Please be patient as the page loads
|
RHG05_HUMAN
|
||||||
| SwissProt Accessions | Q13017 | Gene names | ARHGAP5, RHOGAP5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
GRLF1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.964174 (rank : 3) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9NRY4, Q14452, Q9C0E1 | Gene names | GRLF1, GRF1, KIAA1722 | |||
|
Domain Architecture |

|
|||||
| Description | Glucocorticoid receptor DNA-binding factor 1 (Glucocorticoid receptor repression factor 1) (GRF-1) (Rho GAP p190A) (p190-A). | |||||
|
RHG05_HUMAN
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 158 | |
| SwissProt Accessions | Q13017 | Gene names | ARHGAP5, RHOGAP5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
RHG05_MOUSE
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.995040 (rank : 2) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 139 | |
| SwissProt Accessions | P97393 | Gene names | Arhgap5, Rhogap5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
CHIN_HUMAN
|
||||||
| θ value | 2.05525e-28 (rank : 4) | NC score | 0.712541 (rank : 17) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P15882, Q53SD6, Q53SH5, Q96FB0 | Gene names | CHN1, ARHGAP2, CHN | |||
|
Domain Architecture |

|
|||||
| Description | N-chimaerin (NC) (N-chimerin) (Alpha chimerin) (A-chimaerin) (Rho- GTPase-activating protein 2). | |||||
|
CHIN_MOUSE
|
||||||
| θ value | 3.50572e-28 (rank : 5) | NC score | 0.714138 (rank : 16) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q91V57, Q3UGY3, Q7TQE5, Q8BWU6, Q9D9B3 | Gene names | Chn1, Arhgap2 | |||
|
Domain Architecture |

|
|||||
| Description | N-chimaerin (NC) (N-chimerin) (Alpha chimerin) (A-chimaerin) (Rho- GTPase-activating protein 2). | |||||
|
CHIO_MOUSE
|
||||||
| θ value | 3.28125e-26 (rank : 6) | NC score | 0.742127 (rank : 7) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q80XD1, Q9D9W2, Q9ER57 | Gene names | Chn2, Arhgap3, Bch | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-chimaerin (Beta-chimerin) (Rho-GTPase-activating protein 3). | |||||
|
CHIO_HUMAN
|
||||||
| θ value | 2.12685e-25 (rank : 7) | NC score | 0.710795 (rank : 18) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P52757, Q75MM2 | Gene names | CHN2, ARHGAP3, BCH | |||
|
Domain Architecture |
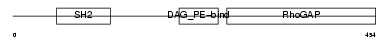
|
|||||
| Description | Beta-chimaerin (Beta-chimerin) (Rho-GTPase-activating protein 3). | |||||
|
ABR_HUMAN
|
||||||
| θ value | 2.06002e-20 (rank : 8) | NC score | 0.684868 (rank : 23) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q12979, Q13693, Q13694 | Gene names | ABR | |||
|
Domain Architecture |

|
|||||
| Description | Active breakpoint cluster region-related protein. | |||||
|
BCR_HUMAN
|
||||||
| θ value | 2.06002e-20 (rank : 9) | NC score | 0.672102 (rank : 30) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P11274, P78501, Q12842 | Gene names | BCR, BCR1 | |||
|
Domain Architecture |

|
|||||
| Description | Breakpoint cluster region protein (EC 2.7.11.1) (NY-REN-26 antigen). | |||||
|
OPHN1_MOUSE
|
||||||
| θ value | 1.74391e-19 (rank : 10) | NC score | 0.725524 (rank : 13) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q99J31, Q544K7 | Gene names | Ophn1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
RHG12_HUMAN
|
||||||
| θ value | 2.97466e-19 (rank : 11) | NC score | 0.756801 (rank : 6) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8IWW6, Q86UB3, Q8IWW7, Q8N3L1, Q9NT76 | Gene names | ARHGAP12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
RHG12_MOUSE
|
||||||
| θ value | 2.97466e-19 (rank : 12) | NC score | 0.757905 (rank : 5) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q8C0D4, Q8BVP8 | Gene names | Arhgap12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
OPHN1_HUMAN
|
||||||
| θ value | 3.88503e-19 (rank : 13) | NC score | 0.727278 (rank : 12) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | O60890, Q5JQ81, Q8WX47 | Gene names | OPHN1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
SRGP2_HUMAN
|
||||||
| θ value | 2.5182e-18 (rank : 14) | NC score | 0.664917 (rank : 32) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | O43295, Q8IX13, Q8IZV8 | Gene names | SRGAP3, ARHGAP14, KIAA0411, MEGAP, SRGAP2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Mental disorder- associated GAP) (Rho-GTPase-activating protein 14). | |||||
|
RHG04_HUMAN
|
||||||
| θ value | 9.56915e-18 (rank : 15) | NC score | 0.673044 (rank : 29) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P98171, Q14144 | Gene names | ARHGAP4, KIAA0131, RGC1, RHOGAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 4 (Rho-GAP hematopoietic protein C1) (p115). | |||||
|
SRGP2_MOUSE
|
||||||
| θ value | 9.56915e-18 (rank : 16) | NC score | 0.658223 (rank : 34) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q812A2, Q80U09, Q8BKP4, Q8BLD0, Q925I2 | Gene names | Srgap3, Arhgap14, Kiaa0411, Srgap2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Rho-GTPase-activating protein 14). | |||||
|
3BP1_HUMAN
|
||||||
| θ value | 6.20254e-17 (rank : 17) | NC score | 0.719086 (rank : 15) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9Y3L3, Q6IBZ2, Q6ZVL9, Q96HQ5, Q9NSQ9 | Gene names | SH3BP1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
CEND2_HUMAN
|
||||||
| θ value | 1.058e-16 (rank : 18) | NC score | 0.611763 (rank : 42) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q96P48, O94879, Q59FI7, Q6PHS3, Q8WU51, Q96HP6, Q96L71 | Gene names | CENTD2, ARAP1, KIAA0782 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-delta 2 (Cnt-d2) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 1). | |||||
|
RHG09_HUMAN
|
||||||
| θ value | 1.058e-16 (rank : 19) | NC score | 0.774564 (rank : 4) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9BRR9, Q8TCJ3, Q8WYR0, Q96EZ2, Q96S74 | Gene names | ARHGAP9 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 9. | |||||
|
MYO9B_HUMAN
|
||||||
| θ value | 1.38178e-16 (rank : 20) | NC score | 0.391274 (rank : 58) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
MYO9B_MOUSE
|
||||||
| θ value | 1.80466e-16 (rank : 21) | NC score | 0.396938 (rank : 57) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9QY06, Q9QY07, Q9QY08, Q9QY09 | Gene names | Myo9b, Myr5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
SRGP1_HUMAN
|
||||||
| θ value | 3.07829e-16 (rank : 22) | NC score | 0.653801 (rank : 36) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q7Z6B7, Q9H8A3, Q9P2P2 | Gene names | SRGAP1, ARHGAP13, KIAA1304 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 1 (srGAP1) (Rho-GTPase- activating protein 13). | |||||
|
RHG26_HUMAN
|
||||||
| θ value | 4.02038e-16 (rank : 23) | NC score | 0.676217 (rank : 26) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9UNA1, O75117, Q9BYS6, Q9BYS7, Q9UJ00 | Gene names | ARHGAP26, GRAF, KIAA0621, OPHN1L | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 26 (Oligophrenin-1-like protein) (GTPase regulator associated with focal adhesion kinase). | |||||
|
RGAP1_HUMAN
|
||||||
| θ value | 5.25075e-16 (rank : 24) | NC score | 0.741197 (rank : 8) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9H0H5, Q6PJ26, Q9NWN2, Q9P250, Q9P2W2 | Gene names | RACGAP1, KIAA1478, MGCRACGAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rac GTPase-activating protein 1 (MgcRacGAP). | |||||
|
CEND3_HUMAN
|
||||||
| θ value | 3.40345e-15 (rank : 25) | NC score | 0.565391 (rank : 48) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8WWN8 | Gene names | CENTD3, ARAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3). | |||||
|
CEND3_MOUSE
|
||||||
| θ value | 3.40345e-15 (rank : 26) | NC score | 0.576708 (rank : 47) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8R5G7, Q5DTN4, Q6NVF1, Q8R5G6 | Gene names | Centd3, Arap3, Drag1, Kiaa4097 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3) (Dual specificity Rho- and Arf-GTPase-activating protein 1). | |||||
|
FNBP2_HUMAN
|
||||||
| θ value | 4.44505e-15 (rank : 27) | NC score | 0.648444 (rank : 38) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | O75044 | Gene names | SRGAP2, FNBP2, KIAA0456 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 2 (srGAP2) (Formin-binding protein 2). | |||||
|
RGAP1_MOUSE
|
||||||
| θ value | 1.68911e-14 (rank : 28) | NC score | 0.736999 (rank : 9) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9WVM1, Q3THR5, Q3TI41, Q3TM81 | Gene names | Racgap1, Mgcracgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rac GTPase-activating protein 1 (MgcRacGAP). | |||||
|
CE005_MOUSE
|
||||||
| θ value | 2.20605e-14 (rank : 29) | NC score | 0.603382 (rank : 45) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q8K2H3 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Protein C5orf5 homolog. | |||||
|
RBP1_HUMAN
|
||||||
| θ value | 3.76295e-14 (rank : 30) | NC score | 0.656637 (rank : 35) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q15311 | Gene names | RALBP1, RLIP1, RLIP76 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (76 kDa Ral-interacting protein) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | 4.91457e-14 (rank : 31) | NC score | 0.612141 (rank : 41) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
TCGAP_MOUSE
|
||||||
| θ value | 4.91457e-14 (rank : 32) | NC score | 0.689424 (rank : 22) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q80YF9, Q6P7W6 | Gene names | Snx26, Tcgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
CE005_HUMAN
|
||||||
| θ value | 8.38298e-14 (rank : 33) | NC score | 0.604787 (rank : 44) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9NYF5, Q6PGQ2, Q9P0I7 | Gene names | C5orf5 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C5orf5 (GAP-like protein N61). | |||||
|
RHG25_MOUSE
|
||||||
| θ value | 1.42992e-13 (rank : 34) | NC score | 0.659236 (rank : 33) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q8BYW1, Q2VPR2, Q3TE04, Q3TVX0, Q3UVB7, Q6A0E0, Q8BX98 | Gene names | Arhgap25, Kiaa0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
CEND1_MOUSE
|
||||||
| θ value | 1.86753e-13 (rank : 35) | NC score | 0.527823 (rank : 53) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8BZ05, Q80TX2, Q8C3T2, Q8VEL6 | Gene names | Centd1, Kiaa0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1). | |||||
|
GMIP_HUMAN
|
||||||
| θ value | 1.86753e-13 (rank : 36) | NC score | 0.725443 (rank : 14) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9P107 | Gene names | GMIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GEM-interacting protein (GMIP). | |||||
|
3BP1_MOUSE
|
||||||
| θ value | 2.43908e-13 (rank : 37) | NC score | 0.727976 (rank : 11) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P55194, Q99KK8 | Gene names | Sh3bp1, 3bp1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
GMIP_MOUSE
|
||||||
| θ value | 2.43908e-13 (rank : 38) | NC score | 0.731869 (rank : 10) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q6PGG2, Q6P9S3 | Gene names | Gmip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GEM-interacting protein (GMIP). | |||||
|
RHG01_HUMAN
|
||||||
| θ value | 7.09661e-13 (rank : 39) | NC score | 0.703387 (rank : 19) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q07960 | Gene names | ARHGAP1, CDC42GAP, RHOGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 1 (GTPase-activating protein rhoOGAP) (Rho-related small GTPase protein activator) (CDC42 GTPase-activating protein) (p50-RhoGAP). | |||||
|
CEND1_HUMAN
|
||||||
| θ value | 9.26847e-13 (rank : 40) | NC score | 0.506706 (rank : 54) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8WZ64, Q4W5D2, Q7Z2L5, Q96L70, Q96P49, Q9Y4E4 | Gene names | CENTD1, ARAP2, KIAA0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 2) (PARX protein). | |||||
|
RHG25_HUMAN
|
||||||
| θ value | 2.0648e-12 (rank : 41) | NC score | 0.637753 (rank : 40) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 550 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | P42331, Q8IXQ2 | Gene names | ARHGAP25, KIAA0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
RBP1_MOUSE
|
||||||
| θ value | 3.52202e-12 (rank : 42) | NC score | 0.665459 (rank : 31) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q62172, Q9CRE5 | Gene names | Ralbp1, Rip1 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
RHG08_HUMAN
|
||||||
| θ value | 1.33837e-11 (rank : 43) | NC score | 0.680465 (rank : 25) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9NSG0, O75983, O95695, Q96RW1, Q96RW2, Q9HA49, Q9HC46, Q9NVX8, Q9NXL1, Q9UH20 | Gene names | ARHGAP8 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 8. | |||||
|
RHG08_MOUSE
|
||||||
| θ value | 1.47974e-10 (rank : 44) | NC score | 0.680950 (rank : 24) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9CXP4, Q99JY7 | Gene names | Arhgap8 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 8. | |||||
|
TAGAP_HUMAN
|
||||||
| θ value | 3.29651e-10 (rank : 45) | NC score | 0.651984 (rank : 37) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8N103, Q2NKM8, Q8NI40, Q96KZ2, Q96QA2 | Gene names | TAGAP, TAGAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell activation Rho GTPase-activating protein (T-cell activation GTPase-activating protein). | |||||
|
TCRG1_HUMAN
|
||||||
| θ value | 5.62301e-10 (rank : 46) | NC score | 0.164713 (rank : 63) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
TCRG1_MOUSE
|
||||||
| θ value | 5.62301e-10 (rank : 47) | NC score | 0.164106 (rank : 64) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
RHG07_HUMAN
|
||||||
| θ value | 9.59137e-10 (rank : 48) | NC score | 0.598652 (rank : 46) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q96QB1, O14868, O43199, Q9C0E0 | Gene names | DLC1, ARHGAP7, KIAA1723, STARD12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 7 (Rho-type GTPase-activating protein 7) (Deleted in liver cancer 1 protein) (Dlc-1) (HP protein) (StAR-related lipid transfer protein 12) (StARD12) (START domain-containing protein 12). | |||||
|
RHG07_MOUSE
|
||||||
| θ value | 9.59137e-10 (rank : 49) | NC score | 0.605662 (rank : 43) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9R0Z9, Q8R541 | Gene names | Dlc1, Arhgap7, Stard12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 7 (Rho-type GTPase-activating protein 7) (Deleted in liver cancer 1 protein homolog) (Dlc-1) (StAR-related lipid transfer protein 12) (StARD12) (START domain-containing protein 12). | |||||
|
RHG06_HUMAN
|
||||||
| θ value | 2.13673e-09 (rank : 50) | NC score | 0.697146 (rank : 20) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O43182, O43437, Q9P1B3, Q9UK81, Q9UK82 | Gene names | ARHGAP6, RHOGAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
RHG18_MOUSE
|
||||||
| θ value | 6.21693e-09 (rank : 51) | NC score | 0.673764 (rank : 28) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8K0Q5, Q8BP03, Q8R196 | Gene names | Arhgap18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho-GTPase-activating protein 18. | |||||
|
RHG06_MOUSE
|
||||||
| θ value | 8.11959e-09 (rank : 52) | NC score | 0.692908 (rank : 21) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O54834, Q9QZL8 | Gene names | Arhgap6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
STA13_HUMAN
|
||||||
| θ value | 1.06045e-08 (rank : 53) | NC score | 0.563566 (rank : 49) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9Y3M8, Q5HYH1, Q5TAE3, Q6UN61, Q86TP6, Q86WQ3, Q86XT1 | Gene names | STARD13, DLC2, GT650 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 13 (StARD13) (START domain- containing protein 13) (46H23.2) (Deleted in liver cancer protein 2) (Rho GTPase-activating protein). | |||||
|
P85A_MOUSE
|
||||||
| θ value | 4.0297e-08 (rank : 54) | NC score | 0.376212 (rank : 60) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P26450 | Gene names | Pik3r1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit alpha (PI3-kinase p85-subunit alpha) (PtdIns-3-kinase p85-alpha) (PI3K). | |||||
|
RHG18_HUMAN
|
||||||
| θ value | 4.0297e-08 (rank : 55) | NC score | 0.674161 (rank : 27) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8N392, Q58EZ3, Q6P679, Q6PJD7, Q96S64 | Gene names | ARHGAP18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho-GTPase-activating protein 18 (MacGAP). | |||||
|
TAGAP_MOUSE
|
||||||
| θ value | 5.26297e-08 (rank : 56) | NC score | 0.639735 (rank : 39) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8K2L9, Q920D6 | Gene names | Tagap, Tagap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell activation Rho GTPase-activating protein (T-cell activation GTPase-activating protein). | |||||
|
P85A_HUMAN
|
||||||
| θ value | 8.97725e-08 (rank : 57) | NC score | 0.379721 (rank : 59) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P27986 | Gene names | PIK3R1, GRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit alpha (PI3-kinase p85-subunit alpha) (PtdIns-3-kinase p85-alpha) (PI3K). | |||||
|
STA13_MOUSE
|
||||||
| θ value | 1.17247e-07 (rank : 58) | NC score | 0.552038 (rank : 52) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q923Q2, Q8K369 | Gene names | Stard13 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 13 (StARD13) (START domain- containing protein 13). | |||||
|
STAR8_HUMAN
|
||||||
| θ value | 1.29631e-06 (rank : 59) | NC score | 0.552230 (rank : 51) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q92502, Q5JST0 | Gene names | STARD8, KIAA0189 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
STAR8_MOUSE
|
||||||
| θ value | 8.40245e-06 (rank : 60) | NC score | 0.556635 (rank : 50) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8K031, Q3UZC7, Q6A0A8, Q6P5E0, Q8R3X8 | Gene names | Stard8, Kiaa0189 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
CTRO_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 61) | NC score | 0.032024 (rank : 133) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
OCRL_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 62) | NC score | 0.304501 (rank : 62) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q01968, O60800, Q15684, Q15774, Q5JQF1, Q5JQF2, Q9UJG5, Q9UMA5 | Gene names | OCRL, INPP5F, OCRL1 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol polyphosphate 5-phosphatase OCRL-1 (EC 3.1.3.36) (Lowe oculocerebrorenal syndrome protein). | |||||
|
I5P2_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 63) | NC score | 0.312426 (rank : 61) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P32019, Q5VSG9, Q5VSH0, Q5VSH1, Q658Q5, Q6P6D4, Q6PD53, Q86YE1 | Gene names | INPP5B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Type II inositol-1,4,5-trisphosphate 5-phosphatase precursor (EC 3.1.3.36) (Phosphoinositide 5-phosphatase) (5PTase) (75 kDa inositol polyphosphate-5-phosphatase). | |||||
|
STK10_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 64) | NC score | 0.014110 (rank : 164) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O94804, Q9UIW4 | Gene names | STK10, LOK | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
PRP40_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 65) | NC score | 0.109320 (rank : 66) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O75400, O43856, O75404, Q8TBQ1, Q9H782, Q9NWU9, Q9P0Q2, Q9Y5A8 | Gene names | PRPF40A, FLAF1, FNBP3, HYPA | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Huntingtin yeast partner A) (Huntingtin-interacting protein HYPA/FBP11) (Fas ligand-associated factor 1) (NY-REN-6 antigen). | |||||
|
PRP40_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 66) | NC score | 0.115148 (rank : 65) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9R1C7, Q61049, Q8BQ76, Q8BRW4, Q8C2U1 | Gene names | Prpf40a, Fbp11, Fnbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Formin-binding protein 11) (FBP-11). | |||||
|
SYNE1_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 67) | NC score | 0.043185 (rank : 121) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
CENPF_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 68) | NC score | 0.047260 (rank : 116) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
MORC4_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 69) | NC score | 0.048074 (rank : 114) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8TE76, Q5JUK7, Q96MZ2, Q9HAI7 | Gene names | MORC4, ZCWCC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 4 (Zinc finger CW-type coiled- coil domain protein 2). | |||||
|
NIN_HUMAN
|
||||||
| θ value | 0.125558 (rank : 70) | NC score | 0.051253 (rank : 106) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
RAD50_MOUSE
|
||||||
| θ value | 0.125558 (rank : 71) | NC score | 0.045534 (rank : 117) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
COG8_HUMAN
|
||||||
| θ value | 0.21417 (rank : 72) | NC score | 0.055133 (rank : 83) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96MW5, Q8WVV6, Q9H6F8 | Gene names | COG8 | |||
|
Domain Architecture |
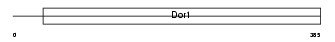
|
|||||
| Description | Conserved oligomeric Golgi complex component 8. | |||||
|
TRIPB_HUMAN
|
||||||
| θ value | 0.21417 (rank : 73) | NC score | 0.042010 (rank : 124) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
KTN1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 74) | NC score | 0.042066 (rank : 123) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q86UP2, Q13999, Q14707, Q15387, Q86W57 | Gene names | KTN1, CG1, KIAA0004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin (Kinesin receptor) (CG-1 antigen). | |||||
|
RAD50_HUMAN
|
||||||
| θ value | 0.365318 (rank : 75) | NC score | 0.044080 (rank : 119) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
MYH1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 76) | NC score | 0.051953 (rank : 99) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
PAWR_HUMAN
|
||||||
| θ value | 0.47712 (rank : 77) | NC score | 0.059676 (rank : 78) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96IZ0, O75796, Q6FHY9, Q8N700 | Gene names | PAWR, PAR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRKC apoptosis WT1 regulator protein (Prostate apoptosis response 4 protein) (Par-4). | |||||
|
SCYL2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 78) | NC score | 0.021553 (rank : 151) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 632 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6P3W7, Q96EF4, Q96ST4, Q9H7V5, Q9NVH3, Q9P2I7 | Gene names | SCYL2, CVAK104, KIAA1360 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCY1-like protein 2 (Coated vesicle-associated kinase of 104 kDa). | |||||
|
BMR1B_HUMAN
|
||||||
| θ value | 0.62314 (rank : 79) | NC score | 0.009095 (rank : 177) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O00238, P78366 | Gene names | BMPR1B | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein receptor type IB precursor (EC 2.7.11.30) (CDw293 antigen). | |||||
|
BMR1B_MOUSE
|
||||||
| θ value | 0.62314 (rank : 80) | NC score | 0.008957 (rank : 179) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 758 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P36898, Q3TRF2 | Gene names | Bmpr1b, Acvrlk6 | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein receptor type IB precursor (EC 2.7.11.30) (Serine/threonine-protein kinase receptor R6) (SKR6) (Activin receptor-like kinase 6) (ALK-6). | |||||
|
CTRO_HUMAN
|
||||||
| θ value | 0.62314 (rank : 81) | NC score | 0.022782 (rank : 147) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
IF3A_HUMAN
|
||||||
| θ value | 0.62314 (rank : 82) | NC score | 0.055275 (rank : 82) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q14152, O00653 | Gene names | EIF3S10, KIAA0139 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a). | |||||
|
MYH2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 83) | NC score | 0.049614 (rank : 112) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
RAB4B_HUMAN
|
||||||
| θ value | 0.62314 (rank : 84) | NC score | 0.017183 (rank : 156) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P61018, P22750, Q7Z514, Q9HBR6 | Gene names | RAB4B | |||
|
Domain Architecture |
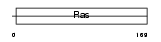
|
|||||
| Description | Ras-related protein Rab-4B. | |||||
|
RAB4B_MOUSE
|
||||||
| θ value | 0.62314 (rank : 85) | NC score | 0.017185 (rank : 155) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q91ZR1 | Gene names | Rab4b, Rab4 | |||
|
Domain Architecture |
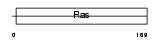
|
|||||
| Description | Ras-related protein Rab-4B. | |||||
|
SCYL2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 86) | NC score | 0.023069 (rank : 145) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CFE4, Q3UT57, Q3UWU9, Q8K0M4 | Gene names | Scyl2, Cvak104, D10Ertd802e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCY1-like protein 2 (Coated vesicle-associated kinase of 104 kDa). | |||||
|
SLK_MOUSE
|
||||||
| θ value | 0.62314 (rank : 87) | NC score | 0.012118 (rank : 171) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1798 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O54988, Q80U65, Q8CAU2, Q8CDW2, Q9WU41 | Gene names | Slk, Kiaa0204, Stk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (mSLK) (Serine/threonine-protein kinase 2) (STE20-related kinase SMAK) (Etk4). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 88) | NC score | 0.043642 (rank : 120) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
K1688_MOUSE
|
||||||
| θ value | 0.813845 (rank : 89) | NC score | 0.431187 (rank : 55) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P59281, Q69ZD4, Q6P9R6 | Gene names | Kiaa1688, D15Wsu169e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1688. | |||||
|
KI13A_HUMAN
|
||||||
| θ value | 0.813845 (rank : 90) | NC score | 0.012348 (rank : 168) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H1H9, Q9H1H8 | Gene names | KIF13A, RBKIN | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF13A (Kinesin-like protein RBKIN). | |||||
|
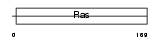
RAB4A_HUMAN
|
||||||
| θ value | 0.813845 (rank : 91) | NC score | 0.015418 (rank : 159) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P20338, Q9BQ44 | Gene names | RAB4A, RAB4 | |||
|
Domain Architecture |
|
|||||
| Description | Ras-related protein Rab-4A. | |||||
|
TACC3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 92) | NC score | 0.038915 (rank : 127) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9JJ11, Q9WVK9 | Gene names | Tacc3, Aint | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ARNT-interacting protein). | |||||
|
LAMA3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 93) | NC score | 0.022871 (rank : 146) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q16787, Q13679, Q13680, Q96TG0 | Gene names | LAMA3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-3 chain precursor (Epiligrin 170 kDa subunit) (E170) (Nicein subunit alpha). | |||||
|
MYH8_HUMAN
|
||||||
| θ value | 1.06291 (rank : 94) | NC score | 0.053344 (rank : 90) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
SLK_HUMAN
|
||||||
| θ value | 1.06291 (rank : 95) | NC score | 0.011186 (rank : 173) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1775 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9H2G2, O00211, Q6P1Z4, Q86WU7, Q86WW1, Q92603, Q9NQL0, Q9NQL1 | Gene names | SLK, KIAA0204, STK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (hSLK) (Serine/threonine-protein kinase 2) (CTCL tumor antigen se20-9). | |||||
|
SPTA1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 96) | NC score | 0.050326 (rank : 110) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P02549, Q15514, Q6LDY5 | Gene names | SPTA1, SPTA | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
IF3A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 97) | NC score | 0.053257 (rank : 91) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P23116, Q60697, Q62162 | Gene names | Eif3s10, Csma, Eif3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a) (p162 protein) (Centrosomin). | |||||
|
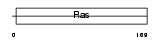
RAB4A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 98) | NC score | 0.014455 (rank : 163) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P56371 | Gene names | Rab4a, Rab4 | |||
|
Domain Architecture |
|
|||||
| Description | Ras-related protein Rab-4A. | |||||
|
AASS_HUMAN
|
||||||
| θ value | 1.81305 (rank : 99) | NC score | 0.026186 (rank : 139) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UDR5, O95462 | Gene names | AASS | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-aminoadipic semialdehyde synthase, mitochondrial precursor (LKR/SDH) [Includes: Lysine ketoglutarate reductase (EC 1.5.1.8) (LOR) (LKR); Saccharopine dehydrogenase (EC 1.5.1.9) (SDH)]. | |||||
|
CCD11_HUMAN
|
||||||
| θ value | 1.81305 (rank : 100) | NC score | 0.042978 (rank : 122) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96M91 | Gene names | CCDC11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 11. | |||||
|
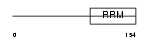
COVA1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 101) | NC score | 0.023562 (rank : 144) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q16206, Q5VTJ1, Q5VTJ2, Q8WUX0, Q9NTP6, Q9UH82 | Gene names | COVA1 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor-associated hydroquinone oxidase (tNOX) (Cytosolic ovarian carcinoma antigen 1) (APK1 antigen) [Includes: Hydroquinone [NADH] oxidase (EC 1.-.-.-); Protein disulfide-thiol oxidoreductase (EC 1.-.-.-)]. | |||||
|
G45IP_HUMAN
|
||||||
| θ value | 1.81305 (rank : 102) | NC score | 0.035465 (rank : 128) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8TAE8, Q8IVM3, Q8TE51, Q969P9, Q9BSM6 | Gene names | GADD45GIP1, PLINP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth arrest and DNA-damage-inducible proteins-interacting protein 1 (CR6-interacting factor 1) (CRIF1) (CKII beta-associating protein) (Papillomavirus L2-interacting nuclear protein 1) (PLINP-1). | |||||
|
MYH4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 103) | NC score | 0.051527 (rank : 102) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
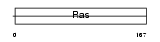
RAB2B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 104) | NC score | 0.012183 (rank : 170) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8WUD1 | Gene names | RAB2B | |||
|
Domain Architecture |
|
|||||
| Description | Ras-related protein Rab-2B. | |||||
|
RAB2B_MOUSE
|
||||||
| θ value | 1.81305 (rank : 105) | NC score | 0.012264 (rank : 169) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P59279, Q3TV67 | Gene names | Rab2b | |||
|
Domain Architecture |
|
|||||
| Description | Ras-related protein Rab-2B. | |||||
|
SPTA1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 106) | NC score | 0.048221 (rank : 113) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P08032, P97502 | Gene names | Spta1, Spna1, Spta | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
CA065_HUMAN
|
||||||
| θ value | 2.36792 (rank : 107) | NC score | 0.041868 (rank : 125) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 469 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8N715, Q8N746, Q8NA93 | Gene names | C1orf65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf65. | |||||
|
DTBP1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 108) | NC score | 0.024877 (rank : 142) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96EV8, Q96NV2, Q9H0U2, Q9H3J5 | Gene names | DTNBP1 | |||
|
Domain Architecture |
|
|||||
| Description | Dystrobrevin-binding protein 1 (Dysbindin) (Hermansky-Pudlak syndrome 7 protein). | |||||
|
MYH8_MOUSE
|
||||||
| θ value | 2.36792 (rank : 109) | NC score | 0.050930 (rank : 108) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
TXLNA_MOUSE
|
||||||
| θ value | 2.36792 (rank : 110) | NC score | 0.033999 (rank : 129) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 846 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6PAM1, Q6P1E5 | Gene names | Txlna, Txln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
ES8L3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 111) | NC score | 0.008897 (rank : 180) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TE67, Q5T8Q6, Q5T8Q7, Q5T8Q8, Q96E47, Q9H719 | Gene names | EPS8L3, EPS8R3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epidermal growth factor receptor kinase substrate 8-like protein 3 (Epidermal growth factor receptor pathway substrate 8-related protein 3) (EPS8-like protein 3). | |||||
|
FAF1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 112) | NC score | 0.032822 (rank : 132) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P54731 | Gene names | Faf1 | |||
|
Domain Architecture |

|
|||||
| Description | FAS-associated factor 1 (Protein FAF1). | |||||
|
K0423_HUMAN
|
||||||
| θ value | 3.0926 (rank : 113) | NC score | 0.027845 (rank : 138) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y4F4, Q68D66, Q6PG27 | Gene names | KIAA0423 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0423. | |||||
|
K0423_MOUSE
|
||||||
| θ value | 3.0926 (rank : 114) | NC score | 0.025085 (rank : 141) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6A070 | Gene names | Kiaa0423 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0423. | |||||
|
MORC4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 115) | NC score | 0.028820 (rank : 136) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BMD7, Q4KMM6, Q8BX95, Q9CS96 | Gene names | Morc4, Zcwcc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 4 (Zinc finger CW-type coiled- coil domain protein 2). | |||||
|
MYH1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 116) | NC score | 0.049937 (rank : 111) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
SIN3A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 117) | NC score | 0.024305 (rank : 143) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q60520, Q60820, Q62139, Q62140, Q7TPU8, Q7TSZ2 | Gene names | Sin3a | |||
|
Domain Architecture |

|
|||||
| Description | Paired amphipathic helix protein Sin3a (Transcriptional corepressor Sin3a) (Histone deacetylase complex subunit Sin3a). | |||||
|
TXLNA_HUMAN
|
||||||
| θ value | 3.0926 (rank : 118) | NC score | 0.032989 (rank : 131) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 982 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P40222, Q66K62, Q86T54, Q86T85, Q86T86, Q86Y86, Q86YW3, Q8N2Y3 | Gene names | TXLNA, TXLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
CI093_HUMAN
|
||||||
| θ value | 4.03905 (rank : 119) | NC score | 0.045310 (rank : 118) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q6TFL3, Q5SU58, Q6P1W1, Q6ZR13, Q8N1Z4, Q8N8L3, Q8TBR2 | Gene names | C9orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf93. | |||||
|
K1688_HUMAN
|
||||||
| θ value | 4.03905 (rank : 120) | NC score | 0.419454 (rank : 56) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9C0H5 | Gene names | KIAA1688 | |||
|
Domain Architecture |

|
|||||
| Description | Protein KIAA1688. | |||||
|
PIGN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 121) | NC score | 0.022524 (rank : 148) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95427, Q7L8F8, Q8TC01, Q9NT05 | Gene names | PIGN, MCD4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GPI ethanolamine phosphate transferase 1 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class N protein) (PIG-N) (MCD4 homolog). | |||||
|
PROX1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 122) | NC score | 0.020418 (rank : 152) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P48437, O88478, Q3UPV1, Q543D8 | Gene names | Prox1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox prospero-like protein PROX1 (PROX 1). | |||||
|
SEN54_MOUSE
|
||||||
| θ value | 4.03905 (rank : 123) | NC score | 0.022512 (rank : 149) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C2A2, Q9DCI5 | Gene names | Tsen54, Sen54 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-splicing endonuclease subunit Sen54 (tRNA-intron endonuclease Sen54). | |||||
|
WDR67_HUMAN
|
||||||
| θ value | 4.03905 (rank : 124) | NC score | 0.025209 (rank : 140) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 782 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96DN5, Q3MIR6, Q8TBP9 | Gene names | WDR67 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 67. | |||||
|
ETV1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 125) | NC score | 0.008875 (rank : 181) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P50549, O75849, Q9UQ71, Q9Y636 | Gene names | ETV1, ER81 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 1 (ER81 protein). | |||||
|
FA48A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 126) | NC score | 0.011504 (rank : 172) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NEM7, Q71RF3, Q9Y6A6 | Gene names | FAM48A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM48A (p38-interacting protein) (p38IP). | |||||
|
FAF1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 127) | NC score | 0.029416 (rank : 135) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UNN5, Q9UF34, Q9UNT3, Q9Y2Z3 | Gene names | FAF1 | |||
|
Domain Architecture |

|
|||||
| Description | FAS-associated factor 1 (Protein FAF1) (hFAF1). | |||||
|
K2C4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 128) | NC score | 0.009620 (rank : 176) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P19013, Q6GTR8, Q96LA7, Q9BTL1 | Gene names | KRT4, CYK4 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 4 (Cytokeratin-4) (CK-4) (Keratin-4) (K4). | |||||
|
MYCN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 129) | NC score | 0.015626 (rank : 158) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P04198, Q6LDT9 | Gene names | MYCN, NMYC | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
MYCN_MOUSE
|
||||||
| θ value | 5.27518 (rank : 130) | NC score | 0.015825 (rank : 157) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P03966, Q61978 | Gene names | Mycn, Nmyc, Nmyc1 | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
PLXB1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 131) | NC score | 0.005742 (rank : 188) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43157, Q6NY20, Q9UIV7, Q9UJ92, Q9UJ93 | Gene names | PLXNB1, KIAA0407, SEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor (Semaphorin receptor SEP). | |||||
|
POLI_HUMAN
|
||||||
| θ value | 5.27518 (rank : 132) | NC score | 0.009075 (rank : 178) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UNA4, Q8N590, Q9H0S1, Q9NYH6 | Gene names | POLI, RAD30B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase iota (EC 2.7.7.7) (RAD30 homolog B) (Eta2). | |||||
|
SIN3A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 133) | NC score | 0.022258 (rank : 150) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96ST3, Q8N8N4, Q8NC83, Q8WV18, Q96L98, Q9UFQ1 | Gene names | SIN3A | |||
|
Domain Architecture |

|
|||||
| Description | Paired amphipathic helix protein Sin3a (Transcriptional corepressor Sin3a) (Histone deacetylase complex subunit Sin3a). | |||||
|
TTLL2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 134) | NC score | 0.012635 (rank : 167) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BWV7, Q7Z6R8, Q86X22 | Gene names | TTLL2, C6orf104 | |||
|
Domain Architecture |

|
|||||
| Description | Tubulin--tyrosine ligase-like protein 2 (Testis-specific protein NYD- TSPG). | |||||
|
CE110_HUMAN
|
||||||
| θ value | 6.88961 (rank : 135) | NC score | 0.013091 (rank : 165) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O43303, O43335, Q68DV9, Q8NE13 | Gene names | CEP110, CP110, KIAA0419 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 110 kDa (Cep110 protein). | |||||
|
ETV1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 136) | NC score | 0.007942 (rank : 187) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P41164 | Gene names | Etv1, Er81, Etsrp81 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 1 (ER81 protein). | |||||
|
EXOC6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 137) | NC score | 0.011046 (rank : 174) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8TAG9, Q9NZ24 | Gene names | EXOC6, SEC15A, SEC15L, SEC15L1 | |||
|
Domain Architecture |

|
|||||
| Description | Exocyst complex component 6 (Exocyst complex component Sec15A) (Sec15- like 1). | |||||
|
JHD3B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 138) | NC score | 0.003023 (rank : 189) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91VY5, Q3UR22, Q6ZQ30, Q99K42 | Gene names | Jmjd2b, Jhdm3b | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3B (EC 1.14.11.-) (Jumonji domain-containing protein 2B). | |||||
|
LMNB2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 139) | NC score | 0.012644 (rank : 166) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P21619, P48680, Q8CGB1 | Gene names | Lmnb2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-B2. | |||||
|
PIGN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 140) | NC score | 0.019556 (rank : 153) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9R1S3, Q3V0S6, Q8VCC3, Q9R1S1, Q9R1S2 | Gene names | Pign | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GPI ethanolamine phosphate transferase 1 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class N protein) (PIG-N). | |||||
|
PTPRZ_HUMAN
|
||||||
| θ value | 6.88961 (rank : 141) | NC score | 0.000700 (rank : 191) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P23471 | Gene names | PTPRZ1, PTPRZ, PTPZ | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase zeta precursor (EC 3.1.3.48) (R-PTP-zeta). | |||||
|
RAB1A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 142) | NC score | 0.008105 (rank : 184) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P62820, P11476, Q96N61, Q9Y3T2 | Gene names | RAB1A, RAB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-related protein Rab-1A (YPT1-related protein). | |||||
|
RAB1A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 143) | NC score | 0.008105 (rank : 185) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P62821, P11476, Q3TX44, Q811M4, Q96N61, Q9Y3T2 | Gene names | Rab1A, Rab1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-related protein Rab-1A (YPT1-related protein). | |||||
|
RAB1B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 144) | NC score | 0.008111 (rank : 183) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H0U4 | Gene names | RAB1B | |||
|
Domain Architecture |
|
|||||
| Description | Ras-related protein Rab-1B. | |||||
|
RAB1B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 145) | NC score | 0.008096 (rank : 186) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9D1G1, Q3U0N1 | Gene names | Rab1b | |||
|
Domain Architecture |
|
|||||
| Description | Ras-related protein Rab-1B. | |||||
|
1433Z_MOUSE
|
||||||
| θ value | 8.99809 (rank : 146) | NC score | 0.002814 (rank : 190) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P63101, P35215, P70197, P97286, Q3TSF1, Q5EBQ1 | Gene names | Ywhaz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 14-3-3 protein zeta/delta (Protein kinase C inhibitor protein 1) (KCIP-1) (SEZ-2). | |||||
|
AN30A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 147) | NC score | 0.017745 (rank : 154) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1119 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9BXX3, Q5W025 | Gene names | ANKRD30A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 30A (NY-BR-1 antigen). | |||||
|
BCAR1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 148) | NC score | 0.030761 (rank : 134) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q61140, Q60869 | Gene names | Bcar1, Cas, Crkas | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
CA048_HUMAN
|
||||||
| θ value | 8.99809 (rank : 149) | NC score | 0.015145 (rank : 161) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96IY1, Q5SY75, Q9H2M5, Q9NRN8, Q9Y415 | Gene names | C1orf48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C1orf48. | |||||
|
CCD46_HUMAN
|
||||||
| θ value | 8.99809 (rank : 150) | NC score | 0.041866 (rank : 126) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
COG8_MOUSE
|
||||||
| θ value | 8.99809 (rank : 151) | NC score | 0.033660 (rank : 130) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JJA2 | Gene names | Cog8 | |||
|
Domain Architecture |

|
|||||
| Description | Conserved oligomeric Golgi complex component 8. | |||||
|
CYLN2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 152) | NC score | 0.028697 (rank : 137) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1004 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9UDT6, O14527, O43611 | Gene names | CYLN2, KIAA0291, WBSCR4, WSCR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115) (Williams-Beuren syndrome chromosome region 4 protein). | |||||
|
GHR_MOUSE
|
||||||
| θ value | 8.99809 (rank : 153) | NC score | 0.010574 (rank : 175) | |||
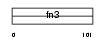
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P16882, P16590, Q61653, Q6DI66, Q80W86, Q8R1M5, Q920Z3, Q9R264 | Gene names | Ghr | |||
|
Domain Architecture |
|
|||||
| Description | Growth hormone receptor precursor (GH receptor) (Somatotropin receptor) [Contains: Growth hormone-binding protein (GH-binding protein) (GHBP) (Serum-binding protein)]. | |||||
|
MYH4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 154) | NC score | 0.047839 (rank : 115) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
NOG1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 155) | NC score | 0.008393 (rank : 182) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BZE4, O95446, Q9NVJ8 | Gene names | GTPBP4, CRFG, NOG1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar GTP-binding protein 1 (Chronic renal failure gene protein) (GTP-binding protein NGB). | |||||
|
NUCB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 156) | NC score | 0.014921 (rank : 162) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q02818, Q15838, Q7Z4J7, Q9BUR1 | Gene names | NUCB1, NUC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleobindin-1 precursor (CALNUC). | |||||
|
SSPO_MOUSE
|
||||||
| θ value | 8.99809 (rank : 157) | NC score | -0.000884 (rank : 192) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 158) | NC score | 0.015324 (rank : 160) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
CENA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.051681 (rank : 101) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75689 | Gene names | CENTA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-alpha 1 (Putative MAPK-activating protein PM25). | |||||
|
CENA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.054121 (rank : 88) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NPF8, Q8N4Q6, Q96SD5 | Gene names | CENTA2 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-alpha 2. | |||||
|
CENB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.070705 (rank : 72) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15027 | Gene names | CENTB1, KIAA0050 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 1 (Cnt-b1). | |||||
|
CENB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.077067 (rank : 68) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q15057, Q9UQR3 | Gene names | CENTB2, KIAA0041 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 2 (Cnt-b2). | |||||
|
CENB5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.071710 (rank : 71) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96P50, Q9BSR9, Q9C0E7 | Gene names | CENTB5, KIAA1716 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 5 (Cnt-b5). | |||||
|
CIP4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.059677 (rank : 77) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q15642, O15184, Q5TZN1 | Gene names | TRIP10, CIP4 | |||
|
Domain Architecture |

|
|||||
| Description | Cdc42-interacting protein 4 (Thyroid receptor-interacting protein 10) (TRIP-10). | |||||
|
DDEF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.050964 (rank : 107) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9QWY8, O08612, Q80TG8, Q80UV6, Q99LV8, Q9Z2B6 | Gene names | Ddef1, Asap1, Kiaa1249, Shag1 | |||
|
Domain Architecture |

|
|||||
| Description | 130 kDa phosphatidylinositol 4,5-biphosphate-dependent ARF1 GTPase- activating protein (PIP2-dependent ARF1 GAP) (ADP-ribosylation factor- directed GTPase-activating protein 1) (ARF GTPase-activating protein 1) (Development and differentiation-enhancing factor 1) (Differentiation-enhancing factor 1) (DEF-1). | |||||
|
DDEF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.055825 (rank : 81) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 599 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O43150 | Gene names | DDEF2, KIAA0400 | |||
|
Domain Architecture |

|
|||||
| Description | Development and differentiation-enhancing factor 2 (Pyk2 C-terminus- associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3). | |||||
|
DDEF2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.054236 (rank : 86) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7SIG6, Q501K1, Q66JN2 | Gene names | Ddef2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor 2 (Pyk2 C-terminus- associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3). | |||||
|
DDFL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.053642 (rank : 89) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8TDY4, Q6P9F4, Q86UY1, Q9NXK2 | Gene names | DDEFL1, UPLC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor-like 1 (Protein up- regulated in liver cancer 1). | |||||
|
FA13A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.069170 (rank : 74) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O94988, Q8NBA3 | Gene names | FAM13A1, KIAA0914 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13A1. | |||||
|
FA13A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.069834 (rank : 73) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BGI4, Q8BZ91 | Gene names | Fam13a1, Precm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13A1 (Precm1 protein). | |||||
|
FA13C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.053137 (rank : 92) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NE31, Q99787 | Gene names | FAM13C1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13C1. | |||||
|
FA13C_MOUSE
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.051751 (rank : 100) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9DBR2 | Gene names | Fam13c1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13C1. | |||||
|
GRAP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.053062 (rank : 94) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13588 | Gene names | GRAP | |||
|
Domain Architecture |
|
|||||
| Description | GRB2-related adapter protein. | |||||
|
GRAP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.050526 (rank : 109) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9CX99, Q3U545 | Gene names | Grap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRB2-related adapter protein. | |||||
|
GRB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.051370 (rank : 103) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P62993, P29354, Q14450, Q63057, Q63059 | Gene names | GRB2, ASH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth factor receptor-bound protein 2 (Adapter protein GRB2) (SH2/SH3 adapter GRB2) (Protein Ash). | |||||
|
GRB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.051362 (rank : 104) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q60631, Q61240 | Gene names | Grb2 | |||
|
Domain Architecture |
|
|||||
| Description | Growth factor receptor-bound protein 2 (Adapter protein GRB2) (SH2/SH3 adapter GRB2). | |||||
|
GRP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.075566 (rank : 70) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8IV61, O94931 | Gene names | RASGRP3, GRP3, KIAA0846 | |||
|
Domain Architecture |

|
|||||
| Description | RAS guanyl-releasing protein 3 (Calcium and DAG-regulated guanine nucleotide exchange factor III) (Guanine nucleotide exchange factor for Rap1). | |||||
|
ITSN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.052573 (rank : 97) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
MYH3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.051318 (rank : 105) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
MYO10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.057826 (rank : 80) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9HD67, Q9NYM7, Q9P110, Q9P111, Q9UHF6 | Gene names | MYO10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin X). | |||||
|
NCK2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.058578 (rank : 79) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O43639, Q9BWN9, Q9UIC3 | Gene names | NCK2 | |||
|
Domain Architecture |
|
|||||
| Description | Cytoplasmic protein NCK2 (NCK adaptor protein 2) (SH2/SH3 adaptor protein NCK-beta) (Nck-2). | |||||
|
P55G_HUMAN
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.060954 (rank : 76) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q92569, O60482 | Gene names | PIK3R3 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit gamma (PI3-kinase p85-subunit gamma) (PtdIns-3-kinase p85-gamma) (p55PIK). | |||||
|
P55G_MOUSE
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.061375 (rank : 75) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q64143 | Gene names | Pik3r3 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit gamma (PI3-kinase p85-subunit gamma) (PtdIns-3-kinase p85-gamma) (p55PIK). | |||||
|
P85B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.102497 (rank : 67) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O00459, Q9UPH9 | Gene names | PIK3R2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit beta (PI3-kinase p85- subunit beta) (PtdIns-3-kinase p85-beta). | |||||
|
P85B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.075566 (rank : 69) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O08908 | Gene names | Pik3r2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit beta (PI3-kinase p85- subunit beta) (PtdIns-3-kinase p85-beta). | |||||
|
PDZD8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.054480 (rank : 84) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8NEN9, Q86WE0, Q86WE5, Q9UFF1 | Gene names | PDZD8, PDZK8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 8 (Sarcoma antigen NY-SAR-84/NY-SAR- 104). | |||||
|
SMAP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.052661 (rank : 95) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IYB5, Q53H70, Q5SYQ2, Q6PK24, Q8NDH4, Q96L38, Q96L39, Q9H8X4 | Gene names | SMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1. | |||||
|
SMAP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.052550 (rank : 98) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91VZ6, Q497I6, Q68EF3 | Gene names | Smap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1. | |||||
|
SMP1L_HUMAN
|
||||||
| θ value | θ > 10 (rank : 189) | NC score | 0.054253 (rank : 85) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WU79, Q5QPL2, Q96C93, Q9NST2, Q9UJL8 | Gene names | SMAP1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1-like. | |||||
|
SMP1L_MOUSE
|
||||||
| θ value | θ > 10 (rank : 190) | NC score | 0.054235 (rank : 87) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TN29, Q3U798 | Gene names | Smap1l, Smap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1-like (Stromal membrane- associated protein 2). | |||||
|
VAV3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 191) | NC score | 0.053087 (rank : 93) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 398 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UKW4, O95230, Q9Y5X8 | Gene names | VAV3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-3. | |||||
|
VAV3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 192) | NC score | 0.052620 (rank : 96) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 404 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9R0C8, Q7TS85 | Gene names | Vav3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-3. | |||||
|
RHG05_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 158 | |
| SwissProt Accessions | Q13017 | Gene names | ARHGAP5, RHOGAP5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
RHG05_MOUSE
|
||||||
| NC score | 0.995040 (rank : 2) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 139 | |
| SwissProt Accessions | P97393 | Gene names | Arhgap5, Rhogap5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
GRLF1_HUMAN
|
||||||
| NC score | 0.964174 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9NRY4, Q14452, Q9C0E1 | Gene names | GRLF1, GRF1, KIAA1722 | |||
|
Domain Architecture |

|
|||||
| Description | Glucocorticoid receptor DNA-binding factor 1 (Glucocorticoid receptor repression factor 1) (GRF-1) (Rho GAP p190A) (p190-A). | |||||
|
RHG09_HUMAN
|
||||||
| NC score | 0.774564 (rank : 4) | θ value | 1.058e-16 (rank : 19) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9BRR9, Q8TCJ3, Q8WYR0, Q96EZ2, Q96S74 | Gene names | ARHGAP9 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 9. | |||||
|
RHG12_MOUSE
|
||||||
| NC score | 0.757905 (rank : 5) | θ value | 2.97466e-19 (rank : 12) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q8C0D4, Q8BVP8 | Gene names | Arhgap12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
RHG12_HUMAN
|
||||||
| NC score | 0.756801 (rank : 6) | θ value | 2.97466e-19 (rank : 11) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8IWW6, Q86UB3, Q8IWW7, Q8N3L1, Q9NT76 | Gene names | ARHGAP12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
CHIO_MOUSE
|
||||||
| NC score | 0.742127 (rank : 7) | θ value | 3.28125e-26 (rank : 6) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q80XD1, Q9D9W2, Q9ER57 | Gene names | Chn2, Arhgap3, Bch | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-chimaerin (Beta-chimerin) (Rho-GTPase-activating protein 3). | |||||
|
RGAP1_HUMAN
|
||||||
| NC score | 0.741197 (rank : 8) | θ value | 5.25075e-16 (rank : 24) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9H0H5, Q6PJ26, Q9NWN2, Q9P250, Q9P2W2 | Gene names | RACGAP1, KIAA1478, MGCRACGAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rac GTPase-activating protein 1 (MgcRacGAP). | |||||
|
RGAP1_MOUSE
|
||||||
| NC score | 0.736999 (rank : 9) | θ value | 1.68911e-14 (rank : 28) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9WVM1, Q3THR5, Q3TI41, Q3TM81 | Gene names | Racgap1, Mgcracgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rac GTPase-activating protein 1 (MgcRacGAP). | |||||
|
GMIP_MOUSE
|
||||||
| NC score | 0.731869 (rank : 10) | θ value | 2.43908e-13 (rank : 38) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q6PGG2, Q6P9S3 | Gene names | Gmip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GEM-interacting protein (GMIP). | |||||
|
3BP1_MOUSE
|
||||||
| NC score | 0.727976 (rank : 11) | θ value | 2.43908e-13 (rank : 37) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P55194, Q99KK8 | Gene names | Sh3bp1, 3bp1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
OPHN1_HUMAN
|
||||||
| NC score | 0.727278 (rank : 12) | θ value | 3.88503e-19 (rank : 13) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | O60890, Q5JQ81, Q8WX47 | Gene names | OPHN1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
OPHN1_MOUSE
|
||||||
| NC score | 0.725524 (rank : 13) | θ value | 1.74391e-19 (rank : 10) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q99J31, Q544K7 | Gene names | Ophn1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
GMIP_HUMAN
|
||||||
| NC score | 0.725443 (rank : 14) | θ value | 1.86753e-13 (rank : 36) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9P107 | Gene names | GMIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GEM-interacting protein (GMIP). | |||||
|
3BP1_HUMAN
|
||||||
| NC score | 0.719086 (rank : 15) | θ value | 6.20254e-17 (rank : 17) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9Y3L3, Q6IBZ2, Q6ZVL9, Q96HQ5, Q9NSQ9 | Gene names | SH3BP1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
CHIN_MOUSE
|
||||||
| NC score | 0.714138 (rank : 16) | θ value | 3.50572e-28 (rank : 5) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q91V57, Q3UGY3, Q7TQE5, Q8BWU6, Q9D9B3 | Gene names | Chn1, Arhgap2 | |||
|
Domain Architecture |

|
|||||
| Description | N-chimaerin (NC) (N-chimerin) (Alpha chimerin) (A-chimaerin) (Rho- GTPase-activating protein 2). | |||||
|
CHIN_HUMAN
|
||||||
| NC score | 0.712541 (rank : 17) | θ value | 2.05525e-28 (rank : 4) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P15882, Q53SD6, Q53SH5, Q96FB0 | Gene names | CHN1, ARHGAP2, CHN | |||
|
Domain Architecture |

|
|||||
| Description | N-chimaerin (NC) (N-chimerin) (Alpha chimerin) (A-chimaerin) (Rho- GTPase-activating protein 2). | |||||
|
CHIO_HUMAN
|
||||||
| NC score | 0.710795 (rank : 18) | θ value | 2.12685e-25 (rank : 7) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P52757, Q75MM2 | Gene names | CHN2, ARHGAP3, BCH | |||
|
Domain Architecture |
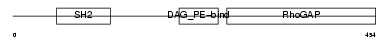
|
|||||
| Description | Beta-chimaerin (Beta-chimerin) (Rho-GTPase-activating protein 3). | |||||
|
RHG01_HUMAN
|
||||||
| NC score | 0.703387 (rank : 19) | θ value | 7.09661e-13 (rank : 39) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q07960 | Gene names | ARHGAP1, CDC42GAP, RHOGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 1 (GTPase-activating protein rhoOGAP) (Rho-related small GTPase protein activator) (CDC42 GTPase-activating protein) (p50-RhoGAP). | |||||
|
RHG06_HUMAN
|
||||||
| NC score | 0.697146 (rank : 20) | θ value | 2.13673e-09 (rank : 50) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O43182, O43437, Q9P1B3, Q9UK81, Q9UK82 | Gene names | ARHGAP6, RHOGAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
RHG06_MOUSE
|
||||||
| NC score | 0.692908 (rank : 21) | θ value | 8.11959e-09 (rank : 52) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O54834, Q9QZL8 | Gene names | Arhgap6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
TCGAP_MOUSE
|
||||||
| NC score | 0.689424 (rank : 22) | θ value | 4.91457e-14 (rank : 32) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q80YF9, Q6P7W6 | Gene names | Snx26, Tcgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
ABR_HUMAN
|
||||||
| NC score | 0.684868 (rank : 23) | θ value | 2.06002e-20 (rank : 8) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q12979, Q13693, Q13694 | Gene names | ABR | |||
|
Domain Architecture |

|
|||||
| Description | Active breakpoint cluster region-related protein. | |||||
|
RHG08_MOUSE
|
||||||
| NC score | 0.680950 (rank : 24) | θ value | 1.47974e-10 (rank : 44) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9CXP4, Q99JY7 | Gene names | Arhgap8 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 8. | |||||
|
RHG08_HUMAN
|
||||||
| NC score | 0.680465 (rank : 25) | θ value | 1.33837e-11 (rank : 43) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9NSG0, O75983, O95695, Q96RW1, Q96RW2, Q9HA49, Q9HC46, Q9NVX8, Q9NXL1, Q9UH20 | Gene names | ARHGAP8 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 8. | |||||
|
RHG26_HUMAN
|
||||||
| NC score | 0.676217 (rank : 26) | θ value | 4.02038e-16 (rank : 23) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9UNA1, O75117, Q9BYS6, Q9BYS7, Q9UJ00 | Gene names | ARHGAP26, GRAF, KIAA0621, OPHN1L | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 26 (Oligophrenin-1-like protein) (GTPase regulator associated with focal adhesion kinase). | |||||
|
RHG18_HUMAN
|
||||||
| NC score | 0.674161 (rank : 27) | θ value | 4.0297e-08 (rank : 55) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8N392, Q58EZ3, Q6P679, Q6PJD7, Q96S64 | Gene names | ARHGAP18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho-GTPase-activating protein 18 (MacGAP). | |||||
|
RHG18_MOUSE
|
||||||
| NC score | 0.673764 (rank : 28) | θ value | 6.21693e-09 (rank : 51) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8K0Q5, Q8BP03, Q8R196 | Gene names | Arhgap18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho-GTPase-activating protein 18. | |||||
|
RHG04_HUMAN
|
||||||
| NC score | 0.673044 (rank : 29) | θ value | 9.56915e-18 (rank : 15) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P98171, Q14144 | Gene names | ARHGAP4, KIAA0131, RGC1, RHOGAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 4 (Rho-GAP hematopoietic protein C1) (p115). | |||||
|
BCR_HUMAN
|
||||||
| NC score | 0.672102 (rank : 30) | θ value | 2.06002e-20 (rank : 9) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P11274, P78501, Q12842 | Gene names | BCR, BCR1 | |||
|
Domain Architecture |

|
|||||
| Description | Breakpoint cluster region protein (EC 2.7.11.1) (NY-REN-26 antigen). | |||||
|
RBP1_MOUSE
|
||||||
| NC score | 0.665459 (rank : 31) | θ value | 3.52202e-12 (rank : 42) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q62172, Q9CRE5 | Gene names | Ralbp1, Rip1 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
SRGP2_HUMAN
|
||||||
| NC score | 0.664917 (rank : 32) | θ value | 2.5182e-18 (rank : 14) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | O43295, Q8IX13, Q8IZV8 | Gene names | SRGAP3, ARHGAP14, KIAA0411, MEGAP, SRGAP2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Mental disorder- associated GAP) (Rho-GTPase-activating protein 14). | |||||
|
RHG25_MOUSE
|
||||||
| NC score | 0.659236 (rank : 33) | θ value | 1.42992e-13 (rank : 34) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q8BYW1, Q2VPR2, Q3TE04, Q3TVX0, Q3UVB7, Q6A0E0, Q8BX98 | Gene names | Arhgap25, Kiaa0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
SRGP2_MOUSE
|
||||||
| NC score | 0.658223 (rank : 34) | θ value | 9.56915e-18 (rank : 16) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q812A2, Q80U09, Q8BKP4, Q8BLD0, Q925I2 | Gene names | Srgap3, Arhgap14, Kiaa0411, Srgap2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Rho-GTPase-activating protein 14). | |||||
|
RBP1_HUMAN
|
||||||
| NC score | 0.656637 (rank : 35) | θ value | 3.76295e-14 (rank : 30) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q15311 | Gene names | RALBP1, RLIP1, RLIP76 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (76 kDa Ral-interacting protein) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
SRGP1_HUMAN
|
||||||
| NC score | 0.653801 (rank : 36) | θ value | 3.07829e-16 (rank : 22) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q7Z6B7, Q9H8A3, Q9P2P2 | Gene names | SRGAP1, ARHGAP13, KIAA1304 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 1 (srGAP1) (Rho-GTPase- activating protein 13). | |||||
|
TAGAP_HUMAN
|
||||||
| NC score | 0.651984 (rank : 37) | θ value | 3.29651e-10 (rank : 45) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8N103, Q2NKM8, Q8NI40, Q96KZ2, Q96QA2 | Gene names | TAGAP, TAGAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell activation Rho GTPase-activating protein (T-cell activation GTPase-activating protein). | |||||
|
FNBP2_HUMAN
|
||||||
| NC score | 0.648444 (rank : 38) | θ value | 4.44505e-15 (rank : 27) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | O75044 | Gene names | SRGAP2, FNBP2, KIAA0456 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 2 (srGAP2) (Formin-binding protein 2). | |||||
|
TAGAP_MOUSE
|
||||||
| NC score | 0.639735 (rank : 39) | θ value | 5.26297e-08 (rank : 56) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8K2L9, Q920D6 | Gene names | Tagap, Tagap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell activation Rho GTPase-activating protein (T-cell activation GTPase-activating protein). | |||||
|
RHG25_HUMAN
|
||||||
| NC score | 0.637753 (rank : 40) | θ value | 2.0648e-12 (rank : 41) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 550 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | P42331, Q8IXQ2 | Gene names | ARHGAP25, KIAA0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.612141 (rank : 41) | θ value | 4.91457e-14 (rank : 31) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
CEND2_HUMAN
|
||||||
| NC score | 0.611763 (rank : 42) | θ value | 1.058e-16 (rank : 18) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q96P48, O94879, Q59FI7, Q6PHS3, Q8WU51, Q96HP6, Q96L71 | Gene names | CENTD2, ARAP1, KIAA0782 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-delta 2 (Cnt-d2) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 1). | |||||
|
RHG07_MOUSE
|
||||||
| NC score | 0.605662 (rank : 43) | θ value | 9.59137e-10 (rank : 49) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9R0Z9, Q8R541 | Gene names | Dlc1, Arhgap7, Stard12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 7 (Rho-type GTPase-activating protein 7) (Deleted in liver cancer 1 protein homolog) (Dlc-1) (StAR-related lipid transfer protein 12) (StARD12) (START domain-containing protein 12). | |||||
|
CE005_HUMAN
|
||||||
| NC score | 0.604787 (rank : 44) | θ value | 8.38298e-14 (rank : 33) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9NYF5, Q6PGQ2, Q9P0I7 | Gene names | C5orf5 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C5orf5 (GAP-like protein N61). | |||||
|
CE005_MOUSE
|
||||||
| NC score | 0.603382 (rank : 45) | θ value | 2.20605e-14 (rank : 29) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q8K2H3 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Protein C5orf5 homolog. | |||||
|
RHG07_HUMAN
|
||||||
| NC score | 0.598652 (rank : 46) | θ value | 9.59137e-10 (rank : 48) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q96QB1, O14868, O43199, Q9C0E0 | Gene names | DLC1, ARHGAP7, KIAA1723, STARD12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 7 (Rho-type GTPase-activating protein 7) (Deleted in liver cancer 1 protein) (Dlc-1) (HP protein) (StAR-related lipid transfer protein 12) (StARD12) (START domain-containing protein 12). | |||||
|
CEND3_MOUSE
|
||||||
| NC score | 0.576708 (rank : 47) | θ value | 3.40345e-15 (rank : 26) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8R5G7, Q5DTN4, Q6NVF1, Q8R5G6 | Gene names | Centd3, Arap3, Drag1, Kiaa4097 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3) (Dual specificity Rho- and Arf-GTPase-activating protein 1). | |||||
|
CEND3_HUMAN
|
||||||
| NC score | 0.565391 (rank : 48) | θ value | 3.40345e-15 (rank : 25) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8WWN8 | Gene names | CENTD3, ARAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3). | |||||
|
STA13_HUMAN
|
||||||
| NC score | 0.563566 (rank : 49) | θ value | 1.06045e-08 (rank : 53) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9Y3M8, Q5HYH1, Q5TAE3, Q6UN61, Q86TP6, Q86WQ3, Q86XT1 | Gene names | STARD13, DLC2, GT650 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 13 (StARD13) (START domain- containing protein 13) (46H23.2) (Deleted in liver cancer protein 2) (Rho GTPase-activating protein). | |||||
|
STAR8_MOUSE
|
||||||
| NC score | 0.556635 (rank : 50) | θ value | 8.40245e-06 (rank : 60) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8K031, Q3UZC7, Q6A0A8, Q6P5E0, Q8R3X8 | Gene names | Stard8, Kiaa0189 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
STAR8_HUMAN
|
||||||
| NC score | 0.552230 (rank : 51) | θ value | 1.29631e-06 (rank : 59) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q92502, Q5JST0 | Gene names | STARD8, KIAA0189 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
STA13_MOUSE
|
||||||
| NC score | 0.552038 (rank : 52) | θ value | 1.17247e-07 (rank : 58) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q923Q2, Q8K369 | Gene names | Stard13 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 13 (StARD13) (START domain- containing protein 13). | |||||
|
CEND1_MOUSE
|
||||||
| NC score | 0.527823 (rank : 53) | θ value | 1.86753e-13 (rank : 35) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8BZ05, Q80TX2, Q8C3T2, Q8VEL6 | Gene names | Centd1, Kiaa0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1). | |||||
|
CEND1_HUMAN
|
||||||
| NC score | 0.506706 (rank : 54) | θ value | 9.26847e-13 (rank : 40) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8WZ64, Q4W5D2, Q7Z2L5, Q96L70, Q96P49, Q9Y4E4 | Gene names | CENTD1, ARAP2, KIAA0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 2) (PARX protein). | |||||
|
K1688_MOUSE
|
||||||
| NC score | 0.431187 (rank : 55) | θ value | 0.813845 (rank : 89) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P59281, Q69ZD4, Q6P9R6 | Gene names | Kiaa1688, D15Wsu169e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1688. | |||||
|
K1688_HUMAN
|
||||||
| NC score | 0.419454 (rank : 56) | θ value | 4.03905 (rank : 120) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9C0H5 | Gene names | KIAA1688 | |||
|
Domain Architecture |

|
|||||
| Description | Protein KIAA1688. | |||||
|
MYO9B_MOUSE
|
||||||
| NC score | 0.396938 (rank : 57) | θ value | 1.80466e-16 (rank : 21) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9QY06, Q9QY07, Q9QY08, Q9QY09 | Gene names | Myo9b, Myr5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
MYO9B_HUMAN
|
||||||
| NC score | 0.391274 (rank : 58) | θ value | 1.38178e-16 (rank : 20) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
P85A_HUMAN
|
||||||
| NC score | 0.379721 (rank : 59) | θ value | 8.97725e-08 (rank : 57) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P27986 | Gene names | PIK3R1, GRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit alpha (PI3-kinase p85-subunit alpha) (PtdIns-3-kinase p85-alpha) (PI3K). | |||||
|
P85A_MOUSE
|
||||||
| NC score | 0.376212 (rank : 60) | θ value | 4.0297e-08 (rank : 54) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P26450 | Gene names | Pik3r1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit alpha (PI3-kinase p85-subunit alpha) (PtdIns-3-kinase p85-alpha) (PI3K). | |||||
|
I5P2_HUMAN
|
||||||
| NC score | 0.312426 (rank : 61) | θ value | 0.00665767 (rank : 63) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P32019, Q5VSG9, Q5VSH0, Q5VSH1, Q658Q5, Q6P6D4, Q6PD53, Q86YE1 | Gene names | INPP5B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Type II inositol-1,4,5-trisphosphate 5-phosphatase precursor (EC 3.1.3.36) (Phosphoinositide 5-phosphatase) (5PTase) (75 kDa inositol polyphosphate-5-phosphatase). | |||||
|
OCRL_HUMAN
|
||||||
| NC score | 0.304501 (rank : 62) | θ value | 0.000786445 (rank : 62) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q01968, O60800, Q15684, Q15774, Q5JQF1, Q5JQF2, Q9UJG5, Q9UMA5 | Gene names | OCRL, INPP5F, OCRL1 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol polyphosphate 5-phosphatase OCRL-1 (EC 3.1.3.36) (Lowe oculocerebrorenal syndrome protein). | |||||
|
TCRG1_HUMAN
|
||||||
| NC score | 0.164713 (rank : 63) | θ value | 5.62301e-10 (rank : 46) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
TCRG1_MOUSE
|
||||||
| NC score | 0.164106 (rank : 64) | θ value | 5.62301e-10 (rank : 47) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
PRP40_MOUSE
|
||||||
| NC score | 0.115148 (rank : 65) | θ value | 0.0148317 (rank : 66) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9R1C7, Q61049, Q8BQ76, Q8BRW4, Q8C2U1 | Gene names | Prpf40a, Fbp11, Fnbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Formin-binding protein 11) (FBP-11). | |||||
|
PRP40_HUMAN
|
||||||
| NC score | 0.109320 (rank : 66) | θ value | 0.0148317 (rank : 65) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O75400, O43856, O75404, Q8TBQ1, Q9H782, Q9NWU9, Q9P0Q2, Q9Y5A8 | Gene names | PRPF40A, FLAF1, FNBP3, HYPA | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Huntingtin yeast partner A) (Huntingtin-interacting protein HYPA/FBP11) (Fas ligand-associated factor 1) (NY-REN-6 antigen). | |||||
|
P85B_HUMAN
|
||||||
| NC score | 0.102497 (rank : 67) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O00459, Q9UPH9 | Gene names | PIK3R2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit beta (PI3-kinase p85- subunit beta) (PtdIns-3-kinase p85-beta). | |||||
|
CENB2_HUMAN
|
||||||
| NC score | 0.077067 (rank : 68) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q15057, Q9UQR3 | Gene names | CENTB2, KIAA0041 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 2 (Cnt-b2). | |||||
|
P85B_MOUSE
|
||||||
| NC score | 0.075566 (rank : 69) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O08908 | Gene names | Pik3r2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit beta (PI3-kinase p85- subunit beta) (PtdIns-3-kinase p85-beta). | |||||
|
GRP3_HUMAN
|
||||||
| NC score | 0.075566 (rank : 70) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8IV61, O94931 | Gene names | RASGRP3, GRP3, KIAA0846 | |||
|
Domain Architecture |

|
|||||
| Description | RAS guanyl-releasing protein 3 (Calcium and DAG-regulated guanine nucleotide exchange factor III) (Guanine nucleotide exchange factor for Rap1). | |||||
|
CENB5_HUMAN
|
||||||
| NC score | 0.071710 (rank : 71) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96P50, Q9BSR9, Q9C0E7 | Gene names | CENTB5, KIAA1716 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 5 (Cnt-b5). | |||||
|
CENB1_HUMAN
|
||||||
| NC score | 0.070705 (rank : 72) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15027 | Gene names | CENTB1, KIAA0050 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 1 (Cnt-b1). | |||||
|
FA13A_MOUSE
|
||||||
| NC score | 0.069834 (rank : 73) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BGI4, Q8BZ91 | Gene names | Fam13a1, Precm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13A1 (Precm1 protein). | |||||
|
FA13A_HUMAN
|
||||||
| NC score | 0.069170 (rank : 74) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O94988, Q8NBA3 | Gene names | FAM13A1, KIAA0914 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13A1. | |||||
|
P55G_MOUSE
|
||||||
| NC score | 0.061375 (rank : 75) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q64143 | Gene names | Pik3r3 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit gamma (PI3-kinase p85-subunit gamma) (PtdIns-3-kinase p85-gamma) (p55PIK). | |||||
|
P55G_HUMAN
|
||||||
| NC score | 0.060954 (rank : 76) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q92569, O60482 | Gene names | PIK3R3 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit gamma (PI3-kinase p85-subunit gamma) (PtdIns-3-kinase p85-gamma) (p55PIK). | |||||
|
CIP4_HUMAN
|
||||||
| NC score | 0.059677 (rank : 77) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q15642, O15184, Q5TZN1 | Gene names | TRIP10, CIP4 | |||
|
Domain Architecture |

|
|||||
| Description | Cdc42-interacting protein 4 (Thyroid receptor-interacting protein 10) (TRIP-10). | |||||
|
PAWR_HUMAN
|
||||||
| NC score | 0.059676 (rank : 78) | θ value | 0.47712 (rank : 77) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96IZ0, O75796, Q6FHY9, Q8N700 | Gene names | PAWR, PAR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PRKC apoptosis WT1 regulator protein (Prostate apoptosis response 4 protein) (Par-4). | |||||
|
NCK2_HUMAN
|
||||||
| NC score | 0.058578 (rank : 79) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O43639, Q9BWN9, Q9UIC3 | Gene names | NCK2 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoplasmic protein NCK2 (NCK adaptor protein 2) (SH2/SH3 adaptor protein NCK-beta) (Nck-2). | |||||
|
MYO10_HUMAN
|
||||||
| NC score | 0.057826 (rank : 80) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9HD67, Q9NYM7, Q9P110, Q9P111, Q9UHF6 | Gene names | MYO10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin X). | |||||
|
DDEF2_HUMAN
|
||||||
| NC score | 0.055825 (rank : 81) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 599 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O43150 | Gene names | DDEF2, KIAA0400 | |||
|
Domain Architecture |

|
|||||
| Description | Development and differentiation-enhancing factor 2 (Pyk2 C-terminus- associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3). | |||||
|
IF3A_HUMAN
|
||||||
| NC score | 0.055275 (rank : 82) | θ value | 0.62314 (rank : 82) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q14152, O00653 | Gene names | EIF3S10, KIAA0139 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a). | |||||
|
COG8_HUMAN
|
||||||
| NC score | 0.055133 (rank : 83) | θ value | 0.21417 (rank : 72) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96MW5, Q8WVV6, Q9H6F8 | Gene names | COG8 | |||
|
Domain Architecture |

|
|||||
| Description | Conserved oligomeric Golgi complex component 8. | |||||
|
PDZD8_HUMAN
|
||||||
| NC score | 0.054480 (rank : 84) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8NEN9, Q86WE0, Q86WE5, Q9UFF1 | Gene names | PDZD8, PDZK8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 8 (Sarcoma antigen NY-SAR-84/NY-SAR- 104). | |||||
|
SMP1L_HUMAN
|
||||||
| NC score | 0.054253 (rank : 85) | θ value | θ > 10 (rank : 189) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WU79, Q5QPL2, Q96C93, Q9NST2, Q9UJL8 | Gene names | SMAP1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1-like. | |||||
|
DDEF2_MOUSE
|
||||||
| NC score | 0.054236 (rank : 86) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7SIG6, Q501K1, Q66JN2 | Gene names | Ddef2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor 2 (Pyk2 C-terminus- associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3). | |||||
|
SMP1L_MOUSE
|
||||||
| NC score | 0.054235 (rank : 87) | θ value | θ > 10 (rank : 190) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TN29, Q3U798 | Gene names | Smap1l, Smap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1-like (Stromal membrane- associated protein 2). | |||||
|
CENA2_HUMAN
|
||||||
| NC score | 0.054121 (rank : 88) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NPF8, Q8N4Q6, Q96SD5 | Gene names | CENTA2 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-alpha 2. | |||||
|
DDFL1_HUMAN
|
||||||
| NC score | 0.053642 (rank : 89) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8TDY4, Q6P9F4, Q86UY1, Q9NXK2 | Gene names | DDEFL1, UPLC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor-like 1 (Protein up- regulated in liver cancer 1). | |||||
|
MYH8_HUMAN
|
||||||
| NC score | 0.053344 (rank : 90) | θ value | 1.06291 (rank : 94) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
IF3A_MOUSE
|
||||||
| NC score | 0.053257 (rank : 91) | θ value | 1.38821 (rank : 97) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P23116, Q60697, Q62162 | Gene names | Eif3s10, Csma, Eif3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a) (p162 protein) (Centrosomin). | |||||
|
FA13C_HUMAN
|
||||||
| NC score | 0.053137 (rank : 92) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NE31, Q99787 | Gene names | FAM13C1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13C1. | |||||
|
VAV3_HUMAN
|
||||||
| NC score | 0.053087 (rank : 93) | θ value | θ > 10 (rank : 191) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 398 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UKW4, O95230, Q9Y5X8 | Gene names | VAV3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-3. | |||||
|
GRAP_HUMAN
|
||||||
| NC score | 0.053062 (rank : 94) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13588 | Gene names | GRAP | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-related adapter protein. | |||||
|
SMAP1_HUMAN
|
||||||
| NC score | 0.052661 (rank : 95) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IYB5, Q53H70, Q5SYQ2, Q6PK24, Q8NDH4, Q96L38, Q96L39, Q9H8X4 | Gene names | SMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1. | |||||
|
VAV3_MOUSE
|
||||||
| NC score | 0.052620 (rank : 96) | θ value | θ > 10 (rank : 192) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 404 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9R0C8, Q7TS85 | Gene names | Vav3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-3. | |||||
|
ITSN1_HUMAN
|
||||||
| NC score | 0.052573 (rank : 97) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
SMAP1_MOUSE
|
||||||
| NC score | 0.052550 (rank : 98) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91VZ6, Q497I6, Q68EF3 | Gene names | Smap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1. | |||||
|
MYH1_MOUSE
|
||||||
| NC score | 0.051953 (rank : 99) | θ value | 0.47712 (rank : 76) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
FA13C_MOUSE
|
||||||
| NC score | 0.051751 (rank : 100) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9DBR2 | Gene names | Fam13c1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13C1. | |||||
|
CENA1_HUMAN
|
||||||
| NC score | 0.051681 (rank : 101) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75689 | Gene names | CENTA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-alpha 1 (Putative MAPK-activating protein PM25). | |||||
|
MYH4_HUMAN
|
||||||
| NC score | 0.051527 (rank : 102) | θ value | 1.81305 (rank : 103) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
GRB2_HUMAN
|
||||||
| NC score | 0.051370 (rank : 103) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P62993, P29354, Q14450, Q63057, Q63059 | Gene names | GRB2, ASH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth factor receptor-bound protein 2 (Adapter protein GRB2) (SH2/SH3 adapter GRB2) (Protein Ash). | |||||
|
GRB2_MOUSE
|
||||||
| NC score | 0.051362 (rank : 104) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q60631, Q61240 | Gene names | Grb2 | |||
|
Domain Architecture |

|
|||||
| Description | Growth factor receptor-bound protein 2 (Adapter protein GRB2) (SH2/SH3 adapter GRB2). | |||||
|
MYH3_HUMAN
|
||||||
| NC score | 0.051318 (rank : 105) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
NIN_HUMAN
|
||||||
| NC score | 0.051253 (rank : 106) | θ value | 0.125558 (rank : 70) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
DDEF1_MOUSE
|
||||||
| NC score | 0.050964 (rank : 107) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9QWY8, O08612, Q80TG8, Q80UV6, Q99LV8, Q9Z2B6 | Gene names | Ddef1, Asap1, Kiaa1249, Shag1 | |||
|
Domain Architecture |

|
|||||
| Description | 130 kDa phosphatidylinositol 4,5-biphosphate-dependent ARF1 GTPase- activating protein (PIP2-dependent ARF1 GAP) (ADP-ribosylation factor- directed GTPase-activating protein 1) (ARF GTPase-activating protein 1) (Development and differentiation-enhancing factor 1) (Differentiation-enhancing factor 1) (DEF-1). | |||||
|
MYH8_MOUSE
|
||||||
| NC score | 0.050930 (rank : 108) | θ value | 2.36792 (rank : 109) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
GRAP_MOUSE
|
||||||
| NC score | 0.050526 (rank : 109) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9CX99, Q3U545 | Gene names | Grap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRB2-related adapter protein. | |||||
|
SPTA1_HUMAN
|
||||||
| NC score | 0.050326 (rank : 110) | θ value | 1.06291 (rank : 96) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P02549, Q15514, Q6LDY5 | Gene names | SPTA1, SPTA | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
MYH1_HUMAN
|
||||||
| NC score | 0.049937 (rank : 111) | θ value | 3.0926 (rank : 116) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
MYH2_HUMAN
|
||||||
| NC score | 0.049614 (rank : 112) | θ value | 0.62314 (rank : 83) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
SPTA1_MOUSE
|
||||||
| NC score | 0.048221 (rank : 113) | θ value | 1.81305 (rank : 106) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P08032, P97502 | Gene names | Spta1, Spna1, Spta | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
MORC4_HUMAN
|
||||||
| NC score | 0.048074 (rank : 114) | θ value | 0.0736092 (rank : 69) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8TE76, Q5JUK7, Q96MZ2, Q9HAI7 | Gene names | MORC4, ZCWCC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 4 (Zinc finger CW-type coiled- coil domain protein 2). | |||||
|
MYH4_MOUSE
|
||||||
| NC score | 0.047839 (rank : 115) | θ value | 8.99809 (rank : 154) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.047260 (rank : 116) | θ value | 0.0736092 (rank : 68) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
RAD50_MOUSE
|
||||||
| NC score | 0.045534 (rank : 117) | θ value | 0.125558 (rank : 71) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
CI093_HUMAN
|
||||||
| NC score | 0.045310 (rank : 118) | θ value | 4.03905 (rank : 119) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q6TFL3, Q5SU58, Q6P1W1, Q6ZR13, Q8N1Z4, Q8N8L3, Q8TBR2 | Gene names | C9orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf93. | |||||
|
RAD50_HUMAN
|
||||||
| NC score | 0.044080 (rank : 119) | θ value | 0.365318 (rank : 75) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.043642 (rank : 120) | θ value | 0.813845 (rank : 88) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
SYNE1_HUMAN
|
||||||
| NC score | 0.043185 (rank : 121) | θ value | 0.0193708 (rank : 67) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
CCD11_HUMAN
|
||||||
| NC score | 0.042978 (rank : 122) | θ value | 1.81305 (rank : 100) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96M91 | Gene names | CCDC11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 11. | |||||
|
KTN1_HUMAN
|
||||||
| NC score | 0.042066 (rank : 123) | θ value | 0.365318 (rank : 74) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q86UP2, Q13999, Q14707, Q15387, Q86W57 | Gene names | KTN1, CG1, KIAA0004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin (Kinesin receptor) (CG-1 antigen). | |||||
|
TRIPB_HUMAN
|
||||||
| NC score | 0.042010 (rank : 124) | θ value | 0.21417 (rank : 73) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
CA065_HUMAN
|
||||||
| NC score | 0.041868 (rank : 125) | θ value | 2.36792 (rank : 107) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 469 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8N715, Q8N746, Q8NA93 | Gene names | C1orf65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf65. | |||||
|
CCD46_HUMAN
|
||||||
| NC score | 0.041866 (rank : 126) | θ value | 8.99809 (rank : 150) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
TACC3_MOUSE
|
||||||
| NC score | 0.038915 (rank : 127) | θ value | 0.813845 (rank : 92) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9JJ11, Q9WVK9 | Gene names | Tacc3, Aint | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ARNT-interacting protein). | |||||
|
G45IP_HUMAN
|
||||||
| NC score | 0.035465 (rank : 128) | θ value | 1.81305 (rank : 102) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8TAE8, Q8IVM3, Q8TE51, Q969P9, Q9BSM6 | Gene names | GADD45GIP1, PLINP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth arrest and DNA-damage-inducible proteins-interacting protein 1 (CR6-interacting factor 1) (CRIF1) (CKII beta-associating protein) (Papillomavirus L2-interacting nuclear protein 1) (PLINP-1). | |||||
|
TXLNA_MOUSE
|
||||||
| NC score | 0.033999 (rank : 129) | θ value | 2.36792 (rank : 110) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 846 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6PAM1, Q6P1E5 | Gene names | Txlna, Txln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
COG8_MOUSE
|
||||||
| NC score | 0.033660 (rank : 130) | θ value | 8.99809 (rank : 151) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JJA2 | Gene names | Cog8 | |||
|
Domain Architecture |

|
|||||
| Description | Conserved oligomeric Golgi complex component 8. | |||||
|
TXLNA_HUMAN
|
||||||
| NC score | 0.032989 (rank : 131) | θ value | 3.0926 (rank : 118) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 982 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P40222, Q66K62, Q86T54, Q86T85, Q86T86, Q86Y86, Q86YW3, Q8N2Y3 | Gene names | TXLNA, TXLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
FAF1_MOUSE
|
||||||
| NC score | 0.032822 (rank : 132) | θ value | 3.0926 (rank : 112) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P54731 | Gene names | Faf1 | |||
|
Domain Architecture |

|
|||||
| Description | FAS-associated factor 1 (Protein FAF1). | |||||
|
CTRO_MOUSE
|
||||||
| NC score | 0.032024 (rank : 133) | θ value | 4.1701e-05 (rank : 61) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
BCAR1_MOUSE
|
||||||
| NC score | 0.030761 (rank : 134) | θ value | 8.99809 (rank : 148) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q61140, Q60869 | Gene names | Bcar1, Cas, Crkas | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
FAF1_HUMAN
|
||||||
| NC score | 0.029416 (rank : 135) | θ value | 5.27518 (rank : 127) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UNN5, Q9UF34, Q9UNT3, Q9Y2Z3 | Gene names | FAF1 | |||
|
Domain Architecture |

|
|||||
| Description | FAS-associated factor 1 (Protein FAF1) (hFAF1). | |||||
|
MORC4_MOUSE
|
||||||
| NC score | 0.028820 (rank : 136) | θ value | 3.0926 (rank : 115) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BMD7, Q4KMM6, Q8BX95, Q9CS96 | Gene names | Morc4, Zcwcc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 4 (Zinc finger CW-type coiled- coil domain protein 2). | |||||
|
CYLN2_HUMAN
|
||||||
| NC score | 0.028697 (rank : 137) | θ value | 8.99809 (rank : 152) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1004 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9UDT6, O14527, O43611 | Gene names | CYLN2, KIAA0291, WBSCR4, WSCR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115) (Williams-Beuren syndrome chromosome region 4 protein). | |||||
|
K0423_HUMAN
|
||||||
| NC score | 0.027845 (rank : 138) | θ value | 3.0926 (rank : 113) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y4F4, Q68D66, Q6PG27 | Gene names | KIAA0423 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0423. | |||||
|
AASS_HUMAN
|
||||||
| NC score | 0.026186 (rank : 139) | θ value | 1.81305 (rank : 99) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UDR5, O95462 | Gene names | AASS | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-aminoadipic semialdehyde synthase, mitochondrial precursor (LKR/SDH) [Includes: Lysine ketoglutarate reductase (EC 1.5.1.8) (LOR) (LKR); Saccharopine dehydrogenase (EC 1.5.1.9) (SDH)]. | |||||
|
WDR67_HUMAN
|
||||||
| NC score | 0.025209 (rank : 140) | θ value | 4.03905 (rank : 124) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 782 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96DN5, Q3MIR6, Q8TBP9 | Gene names | WDR67 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 67. | |||||
|
K0423_MOUSE
|
||||||
| NC score | 0.025085 (rank : 141) | θ value | 3.0926 (rank : 114) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6A070 | Gene names | Kiaa0423 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0423. | |||||
|
DTBP1_HUMAN
|
||||||
| NC score | 0.024877 (rank : 142) | θ value | 2.36792 (rank : 108) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96EV8, Q96NV2, Q9H0U2, Q9H3J5 | Gene names | DTNBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dystrobrevin-binding protein 1 (Dysbindin) (Hermansky-Pudlak syndrome 7 protein). | |||||
|
SIN3A_MOUSE
|
||||||
| NC score | 0.024305 (rank : 143) | θ value | 3.0926 (rank : 117) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q60520, Q60820, Q62139, Q62140, Q7TPU8, Q7TSZ2 | Gene names | Sin3a | |||
|
Domain Architecture |

|
|||||
| Description | Paired amphipathic helix protein Sin3a (Transcriptional corepressor Sin3a) (Histone deacetylase complex subunit Sin3a). | |||||
|
COVA1_HUMAN
|
||||||
| NC score | 0.023562 (rank : 144) | θ value | 1.81305 (rank : 101) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q16206, Q5VTJ1, Q5VTJ2, Q8WUX0, Q9NTP6, Q9UH82 | Gene names | COVA1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor-associated hydroquinone oxidase (tNOX) (Cytosolic ovarian carcinoma antigen 1) (APK1 antigen) [Includes: Hydroquinone [NADH] oxidase (EC 1.-.-.-); Protein disulfide-thiol oxidoreductase (EC 1.-.-.-)]. | |||||
|
SCYL2_MOUSE
|
||||||
| NC score | 0.023069 (rank : 145) | θ value | 0.62314 (rank : 86) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CFE4, Q3UT57, Q3UWU9, Q8K0M4 | Gene names | Scyl2, Cvak104, D10Ertd802e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCY1-like protein 2 (Coated vesicle-associated kinase of 104 kDa). | |||||
|
LAMA3_HUMAN
|
||||||
| NC score | 0.022871 (rank : 146) | θ value | 1.06291 (rank : 93) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q16787, Q13679, Q13680, Q96TG0 | Gene names | LAMA3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-3 chain precursor (Epiligrin 170 kDa subunit) (E170) (Nicein subunit alpha). | |||||
|
CTRO_HUMAN
|
||||||
| NC score | 0.022782 (rank : 147) | θ value | 0.62314 (rank : 81) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
PIGN_HUMAN
|
||||||
| NC score | 0.022524 (rank : 148) | θ value | 4.03905 (rank : 121) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95427, Q7L8F8, Q8TC01, Q9NT05 | Gene names | PIGN, MCD4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GPI ethanolamine phosphate transferase 1 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class N protein) (PIG-N) (MCD4 homolog). | |||||
|
SEN54_MOUSE
|
||||||
| NC score | 0.022512 (rank : 149) | θ value | 4.03905 (rank : 123) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C2A2, Q9DCI5 | Gene names | Tsen54, Sen54 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-splicing endonuclease subunit Sen54 (tRNA-intron endonuclease Sen54). | |||||
|
SIN3A_HUMAN
|
||||||
| NC score | 0.022258 (rank : 150) | θ value | 5.27518 (rank : 133) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96ST3, Q8N8N4, Q8NC83, Q8WV18, Q96L98, Q9UFQ1 | Gene names | SIN3A | |||
|
Domain Architecture |

|
|||||
| Description | Paired amphipathic helix protein Sin3a (Transcriptional corepressor Sin3a) (Histone deacetylase complex subunit Sin3a). | |||||
|
SCYL2_HUMAN
|
||||||
| NC score | 0.021553 (rank : 151) | θ value | 0.47712 (rank : 78) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 632 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6P3W7, Q96EF4, Q96ST4, Q9H7V5, Q9NVH3, Q9P2I7 | Gene names | SCYL2, CVAK104, KIAA1360 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCY1-like protein 2 (Coated vesicle-associated kinase of 104 kDa). | |||||
|
PROX1_MOUSE
|
||||||
| NC score | 0.020418 (rank : 152) | θ value | 4.03905 (rank : 122) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P48437, O88478, Q3UPV1, Q543D8 | Gene names | Prox1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox prospero-like protein PROX1 (PROX 1). | |||||
|
PIGN_MOUSE
|
||||||
| NC score | 0.019556 (rank : 153) | θ value | 6.88961 (rank : 140) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9R1S3, Q3V0S6, Q8VCC3, Q9R1S1, Q9R1S2 | Gene names | Pign | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GPI ethanolamine phosphate transferase 1 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class N protein) (PIG-N). | |||||
|
AN30A_HUMAN
|
||||||
| NC score | 0.017745 (rank : 154) | θ value | 8.99809 (rank : 147) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1119 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9BXX3, Q5W025 | Gene names | ANKRD30A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 30A (NY-BR-1 antigen). | |||||
|
RAB4B_MOUSE
|
||||||
| NC score | 0.017185 (rank : 155) | θ value | 0.62314 (rank : 85) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q91ZR1 | Gene names | Rab4b, Rab4 | |||
|
Domain Architecture |

|
|||||
| Description | Ras-related protein Rab-4B. | |||||
|
RAB4B_HUMAN
|
||||||
| NC score | 0.017183 (rank : 156) | θ value | 0.62314 (rank : 84) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P61018, P22750, Q7Z514, Q9HBR6 | Gene names | RAB4B | |||
|
Domain Architecture |

|
|||||
| Description | Ras-related protein Rab-4B. | |||||
|
MYCN_MOUSE
|
||||||
| NC score | 0.015825 (rank : 157) | θ value | 5.27518 (rank : 130) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P03966, Q61978 | Gene names | Mycn, Nmyc, Nmyc1 | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
MYCN_HUMAN
|
||||||
| NC score | 0.015626 (rank : 158) | θ value | 5.27518 (rank : 129) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P04198, Q6LDT9 | Gene names | MYCN, NMYC | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
RAB4A_HUMAN
|
||||||
| NC score | 0.015418 (rank : 159) | θ value | 0.813845 (rank : 91) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P20338, Q9BQ44 | Gene names | RAB4A, RAB4 | |||
|
Domain Architecture |

|
|||||
| Description | Ras-related protein Rab-4A. | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.015324 (rank : 160) | θ value | 8.99809 (rank : 158) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
CA048_HUMAN
|
||||||
| NC score | 0.015145 (rank : 161) | θ value | 8.99809 (rank : 149) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96IY1, Q5SY75, Q9H2M5, Q9NRN8, Q9Y415 | Gene names | C1orf48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C1orf48. | |||||
|
NUCB1_HUMAN
|
||||||
| NC score | 0.014921 (rank : 162) | θ value | 8.99809 (rank : 156) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q02818, Q15838, Q7Z4J7, Q9BUR1 | Gene names | NUCB1, NUC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleobindin-1 precursor (CALNUC). | |||||
|
RAB4A_MOUSE
|
||||||
| NC score | 0.014455 (rank : 163) | θ value | 1.38821 (rank : 98) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P56371 | Gene names | Rab4a, Rab4 | |||
|
Domain Architecture |

|
|||||
| Description | Ras-related protein Rab-4A. | |||||
|
STK10_HUMAN
|
||||||
| NC score | 0.014110 (rank : 164) | θ value | 0.00869519 (rank : 64) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O94804, Q9UIW4 | Gene names | STK10, LOK | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
CE110_HUMAN
|
||||||
| NC score | 0.013091 (rank : 165) | θ value | 6.88961 (rank : 135) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O43303, O43335, Q68DV9, Q8NE13 | Gene names | CEP110, CP110, KIAA0419 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 110 kDa (Cep110 protein). | |||||
|
LMNB2_MOUSE
|
||||||
| NC score | 0.012644 (rank : 166) | θ value | 6.88961 (rank : 139) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P21619, P48680, Q8CGB1 | Gene names | Lmnb2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-B2. | |||||
|
TTLL2_HUMAN
|
||||||
| NC score | 0.012635 (rank : 167) | θ value | 5.27518 (rank : 134) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BWV7, Q7Z6R8, Q86X22 | Gene names | TTLL2, C6orf104 | |||
|
Domain Architecture |

|
|||||
| Description | Tubulin--tyrosine ligase-like protein 2 (Testis-specific protein NYD- TSPG). | |||||
|
KI13A_HUMAN
|
||||||
| NC score | 0.012348 (rank : 168) | θ value | 0.813845 (rank : 90) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H1H9, Q9H1H8 | Gene names | KIF13A, RBKIN | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF13A (Kinesin-like protein RBKIN). | |||||
|
RAB2B_MOUSE
|
||||||
| NC score | 0.012264 (rank : 169) | θ value | 1.81305 (rank : 105) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P59279, Q3TV67 | Gene names | Rab2b | |||
|
Domain Architecture |

|
|||||
| Description | Ras-related protein Rab-2B. | |||||
|
RAB2B_HUMAN
|
||||||
| NC score | 0.012183 (rank : 170) | θ value | 1.81305 (rank : 104) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8WUD1 | Gene names | RAB2B | |||
|
Domain Architecture |

|
|||||
| Description | Ras-related protein Rab-2B. | |||||
|
SLK_MOUSE
|
||||||
| NC score | 0.012118 (rank : 171) | θ value | 0.62314 (rank : 87) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1798 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O54988, Q80U65, Q8CAU2, Q8CDW2, Q9WU41 | Gene names | Slk, Kiaa0204, Stk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (mSLK) (Serine/threonine-protein kinase 2) (STE20-related kinase SMAK) (Etk4). | |||||
|
FA48A_HUMAN
|
||||||
| NC score | 0.011504 (rank : 172) | θ value | 5.27518 (rank : 126) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NEM7, Q71RF3, Q9Y6A6 | Gene names | FAM48A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM48A (p38-interacting protein) (p38IP). | |||||
|
SLK_HUMAN
|
||||||
| NC score | 0.011186 (rank : 173) | θ value | 1.06291 (rank : 95) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1775 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9H2G2, O00211, Q6P1Z4, Q86WU7, Q86WW1, Q92603, Q9NQL0, Q9NQL1 | Gene names | SLK, KIAA0204, STK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (hSLK) (Serine/threonine-protein kinase 2) (CTCL tumor antigen se20-9). | |||||
|
EXOC6_HUMAN
|
||||||
| NC score | 0.011046 (rank : 174) | θ value | 6.88961 (rank : 137) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8TAG9, Q9NZ24 | Gene names | EXOC6, SEC15A, SEC15L, SEC15L1 | |||
|
Domain Architecture |

|
|||||
| Description | Exocyst complex component 6 (Exocyst complex component Sec15A) (Sec15- like 1). | |||||
|
GHR_MOUSE
|
||||||
| NC score | 0.010574 (rank : 175) | θ value | 8.99809 (rank : 153) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P16882, P16590, Q61653, Q6DI66, Q80W86, Q8R1M5, Q920Z3, Q9R264 | Gene names | Ghr | |||
|
Domain Architecture |

|
|||||
| Description | Growth hormone receptor precursor (GH receptor) (Somatotropin receptor) [Contains: Growth hormone-binding protein (GH-binding protein) (GHBP) (Serum-binding protein)]. | |||||
|
K2C4_HUMAN
|
||||||
| NC score | 0.009620 (rank : 176) | θ value | 5.27518 (rank : 128) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P19013, Q6GTR8, Q96LA7, Q9BTL1 | Gene names | KRT4, CYK4 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 4 (Cytokeratin-4) (CK-4) (Keratin-4) (K4). | |||||
|
BMR1B_HUMAN
|
||||||
| NC score | 0.009095 (rank : 177) | θ value | 0.62314 (rank : 79) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O00238, P78366 | Gene names | BMPR1B | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein receptor type IB precursor (EC 2.7.11.30) (CDw293 antigen). | |||||
|
POLI_HUMAN
|
||||||
| NC score | 0.009075 (rank : 178) | θ value | 5.27518 (rank : 132) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UNA4, Q8N590, Q9H0S1, Q9NYH6 | Gene names | POLI, RAD30B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase iota (EC 2.7.7.7) (RAD30 homolog B) (Eta2). | |||||
|
BMR1B_MOUSE
|
||||||
| NC score | 0.008957 (rank : 179) | θ value | 0.62314 (rank : 80) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 758 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P36898, Q3TRF2 | Gene names | Bmpr1b, Acvrlk6 | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein receptor type IB precursor (EC 2.7.11.30) (Serine/threonine-protein kinase receptor R6) (SKR6) (Activin receptor-like kinase 6) (ALK-6). | |||||
|
ES8L3_HUMAN
|
||||||
| NC score | 0.008897 (rank : 180) | θ value | 3.0926 (rank : 111) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TE67, Q5T8Q6, Q5T8Q7, Q5T8Q8, Q96E47, Q9H719 | Gene names | EPS8L3, EPS8R3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epidermal growth factor receptor kinase substrate 8-like protein 3 (Epidermal growth factor receptor pathway substrate 8-related protein 3) (EPS8-like protein 3). | |||||
|
ETV1_HUMAN
|
||||||
| NC score | 0.008875 (rank : 181) | θ value | 5.27518 (rank : 125) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P50549, O75849, Q9UQ71, Q9Y636 | Gene names | ETV1, ER81 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 1 (ER81 protein). | |||||
|
NOG1_HUMAN
|
||||||
| NC score | 0.008393 (rank : 182) | θ value | 8.99809 (rank : 155) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BZE4, O95446, Q9NVJ8 | Gene names | GTPBP4, CRFG, NOG1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar GTP-binding protein 1 (Chronic renal failure gene protein) (GTP-binding protein NGB). | |||||
|
RAB1B_HUMAN
|
||||||
| NC score | 0.008111 (rank : 183) | θ value | 6.88961 (rank : 144) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H0U4 | Gene names | RAB1B | |||
|
Domain Architecture |

|
|||||
| Description | Ras-related protein Rab-1B. | |||||
|
RAB1A_HUMAN
|
||||||
| NC score | 0.008105 (rank : 184) | θ value | 6.88961 (rank : 142) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P62820, P11476, Q96N61, Q9Y3T2 | Gene names | RAB1A, RAB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-related protein Rab-1A (YPT1-related protein). | |||||
|
RAB1A_MOUSE
|
||||||
| NC score | 0.008105 (rank : 185) | θ value | 6.88961 (rank : 143) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P62821, P11476, Q3TX44, Q811M4, Q96N61, Q9Y3T2 | Gene names | Rab1A, Rab1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-related protein Rab-1A (YPT1-related protein). | |||||
|
RAB1B_MOUSE
|
||||||
| NC score | 0.008096 (rank : 186) | θ value | 6.88961 (rank : 145) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9D1G1, Q3U0N1 | Gene names | Rab1b | |||
|
Domain Architecture |

|
|||||
| Description | Ras-related protein Rab-1B. | |||||
|
ETV1_MOUSE
|
||||||
| NC score | 0.007942 (rank : 187) | θ value | 6.88961 (rank : 136) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P41164 | Gene names | Etv1, Er81, Etsrp81 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 1 (ER81 protein). | |||||
|
PLXB1_HUMAN
|
||||||
| NC score | 0.005742 (rank : 188) | θ value | 5.27518 (rank : 131) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43157, Q6NY20, Q9UIV7, Q9UJ92, Q9UJ93 | Gene names | PLXNB1, KIAA0407, SEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor (Semaphorin receptor SEP). | |||||
|
JHD3B_MOUSE
|
||||||
| NC score | 0.003023 (rank : 189) | θ value | 6.88961 (rank : 138) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91VY5, Q3UR22, Q6ZQ30, Q99K42 | Gene names | Jmjd2b, Jhdm3b | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3B (EC 1.14.11.-) (Jumonji domain-containing protein 2B). | |||||
|
1433Z_MOUSE
|
||||||
| NC score | 0.002814 (rank : 190) | θ value | 8.99809 (rank : 146) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P63101, P35215, P70197, P97286, Q3TSF1, Q5EBQ1 | Gene names | Ywhaz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 14-3-3 protein zeta/delta (Protein kinase C inhibitor protein 1) (KCIP-1) (SEZ-2). | |||||
|
PTPRZ_HUMAN
|
||||||
| NC score | 0.000700 (rank : 191) | θ value | 6.88961 (rank : 141) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P23471 | Gene names | PTPRZ1, PTPRZ, PTPZ | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase zeta precursor (EC 3.1.3.48) (R-PTP-zeta). | |||||
|
SSPO_MOUSE
|
||||||
| NC score | -0.000884 (rank : 192) | θ value | 8.99809 (rank : 157) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||