Please be patient as the page loads
|
ALMS1_MOUSE
|
||||||
| SwissProt Accessions | Q8K4E0, Q6A084, Q8C9N9 | Gene names | Alms1, Kiaa0328 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alstrom syndrome protein 1 homolog. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ALMS1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.895643 (rank : 2) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8TCU4, Q53S05, Q580Q8, Q86VP9, Q9Y4G4 | Gene names | ALMS1, KIAA0328 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alstrom syndrome protein 1. | |||||
|
ALMS1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 181 | |
| SwissProt Accessions | Q8K4E0, Q6A084, Q8C9N9 | Gene names | Alms1, Kiaa0328 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alstrom syndrome protein 1 homolog. | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 3) | NC score | 0.128256 (rank : 3) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 4) | NC score | 0.123368 (rank : 4) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
AFF4_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 5) | NC score | 0.073329 (rank : 12) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
ANK3_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 6) | NC score | 0.027805 (rank : 90) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
NFRKB_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 7) | NC score | 0.076643 (rank : 11) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6P4R8, Q12869, Q15312, Q9H048 | Gene names | NFRKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor related to kappa-B-binding protein (DNA-binding protein R kappa-B). | |||||
|
CBP_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 8) | NC score | 0.079699 (rank : 9) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
SSH2_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 9) | NC score | 0.041726 (rank : 48) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5SW75, Q3TDK8, Q3TYP8, Q3U2K3, Q5F268, Q5SW74, Q69ZC3, Q76I78 | Gene names | Ssh2, Kiaa1725, Ssh2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 2 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-2L) (mSSH-2L). | |||||
|
KCNQ2_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 10) | NC score | 0.029255 (rank : 85) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z351, Q8R498, Q9QWN9, Q9Z342, Q9Z343, Q9Z344, Q9Z345, Q9Z346, Q9Z347, Q9Z348, Q9Z349, Q9Z350 | Gene names | Kcnq2, Kqt2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
AKA12_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 11) | NC score | 0.069852 (rank : 13) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
EP300_HUMAN
|
||||||
| θ value | 0.163984 (rank : 12) | NC score | 0.094940 (rank : 6) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
CBP_HUMAN
|
||||||
| θ value | 0.21417 (rank : 13) | NC score | 0.084270 (rank : 8) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
MAP4_HUMAN
|
||||||
| θ value | 0.279714 (rank : 14) | NC score | 0.068674 (rank : 14) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
PDZD2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 15) | NC score | 0.048744 (rank : 31) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
RBMX2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 16) | NC score | 0.030407 (rank : 79) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R0F5, Q3TI42 | Gene names | Rbmx2 | |||
|
Domain Architecture |
|
|||||
| Description | RNA-binding motif protein, X-linked 2. | |||||
|
VPS72_HUMAN
|
||||||
| θ value | 0.279714 (rank : 17) | NC score | 0.067696 (rank : 15) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q15906, Q53GJ2, Q5U0R4 | Gene names | VPS72, TCFL1, YL1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 72 homolog (Transcription factor-like 1) (Protein YL-1). | |||||
|
VPS72_MOUSE
|
||||||
| θ value | 0.365318 (rank : 18) | NC score | 0.065519 (rank : 17) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q62481, Q3U2N4, Q810A9, Q99K81 | Gene names | Vps72, Tcfl1, Yl1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 72 homolog (Transcription factor-like 1) (Protein YL-1). | |||||
|
ARI1A_HUMAN
|
||||||
| θ value | 0.47712 (rank : 19) | NC score | 0.063302 (rank : 19) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
K0146_HUMAN
|
||||||
| θ value | 0.47712 (rank : 20) | NC score | 0.044151 (rank : 43) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14159, Q96BI5 | Gene names | KIAA0146 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0146. | |||||
|
MAGC1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 21) | NC score | 0.038366 (rank : 60) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
MAML1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 22) | NC score | 0.045694 (rank : 40) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q92585, Q9NZ12 | Gene names | MAML1, KIAA0200 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 1 (Mam-1). | |||||
|
TMUB1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 23) | NC score | 0.050237 (rank : 30) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BVT8 | Gene names | TMUB1, C7orf21 | |||
|
Domain Architecture |
|
|||||
| Description | Transmembrane and ubiquitin-like domain-containing protein 1 (Ubiquitin-like protein SB144). | |||||
|
CJ084_HUMAN
|
||||||
| θ value | 0.62314 (rank : 24) | NC score | 0.054316 (rank : 23) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H8W3, Q9H5V5 | Gene names | C10orf84 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf84. | |||||
|
KIF17_MOUSE
|
||||||
| θ value | 0.62314 (rank : 25) | NC score | 0.017469 (rank : 122) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99PW8 | Gene names | Kif17 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF17 (MmKIF17). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 26) | NC score | 0.098836 (rank : 5) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
TPR_HUMAN
|
||||||
| θ value | 0.62314 (rank : 27) | NC score | 0.019198 (rank : 115) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
XKR5_HUMAN
|
||||||
| θ value | 0.62314 (rank : 28) | NC score | 0.043809 (rank : 46) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6UX68, Q5GH74 | Gene names | XKR5, XRG5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | XK-related protein 5. | |||||
|
DSCL1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 29) | NC score | 0.011836 (rank : 150) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8TD84, Q76MU9, Q8IZY3, Q8IZY4, Q8WXU7, Q9ULT7 | Gene names | DSCAML1, DSCAM2, KIAA1132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Down syndrome cell adhesion molecule-like protein 1 precursor (Down syndrome cell adhesion molecule 2). | |||||
|
FGD1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 30) | NC score | 0.021639 (rank : 107) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P52734 | Gene names | Fgd1 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein homolog) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
TFAM_MOUSE
|
||||||
| θ value | 0.813845 (rank : 31) | NC score | 0.078768 (rank : 10) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P40630, P97894, P97906, Q543I8, Q9DBM9 | Gene names | Tfam, Hmgts | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor A, mitochondrial precursor (mtTFA) (Testis- specific high mobility group protein) (TS-HMG). | |||||
|
ATX7_HUMAN
|
||||||
| θ value | 1.06291 (rank : 32) | NC score | 0.042162 (rank : 47) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O15265, O75328, O75329, Q9Y6P8 | Gene names | ATXN7, SCA7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein). | |||||
|
FA21C_HUMAN
|
||||||
| θ value | 1.06291 (rank : 33) | NC score | 0.046981 (rank : 35) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y4E1, Q5SQU4, Q5SQU5, Q7L521, Q9UG79 | Gene names | FAM21C, KIAA0592 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM21C. | |||||
|
HNRPC_MOUSE
|
||||||
| θ value | 1.06291 (rank : 34) | NC score | 0.029638 (rank : 84) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z204, Q3TLB5, Q501Q3, Q8C2G5, Q99KE2, Q9CQT3, Q9CY83 | Gene names | Hnrpc, Hnrnpc | |||
|
Domain Architecture |
|
|||||
| Description | Heterogeneous nuclear ribonucleoproteins C1/C2 (hnRNP C1 / hnRNP C2). | |||||
|
HXA11_HUMAN
|
||||||
| θ value | 1.06291 (rank : 35) | NC score | 0.018750 (rank : 116) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P31270 | Gene names | HOXA11, HOX1I | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A11 (Hox-1I). | |||||
|
HXA11_MOUSE
|
||||||
| θ value | 1.06291 (rank : 36) | NC score | 0.014956 (rank : 130) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P31311, Q3V026 | Gene names | Hoxa11, Hox-1.9, Hoxa-11 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A11 (Hox-1.9). | |||||
|
PENK_HUMAN
|
||||||
| θ value | 1.06291 (rank : 37) | NC score | 0.038849 (rank : 57) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P01210 | Gene names | PENK | |||
|
Domain Architecture |

|
|||||
| Description | Proenkephalin A precursor [Contains: Synenkephalin; Met-enkephalin (Opioid growth factor) (OGF); Met-enkephalin-Arg-Gly-Leu; Leu- enkephalin; Met-enkephalin-Arg-Phe]. | |||||
|
PRGC1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 38) | NC score | 0.036254 (rank : 64) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UBK2, Q9UN32 | Gene names | PPARGC1A, LEM6, PGC1, PGC1A, PPARGC1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PPAR gamma coactivator 1-alpha) (PPARGC-1-alpha) (PGC-1-alpha) (Ligand effect modulator 6). | |||||
|
ST5_MOUSE
|
||||||
| θ value | 1.06291 (rank : 39) | NC score | 0.023102 (rank : 102) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q924W7, Q78H54, Q8K2P3, Q924W8 | Gene names | St5, Dennd2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppression of tumorigenicity 5 (DENN domain-containing protein 2B). | |||||
|
SYTL2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 40) | NC score | 0.021435 (rank : 108) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9HCH5, Q2YDA7, Q8ND34, Q96BJ2, Q9H768, Q9NXM1 | Gene names | SYTL2, KIAA1597, SLP2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
ZAN_MOUSE
|
||||||
| θ value | 1.06291 (rank : 41) | NC score | 0.041520 (rank : 50) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
A4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 42) | NC score | 0.041069 (rank : 52) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P05067, P09000, P78438, Q13764, Q13778, Q13793, Q16011, Q16014, Q16019, Q16020, Q9BT38, Q9UCA9, Q9UCB6, Q9UCC8, Q9UCD1, Q9UQ58 | Gene names | APP, A4, AD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein) (Cerebral vascular amyloid peptide) (CVAP) (Protease nexin-II) (PN-II) (APPI) (PreA4) [Contains: Soluble APP-alpha (S-APP- alpha); Soluble APP-beta (S-APP-beta); C99; Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)); Gamma-CTF(57) (Gamma- secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
APLP2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 43) | NC score | 0.046997 (rank : 34) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q06481 | Gene names | APLP2, APPL2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 2 precursor (Amyloid protein homolog) (APPH) (CDEI box-binding protein) (CDEBP). | |||||
|
BPAEA_MOUSE
|
||||||
| θ value | 1.38821 (rank : 44) | NC score | 0.013265 (rank : 141) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
ELK3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 45) | NC score | 0.017051 (rank : 125) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P41971, P97747, Q62346 | Gene names | Elk3, Erp, Net | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-3 (ETS-related protein NET) (ETS- related protein ERP). | |||||
|
FMNL_HUMAN
|
||||||
| θ value | 1.38821 (rank : 46) | NC score | 0.028307 (rank : 89) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O95466, Q6DKG5, Q6IBP3, Q86UH1, Q8N671, Q8TDH1, Q96H10 | Gene names | FMNL1, C17orf1, C17orf1B, FMNL | |||
|
Domain Architecture |

|
|||||
| Description | Formin-like protein 1 (Leukocyte formin) (CLL-associated antigen KW- 13). | |||||
|
HUWE1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 47) | NC score | 0.020928 (rank : 110) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
MYCB2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 48) | NC score | 0.040286 (rank : 55) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O75592, Q5JSX8, Q5VZN6, Q6PIB6, Q9UQ11, Q9Y6E4 | Gene names | MYCBP2, KIAA0916, PAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
NTHL1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 49) | NC score | 0.045760 (rank : 39) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35980 | Gene names | Nthl1, Nth1 | |||
|
Domain Architecture |
|
|||||
| Description | Endonuclease III-like protein 1 (EC 4.2.99.18). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 1.38821 (rank : 50) | NC score | 0.046922 (rank : 37) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
PDXL2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 51) | NC score | 0.053705 (rank : 26) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8CAE9, Q8CFW3 | Gene names | Podxl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Podocalyxin-like protein 2 precursor (Endoglycan). | |||||
|
PHAR2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 52) | NC score | 0.040537 (rank : 54) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O75167, Q68DM2 | Gene names | PHACTR2, C6orf56, KIAA0680 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatase and actin regulator 2. | |||||
|
SETB1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 53) | NC score | 0.029786 (rank : 83) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O88974, Q922K1 | Gene names | Setdb1, Eset | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 4 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 4) (H3-K9-HMTase 4) (SET domain bifurcated 1) (ERG-associated protein with SET domain) (ESET). | |||||
|
TLR2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 54) | NC score | 0.013531 (rank : 139) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60603, O15454, Q8NI00 | Gene names | TLR2, TIL4 | |||
|
Domain Architecture |

|
|||||
| Description | Toll-like receptor 2 precursor (Toll/interleukin 1 receptor-like protein 4) (CD282 antigen). | |||||
|
TREF1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 55) | NC score | 0.023674 (rank : 99) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BXJ2, Q6PCQ4, Q80X25, Q810H8, Q8BY31 | Gene names | Trerf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132). | |||||
|
3BP2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 56) | NC score | 0.030837 (rank : 78) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q06649 | Gene names | Sh3bp2, 3bp2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
BMP2K_HUMAN
|
||||||
| θ value | 1.81305 (rank : 57) | NC score | 0.010697 (rank : 156) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NSY1, O94791, Q8IYF2, Q8N2G7, Q8NHG9, Q9NTG8 | Gene names | BMP2K, BIKE | |||
|
Domain Architecture |
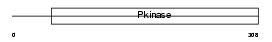
|
|||||
| Description | BMP-2-inducible protein kinase (EC 2.7.11.1) (BIKe). | |||||
|
CC47_MOUSE
|
||||||
| θ value | 1.81305 (rank : 58) | NC score | 0.030927 (rank : 77) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9D024, Q3TSB5, Q3V1S7, Q8C5D9, Q920S6 | Gene names | Ccdc47, Asp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 47 precursor (Adipocyte-specific protein 4). | |||||
|
LR37B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 59) | NC score | 0.025728 (rank : 94) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96QE4, Q5YKG6 | Gene names | LRRC37B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37B precursor (C66 SLIT-like testicular protein). | |||||
|
MARK2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 60) | NC score | 0.006182 (rank : 178) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1056 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q7KZI7, Q15449, Q15524, Q5XGA3, Q68A18, Q96HB3, Q96RG0 | Gene names | MARK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MARK2 (EC 2.7.11.1) (MAP/microtubule affinity-regulating kinase 2) (ELKL motif kinase) (EMK1) (PAR1 homolog). | |||||
|
PGCA_HUMAN
|
||||||
| θ value | 1.81305 (rank : 61) | NC score | 0.037814 (rank : 61) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
PRG4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 62) | NC score | 0.045118 (rank : 42) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
T22D2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 63) | NC score | 0.047723 (rank : 32) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O75157, Q6PI50, Q9H2Z6, Q9H2Z7, Q9H2Z8 | Gene names | TSC22D2, KIAA0669, TILZ4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TSC22 domain family protein 2 (TSC22-related-inducible leucine zipper protein 4). | |||||
|
ZN551_HUMAN
|
||||||
| θ value | 1.81305 (rank : 64) | NC score | 0.001394 (rank : 187) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7Z340, P17034, Q8N246, Q9BRY1 | Gene names | ZNF551, KOX23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 551 (Zinc finger protein KOX23). | |||||
|
ARID2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 65) | NC score | 0.043830 (rank : 45) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q68CP9, Q5EB51, Q645I3, Q6ZRY5, Q7Z3I5, Q86T28, Q96SJ6, Q9HCL5 | Gene names | ARID2, KIAA1557 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 2 (ARID domain- containing protein 2) (BRG1-associated factor 200) (BAF200). | |||||
|
CNOT3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 66) | NC score | 0.038676 (rank : 59) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O75175, Q9NZN7, Q9UF76 | Gene names | CNOT3, KIAA0691, NOT3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
CO7A1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 67) | NC score | 0.014967 (rank : 129) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
DCP1A_MOUSE
|
||||||
| θ value | 2.36792 (rank : 68) | NC score | 0.046809 (rank : 38) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q91YD3, Q6NZE3 | Gene names | Dcp1a, Mitc1, Smif | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA decapping enzyme 1A (EC 3.-.-.-) (Transcription factor SMIF) (MAD homolog 4-interacting transcription coactivator 1) (Smad4-interacting transcriptional co-activator). | |||||
|
FXL19_HUMAN
|
||||||
| θ value | 2.36792 (rank : 69) | NC score | 0.023541 (rank : 100) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6PCT2, Q8N789, Q9NT14 | Gene names | FBXL19, FBL19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box/LRR-repeat protein 19 (F-box and leucine-rich repeat protein 19). | |||||
|
K2027_MOUSE
|
||||||
| θ value | 2.36792 (rank : 70) | NC score | 0.036045 (rank : 65) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BWS5, Q69Z30 | Gene names | Kiaa2027 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA2027. | |||||
|
KI67_HUMAN
|
||||||
| θ value | 2.36792 (rank : 71) | NC score | 0.052716 (rank : 27) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
MACF1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 72) | NC score | 0.011517 (rank : 152) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
MAFB_MOUSE
|
||||||
| θ value | 2.36792 (rank : 73) | NC score | 0.021235 (rank : 109) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P54841 | Gene names | Mafb, Krml, Maf1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafB (V-maf musculoaponeurotic fibrosarcoma oncogene homolog B) (Transcription factor MAF1) (Segmentation protein KR) (Kreisler). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 74) | NC score | 0.040686 (rank : 53) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
NCOAT_MOUSE
|
||||||
| θ value | 2.36792 (rank : 75) | NC score | 0.026709 (rank : 93) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EQQ9, Q3ULY7, Q6ZQ71, Q8BK05, Q8BTT2, Q8CFX2, Q9CSJ4, Q9CUR7 | Gene names | Mgea5, Hexc, Kiaa0679 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional protein NCOAT (Nuclear cytoplasmic O-GlcNAcase and acetyltransferase) (Meningioma-expressed antigen 5) [Includes: Beta- hexosaminidase (EC 3.2.1.52) (N-acetyl-beta-glucosaminidase) (Beta-N- acetylhexosaminidase) (Hexosaminidase C) (N-acetyl-beta-D- glucosaminidase) (O-GlcNAcase); Histone acetyltransferase (EC 2.3.1.48) (HAT)]. | |||||
|
PDPN_HUMAN
|
||||||
| θ value | 2.36792 (rank : 76) | NC score | 0.050340 (rank : 29) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86YL7, O60836, O95128, Q7L375, Q8NBQ8, Q8NBR3 | Gene names | PDPN, GP36 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Podoplanin precursor (Glycoprotein 36) (Gp36) (PA2.26 antigen) (T1A) (T1-alpha) (Aggrus). | |||||
|
PPR3A_MOUSE
|
||||||
| θ value | 2.36792 (rank : 77) | NC score | 0.027113 (rank : 92) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99MR9, Q8BUJ4, Q8BUL0 | Gene names | Ppp1r3a, Pp1g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase 1 regulatory subunit 3A (Protein phosphatase 1 glycogen-associated regulatory subunit) (Protein phosphatase type-1 glycogen targeting subunit) (RGL). | |||||
|
SKI_MOUSE
|
||||||
| θ value | 2.36792 (rank : 78) | NC score | 0.020610 (rank : 111) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q60698, Q8VIL5 | Gene names | Ski | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ski oncogene (C-ski). | |||||
|
TENS1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 79) | NC score | 0.033912 (rank : 71) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9HBL0 | Gene names | TNS1, TNS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-1. | |||||
|
TRI36_HUMAN
|
||||||
| θ value | 2.36792 (rank : 80) | NC score | 0.011832 (rank : 151) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NQ86 | Gene names | TRIM36, RBCC728, RNF98 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 36 (Zinc-binding protein Rbcc728) (RING finger protein 98). | |||||
|
WTAP_MOUSE
|
||||||
| θ value | 2.36792 (rank : 81) | NC score | 0.034381 (rank : 70) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9ER69 | Gene names | Wtap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wilms' tumor 1-associating protein (WT1-associated protein) (Putative pre-mRNA-splicing regulator female-lethal(2D) homolog). | |||||
|
APLP2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | 0.039708 (rank : 56) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q06335 | Gene names | Aplp2 | |||
|
Domain Architecture |
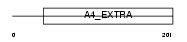
|
|||||
| Description | Amyloid-like protein 2 precursor (CDEI box-binding protein) (CDEBP). | |||||
|
CO3A1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 83) | NC score | 0.017875 (rank : 120) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P02461, Q15112, Q16403, Q6LDB3, Q6LDJ2, Q6LDJ3, Q7KZ56 | Gene names | COL3A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
DOCK2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 84) | NC score | 0.012758 (rank : 144) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C3J5, Q99M79 | Gene names | Dock2 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 2 (Hch protein). | |||||
|
GLI1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 85) | NC score | 0.005442 (rank : 179) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P47806, Q9QYK1 | Gene names | Gli1, Gli | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI1 (Glioma-associated oncogene homolog). | |||||
|
MUC1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 86) | NC score | 0.090532 (rank : 7) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 87) | NC score | 0.046953 (rank : 36) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
RFX1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 88) | NC score | 0.028460 (rank : 88) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P22670 | Gene names | RFX1 | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II regulatory factor RFX1 (RFX) (Enhancer factor C) (EF-C). | |||||
|
SSH2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 89) | NC score | 0.031035 (rank : 76) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q76I76, Q8TDB5, Q8WYL1, Q8WYL2, Q96F40, Q96H36, Q9C0D8 | Gene names | SSH2, KIAA1725, SSH2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 2 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-2L) (hSSH-2L). | |||||
|
STA13_MOUSE
|
||||||
| θ value | 3.0926 (rank : 90) | NC score | 0.012483 (rank : 146) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q923Q2, Q8K369 | Gene names | Stard13 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 13 (StARD13) (START domain- containing protein 13). | |||||
|
CENPC_HUMAN
|
||||||
| θ value | 4.03905 (rank : 91) | NC score | 0.028543 (rank : 87) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q03188, Q9P0M5 | Gene names | CENPC1, CENPC, ICEN7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein C 1 (CENP-C) (Centromere autoantigen C) (Interphase centromere complex protein 7). | |||||
|
CEP63_HUMAN
|
||||||
| θ value | 4.03905 (rank : 92) | NC score | 0.010904 (rank : 155) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1005 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96MT8, Q96CR0, Q9H8F5, Q9H8N0 | Gene names | CEP63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 63 kDa (Cep63 protein). | |||||
|
CHD8_HUMAN
|
||||||
| θ value | 4.03905 (rank : 93) | NC score | 0.010530 (rank : 159) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9HCK8, Q68DQ0, Q8N3Z9, Q8NCY4, Q8TBR9, Q96F26 | Gene names | CHD8, HELSNF1, KIAA1564 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 8 (EC 3.6.1.-) (ATP- dependent helicase CHD8) (CHD-8) (Helicase with SNF2 domain 1). | |||||
|
CIC_MOUSE
|
||||||
| θ value | 4.03905 (rank : 94) | NC score | 0.032427 (rank : 73) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
FGD1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 95) | NC score | 0.017735 (rank : 121) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 367 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P98174, Q8N4D9 | Gene names | FGD1, ZFYVE3 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
GAK_MOUSE
|
||||||
| θ value | 4.03905 (rank : 96) | NC score | 0.008422 (rank : 169) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99KY4, Q6P1I8, Q6P9S5, Q8BM74, Q8K0Q4 | Gene names | Gak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||
|
GLI2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 97) | NC score | 0.006712 (rank : 175) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 971 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P10070, O60252, O60253, O60254, O60255, Q15590, Q15591 | Gene names | GLI2, THP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI2 (Tax helper protein). | |||||
|
GLIS3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 98) | NC score | 0.004508 (rank : 182) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 758 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8NEA6 | Gene names | GLIS3, ZNF515 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLIS3 (GLI-similar 3) (Zinc finger protein 515). | |||||
|
INPP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 99) | NC score | 0.023106 (rank : 101) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49441 | Gene names | INPP1 | |||
|
Domain Architecture |
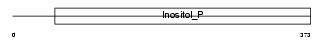
|
|||||
| Description | Inositol polyphosphate 1-phosphatase (EC 3.1.3.57) (IPPase) (IPP). | |||||
|
MAP4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 100) | NC score | 0.059472 (rank : 21) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
MBD5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 101) | NC score | 0.035113 (rank : 66) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9P267, Q9NUV6 | Gene names | MBD5, KIAA1461 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 5 (Methyl-CpG-binding protein MBD5). | |||||
|
MK07_MOUSE
|
||||||
| θ value | 4.03905 (rank : 102) | NC score | 0.006222 (rank : 177) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1054 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9WVS8 | Gene names | Mapk7, Erk5 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase 7 (EC 2.7.11.24) (Extracellular signal-regulated kinase 5) (ERK-5) (BMK1 kinase). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 103) | NC score | 0.043965 (rank : 44) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
P73L_HUMAN
|
||||||
| θ value | 4.03905 (rank : 104) | NC score | 0.020056 (rank : 113) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H3D4, O75080, O75195, O75922, O76078, Q6VEG2, Q6VEG3, Q6VEG4, Q6VFJ1, Q6VFJ2, Q6VFJ3, Q6VH20, Q7LDI3, Q7LDI4, Q7LDI5, Q96KR0, Q9H3D2, Q9H3D3, Q9H3P8, Q9NPH7, Q9P1B4, Q9P1B5, Q9P1B6, Q9P1B7, Q9UBV9, Q9UE10, Q9UP26, Q9UP27, Q9UP28, Q9UP74 | Gene names | TP73L, KET, P63, P73H, P73L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor protein p73-like (p73L) (p63) (Tumor protein 63) (TP63) (p51) (p40) (Keratinocyte transcription factor KET) (Chronic ulcerative stomatitis protein) (CUSP). | |||||
|
PCDA6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 105) | NC score | 0.002887 (rank : 184) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UN73, O75283, Q9NRT8 | Gene names | PCDHA6 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 6 precursor (PCDH-alpha6). | |||||
|
PO121_MOUSE
|
||||||
| θ value | 4.03905 (rank : 106) | NC score | 0.047145 (rank : 33) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
WTAP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 107) | NC score | 0.030204 (rank : 81) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15007, Q96T28, Q9BYJ7, Q9H4E2 | Gene names | WTAP, KIAA0105 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wilms' tumor 1-associating protein (WT1-associated protein) (Putative pre-mRNA-splicing regulator female-lethal(2D) homolog). | |||||
|
ZBT40_HUMAN
|
||||||
| θ value | 4.03905 (rank : 108) | NC score | 0.002845 (rank : 185) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NUA8, O75066, Q5TFU5, Q8N1R1 | Gene names | ZBTB40, KIAA0478 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 40. | |||||
|
A4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 109) | NC score | 0.034663 (rank : 68) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P12023, P97487, P97942, Q99K32 | Gene names | App | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein homolog) (Amyloidogenic glycoprotein) (AG) [Contains: Soluble APP-alpha (S-APP-alpha); Soluble APP-beta (S-APP-beta); C99 (APP-C99); Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)) (APP-C59); Gamma-CTF(57) (Gamma-secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)) (APP-C57); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
AB1IP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 110) | NC score | 0.018413 (rank : 119) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8R5A3, O35329, Q8BRU0, Q99KV8 | Gene names | Apbb1ip, Prel1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 1- interacting protein (APBB1-interacting protein 1) (Proline-rich EVH1 ligand 1) (PREL-1) (Proline-rich protein 48). | |||||
|
ABP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 111) | NC score | 0.012744 (rank : 145) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P19801, Q16683, Q16684, Q56II4, Q6GU42 | Gene names | ABP1, AOC1, DAO1 | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive amine oxidase [copper-containing] precursor (EC 1.4.3.6) (Diamine oxidase) (DAO) (Amiloride-binding protein) (ABP) (Histaminase) (Kidney amine oxidase) (KAO). | |||||
|
AFF2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 112) | NC score | 0.041290 (rank : 51) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P51816, O43786, O60215, P78407, Q13521, Q14323, Q9UNA5 | Gene names | AFF2, FMR2, OX19 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 2 (Fragile X mental retardation 2 protein) (Protein FMR-2) (FMR2P) (Protein Ox19) (Fragile X E mental retardation syndrome protein). | |||||
|
ANR35_HUMAN
|
||||||
| θ value | 5.27518 (rank : 113) | NC score | 0.010109 (rank : 162) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8N283, Q3MJ10, Q96LS3 | Gene names | ANKRD35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 35. | |||||
|
BMP2K_MOUSE
|
||||||
| θ value | 5.27518 (rank : 114) | NC score | 0.009471 (rank : 166) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q91Z96, Q8C8L7 | Gene names | Bmp2k, Bike | |||
|
Domain Architecture |

|
|||||
| Description | BMP-2-inducible protein kinase (EC 2.7.11.1) (BIKe). | |||||
|
CD2L2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 115) | NC score | 0.005305 (rank : 180) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
CG1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 116) | NC score | 0.045226 (rank : 41) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q13495 | Gene names | CXorf6, CG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CG1 protein (F18). | |||||
|
COHA1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 117) | NC score | 0.023878 (rank : 98) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UMD9, Q02802, Q99018, Q9NQK9, Q9UC14 | Gene names | COL17A1, BP180, BPAG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XVII) chain (Bullous pemphigoid antigen 2) (180 kDa bullous pemphigoid antigen 2). | |||||
|
FXL19_MOUSE
|
||||||
| θ value | 5.27518 (rank : 118) | NC score | 0.020030 (rank : 114) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6PB97, Q7TSH0, Q8BIB4 | Gene names | Fbxl19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box/LRR-repeat protein 19 (F-box and leucine-rich repeat protein 19). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 5.27518 (rank : 119) | NC score | 0.034777 (rank : 67) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
OSTP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 120) | NC score | 0.038791 (rank : 58) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P10451, Q15681, Q15682, Q15683, Q8NBK2, Q96IZ1 | Gene names | SPP1, OPN | |||
|
Domain Architecture |

|
|||||
| Description | Osteopontin precursor (Bone sialoprotein-1) (Secreted phosphoprotein 1) (SPP-1) (Urinary stone protein) (Nephropontin) (Uropontin). | |||||
|
PIWL1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 121) | NC score | 0.013709 (rank : 136) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96J94, O95404, Q8NA60, Q8TBY5, Q96JD5 | Gene names | PIWIL1, HIWI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Piwi-like protein 1. | |||||
|
PIWL1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 122) | NC score | 0.013779 (rank : 135) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JMB7 | Gene names | Piwil1, Miwi | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Piwi-like protein 1. | |||||
|
PLK4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 123) | NC score | 0.001687 (rank : 186) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 879 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q64702, Q6PEP6, Q78EG6, Q8R0I5, Q9CVR6, Q9CVU6 | Gene names | Plk4, Sak, Stk18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase PLK4 (EC 2.7.11.21) (Polo-like kinase 4) (PLK-4) (Serine/threonine-protein kinase Sak) (Serine/threonine- protein kinase 18). | |||||
|
RTN1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 124) | NC score | 0.017207 (rank : 123) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8K0T0, Q8K4S4 | Gene names | Rtn1, Nsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulon-1 (Neuroendocrine-specific protein). | |||||
|
TACC3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 125) | NC score | 0.022726 (rank : 103) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Y6A5, Q9UMQ1 | Gene names | TACC3, ERIC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ERIC-1). | |||||
|
AFF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 126) | NC score | 0.066754 (rank : 16) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
ATF7_MOUSE
|
||||||
| θ value | 6.88961 (rank : 127) | NC score | 0.013660 (rank : 137) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8R0S1 | Gene names | Atf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
BRD4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 128) | NC score | 0.034580 (rank : 69) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
CCD60_HUMAN
|
||||||
| θ value | 6.88961 (rank : 129) | NC score | 0.017180 (rank : 124) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IWA6 | Gene names | CCDC60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 60. | |||||
|
CIC_HUMAN
|
||||||
| θ value | 6.88961 (rank : 130) | NC score | 0.032486 (rank : 72) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CN044_HUMAN
|
||||||
| θ value | 6.88961 (rank : 131) | NC score | 0.020337 (rank : 112) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96MY7 | Gene names | C14orf44 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf44. | |||||
|
CO039_MOUSE
|
||||||
| θ value | 6.88961 (rank : 132) | NC score | 0.030100 (rank : 82) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q3TEI4, Q3TMF2, Q3U253, Q3UM75, Q9DA93 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39 homolog. | |||||
|
COHA1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 133) | NC score | 0.024602 (rank : 97) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q07563, Q99LK8 | Gene names | Col17a1, Bp180, Bpag2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVII) chain (Bullous pemphigoid antigen 2) (180 kDa bullous pemphigoid antigen 2). | |||||
|
CP135_HUMAN
|
||||||
| θ value | 6.88961 (rank : 134) | NC score | 0.008081 (rank : 171) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
EYA3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 135) | NC score | 0.013572 (rank : 138) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99504, O95463, Q99813 | Gene names | EYA3 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 3 (EC 3.1.3.48). | |||||
|
EYA3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 136) | NC score | 0.013291 (rank : 140) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97480, P97768 | Gene names | Eya3 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 3 (EC 3.1.3.48). | |||||
|
FOLC_MOUSE
|
||||||
| θ value | 6.88961 (rank : 137) | NC score | 0.016482 (rank : 126) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P48760 | Gene names | Fpgs | |||
|
Domain Architecture |
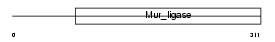
|
|||||
| Description | Folylpolyglutamate synthase, mitochondrial precursor (EC 6.3.2.17) (Folylpoly-gamma-glutamate synthetase) (FPGS) (Tetrahydrofolate synthase) (Tetrahydrofolylpolyglutamate synthase). | |||||
|
GRIP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 138) | NC score | 0.009710 (rank : 165) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q925T6, Q8BLQ3, Q8C0T3, Q925T5, Q925T7 | Gene names | Grip1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor-interacting protein 1 (GRIP1 protein). | |||||
|
GTSE1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 139) | NC score | 0.029193 (rank : 86) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8R080, O89015, Q9CSG9 | Gene names | Gtse1, B99 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G2 and S phase expressed protein 1 (Gtse-1) (B99 protein). | |||||
|
ITB4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 140) | NC score | 0.006923 (rank : 173) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P16144, O14690, O14691, O15339, O15340, O15341, Q9UIQ4 | Gene names | ITGB4 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-4 precursor (GP150) (CD104 antigen). | |||||
|
KI13B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 141) | NC score | 0.012382 (rank : 147) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NQT8, O75134, Q9BYJ6 | Gene names | KIF13B, GAKIN, KIAA0639 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF13B (Kinesin-like protein GAKIN). | |||||
|
KIF1C_HUMAN
|
||||||
| θ value | 6.88961 (rank : 142) | NC score | 0.011112 (rank : 153) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43896, O75186 | Gene names | KIF1C, KIAA0706 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF1C. | |||||
|
KIF1C_MOUSE
|
||||||
| θ value | 6.88961 (rank : 143) | NC score | 0.010955 (rank : 154) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35071, Q5SX62 | Gene names | Kif1c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF1C. | |||||
|
MTSS1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 144) | NC score | 0.027395 (rank : 91) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8R1S4, Q8BMM3, Q99LB3 | Gene names | Mtss1, Mim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein). | |||||
|
MUC5B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 145) | NC score | 0.041718 (rank : 49) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
MYPC3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 146) | NC score | 0.007822 (rank : 172) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O70468, O88997 | Gene names | Mybpc3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-binding protein C, cardiac-type (Cardiac MyBP-C) (C-protein, cardiac muscle isoform). | |||||
|
NCOA3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 147) | NC score | 0.011913 (rank : 149) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y6Q9, Q5JYD9, Q5JYE0, Q9BR49, Q9UPC9, Q9UPG4, Q9UPG7 | Gene names | NCOA3, AIB1, RAC3, TRAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP-interacting protein) (pCIP). | |||||
|
NTHL1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 148) | NC score | 0.031790 (rank : 75) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P78549, Q99566, Q99794, Q9BPX2 | Gene names | NTHL1, NTH1, OCTS3 | |||
|
Domain Architecture |
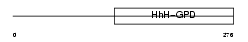
|
|||||
| Description | Endonuclease III-like protein 1 (EC 4.2.99.18). | |||||
|
OBFC1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 149) | NC score | 0.018467 (rank : 118) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K2X3, Q8C6I6 | Gene names | Obfc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oligonucleotide/oligosaccharide-binding fold-containing protein 1. | |||||
|
PCX1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 150) | NC score | 0.025167 (rank : 96) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96RV3, O94897, Q96AI7, Q9Y2J9 | Gene names | PCNX, KIAA0805, KIAA0995, PCNXL1 | |||
|
Domain Architecture |

|
|||||
| Description | Pecanex-like protein 1 (Pecanex homolog). | |||||
|
PER3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 151) | NC score | 0.009930 (rank : 163) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O70361 | Gene names | Per3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (mPER3). | |||||
|
PHLPP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 152) | NC score | 0.010621 (rank : 158) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
PHLPP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 153) | NC score | 0.008948 (rank : 167) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CHE4, Q6PCN0, Q8QZU8, Q8R5E5, Q99KL6 | Gene names | Phlpp, Kiaa0606, Plekhe1, Scop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein). | |||||
|
PKHA3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 154) | NC score | 0.014156 (rank : 133) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HB20, Q86TQ1, Q9NXT3 | Gene names | PLEKHA3, FAPP1 | |||
|
Domain Architecture |
|
|||||
| Description | Pleckstrin homology domain-containing family A member 3 (Phosphoinositol 4-phosphate adaptor protein 1) (FAPP-1). | |||||
|
PPRB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 155) | NC score | 0.037490 (rank : 62) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q15648, O43810, O75447, Q9HD39 | Gene names | PPARBP, ARC205, DRIP205, TRAP220, TRIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2) (p53 regulatory protein RB18A) (Vitamin D receptor-interacting protein complex component DRIP205) (Activator- recruited cofactor 205 kDa component) (ARC205). | |||||
|
SOS1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 156) | NC score | 0.012920 (rank : 143) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q62245, Q62244 | Gene names | Sos1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1) (mSOS-1). | |||||
|
TAU_HUMAN
|
||||||
| θ value | 6.88961 (rank : 157) | NC score | 0.037254 (rank : 63) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P10636, P18518, Q14799, Q15549, Q15550, Q15551, Q9UDJ3, Q9UMH0, Q9UQ96 | Gene names | MAPT, MTBT1, TAU | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
TF3C2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 158) | NC score | 0.016059 (rank : 128) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BL74, Q3U7Z9, Q80XQ7, Q8BJJ4, Q8BLP3, Q8K1J1, Q9CSS0, Q9CVE6, Q9DBK6 | Gene names | Gtf3c2, Kiaa0011 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor 3C polypeptide 2 (Transcription factor IIIC-subunit beta) (TF3C-beta) (TFIIIC 110 kDa subunit) (TFIIIC110). | |||||
|
THB1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 159) | NC score | 0.003631 (rank : 183) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P37242 | Gene names | Thrb, C-erba-beta, Nr1a2 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid hormone receptor beta-1. | |||||
|
AGGF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 160) | NC score | 0.013116 (rank : 142) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8N302, O00581, Q53YS3, Q9BU84, Q9NW66 | Gene names | AGGF1, VG5Q | |||
|
Domain Architecture |

|
|||||
| Description | Angiogenic factor with G patch and FHA domains 1 (Angiogenic factor VG5Q) (Vasculogenesis gene on 5q protein) (hVG5Q). | |||||
|
ARI4A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 161) | NC score | 0.022331 (rank : 105) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P29374, Q15991, Q15992, Q15993 | Gene names | ARID4A, RBBP1, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 4A (ARID domain- containing protein 4A) (Retinoblastoma-binding protein 1) (RBBP-1). | |||||
|
BSND_MOUSE
|
||||||
| θ value | 8.99809 (rank : 162) | NC score | 0.016260 (rank : 127) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VIM4, Q8C740, Q8CHY0 | Gene names | Bsnd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Barttin. | |||||
|
CB013_MOUSE
|
||||||
| θ value | 8.99809 (rank : 163) | NC score | 0.014700 (rank : 131) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D842, Q8BZL5, Q99LX6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C2orf13 homolog. | |||||
|
CENPC_MOUSE
|
||||||
| θ value | 8.99809 (rank : 164) | NC score | 0.025172 (rank : 95) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P49452 | Gene names | Cenpc1, Cenpc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein C 1 (CENP-C) (Centromere autoantigen C). | |||||
|
CJ047_MOUSE
|
||||||
| θ value | 8.99809 (rank : 165) | NC score | 0.021827 (rank : 106) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8C5R2, Q8R017, Q8R2Z9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf47 homolog. | |||||
|
CN044_MOUSE
|
||||||
| θ value | 8.99809 (rank : 166) | NC score | 0.022531 (rank : 104) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CB59, Q8BQX8, Q8BSL9, Q8CBZ3 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf44 homolog. | |||||
|
ELL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 167) | NC score | 0.014357 (rank : 132) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P55199 | Gene names | ELL, C19orf17 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II elongation factor ELL (Eleven-nineteen lysine-rich leukemia protein). | |||||
|
FOXP4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 168) | NC score | 0.006348 (rank : 176) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9DBY0, Q80V92, Q8CG10, Q8CIS1 | Gene names | Foxp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Forkhead box protein P4 (Fork head-related protein like A) (mFKHLA). | |||||
|
LIMA1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 169) | NC score | 0.010638 (rank : 157) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UHB6, Q2TAN7, Q9BVF2, Q9H8J1, Q9HBN5, Q9NX96, Q9NXC3, Q9NXU6, Q9P0H8, Q9UHB5 | Gene names | LIMA1, EPLIN, SREBP3 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain and actin-binding protein 1 (Epithelial protein lost in neoplasm). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 170) | NC score | 0.030383 (rank : 80) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 171) | NC score | 0.032075 (rank : 74) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
MYLK_MOUSE
|
||||||
| θ value | 8.99809 (rank : 172) | NC score | 0.004654 (rank : 181) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1489 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6PDN3, Q3TSJ7, Q80UX0, Q80YN7, Q80YN8, Q8K026, Q924D2, Q9ERD3 | Gene names | Mylk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin light chain kinase, smooth muscle (EC 2.7.11.18) (MLCK) (Telokin) (Kinase-related protein) (KRP). | |||||
|
NFIB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 173) | NC score | 0.008189 (rank : 170) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O00712, O00166, Q12858, Q96J45 | Gene names | NFIB | |||
|
Domain Architecture |
|
|||||
| Description | Nuclear factor 1 B-type (Nuclear factor 1/B) (NF1-B) (NFI-B) (NF-I/B) (CCAAT-box-binding transcription factor) (CTF) (TGGCA-binding protein). | |||||
|
PPE2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 174) | NC score | 0.006908 (rank : 174) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35385 | Gene names | Ppef2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase with EF-hands 2 (EC 3.1.3.16) (PPEF-2). | |||||
|
PPGB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 175) | NC score | 0.012033 (rank : 148) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P10619, Q5JZH1, Q96KJ2, Q9BR08, Q9BW68 | Gene names | PPGB | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal protective protein precursor (EC 3.4.16.5) (Cathepsin A) (Carboxypeptidase C) (Protective protein for beta-galactosidase) [Contains: Lysosomal protective protein 32 kDa chain; Lysosomal protective protein 20 kDa chain]. | |||||
|
PRDM2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 176) | NC score | 0.008436 (rank : 168) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
SAMD4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 177) | NC score | 0.013862 (rank : 134) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UPU9 | Gene names | SAMD4, KIAA1053 | |||
|
Domain Architecture |

|
|||||
| Description | Sterile alpha motif domain-containing protein 4. | |||||
|
SMC3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 178) | NC score | 0.010387 (rank : 161) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1186 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UQE7, O60464 | Gene names | CSPG6, BAM, BMH, SMC3, SMC3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome-associated polypeptide) (hCAP) (Bamacan) (Basement membrane-associated chondroitin proteoglycan). | |||||
|
SMC3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 179) | NC score | 0.010432 (rank : 160) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1187 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9CW03, O35667, Q9QUS3 | Gene names | Cspg6, Bam, Bmh, Mmip1, Smc3, Smc3l1, Smcd | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome segregation protein SmcD) (Bamacan) (Basement membrane-associated chondroitin proteoglycan) (Mad member- interacting protein 1). | |||||
|
TCAL5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 180) | NC score | 0.018504 (rank : 117) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
TTLL4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 181) | NC score | 0.009917 (rank : 164) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14679, Q8WW29 | Gene names | TTLL4, KIAA0173 | |||
|
Domain Architecture |

|
|||||
| Description | Tubulin--tyrosine ligase-like protein 4. | |||||
|
MAML3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.060989 (rank : 20) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96JK9 | Gene names | MAML3, KIAA1816 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 3 (Mam-3). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.053964 (rank : 25) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PRG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.064257 (rank : 18) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
SELPL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.054104 (rank : 24) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
VCX3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.052211 (rank : 28) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NNX9, Q9P0H4 | Gene names | VCX3A, VCX3, VCX8R, VCXA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 3 (VCX-A protein) (Variably charged protein X-A) (Variable charge protein on X with eight repeats) (VCX- 8r). | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.056483 (rank : 22) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
ALMS1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 181 | |
| SwissProt Accessions | Q8K4E0, Q6A084, Q8C9N9 | Gene names | Alms1, Kiaa0328 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alstrom syndrome protein 1 homolog. | |||||
|
ALMS1_HUMAN
|
||||||
| NC score | 0.895643 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8TCU4, Q53S05, Q580Q8, Q86VP9, Q9Y4G4 | Gene names | ALMS1, KIAA0328 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alstrom syndrome protein 1. | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.128256 (rank : 3) | θ value | 5.44631e-05 (rank : 3) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.123368 (rank : 4) | θ value | 9.29e-05 (rank : 4) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.098836 (rank : 5) | θ value | 0.62314 (rank : 26) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
EP300_HUMAN
|
||||||
| NC score | 0.094940 (rank : 6) | θ value | 0.163984 (rank : 12) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
MUC1_MOUSE
|
||||||
| NC score | 0.090532 (rank : 7) | θ value | 3.0926 (rank : 86) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.084270 (rank : 8) | θ value | 0.21417 (rank : 13) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.079699 (rank : 9) | θ value | 0.0431538 (rank : 8) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
TFAM_MOUSE
|
||||||
| NC score | 0.078768 (rank : 10) | θ value | 0.813845 (rank : 31) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P40630, P97894, P97906, Q543I8, Q9DBM9 | Gene names | Tfam, Hmgts | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor A, mitochondrial precursor (mtTFA) (Testis- specific high mobility group protein) (TS-HMG). | |||||
|
NFRKB_HUMAN
|
||||||
| NC score | 0.076643 (rank : 11) | θ value | 0.0148317 (rank : 7) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6P4R8, Q12869, Q15312, Q9H048 | Gene names | NFRKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor related to kappa-B-binding protein (DNA-binding protein R kappa-B). | |||||
|
AFF4_MOUSE
|
||||||
| NC score | 0.073329 (rank : 12) | θ value | 0.00509761 (rank : 5) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.069852 (rank : 13) | θ value | 0.0961366 (rank : 11) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
MAP4_HUMAN
|
||||||
| NC score | 0.068674 (rank : 14) | θ value | 0.279714 (rank : 14) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
VPS72_HUMAN
|
||||||
| NC score | 0.067696 (rank : 15) | θ value | 0.279714 (rank : 17) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q15906, Q53GJ2, Q5U0R4 | Gene names | VPS72, TCFL1, YL1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 72 homolog (Transcription factor-like 1) (Protein YL-1). | |||||
|
AFF1_HUMAN
|
||||||
| NC score | 0.066754 (rank : 16) | θ value | 6.88961 (rank : 126) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
VPS72_MOUSE
|
||||||
| NC score | 0.065519 (rank : 17) | θ value | 0.365318 (rank : 18) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q62481, Q3U2N4, Q810A9, Q99K81 | Gene names | Vps72, Tcfl1, Yl1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 72 homolog (Transcription factor-like 1) (Protein YL-1). | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.064257 (rank : 18) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
ARI1A_HUMAN
|
||||||
| NC score | 0.063302 (rank : 19) | θ value | 0.47712 (rank : 19) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
MAML3_HUMAN
|
||||||
| NC score | 0.060989 (rank : 20) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96JK9 | Gene names | MAML3, KIAA1816 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 3 (Mam-3). | |||||
|
MAP4_MOUSE
|
||||||
| NC score | 0.059472 (rank : 21) | θ value | 4.03905 (rank : 100) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.056483 (rank : 22) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
CJ084_HUMAN
|
||||||
| NC score | 0.054316 (rank : 23) | θ value | 0.62314 (rank : 24) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H8W3, Q9H5V5 | Gene names | C10orf84 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf84. | |||||
|
SELPL_MOUSE
|
||||||
| NC score | 0.054104 (rank : 24) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.053964 (rank : 25) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PDXL2_MOUSE
|
||||||
| NC score | 0.053705 (rank : 26) | θ value | 1.38821 (rank : 51) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8CAE9, Q8CFW3 | Gene names | Podxl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Podocalyxin-like protein 2 precursor (Endoglycan). | |||||
|
KI67_HUMAN
|
||||||
| NC score | 0.052716 (rank : 27) | θ value | 2.36792 (rank : 71) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
VCX3_HUMAN
|
||||||
| NC score | 0.052211 (rank : 28) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NNX9, Q9P0H4 | Gene names | VCX3A, VCX3, VCX8R, VCXA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 3 (VCX-A protein) (Variably charged protein X-A) (Variable charge protein on X with eight repeats) (VCX- 8r). | |||||
|
PDPN_HUMAN
|
||||||
| NC score | 0.050340 (rank : 29) | θ value | 2.36792 (rank : 76) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86YL7, O60836, O95128, Q7L375, Q8NBQ8, Q8NBR3 | Gene names | PDPN, GP36 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Podoplanin precursor (Glycoprotein 36) (Gp36) (PA2.26 antigen) (T1A) (T1-alpha) (Aggrus). | |||||
|
TMUB1_HUMAN
|
||||||
| NC score | 0.050237 (rank : 30) | θ value | 0.47712 (rank : 23) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BVT8 | Gene names | TMUB1, C7orf21 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane and ubiquitin-like domain-containing protein 1 (Ubiquitin-like protein SB144). | |||||
|
PDZD2_HUMAN
|
||||||
| NC score | 0.048744 (rank : 31) | θ value | 0.279714 (rank : 15) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
T22D2_HUMAN
|
||||||
| NC score | 0.047723 (rank : 32) | θ value | 1.81305 (rank : 63) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O75157, Q6PI50, Q9H2Z6, Q9H2Z7, Q9H2Z8 | Gene names | TSC22D2, KIAA0669, TILZ4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TSC22 domain family protein 2 (TSC22-related-inducible leucine zipper protein 4). | |||||
|
PO121_MOUSE
|
||||||
| NC score | 0.047145 (rank : 33) | θ value | 4.03905 (rank : 106) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
APLP2_HUMAN
|
||||||
| NC score | 0.046997 (rank : 34) | θ value | 1.38821 (rank : 43) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q06481 | Gene names | APLP2, APPL2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 2 precursor (Amyloid protein homolog) (APPH) (CDEI box-binding protein) (CDEBP). | |||||
|
FA21C_HUMAN
|
||||||
| NC score | 0.046981 (rank : 35) | θ value | 1.06291 (rank : 33) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y4E1, Q5SQU4, Q5SQU5, Q7L521, Q9UG79 | Gene names | FAM21C, KIAA0592 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM21C. | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.046953 (rank : 36) | θ value | 3.0926 (rank : 87) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.046922 (rank : 37) | θ value | 1.38821 (rank : 50) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
DCP1A_MOUSE
|
||||||
| NC score | 0.046809 (rank : 38) | θ value | 2.36792 (rank : 68) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q91YD3, Q6NZE3 | Gene names | Dcp1a, Mitc1, Smif | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA decapping enzyme 1A (EC 3.-.-.-) (Transcription factor SMIF) (MAD homolog 4-interacting transcription coactivator 1) (Smad4-interacting transcriptional co-activator). | |||||
|
NTHL1_MOUSE
|
||||||
| NC score | 0.045760 (rank : 39) | θ value | 1.38821 (rank : 49) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35980 | Gene names | Nthl1, Nth1 | |||
|
Domain Architecture |

|
|||||
| Description | Endonuclease III-like protein 1 (EC 4.2.99.18). | |||||
|
MAML1_HUMAN
|
||||||
| NC score | 0.045694 (rank : 40) | θ value | 0.47712 (rank : 22) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q92585, Q9NZ12 | Gene names | MAML1, KIAA0200 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 1 (Mam-1). | |||||
|
CG1_HUMAN
|
||||||
| NC score | 0.045226 (rank : 41) | θ value | 5.27518 (rank : 116) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q13495 | Gene names | CXorf6, CG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CG1 protein (F18). | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.045118 (rank : 42) | θ value | 1.81305 (rank : 62) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
K0146_HUMAN
|
||||||
| NC score | 0.044151 (rank : 43) | θ value | 0.47712 (rank : 20) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14159, Q96BI5 | Gene names | KIAA0146 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0146. | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.043965 (rank : 44) | θ value | 4.03905 (rank : 103) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
ARID2_HUMAN
|
||||||
| NC score | 0.043830 (rank : 45) | θ value | 2.36792 (rank : 65) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q68CP9, Q5EB51, Q645I3, Q6ZRY5, Q7Z3I5, Q86T28, Q96SJ6, Q9HCL5 | Gene names | ARID2, KIAA1557 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 2 (ARID domain- containing protein 2) (BRG1-associated factor 200) (BAF200). | |||||
|
XKR5_HUMAN
|
||||||
| NC score | 0.043809 (rank : 46) | θ value | 0.62314 (rank : 28) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6UX68, Q5GH74 | Gene names | XKR5, XRG5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | XK-related protein 5. | |||||
|
ATX7_HUMAN
|
||||||
| NC score | 0.042162 (rank : 47) | θ value | 1.06291 (rank : 32) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O15265, O75328, O75329, Q9Y6P8 | Gene names | ATXN7, SCA7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein). | |||||
|
SSH2_MOUSE
|
||||||
| NC score | 0.041726 (rank : 48) | θ value | 0.0431538 (rank : 9) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5SW75, Q3TDK8, Q3TYP8, Q3U2K3, Q5F268, Q5SW74, Q69ZC3, Q76I78 | Gene names | Ssh2, Kiaa1725, Ssh2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 2 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-2L) (mSSH-2L). | |||||
|
MUC5B_HUMAN
|
||||||
| NC score | 0.041718 (rank : 49) | θ value | 6.88961 (rank : 145) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
ZAN_MOUSE
|
||||||
| NC score | 0.041520 (rank : 50) | θ value | 1.06291 (rank : 41) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
AFF2_HUMAN
|
||||||
| NC score | 0.041290 (rank : 51) | θ value | 5.27518 (rank : 112) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P51816, O43786, O60215, P78407, Q13521, Q14323, Q9UNA5 | Gene names | AFF2, FMR2, OX19 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 2 (Fragile X mental retardation 2 protein) (Protein FMR-2) (FMR2P) (Protein Ox19) (Fragile X E mental retardation syndrome protein). | |||||
|
A4_HUMAN
|
||||||
| NC score | 0.041069 (rank : 52) | θ value | 1.38821 (rank : 42) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P05067, P09000, P78438, Q13764, Q13778, Q13793, Q16011, Q16014, Q16019, Q16020, Q9BT38, Q9UCA9, Q9UCB6, Q9UCC8, Q9UCD1, Q9UQ58 | Gene names | APP, A4, AD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein) (Cerebral vascular amyloid peptide) (CVAP) (Protease nexin-II) (PN-II) (APPI) (PreA4) [Contains: Soluble APP-alpha (S-APP- alpha); Soluble APP-beta (S-APP-beta); C99; Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)); Gamma-CTF(57) (Gamma- secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.040686 (rank : 53) | θ value | 2.36792 (rank : 74) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
PHAR2_HUMAN
|
||||||
| NC score | 0.040537 (rank : 54) | θ value | 1.38821 (rank : 52) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O75167, Q68DM2 | Gene names | PHACTR2, C6orf56, KIAA0680 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatase and actin regulator 2. | |||||
|
MYCB2_HUMAN
|
||||||
| NC score | 0.040286 (rank : 55) | θ value | 1.38821 (rank : 48) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O75592, Q5JSX8, Q5VZN6, Q6PIB6, Q9UQ11, Q9Y6E4 | Gene names | MYCBP2, KIAA0916, PAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
APLP2_MOUSE
|
||||||
| NC score | 0.039708 (rank : 56) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q06335 | Gene names | Aplp2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 2 precursor (CDEI box-binding protein) (CDEBP). | |||||
|
PENK_HUMAN
|
||||||
| NC score | 0.038849 (rank : 57) | θ value | 1.06291 (rank : 37) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P01210 | Gene names | PENK | |||
|
Domain Architecture |

|
|||||
| Description | Proenkephalin A precursor [Contains: Synenkephalin; Met-enkephalin (Opioid growth factor) (OGF); Met-enkephalin-Arg-Gly-Leu; Leu- enkephalin; Met-enkephalin-Arg-Phe]. | |||||
|
OSTP_HUMAN
|
||||||
| NC score | 0.038791 (rank : 58) | θ value | 5.27518 (rank : 120) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P10451, Q15681, Q15682, Q15683, Q8NBK2, Q96IZ1 | Gene names | SPP1, OPN | |||
|
Domain Architecture |

|
|||||
| Description | Osteopontin precursor (Bone sialoprotein-1) (Secreted phosphoprotein 1) (SPP-1) (Urinary stone protein) (Nephropontin) (Uropontin). | |||||
|
CNOT3_HUMAN
|
||||||
| NC score | 0.038676 (rank : 59) | θ value | 2.36792 (rank : 66) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O75175, Q9NZN7, Q9UF76 | Gene names | CNOT3, KIAA0691, NOT3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
MAGC1_HUMAN
|
||||||
| NC score | 0.038366 (rank : 60) | θ value | 0.47712 (rank : 21) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
PGCA_HUMAN
|
||||||
| NC score | 0.037814 (rank : 61) | θ value | 1.81305 (rank : 61) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
PPRB_HUMAN
|
||||||
| NC score | 0.037490 (rank : 62) | θ value | 6.88961 (rank : 155) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q15648, O43810, O75447, Q9HD39 | Gene names | PPARBP, ARC205, DRIP205, TRAP220, TRIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2) (p53 regulatory protein RB18A) (Vitamin D receptor-interacting protein complex component DRIP205) (Activator- recruited cofactor 205 kDa component) (ARC205). | |||||
|
TAU_HUMAN
|
||||||
| NC score | 0.037254 (rank : 63) | θ value | 6.88961 (rank : 157) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P10636, P18518, Q14799, Q15549, Q15550, Q15551, Q9UDJ3, Q9UMH0, Q9UQ96 | Gene names | MAPT, MTBT1, TAU | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
PRGC1_HUMAN
|
||||||
| NC score | 0.036254 (rank : 64) | θ value | 1.06291 (rank : 38) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UBK2, Q9UN32 | Gene names | PPARGC1A, LEM6, PGC1, PGC1A, PPARGC1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PPAR gamma coactivator 1-alpha) (PPARGC-1-alpha) (PGC-1-alpha) (Ligand effect modulator 6). | |||||
|
K2027_MOUSE
|
||||||
| NC score | 0.036045 (rank : 65) | θ value | 2.36792 (rank : 70) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BWS5, Q69Z30 | Gene names | Kiaa2027 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA2027. | |||||
|
MBD5_HUMAN
|
||||||
| NC score | 0.035113 (rank : 66) | θ value | 4.03905 (rank : 101) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9P267, Q9NUV6 | Gene names | MBD5, KIAA1461 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 5 (Methyl-CpG-binding protein MBD5). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.034777 (rank : 67) | θ value | 5.27518 (rank : 119) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
A4_MOUSE
|
||||||
| NC score | 0.034663 (rank : 68) | θ value | 5.27518 (rank : 109) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P12023, P97487, P97942, Q99K32 | Gene names | App | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein homolog) (Amyloidogenic glycoprotein) (AG) [Contains: Soluble APP-alpha (S-APP-alpha); Soluble APP-beta (S-APP-beta); C99 (APP-C99); Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)) (APP-C59); Gamma-CTF(57) (Gamma-secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)) (APP-C57); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
BRD4_HUMAN
|
||||||
| NC score | 0.034580 (rank : 69) | θ value | 6.88961 (rank : 128) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
WTAP_MOUSE
|
||||||
| NC score | 0.034381 (rank : 70) | θ value | 2.36792 (rank : 81) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9ER69 | Gene names | Wtap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wilms' tumor 1-associating protein (WT1-associated protein) (Putative pre-mRNA-splicing regulator female-lethal(2D) homolog). | |||||
|
TENS1_HUMAN
|
||||||
| NC score | 0.033912 (rank : 71) | θ value | 2.36792 (rank : 79) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9HBL0 | Gene names | TNS1, TNS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-1. | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.032486 (rank : 72) | θ value | 6.88961 (rank : 130) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CIC_MOUSE
|
||||||
| NC score | 0.032427 (rank : 73) | θ value | 4.03905 (rank : 94) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.032075 (rank : 74) | θ value | 8.99809 (rank : 171) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
NTHL1_HUMAN
|
||||||
| NC score | 0.031790 (rank : 75) | θ value | 6.88961 (rank : 148) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P78549, Q99566, Q99794, Q9BPX2 | Gene names | NTHL1, NTH1, OCTS3 | |||
|
Domain Architecture |

|
|||||
| Description | Endonuclease III-like protein 1 (EC 4.2.99.18). | |||||
|
SSH2_HUMAN
|
||||||
| NC score | 0.031035 (rank : 76) | θ value | 3.0926 (rank : 89) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q76I76, Q8TDB5, Q8WYL1, Q8WYL2, Q96F40, Q96H36, Q9C0D8 | Gene names | SSH2, KIAA1725, SSH2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 2 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-2L) (hSSH-2L). | |||||
|
CC47_MOUSE
|
||||||
| NC score | 0.030927 (rank : 77) | θ value | 1.81305 (rank : 58) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9D024, Q3TSB5, Q3V1S7, Q8C5D9, Q920S6 | Gene names | Ccdc47, Asp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 47 precursor (Adipocyte-specific protein 4). | |||||
|
3BP2_MOUSE
|
||||||
| NC score | 0.030837 (rank : 78) | θ value | 1.81305 (rank : 56) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q06649 | Gene names | Sh3bp2, 3bp2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
RBMX2_MOUSE
|
||||||
| NC score | 0.030407 (rank : 79) | θ value | 0.279714 (rank : 16) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R0F5, Q3TI42 | Gene names | Rbmx2 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding motif protein, X-linked 2. | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.030383 (rank : 80) | θ value | 8.99809 (rank : 170) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
WTAP_HUMAN
|
||||||
| NC score | 0.030204 (rank : 81) | θ value | 4.03905 (rank : 107) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15007, Q96T28, Q9BYJ7, Q9H4E2 | Gene names | WTAP, KIAA0105 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wilms' tumor 1-associating protein (WT1-associated protein) (Putative pre-mRNA-splicing regulator female-lethal(2D) homolog). | |||||
|
CO039_MOUSE
|
||||||
| NC score | 0.030100 (rank : 82) | θ value | 6.88961 (rank : 132) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q3TEI4, Q3TMF2, Q3U253, Q3UM75, Q9DA93 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39 homolog. | |||||
|
SETB1_MOUSE
|
||||||
| NC score | 0.029786 (rank : 83) | θ value | 1.38821 (rank : 53) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O88974, Q922K1 | Gene names | Setdb1, Eset | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 4 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 4) (H3-K9-HMTase 4) (SET domain bifurcated 1) (ERG-associated protein with SET domain) (ESET). | |||||
|
HNRPC_MOUSE
|
||||||
| NC score | 0.029638 (rank : 84) | θ value | 1.06291 (rank : 34) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z204, Q3TLB5, Q501Q3, Q8C2G5, Q99KE2, Q9CQT3, Q9CY83 | Gene names | Hnrpc, Hnrnpc | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoproteins C1/C2 (hnRNP C1 / hnRNP C2). | |||||
|
KCNQ2_MOUSE
|
||||||
| NC score | 0.029255 (rank : 85) | θ value | 0.0736092 (rank : 10) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z351, Q8R498, Q9QWN9, Q9Z342, Q9Z343, Q9Z344, Q9Z345, Q9Z346, Q9Z347, Q9Z348, Q9Z349, Q9Z350 | Gene names | Kcnq2, Kqt2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
GTSE1_MOUSE
|
||||||
| NC score | 0.029193 (rank : 86) | θ value | 6.88961 (rank : 139) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8R080, O89015, Q9CSG9 | Gene names | Gtse1, B99 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G2 and S phase expressed protein 1 (Gtse-1) (B99 protein). | |||||
|
CENPC_HUMAN
|
||||||
| NC score | 0.028543 (rank : 87) | θ value | 4.03905 (rank : 91) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q03188, Q9P0M5 | Gene names | CENPC1, CENPC, ICEN7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein C 1 (CENP-C) (Centromere autoantigen C) (Interphase centromere complex protein 7). | |||||
|
RFX1_HUMAN
|
||||||
| NC score | 0.028460 (rank : 88) | θ value | 3.0926 (rank : 88) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P22670 | Gene names | RFX1 | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II regulatory factor RFX1 (RFX) (Enhancer factor C) (EF-C). | |||||
|
FMNL_HUMAN
|
||||||
| NC score | 0.028307 (rank : 89) | θ value | 1.38821 (rank : 46) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O95466, Q6DKG5, Q6IBP3, Q86UH1, Q8N671, Q8TDH1, Q96H10 | Gene names | FMNL1, C17orf1, C17orf1B, FMNL | |||
|
Domain Architecture |

|
|||||
| Description | Formin-like protein 1 (Leukocyte formin) (CLL-associated antigen KW- 13). | |||||
|
ANK3_HUMAN
|
||||||
| NC score | 0.027805 (rank : 90) | θ value | 0.00509761 (rank : 6) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
MTSS1_MOUSE
|
||||||
| NC score | 0.027395 (rank : 91) | θ value | 6.88961 (rank : 144) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8R1S4, Q8BMM3, Q99LB3 | Gene names | Mtss1, Mim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein). | |||||
|
PPR3A_MOUSE
|
||||||
| NC score | 0.027113 (rank : 92) | θ value | 2.36792 (rank : 77) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99MR9, Q8BUJ4, Q8BUL0 | Gene names | Ppp1r3a, Pp1g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase 1 regulatory subunit 3A (Protein phosphatase 1 glycogen-associated regulatory subunit) (Protein phosphatase type-1 glycogen targeting subunit) (RGL). | |||||
|
NCOAT_MOUSE
|
||||||
| NC score | 0.026709 (rank : 93) | θ value | 2.36792 (rank : 75) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EQQ9, Q3ULY7, Q6ZQ71, Q8BK05, Q8BTT2, Q8CFX2, Q9CSJ4, Q9CUR7 | Gene names | Mgea5, Hexc, Kiaa0679 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional protein NCOAT (Nuclear cytoplasmic O-GlcNAcase and acetyltransferase) (Meningioma-expressed antigen 5) [Includes: Beta- hexosaminidase (EC 3.2.1.52) (N-acetyl-beta-glucosaminidase) (Beta-N- acetylhexosaminidase) (Hexosaminidase C) (N-acetyl-beta-D- glucosaminidase) (O-GlcNAcase); Histone acetyltransferase (EC 2.3.1.48) (HAT)]. | |||||
|
LR37B_HUMAN
|
||||||
| NC score | 0.025728 (rank : 94) | θ value | 1.81305 (rank : 59) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96QE4, Q5YKG6 | Gene names | LRRC37B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37B precursor (C66 SLIT-like testicular protein). | |||||
|
CENPC_MOUSE
|
||||||
| NC score | 0.025172 (rank : 95) | θ value | 8.99809 (rank : 164) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P49452 | Gene names | Cenpc1, Cenpc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein C 1 (CENP-C) (Centromere autoantigen C). | |||||
|
PCX1_HUMAN
|
||||||
| NC score | 0.025167 (rank : 96) | θ value | 6.88961 (rank : 150) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96RV3, O94897, Q96AI7, Q9Y2J9 | Gene names | PCNX, KIAA0805, KIAA0995, PCNXL1 | |||
|
Domain Architecture |

|
|||||
| Description | Pecanex-like protein 1 (Pecanex homolog). | |||||
|
COHA1_MOUSE
|
||||||
| NC score | 0.024602 (rank : 97) | θ value | 6.88961 (rank : 133) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q07563, Q99LK8 | Gene names | Col17a1, Bp180, Bpag2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVII) chain (Bullous pemphigoid antigen 2) (180 kDa bullous pemphigoid antigen 2). | |||||
|
COHA1_HUMAN
|
||||||
| NC score | 0.023878 (rank : 98) | θ value | 5.27518 (rank : 117) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UMD9, Q02802, Q99018, Q9NQK9, Q9UC14 | Gene names | COL17A1, BP180, BPAG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XVII) chain (Bullous pemphigoid antigen 2) (180 kDa bullous pemphigoid antigen 2). | |||||
|
TREF1_MOUSE
|
||||||
| NC score | 0.023674 (rank : 99) | θ value | 1.38821 (rank : 55) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BXJ2, Q6PCQ4, Q80X25, Q810H8, Q8BY31 | Gene names | Trerf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132). | |||||
|
FXL19_HUMAN
|
||||||
| NC score | 0.023541 (rank : 100) | θ value | 2.36792 (rank : 69) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6PCT2, Q8N789, Q9NT14 | Gene names | FBXL19, FBL19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box/LRR-repeat protein 19 (F-box and leucine-rich repeat protein 19). | |||||
|
INPP_HUMAN
|
||||||
| NC score | 0.023106 (rank : 101) | θ value | 4.03905 (rank : 99) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49441 | Gene names | INPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol polyphosphate 1-phosphatase (EC 3.1.3.57) (IPPase) (IPP). | |||||
|
ST5_MOUSE
|
||||||
| NC score | 0.023102 (rank : 102) | θ value | 1.06291 (rank : 39) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q924W7, Q78H54, Q8K2P3, Q924W8 | Gene names | St5, Dennd2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppression of tumorigenicity 5 (DENN domain-containing protein 2B). | |||||
|
TACC3_HUMAN
|
||||||
| NC score | 0.022726 (rank : 103) | θ value | 5.27518 (rank : 125) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Y6A5, Q9UMQ1 | Gene names | TACC3, ERIC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ERIC-1). | |||||
|
CN044_MOUSE
|
||||||
| NC score | 0.022531 (rank : 104) | θ value | 8.99809 (rank : 166) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CB59, Q8BQX8, Q8BSL9, Q8CBZ3 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf44 homolog. | |||||
|
ARI4A_HUMAN
|
||||||
| NC score | 0.022331 (rank : 105) | θ value | 8.99809 (rank : 161) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P29374, Q15991, Q15992, Q15993 | Gene names | ARID4A, RBBP1, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 4A (ARID domain- containing protein 4A) (Retinoblastoma-binding protein 1) (RBBP-1). | |||||
|
CJ047_MOUSE
|
||||||
| NC score | 0.021827 (rank : 106) | θ value | 8.99809 (rank : 165) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8C5R2, Q8R017, Q8R2Z9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf47 homolog. | |||||
|
FGD1_MOUSE
|
||||||
| NC score | 0.021639 (rank : 107) | θ value | 0.813845 (rank : 30) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P52734 | Gene names | Fgd1 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein homolog) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
SYTL2_HUMAN
|
||||||
| NC score | 0.021435 (rank : 108) | θ value | 1.06291 (rank : 40) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9HCH5, Q2YDA7, Q8ND34, Q96BJ2, Q9H768, Q9NXM1 | Gene names | SYTL2, KIAA1597, SLP2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
MAFB_MOUSE
|
||||||
| NC score | 0.021235 (rank : 109) | θ value | 2.36792 (rank : 73) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P54841 | Gene names | Mafb, Krml, Maf1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafB (V-maf musculoaponeurotic fibrosarcoma oncogene homolog B) (Transcription factor MAF1) (Segmentation protein KR) (Kreisler). | |||||
|
HUWE1_HUMAN
|
||||||
| NC score | 0.020928 (rank : 110) | θ value | 1.38821 (rank : 47) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
SKI_MOUSE
|
||||||
| NC score | 0.020610 (rank : 111) | θ value | 2.36792 (rank : 78) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q60698, Q8VIL5 | Gene names | Ski | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ski oncogene (C-ski). | |||||
|
CN044_HUMAN
|
||||||
| NC score | 0.020337 (rank : 112) | θ value | 6.88961 (rank : 131) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96MY7 | Gene names | C14orf44 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf44. | |||||
|
P73L_HUMAN
|
||||||
| NC score | 0.020056 (rank : 113) | θ value | 4.03905 (rank : 104) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H3D4, O75080, O75195, O75922, O76078, Q6VEG2, Q6VEG3, Q6VEG4, Q6VFJ1, Q6VFJ2, Q6VFJ3, Q6VH20, Q7LDI3, Q7LDI4, Q7LDI5, Q96KR0, Q9H3D2, Q9H3D3, Q9H3P8, Q9NPH7, Q9P1B4, Q9P1B5, Q9P1B6, Q9P1B7, Q9UBV9, Q9UE10, Q9UP26, Q9UP27, Q9UP28, Q9UP74 | Gene names | TP73L, KET, P63, P73H, P73L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor protein p73-like (p73L) (p63) (Tumor protein 63) (TP63) (p51) (p40) (Keratinocyte transcription factor KET) (Chronic ulcerative stomatitis protein) (CUSP). | |||||
|
FXL19_MOUSE
|
||||||
| NC score | 0.020030 (rank : 114) | θ value | 5.27518 (rank : 118) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6PB97, Q7TSH0, Q8BIB4 | Gene names | Fbxl19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box/LRR-repeat protein 19 (F-box and leucine-rich repeat protein 19). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.019198 (rank : 115) | θ value | 0.62314 (rank : 27) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
HXA11_HUMAN
|
||||||
| NC score | 0.018750 (rank : 116) | θ value | 1.06291 (rank : 35) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P31270 | Gene names | HOXA11, HOX1I | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A11 (Hox-1I). | |||||
|
TCAL5_HUMAN
|
||||||
| NC score | 0.018504 (rank : 117) | θ value | 8.99809 (rank : 180) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
OBFC1_MOUSE
|
||||||
| NC score | 0.018467 (rank : 118) | θ value | 6.88961 (rank : 149) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K2X3, Q8C6I6 | Gene names | Obfc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oligonucleotide/oligosaccharide-binding fold-containing protein 1. | |||||
|
AB1IP_MOUSE
|
||||||
| NC score | 0.018413 (rank : 119) | θ value | 5.27518 (rank : 110) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8R5A3, O35329, Q8BRU0, Q99KV8 | Gene names | Apbb1ip, Prel1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 1- interacting protein (APBB1-interacting protein 1) (Proline-rich EVH1 ligand 1) (PREL-1) (Proline-rich protein 48). | |||||
|
CO3A1_HUMAN
|
||||||
| NC score | 0.017875 (rank : 120) | θ value | 3.0926 (rank : 83) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P02461, Q15112, Q16403, Q6LDB3, Q6LDJ2, Q6LDJ3, Q7KZ56 | Gene names | COL3A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
FGD1_HUMAN
|
||||||
| NC score | 0.017735 (rank : 121) | θ value | 4.03905 (rank : 95) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 367 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P98174, Q8N4D9 | Gene names | FGD1, ZFYVE3 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
KIF17_MOUSE
|
||||||
| NC score | 0.017469 (rank : 122) | θ value | 0.62314 (rank : 25) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99PW8 | Gene names | Kif17 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF17 (MmKIF17). | |||||
|
RTN1_MOUSE
|
||||||
| NC score | 0.017207 (rank : 123) | θ value | 5.27518 (rank : 124) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8K0T0, Q8K4S4 | Gene names | Rtn1, Nsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulon-1 (Neuroendocrine-specific protein). | |||||
|
CCD60_HUMAN
|
||||||
| NC score | 0.017180 (rank : 124) | θ value | 6.88961 (rank : 129) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IWA6 | Gene names | CCDC60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 60. | |||||
|
ELK3_MOUSE
|
||||||
| NC score | 0.017051 (rank : 125) | θ value | 1.38821 (rank : 45) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P41971, P97747, Q62346 | Gene names | Elk3, Erp, Net | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-3 (ETS-related protein NET) (ETS- related protein ERP). | |||||
|
FOLC_MOUSE
|
||||||
| NC score | 0.016482 (rank : 126) | θ value | 6.88961 (rank : 137) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P48760 | Gene names | Fpgs | |||
|
Domain Architecture |

|
|||||
| Description | Folylpolyglutamate synthase, mitochondrial precursor (EC 6.3.2.17) (Folylpoly-gamma-glutamate synthetase) (FPGS) (Tetrahydrofolate synthase) (Tetrahydrofolylpolyglutamate synthase). | |||||
|
BSND_MOUSE
|
||||||
| NC score | 0.016260 (rank : 127) | θ value | 8.99809 (rank : 162) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VIM4, Q8C740, Q8CHY0 | Gene names | Bsnd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Barttin. | |||||
|
TF3C2_MOUSE
|
||||||
| NC score | 0.016059 (rank : 128) | θ value | 6.88961 (rank : 158) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BL74, Q3U7Z9, Q80XQ7, Q8BJJ4, Q8BLP3, Q8K1J1, Q9CSS0, Q9CVE6, Q9DBK6 | Gene names | Gtf3c2, Kiaa0011 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor 3C polypeptide 2 (Transcription factor IIIC-subunit beta) (TF3C-beta) (TFIIIC 110 kDa subunit) (TFIIIC110). | |||||
|
CO7A1_HUMAN
|
||||||
| NC score | 0.014967 (rank : 129) | θ value | 2.36792 (rank : 67) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
HXA11_MOUSE
|
||||||
| NC score | 0.014956 (rank : 130) | θ value | 1.06291 (rank : 36) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P31311, Q3V026 | Gene names | Hoxa11, Hox-1.9, Hoxa-11 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A11 (Hox-1.9). | |||||
|
CB013_MOUSE
|
||||||
| NC score | 0.014700 (rank : 131) | θ value | 8.99809 (rank : 163) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D842, Q8BZL5, Q99LX6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C2orf13 homolog. | |||||
|
ELL_HUMAN
|
||||||
| NC score | 0.014357 (rank : 132) | θ value | 8.99809 (rank : 167) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P55199 | Gene names | ELL, C19orf17 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II elongation factor ELL (Eleven-nineteen lysine-rich leukemia protein). | |||||
|
PKHA3_HUMAN
|
||||||
| NC score | 0.014156 (rank : 133) | θ value | 6.88961 (rank : 154) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HB20, Q86TQ1, Q9NXT3 | Gene names | PLEKHA3, FAPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 3 (Phosphoinositol 4-phosphate adaptor protein 1) (FAPP-1). | |||||
|
SAMD4_HUMAN
|
||||||
| NC score | 0.013862 (rank : 134) | θ value | 8.99809 (rank : 177) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UPU9 | Gene names | SAMD4, KIAA1053 | |||
|
Domain Architecture |

|
|||||
| Description | Sterile alpha motif domain-containing protein 4. | |||||
|
PIWL1_MOUSE
|
||||||
| NC score | 0.013779 (rank : 135) | θ value | 5.27518 (rank : 122) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JMB7 | Gene names | Piwil1, Miwi | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Piwi-like protein 1. | |||||
|
PIWL1_HUMAN
|
||||||
| NC score | 0.013709 (rank : 136) | θ value | 5.27518 (rank : 121) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96J94, O95404, Q8NA60, Q8TBY5, Q96JD5 | Gene names | PIWIL1, HIWI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Piwi-like protein 1. | |||||
|
ATF7_MOUSE
|
||||||
| NC score | 0.013660 (rank : 137) | θ value | 6.88961 (rank : 127) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8R0S1 | Gene names | Atf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
EYA3_HUMAN
|
||||||
| NC score | 0.013572 (rank : 138) | θ value | 6.88961 (rank : 135) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99504, O95463, Q99813 | Gene names | EYA3 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 3 (EC 3.1.3.48). | |||||
|
TLR2_HUMAN
|
||||||
| NC score | 0.013531 (rank : 139) | θ value | 1.38821 (rank : 54) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60603, O15454, Q8NI00 | Gene names | TLR2, TIL4 | |||
|
Domain Architecture |

|
|||||
| Description | Toll-like receptor 2 precursor (Toll/interleukin 1 receptor-like protein 4) (CD282 antigen). | |||||
|
EYA3_MOUSE
|
||||||
| NC score | 0.013291 (rank : 140) | θ value | 6.88961 (rank : 136) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97480, P97768 | Gene names | Eya3 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 3 (EC 3.1.3.48). | |||||
|
BPAEA_MOUSE
|
||||||
| NC score | 0.013265 (rank : 141) | θ value | 1.38821 (rank : 44) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
AGGF1_HUMAN
|
||||||
| NC score | 0.013116 (rank : 142) | θ value | 8.99809 (rank : 160) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8N302, O00581, Q53YS3, Q9BU84, Q9NW66 | Gene names | AGGF1, VG5Q | |||
|
Domain Architecture |

|
|||||
| Description | Angiogenic factor with G patch and FHA domains 1 (Angiogenic factor VG5Q) (Vasculogenesis gene on 5q protein) (hVG5Q). | |||||
|
SOS1_MOUSE
|
||||||
| NC score | 0.012920 (rank : 143) | θ value | 6.88961 (rank : 156) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q62245, Q62244 | Gene names | Sos1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1) (mSOS-1). | |||||
|
DOCK2_MOUSE
|
||||||
| NC score | 0.012758 (rank : 144) | θ value | 3.0926 (rank : 84) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C3J5, Q99M79 | Gene names | Dock2 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 2 (Hch protein). | |||||
|
ABP1_HUMAN
|
||||||
| NC score | 0.012744 (rank : 145) | θ value | 5.27518 (rank : 111) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P19801, Q16683, Q16684, Q56II4, Q6GU42 | Gene names | ABP1, AOC1, DAO1 | |||
|
Domain Architecture |

|
|||||
| Description | Amiloride-sensitive amine oxidase [copper-containing] precursor (EC 1.4.3.6) (Diamine oxidase) (DAO) (Amiloride-binding protein) (ABP) (Histaminase) (Kidney amine oxidase) (KAO). | |||||
|
STA13_MOUSE
|
||||||
| NC score | 0.012483 (rank : 146) | θ value | 3.0926 (rank : 90) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q923Q2, Q8K369 | Gene names | Stard13 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 13 (StARD13) (START domain- containing protein 13). | |||||
|
KI13B_HUMAN
|
||||||
| NC score | 0.012382 (rank : 147) | θ value | 6.88961 (rank : 141) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NQT8, O75134, Q9BYJ6 | Gene names | KIF13B, GAKIN, KIAA0639 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF13B (Kinesin-like protein GAKIN). | |||||
|
PPGB_HUMAN
|
||||||
| NC score | 0.012033 (rank : 148) | θ value | 8.99809 (rank : 175) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P10619, Q5JZH1, Q96KJ2, Q9BR08, Q9BW68 | Gene names | PPGB | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal protective protein precursor (EC 3.4.16.5) (Cathepsin A) (Carboxypeptidase C) (Protective protein for beta-galactosidase) [Contains: Lysosomal protective protein 32 kDa chain; Lysosomal protective protein 20 kDa chain]. | |||||
|
NCOA3_HUMAN
|
||||||
| NC score | 0.011913 (rank : 149) | θ value | 6.88961 (rank : 147) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y6Q9, Q5JYD9, Q5JYE0, Q9BR49, Q9UPC9, Q9UPG4, Q9UPG7 | Gene names | NCOA3, AIB1, RAC3, TRAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP-interacting protein) (pCIP). | |||||
|
DSCL1_HUMAN
|
||||||
| NC score | 0.011836 (rank : 150) | θ value | 0.813845 (rank : 29) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8TD84, Q76MU9, Q8IZY3, Q8IZY4, Q8WXU7, Q9ULT7 | Gene names | DSCAML1, DSCAM2, KIAA1132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Down syndrome cell adhesion molecule-like protein 1 precursor (Down syndrome cell adhesion molecule 2). | |||||
|
TRI36_HUMAN
|
||||||
| NC score | 0.011832 (rank : 151) | θ value | 2.36792 (rank : 80) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NQ86 | Gene names | TRIM36, RBCC728, RNF98 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 36 (Zinc-binding protein Rbcc728) (RING finger protein 98). | |||||
|
MACF1_MOUSE
|
||||||
| NC score | 0.011517 (rank : 152) | θ value | 2.36792 (rank : 72) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
KIF1C_HUMAN
|
||||||
| NC score | 0.011112 (rank : 153) | θ value | 6.88961 (rank : 142) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43896, O75186 | Gene names | KIF1C, KIAA0706 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF1C. | |||||
|
KIF1C_MOUSE
|
||||||
| NC score | 0.010955 (rank : 154) | θ value | 6.88961 (rank : 143) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35071, Q5SX62 | Gene names | Kif1c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF1C. | |||||
|
CEP63_HUMAN
|
||||||
| NC score | 0.010904 (rank : 155) | θ value | 4.03905 (rank : 92) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1005 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96MT8, Q96CR0, Q9H8F5, Q9H8N0 | Gene names | CEP63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 63 kDa (Cep63 protein). | |||||
|
BMP2K_HUMAN
|
||||||
| NC score | 0.010697 (rank : 156) | θ value | 1.81305 (rank : 57) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NSY1, O94791, Q8IYF2, Q8N2G7, Q8NHG9, Q9NTG8 | Gene names | BMP2K, BIKE | |||
|
Domain Architecture |

|
|||||
| Description | BMP-2-inducible protein kinase (EC 2.7.11.1) (BIKe). | |||||
|
LIMA1_HUMAN
|
||||||
| NC score | 0.010638 (rank : 157) | θ value | 8.99809 (rank : 169) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UHB6, Q2TAN7, Q9BVF2, Q9H8J1, Q9HBN5, Q9NX96, Q9NXC3, Q9NXU6, Q9P0H8, Q9UHB5 | Gene names | LIMA1, EPLIN, SREBP3 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain and actin-binding protein 1 (Epithelial protein lost in neoplasm). | |||||
|
PHLPP_HUMAN
|
||||||
| NC score | 0.010621 (rank : 158) | θ value | 6.88961 (rank : 152) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
CHD8_HUMAN
|
||||||
| NC score | 0.010530 (rank : 159) | θ value | 4.03905 (rank : 93) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9HCK8, Q68DQ0, Q8N3Z9, Q8NCY4, Q8TBR9, Q96F26 | Gene names | CHD8, HELSNF1, KIAA1564 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 8 (EC 3.6.1.-) (ATP- dependent helicase CHD8) (CHD-8) (Helicase with SNF2 domain 1). | |||||
|
SMC3_MOUSE
|
||||||
| NC score | 0.010432 (rank : 160) | θ value | 8.99809 (rank : 179) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1187 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9CW03, O35667, Q9QUS3 | Gene names | Cspg6, Bam, Bmh, Mmip1, Smc3, Smc3l1, Smcd | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome segregation protein SmcD) (Bamacan) (Basement membrane-associated chondroitin proteoglycan) (Mad member- interacting protein 1). | |||||
|
SMC3_HUMAN
|
||||||
| NC score | 0.010387 (rank : 161) | θ value | 8.99809 (rank : 178) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1186 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UQE7, O60464 | Gene names | CSPG6, BAM, BMH, SMC3, SMC3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome-associated polypeptide) (hCAP) (Bamacan) (Basement membrane-associated chondroitin proteoglycan). | |||||
|
ANR35_HUMAN
|
||||||
| NC score | 0.010109 (rank : 162) | θ value | 5.27518 (rank : 113) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8N283, Q3MJ10, Q96LS3 | Gene names | ANKRD35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 35. | |||||
|
PER3_MOUSE
|
||||||
| NC score | 0.009930 (rank : 163) | θ value | 6.88961 (rank : 151) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O70361 | Gene names | Per3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (mPER3). | |||||
|
TTLL4_HUMAN
|
||||||
| NC score | 0.009917 (rank : 164) | θ value | 8.99809 (rank : 181) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14679, Q8WW29 | Gene names | TTLL4, KIAA0173 | |||
|
Domain Architecture |

|
|||||
| Description | Tubulin--tyrosine ligase-like protein 4. | |||||
|
GRIP1_MOUSE
|
||||||
| NC score | 0.009710 (rank : 165) | θ value | 6.88961 (rank : 138) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q925T6, Q8BLQ3, Q8C0T3, Q925T5, Q925T7 | Gene names | Grip1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor-interacting protein 1 (GRIP1 protein). | |||||
|
BMP2K_MOUSE
|
||||||
| NC score | 0.009471 (rank : 166) | θ value | 5.27518 (rank : 114) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q91Z96, Q8C8L7 | Gene names | Bmp2k, Bike | |||
|
Domain Architecture |

|
|||||
| Description | BMP-2-inducible protein kinase (EC 2.7.11.1) (BIKe). | |||||
|
PHLPP_MOUSE
|
||||||
| NC score | 0.008948 (rank : 167) | θ value | 6.88961 (rank : 153) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CHE4, Q6PCN0, Q8QZU8, Q8R5E5, Q99KL6 | Gene names | Phlpp, Kiaa0606, Plekhe1, Scop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein). | |||||
|
PRDM2_HUMAN
|
||||||
| NC score | 0.008436 (rank : 168) | θ value | 8.99809 (rank : 176) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
GAK_MOUSE
|
||||||
| NC score | 0.008422 (rank : 169) | θ value | 4.03905 (rank : 96) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99KY4, Q6P1I8, Q6P9S5, Q8BM74, Q8K0Q4 | Gene names | Gak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||
|
NFIB_HUMAN
|
||||||
| NC score | 0.008189 (rank : 170) | θ value | 8.99809 (rank : 173) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O00712, O00166, Q12858, Q96J45 | Gene names | NFIB | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor 1 B-type (Nuclear factor 1/B) (NF1-B) (NFI-B) (NF-I/B) (CCAAT-box-binding transcription factor) (CTF) (TGGCA-binding protein). | |||||
|
CP135_HUMAN
|
||||||
| NC score | 0.008081 (rank : 171) | θ value | 6.88961 (rank : 134) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
MYPC3_MOUSE
|
||||||
| NC score | 0.007822 (rank : 172) | θ value | 6.88961 (rank : 146) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O70468, O88997 | Gene names | Mybpc3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-binding protein C, cardiac-type (Cardiac MyBP-C) (C-protein, cardiac muscle isoform). | |||||
|
ITB4_HUMAN
|
||||||
| NC score | 0.006923 (rank : 173) | θ value | 6.88961 (rank : 140) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P16144, O14690, O14691, O15339, O15340, O15341, Q9UIQ4 | Gene names | ITGB4 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-4 precursor (GP150) (CD104 antigen). | |||||
|
PPE2_MOUSE
|
||||||
| NC score | 0.006908 (rank : 174) | θ value | 8.99809 (rank : 174) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35385 | Gene names | Ppef2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase with EF-hands 2 (EC 3.1.3.16) (PPEF-2). | |||||
|
GLI2_HUMAN
|
||||||
| NC score | 0.006712 (rank : 175) | θ value | 4.03905 (rank : 97) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 971 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P10070, O60252, O60253, O60254, O60255, Q15590, Q15591 | Gene names | GLI2, THP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI2 (Tax helper protein). | |||||
|
FOXP4_MOUSE
|
||||||
| NC score | 0.006348 (rank : 176) | θ value | 8.99809 (rank : 168) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9DBY0, Q80V92, Q8CG10, Q8CIS1 | Gene names | Foxp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Forkhead box protein P4 (Fork head-related protein like A) (mFKHLA). | |||||
|
MK07_MOUSE
|
||||||
| NC score | 0.006222 (rank : 177) | θ value | 4.03905 (rank : 102) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1054 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9WVS8 | Gene names | Mapk7, Erk5 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase 7 (EC 2.7.11.24) (Extracellular signal-regulated kinase 5) (ERK-5) (BMK1 kinase). | |||||
|
MARK2_HUMAN
|
||||||
| NC score | 0.006182 (rank : 178) | θ value | 1.81305 (rank : 60) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1056 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q7KZI7, Q15449, Q15524, Q5XGA3, Q68A18, Q96HB3, Q96RG0 | Gene names | MARK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MARK2 (EC 2.7.11.1) (MAP/microtubule affinity-regulating kinase 2) (ELKL motif kinase) (EMK1) (PAR1 homolog). | |||||
|
GLI1_MOUSE
|
||||||
| NC score | 0.005442 (rank : 179) | θ value | 3.0926 (rank : 85) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P47806, Q9QYK1 | Gene names | Gli1, Gli | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI1 (Glioma-associated oncogene homolog). | |||||
|
CD2L2_HUMAN
|
||||||
| NC score | 0.005305 (rank : 180) | θ value | 5.27518 (rank : 115) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
MYLK_MOUSE
|
||||||
| NC score | 0.004654 (rank : 181) | θ value | 8.99809 (rank : 172) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1489 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6PDN3, Q3TSJ7, Q80UX0, Q80YN7, Q80YN8, Q8K026, Q924D2, Q9ERD3 | Gene names | Mylk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin light chain kinase, smooth muscle (EC 2.7.11.18) (MLCK) (Telokin) (Kinase-related protein) (KRP). | |||||
|
GLIS3_HUMAN
|
||||||
| NC score | 0.004508 (rank : 182) | θ value | 4.03905 (rank : 98) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 758 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8NEA6 | Gene names | GLIS3, ZNF515 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLIS3 (GLI-similar 3) (Zinc finger protein 515). | |||||
|
THB1_MOUSE
|
||||||
| NC score | 0.003631 (rank : 183) | θ value | 6.88961 (rank : 159) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P37242 | Gene names | Thrb, C-erba-beta, Nr1a2 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid hormone receptor beta-1. | |||||
|
PCDA6_HUMAN
|
||||||
| NC score | 0.002887 (rank : 184) | θ value | 4.03905 (rank : 105) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UN73, O75283, Q9NRT8 | Gene names | PCDHA6 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 6 precursor (PCDH-alpha6). | |||||
|
ZBT40_HUMAN
|
||||||
| NC score | 0.002845 (rank : 185) | θ value | 4.03905 (rank : 108) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NUA8, O75066, Q5TFU5, Q8N1R1 | Gene names | ZBTB40, KIAA0478 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 40. | |||||
|
PLK4_MOUSE
|
||||||
| NC score | 0.001687 (rank : 186) | θ value | 5.27518 (rank : 123) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 879 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q64702, Q6PEP6, Q78EG6, Q8R0I5, Q9CVR6, Q9CVU6 | Gene names | Plk4, Sak, Stk18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase PLK4 (EC 2.7.11.21) (Polo-like kinase 4) (PLK-4) (Serine/threonine-protein kinase Sak) (Serine/threonine- protein kinase 18). | |||||
|
ZN551_HUMAN
|
||||||
| NC score | 0.001394 (rank : 187) | θ value | 1.81305 (rank : 64) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7Z340, P17034, Q8N246, Q9BRY1 | Gene names | ZNF551, KOX23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 551 (Zinc finger protein KOX23). | |||||