Please be patient as the page loads
|
PCTL_HUMAN
|
||||||
| SwissProt Accessions | Q9Y365, O60532 | Gene names | STARD10, SDCCAG28 | |||
|
Domain Architecture |
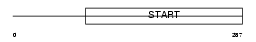
|
|||||
| Description | PCTP-like protein (PCTP-L) (StAR-related lipid transfer protein 10) (StARD10) (START domain-containing protein 10) (Serologically defined colon cancer antigen 28) (Antigen NY-CO-28). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PCTL_HUMAN
|
||||||
| θ value | 6.59259e-152 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Y365, O60532 | Gene names | STARD10, SDCCAG28 | |||
|
Domain Architecture |

|
|||||
| Description | PCTP-like protein (PCTP-L) (StAR-related lipid transfer protein 10) (StARD10) (START domain-containing protein 10) (Serologically defined colon cancer antigen 28) (Antigen NY-CO-28). | |||||
|
PCTL_MOUSE
|
||||||
| θ value | 1.51984e-148 (rank : 2) | NC score | 0.999416 (rank : 2) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9JMD3 | Gene names | Stard10, Pctpl, Sdccag28, Sdccagg28 | |||
|
Domain Architecture |
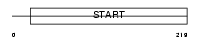
|
|||||
| Description | PCTP-like protein (PCTP-L) (StAR-related lipid transfer protein 10) (StARD10) (START domain-containing protein 10) (Serologically defined colon cancer antigen 28 homolog). | |||||
|
PPCT_MOUSE
|
||||||
| θ value | 8.95645e-16 (rank : 3) | NC score | 0.629968 (rank : 3) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P53808, Q9QZX0, Q9R058 | Gene names | Pctp, Stard2 | |||
|
Domain Architecture |
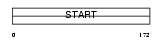
|
|||||
| Description | Phosphatidylcholine transfer protein (PC-TP) (StAR-related lipid transfer protein 2) (StARD2) (START domain-containing protein 2). | |||||
|
PPCT_HUMAN
|
||||||
| θ value | 1.2105e-12 (rank : 4) | NC score | 0.599742 (rank : 4) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UKL6, Q9UIT3, Q9UKW7 | Gene names | PCTP, STARD2 | |||
|
Domain Architecture |
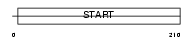
|
|||||
| Description | Phosphatidylcholine transfer protein (PC-TP) (StAR-related lipid transfer protein 2) (StARD2) (START domain-containing protein 2). | |||||
|
RHG07_HUMAN
|
||||||
| θ value | 1.38499e-08 (rank : 5) | NC score | 0.323740 (rank : 8) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96QB1, O14868, O43199, Q9C0E0 | Gene names | DLC1, ARHGAP7, KIAA1723, STARD12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 7 (Rho-type GTPase-activating protein 7) (Deleted in liver cancer 1 protein) (Dlc-1) (HP protein) (StAR-related lipid transfer protein 12) (StARD12) (START domain-containing protein 12). | |||||
|
ACO12_MOUSE
|
||||||
| θ value | 7.59969e-07 (rank : 6) | NC score | 0.256569 (rank : 15) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9DBK0, Q544M5, Q8R108 | Gene names | Acot12, Cach, Cach1 | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A thioesterase 12 (EC 3.1.2.1) (Acyl-CoA thioesterase 12) (Acyl-CoA thioester hydrolase 12) (Cytoplasmic acetyl-CoA hydrolase 1) (CACH-1) (mCACH-1). | |||||
|
RHG07_MOUSE
|
||||||
| θ value | 9.92553e-07 (rank : 7) | NC score | 0.309791 (rank : 12) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9R0Z9, Q8R541 | Gene names | Dlc1, Arhgap7, Stard12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 7 (Rho-type GTPase-activating protein 7) (Deleted in liver cancer 1 protein homolog) (Dlc-1) (StAR-related lipid transfer protein 12) (StARD12) (START domain-containing protein 12). | |||||
|
STAR7_MOUSE
|
||||||
| θ value | 1.29631e-06 (rank : 8) | NC score | 0.491066 (rank : 6) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8R1R3 | Gene names | Stard7 | |||
|
Domain Architecture |
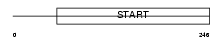
|
|||||
| Description | StAR-related lipid transfer protein 7 (StARD7) (START domain- containing protein 7). | |||||
|
STAR7_HUMAN
|
||||||
| θ value | 1.69304e-06 (rank : 9) | NC score | 0.494885 (rank : 5) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NQZ5, Q6GU43, Q969M6 | Gene names | STARD7 | |||
|
Domain Architecture |
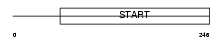
|
|||||
| Description | StAR-related lipid transfer protein 7 (StARD7) (START domain- containing protein 7) (Protein GTT1). | |||||
|
STA13_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 10) | NC score | 0.323388 (rank : 9) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y3M8, Q5HYH1, Q5TAE3, Q6UN61, Q86TP6, Q86WQ3, Q86XT1 | Gene names | STARD13, DLC2, GT650 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 13 (StARD13) (START domain- containing protein 13) (46H23.2) (Deleted in liver cancer protein 2) (Rho GTPase-activating protein). | |||||
|
STA13_MOUSE
|
||||||
| θ value | 8.40245e-06 (rank : 11) | NC score | 0.324688 (rank : 7) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q923Q2, Q8K369 | Gene names | Stard13 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 13 (StARD13) (START domain- containing protein 13). | |||||
|
C43BP_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 12) | NC score | 0.262074 (rank : 13) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9EQG9, Q91WB1, Q9CU52, Q9EQG8 | Gene names | Col4a3bp, Stard11 | |||
|
Domain Architecture |

|
|||||
| Description | Goodpasture antigen-binding protein (EC 2.7.11.9) (GPBP) (Collagen type IV alpha-3-binding protein) (StAR-related lipid transfer protein 11) (StARD11) (START domain-containing protein 11). | |||||
|
STAR8_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 13) | NC score | 0.311667 (rank : 11) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q92502, Q5JST0 | Gene names | STARD8, KIAA0189 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
STAR8_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 14) | NC score | 0.314960 (rank : 10) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8K031, Q3UZC7, Q6A0A8, Q6P5E0, Q8R3X8 | Gene names | Stard8, Kiaa0189 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
C43BP_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 15) | NC score | 0.259311 (rank : 14) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y5P4, Q96Q85, Q96Q88, Q9H2S7, Q9H2S8 | Gene names | COL4A3BP, STARD11 | |||
|
Domain Architecture |

|
|||||
| Description | Goodpasture antigen-binding protein (EC 2.7.11.9) (GPBP) (Collagen type IV alpha-3-binding protein) (StAR-related lipid transfer protein 11) (StARD11) (START domain-containing protein 11). | |||||
|
ACO12_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 16) | NC score | 0.224973 (rank : 16) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8WYK0 | Gene names | ACOT12, CACH, CACH1, STARD12 | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A thioesterase 12 (EC 3.1.2.1) (Acyl-CoA thioesterase 12) (Acyl-CoA thioester hydrolase 12) (Cytoplasmic acetyl-CoA hydrolase 1) (CACH-1) (hCACH-1) (START domain-containing protein 12) (StARD12). | |||||
|
STAR3_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 17) | NC score | 0.153404 (rank : 20) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14849, Q53Y53, Q96HM9 | Gene names | STARD3, CAB1, MLN64 | |||
|
Domain Architecture |
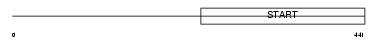
|
|||||
| Description | StAR-related lipid transfer protein 3 (StARD3) (START domain- containing protein 3) (Metastatic lymph node protein 64) (Protein MLN 64) (Protein CAB1). | |||||
|
STAR6_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 18) | NC score | 0.190537 (rank : 19) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P59096 | Gene names | Stard6 | |||
|
Domain Architecture |
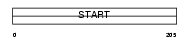
|
|||||
| Description | StAR-related lipid transfer protein 6 (StARD6) (START domain- containing protein 6). | |||||
|
STAR_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 19) | NC score | 0.195438 (rank : 18) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P51557, Q543A5, Q924Y5, Q9D2G1 | Gene names | Star | |||
|
Domain Architecture |
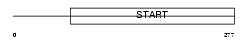
|
|||||
| Description | Steroidogenic acute regulatory protein, mitochondrial precursor (StAR) (StARD1) (Luteinizing hormone-induced protein). | |||||
|
STAR_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 20) | NC score | 0.195509 (rank : 17) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P49675, Q16396 | Gene names | STAR | |||
|
Domain Architecture |
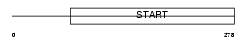
|
|||||
| Description | Steroidogenic acute regulatory protein, mitochondrial precursor (StAR) (StARD1). | |||||
|
STAR6_HUMAN
|
||||||
| θ value | 1.06291 (rank : 21) | NC score | 0.147756 (rank : 21) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P59095 | Gene names | STARD6 | |||
|
Domain Architecture |
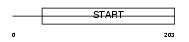
|
|||||
| Description | StAR-related lipid transfer protein 6 (StARD6) (START domain- containing protein 6). | |||||
|
AP3D1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 22) | NC score | 0.020109 (rank : 71) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14617, O00202, O75262, Q59HF5, Q96G11, Q9H3C6 | Gene names | AP3D1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin). | |||||
|
AP3D1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 23) | NC score | 0.020815 (rank : 70) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54774 | Gene names | Ap3d1, Ap3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin) (mBLVR1). | |||||
|
MKL1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 24) | NC score | 0.022513 (rank : 69) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q969V6, Q8TCL1, Q96SC5, Q96SC6, Q9P2B0 | Gene names | MKL1, KIAA1438, MAL | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein) (Megakaryocytic acute leukemia protein). | |||||
|
SNX24_HUMAN
|
||||||
| θ value | 8.99809 (rank : 25) | NC score | 0.028962 (rank : 67) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y343, Q6UY33 | Gene names | SNX24 | |||
|
Domain Architecture |
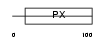
|
|||||
| Description | Sorting nexin-24. | |||||
|
SNX24_MOUSE
|
||||||
| θ value | 8.99809 (rank : 26) | NC score | 0.028881 (rank : 68) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CRB0 | Gene names | Snx24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorting nexin-24. | |||||
|
3BP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 27) | NC score | 0.074413 (rank : 30) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y3L3, Q6IBZ2, Q6ZVL9, Q96HQ5, Q9NSQ9 | Gene names | SH3BP1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
3BP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 28) | NC score | 0.075475 (rank : 29) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P55194, Q99KK8 | Gene names | Sh3bp1, 3bp1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
ABR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 29) | NC score | 0.057432 (rank : 46) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q12979, Q13693, Q13694 | Gene names | ABR | |||
|
Domain Architecture |

|
|||||
| Description | Active breakpoint cluster region-related protein. | |||||
|
ACO11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 30) | NC score | 0.117859 (rank : 22) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8WXI4, O75187, Q96DI1, Q9H883 | Gene names | ACOT11, BFIT, KIAA0707, THEA | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A thioesterase 11 (EC 3.1.2.-) (Acyl-CoA thioesterase 11) (Acyl-CoA thioester hydrolase 11) (Brown fat-inducible thioesterase) (BFIT) (Adipose-associated thioesterase). | |||||
|
ACO11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 31) | NC score | 0.110534 (rank : 23) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VHQ9 | Gene names | Acot11, Bfit, Thea | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A thioesterase 11 (EC 3.1.2.-) (Acyl-CoA thioesterase 11) (Acyl-CoA thioester hydrolase 11) (Brown fat-inducible thioesterase) (BFIT) (Adipose-associated thioesterase). | |||||
|
BACH_HUMAN
|
||||||
| θ value | θ > 10 (rank : 32) | NC score | 0.050690 (rank : 63) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00154, O43703, Q5JYL4, Q9UJM9, Q9Y539, Q9Y540 | Gene names | ACOT7, BACH | |||
|
Domain Architecture |

|
|||||
| Description | Cytosolic acyl coenzyme A thioester hydrolase (EC 3.1.2.2) (Long chain acyl-CoA thioester hydrolase) (CTE-II) (CTE-IIa) (Brain acyl-CoA hydrolase) (Acyl-CoA thioesterase 7). | |||||
|
BACH_MOUSE
|
||||||
| θ value | θ > 10 (rank : 33) | NC score | 0.050228 (rank : 66) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91V12, Q59HQ2 | Gene names | Acot7, Bach | |||
|
Domain Architecture |

|
|||||
| Description | Cytosolic acyl coenzyme A thioester hydrolase (EC 3.1.2.2) (Long chain acyl-CoA thioester hydrolase) (CTE-II) (CTE-IIa) (Brain acyl-CoA hydrolase) (Acyl-CoA thioesterase 7). | |||||
|
BCR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 34) | NC score | 0.055172 (rank : 51) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P11274, P78501, Q12842 | Gene names | BCR, BCR1 | |||
|
Domain Architecture |

|
|||||
| Description | Breakpoint cluster region protein (EC 2.7.11.1) (NY-REN-26 antigen). | |||||
|
CE005_HUMAN
|
||||||
| θ value | θ > 10 (rank : 35) | NC score | 0.056596 (rank : 49) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NYF5, Q6PGQ2, Q9P0I7 | Gene names | C5orf5 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C5orf5 (GAP-like protein N61). | |||||
|
CE005_MOUSE
|
||||||
| θ value | θ > 10 (rank : 36) | NC score | 0.056857 (rank : 47) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K2H3 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Protein C5orf5 homolog. | |||||
|
CEND2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 37) | NC score | 0.052164 (rank : 58) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96P48, O94879, Q59FI7, Q6PHS3, Q8WU51, Q96HP6, Q96L71 | Gene names | CENTD2, ARAP1, KIAA0782 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-delta 2 (Cnt-d2) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 1). | |||||
|
CEND3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 38) | NC score | 0.052259 (rank : 57) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R5G7, Q5DTN4, Q6NVF1, Q8R5G6 | Gene names | Centd3, Arap3, Drag1, Kiaa4097 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3) (Dual specificity Rho- and Arf-GTPase-activating protein 1). | |||||
|
FNBP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 39) | NC score | 0.052694 (rank : 56) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75044 | Gene names | SRGAP2, FNBP2, KIAA0456 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 2 (srGAP2) (Formin-binding protein 2). | |||||
|
GMIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 40) | NC score | 0.059158 (rank : 45) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P107 | Gene names | GMIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GEM-interacting protein (GMIP). | |||||
|
GMIP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 41) | NC score | 0.059858 (rank : 44) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6PGG2, Q6P9S3 | Gene names | Gmip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GEM-interacting protein (GMIP). | |||||
|
GRLF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 42) | NC score | 0.067654 (rank : 34) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NRY4, Q14452, Q9C0E1 | Gene names | GRLF1, GRF1, KIAA1722 | |||
|
Domain Architecture |

|
|||||
| Description | Glucocorticoid receptor DNA-binding factor 1 (Glucocorticoid receptor repression factor 1) (GRF-1) (Rho GAP p190A) (p190-A). | |||||
|
OPHN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 43) | NC score | 0.050498 (rank : 65) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60890, Q5JQ81, Q8WX47 | Gene names | OPHN1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
OPHN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 44) | NC score | 0.052136 (rank : 59) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99J31, Q544K7 | Gene names | Ophn1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
RBP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 45) | NC score | 0.067020 (rank : 35) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15311 | Gene names | RALBP1, RLIP1, RLIP76 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (76 kDa Ral-interacting protein) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
RBP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 46) | NC score | 0.071221 (rank : 33) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62172, Q9CRE5 | Gene names | Ralbp1, Rip1 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
RGAP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 47) | NC score | 0.053819 (rank : 54) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H0H5, Q6PJ26, Q9NWN2, Q9P250, Q9P2W2 | Gene names | RACGAP1, KIAA1478, MGCRACGAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rac GTPase-activating protein 1 (MgcRacGAP). | |||||
|
RHG01_HUMAN
|
||||||
| θ value | θ > 10 (rank : 48) | NC score | 0.066555 (rank : 36) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q07960 | Gene names | ARHGAP1, CDC42GAP, RHOGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 1 (GTPase-activating protein rhoOGAP) (Rho-related small GTPase protein activator) (CDC42 GTPase-activating protein) (p50-RhoGAP). | |||||
|
RHG04_HUMAN
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.064162 (rank : 39) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P98171, Q14144 | Gene names | ARHGAP4, KIAA0131, RGC1, RHOGAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 4 (Rho-GAP hematopoietic protein C1) (p115). | |||||
|
RHG06_HUMAN
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.082208 (rank : 28) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43182, O43437, Q9P1B3, Q9UK81, Q9UK82 | Gene names | ARHGAP6, RHOGAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
RHG06_MOUSE
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.083454 (rank : 27) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O54834, Q9QZL8 | Gene names | Arhgap6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
RHG08_HUMAN
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.051306 (rank : 61) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NSG0, O75983, O95695, Q96RW1, Q96RW2, Q9HA49, Q9HC46, Q9NVX8, Q9NXL1, Q9UH20 | Gene names | ARHGAP8 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 8. | |||||
|
RHG08_MOUSE
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.054767 (rank : 52) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CXP4, Q99JY7 | Gene names | Arhgap8 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 8. | |||||
|
RHG09_HUMAN
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.054161 (rank : 53) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BRR9, Q8TCJ3, Q8WYR0, Q96EZ2, Q96S74 | Gene names | ARHGAP9 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 9. | |||||
|
RHG12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.061212 (rank : 41) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IWW6, Q86UB3, Q8IWW7, Q8N3L1, Q9NT76 | Gene names | ARHGAP12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
RHG12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.060366 (rank : 42) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C0D4, Q8BVP8 | Gene names | Arhgap12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
RHG18_HUMAN
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.093774 (rank : 25) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N392, Q58EZ3, Q6P679, Q6PJD7, Q96S64 | Gene names | ARHGAP18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho-GTPase-activating protein 18 (MacGAP). | |||||
|
RHG18_MOUSE
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.099299 (rank : 24) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K0Q5, Q8BP03, Q8R196 | Gene names | Arhgap18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho-GTPase-activating protein 18. | |||||
|
RHG25_MOUSE
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.050654 (rank : 64) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BYW1, Q2VPR2, Q3TE04, Q3TVX0, Q3UVB7, Q6A0E0, Q8BX98 | Gene names | Arhgap25, Kiaa0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
RHG26_HUMAN
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.052047 (rank : 60) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UNA1, O75117, Q9BYS6, Q9BYS7, Q9UJ00 | Gene names | ARHGAP26, GRAF, KIAA0621, OPHN1L | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 26 (Oligophrenin-1-like protein) (GTPase regulator associated with focal adhesion kinase). | |||||
|
SRGP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.050727 (rank : 62) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7Z6B7, Q9H8A3, Q9P2P2 | Gene names | SRGAP1, ARHGAP13, KIAA1304 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 1 (srGAP1) (Rho-GTPase- activating protein 13). | |||||
|
SRGP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.055323 (rank : 50) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43295, Q8IX13, Q8IZV8 | Gene names | SRGAP3, ARHGAP14, KIAA0411, MEGAP, SRGAP2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Mental disorder- associated GAP) (Rho-GTPase-activating protein 14). | |||||
|
SRGP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.053104 (rank : 55) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q812A2, Q80U09, Q8BKP4, Q8BLD0, Q925I2 | Gene names | Srgap3, Arhgap14, Kiaa0411, Srgap2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Rho-GTPase-activating protein 14). | |||||
|
STAR3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.085669 (rank : 26) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61542 | Gene names | Stard3, Es64, Mln64 | |||
|
Domain Architecture |
|
|||||
| Description | StAR-related lipid transfer protein 3 (StARD3) (START domain- containing protein 3) (Protein MLN 64) (Protein ES 64). | |||||
|
STAR4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.065005 (rank : 38) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96DR4 | Gene names | STARD4 | |||
|
Domain Architecture |
|
|||||
| Description | StAR-related lipid transfer protein 4 (StARD4) (START domain- containing protein 4). | |||||
|
STAR4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.073317 (rank : 31) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99JV5 | Gene names | Stard4 | |||
|
Domain Architecture |
|
|||||
| Description | StAR-related lipid transfer protein 4 (StARD4) (START domain- containing protein 4). | |||||
|
STAR5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.066447 (rank : 37) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NSY2, P59094 | Gene names | STARD5 | |||
|
Domain Architecture |
|
|||||
| Description | StAR-related lipid transfer protein 5 (StARD5) (START domain- containing protein 5). | |||||
|
STAR5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.071437 (rank : 32) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9EPQ7, Q9CQY0, Q9EPQ6 | Gene names | Stard5 | |||
|
Domain Architecture |
|
|||||
| Description | StAR-related lipid transfer protein 5 (StARD5) (START domain- containing protein 5). | |||||
|
TAGAP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.063341 (rank : 40) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N103, Q2NKM8, Q8NI40, Q96KZ2, Q96QA2 | Gene names | TAGAP, TAGAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell activation Rho GTPase-activating protein (T-cell activation GTPase-activating protein). | |||||
|
TAGAP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.060054 (rank : 43) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K2L9, Q920D6 | Gene names | Tagap, Tagap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell activation Rho GTPase-activating protein (T-cell activation GTPase-activating protein). | |||||
|
TCGAP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.056599 (rank : 48) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80YF9, Q6P7W6 | Gene names | Snx26, Tcgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
PCTL_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 6.59259e-152 (rank : 1) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Y365, O60532 | Gene names | STARD10, SDCCAG28 | |||
|
Domain Architecture |

|
|||||
| Description | PCTP-like protein (PCTP-L) (StAR-related lipid transfer protein 10) (StARD10) (START domain-containing protein 10) (Serologically defined colon cancer antigen 28) (Antigen NY-CO-28). | |||||
|
PCTL_MOUSE
|
||||||
| NC score | 0.999416 (rank : 2) | θ value | 1.51984e-148 (rank : 2) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9JMD3 | Gene names | Stard10, Pctpl, Sdccag28, Sdccagg28 | |||
|
Domain Architecture |

|
|||||
| Description | PCTP-like protein (PCTP-L) (StAR-related lipid transfer protein 10) (StARD10) (START domain-containing protein 10) (Serologically defined colon cancer antigen 28 homolog). | |||||
|
PPCT_MOUSE
|
||||||
| NC score | 0.629968 (rank : 3) | θ value | 8.95645e-16 (rank : 3) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P53808, Q9QZX0, Q9R058 | Gene names | Pctp, Stard2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylcholine transfer protein (PC-TP) (StAR-related lipid transfer protein 2) (StARD2) (START domain-containing protein 2). | |||||
|
PPCT_HUMAN
|
||||||
| NC score | 0.599742 (rank : 4) | θ value | 1.2105e-12 (rank : 4) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UKL6, Q9UIT3, Q9UKW7 | Gene names | PCTP, STARD2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylcholine transfer protein (PC-TP) (StAR-related lipid transfer protein 2) (StARD2) (START domain-containing protein 2). | |||||
|
STAR7_HUMAN
|
||||||
| NC score | 0.494885 (rank : 5) | θ value | 1.69304e-06 (rank : 9) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NQZ5, Q6GU43, Q969M6 | Gene names | STARD7 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 7 (StARD7) (START domain- containing protein 7) (Protein GTT1). | |||||
|
STAR7_MOUSE
|
||||||
| NC score | 0.491066 (rank : 6) | θ value | 1.29631e-06 (rank : 8) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8R1R3 | Gene names | Stard7 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 7 (StARD7) (START domain- containing protein 7). | |||||
|
STA13_MOUSE
|
||||||
| NC score | 0.324688 (rank : 7) | θ value | 8.40245e-06 (rank : 11) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q923Q2, Q8K369 | Gene names | Stard13 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 13 (StARD13) (START domain- containing protein 13). | |||||
|
RHG07_HUMAN
|
||||||
| NC score | 0.323740 (rank : 8) | θ value | 1.38499e-08 (rank : 5) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96QB1, O14868, O43199, Q9C0E0 | Gene names | DLC1, ARHGAP7, KIAA1723, STARD12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 7 (Rho-type GTPase-activating protein 7) (Deleted in liver cancer 1 protein) (Dlc-1) (HP protein) (StAR-related lipid transfer protein 12) (StARD12) (START domain-containing protein 12). | |||||
|
STA13_HUMAN
|
||||||
| NC score | 0.323388 (rank : 9) | θ value | 6.43352e-06 (rank : 10) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y3M8, Q5HYH1, Q5TAE3, Q6UN61, Q86TP6, Q86WQ3, Q86XT1 | Gene names | STARD13, DLC2, GT650 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 13 (StARD13) (START domain- containing protein 13) (46H23.2) (Deleted in liver cancer protein 2) (Rho GTPase-activating protein). | |||||
|
STAR8_MOUSE
|
||||||
| NC score | 0.314960 (rank : 10) | θ value | 4.1701e-05 (rank : 14) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8K031, Q3UZC7, Q6A0A8, Q6P5E0, Q8R3X8 | Gene names | Stard8, Kiaa0189 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
STAR8_HUMAN
|
||||||
| NC score | 0.311667 (rank : 11) | θ value | 4.1701e-05 (rank : 13) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q92502, Q5JST0 | Gene names | STARD8, KIAA0189 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
RHG07_MOUSE
|
||||||
| NC score | 0.309791 (rank : 12) | θ value | 9.92553e-07 (rank : 7) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9R0Z9, Q8R541 | Gene names | Dlc1, Arhgap7, Stard12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 7 (Rho-type GTPase-activating protein 7) (Deleted in liver cancer 1 protein homolog) (Dlc-1) (StAR-related lipid transfer protein 12) (StARD12) (START domain-containing protein 12). | |||||
|
C43BP_MOUSE
|
||||||
| NC score | 0.262074 (rank : 13) | θ value | 4.1701e-05 (rank : 12) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9EQG9, Q91WB1, Q9CU52, Q9EQG8 | Gene names | Col4a3bp, Stard11 | |||
|
Domain Architecture |

|
|||||
| Description | Goodpasture antigen-binding protein (EC 2.7.11.9) (GPBP) (Collagen type IV alpha-3-binding protein) (StAR-related lipid transfer protein 11) (StARD11) (START domain-containing protein 11). | |||||
|
C43BP_HUMAN
|
||||||
| NC score | 0.259311 (rank : 14) | θ value | 9.29e-05 (rank : 15) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y5P4, Q96Q85, Q96Q88, Q9H2S7, Q9H2S8 | Gene names | COL4A3BP, STARD11 | |||
|
Domain Architecture |

|
|||||
| Description | Goodpasture antigen-binding protein (EC 2.7.11.9) (GPBP) (Collagen type IV alpha-3-binding protein) (StAR-related lipid transfer protein 11) (StARD11) (START domain-containing protein 11). | |||||
|
ACO12_MOUSE
|
||||||
| NC score | 0.256569 (rank : 15) | θ value | 7.59969e-07 (rank : 6) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9DBK0, Q544M5, Q8R108 | Gene names | Acot12, Cach, Cach1 | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A thioesterase 12 (EC 3.1.2.1) (Acyl-CoA thioesterase 12) (Acyl-CoA thioester hydrolase 12) (Cytoplasmic acetyl-CoA hydrolase 1) (CACH-1) (mCACH-1). | |||||
|
ACO12_HUMAN
|
||||||
| NC score | 0.224973 (rank : 16) | θ value | 0.000121331 (rank : 16) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8WYK0 | Gene names | ACOT12, CACH, CACH1, STARD12 | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A thioesterase 12 (EC 3.1.2.1) (Acyl-CoA thioesterase 12) (Acyl-CoA thioester hydrolase 12) (Cytoplasmic acetyl-CoA hydrolase 1) (CACH-1) (hCACH-1) (START domain-containing protein 12) (StARD12). | |||||
|
STAR_HUMAN
|
||||||
| NC score | 0.195509 (rank : 17) | θ value | 0.0330416 (rank : 20) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P49675, Q16396 | Gene names | STAR | |||
|
Domain Architecture |

|
|||||
| Description | Steroidogenic acute regulatory protein, mitochondrial precursor (StAR) (StARD1). | |||||
|
STAR_MOUSE
|
||||||
| NC score | 0.195438 (rank : 18) | θ value | 0.0193708 (rank : 19) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P51557, Q543A5, Q924Y5, Q9D2G1 | Gene names | Star | |||
|
Domain Architecture |

|
|||||
| Description | Steroidogenic acute regulatory protein, mitochondrial precursor (StAR) (StARD1) (Luteinizing hormone-induced protein). | |||||
|
STAR6_MOUSE
|
||||||
| NC score | 0.190537 (rank : 19) | θ value | 0.00665767 (rank : 18) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P59096 | Gene names | Stard6 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 6 (StARD6) (START domain- containing protein 6). | |||||
|
STAR3_HUMAN
|
||||||
| NC score | 0.153404 (rank : 20) | θ value | 0.00509761 (rank : 17) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14849, Q53Y53, Q96HM9 | Gene names | STARD3, CAB1, MLN64 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 3 (StARD3) (START domain- containing protein 3) (Metastatic lymph node protein 64) (Protein MLN 64) (Protein CAB1). | |||||
|
STAR6_HUMAN
|
||||||
| NC score | 0.147756 (rank : 21) | θ value | 1.06291 (rank : 21) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P59095 | Gene names | STARD6 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 6 (StARD6) (START domain- containing protein 6). | |||||
|
ACO11_HUMAN
|
||||||
| NC score | 0.117859 (rank : 22) | θ value | θ > 10 (rank : 30) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8WXI4, O75187, Q96DI1, Q9H883 | Gene names | ACOT11, BFIT, KIAA0707, THEA | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A thioesterase 11 (EC 3.1.2.-) (Acyl-CoA thioesterase 11) (Acyl-CoA thioester hydrolase 11) (Brown fat-inducible thioesterase) (BFIT) (Adipose-associated thioesterase). | |||||
|
ACO11_MOUSE
|
||||||
| NC score | 0.110534 (rank : 23) | θ value | θ > 10 (rank : 31) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VHQ9 | Gene names | Acot11, Bfit, Thea | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A thioesterase 11 (EC 3.1.2.-) (Acyl-CoA thioesterase 11) (Acyl-CoA thioester hydrolase 11) (Brown fat-inducible thioesterase) (BFIT) (Adipose-associated thioesterase). | |||||
|
RHG18_MOUSE
|
||||||
| NC score | 0.099299 (rank : 24) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K0Q5, Q8BP03, Q8R196 | Gene names | Arhgap18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho-GTPase-activating protein 18. | |||||
|
RHG18_HUMAN
|
||||||
| NC score | 0.093774 (rank : 25) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N392, Q58EZ3, Q6P679, Q6PJD7, Q96S64 | Gene names | ARHGAP18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho-GTPase-activating protein 18 (MacGAP). | |||||
|
STAR3_MOUSE
|
||||||
| NC score | 0.085669 (rank : 26) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61542 | Gene names | Stard3, Es64, Mln64 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 3 (StARD3) (START domain- containing protein 3) (Protein MLN 64) (Protein ES 64). | |||||
|
RHG06_MOUSE
|
||||||
| NC score | 0.083454 (rank : 27) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O54834, Q9QZL8 | Gene names | Arhgap6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
RHG06_HUMAN
|
||||||
| NC score | 0.082208 (rank : 28) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43182, O43437, Q9P1B3, Q9UK81, Q9UK82 | Gene names | ARHGAP6, RHOGAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
3BP1_MOUSE
|
||||||
| NC score | 0.075475 (rank : 29) | θ value | θ > 10 (rank : 28) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P55194, Q99KK8 | Gene names | Sh3bp1, 3bp1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
3BP1_HUMAN
|
||||||
| NC score | 0.074413 (rank : 30) | θ value | θ > 10 (rank : 27) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y3L3, Q6IBZ2, Q6ZVL9, Q96HQ5, Q9NSQ9 | Gene names | SH3BP1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
STAR4_MOUSE
|
||||||
| NC score | 0.073317 (rank : 31) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99JV5 | Gene names | Stard4 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 4 (StARD4) (START domain- containing protein 4). | |||||
|
STAR5_MOUSE
|
||||||
| NC score | 0.071437 (rank : 32) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9EPQ7, Q9CQY0, Q9EPQ6 | Gene names | Stard5 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 5 (StARD5) (START domain- containing protein 5). | |||||
|
RBP1_MOUSE
|
||||||
| NC score | 0.071221 (rank : 33) | θ value | θ > 10 (rank : 46) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62172, Q9CRE5 | Gene names | Ralbp1, Rip1 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
GRLF1_HUMAN
|
||||||
| NC score | 0.067654 (rank : 34) | θ value | θ > 10 (rank : 42) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NRY4, Q14452, Q9C0E1 | Gene names | GRLF1, GRF1, KIAA1722 | |||
|
Domain Architecture |

|
|||||
| Description | Glucocorticoid receptor DNA-binding factor 1 (Glucocorticoid receptor repression factor 1) (GRF-1) (Rho GAP p190A) (p190-A). | |||||
|
RBP1_HUMAN
|
||||||
| NC score | 0.067020 (rank : 35) | θ value | θ > 10 (rank : 45) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15311 | Gene names | RALBP1, RLIP1, RLIP76 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (76 kDa Ral-interacting protein) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
RHG01_HUMAN
|
||||||
| NC score | 0.066555 (rank : 36) | θ value | θ > 10 (rank : 48) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q07960 | Gene names | ARHGAP1, CDC42GAP, RHOGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 1 (GTPase-activating protein rhoOGAP) (Rho-related small GTPase protein activator) (CDC42 GTPase-activating protein) (p50-RhoGAP). | |||||
|
STAR5_HUMAN
|
||||||
| NC score | 0.066447 (rank : 37) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NSY2, P59094 | Gene names | STARD5 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 5 (StARD5) (START domain- containing protein 5). | |||||
|
STAR4_HUMAN
|
||||||
| NC score | 0.065005 (rank : 38) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96DR4 | Gene names | STARD4 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 4 (StARD4) (START domain- containing protein 4). | |||||
|
RHG04_HUMAN
|
||||||
| NC score | 0.064162 (rank : 39) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P98171, Q14144 | Gene names | ARHGAP4, KIAA0131, RGC1, RHOGAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 4 (Rho-GAP hematopoietic protein C1) (p115). | |||||
|
TAGAP_HUMAN
|
||||||
| NC score | 0.063341 (rank : 40) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N103, Q2NKM8, Q8NI40, Q96KZ2, Q96QA2 | Gene names | TAGAP, TAGAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell activation Rho GTPase-activating protein (T-cell activation GTPase-activating protein). | |||||
|
RHG12_HUMAN
|
||||||
| NC score | 0.061212 (rank : 41) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IWW6, Q86UB3, Q8IWW7, Q8N3L1, Q9NT76 | Gene names | ARHGAP12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
RHG12_MOUSE
|
||||||
| NC score | 0.060366 (rank : 42) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C0D4, Q8BVP8 | Gene names | Arhgap12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
TAGAP_MOUSE
|
||||||
| NC score | 0.060054 (rank : 43) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K2L9, Q920D6 | Gene names | Tagap, Tagap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell activation Rho GTPase-activating protein (T-cell activation GTPase-activating protein). | |||||
|
GMIP_MOUSE
|
||||||
| NC score | 0.059858 (rank : 44) | θ value | θ > 10 (rank : 41) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6PGG2, Q6P9S3 | Gene names | Gmip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GEM-interacting protein (GMIP). | |||||
|
GMIP_HUMAN
|
||||||
| NC score | 0.059158 (rank : 45) | θ value | θ > 10 (rank : 40) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P107 | Gene names | GMIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GEM-interacting protein (GMIP). | |||||
|
ABR_HUMAN
|
||||||
| NC score | 0.057432 (rank : 46) | θ value | θ > 10 (rank : 29) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q12979, Q13693, Q13694 | Gene names | ABR | |||
|
Domain Architecture |

|
|||||
| Description | Active breakpoint cluster region-related protein. | |||||
|
CE005_MOUSE
|
||||||
| NC score | 0.056857 (rank : 47) | θ value | θ > 10 (rank : 36) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K2H3 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Protein C5orf5 homolog. | |||||
|
TCGAP_MOUSE
|
||||||
| NC score | 0.056599 (rank : 48) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80YF9, Q6P7W6 | Gene names | Snx26, Tcgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
CE005_HUMAN
|
||||||
| NC score | 0.056596 (rank : 49) | θ value | θ > 10 (rank : 35) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NYF5, Q6PGQ2, Q9P0I7 | Gene names | C5orf5 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C5orf5 (GAP-like protein N61). | |||||
|
SRGP2_HUMAN
|
||||||
| NC score | 0.055323 (rank : 50) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43295, Q8IX13, Q8IZV8 | Gene names | SRGAP3, ARHGAP14, KIAA0411, MEGAP, SRGAP2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Mental disorder- associated GAP) (Rho-GTPase-activating protein 14). | |||||
|
BCR_HUMAN
|
||||||
| NC score | 0.055172 (rank : 51) | θ value | θ > 10 (rank : 34) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P11274, P78501, Q12842 | Gene names | BCR, BCR1 | |||
|
Domain Architecture |

|
|||||
| Description | Breakpoint cluster region protein (EC 2.7.11.1) (NY-REN-26 antigen). | |||||
|
RHG08_MOUSE
|
||||||
| NC score | 0.054767 (rank : 52) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CXP4, Q99JY7 | Gene names | Arhgap8 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 8. | |||||
|
RHG09_HUMAN
|
||||||
| NC score | 0.054161 (rank : 53) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BRR9, Q8TCJ3, Q8WYR0, Q96EZ2, Q96S74 | Gene names | ARHGAP9 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 9. | |||||
|
RGAP1_HUMAN
|
||||||
| NC score | 0.053819 (rank : 54) | θ value | θ > 10 (rank : 47) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H0H5, Q6PJ26, Q9NWN2, Q9P250, Q9P2W2 | Gene names | RACGAP1, KIAA1478, MGCRACGAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rac GTPase-activating protein 1 (MgcRacGAP). | |||||
|
SRGP2_MOUSE
|
||||||
| NC score | 0.053104 (rank : 55) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q812A2, Q80U09, Q8BKP4, Q8BLD0, Q925I2 | Gene names | Srgap3, Arhgap14, Kiaa0411, Srgap2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Rho-GTPase-activating protein 14). | |||||
|
FNBP2_HUMAN
|
||||||
| NC score | 0.052694 (rank : 56) | θ value | θ > 10 (rank : 39) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75044 | Gene names | SRGAP2, FNBP2, KIAA0456 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 2 (srGAP2) (Formin-binding protein 2). | |||||
|
CEND3_MOUSE
|
||||||
| NC score | 0.052259 (rank : 57) | θ value | θ > 10 (rank : 38) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R5G7, Q5DTN4, Q6NVF1, Q8R5G6 | Gene names | Centd3, Arap3, Drag1, Kiaa4097 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3) (Dual specificity Rho- and Arf-GTPase-activating protein 1). | |||||
|
CEND2_HUMAN
|
||||||
| NC score | 0.052164 (rank : 58) | θ value | θ > 10 (rank : 37) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96P48, O94879, Q59FI7, Q6PHS3, Q8WU51, Q96HP6, Q96L71 | Gene names | CENTD2, ARAP1, KIAA0782 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-delta 2 (Cnt-d2) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 1). | |||||
|
OPHN1_MOUSE
|
||||||
| NC score | 0.052136 (rank : 59) | θ value | θ > 10 (rank : 44) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99J31, Q544K7 | Gene names | Ophn1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
RHG26_HUMAN
|
||||||
| NC score | 0.052047 (rank : 60) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UNA1, O75117, Q9BYS6, Q9BYS7, Q9UJ00 | Gene names | ARHGAP26, GRAF, KIAA0621, OPHN1L | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 26 (Oligophrenin-1-like protein) (GTPase regulator associated with focal adhesion kinase). | |||||
|
RHG08_HUMAN
|
||||||
| NC score | 0.051306 (rank : 61) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NSG0, O75983, O95695, Q96RW1, Q96RW2, Q9HA49, Q9HC46, Q9NVX8, Q9NXL1, Q9UH20 | Gene names | ARHGAP8 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 8. | |||||
|
SRGP1_HUMAN
|
||||||
| NC score | 0.050727 (rank : 62) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7Z6B7, Q9H8A3, Q9P2P2 | Gene names | SRGAP1, ARHGAP13, KIAA1304 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 1 (srGAP1) (Rho-GTPase- activating protein 13). | |||||
|
BACH_HUMAN
|
||||||
| NC score | 0.050690 (rank : 63) | θ value | θ > 10 (rank : 32) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00154, O43703, Q5JYL4, Q9UJM9, Q9Y539, Q9Y540 | Gene names | ACOT7, BACH | |||
|
Domain Architecture |

|
|||||
| Description | Cytosolic acyl coenzyme A thioester hydrolase (EC 3.1.2.2) (Long chain acyl-CoA thioester hydrolase) (CTE-II) (CTE-IIa) (Brain acyl-CoA hydrolase) (Acyl-CoA thioesterase 7). | |||||
|
RHG25_MOUSE
|
||||||
| NC score | 0.050654 (rank : 64) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BYW1, Q2VPR2, Q3TE04, Q3TVX0, Q3UVB7, Q6A0E0, Q8BX98 | Gene names | Arhgap25, Kiaa0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
OPHN1_HUMAN
|
||||||
| NC score | 0.050498 (rank : 65) | θ value | θ > 10 (rank : 43) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60890, Q5JQ81, Q8WX47 | Gene names | OPHN1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
BACH_MOUSE
|
||||||
| NC score | 0.050228 (rank : 66) | θ value | θ > 10 (rank : 33) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91V12, Q59HQ2 | Gene names | Acot7, Bach | |||
|
Domain Architecture |

|
|||||
| Description | Cytosolic acyl coenzyme A thioester hydrolase (EC 3.1.2.2) (Long chain acyl-CoA thioester hydrolase) (CTE-II) (CTE-IIa) (Brain acyl-CoA hydrolase) (Acyl-CoA thioesterase 7). | |||||
|
SNX24_HUMAN
|
||||||
| NC score | 0.028962 (rank : 67) | θ value | 8.99809 (rank : 25) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y343, Q6UY33 | Gene names | SNX24 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-24. | |||||
|
SNX24_MOUSE
|
||||||
| NC score | 0.028881 (rank : 68) | θ value | 8.99809 (rank : 26) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CRB0 | Gene names | Snx24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorting nexin-24. | |||||
|
MKL1_HUMAN
|
||||||
| NC score | 0.022513 (rank : 69) | θ value | 5.27518 (rank : 24) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q969V6, Q8TCL1, Q96SC5, Q96SC6, Q9P2B0 | Gene names | MKL1, KIAA1438, MAL | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein) (Megakaryocytic acute leukemia protein). | |||||
|
AP3D1_MOUSE
|
||||||
| NC score | 0.020815 (rank : 70) | θ value | 5.27518 (rank : 23) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54774 | Gene names | Ap3d1, Ap3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin) (mBLVR1). | |||||
|
AP3D1_HUMAN
|
||||||
| NC score | 0.020109 (rank : 71) | θ value | 5.27518 (rank : 22) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14617, O00202, O75262, Q59HF5, Q96G11, Q9H3C6 | Gene names | AP3D1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin). | |||||