Please be patient as the page loads
|
STA13_MOUSE
|
||||||
| SwissProt Accessions | Q923Q2, Q8K369 | Gene names | Stard13 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 13 (StARD13) (START domain- containing protein 13). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
RHG07_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.981808 (rank : 3) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q96QB1, O14868, O43199, Q9C0E0 | Gene names | DLC1, ARHGAP7, KIAA1723, STARD12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 7 (Rho-type GTPase-activating protein 7) (Deleted in liver cancer 1 protein) (Dlc-1) (HP protein) (StAR-related lipid transfer protein 12) (StARD12) (START domain-containing protein 12). | |||||
|
RHG07_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.981435 (rank : 4) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9R0Z9, Q8R541 | Gene names | Dlc1, Arhgap7, Stard12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 7 (Rho-type GTPase-activating protein 7) (Deleted in liver cancer 1 protein homolog) (Dlc-1) (StAR-related lipid transfer protein 12) (StARD12) (START domain-containing protein 12). | |||||
|
STA13_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.997673 (rank : 2) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9Y3M8, Q5HYH1, Q5TAE3, Q6UN61, Q86TP6, Q86WQ3, Q86XT1 | Gene names | STARD13, DLC2, GT650 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 13 (StARD13) (START domain- containing protein 13) (46H23.2) (Deleted in liver cancer protein 2) (Rho GTPase-activating protein). | |||||
|
STA13_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q923Q2, Q8K369 | Gene names | Stard13 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 13 (StARD13) (START domain- containing protein 13). | |||||
|
STAR8_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.960890 (rank : 6) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q92502, Q5JST0 | Gene names | STARD8, KIAA0189 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
STAR8_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.971905 (rank : 5) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8K031, Q3UZC7, Q6A0A8, Q6P5E0, Q8R3X8 | Gene names | Stard8, Kiaa0189 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
RHG18_MOUSE
|
||||||
| θ value | 2.7842e-17 (rank : 7) | NC score | 0.715514 (rank : 7) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8K0Q5, Q8BP03, Q8R196 | Gene names | Arhgap18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho-GTPase-activating protein 18. | |||||
|
RHG04_HUMAN
|
||||||
| θ value | 5.25075e-16 (rank : 8) | NC score | 0.599673 (rank : 21) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P98171, Q14144 | Gene names | ARHGAP4, KIAA0131, RGC1, RHOGAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 4 (Rho-GAP hematopoietic protein C1) (p115). | |||||
|
3BP1_HUMAN
|
||||||
| θ value | 3.40345e-15 (rank : 9) | NC score | 0.652780 (rank : 12) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9Y3L3, Q6IBZ2, Q6ZVL9, Q96HQ5, Q9NSQ9 | Gene names | SH3BP1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
MYO9B_MOUSE
|
||||||
| θ value | 3.40345e-15 (rank : 10) | NC score | 0.341567 (rank : 56) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9QY06, Q9QY07, Q9QY08, Q9QY09 | Gene names | Myo9b, Myr5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
RHG06_MOUSE
|
||||||
| θ value | 7.58209e-15 (rank : 11) | NC score | 0.686960 (rank : 9) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O54834, Q9QZL8 | Gene names | Arhgap6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
MYO9B_HUMAN
|
||||||
| θ value | 9.90251e-15 (rank : 12) | NC score | 0.336222 (rank : 58) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
GRLF1_HUMAN
|
||||||
| θ value | 1.68911e-14 (rank : 13) | NC score | 0.630036 (rank : 13) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9NRY4, Q14452, Q9C0E1 | Gene names | GRLF1, GRF1, KIAA1722 | |||
|
Domain Architecture |

|
|||||
| Description | Glucocorticoid receptor DNA-binding factor 1 (Glucocorticoid receptor repression factor 1) (GRF-1) (Rho GAP p190A) (p190-A). | |||||
|
RHG06_HUMAN
|
||||||
| θ value | 3.76295e-14 (rank : 14) | NC score | 0.683730 (rank : 10) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O43182, O43437, Q9P1B3, Q9UK81, Q9UK82 | Gene names | ARHGAP6, RHOGAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
RHG18_HUMAN
|
||||||
| θ value | 6.41864e-14 (rank : 15) | NC score | 0.704890 (rank : 8) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8N392, Q58EZ3, Q6P679, Q6PJD7, Q96S64 | Gene names | ARHGAP18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho-GTPase-activating protein 18 (MacGAP). | |||||
|
RBP1_MOUSE
|
||||||
| θ value | 1.42992e-13 (rank : 16) | NC score | 0.606224 (rank : 19) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q62172, Q9CRE5 | Gene names | Ralbp1, Rip1 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
SRGP2_HUMAN
|
||||||
| θ value | 1.42992e-13 (rank : 17) | NC score | 0.572472 (rank : 26) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O43295, Q8IX13, Q8IZV8 | Gene names | SRGAP3, ARHGAP14, KIAA0411, MEGAP, SRGAP2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Mental disorder- associated GAP) (Rho-GTPase-activating protein 14). | |||||
|
3BP1_MOUSE
|
||||||
| θ value | 3.18553e-13 (rank : 18) | NC score | 0.666379 (rank : 11) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P55194, Q99KK8 | Gene names | Sh3bp1, 3bp1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
RBP1_HUMAN
|
||||||
| θ value | 1.2105e-12 (rank : 19) | NC score | 0.583832 (rank : 22) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q15311 | Gene names | RALBP1, RLIP1, RLIP76 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (76 kDa Ral-interacting protein) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
RHG12_HUMAN
|
||||||
| θ value | 1.2105e-12 (rank : 20) | NC score | 0.623281 (rank : 14) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8IWW6, Q86UB3, Q8IWW7, Q8N3L1, Q9NT76 | Gene names | ARHGAP12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
RHG12_MOUSE
|
||||||
| θ value | 1.2105e-12 (rank : 21) | NC score | 0.619162 (rank : 16) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8C0D4, Q8BVP8 | Gene names | Arhgap12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
SRGP2_MOUSE
|
||||||
| θ value | 1.58096e-12 (rank : 22) | NC score | 0.563210 (rank : 33) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q812A2, Q80U09, Q8BKP4, Q8BLD0, Q925I2 | Gene names | Srgap3, Arhgap14, Kiaa0411, Srgap2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Rho-GTPase-activating protein 14). | |||||
|
SRGP1_HUMAN
|
||||||
| θ value | 7.84624e-12 (rank : 23) | NC score | 0.552167 (rank : 39) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q7Z6B7, Q9H8A3, Q9P2P2 | Gene names | SRGAP1, ARHGAP13, KIAA1304 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 1 (srGAP1) (Rho-GTPase- activating protein 13). | |||||
|
FNBP2_HUMAN
|
||||||
| θ value | 1.02475e-11 (rank : 24) | NC score | 0.556022 (rank : 36) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O75044 | Gene names | SRGAP2, FNBP2, KIAA0456 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 2 (srGAP2) (Formin-binding protein 2). | |||||
|
RHG01_HUMAN
|
||||||
| θ value | 1.74796e-11 (rank : 25) | NC score | 0.621758 (rank : 15) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q07960 | Gene names | ARHGAP1, CDC42GAP, RHOGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 1 (GTPase-activating protein rhoOGAP) (Rho-related small GTPase protein activator) (CDC42 GTPase-activating protein) (p50-RhoGAP). | |||||
|
ABR_HUMAN
|
||||||
| θ value | 6.64225e-11 (rank : 26) | NC score | 0.566544 (rank : 29) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q12979, Q13693, Q13694 | Gene names | ABR | |||
|
Domain Architecture |

|
|||||
| Description | Active breakpoint cluster region-related protein. | |||||
|
CEND2_HUMAN
|
||||||
| θ value | 1.133e-10 (rank : 27) | NC score | 0.492132 (rank : 50) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q96P48, O94879, Q59FI7, Q6PHS3, Q8WU51, Q96HP6, Q96L71 | Gene names | CENTD2, ARAP1, KIAA0782 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-delta 2 (Cnt-d2) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 1). | |||||
|
CEND3_MOUSE
|
||||||
| θ value | 1.47974e-10 (rank : 28) | NC score | 0.484667 (rank : 51) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8R5G7, Q5DTN4, Q6NVF1, Q8R5G6 | Gene names | Centd3, Arap3, Drag1, Kiaa4097 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3) (Dual specificity Rho- and Arf-GTPase-activating protein 1). | |||||
|
CEND3_HUMAN
|
||||||
| θ value | 5.62301e-10 (rank : 29) | NC score | 0.473536 (rank : 52) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8WWN8 | Gene names | CENTD3, ARAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3). | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | 5.62301e-10 (rank : 30) | NC score | 0.515776 (rank : 46) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
TCGAP_MOUSE
|
||||||
| θ value | 5.62301e-10 (rank : 31) | NC score | 0.581081 (rank : 24) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q80YF9, Q6P7W6 | Gene names | Snx26, Tcgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
BCR_HUMAN
|
||||||
| θ value | 2.13673e-09 (rank : 32) | NC score | 0.553312 (rank : 38) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P11274, P78501, Q12842 | Gene names | BCR, BCR1 | |||
|
Domain Architecture |

|
|||||
| Description | Breakpoint cluster region protein (EC 2.7.11.1) (NY-REN-26 antigen). | |||||
|
GMIP_MOUSE
|
||||||
| θ value | 8.11959e-09 (rank : 33) | NC score | 0.609660 (rank : 18) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q6PGG2, Q6P9S3 | Gene names | Gmip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GEM-interacting protein (GMIP). | |||||
|
RHG05_MOUSE
|
||||||
| θ value | 1.06045e-08 (rank : 34) | NC score | 0.563240 (rank : 32) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P97393 | Gene names | Arhgap5, Rhogap5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
CHIN_HUMAN
|
||||||
| θ value | 1.80886e-08 (rank : 35) | NC score | 0.540605 (rank : 42) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P15882, Q53SD6, Q53SH5, Q96FB0 | Gene names | CHN1, ARHGAP2, CHN | |||
|
Domain Architecture |

|
|||||
| Description | N-chimaerin (NC) (N-chimerin) (Alpha chimerin) (A-chimaerin) (Rho- GTPase-activating protein 2). | |||||
|
GMIP_HUMAN
|
||||||
| θ value | 3.08544e-08 (rank : 36) | NC score | 0.603612 (rank : 20) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9P107 | Gene names | GMIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GEM-interacting protein (GMIP). | |||||
|
CHIO_MOUSE
|
||||||
| θ value | 6.87365e-08 (rank : 37) | NC score | 0.563855 (rank : 31) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q80XD1, Q9D9W2, Q9ER57 | Gene names | Chn2, Arhgap3, Bch | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-chimaerin (Beta-chimerin) (Rho-GTPase-activating protein 3). | |||||
|
OPHN1_MOUSE
|
||||||
| θ value | 6.87365e-08 (rank : 38) | NC score | 0.564423 (rank : 30) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q99J31, Q544K7 | Gene names | Ophn1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
RHG08_HUMAN
|
||||||
| θ value | 8.97725e-08 (rank : 39) | NC score | 0.570049 (rank : 27) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9NSG0, O75983, O95695, Q96RW1, Q96RW2, Q9HA49, Q9HC46, Q9NVX8, Q9NXL1, Q9UH20 | Gene names | ARHGAP8 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 8. | |||||
|
RHG08_MOUSE
|
||||||
| θ value | 8.97725e-08 (rank : 40) | NC score | 0.579538 (rank : 25) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9CXP4, Q99JY7 | Gene names | Arhgap8 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 8. | |||||
|
CHIN_MOUSE
|
||||||
| θ value | 1.17247e-07 (rank : 41) | NC score | 0.535547 (rank : 43) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q91V57, Q3UGY3, Q7TQE5, Q8BWU6, Q9D9B3 | Gene names | Chn1, Arhgap2 | |||
|
Domain Architecture |

|
|||||
| Description | N-chimaerin (NC) (N-chimerin) (Alpha chimerin) (A-chimaerin) (Rho- GTPase-activating protein 2). | |||||
|
RHG05_HUMAN
|
||||||
| θ value | 1.17247e-07 (rank : 42) | NC score | 0.552038 (rank : 40) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q13017 | Gene names | ARHGAP5, RHOGAP5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
CHIO_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 43) | NC score | 0.534440 (rank : 44) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P52757, Q75MM2 | Gene names | CHN2, ARHGAP3, BCH | |||
|
Domain Architecture |
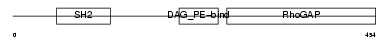
|
|||||
| Description | Beta-chimaerin (Beta-chimerin) (Rho-GTPase-activating protein 3). | |||||
|
RHG26_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 44) | NC score | 0.541627 (rank : 41) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9UNA1, O75117, Q9BYS6, Q9BYS7, Q9UJ00 | Gene names | ARHGAP26, GRAF, KIAA0621, OPHN1L | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 26 (Oligophrenin-1-like protein) (GTPase regulator associated with focal adhesion kinase). | |||||
|
OPHN1_HUMAN
|
||||||
| θ value | 1.99992e-07 (rank : 45) | NC score | 0.560349 (rank : 34) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O60890, Q5JQ81, Q8WX47 | Gene names | OPHN1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
RHG09_HUMAN
|
||||||
| θ value | 7.59969e-07 (rank : 46) | NC score | 0.610885 (rank : 17) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9BRR9, Q8TCJ3, Q8WYR0, Q96EZ2, Q96S74 | Gene names | ARHGAP9 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 9. | |||||
|
RHG25_MOUSE
|
||||||
| θ value | 2.21117e-06 (rank : 47) | NC score | 0.526916 (rank : 45) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8BYW1, Q2VPR2, Q3TE04, Q3TVX0, Q3UVB7, Q6A0E0, Q8BX98 | Gene names | Arhgap25, Kiaa0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
PCTL_MOUSE
|
||||||
| θ value | 4.92598e-06 (rank : 48) | NC score | 0.327588 (rank : 59) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JMD3 | Gene names | Stard10, Pctpl, Sdccag28, Sdccagg28 | |||
|
Domain Architecture |
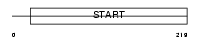
|
|||||
| Description | PCTP-like protein (PCTP-L) (StAR-related lipid transfer protein 10) (StARD10) (START domain-containing protein 10) (Serologically defined colon cancer antigen 28 homolog). | |||||
|
RHG25_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 49) | NC score | 0.511454 (rank : 47) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 550 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P42331, Q8IXQ2 | Gene names | ARHGAP25, KIAA0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
RGAP1_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 50) | NC score | 0.582236 (rank : 23) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9H0H5, Q6PJ26, Q9NWN2, Q9P250, Q9P2W2 | Gene names | RACGAP1, KIAA1478, MGCRACGAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rac GTPase-activating protein 1 (MgcRacGAP). | |||||
|
PCTL_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 51) | NC score | 0.324688 (rank : 60) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y365, O60532 | Gene names | STARD10, SDCCAG28 | |||
|
Domain Architecture |
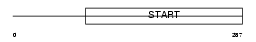
|
|||||
| Description | PCTP-like protein (PCTP-L) (StAR-related lipid transfer protein 10) (StARD10) (START domain-containing protein 10) (Serologically defined colon cancer antigen 28) (Antigen NY-CO-28). | |||||
|
TAGAP_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 52) | NC score | 0.567988 (rank : 28) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8N103, Q2NKM8, Q8NI40, Q96KZ2, Q96QA2 | Gene names | TAGAP, TAGAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell activation Rho GTPase-activating protein (T-cell activation GTPase-activating protein). | |||||
|
CE005_MOUSE
|
||||||
| θ value | 0.000121331 (rank : 53) | NC score | 0.494124 (rank : 49) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8K2H3 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Protein C5orf5 homolog. | |||||
|
CE005_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 54) | NC score | 0.494421 (rank : 48) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9NYF5, Q6PGQ2, Q9P0I7 | Gene names | C5orf5 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C5orf5 (GAP-like protein N61). | |||||
|
TAGAP_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 55) | NC score | 0.557850 (rank : 35) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8K2L9, Q920D6 | Gene names | Tagap, Tagap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell activation Rho GTPase-activating protein (T-cell activation GTPase-activating protein). | |||||
|
RGAP1_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 56) | NC score | 0.553601 (rank : 37) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9WVM1, Q3THR5, Q3TI41, Q3TM81 | Gene names | Racgap1, Mgcracgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rac GTPase-activating protein 1 (MgcRacGAP). | |||||
|
CEND1_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 57) | NC score | 0.352427 (rank : 55) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8BZ05, Q80TX2, Q8C3T2, Q8VEL6 | Gene names | Centd1, Kiaa0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1). | |||||
|
STAR7_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 58) | NC score | 0.175531 (rank : 65) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NQZ5, Q6GU43, Q969M6 | Gene names | STARD7 | |||
|
Domain Architecture |
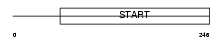
|
|||||
| Description | StAR-related lipid transfer protein 7 (StARD7) (START domain- containing protein 7) (Protein GTT1). | |||||
|
CEND1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 59) | NC score | 0.336697 (rank : 57) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8WZ64, Q4W5D2, Q7Z2L5, Q96L70, Q96P49, Q9Y4E4 | Gene names | CENTD1, ARAP2, KIAA0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 2) (PARX protein). | |||||
|
K1688_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 60) | NC score | 0.392066 (rank : 53) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P59281, Q69ZD4, Q6P9R6 | Gene names | Kiaa1688, D15Wsu169e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1688. | |||||
|
STAR7_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 61) | NC score | 0.157085 (rank : 66) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8R1R3 | Gene names | Stard7 | |||
|
Domain Architecture |
|
|||||
| Description | StAR-related lipid transfer protein 7 (StARD7) (START domain- containing protein 7). | |||||
|
K1688_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 62) | NC score | 0.380809 (rank : 54) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9C0H5 | Gene names | KIAA1688 | |||
|
Domain Architecture |

|
|||||
| Description | Protein KIAA1688. | |||||
|
STAR_HUMAN
|
||||||
| θ value | 0.125558 (rank : 63) | NC score | 0.075531 (rank : 67) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P49675, Q16396 | Gene names | STAR | |||
|
Domain Architecture |
|
|||||
| Description | Steroidogenic acute regulatory protein, mitochondrial precursor (StAR) (StARD1). | |||||
|
STAR_MOUSE
|
||||||
| θ value | 0.163984 (rank : 64) | NC score | 0.067830 (rank : 68) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P51557, Q543A5, Q924Y5, Q9D2G1 | Gene names | Star | |||
|
Domain Architecture |
|
|||||
| Description | Steroidogenic acute regulatory protein, mitochondrial precursor (StAR) (StARD1) (Luteinizing hormone-induced protein). | |||||
|
STAT2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 65) | NC score | 0.018647 (rank : 77) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WVL2, Q64188, Q64189, Q64250 | Gene names | Stat2 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 2. | |||||
|
MEPE_HUMAN
|
||||||
| θ value | 0.813845 (rank : 66) | NC score | 0.029390 (rank : 73) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NQ76 | Gene names | MEPE | |||
|
Domain Architecture |
|
|||||
| Description | Matrix extracellular phosphoglycoprotein precursor (Osteoblast/osteocyte factor 45) (OF45). | |||||
|
TREF1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 67) | NC score | 0.021127 (rank : 74) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BXJ2, Q6PCQ4, Q80X25, Q810H8, Q8BY31 | Gene names | Trerf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132). | |||||
|
ACO11_HUMAN
|
||||||
| θ value | 1.38821 (rank : 68) | NC score | 0.043468 (rank : 71) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WXI4, O75187, Q96DI1, Q9H883 | Gene names | ACOT11, BFIT, KIAA0707, THEA | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A thioesterase 11 (EC 3.1.2.-) (Acyl-CoA thioesterase 11) (Acyl-CoA thioester hydrolase 11) (Brown fat-inducible thioesterase) (BFIT) (Adipose-associated thioesterase). | |||||
|
ACO12_MOUSE
|
||||||
| θ value | 1.38821 (rank : 69) | NC score | 0.061870 (rank : 69) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9DBK0, Q544M5, Q8R108 | Gene names | Acot12, Cach, Cach1 | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A thioesterase 12 (EC 3.1.2.1) (Acyl-CoA thioesterase 12) (Acyl-CoA thioester hydrolase 12) (Cytoplasmic acetyl-CoA hydrolase 1) (CACH-1) (mCACH-1). | |||||
|
TREF1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 70) | NC score | 0.016518 (rank : 78) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96PN7, Q7Z6T2, Q7Z6T3, Q9NQ72, Q9NQ73, Q9NUN9 | Gene names | TRERF1, RAPA, TREP132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132) (Zinc finger protein rapa). | |||||
|
HIRA_MOUSE
|
||||||
| θ value | 2.36792 (rank : 71) | NC score | 0.011689 (rank : 81) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61666, O08845, Q62365 | Gene names | Hira, Tuple1 | |||
|
Domain Architecture |
|
|||||
| Description | HIRA protein (TUP1-like enhancer of split protein 1). | |||||
|
KCNH1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 72) | NC score | 0.005760 (rank : 85) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q60603 | Gene names | Kcnh1, Eag | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 1 (Voltage-gated potassium channel subunit Kv10.1) (Ether-a-go-go potassium channel 1) (EAG1) (m-eag). | |||||
|
SFRIP_HUMAN
|
||||||
| θ value | 2.36792 (rank : 73) | NC score | 0.020114 (rank : 76) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
ALMS1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 74) | NC score | 0.012483 (rank : 79) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K4E0, Q6A084, Q8C9N9 | Gene names | Alms1, Kiaa0328 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alstrom syndrome protein 1 homolog. | |||||
|
ILRL2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 75) | NC score | 0.006384 (rank : 84) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HB29, Q13525, Q45H74 | Gene names | IL1RL2, IL1RRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-1 receptor-like 2 precursor (IL-1Rrp2) (Interleukin-1 receptor-related protein 2) (IL1R-rp2). | |||||
|
OSTP_MOUSE
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | 0.032678 (rank : 72) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P10923, P19008 | Gene names | Spp1, Eta-1, Op, Spp-1 | |||
|
Domain Architecture |

|
|||||
| Description | Osteopontin precursor (Bone sialoprotein-1) (Secreted phosphoprotein 1) (SPP-1) (Minopontin) (Early T-lymphocyte activation 1 protein) (2AR) (Calcium oxalate crystal growth inhibitor protein). | |||||
|
SYCP2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.012122 (rank : 80) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BX26, O75763, Q5JX11, Q9NTX8, Q9UG27 | Gene names | SYCP2, SCP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptonemal complex protein 2 (SCP-2) (Synaptonemal complex lateral element protein) (hsSCP2). | |||||
|
ATS13_MOUSE
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.002334 (rank : 90) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q769J6, Q76LW1 | Gene names | Adamts13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAMTS-13 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 13) (ADAM-TS 13) (ADAM-TS13) (von Willebrand factor-cleaving protease) (vWF-cleaving protease) (vWF-CP). | |||||
|
HIRA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.010230 (rank : 82) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P54198, Q8IXN2 | Gene names | HIRA, DGCR1, HIR, TUPLE1 | |||
|
Domain Architecture |
|
|||||
| Description | HIRA protein (TUP1-like enhancer of split protein 1). | |||||
|
OSTP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.020269 (rank : 75) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P10451, Q15681, Q15682, Q15683, Q8NBK2, Q96IZ1 | Gene names | SPP1, OPN | |||
|
Domain Architecture |

|
|||||
| Description | Osteopontin precursor (Bone sialoprotein-1) (Secreted phosphoprotein 1) (SPP-1) (Urinary stone protein) (Nephropontin) (Uropontin). | |||||
|
TEX14_HUMAN
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | -0.000593 (rank : 92) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1103 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IWB6, Q7RTP3, Q8ND97, Q9BXT9 | Gene names | TEX14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed protein 14 (Testis-expressed sequence 14). | |||||
|
B3GN1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.009655 (rank : 83) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z222, Q91V18 | Gene names | B3gnt1, Beta3gnt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1 (EC 2.4.1.-) (Beta3Gn-T1) (BGnT-1) (Beta-1,3-galactosyltransferase 7) (Beta-1,3-GalTase 7) (Beta3Gal-T7) (b3Gal-T7) (UDP-galactose:beta-N- acetylglucosamine beta-1,3-galactosyltransferase 7) (UDP-Gal:beta- GlcNAc beta-1,3-galactosyltransferase 7) (Beta-3-Gx-T7). | |||||
|
FRMD5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.003127 (rank : 89) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P5H6, Q6PFH6, Q8C0K0 | Gene names | Frmd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 5. | |||||
|
GOGA6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.004181 (rank : 87) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NYA3, Q9NYA7 | Gene names | GOLGA6, GLP | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 6 (Golgin linked to PML) (Golgin-like protein). | |||||
|
MK10_HUMAN
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | -0.004110 (rank : 93) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 836 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P53779, Q15707 | Gene names | MAPK10, JNK3, JNK3A, PRKM10 | |||
|
Domain Architecture |
|
|||||
| Description | Mitogen-activated protein kinase 10 (EC 2.7.11.24) (Stress-activated protein kinase JNK3) (c-Jun N-terminal kinase 3) (MAP kinase p49 3F12). | |||||
|
MK10_MOUSE
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | -0.004117 (rank : 94) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 849 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61831, Q9R0U6 | Gene names | Mapk10, Jnk3, Prkm10, Serk2 | |||
|
Domain Architecture |
|
|||||
| Description | Mitogen-activated protein kinase 10 (EC 2.7.11.24) (Stress-activated protein kinase JNK3) (c-Jun N-terminal kinase 3) (MAP kinase p49 3F12). | |||||
|
PARC_HUMAN
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.003766 (rank : 88) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IWT3, O75188, Q68CP2, Q68D92, Q8N3W9, Q9BU56 | Gene names | PARC, H7AP1, KIAA0708 | |||
|
Domain Architecture |

|
|||||
| Description | p53-associated parkin-like cytoplasmic protein (UbcH7-associated protein 1). | |||||
|
SYNE2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.000172 (rank : 91) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
TRRAP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.005512 (rank : 86) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y4A5, O75218, Q9Y631, Q9Y6H4 | Gene names | TRRAP, PAF400 | |||
|
Domain Architecture |

|
|||||
| Description | Transformation/transcription domain-associated protein (350/400 kDa PCAF-associated factor) (PAF350/400) (STAF40) (Tra1 homolog). | |||||
|
CCD48_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.051214 (rank : 70) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HA90 | Gene names | CCDC48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 48. | |||||
|
I5P2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.214569 (rank : 63) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P32019, Q5VSG9, Q5VSH0, Q5VSH1, Q658Q5, Q6P6D4, Q6PD53, Q86YE1 | Gene names | INPP5B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Type II inositol-1,4,5-trisphosphate 5-phosphatase precursor (EC 3.1.3.36) (Phosphoinositide 5-phosphatase) (5PTase) (75 kDa inositol polyphosphate-5-phosphatase). | |||||
|
OCRL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.184881 (rank : 64) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q01968, O60800, Q15684, Q15774, Q5JQF1, Q5JQF2, Q9UJG5, Q9UMA5 | Gene names | OCRL, INPP5F, OCRL1 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol polyphosphate 5-phosphatase OCRL-1 (EC 3.1.3.36) (Lowe oculocerebrorenal syndrome protein). | |||||
|
P85A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.223616 (rank : 61) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P27986 | Gene names | PIK3R1, GRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit alpha (PI3-kinase p85-subunit alpha) (PtdIns-3-kinase p85-alpha) (PI3K). | |||||
|
P85A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.220607 (rank : 62) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P26450 | Gene names | Pik3r1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit alpha (PI3-kinase p85-subunit alpha) (PtdIns-3-kinase p85-alpha) (PI3K). | |||||
|
STA13_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q923Q2, Q8K369 | Gene names | Stard13 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 13 (StARD13) (START domain- containing protein 13). | |||||
|
STA13_HUMAN
|
||||||
| NC score | 0.997673 (rank : 2) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9Y3M8, Q5HYH1, Q5TAE3, Q6UN61, Q86TP6, Q86WQ3, Q86XT1 | Gene names | STARD13, DLC2, GT650 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 13 (StARD13) (START domain- containing protein 13) (46H23.2) (Deleted in liver cancer protein 2) (Rho GTPase-activating protein). | |||||
|
RHG07_HUMAN
|
||||||
| NC score | 0.981808 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q96QB1, O14868, O43199, Q9C0E0 | Gene names | DLC1, ARHGAP7, KIAA1723, STARD12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 7 (Rho-type GTPase-activating protein 7) (Deleted in liver cancer 1 protein) (Dlc-1) (HP protein) (StAR-related lipid transfer protein 12) (StARD12) (START domain-containing protein 12). | |||||
|
RHG07_MOUSE
|
||||||
| NC score | 0.981435 (rank : 4) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9R0Z9, Q8R541 | Gene names | Dlc1, Arhgap7, Stard12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 7 (Rho-type GTPase-activating protein 7) (Deleted in liver cancer 1 protein homolog) (Dlc-1) (StAR-related lipid transfer protein 12) (StARD12) (START domain-containing protein 12). | |||||
|
STAR8_MOUSE
|
||||||
| NC score | 0.971905 (rank : 5) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8K031, Q3UZC7, Q6A0A8, Q6P5E0, Q8R3X8 | Gene names | Stard8, Kiaa0189 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
STAR8_HUMAN
|
||||||
| NC score | 0.960890 (rank : 6) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q92502, Q5JST0 | Gene names | STARD8, KIAA0189 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
RHG18_MOUSE
|
||||||
| NC score | 0.715514 (rank : 7) | θ value | 2.7842e-17 (rank : 7) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8K0Q5, Q8BP03, Q8R196 | Gene names | Arhgap18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho-GTPase-activating protein 18. | |||||
|
RHG18_HUMAN
|
||||||
| NC score | 0.704890 (rank : 8) | θ value | 6.41864e-14 (rank : 15) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8N392, Q58EZ3, Q6P679, Q6PJD7, Q96S64 | Gene names | ARHGAP18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho-GTPase-activating protein 18 (MacGAP). | |||||
|
RHG06_MOUSE
|
||||||
| NC score | 0.686960 (rank : 9) | θ value | 7.58209e-15 (rank : 11) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O54834, Q9QZL8 | Gene names | Arhgap6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
RHG06_HUMAN
|
||||||
| NC score | 0.683730 (rank : 10) | θ value | 3.76295e-14 (rank : 14) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O43182, O43437, Q9P1B3, Q9UK81, Q9UK82 | Gene names | ARHGAP6, RHOGAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
3BP1_MOUSE
|
||||||
| NC score | 0.666379 (rank : 11) | θ value | 3.18553e-13 (rank : 18) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P55194, Q99KK8 | Gene names | Sh3bp1, 3bp1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
3BP1_HUMAN
|
||||||
| NC score | 0.652780 (rank : 12) | θ value | 3.40345e-15 (rank : 9) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9Y3L3, Q6IBZ2, Q6ZVL9, Q96HQ5, Q9NSQ9 | Gene names | SH3BP1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
GRLF1_HUMAN
|
||||||
| NC score | 0.630036 (rank : 13) | θ value | 1.68911e-14 (rank : 13) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9NRY4, Q14452, Q9C0E1 | Gene names | GRLF1, GRF1, KIAA1722 | |||
|
Domain Architecture |

|
|||||
| Description | Glucocorticoid receptor DNA-binding factor 1 (Glucocorticoid receptor repression factor 1) (GRF-1) (Rho GAP p190A) (p190-A). | |||||
|
RHG12_HUMAN
|
||||||
| NC score | 0.623281 (rank : 14) | θ value | 1.2105e-12 (rank : 20) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8IWW6, Q86UB3, Q8IWW7, Q8N3L1, Q9NT76 | Gene names | ARHGAP12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
RHG01_HUMAN
|
||||||
| NC score | 0.621758 (rank : 15) | θ value | 1.74796e-11 (rank : 25) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q07960 | Gene names | ARHGAP1, CDC42GAP, RHOGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 1 (GTPase-activating protein rhoOGAP) (Rho-related small GTPase protein activator) (CDC42 GTPase-activating protein) (p50-RhoGAP). | |||||
|
RHG12_MOUSE
|
||||||
| NC score | 0.619162 (rank : 16) | θ value | 1.2105e-12 (rank : 21) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8C0D4, Q8BVP8 | Gene names | Arhgap12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
RHG09_HUMAN
|
||||||
| NC score | 0.610885 (rank : 17) | θ value | 7.59969e-07 (rank : 46) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9BRR9, Q8TCJ3, Q8WYR0, Q96EZ2, Q96S74 | Gene names | ARHGAP9 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 9. | |||||
|
GMIP_MOUSE
|
||||||
| NC score | 0.609660 (rank : 18) | θ value | 8.11959e-09 (rank : 33) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q6PGG2, Q6P9S3 | Gene names | Gmip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GEM-interacting protein (GMIP). | |||||
|
RBP1_MOUSE
|
||||||
| NC score | 0.606224 (rank : 19) | θ value | 1.42992e-13 (rank : 16) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q62172, Q9CRE5 | Gene names | Ralbp1, Rip1 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
GMIP_HUMAN
|
||||||
| NC score | 0.603612 (rank : 20) | θ value | 3.08544e-08 (rank : 36) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9P107 | Gene names | GMIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GEM-interacting protein (GMIP). | |||||
|
RHG04_HUMAN
|
||||||
| NC score | 0.599673 (rank : 21) | θ value | 5.25075e-16 (rank : 8) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P98171, Q14144 | Gene names | ARHGAP4, KIAA0131, RGC1, RHOGAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 4 (Rho-GAP hematopoietic protein C1) (p115). | |||||
|
RBP1_HUMAN
|
||||||
| NC score | 0.583832 (rank : 22) | θ value | 1.2105e-12 (rank : 19) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q15311 | Gene names | RALBP1, RLIP1, RLIP76 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (76 kDa Ral-interacting protein) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
RGAP1_HUMAN
|
||||||
| NC score | 0.582236 (rank : 23) | θ value | 6.43352e-06 (rank : 50) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9H0H5, Q6PJ26, Q9NWN2, Q9P250, Q9P2W2 | Gene names | RACGAP1, KIAA1478, MGCRACGAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rac GTPase-activating protein 1 (MgcRacGAP). | |||||
|
TCGAP_MOUSE
|
||||||
| NC score | 0.581081 (rank : 24) | θ value | 5.62301e-10 (rank : 31) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q80YF9, Q6P7W6 | Gene names | Snx26, Tcgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
RHG08_MOUSE
|
||||||
| NC score | 0.579538 (rank : 25) | θ value | 8.97725e-08 (rank : 40) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9CXP4, Q99JY7 | Gene names | Arhgap8 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 8. | |||||
|
SRGP2_HUMAN
|
||||||
| NC score | 0.572472 (rank : 26) | θ value | 1.42992e-13 (rank : 17) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O43295, Q8IX13, Q8IZV8 | Gene names | SRGAP3, ARHGAP14, KIAA0411, MEGAP, SRGAP2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Mental disorder- associated GAP) (Rho-GTPase-activating protein 14). | |||||
|
RHG08_HUMAN
|
||||||
| NC score | 0.570049 (rank : 27) | θ value | 8.97725e-08 (rank : 39) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9NSG0, O75983, O95695, Q96RW1, Q96RW2, Q9HA49, Q9HC46, Q9NVX8, Q9NXL1, Q9UH20 | Gene names | ARHGAP8 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 8. | |||||
|
TAGAP_HUMAN
|
||||||
| NC score | 0.567988 (rank : 28) | θ value | 5.44631e-05 (rank : 52) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8N103, Q2NKM8, Q8NI40, Q96KZ2, Q96QA2 | Gene names | TAGAP, TAGAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell activation Rho GTPase-activating protein (T-cell activation GTPase-activating protein). | |||||
|
ABR_HUMAN
|
||||||
| NC score | 0.566544 (rank : 29) | θ value | 6.64225e-11 (rank : 26) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q12979, Q13693, Q13694 | Gene names | ABR | |||
|
Domain Architecture |

|
|||||
| Description | Active breakpoint cluster region-related protein. | |||||
|
OPHN1_MOUSE
|
||||||
| NC score | 0.564423 (rank : 30) | θ value | 6.87365e-08 (rank : 38) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q99J31, Q544K7 | Gene names | Ophn1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
CHIO_MOUSE
|
||||||
| NC score | 0.563855 (rank : 31) | θ value | 6.87365e-08 (rank : 37) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q80XD1, Q9D9W2, Q9ER57 | Gene names | Chn2, Arhgap3, Bch | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-chimaerin (Beta-chimerin) (Rho-GTPase-activating protein 3). | |||||
|
RHG05_MOUSE
|
||||||
| NC score | 0.563240 (rank : 32) | θ value | 1.06045e-08 (rank : 34) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P97393 | Gene names | Arhgap5, Rhogap5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
SRGP2_MOUSE
|
||||||
| NC score | 0.563210 (rank : 33) | θ value | 1.58096e-12 (rank : 22) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q812A2, Q80U09, Q8BKP4, Q8BLD0, Q925I2 | Gene names | Srgap3, Arhgap14, Kiaa0411, Srgap2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Rho-GTPase-activating protein 14). | |||||
|
OPHN1_HUMAN
|
||||||
| NC score | 0.560349 (rank : 34) | θ value | 1.99992e-07 (rank : 45) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O60890, Q5JQ81, Q8WX47 | Gene names | OPHN1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
TAGAP_MOUSE
|
||||||
| NC score | 0.557850 (rank : 35) | θ value | 0.000786445 (rank : 55) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8K2L9, Q920D6 | Gene names | Tagap, Tagap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell activation Rho GTPase-activating protein (T-cell activation GTPase-activating protein). | |||||
|
FNBP2_HUMAN
|
||||||
| NC score | 0.556022 (rank : 36) | θ value | 1.02475e-11 (rank : 24) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O75044 | Gene names | SRGAP2, FNBP2, KIAA0456 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 2 (srGAP2) (Formin-binding protein 2). | |||||
|
RGAP1_MOUSE
|
||||||
| NC score | 0.553601 (rank : 37) | θ value | 0.00228821 (rank : 56) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9WVM1, Q3THR5, Q3TI41, Q3TM81 | Gene names | Racgap1, Mgcracgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rac GTPase-activating protein 1 (MgcRacGAP). | |||||
|
BCR_HUMAN
|
||||||
| NC score | 0.553312 (rank : 38) | θ value | 2.13673e-09 (rank : 32) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P11274, P78501, Q12842 | Gene names | BCR, BCR1 | |||
|
Domain Architecture |

|
|||||
| Description | Breakpoint cluster region protein (EC 2.7.11.1) (NY-REN-26 antigen). | |||||
|
SRGP1_HUMAN
|
||||||
| NC score | 0.552167 (rank : 39) | θ value | 7.84624e-12 (rank : 23) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q7Z6B7, Q9H8A3, Q9P2P2 | Gene names | SRGAP1, ARHGAP13, KIAA1304 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 1 (srGAP1) (Rho-GTPase- activating protein 13). | |||||
|
RHG05_HUMAN
|
||||||
| NC score | 0.552038 (rank : 40) | θ value | 1.17247e-07 (rank : 42) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q13017 | Gene names | ARHGAP5, RHOGAP5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
RHG26_HUMAN
|
||||||
| NC score | 0.541627 (rank : 41) | θ value | 1.53129e-07 (rank : 44) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9UNA1, O75117, Q9BYS6, Q9BYS7, Q9UJ00 | Gene names | ARHGAP26, GRAF, KIAA0621, OPHN1L | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 26 (Oligophrenin-1-like protein) (GTPase regulator associated with focal adhesion kinase). | |||||
|
CHIN_HUMAN
|
||||||
| NC score | 0.540605 (rank : 42) | θ value | 1.80886e-08 (rank : 35) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P15882, Q53SD6, Q53SH5, Q96FB0 | Gene names | CHN1, ARHGAP2, CHN | |||
|
Domain Architecture |

|
|||||
| Description | N-chimaerin (NC) (N-chimerin) (Alpha chimerin) (A-chimaerin) (Rho- GTPase-activating protein 2). | |||||
|
CHIN_MOUSE
|
||||||
| NC score | 0.535547 (rank : 43) | θ value | 1.17247e-07 (rank : 41) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q91V57, Q3UGY3, Q7TQE5, Q8BWU6, Q9D9B3 | Gene names | Chn1, Arhgap2 | |||
|
Domain Architecture |

|
|||||
| Description | N-chimaerin (NC) (N-chimerin) (Alpha chimerin) (A-chimaerin) (Rho- GTPase-activating protein 2). | |||||
|
CHIO_HUMAN
|
||||||
| NC score | 0.534440 (rank : 44) | θ value | 1.53129e-07 (rank : 43) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P52757, Q75MM2 | Gene names | CHN2, ARHGAP3, BCH | |||
|
Domain Architecture |
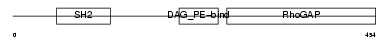
|
|||||
| Description | Beta-chimaerin (Beta-chimerin) (Rho-GTPase-activating protein 3). | |||||
|
RHG25_MOUSE
|
||||||
| NC score | 0.526916 (rank : 45) | θ value | 2.21117e-06 (rank : 47) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8BYW1, Q2VPR2, Q3TE04, Q3TVX0, Q3UVB7, Q6A0E0, Q8BX98 | Gene names | Arhgap25, Kiaa0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.515776 (rank : 46) | θ value | 5.62301e-10 (rank : 30) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
RHG25_HUMAN
|
||||||
| NC score | 0.511454 (rank : 47) | θ value | 4.92598e-06 (rank : 49) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 550 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P42331, Q8IXQ2 | Gene names | ARHGAP25, KIAA0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
CE005_HUMAN
|
||||||
| NC score | 0.494421 (rank : 48) | θ value | 0.00020696 (rank : 54) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9NYF5, Q6PGQ2, Q9P0I7 | Gene names | C5orf5 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C5orf5 (GAP-like protein N61). | |||||
|
CE005_MOUSE
|
||||||
| NC score | 0.494124 (rank : 49) | θ value | 0.000121331 (rank : 53) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8K2H3 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Protein C5orf5 homolog. | |||||
|
CEND2_HUMAN
|
||||||
| NC score | 0.492132 (rank : 50) | θ value | 1.133e-10 (rank : 27) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q96P48, O94879, Q59FI7, Q6PHS3, Q8WU51, Q96HP6, Q96L71 | Gene names | CENTD2, ARAP1, KIAA0782 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-delta 2 (Cnt-d2) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 1). | |||||
|
CEND3_MOUSE
|
||||||
| NC score | 0.484667 (rank : 51) | θ value | 1.47974e-10 (rank : 28) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8R5G7, Q5DTN4, Q6NVF1, Q8R5G6 | Gene names | Centd3, Arap3, Drag1, Kiaa4097 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3) (Dual specificity Rho- and Arf-GTPase-activating protein 1). | |||||
|
CEND3_HUMAN
|
||||||
| NC score | 0.473536 (rank : 52) | θ value | 5.62301e-10 (rank : 29) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8WWN8 | Gene names | CENTD3, ARAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3). | |||||
|
K1688_MOUSE
|
||||||
| NC score | 0.392066 (rank : 53) | θ value | 0.0736092 (rank : 60) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P59281, Q69ZD4, Q6P9R6 | Gene names | Kiaa1688, D15Wsu169e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1688. | |||||
|
K1688_HUMAN
|
||||||
| NC score | 0.380809 (rank : 54) | θ value | 0.0961366 (rank : 62) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9C0H5 | Gene names | KIAA1688 | |||
|
Domain Architecture |

|
|||||
| Description | Protein KIAA1688. | |||||
|
CEND1_MOUSE
|
||||||
| NC score | 0.352427 (rank : 55) | θ value | 0.0193708 (rank : 57) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8BZ05, Q80TX2, Q8C3T2, Q8VEL6 | Gene names | Centd1, Kiaa0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1). | |||||
|
MYO9B_MOUSE
|
||||||
| NC score | 0.341567 (rank : 56) | θ value | 3.40345e-15 (rank : 10) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9QY06, Q9QY07, Q9QY08, Q9QY09 | Gene names | Myo9b, Myr5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
CEND1_HUMAN
|
||||||
| NC score | 0.336697 (rank : 57) | θ value | 0.0431538 (rank : 59) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8WZ64, Q4W5D2, Q7Z2L5, Q96L70, Q96P49, Q9Y4E4 | Gene names | CENTD1, ARAP2, KIAA0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 2) (PARX protein). | |||||
|
MYO9B_HUMAN
|
||||||
| NC score | 0.336222 (rank : 58) | θ value | 9.90251e-15 (rank : 12) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
PCTL_MOUSE
|
||||||
| NC score | 0.327588 (rank : 59) | θ value | 4.92598e-06 (rank : 48) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JMD3 | Gene names | Stard10, Pctpl, Sdccag28, Sdccagg28 | |||
|
Domain Architecture |

|
|||||
| Description | PCTP-like protein (PCTP-L) (StAR-related lipid transfer protein 10) (StARD10) (START domain-containing protein 10) (Serologically defined colon cancer antigen 28 homolog). | |||||
|
PCTL_HUMAN
|
||||||
| NC score | 0.324688 (rank : 60) | θ value | 8.40245e-06 (rank : 51) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y365, O60532 | Gene names | STARD10, SDCCAG28 | |||
|
Domain Architecture |

|
|||||
| Description | PCTP-like protein (PCTP-L) (StAR-related lipid transfer protein 10) (StARD10) (START domain-containing protein 10) (Serologically defined colon cancer antigen 28) (Antigen NY-CO-28). | |||||
|
P85A_HUMAN
|
||||||
| NC score | 0.223616 (rank : 61) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P27986 | Gene names | PIK3R1, GRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit alpha (PI3-kinase p85-subunit alpha) (PtdIns-3-kinase p85-alpha) (PI3K). | |||||
|
P85A_MOUSE
|
||||||
| NC score | 0.220607 (rank : 62) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P26450 | Gene names | Pik3r1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit alpha (PI3-kinase p85-subunit alpha) (PtdIns-3-kinase p85-alpha) (PI3K). | |||||
|
I5P2_HUMAN
|
||||||
| NC score | 0.214569 (rank : 63) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P32019, Q5VSG9, Q5VSH0, Q5VSH1, Q658Q5, Q6P6D4, Q6PD53, Q86YE1 | Gene names | INPP5B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Type II inositol-1,4,5-trisphosphate 5-phosphatase precursor (EC 3.1.3.36) (Phosphoinositide 5-phosphatase) (5PTase) (75 kDa inositol polyphosphate-5-phosphatase). | |||||
|
OCRL_HUMAN
|
||||||
| NC score | 0.184881 (rank : 64) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q01968, O60800, Q15684, Q15774, Q5JQF1, Q5JQF2, Q9UJG5, Q9UMA5 | Gene names | OCRL, INPP5F, OCRL1 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol polyphosphate 5-phosphatase OCRL-1 (EC 3.1.3.36) (Lowe oculocerebrorenal syndrome protein). | |||||
|
STAR7_HUMAN
|
||||||
| NC score | 0.175531 (rank : 65) | θ value | 0.0252991 (rank : 58) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NQZ5, Q6GU43, Q969M6 | Gene names | STARD7 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 7 (StARD7) (START domain- containing protein 7) (Protein GTT1). | |||||
|
STAR7_MOUSE
|
||||||
| NC score | 0.157085 (rank : 66) | θ value | 0.0736092 (rank : 61) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8R1R3 | Gene names | Stard7 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 7 (StARD7) (START domain- containing protein 7). | |||||
|
STAR_HUMAN
|
||||||
| NC score | 0.075531 (rank : 67) | θ value | 0.125558 (rank : 63) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P49675, Q16396 | Gene names | STAR | |||
|
Domain Architecture |

|
|||||
| Description | Steroidogenic acute regulatory protein, mitochondrial precursor (StAR) (StARD1). | |||||
|
STAR_MOUSE
|
||||||
| NC score | 0.067830 (rank : 68) | θ value | 0.163984 (rank : 64) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P51557, Q543A5, Q924Y5, Q9D2G1 | Gene names | Star | |||
|
Domain Architecture |

|
|||||
| Description | Steroidogenic acute regulatory protein, mitochondrial precursor (StAR) (StARD1) (Luteinizing hormone-induced protein). | |||||
|
ACO12_MOUSE
|
||||||
| NC score | 0.061870 (rank : 69) | θ value | 1.38821 (rank : 69) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9DBK0, Q544M5, Q8R108 | Gene names | Acot12, Cach, Cach1 | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A thioesterase 12 (EC 3.1.2.1) (Acyl-CoA thioesterase 12) (Acyl-CoA thioester hydrolase 12) (Cytoplasmic acetyl-CoA hydrolase 1) (CACH-1) (mCACH-1). | |||||
|
CCD48_HUMAN
|
||||||
| NC score | 0.051214 (rank : 70) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HA90 | Gene names | CCDC48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 48. | |||||
|
ACO11_HUMAN
|
||||||
| NC score | 0.043468 (rank : 71) | θ value | 1.38821 (rank : 68) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WXI4, O75187, Q96DI1, Q9H883 | Gene names | ACOT11, BFIT, KIAA0707, THEA | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A thioesterase 11 (EC 3.1.2.-) (Acyl-CoA thioesterase 11) (Acyl-CoA thioester hydrolase 11) (Brown fat-inducible thioesterase) (BFIT) (Adipose-associated thioesterase). | |||||
|
OSTP_MOUSE
|
||||||
| NC score | 0.032678 (rank : 72) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P10923, P19008 | Gene names | Spp1, Eta-1, Op, Spp-1 | |||
|
Domain Architecture |

|
|||||
| Description | Osteopontin precursor (Bone sialoprotein-1) (Secreted phosphoprotein 1) (SPP-1) (Minopontin) (Early T-lymphocyte activation 1 protein) (2AR) (Calcium oxalate crystal growth inhibitor protein). | |||||
|
MEPE_HUMAN
|
||||||
| NC score | 0.029390 (rank : 73) | θ value | 0.813845 (rank : 66) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NQ76 | Gene names | MEPE | |||
|
Domain Architecture |

|
|||||
| Description | Matrix extracellular phosphoglycoprotein precursor (Osteoblast/osteocyte factor 45) (OF45). | |||||
|
TREF1_MOUSE
|
||||||
| NC score | 0.021127 (rank : 74) | θ value | 1.06291 (rank : 67) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BXJ2, Q6PCQ4, Q80X25, Q810H8, Q8BY31 | Gene names | Trerf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132). | |||||
|
OSTP_HUMAN
|
||||||
| NC score | 0.020269 (rank : 75) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P10451, Q15681, Q15682, Q15683, Q8NBK2, Q96IZ1 | Gene names | SPP1, OPN | |||
|
Domain Architecture |

|
|||||
| Description | Osteopontin precursor (Bone sialoprotein-1) (Secreted phosphoprotein 1) (SPP-1) (Urinary stone protein) (Nephropontin) (Uropontin). | |||||
|
SFRIP_HUMAN
|
||||||
| NC score | 0.020114 (rank : 76) | θ value | 2.36792 (rank : 73) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
STAT2_MOUSE
|
||||||
| NC score | 0.018647 (rank : 77) | θ value | 0.62314 (rank : 65) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WVL2, Q64188, Q64189, Q64250 | Gene names | Stat2 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 2. | |||||
|
TREF1_HUMAN
|
||||||
| NC score | 0.016518 (rank : 78) | θ value | 1.38821 (rank : 70) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96PN7, Q7Z6T2, Q7Z6T3, Q9NQ72, Q9NQ73, Q9NUN9 | Gene names | TRERF1, RAPA, TREP132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132) (Zinc finger protein rapa). | |||||
|
ALMS1_MOUSE
|
||||||
| NC score | 0.012483 (rank : 79) | θ value | 3.0926 (rank : 74) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K4E0, Q6A084, Q8C9N9 | Gene names | Alms1, Kiaa0328 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alstrom syndrome protein 1 homolog. | |||||
|
SYCP2_HUMAN
|
||||||
| NC score | 0.012122 (rank : 80) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BX26, O75763, Q5JX11, Q9NTX8, Q9UG27 | Gene names | SYCP2, SCP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptonemal complex protein 2 (SCP-2) (Synaptonemal complex lateral element protein) (hsSCP2). | |||||
|
HIRA_MOUSE
|
||||||
| NC score | 0.011689 (rank : 81) | θ value | 2.36792 (rank : 71) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61666, O08845, Q62365 | Gene names | Hira, Tuple1 | |||
|
Domain Architecture |

|
|||||
| Description | HIRA protein (TUP1-like enhancer of split protein 1). | |||||
|
HIRA_HUMAN
|
||||||
| NC score | 0.010230 (rank : 82) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P54198, Q8IXN2 | Gene names | HIRA, DGCR1, HIR, TUPLE1 | |||
|
Domain Architecture |

|
|||||
| Description | HIRA protein (TUP1-like enhancer of split protein 1). | |||||
|
B3GN1_MOUSE
|
||||||
| NC score | 0.009655 (rank : 83) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z222, Q91V18 | Gene names | B3gnt1, Beta3gnt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1 (EC 2.4.1.-) (Beta3Gn-T1) (BGnT-1) (Beta-1,3-galactosyltransferase 7) (Beta-1,3-GalTase 7) (Beta3Gal-T7) (b3Gal-T7) (UDP-galactose:beta-N- acetylglucosamine beta-1,3-galactosyltransferase 7) (UDP-Gal:beta- GlcNAc beta-1,3-galactosyltransferase 7) (Beta-3-Gx-T7). | |||||
|
ILRL2_HUMAN
|
||||||
| NC score | 0.006384 (rank : 84) | θ value | 4.03905 (rank : 75) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HB29, Q13525, Q45H74 | Gene names | IL1RL2, IL1RRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-1 receptor-like 2 precursor (IL-1Rrp2) (Interleukin-1 receptor-related protein 2) (IL1R-rp2). | |||||
|
KCNH1_MOUSE
|
||||||
| NC score | 0.005760 (rank : 85) | θ value | 2.36792 (rank : 72) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q60603 | Gene names | Kcnh1, Eag | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 1 (Voltage-gated potassium channel subunit Kv10.1) (Ether-a-go-go potassium channel 1) (EAG1) (m-eag). | |||||
|
TRRAP_HUMAN
|
||||||
| NC score | 0.005512 (rank : 86) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y4A5, O75218, Q9Y631, Q9Y6H4 | Gene names | TRRAP, PAF400 | |||
|
Domain Architecture |

|
|||||
| Description | Transformation/transcription domain-associated protein (350/400 kDa PCAF-associated factor) (PAF350/400) (STAF40) (Tra1 homolog). | |||||
|
GOGA6_HUMAN
|
||||||
| NC score | 0.004181 (rank : 87) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NYA3, Q9NYA7 | Gene names | GOLGA6, GLP | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 6 (Golgin linked to PML) (Golgin-like protein). | |||||
|
PARC_HUMAN
|
||||||
| NC score | 0.003766 (rank : 88) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IWT3, O75188, Q68CP2, Q68D92, Q8N3W9, Q9BU56 | Gene names | PARC, H7AP1, KIAA0708 | |||
|
Domain Architecture |

|
|||||
| Description | p53-associated parkin-like cytoplasmic protein (UbcH7-associated protein 1). | |||||
|
FRMD5_MOUSE
|
||||||
| NC score | 0.003127 (rank : 89) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P5H6, Q6PFH6, Q8C0K0 | Gene names | Frmd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 5. | |||||
|
ATS13_MOUSE
|
||||||
| NC score | 0.002334 (rank : 90) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q769J6, Q76LW1 | Gene names | Adamts13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAMTS-13 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 13) (ADAM-TS 13) (ADAM-TS13) (von Willebrand factor-cleaving protease) (vWF-cleaving protease) (vWF-CP). | |||||
|
SYNE2_HUMAN
|
||||||
| NC score | 0.000172 (rank : 91) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
TEX14_HUMAN
|
||||||
| NC score | -0.000593 (rank : 92) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1103 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IWB6, Q7RTP3, Q8ND97, Q9BXT9 | Gene names | TEX14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed protein 14 (Testis-expressed sequence 14). | |||||
|
MK10_HUMAN
|
||||||
| NC score | -0.004110 (rank : 93) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 836 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P53779, Q15707 | Gene names | MAPK10, JNK3, JNK3A, PRKM10 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase 10 (EC 2.7.11.24) (Stress-activated protein kinase JNK3) (c-Jun N-terminal kinase 3) (MAP kinase p49 3F12). | |||||
|
MK10_MOUSE
|
||||||
| NC score | -0.004117 (rank : 94) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 849 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61831, Q9R0U6 | Gene names | Mapk10, Jnk3, Prkm10, Serk2 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase 10 (EC 2.7.11.24) (Stress-activated protein kinase JNK3) (c-Jun N-terminal kinase 3) (MAP kinase p49 3F12). | |||||