Please be patient as the page loads
|
NCOAT_MOUSE
|
||||||
| SwissProt Accessions | Q9EQQ9, Q3ULY7, Q6ZQ71, Q8BK05, Q8BTT2, Q8CFX2, Q9CSJ4, Q9CUR7 | Gene names | Mgea5, Hexc, Kiaa0679 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional protein NCOAT (Nuclear cytoplasmic O-GlcNAcase and acetyltransferase) (Meningioma-expressed antigen 5) [Includes: Beta- hexosaminidase (EC 3.2.1.52) (N-acetyl-beta-glucosaminidase) (Beta-N- acetylhexosaminidase) (Hexosaminidase C) (N-acetyl-beta-D- glucosaminidase) (O-GlcNAcase); Histone acetyltransferase (EC 2.3.1.48) (HAT)]. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NCOAT_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.993839 (rank : 2) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O60502, O75166, Q86WV0, Q8IV98, Q9BVA5, Q9HAR0 | Gene names | MGEA5, HEXC, KIAA0679, MEA5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional protein NCOAT (Nuclear cytoplasmic O-GlcNAcase and acetyltransferase) (Meningioma-expressed antigen 5) [Includes: Beta- hexosaminidase (EC 3.2.1.52) (N-acetyl-beta-glucosaminidase) (Beta-N- acetylhexosaminidase) (Hexosaminidase C) (N-acetyl-beta-D- glucosaminidase) (O-GlcNAcase); Histone acetyltransferase (EC 2.3.1.48) (HAT)]. | |||||
|
NCOAT_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9EQQ9, Q3ULY7, Q6ZQ71, Q8BK05, Q8BTT2, Q8CFX2, Q9CSJ4, Q9CUR7 | Gene names | Mgea5, Hexc, Kiaa0679 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional protein NCOAT (Nuclear cytoplasmic O-GlcNAcase and acetyltransferase) (Meningioma-expressed antigen 5) [Includes: Beta- hexosaminidase (EC 3.2.1.52) (N-acetyl-beta-glucosaminidase) (Beta-N- acetylhexosaminidase) (Hexosaminidase C) (N-acetyl-beta-D- glucosaminidase) (O-GlcNAcase); Histone acetyltransferase (EC 2.3.1.48) (HAT)]. | |||||
|
EF1G_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 3) | NC score | 0.137852 (rank : 3) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P26641, Q6PJ62, Q6PK31, Q96CU2, Q9P196 | Gene names | EEF1G, EF1G | |||
|
Domain Architecture |
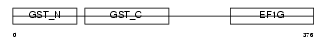
|
|||||
| Description | Elongation factor 1-gamma (EF-1-gamma) (eEF-1B gamma). | |||||
|
EF1G_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 4) | NC score | 0.130262 (rank : 4) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D8N0, Q920C5, Q9CRT5, Q9CSU3, Q9D004 | Gene names | Eef1g | |||
|
Domain Architecture |
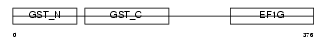
|
|||||
| Description | Elongation factor 1-gamma (EF-1-gamma) (eEF-1B gamma). | |||||
|
MYST4_HUMAN
|
||||||
| θ value | 0.279714 (rank : 5) | NC score | 0.050698 (rank : 7) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
NFM_HUMAN
|
||||||
| θ value | 0.279714 (rank : 6) | NC score | 0.019192 (rank : 25) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
F51A1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 7) | NC score | 0.095148 (rank : 5) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NWZ8 | Gene names | FAM51A1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM51A1. | |||||
|
MYST3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 8) | NC score | 0.044778 (rank : 9) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
HPS5_HUMAN
|
||||||
| θ value | 0.813845 (rank : 9) | NC score | 0.056944 (rank : 6) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UPZ3, O95942, Q8N4U0 | Gene names | HPS5, AIBP63, KIAA1017 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hermansky-Pudlak syndrome 5 protein (Alpha-integrin-binding protein 63) (Ruby-eye protein 2 homolog) (Ru2). | |||||
|
FBXW7_HUMAN
|
||||||
| θ value | 1.06291 (rank : 10) | NC score | 0.011157 (rank : 36) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q969H0, Q96A16, Q96LE0, Q96RI2, Q9NUX6 | Gene names | FBXW7, FBW7, FBX30, SEL10 | |||
|
Domain Architecture |

|
|||||
| Description | F-box/WD repeat protein 7 (F-box and WD-40 domain protein 7) (F-box protein FBX30) (hCdc4) (Archipelago homolog) (hAgo) (SEL-10). | |||||
|
SC10A_HUMAN
|
||||||
| θ value | 1.06291 (rank : 11) | NC score | 0.019677 (rank : 24) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y5Y9 | Gene names | SCN10A, PN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (hPN3). | |||||
|
CCD93_HUMAN
|
||||||
| θ value | 1.81305 (rank : 12) | NC score | 0.025199 (rank : 18) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 600 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q567U6, Q4LE78, Q53TJ2, Q8TBX5, Q9H6R5, Q9NV15 | Gene names | CCDC93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 93. | |||||
|
SDC3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 13) | NC score | 0.035290 (rank : 11) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75056, Q5T1Z6, Q5T1Z7, Q96CT3, Q96PR8 | Gene names | SDC3, KIAA0468 | |||
|
Domain Architecture |
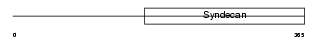
|
|||||
| Description | Syndecan-3 (SYND3). | |||||
|
TRHY_HUMAN
|
||||||
| θ value | 1.81305 (rank : 14) | NC score | 0.021987 (rank : 21) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
ZN326_HUMAN
|
||||||
| θ value | 1.81305 (rank : 15) | NC score | 0.050191 (rank : 8) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5BKZ1, Q5VW93, Q5VW94, Q5VW96, Q5VW97, Q6NSA2, Q7Z638, Q7Z6C2 | Gene names | ZNF326 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 326. | |||||
|
ALMS1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 16) | NC score | 0.026709 (rank : 14) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K4E0, Q6A084, Q8C9N9 | Gene names | Alms1, Kiaa0328 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alstrom syndrome protein 1 homolog. | |||||
|
CP250_MOUSE
|
||||||
| θ value | 2.36792 (rank : 17) | NC score | 0.013330 (rank : 30) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
MYT1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 18) | NC score | 0.024540 (rank : 19) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q01538, O94922, Q9UPV2 | Gene names | MYT1, KIAA0835, KIAA1050, MTF1, MYTI, PLPB1 | |||
|
Domain Architecture |

|
|||||
| Description | Myelin transcription factor 1 (MyT1) (MyTI) (Proteolipid protein- binding protein) (PLPB1). | |||||
|
SL9A3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 19) | NC score | 0.025628 (rank : 15) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P48764 | Gene names | SLC9A3, NHE3 | |||
|
Domain Architecture |
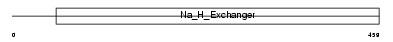
|
|||||
| Description | Sodium/hydrogen exchanger 3 (Na(+)/H(+) exchanger 3) (NHE-3) (Solute carrier family 9 member 3). | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | 4.03905 (rank : 20) | NC score | 0.013082 (rank : 31) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
ANK3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 21) | NC score | 0.007523 (rank : 40) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
GTPB2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 22) | NC score | 0.025411 (rank : 17) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BX10, Q5T7E8, Q8ND84, Q8TAH7, Q8WUA5, Q9HCS9, Q9NRU4, Q9NX60 | Gene names | GTPBP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTP-binding protein 2. | |||||
|
GTPB2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 23) | NC score | 0.025416 (rank : 16) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3UJK4, Q9EST7, Q9JIX7 | Gene names | Gtpbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTP-binding protein 2 (GTP-binding-like protein 2). | |||||
|
LR37A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 24) | NC score | 0.020018 (rank : 23) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O60309, Q49A01, Q49A80, Q8NB33 | Gene names | LRRC37A, KIAA0563 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37A. | |||||
|
PHC1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 25) | NC score | 0.017407 (rank : 26) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q64028, P70359, Q64307, Q8BZ80 | Gene names | Phc1, Edr, Edr1, Rae28 | |||
|
Domain Architecture |

|
|||||
| Description | Polyhomeotic-like protein 1 (mPH1) (Early development regulatory protein 1) (RAE-28). | |||||
|
PININ_MOUSE
|
||||||
| θ value | 4.03905 (rank : 26) | NC score | 0.028190 (rank : 13) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
TAXB1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 27) | NC score | 0.016080 (rank : 28) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q3UKC1, Q91YT6, Q9CVF0, Q9DC45 | Gene names | Tax1bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 1 homolog. | |||||
|
ARHGC_HUMAN
|
||||||
| θ value | 5.27518 (rank : 28) | NC score | 0.020267 (rank : 22) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NZN5, O15086 | Gene names | ARHGEF12, KIAA0382, LARG | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 12 (Leukemia-associated RhoGEF). | |||||
|
CCD21_HUMAN
|
||||||
| θ value | 5.27518 (rank : 29) | NC score | 0.013410 (rank : 29) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
CSPG3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 30) | NC score | 0.011864 (rank : 34) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P55066, Q6P1E3, Q8C4F8 | Gene names | Cspg3, Ncan | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
EVC_MOUSE
|
||||||
| θ value | 5.27518 (rank : 31) | NC score | 0.031799 (rank : 12) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P57680 | Gene names | Evc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ellis-van Creveld syndrome protein homolog. | |||||
|
MAP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 32) | NC score | 0.017360 (rank : 27) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P11137, Q99975, Q99976 | Gene names | MAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2) (MAP-2). | |||||
|
MYST3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 33) | NC score | 0.039595 (rank : 10) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
PDK1L_HUMAN
|
||||||
| θ value | 5.27518 (rank : 34) | NC score | 0.006894 (rank : 41) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N165, Q5T2I0, Q8NDB3 | Gene names | PDIK1L, CLIK1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase PDIK1L (EC 2.7.11.1) (PDLIM1- interacting kinase 1-like). | |||||
|
PDK1L_MOUSE
|
||||||
| θ value | 5.27518 (rank : 35) | NC score | 0.006867 (rank : 42) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8QZR7, Q3URF0, Q8BKB3 | Gene names | Pdik1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase PDIK1L (EC 2.7.11.1) (PDLIM1- interacting kinase 1-like). | |||||
|
ZAN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 36) | NC score | 0.010990 (rank : 37) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
FYV1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 37) | NC score | 0.011901 (rank : 33) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y2I7, Q8NB67 | Gene names | PIP5K3, KIAA0981, PIKFYVE | |||
|
Domain Architecture |

|
|||||
| Description | FYVE finger-containing phosphoinositide kinase (EC 2.7.1.68) (1- phosphatidylinositol-4-phosphate 5-kinase) (Phosphatidylinositol-3- phosphate 5-kinase type III) (PIP5K) (PtdIns(4)P-5-kinase) (PIKfyve) (p235). | |||||
|
URFB1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 38) | NC score | 0.022570 (rank : 20) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6BDS2, Q9NXE0 | Gene names | UHRF1BP1, C6orf107 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UHRF1-binding protein 1 (Ubiquitin-like containing PHD and RING finger domains 1-binding protein 1) (ICBP90-binding protein 1). | |||||
|
CASP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 39) | NC score | 0.010330 (rank : 38) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 1152 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P70403, Q7TQJ4, Q91WN6 | Gene names | Cutl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CUTL1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 40) | NC score | 0.010080 (rank : 39) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
FMN1B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 41) | NC score | 0.012232 (rank : 32) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q05859 | Gene names | Fmn, Ld | |||
|
Domain Architecture |

|
|||||
| Description | Formin-1 isoform IV (Limb deformity protein). | |||||
|
NP1L3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 42) | NC score | 0.011374 (rank : 35) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q794H2, O54802 | Gene names | Nap1l3, Mb20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome assembly protein 1-like 3 (Brain-specific protein MB20). | |||||
|
SUHW4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 43) | NC score | 0.003097 (rank : 43) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 737 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q68FE8, Q3T9B1, Q8BI82 | Gene names | Suhw4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppressor of hairy wing homolog 4. | |||||
|
NCOAT_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9EQQ9, Q3ULY7, Q6ZQ71, Q8BK05, Q8BTT2, Q8CFX2, Q9CSJ4, Q9CUR7 | Gene names | Mgea5, Hexc, Kiaa0679 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional protein NCOAT (Nuclear cytoplasmic O-GlcNAcase and acetyltransferase) (Meningioma-expressed antigen 5) [Includes: Beta- hexosaminidase (EC 3.2.1.52) (N-acetyl-beta-glucosaminidase) (Beta-N- acetylhexosaminidase) (Hexosaminidase C) (N-acetyl-beta-D- glucosaminidase) (O-GlcNAcase); Histone acetyltransferase (EC 2.3.1.48) (HAT)]. | |||||
|
NCOAT_HUMAN
|
||||||
| NC score | 0.993839 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O60502, O75166, Q86WV0, Q8IV98, Q9BVA5, Q9HAR0 | Gene names | MGEA5, HEXC, KIAA0679, MEA5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional protein NCOAT (Nuclear cytoplasmic O-GlcNAcase and acetyltransferase) (Meningioma-expressed antigen 5) [Includes: Beta- hexosaminidase (EC 3.2.1.52) (N-acetyl-beta-glucosaminidase) (Beta-N- acetylhexosaminidase) (Hexosaminidase C) (N-acetyl-beta-D- glucosaminidase) (O-GlcNAcase); Histone acetyltransferase (EC 2.3.1.48) (HAT)]. | |||||
|
EF1G_HUMAN
|
||||||
| NC score | 0.137852 (rank : 3) | θ value | 0.0330416 (rank : 3) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P26641, Q6PJ62, Q6PK31, Q96CU2, Q9P196 | Gene names | EEF1G, EF1G | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor 1-gamma (EF-1-gamma) (eEF-1B gamma). | |||||
|
EF1G_MOUSE
|
||||||
| NC score | 0.130262 (rank : 4) | θ value | 0.0736092 (rank : 4) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D8N0, Q920C5, Q9CRT5, Q9CSU3, Q9D004 | Gene names | Eef1g | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor 1-gamma (EF-1-gamma) (eEF-1B gamma). | |||||
|
F51A1_HUMAN
|
||||||
| NC score | 0.095148 (rank : 5) | θ value | 0.47712 (rank : 7) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NWZ8 | Gene names | FAM51A1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM51A1. | |||||
|
HPS5_HUMAN
|
||||||
| NC score | 0.056944 (rank : 6) | θ value | 0.813845 (rank : 9) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UPZ3, O95942, Q8N4U0 | Gene names | HPS5, AIBP63, KIAA1017 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hermansky-Pudlak syndrome 5 protein (Alpha-integrin-binding protein 63) (Ruby-eye protein 2 homolog) (Ru2). | |||||
|
MYST4_HUMAN
|
||||||
| NC score | 0.050698 (rank : 7) | θ value | 0.279714 (rank : 5) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
ZN326_HUMAN
|
||||||
| NC score | 0.050191 (rank : 8) | θ value | 1.81305 (rank : 15) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5BKZ1, Q5VW93, Q5VW94, Q5VW96, Q5VW97, Q6NSA2, Q7Z638, Q7Z6C2 | Gene names | ZNF326 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 326. | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.044778 (rank : 9) | θ value | 0.47712 (rank : 8) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
MYST3_MOUSE
|
||||||
| NC score | 0.039595 (rank : 10) | θ value | 5.27518 (rank : 33) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
SDC3_HUMAN
|
||||||
| NC score | 0.035290 (rank : 11) | θ value | 1.81305 (rank : 13) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75056, Q5T1Z6, Q5T1Z7, Q96CT3, Q96PR8 | Gene names | SDC3, KIAA0468 | |||
|
Domain Architecture |

|
|||||
| Description | Syndecan-3 (SYND3). | |||||
|
EVC_MOUSE
|
||||||
| NC score | 0.031799 (rank : 12) | θ value | 5.27518 (rank : 31) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P57680 | Gene names | Evc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ellis-van Creveld syndrome protein homolog. | |||||
|
PININ_MOUSE
|
||||||
| NC score | 0.028190 (rank : 13) | θ value | 4.03905 (rank : 26) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
ALMS1_MOUSE
|
||||||
| NC score | 0.026709 (rank : 14) | θ value | 2.36792 (rank : 16) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K4E0, Q6A084, Q8C9N9 | Gene names | Alms1, Kiaa0328 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alstrom syndrome protein 1 homolog. | |||||
|
SL9A3_HUMAN
|
||||||
| NC score | 0.025628 (rank : 15) | θ value | 3.0926 (rank : 19) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P48764 | Gene names | SLC9A3, NHE3 | |||
|
Domain Architecture |
|
|||||
| Description | Sodium/hydrogen exchanger 3 (Na(+)/H(+) exchanger 3) (NHE-3) (Solute carrier family 9 member 3). | |||||
|
GTPB2_MOUSE
|
||||||
| NC score | 0.025416 (rank : 16) | θ value | 4.03905 (rank : 23) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3UJK4, Q9EST7, Q9JIX7 | Gene names | Gtpbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTP-binding protein 2 (GTP-binding-like protein 2). | |||||
|
GTPB2_HUMAN
|
||||||
| NC score | 0.025411 (rank : 17) | θ value | 4.03905 (rank : 22) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BX10, Q5T7E8, Q8ND84, Q8TAH7, Q8WUA5, Q9HCS9, Q9NRU4, Q9NX60 | Gene names | GTPBP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTP-binding protein 2. | |||||
|
CCD93_HUMAN
|
||||||
| NC score | 0.025199 (rank : 18) | θ value | 1.81305 (rank : 12) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 600 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q567U6, Q4LE78, Q53TJ2, Q8TBX5, Q9H6R5, Q9NV15 | Gene names | CCDC93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 93. | |||||
|
MYT1_HUMAN
|
||||||
| NC score | 0.024540 (rank : 19) | θ value | 2.36792 (rank : 18) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q01538, O94922, Q9UPV2 | Gene names | MYT1, KIAA0835, KIAA1050, MTF1, MYTI, PLPB1 | |||
|
Domain Architecture |

|
|||||
| Description | Myelin transcription factor 1 (MyT1) (MyTI) (Proteolipid protein- binding protein) (PLPB1). | |||||
|
URFB1_HUMAN
|
||||||
| NC score | 0.022570 (rank : 20) | θ value | 6.88961 (rank : 38) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6BDS2, Q9NXE0 | Gene names | UHRF1BP1, C6orf107 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UHRF1-binding protein 1 (Ubiquitin-like containing PHD and RING finger domains 1-binding protein 1) (ICBP90-binding protein 1). | |||||
|
TRHY_HUMAN
|
||||||
| NC score | 0.021987 (rank : 21) | θ value | 1.81305 (rank : 14) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
ARHGC_HUMAN
|
||||||
| NC score | 0.020267 (rank : 22) | θ value | 5.27518 (rank : 28) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NZN5, O15086 | Gene names | ARHGEF12, KIAA0382, LARG | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 12 (Leukemia-associated RhoGEF). | |||||
|
LR37A_HUMAN
|
||||||
| NC score | 0.020018 (rank : 23) | θ value | 4.03905 (rank : 24) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O60309, Q49A01, Q49A80, Q8NB33 | Gene names | LRRC37A, KIAA0563 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37A. | |||||
|
SC10A_HUMAN
|
||||||
| NC score | 0.019677 (rank : 24) | θ value | 1.06291 (rank : 11) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y5Y9 | Gene names | SCN10A, PN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (hPN3). | |||||
|
NFM_HUMAN
|
||||||
| NC score | 0.019192 (rank : 25) | θ value | 0.279714 (rank : 6) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
PHC1_MOUSE
|
||||||
| NC score | 0.017407 (rank : 26) | θ value | 4.03905 (rank : 25) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q64028, P70359, Q64307, Q8BZ80 | Gene names | Phc1, Edr, Edr1, Rae28 | |||
|
Domain Architecture |

|
|||||
| Description | Polyhomeotic-like protein 1 (mPH1) (Early development regulatory protein 1) (RAE-28). | |||||
|
MAP2_HUMAN
|
||||||
| NC score | 0.017360 (rank : 27) | θ value | 5.27518 (rank : 32) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P11137, Q99975, Q99976 | Gene names | MAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2) (MAP-2). | |||||
|
TAXB1_MOUSE
|
||||||
| NC score | 0.016080 (rank : 28) | θ value | 4.03905 (rank : 27) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q3UKC1, Q91YT6, Q9CVF0, Q9DC45 | Gene names | Tax1bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 1 homolog. | |||||
|
CCD21_HUMAN
|
||||||
| NC score | 0.013410 (rank : 29) | θ value | 5.27518 (rank : 29) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
CP250_MOUSE
|
||||||
| NC score | 0.013330 (rank : 30) | θ value | 2.36792 (rank : 17) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | 0.013082 (rank : 31) | θ value | 4.03905 (rank : 20) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
FMN1B_MOUSE
|
||||||
| NC score | 0.012232 (rank : 32) | θ value | 8.99809 (rank : 41) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q05859 | Gene names | Fmn, Ld | |||
|
Domain Architecture |

|
|||||
| Description | Formin-1 isoform IV (Limb deformity protein). | |||||
|
FYV1_HUMAN
|
||||||
| NC score | 0.011901 (rank : 33) | θ value | 6.88961 (rank : 37) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y2I7, Q8NB67 | Gene names | PIP5K3, KIAA0981, PIKFYVE | |||
|
Domain Architecture |

|
|||||
| Description | FYVE finger-containing phosphoinositide kinase (EC 2.7.1.68) (1- phosphatidylinositol-4-phosphate 5-kinase) (Phosphatidylinositol-3- phosphate 5-kinase type III) (PIP5K) (PtdIns(4)P-5-kinase) (PIKfyve) (p235). | |||||
|
CSPG3_MOUSE
|
||||||
| NC score | 0.011864 (rank : 34) | θ value | 5.27518 (rank : 30) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P55066, Q6P1E3, Q8C4F8 | Gene names | Cspg3, Ncan | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
NP1L3_MOUSE
|
||||||
| NC score | 0.011374 (rank : 35) | θ value | 8.99809 (rank : 42) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q794H2, O54802 | Gene names | Nap1l3, Mb20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome assembly protein 1-like 3 (Brain-specific protein MB20). | |||||
|
FBXW7_HUMAN
|
||||||
| NC score | 0.011157 (rank : 36) | θ value | 1.06291 (rank : 10) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q969H0, Q96A16, Q96LE0, Q96RI2, Q9NUX6 | Gene names | FBXW7, FBW7, FBX30, SEL10 | |||
|
Domain Architecture |

|
|||||
| Description | F-box/WD repeat protein 7 (F-box and WD-40 domain protein 7) (F-box protein FBX30) (hCdc4) (Archipelago homolog) (hAgo) (SEL-10). | |||||
|
ZAN_HUMAN
|
||||||
| NC score | 0.010990 (rank : 37) | θ value | 5.27518 (rank : 36) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
CASP_MOUSE
|
||||||
| NC score | 0.010330 (rank : 38) | θ value | 8.99809 (rank : 39) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 1152 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P70403, Q7TQJ4, Q91WN6 | Gene names | Cutl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CUTL1_MOUSE
|
||||||
| NC score | 0.010080 (rank : 39) | θ value | 8.99809 (rank : 40) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
ANK3_HUMAN
|
||||||
| NC score | 0.007523 (rank : 40) | θ value | 4.03905 (rank : 21) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
PDK1L_HUMAN
|
||||||
| NC score | 0.006894 (rank : 41) | θ value | 5.27518 (rank : 34) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N165, Q5T2I0, Q8NDB3 | Gene names | PDIK1L, CLIK1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase PDIK1L (EC 2.7.11.1) (PDLIM1- interacting kinase 1-like). | |||||
|
PDK1L_MOUSE
|
||||||
| NC score | 0.006867 (rank : 42) | θ value | 5.27518 (rank : 35) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8QZR7, Q3URF0, Q8BKB3 | Gene names | Pdik1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase PDIK1L (EC 2.7.11.1) (PDLIM1- interacting kinase 1-like). | |||||
|
SUHW4_MOUSE
|
||||||
| NC score | 0.003097 (rank : 43) | θ value | 8.99809 (rank : 43) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 737 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q68FE8, Q3T9B1, Q8BI82 | Gene names | Suhw4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppressor of hairy wing homolog 4. | |||||