP-50) (56 kDa autoantigen). | |||||
|
EMIL2_MOUSE
|
||||||
| NC score | 0.017214 (rank : 129) | θ value | 5.27518 (rank : 105) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8K482 | Gene names | Emilin2 | |||
|
Domain Architecture |

|
|||||
| Description | EMILIN-2 precursor (Elastin microfibril interface-located protein 2) (Elastin microfibril interfacer 2) (Basilin). | |||||
|
ANX11_MOUSE
|
||||||
| NC score | 0.017157 (rank : 130) | θ value | 5.27518 (rank : 100) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P97384 | Gene names | Anxa11, Anx11 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A11 (Annexin XI) (Calcyclin-associated annexin 50) (CAP-50). | |||||
|
GLI3_MOUSE
|
||||||
| NC score | 0.017140 (rank : 131) | θ value | 0.125558 (rank : 9) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61602 | Gene names | Gli3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI3. | |||||
|
CO2A1_MOUSE
|
||||||
| NC score | 0.016671 (rank : 132) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 483 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P28481 | Gene names | Col2a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(II) chain precursor [Contains: Chondrocalcin]. | |||||
|
GLI3_HUMAN
|
||||||
| NC score | 0.016638 (rank : 133) | θ value | 1.06291 (rank : 35) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P10071, O75219, Q9UDT5, Q9UJ39 | Gene names | GLI3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI3. | |||||
|
FBLI1_MOUSE
|
||||||
| NC score | 0.015447 (rank : 134) | θ value | 4.03905 (rank : 89) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q71FD7, Q99J35 | Gene names | Fblim1, Cal | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-binding LIM protein 1 (CSX-associated LIM). | |||||
|
GLU2B_HUMAN
|
||||||
| NC score | 0.015057 (rank : 135) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P14314, Q96BU9, Q96D06, Q9P0W9 | Gene names | PRKCSH, G19P1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glucosidase 2 subunit beta precursor (Glucosidase II subunit beta) (Protein kinase C substrate, 60.1 kDa protein, heavy chain) (PKCSH) (80K-H protein). | |||||
|
MMP15_MOUSE
|
||||||
| NC score | 0.013929 (rank : 136) | θ value | 1.38821 (rank : 43) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O54732 | Gene names | Mmp15 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-15 precursor (EC 3.4.24.-) (MMP-15) (Membrane-type matrix metalloproteinase 2) (MT-MMP 2) (MTMMP2) (Membrane-type-2 matrix metalloproteinase) (MT2-MMP) (MT2MMP). | |||||
|
ESX1L_HUMAN
|
||||||
| NC score | 0.013067 (rank : 137) | θ value | 4.03905 (rank : 88) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8N693, Q7Z6K7 | Gene names | ESX1L, ESX1R | |||
|
Domain Architecture |

|
|||||
| Description | Extraembryonic, spermatogenesis, homeobox 1-like protein. | |||||
|
WNK2_HUMAN
|
||||||
| NC score | 0.012571 (rank : 138) | θ value | 2.36792 (rank : 62) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
QSCN6_HUMAN
|
||||||
| NC score | 0.012074 (rank : 139) | θ value | 8.99809 (rank : 138) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O00391, Q59G29, Q5T2X0, Q8TDL6, Q8WVP4 | Gene names | QSCN6, QSOX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sulfhydryl oxidase 1 precursor (EC 1.8.3.2) (Quiescin Q6) (hQSOX). | |||||
|
SREC2_HUMAN
|
||||||
| NC score | 0.012065 (rank : 140) | θ value | 4.03905 (rank : 97) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96GP6, Q8IXF3, Q9BW74 | Gene names | SCARF2, SREC2, SREPCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II) (SRECRP-1). | |||||
|
NOTC1_HUMAN
|
||||||
| NC score | 0.011642 (rank : 141) | θ value | 2.36792 (rank : 57) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P46531 | Gene names | NOTCH1, TAN1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (hN1) (Translocation-associated notch protein TAN-1) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
RHG18_HUMAN
|
||||||
| NC score | 0.011591 (rank : 142) | θ value | 8.99809 (rank : 140) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N392, Q58EZ3, Q6P679, Q6PJD7, Q96S64 | Gene names | ARHGAP18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho-GTPase-activating protein 18 (MacGAP). | |||||
|
KRHB6_HUMAN
|
||||||
| NC score | 0.010567 (rank : 143) | θ value | 5.27518 (rank : 109) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O43790, P78387 | Gene names | KRTHB6 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cuticular Hb6 (Hair keratin, type II Hb6) (ghHb6). | |||||
|
KRHB1_HUMAN
|
||||||
| NC score | 0.010422 (rank : 144) | θ value | 5.27518 (rank : 108) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14533, Q14846, Q16274, Q8WU52, Q9BR74 | Gene names | KRTHB1, MLN137 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cuticular Hb1 (Hair keratin, type II Hb1) (ghHKb1) (ghHb1) (MLN 137). | |||||
|
GABP2_HUMAN
|
||||||
| NC score | 0.009633 (rank : 145) | θ value | 6.88961 (rank : 123) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q06547, Q06545, Q12940, Q12941, Q12942, Q8IYD0 | Gene names | GABPB2, E4TF1B, GABPB, GABPB1 | |||
|
Domain Architecture |

|
|||||
| Description | GA-binding protein beta chain (GABP subunit beta-2) (GABP-2) (GABP subunit beta-1) (GABPB-1) (Transcription factor E4TF1-53) (Transcription factor E4TF1-47) (Nuclear respiratory factor 2). | |||||
|
KRHB2_HUMAN
|
||||||
| NC score | 0.009341 (rank : 146) | θ value | 8.99809 (rank : 136) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NSB4 | Gene names | KRTHB2 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cuticular Hb2 (Hair keratin, type II Hb2). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.007800 (rank : 147) | θ value | 2.36792 (rank : 61) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
M3K14_HUMAN
|
||||||
| NC score | 0.007459 (rank : 148) | θ value | 2.36792 (rank : 55) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 887 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99558, Q8IYN1 | Gene names | MAP3K14, NIK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 14 (EC 2.7.11.25) (NF- kappa beta-inducing kinase) (Serine/threonine-protein kinase NIK) (HsNIK). | |||||
|
WNK4_HUMAN
|
||||||
| NC score | 0.007263 (rank : 149) | θ value | 6.88961 (rank : 129) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1043 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96J92, Q8N8X3, Q8N8Z2, Q96DT8, Q9BYS5 | Gene names | WNK4, PRKWNK4 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK4 (EC 2.7.11.1) (Protein kinase with no lysine 4) (Protein kinase, lysine-deficient 4). | |||||
|
ZDHC9_HUMAN
|
||||||
| NC score | 0.006760 (rank : 150) | θ value | 8.99809 (rank : 147) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y397, Q59EK4, Q5JSW5, Q8WWS7, Q9BPY4, Q9NSP0, Q9NVL0, Q9NVR6 | Gene names | ZDHHC9, ZNF379 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC9 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 9) (DHHC-9) (DHHC9) (Zinc finger protein 379). | |||||
|
PGCB_MOUSE
|
||||||
| NC score | 0.006379 (rank : 151) | θ value | 8.99809 (rank : 137) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61361 | Gene names | Bcan | |||
|
Domain Architecture |

|
|||||
| Description | Brevican core protein precursor. | |||||
|
FZD8_HUMAN
|
||||||
| NC score | 0.006155 (rank : 152) | θ value | 6.88961 (rank : 122) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H461 | Gene names | FZD8 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-8 precursor (Fz-8) (hFz8). | |||||
|
MK07_HUMAN
|
||||||
| NC score | 0.005516 (rank : 153) | θ value | 4.03905 (rank : 90) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1087 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13164, Q16634 | Gene names | MAPK7, ERK4, ERK5, PRKM7 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase 7 (EC 2.7.11.24) (Extracellular signal-regulated kinase 5) (ERK-5) (ERK4) (BMK1 kinase). | |||||
|
KIF3A_HUMAN
|
||||||
| NC score | 0.005472 (rank : 154) | θ value | 8.99809 (rank : 135) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y496, Q86XE9, Q9Y6V4 | Gene names | KIF3A, KIF3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3A (Microtubule plus end-directed kinesin motor 3A). | |||||
|
MAZ_HUMAN
|
||||||
| NC score | 0.005361 (rank : 155) | θ value | 3.0926 (rank : 74) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 757 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P56270, Q15703, Q99443 | Gene names | MAZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myc-associated zinc finger protein (MAZI) (Purine-binding transcription factor) (Pur-1) (ZF87) (ZIF87). | |||||
|
HIC1_MOUSE
|
||||||
| NC score | 0.005338 (rank : 156) | θ value | 3.0926 (rank : 70) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 863 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9R1Y5, Q9R1Y6, Q9R2B0 | Gene names | Hic1 | |||
|
Domain Architecture |

|
|||||
| Description | Hypermethylated in cancer 1 protein (Hic-1). | |||||
|
CTCF_MOUSE
|
||||||
| NC score | 0.003752 (rank : 157) | θ value | 5.27518 (rank : 104) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 796 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61164 | Gene names | Ctcf | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional repressor CTCF (CCCTC-binding factor) (CTCFL paralog) (11-zinc finger protein). | |||||
|
TAOK2_HUMAN
|
||||||
| NC score | 0.002757 (rank : 158) | θ value | 8.99809 (rank : 143) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1172 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UL54, O94957, Q6UW73, Q7LC09, Q9NSW2 | Gene names | TAOK2, KIAA0881, MAP3K17, PSK, PSK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO2 (EC 2.7.11.1) (Thousand and one amino acid protein 2) (Prostate-derived STE20-like kinase 1) (PSK-1) (Kinase from chicken homolog C) (hKFC-C). | |||||
|
ZN202_HUMAN
|
||||||
| NC score | 0.002129 (rank : 159) | θ value | 5.27518 (rank : 118) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 756 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O95125, Q4JG21, Q9H1B9, Q9NSM4 | Gene names | ZNF202 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 202. | |||||
|
M3K12_HUMAN
|
||||||
| NC score | 0.001908 (rank : 160) | θ value | 6.88961 (rank : 124) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q12852, Q86VQ5, Q8WY25 | Gene names | MAP3K12, ZPK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 12 (EC 2.7.11.25) (Mixed lineage kinase) (Leucine-zipper protein kinase) (ZPK) (Dual leucine zipper bearing kinase) (DLK) (MAPK-upstream kinase) (MUK). | |||||
|
TRIB1_HUMAN
|
||||||
| NC score | 0.001755 (rank : 161) | θ value | 8.99809 (rank : 144) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 598 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96RU8, O15180, Q9H2Y8 | Gene names | TRIB1, C8FW, GIG2, TRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tribbles homolog 1 (TRB-1) (SKIP1) (G-protein-coupled receptor-induced protein 2) (GIG-2). | |||||