Please be patient as the page loads
|
I5P2_HUMAN
|
||||||
| SwissProt Accessions | P32019, Q5VSG9, Q5VSH0, Q5VSH1, Q658Q5, Q6P6D4, Q6PD53, Q86YE1 | Gene names | INPP5B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Type II inositol-1,4,5-trisphosphate 5-phosphatase precursor (EC 3.1.3.36) (Phosphoinositide 5-phosphatase) (5PTase) (75 kDa inositol polyphosphate-5-phosphatase). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
I5P2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P32019, Q5VSG9, Q5VSH0, Q5VSH1, Q658Q5, Q6P6D4, Q6PD53, Q86YE1 | Gene names | INPP5B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Type II inositol-1,4,5-trisphosphate 5-phosphatase precursor (EC 3.1.3.36) (Phosphoinositide 5-phosphatase) (5PTase) (75 kDa inositol polyphosphate-5-phosphatase). | |||||
|
OCRL_HUMAN
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.975892 (rank : 2) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q01968, O60800, Q15684, Q15774, Q5JQF1, Q5JQF2, Q9UJG5, Q9UMA5 | Gene names | OCRL, INPP5F, OCRL1 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol polyphosphate 5-phosphatase OCRL-1 (EC 3.1.3.36) (Lowe oculocerebrorenal syndrome protein). | |||||
|
SYNJ1_MOUSE
|
||||||
| θ value | 1.15626e-55 (rank : 3) | NC score | 0.690844 (rank : 11) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
SYNJ1_HUMAN
|
||||||
| θ value | 1.2784e-54 (rank : 4) | NC score | 0.691745 (rank : 10) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O43426, O43425, O94984 | Gene names | SYNJ1, KIAA0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
SYNJ2_HUMAN
|
||||||
| θ value | 4.11246e-53 (rank : 5) | NC score | 0.751462 (rank : 6) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O15056, Q5TA13, Q5TA16, Q5TA19, Q86XK0, Q8IZA8, Q9H226 | Gene names | SYNJ2, KIAA0348 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptojanin-2 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 2). | |||||
|
SYNJ2_MOUSE
|
||||||
| θ value | 1.32293e-51 (rank : 6) | NC score | 0.758213 (rank : 5) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9D2G5, O35404, O88399, O88400, O88401, O88402, O88403, O88404 | Gene names | Synj2, Kiaa0348 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-2 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 2). | |||||
|
PI5PA_MOUSE
|
||||||
| θ value | 1.15895e-47 (rank : 7) | NC score | 0.733109 (rank : 8) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P59644 | Gene names | Pib5pa | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
PI5PA_HUMAN
|
||||||
| θ value | 1.51363e-47 (rank : 8) | NC score | 0.663022 (rank : 12) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
SKIP_HUMAN
|
||||||
| θ value | 9.18288e-45 (rank : 9) | NC score | 0.799809 (rank : 3) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BT40, Q15733, Q9NPJ5, Q9P2R5 | Gene names | SKIP, PPS | |||
|
Domain Architecture |

|
|||||
| Description | Skeletal muscle and kidney-enriched inositol phosphatase (EC 3.1.3.56). | |||||
|
SKIP_MOUSE
|
||||||
| θ value | 7.77379e-44 (rank : 10) | NC score | 0.799139 (rank : 4) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8C5L6, O09040 | Gene names | Skip, Pps | |||
|
Domain Architecture |
|
|||||
| Description | Skeletal muscle and kidney-enriched inositol phosphatase (EC 3.1.3.56). | |||||
|
INP5E_MOUSE
|
||||||
| θ value | 5.22648e-32 (rank : 11) | NC score | 0.737969 (rank : 7) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JII1, Q3TCC9 | Gene names | Inpp5e | |||
|
Domain Architecture |

|
|||||
| Description | 72 kDa inositol polyphosphate 5-phosphatase (EC 3.1.3.36) (Phosphatidylinositol-4,5-bisphosphate 5-phosphatase) (Phosphatidylinositol polyphosphate 5-phosphatase type IV). | |||||
|
INP5E_HUMAN
|
||||||
| θ value | 4.14116e-29 (rank : 12) | NC score | 0.712754 (rank : 9) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NRR6, Q15734 | Gene names | INPP5E | |||
|
Domain Architecture |

|
|||||
| Description | 72 kDa inositol polyphosphate 5-phosphatase (EC 3.1.3.36) (Phosphatidylinositol-4,5-bisphosphate 5-phosphatase) (Phosphatidylinositol polyphosphate 5-phosphatase type IV). | |||||
|
RHG25_MOUSE
|
||||||
| θ value | 6.00763e-12 (rank : 13) | NC score | 0.385581 (rank : 15) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8BYW1, Q2VPR2, Q3TE04, Q3TVX0, Q3UVB7, Q6A0E0, Q8BX98 | Gene names | Arhgap25, Kiaa0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
RHG25_HUMAN
|
||||||
| θ value | 9.59137e-10 (rank : 14) | NC score | 0.368697 (rank : 18) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 550 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P42331, Q8IXQ2 | Gene names | ARHGAP25, KIAA0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
RHG08_HUMAN
|
||||||
| θ value | 1.06045e-08 (rank : 15) | NC score | 0.393435 (rank : 13) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9NSG0, O75983, O95695, Q96RW1, Q96RW2, Q9HA49, Q9HC46, Q9NVX8, Q9NXL1, Q9UH20 | Gene names | ARHGAP8 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 8. | |||||
|
RHG08_MOUSE
|
||||||
| θ value | 3.08544e-08 (rank : 16) | NC score | 0.392125 (rank : 14) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9CXP4, Q99JY7 | Gene names | Arhgap8 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 8. | |||||
|
FNBP2_HUMAN
|
||||||
| θ value | 4.0297e-08 (rank : 17) | NC score | 0.335023 (rank : 29) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O75044 | Gene names | SRGAP2, FNBP2, KIAA0456 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 2 (srGAP2) (Formin-binding protein 2). | |||||
|
SRGP2_HUMAN
|
||||||
| θ value | 4.45536e-07 (rank : 18) | NC score | 0.341083 (rank : 24) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O43295, Q8IX13, Q8IZV8 | Gene names | SRGAP3, ARHGAP14, KIAA0411, MEGAP, SRGAP2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Mental disorder- associated GAP) (Rho-GTPase-activating protein 14). | |||||
|
CHIO_HUMAN
|
||||||
| θ value | 5.81887e-07 (rank : 19) | NC score | 0.334800 (rank : 30) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P52757, Q75MM2 | Gene names | CHN2, ARHGAP3, BCH | |||
|
Domain Architecture |
|
|||||
| Description | Beta-chimaerin (Beta-chimerin) (Rho-GTPase-activating protein 3). | |||||
|
CHIO_MOUSE
|
||||||
| θ value | 5.81887e-07 (rank : 20) | NC score | 0.348004 (rank : 22) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q80XD1, Q9D9W2, Q9ER57 | Gene names | Chn2, Arhgap3, Bch | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-chimaerin (Beta-chimerin) (Rho-GTPase-activating protein 3). | |||||
|
RHG01_HUMAN
|
||||||
| θ value | 5.81887e-07 (rank : 21) | NC score | 0.373915 (rank : 16) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q07960 | Gene names | ARHGAP1, CDC42GAP, RHOGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 1 (GTPase-activating protein rhoOGAP) (Rho-related small GTPase protein activator) (CDC42 GTPase-activating protein) (p50-RhoGAP). | |||||
|
SRGP2_MOUSE
|
||||||
| θ value | 1.69304e-06 (rank : 22) | NC score | 0.337449 (rank : 27) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q812A2, Q80U09, Q8BKP4, Q8BLD0, Q925I2 | Gene names | Srgap3, Arhgap14, Kiaa0411, Srgap2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Rho-GTPase-activating protein 14). | |||||
|
3BP1_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 23) | NC score | 0.370565 (rank : 17) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9Y3L3, Q6IBZ2, Q6ZVL9, Q96HQ5, Q9NSQ9 | Gene names | SH3BP1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
RHG04_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 24) | NC score | 0.329730 (rank : 32) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P98171, Q14144 | Gene names | ARHGAP4, KIAA0131, RGC1, RHOGAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 4 (Rho-GAP hematopoietic protein C1) (p115). | |||||
|
CE005_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 25) | NC score | 0.309565 (rank : 43) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8K2H3 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Protein C5orf5 homolog. | |||||
|
GMIP_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 26) | NC score | 0.364019 (rank : 19) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9P107 | Gene names | GMIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GEM-interacting protein (GMIP). | |||||
|
K1688_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 27) | NC score | 0.321949 (rank : 39) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P59281, Q69ZD4, Q6P9R6 | Gene names | Kiaa1688, D15Wsu169e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1688. | |||||
|
K1688_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 28) | NC score | 0.319245 (rank : 40) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9C0H5 | Gene names | KIAA1688 | |||
|
Domain Architecture |

|
|||||
| Description | Protein KIAA1688. | |||||
|
RHG12_MOUSE
|
||||||
| θ value | 1.87187e-05 (rank : 29) | NC score | 0.347289 (rank : 23) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8C0D4, Q8BVP8 | Gene names | Arhgap12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
CE005_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 30) | NC score | 0.308982 (rank : 44) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9NYF5, Q6PGQ2, Q9P0I7 | Gene names | C5orf5 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C5orf5 (GAP-like protein N61). | |||||
|
P85A_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 31) | NC score | 0.203187 (rank : 67) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P27986 | Gene names | PIK3R1, GRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit alpha (PI3-kinase p85-subunit alpha) (PtdIns-3-kinase p85-alpha) (PI3K). | |||||
|
P85A_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 32) | NC score | 0.204564 (rank : 66) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P26450 | Gene names | Pik3r1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit alpha (PI3-kinase p85-subunit alpha) (PtdIns-3-kinase p85-alpha) (PI3K). | |||||
|
CHIN_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 33) | NC score | 0.324747 (rank : 37) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P15882, Q53SD6, Q53SH5, Q96FB0 | Gene names | CHN1, ARHGAP2, CHN | |||
|
Domain Architecture |

|
|||||
| Description | N-chimaerin (NC) (N-chimerin) (Alpha chimerin) (A-chimaerin) (Rho- GTPase-activating protein 2). | |||||
|
CHIN_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 34) | NC score | 0.323822 (rank : 38) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q91V57, Q3UGY3, Q7TQE5, Q8BWU6, Q9D9B3 | Gene names | Chn1, Arhgap2 | |||
|
Domain Architecture |

|
|||||
| Description | N-chimaerin (NC) (N-chimerin) (Alpha chimerin) (A-chimaerin) (Rho- GTPase-activating protein 2). | |||||
|
RHG12_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 35) | NC score | 0.329635 (rank : 33) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8IWW6, Q86UB3, Q8IWW7, Q8N3L1, Q9NT76 | Gene names | ARHGAP12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
RHG05_MOUSE
|
||||||
| θ value | 7.1131e-05 (rank : 36) | NC score | 0.324749 (rank : 36) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P97393 | Gene names | Arhgap5, Rhogap5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 37) | NC score | 0.307655 (rank : 45) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
TCGAP_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 38) | NC score | 0.339603 (rank : 26) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q80YF9, Q6P7W6 | Gene names | Snx26, Tcgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
SRGP1_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 39) | NC score | 0.326870 (rank : 35) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q7Z6B7, Q9H8A3, Q9P2P2 | Gene names | SRGAP1, ARHGAP13, KIAA1304 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 1 (srGAP1) (Rho-GTPase- activating protein 13). | |||||
|
3BP1_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 40) | NC score | 0.361254 (rank : 20) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P55194, Q99KK8 | Gene names | Sh3bp1, 3bp1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
GMIP_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 41) | NC score | 0.353932 (rank : 21) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q6PGG2, Q6P9S3 | Gene names | Gmip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GEM-interacting protein (GMIP). | |||||
|
GRLF1_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 42) | NC score | 0.329099 (rank : 34) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9NRY4, Q14452, Q9C0E1 | Gene names | GRLF1, GRF1, KIAA1722 | |||
|
Domain Architecture |

|
|||||
| Description | Glucocorticoid receptor DNA-binding factor 1 (Glucocorticoid receptor repression factor 1) (GRF-1) (Rho GAP p190A) (p190-A). | |||||
|
RHG07_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 43) | NC score | 0.265788 (rank : 56) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q96QB1, O14868, O43199, Q9C0E0 | Gene names | DLC1, ARHGAP7, KIAA1723, STARD12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 7 (Rho-type GTPase-activating protein 7) (Deleted in liver cancer 1 protein) (Dlc-1) (HP protein) (StAR-related lipid transfer protein 12) (StARD12) (START domain-containing protein 12). | |||||
|
BCR_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 44) | NC score | 0.292456 (rank : 48) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P11274, P78501, Q12842 | Gene names | BCR, BCR1 | |||
|
Domain Architecture |

|
|||||
| Description | Breakpoint cluster region protein (EC 2.7.11.1) (NY-REN-26 antigen). | |||||
|
MYO9B_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 45) | NC score | 0.174597 (rank : 70) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
RHG07_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 46) | NC score | 0.267366 (rank : 55) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9R0Z9, Q8R541 | Gene names | Dlc1, Arhgap7, Stard12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 7 (Rho-type GTPase-activating protein 7) (Deleted in liver cancer 1 protein homolog) (Dlc-1) (StAR-related lipid transfer protein 12) (StARD12) (START domain-containing protein 12). | |||||
|
RHG18_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 47) | NC score | 0.305335 (rank : 46) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8N392, Q58EZ3, Q6P679, Q6PJD7, Q96S64 | Gene names | ARHGAP18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho-GTPase-activating protein 18 (MacGAP). | |||||
|
RHG05_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 48) | NC score | 0.312426 (rank : 42) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q13017 | Gene names | ARHGAP5, RHOGAP5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
RHG09_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 49) | NC score | 0.340595 (rank : 25) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9BRR9, Q8TCJ3, Q8WYR0, Q96EZ2, Q96S74 | Gene names | ARHGAP9 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 9. | |||||
|
RHG18_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 50) | NC score | 0.313786 (rank : 41) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8K0Q5, Q8BP03, Q8R196 | Gene names | Arhgap18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho-GTPase-activating protein 18. | |||||
|
ABR_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 51) | NC score | 0.294687 (rank : 47) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q12979, Q13693, Q13694 | Gene names | ABR | |||
|
Domain Architecture |

|
|||||
| Description | Active breakpoint cluster region-related protein. | |||||
|
PRIO_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 52) | NC score | 0.088006 (rank : 76) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P04925 | Gene names | Prnp, Prn-p, Prp | |||
|
Domain Architecture |
|
|||||
| Description | Major prion protein precursor (PrP) (PrP27-30) (PrP33-35C) (CD230 antigen). | |||||
|
RHG06_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 53) | NC score | 0.336942 (rank : 28) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O43182, O43437, Q9P1B3, Q9UK81, Q9UK82 | Gene names | ARHGAP6, RHOGAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
MYO9B_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 54) | NC score | 0.167312 (rank : 73) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9QY06, Q9QY07, Q9QY08, Q9QY09 | Gene names | Myo9b, Myr5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
EVL_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 55) | NC score | 0.040580 (rank : 87) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70429, Q9ERU8 | Gene names | Evl | |||
|
Domain Architecture |

|
|||||
| Description | Ena/VASP-like protein (Ena/vasodilator-stimulated phosphoprotein- like). | |||||
|
RHG06_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 56) | NC score | 0.331914 (rank : 31) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O54834, Q9QZL8 | Gene names | Arhgap6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
STAR8_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 57) | NC score | 0.236760 (rank : 60) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q92502, Q5JST0 | Gene names | STARD8, KIAA0189 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
I5P1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 58) | NC score | 0.207708 (rank : 65) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14642, Q14640 | Gene names | INPP5A | |||
|
Domain Architecture |

|
|||||
| Description | Type I inositol-1,4,5-trisphosphate 5-phosphatase (EC 3.1.3.56) (5PTase). | |||||
|
STAR8_MOUSE
|
||||||
| θ value | 0.279714 (rank : 59) | NC score | 0.235827 (rank : 61) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8K031, Q3UZC7, Q6A0A8, Q6P5E0, Q8R3X8 | Gene names | Stard8, Kiaa0189 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
PRIO_HUMAN
|
||||||
| θ value | 0.813845 (rank : 60) | NC score | 0.063056 (rank : 78) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P04156, O60489, P78446, Q15216, Q15221, Q8TBG0, Q96E70, Q9UP19 | Gene names | PRNP, PRP | |||
|
Domain Architecture |
|
|||||
| Description | Major prion protein precursor (PrP) (PrP27-30) (PrP33-35C) (ASCR) (CD230 antigen). | |||||
|
CEND2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 61) | NC score | 0.210254 (rank : 64) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q96P48, O94879, Q59FI7, Q6PHS3, Q8WU51, Q96HP6, Q96L71 | Gene names | CENTD2, ARAP1, KIAA0782 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-delta 2 (Cnt-d2) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 1). | |||||
|
EVL_HUMAN
|
||||||
| θ value | 1.06291 (rank : 62) | NC score | 0.036641 (rank : 88) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UI08, O95884, Q7Z522, Q8TBV1, Q9UF25, Q9UIC2 | Gene names | EVL, RNB6 | |||
|
Domain Architecture |

|
|||||
| Description | Ena/VASP-like protein (Ena/vasodilator-stimulated phosphoprotein- like). | |||||
|
HS90B_MOUSE
|
||||||
| θ value | 1.38821 (rank : 63) | NC score | 0.014264 (rank : 93) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P11499, O89078 | Gene names | Hsp90ab1, Hsp84, Hsp84-1, Hspcb | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-beta (HSP 84) (Tumor-specific transplantation 84 kDa antigen) (TSTA). | |||||
|
TAGAP_HUMAN
|
||||||
| θ value | 1.81305 (rank : 64) | NC score | 0.274059 (rank : 54) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8N103, Q2NKM8, Q8NI40, Q96KZ2, Q96QA2 | Gene names | TAGAP, TAGAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell activation Rho GTPase-activating protein (T-cell activation GTPase-activating protein). | |||||
|
ACADS_MOUSE
|
||||||
| θ value | 2.36792 (rank : 65) | NC score | 0.009588 (rank : 98) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q07417 | Gene names | Acads | |||
|
Domain Architecture |
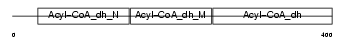
|
|||||
| Description | Short-chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.2) (SCAD) (Butyryl-CoA dehydrogenase). | |||||
|
CN037_HUMAN
|
||||||
| θ value | 2.36792 (rank : 66) | NC score | 0.020010 (rank : 90) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86TY3, Q6P5Q1, Q86TY1 | Gene names | C14orf37 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf37 precursor. | |||||
|
MLL4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 67) | NC score | 0.022899 (rank : 89) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
HORN_HUMAN
|
||||||
| θ value | 3.0926 (rank : 68) | NC score | 0.011495 (rank : 96) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86YZ3 | Gene names | HRNR, S100A18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hornerin. | |||||
|
AK1D1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.008396 (rank : 99) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VCX1 | Gene names | Akr1d1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 3-oxo-5-beta-steroid 4-dehydrogenase (EC 1.3.99.6) (Delta(4)-3- ketosteroid 5-beta-reductase) (Aldo-keto reductase family 1 member D1). | |||||
|
GPR12_MOUSE
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.000238 (rank : 105) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 731 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P35412 | Gene names | Gpr12, Gpcr12 | |||
|
Domain Architecture |
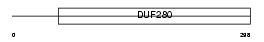
|
|||||
| Description | Probable G-protein coupled receptor 12 (GPCR01). | |||||
|
IRX3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.007514 (rank : 100) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P81067 | Gene names | Irx3, Irxb1 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-3 (Iroquois homeobox protein 3) (Homeodomain protein IRXB1). | |||||
|
DLG7_MOUSE
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.013344 (rank : 94) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K4R9, Q6ZQL3, Q80VS7, Q8BUC8, Q8BZ79 | Gene names | Dlg7, Kiaa0008 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Discs large homolog 7 (Hepatoma up-regulated protein homolog) (HURP). | |||||
|
DPOE1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.011659 (rank : 95) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q07864, Q13533, Q86VH9 | Gene names | POLE, POLE1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase epsilon, catalytic subunit A (EC 2.7.7.7) (DNA polymerase II subunit A). | |||||
|
LYSM1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.015178 (rank : 92) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D0E3 | Gene names | Lysmd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LysM and putative peptidoglycan-binding domain-containing protein 1. | |||||
|
M3K12_HUMAN
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | -0.003531 (rank : 107) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12852, Q86VQ5, Q8WY25 | Gene names | MAP3K12, ZPK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 12 (EC 2.7.11.25) (Mixed lineage kinase) (Leucine-zipper protein kinase) (ZPK) (Dual leucine zipper bearing kinase) (DLK) (MAPK-upstream kinase) (MUK). | |||||
|
OPHN1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.276923 (rank : 52) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q99J31, Q544K7 | Gene names | Ophn1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
RHG26_HUMAN
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.249299 (rank : 57) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9UNA1, O75117, Q9BYS6, Q9BYS7, Q9UJ00 | Gene names | ARHGAP26, GRAF, KIAA0621, OPHN1L | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 26 (Oligophrenin-1-like protein) (GTPase regulator associated with focal adhesion kinase). | |||||
|
TAGAP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.274182 (rank : 53) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8K2L9, Q920D6 | Gene names | Tagap, Tagap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell activation Rho GTPase-activating protein (T-cell activation GTPase-activating protein). | |||||
|
OPHN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.278473 (rank : 50) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O60890, Q5JQ81, Q8WX47 | Gene names | OPHN1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
AK1D1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.006579 (rank : 102) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P51857 | Gene names | AKR1D1, SRD5B1 | |||
|
Domain Architecture |
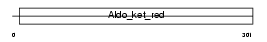
|
|||||
| Description | 3-oxo-5-beta-steroid 4-dehydrogenase (EC 1.3.1.3) (Delta(4)-3- ketosteroid 5-beta-reductase) (Aldo-keto reductase family 1 member D1). | |||||
|
ANR12_HUMAN
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | -0.000359 (rank : 106) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6UB98, O94951, Q658K1, Q6QMF7, Q9H231, Q9H784 | Gene names | ANKRD12, ANCO2, KIAA0874 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 12 (Ankyrin repeat-containing cofactor 2) (GAC-1 protein). | |||||
|
C8AP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.009996 (rank : 97) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 548 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UKL3, Q6PH76, Q7LCQ7, Q86YD9, Q9NUQ4, Q9NZV9, Q9P2N1, Q9Y563 | Gene names | CASP8AP2, FLASH, KIAA1315, RIP25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
DMP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.004562 (rank : 104) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
EPIPL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.005799 (rank : 103) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P58107, Q76E58 | Gene names | EPPK1, EPIPL | |||
|
Domain Architecture |

|
|||||
| Description | Epiplakin (450 kDa epidermal antigen). | |||||
|
HDGR3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.007386 (rank : 101) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JMG7, Q3TRX2, Q8BQ69, Q8BR62, Q9D2M7 | Gene names | Hdgfrp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatoma-derived growth factor-related protein 3 (HRP-3). | |||||
|
STAM2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.015306 (rank : 91) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O88811, Q8C8Y4 | Gene names | Stam2, Hbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 2 (STAM-2) (Hrs-binding protein). | |||||
|
CEND1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.147613 (rank : 75) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8WZ64, Q4W5D2, Q7Z2L5, Q96L70, Q96P49, Q9Y4E4 | Gene names | CENTD1, ARAP2, KIAA0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 2) (PARX protein). | |||||
|
CEND1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.155015 (rank : 74) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8BZ05, Q80TX2, Q8C3T2, Q8VEL6 | Gene names | Centd1, Kiaa0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1). | |||||
|
CEND3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.167990 (rank : 72) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8WWN8 | Gene names | CENTD3, ARAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3). | |||||
|
CEND3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.172722 (rank : 71) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8R5G7, Q5DTN4, Q6NVF1, Q8R5G6 | Gene names | Centd3, Arap3, Drag1, Kiaa4097 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3) (Dual specificity Rho- and Arf-GTPase-activating protein 1). | |||||
|
CF155_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.063566 (rank : 77) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H8W2, Q5TG13 | Gene names | C6orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf155. | |||||
|
CK024_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.055224 (rank : 84) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
CO039_MOUSE
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.053762 (rank : 85) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q3TEI4, Q3TMF2, Q3U253, Q3UM75, Q9DA93 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39 homolog. | |||||
|
MDC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.055519 (rank : 83) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.057862 (rank : 81) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
NACAM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.056641 (rank : 82) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.050366 (rank : 86) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
P85B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.058722 (rank : 80) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O00459, Q9UPH9 | Gene names | PIK3R2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit beta (PI3-kinase p85- subunit beta) (PtdIns-3-kinase p85-beta). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.060211 (rank : 79) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
RBP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.240292 (rank : 59) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q15311 | Gene names | RALBP1, RLIP1, RLIP76 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (76 kDa Ral-interacting protein) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
RBP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.245931 (rank : 58) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q62172, Q9CRE5 | Gene names | Ralbp1, Rip1 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
RGAP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.277160 (rank : 51) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9H0H5, Q6PJ26, Q9NWN2, Q9P250, Q9P2W2 | Gene names | RACGAP1, KIAA1478, MGCRACGAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rac GTPase-activating protein 1 (MgcRacGAP). | |||||
|
RGAP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.278712 (rank : 49) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9WVM1, Q3THR5, Q3TI41, Q3TM81 | Gene names | Racgap1, Mgcracgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rac GTPase-activating protein 1 (MgcRacGAP). | |||||
|
SAC3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.200568 (rank : 69) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92562, Q53H49, Q5TCS6 | Gene names | SAC3, KIAA0274 | |||
|
Domain Architecture |
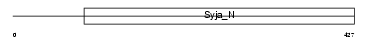
|
|||||
| Description | SAC domain-containing protein 3. | |||||
|
SAC3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.201871 (rank : 68) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91WF7, Q6A092 | Gene names | Sac3, Kiaa0274 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAC domain-containing protein 3. | |||||
|
STA13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.219801 (rank : 62) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9Y3M8, Q5HYH1, Q5TAE3, Q6UN61, Q86TP6, Q86WQ3, Q86XT1 | Gene names | STARD13, DLC2, GT650 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 13 (StARD13) (START domain- containing protein 13) (46H23.2) (Deleted in liver cancer protein 2) (Rho GTPase-activating protein). | |||||
|
STA13_MOUSE
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.214569 (rank : 63) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q923Q2, Q8K369 | Gene names | Stard13 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 13 (StARD13) (START domain- containing protein 13). | |||||
|
I5P2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P32019, Q5VSG9, Q5VSH0, Q5VSH1, Q658Q5, Q6P6D4, Q6PD53, Q86YE1 | Gene names | INPP5B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Type II inositol-1,4,5-trisphosphate 5-phosphatase precursor (EC 3.1.3.36) (Phosphoinositide 5-phosphatase) (5PTase) (75 kDa inositol polyphosphate-5-phosphatase). | |||||
|
OCRL_HUMAN
|
||||||
| NC score | 0.975892 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q01968, O60800, Q15684, Q15774, Q5JQF1, Q5JQF2, Q9UJG5, Q9UMA5 | Gene names | OCRL, INPP5F, OCRL1 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol polyphosphate 5-phosphatase OCRL-1 (EC 3.1.3.36) (Lowe oculocerebrorenal syndrome protein). | |||||
|
SKIP_HUMAN
|
||||||
| NC score | 0.799809 (rank : 3) | θ value | 9.18288e-45 (rank : 9) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BT40, Q15733, Q9NPJ5, Q9P2R5 | Gene names | SKIP, PPS | |||
|
Domain Architecture |

|
|||||
| Description | Skeletal muscle and kidney-enriched inositol phosphatase (EC 3.1.3.56). | |||||
|
SKIP_MOUSE
|
||||||
| NC score | 0.799139 (rank : 4) | θ value | 7.77379e-44 (rank : 10) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8C5L6, O09040 | Gene names | Skip, Pps | |||
|
Domain Architecture |
|
|||||
| Description | Skeletal muscle and kidney-enriched inositol phosphatase (EC 3.1.3.56). | |||||
|
SYNJ2_MOUSE
|
||||||
| NC score | 0.758213 (rank : 5) | θ value | 1.32293e-51 (rank : 6) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9D2G5, O35404, O88399, O88400, O88401, O88402, O88403, O88404 | Gene names | Synj2, Kiaa0348 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-2 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 2). | |||||
|
SYNJ2_HUMAN
|
||||||
| NC score | 0.751462 (rank : 6) | θ value | 4.11246e-53 (rank : 5) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O15056, Q5TA13, Q5TA16, Q5TA19, Q86XK0, Q8IZA8, Q9H226 | Gene names | SYNJ2, KIAA0348 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptojanin-2 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 2). | |||||
|
INP5E_MOUSE
|
||||||
| NC score | 0.737969 (rank : 7) | θ value | 5.22648e-32 (rank : 11) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JII1, Q3TCC9 | Gene names | Inpp5e | |||
|
Domain Architecture |

|
|||||
| Description | 72 kDa inositol polyphosphate 5-phosphatase (EC 3.1.3.36) (Phosphatidylinositol-4,5-bisphosphate 5-phosphatase) (Phosphatidylinositol polyphosphate 5-phosphatase type IV). | |||||
|
PI5PA_MOUSE
|
||||||
| NC score | 0.733109 (rank : 8) | θ value | 1.15895e-47 (rank : 7) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P59644 | Gene names | Pib5pa | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
INP5E_HUMAN
|
||||||
| NC score | 0.712754 (rank : 9) | θ value | 4.14116e-29 (rank : 12) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NRR6, Q15734 | Gene names | INPP5E | |||
|
Domain Architecture |

|
|||||
| Description | 72 kDa inositol polyphosphate 5-phosphatase (EC 3.1.3.36) (Phosphatidylinositol-4,5-bisphosphate 5-phosphatase) (Phosphatidylinositol polyphosphate 5-phosphatase type IV). | |||||
|
SYNJ1_HUMAN
|
||||||
| NC score | 0.691745 (rank : 10) | θ value | 1.2784e-54 (rank : 4) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O43426, O43425, O94984 | Gene names | SYNJ1, KIAA0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
SYNJ1_MOUSE
|
||||||
| NC score | 0.690844 (rank : 11) | θ value | 1.15626e-55 (rank : 3) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
PI5PA_HUMAN
|
||||||
| NC score | 0.663022 (rank : 12) | θ value | 1.51363e-47 (rank : 8) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
RHG08_HUMAN
|
||||||
| NC score | 0.393435 (rank : 13) | θ value | 1.06045e-08 (rank : 15) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9NSG0, O75983, O95695, Q96RW1, Q96RW2, Q9HA49, Q9HC46, Q9NVX8, Q9NXL1, Q9UH20 | Gene names | ARHGAP8 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 8. | |||||
|
RHG08_MOUSE
|
||||||
| NC score | 0.392125 (rank : 14) | θ value | 3.08544e-08 (rank : 16) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9CXP4, Q99JY7 | Gene names | Arhgap8 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 8. | |||||
|
RHG25_MOUSE
|
||||||
| NC score | 0.385581 (rank : 15) | θ value | 6.00763e-12 (rank : 13) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8BYW1, Q2VPR2, Q3TE04, Q3TVX0, Q3UVB7, Q6A0E0, Q8BX98 | Gene names | Arhgap25, Kiaa0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
RHG01_HUMAN
|
||||||
| NC score | 0.373915 (rank : 16) | θ value | 5.81887e-07 (rank : 21) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q07960 | Gene names | ARHGAP1, CDC42GAP, RHOGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 1 (GTPase-activating protein rhoOGAP) (Rho-related small GTPase protein activator) (CDC42 GTPase-activating protein) (p50-RhoGAP). | |||||
|
3BP1_HUMAN
|
||||||
| NC score | 0.370565 (rank : 17) | θ value | 4.92598e-06 (rank : 23) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9Y3L3, Q6IBZ2, Q6ZVL9, Q96HQ5, Q9NSQ9 | Gene names | SH3BP1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
RHG25_HUMAN
|
||||||
| NC score | 0.368697 (rank : 18) | θ value | 9.59137e-10 (rank : 14) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 550 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P42331, Q8IXQ2 | Gene names | ARHGAP25, KIAA0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
GMIP_HUMAN
|
||||||
| NC score | 0.364019 (rank : 19) | θ value | 1.43324e-05 (rank : 26) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9P107 | Gene names | GMIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GEM-interacting protein (GMIP). | |||||
|
3BP1_MOUSE
|
||||||
| NC score | 0.361254 (rank : 20) | θ value | 0.00035302 (rank : 40) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P55194, Q99KK8 | Gene names | Sh3bp1, 3bp1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
GMIP_MOUSE
|
||||||
| NC score | 0.353932 (rank : 21) | θ value | 0.00035302 (rank : 41) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q6PGG2, Q6P9S3 | Gene names | Gmip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GEM-interacting protein (GMIP). | |||||
|
CHIO_MOUSE
|
||||||
| NC score | 0.348004 (rank : 22) | θ value | 5.81887e-07 (rank : 20) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q80XD1, Q9D9W2, Q9ER57 | Gene names | Chn2, Arhgap3, Bch | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-chimaerin (Beta-chimerin) (Rho-GTPase-activating protein 3). | |||||
|
RHG12_MOUSE
|
||||||
| NC score | 0.347289 (rank : 23) | θ value | 1.87187e-05 (rank : 29) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8C0D4, Q8BVP8 | Gene names | Arhgap12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
SRGP2_HUMAN
|
||||||
| NC score | 0.341083 (rank : 24) | θ value | 4.45536e-07 (rank : 18) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O43295, Q8IX13, Q8IZV8 | Gene names | SRGAP3, ARHGAP14, KIAA0411, MEGAP, SRGAP2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Mental disorder- associated GAP) (Rho-GTPase-activating protein 14). | |||||
|
RHG09_HUMAN
|
||||||
| NC score | 0.340595 (rank : 25) | θ value | 0.00869519 (rank : 49) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9BRR9, Q8TCJ3, Q8WYR0, Q96EZ2, Q96S74 | Gene names | ARHGAP9 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 9. | |||||
|
TCGAP_MOUSE
|
||||||
| NC score | 0.339603 (rank : 26) | θ value | 9.29e-05 (rank : 38) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q80YF9, Q6P7W6 | Gene names | Snx26, Tcgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
SRGP2_MOUSE
|
||||||
| NC score | 0.337449 (rank : 27) | θ value | 1.69304e-06 (rank : 22) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q812A2, Q80U09, Q8BKP4, Q8BLD0, Q925I2 | Gene names | Srgap3, Arhgap14, Kiaa0411, Srgap2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Rho-GTPase-activating protein 14). | |||||
|
RHG06_HUMAN
|
||||||
| NC score | 0.336942 (rank : 28) | θ value | 0.0113563 (rank : 53) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O43182, O43437, Q9P1B3, Q9UK81, Q9UK82 | Gene names | ARHGAP6, RHOGAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
FNBP2_HUMAN
|
||||||
| NC score | 0.335023 (rank : 29) | θ value | 4.0297e-08 (rank : 17) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O75044 | Gene names | SRGAP2, FNBP2, KIAA0456 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 2 (srGAP2) (Formin-binding protein 2). | |||||
|
CHIO_HUMAN
|
||||||
| NC score | 0.334800 (rank : 30) | θ value | 5.81887e-07 (rank : 19) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P52757, Q75MM2 | Gene names | CHN2, ARHGAP3, BCH | |||
|
Domain Architecture |
|
|||||
| Description | Beta-chimaerin (Beta-chimerin) (Rho-GTPase-activating protein 3). | |||||
|
RHG06_MOUSE
|
||||||
| NC score | 0.331914 (rank : 31) | θ value | 0.0563607 (rank : 56) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O54834, Q9QZL8 | Gene names | Arhgap6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
RHG04_HUMAN
|
||||||
| NC score | 0.329730 (rank : 32) | θ value | 6.43352e-06 (rank : 24) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P98171, Q14144 | Gene names | ARHGAP4, KIAA0131, RGC1, RHOGAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 4 (Rho-GAP hematopoietic protein C1) (p115). | |||||
|
RHG12_HUMAN
|
||||||
| NC score | 0.329635 (rank : 33) | θ value | 4.1701e-05 (rank : 35) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8IWW6, Q86UB3, Q8IWW7, Q8N3L1, Q9NT76 | Gene names | ARHGAP12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
GRLF1_HUMAN
|
||||||
| NC score | 0.329099 (rank : 34) | θ value | 0.00035302 (rank : 42) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9NRY4, Q14452, Q9C0E1 | Gene names | GRLF1, GRF1, KIAA1722 | |||
|
Domain Architecture |

|
|||||
| Description | Glucocorticoid receptor DNA-binding factor 1 (Glucocorticoid receptor repression factor 1) (GRF-1) (Rho GAP p190A) (p190-A). | |||||
|
SRGP1_HUMAN
|
||||||
| NC score | 0.326870 (rank : 35) | θ value | 0.000121331 (rank : 39) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q7Z6B7, Q9H8A3, Q9P2P2 | Gene names | SRGAP1, ARHGAP13, KIAA1304 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 1 (srGAP1) (Rho-GTPase- activating protein 13). | |||||
|
RHG05_MOUSE
|
||||||
| NC score | 0.324749 (rank : 36) | θ value | 7.1131e-05 (rank : 36) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P97393 | Gene names | Arhgap5, Rhogap5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
CHIN_HUMAN
|
||||||
| NC score | 0.324747 (rank : 37) | θ value | 4.1701e-05 (rank : 33) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P15882, Q53SD6, Q53SH5, Q96FB0 | Gene names | CHN1, ARHGAP2, CHN | |||
|
Domain Architecture |

|
|||||
| Description | N-chimaerin (NC) (N-chimerin) (Alpha chimerin) (A-chimaerin) (Rho- GTPase-activating protein 2). | |||||
|
CHIN_MOUSE
|
||||||
| NC score | 0.323822 (rank : 38) | θ value | 4.1701e-05 (rank : 34) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q91V57, Q3UGY3, Q7TQE5, Q8BWU6, Q9D9B3 | Gene names | Chn1, Arhgap2 | |||
|
Domain Architecture |

|
|||||
| Description | N-chimaerin (NC) (N-chimerin) (Alpha chimerin) (A-chimaerin) (Rho- GTPase-activating protein 2). | |||||
|
K1688_MOUSE
|
||||||
| NC score | 0.321949 (rank : 39) | θ value | 1.43324e-05 (rank : 27) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P59281, Q69ZD4, Q6P9R6 | Gene names | Kiaa1688, D15Wsu169e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1688. | |||||
|
K1688_HUMAN
|
||||||
| NC score | 0.319245 (rank : 40) | θ value | 1.87187e-05 (rank : 28) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9C0H5 | Gene names | KIAA1688 | |||
|
Domain Architecture |

|
|||||
| Description | Protein KIAA1688. | |||||
|
RHG18_MOUSE
|
||||||
| NC score | 0.313786 (rank : 41) | θ value | 0.00869519 (rank : 50) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8K0Q5, Q8BP03, Q8R196 | Gene names | Arhgap18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho-GTPase-activating protein 18. | |||||
|
RHG05_HUMAN
|
||||||
| NC score | 0.312426 (rank : 42) | θ value | 0.00665767 (rank : 48) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q13017 | Gene names | ARHGAP5, RHOGAP5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
CE005_MOUSE
|
||||||
| NC score | 0.309565 (rank : 43) | θ value | 1.43324e-05 (rank : 25) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8K2H3 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Protein C5orf5 homolog. | |||||
|
CE005_HUMAN
|
||||||
| NC score | 0.308982 (rank : 44) | θ value | 2.44474e-05 (rank : 30) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9NYF5, Q6PGQ2, Q9P0I7 | Gene names | C5orf5 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C5orf5 (GAP-like protein N61). | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.307655 (rank : 45) | θ value | 9.29e-05 (rank : 37) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
RHG18_HUMAN
|
||||||
| NC score | 0.305335 (rank : 46) | θ value | 0.00390308 (rank : 47) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8N392, Q58EZ3, Q6P679, Q6PJD7, Q96S64 | Gene names | ARHGAP18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho-GTPase-activating protein 18 (MacGAP). | |||||
|
ABR_HUMAN
|
||||||
| NC score | 0.294687 (rank : 47) | θ value | 0.0113563 (rank : 51) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q12979, Q13693, Q13694 | Gene names | ABR | |||
|
Domain Architecture |

|
|||||
| Description | Active breakpoint cluster region-related protein. | |||||
|
BCR_HUMAN
|
||||||
| NC score | 0.292456 (rank : 48) | θ value | 0.00228821 (rank : 44) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P11274, P78501, Q12842 | Gene names | BCR, BCR1 | |||
|
Domain Architecture |

|
|||||
| Description | Breakpoint cluster region protein (EC 2.7.11.1) (NY-REN-26 antigen). | |||||
|
RGAP1_MOUSE
|
||||||
| NC score | 0.278712 (rank : 49) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9WVM1, Q3THR5, Q3TI41, Q3TM81 | Gene names | Racgap1, Mgcracgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rac GTPase-activating protein 1 (MgcRacGAP). | |||||
|
OPHN1_HUMAN
|
||||||
| NC score | 0.278473 (rank : 50) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O60890, Q5JQ81, Q8WX47 | Gene names | OPHN1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
RGAP1_HUMAN
|
||||||
| NC score | 0.277160 (rank : 51) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9H0H5, Q6PJ26, Q9NWN2, Q9P250, Q9P2W2 | Gene names | RACGAP1, KIAA1478, MGCRACGAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rac GTPase-activating protein 1 (MgcRacGAP). | |||||
|
OPHN1_MOUSE
|
||||||
| NC score | 0.276923 (rank : 52) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q99J31, Q544K7 | Gene names | Ophn1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
TAGAP_MOUSE
|
||||||
| NC score | 0.274182 (rank : 53) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8K2L9, Q920D6 | Gene names | Tagap, Tagap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell activation Rho GTPase-activating protein (T-cell activation GTPase-activating protein). | |||||
|
TAGAP_HUMAN
|
||||||
| NC score | 0.274059 (rank : 54) | θ value | 1.81305 (rank : 64) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8N103, Q2NKM8, Q8NI40, Q96KZ2, Q96QA2 | Gene names | TAGAP, TAGAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell activation Rho GTPase-activating protein (T-cell activation GTPase-activating protein). | |||||
|
RHG07_MOUSE
|
||||||
| NC score | 0.267366 (rank : 55) | θ value | 0.00390308 (rank : 46) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9R0Z9, Q8R541 | Gene names | Dlc1, Arhgap7, Stard12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 7 (Rho-type GTPase-activating protein 7) (Deleted in liver cancer 1 protein homolog) (Dlc-1) (StAR-related lipid transfer protein 12) (StARD12) (START domain-containing protein 12). | |||||
|
RHG07_HUMAN
|
||||||
| NC score | 0.265788 (rank : 56) | θ value | 0.00175202 (rank : 43) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q96QB1, O14868, O43199, Q9C0E0 | Gene names | DLC1, ARHGAP7, KIAA1723, STARD12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 7 (Rho-type GTPase-activating protein 7) (Deleted in liver cancer 1 protein) (Dlc-1) (HP protein) (StAR-related lipid transfer protein 12) (StARD12) (START domain-containing protein 12). | |||||
|
RHG26_HUMAN
|
||||||
| NC score | 0.249299 (rank : 57) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9UNA1, O75117, Q9BYS6, Q9BYS7, Q9UJ00 | Gene names | ARHGAP26, GRAF, KIAA0621, OPHN1L | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 26 (Oligophrenin-1-like protein) (GTPase regulator associated with focal adhesion kinase). | |||||
|
RBP1_MOUSE
|
||||||
| NC score | 0.245931 (rank : 58) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q62172, Q9CRE5 | Gene names | Ralbp1, Rip1 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
RBP1_HUMAN
|
||||||
| NC score | 0.240292 (rank : 59) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q15311 | Gene names | RALBP1, RLIP1, RLIP76 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (76 kDa Ral-interacting protein) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
STAR8_HUMAN
|
||||||
| NC score | 0.236760 (rank : 60) | θ value | 0.0736092 (rank : 57) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q92502, Q5JST0 | Gene names | STARD8, KIAA0189 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
STAR8_MOUSE
|
||||||
| NC score | 0.235827 (rank : 61) | θ value | 0.279714 (rank : 59) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8K031, Q3UZC7, Q6A0A8, Q6P5E0, Q8R3X8 | Gene names | Stard8, Kiaa0189 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
STA13_HUMAN
|
||||||
| NC score | 0.219801 (rank : 62) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9Y3M8, Q5HYH1, Q5TAE3, Q6UN61, Q86TP6, Q86WQ3, Q86XT1 | Gene names | STARD13, DLC2, GT650 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 13 (StARD13) (START domain- containing protein 13) (46H23.2) (Deleted in liver cancer protein 2) (Rho GTPase-activating protein). | |||||
|
STA13_MOUSE
|
||||||
| NC score | 0.214569 (rank : 63) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q923Q2, Q8K369 | Gene names | Stard13 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 13 (StARD13) (START domain- containing protein 13). | |||||
|
CEND2_HUMAN
|
||||||
| NC score | 0.210254 (rank : 64) | θ value | 1.06291 (rank : 61) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q96P48, O94879, Q59FI7, Q6PHS3, Q8WU51, Q96HP6, Q96L71 | Gene names | CENTD2, ARAP1, KIAA0782 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-delta 2 (Cnt-d2) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 1). | |||||
|
I5P1_HUMAN
|
||||||
| NC score | 0.207708 (rank : 65) | θ value | 0.0961366 (rank : 58) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14642, Q14640 | Gene names | INPP5A | |||
|
Domain Architecture |

|
|||||
| Description | Type I inositol-1,4,5-trisphosphate 5-phosphatase (EC 3.1.3.56) (5PTase). | |||||
|
P85A_MOUSE
|
||||||
| NC score | 0.204564 (rank : 66) | θ value | 3.19293e-05 (rank : 32) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P26450 | Gene names | Pik3r1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit alpha (PI3-kinase p85-subunit alpha) (PtdIns-3-kinase p85-alpha) (PI3K). | |||||
|
P85A_HUMAN
|
||||||
| NC score | 0.203187 (rank : 67) | θ value | 3.19293e-05 (rank : 31) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P27986 | Gene names | PIK3R1, GRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit alpha (PI3-kinase p85-subunit alpha) (PtdIns-3-kinase p85-alpha) (PI3K). | |||||
|
SAC3_MOUSE
|
||||||
| NC score | 0.201871 (rank : 68) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91WF7, Q6A092 | Gene names | Sac3, Kiaa0274 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAC domain-containing protein 3. | |||||
|
SAC3_HUMAN
|
||||||
| NC score | 0.200568 (rank : 69) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92562, Q53H49, Q5TCS6 | Gene names | SAC3, KIAA0274 | |||
|
Domain Architecture |

|
|||||
| Description | SAC domain-containing protein 3. | |||||
|
MYO9B_HUMAN
|
||||||
| NC score | 0.174597 (rank : 70) | θ value | 0.00390308 (rank : 45) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
CEND3_MOUSE
|
||||||
| NC score | 0.172722 (rank : 71) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8R5G7, Q5DTN4, Q6NVF1, Q8R5G6 | Gene names | Centd3, Arap3, Drag1, Kiaa4097 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3) (Dual specificity Rho- and Arf-GTPase-activating protein 1). | |||||
|
CEND3_HUMAN
|
||||||
| NC score | 0.167990 (rank : 72) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8WWN8 | Gene names | CENTD3, ARAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3). | |||||
|
MYO9B_MOUSE
|
||||||
| NC score | 0.167312 (rank : 73) | θ value | 0.0252991 (rank : 54) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9QY06, Q9QY07, Q9QY08, Q9QY09 | Gene names | Myo9b, Myr5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
CEND1_MOUSE
|
||||||
| NC score | 0.155015 (rank : 74) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8BZ05, Q80TX2, Q8C3T2, Q8VEL6 | Gene names | Centd1, Kiaa0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1). | |||||
|
CEND1_HUMAN
|
||||||
| NC score | 0.147613 (rank : 75) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8WZ64, Q4W5D2, Q7Z2L5, Q96L70, Q96P49, Q9Y4E4 | Gene names | CENTD1, ARAP2, KIAA0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 2) (PARX protein). | |||||
|
PRIO_MOUSE
|
||||||
| NC score | 0.088006 (rank : 76) | θ value | 0.0113563 (rank : 52) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P04925 | Gene names | Prnp, Prn-p, Prp | |||
|
Domain Architecture |

|
|||||
| Description | Major prion protein precursor (PrP) (PrP27-30) (PrP33-35C) (CD230 antigen). | |||||
|
CF155_HUMAN
|
||||||
| NC score | 0.063566 (rank : 77) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H8W2, Q5TG13 | Gene names | C6orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf155. | |||||
|
PRIO_HUMAN
|
||||||
| NC score | 0.063056 (rank : 78) | θ value | 0.813845 (rank : 60) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P04156, O60489, P78446, Q15216, Q15221, Q8TBG0, Q96E70, Q9UP19 | Gene names | PRNP, PRP | |||
|
Domain Architecture |

|
|||||
| Description | Major prion protein precursor (PrP) (PrP27-30) (PrP33-35C) (ASCR) (CD230 antigen). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.060211 (rank : 79) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
P85B_HUMAN
|
||||||
| NC score | 0.058722 (rank : 80) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O00459, Q9UPH9 | Gene names | PIK3R2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit beta (PI3-kinase p85- subunit beta) (PtdIns-3-kinase p85-beta). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.057862 (rank : 81) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.056641 (rank : 82) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.055519 (rank : 83) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
CK024_HUMAN
|
||||||
| NC score | 0.055224 (rank : 84) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
CO039_MOUSE
|
||||||
| NC score | 0.053762 (rank : 85) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q3TEI4, Q3TMF2, Q3U253, Q3UM75, Q9DA93 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39 homolog. | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.050366 (rank : 86) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
EVL_MOUSE
|
||||||
| NC score | 0.040580 (rank : 87) | θ value | 0.0563607 (rank : 55) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70429, Q9ERU8 | Gene names | Evl | |||
|
Domain Architecture |

|
|||||
| Description | Ena/VASP-like protein (Ena/vasodilator-stimulated phosphoprotein- like). | |||||
|
EVL_HUMAN
|
||||||
| NC score | 0.036641 (rank : 88) | θ value | 1.06291 (rank : 62) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UI08, O95884, Q7Z522, Q8TBV1, Q9UF25, Q9UIC2 | Gene names | EVL, RNB6 | |||
|
Domain Architecture |

|
|||||
| Description | Ena/VASP-like protein (Ena/vasodilator-stimulated phosphoprotein- like). | |||||
|
MLL4_HUMAN
|
||||||
| NC score | 0.022899 (rank : 89) | θ value | 2.36792 (rank : 67) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
CN037_HUMAN
|
||||||
| NC score | 0.020010 (rank : 90) | θ value | 2.36792 (rank : 66) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86TY3, Q6P5Q1, Q86TY1 | Gene names | C14orf37 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf37 precursor. | |||||
|
STAM2_MOUSE
|
||||||
| NC score | 0.015306 (rank : 91) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O88811, Q8C8Y4 | Gene names | Stam2, Hbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 2 (STAM-2) (Hrs-binding protein). | |||||
|
LYSM1_MOUSE
|
||||||
| NC score | 0.015178 (rank : 92) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D0E3 | Gene names | Lysmd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LysM and putative peptidoglycan-binding domain-containing protein 1. | |||||
|
HS90B_MOUSE
|
||||||
| NC score | 0.014264 (rank : 93) | θ value | 1.38821 (rank : 63) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P11499, O89078 | Gene names | Hsp90ab1, Hsp84, Hsp84-1, Hspcb | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-beta (HSP 84) (Tumor-specific transplantation 84 kDa antigen) (TSTA). | |||||
|
DLG7_MOUSE
|
||||||
| NC score | 0.013344 (rank : 94) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K4R9, Q6ZQL3, Q80VS7, Q8BUC8, Q8BZ79 | Gene names | Dlg7, Kiaa0008 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Discs large homolog 7 (Hepatoma up-regulated protein homolog) (HURP). | |||||
|
DPOE1_HUMAN
|
||||||
| NC score | 0.011659 (rank : 95) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q07864, Q13533, Q86VH9 | Gene names | POLE, POLE1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase epsilon, catalytic subunit A (EC 2.7.7.7) (DNA polymerase II subunit A). | |||||
|
HORN_HUMAN
|
||||||
| NC score | 0.011495 (rank : 96) | θ value | 3.0926 (rank : 68) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86YZ3 | Gene names | HRNR, S100A18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hornerin. | |||||
|
C8AP2_HUMAN
|
||||||
| NC score | 0.009996 (rank : 97) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 548 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UKL3, Q6PH76, Q7LCQ7, Q86YD9, Q9NUQ4, Q9NZV9, Q9P2N1, Q9Y563 | Gene names | CASP8AP2, FLASH, KIAA1315, RIP25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
ACADS_MOUSE
|
||||||
| NC score | 0.009588 (rank : 98) | θ value | 2.36792 (rank : 65) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q07417 | Gene names | Acads | |||
|
Domain Architecture |
|
|||||
| Description | Short-chain specific acyl-CoA dehydrogenase, mitochondrial precursor (EC 1.3.99.2) (SCAD) (Butyryl-CoA dehydrogenase). | |||||
|
AK1D1_MOUSE
|
||||||
| NC score | 0.008396 (rank : 99) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VCX1 | Gene names | Akr1d1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 3-oxo-5-beta-steroid 4-dehydrogenase (EC 1.3.99.6) (Delta(4)-3- ketosteroid 5-beta-reductase) (Aldo-keto reductase family 1 member D1). | |||||
|
IRX3_MOUSE
|
||||||
| NC score | 0.007514 (rank : 100) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P81067 | Gene names | Irx3, Irxb1 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-3 (Iroquois homeobox protein 3) (Homeodomain protein IRXB1). | |||||
|
HDGR3_MOUSE
|
||||||
| NC score | 0.007386 (rank : 101) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JMG7, Q3TRX2, Q8BQ69, Q8BR62, Q9D2M7 | Gene names | Hdgfrp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatoma-derived growth factor-related protein 3 (HRP-3). | |||||
|
AK1D1_HUMAN
|
||||||
| NC score | 0.006579 (rank : 102) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P51857 | Gene names | AKR1D1, SRD5B1 | |||
|
Domain Architecture |
|
|||||
| Description | 3-oxo-5-beta-steroid 4-dehydrogenase (EC 1.3.1.3) (Delta(4)-3- ketosteroid 5-beta-reductase) (Aldo-keto reductase family 1 member D1). | |||||
|
EPIPL_HUMAN
|
||||||
| NC score | 0.005799 (rank : 103) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P58107, Q76E58 | Gene names | EPPK1, EPIPL | |||
|
Domain Architecture |

|
|||||
| Description | Epiplakin (450 kDa epidermal antigen). | |||||
|
DMP1_HUMAN
|
||||||
| NC score | 0.004562 (rank : 104) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
GPR12_MOUSE
|
||||||
| NC score | 0.000238 (rank : 105) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 731 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P35412 | Gene names | Gpr12, Gpcr12 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 12 (GPCR01). | |||||
|
ANR12_HUMAN
|
||||||
| NC score | -0.000359 (rank : 106) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6UB98, O94951, Q658K1, Q6QMF7, Q9H231, Q9H784 | Gene names | ANKRD12, ANCO2, KIAA0874 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 12 (Ankyrin repeat-containing cofactor 2) (GAC-1 protein). | |||||
|
M3K12_HUMAN
|
||||||
| NC score | -0.003531 (rank : 107) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12852, Q86VQ5, Q8WY25 | Gene names | MAP3K12, ZPK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 12 (EC 2.7.11.25) (Mixed lineage kinase) (Leucine-zipper protein kinase) (ZPK) (Dual leucine zipper bearing kinase) (DLK) (MAPK-upstream kinase) (MUK). | |||||