Please be patient as the page loads
|
ZN326_HUMAN
|
||||||
| SwissProt Accessions | Q5BKZ1, Q5VW93, Q5VW94, Q5VW96, Q5VW97, Q6NSA2, Q7Z638, Q7Z6C2 | Gene names | ZNF326 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 326. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ZN326_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q5BKZ1, Q5VW93, Q5VW94, Q5VW96, Q5VW97, Q6NSA2, Q7Z638, Q7Z6C2 | Gene names | ZNF326 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 326. | |||||
|
ZN326_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.972274 (rank : 2) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O88291, Q8BSJ5, Q8K1X9, Q9CYG9 | Gene names | Znf326, Zan75, Zfp326 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 326 (Zinc finger protein-associated with nuclear matrix of 75 kDa). | |||||
|
AKP8L_MOUSE
|
||||||
| θ value | 4.42448e-31 (rank : 3) | NC score | 0.741762 (rank : 4) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9R0L7 | Gene names | Akap8l, Nakap95 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 8-like (AKAP8-like protein) (Neighbor of A- kinase-anchoring protein 95) (Neighbor of AKAP95). | |||||
|
AKP8L_HUMAN
|
||||||
| θ value | 1.08979e-29 (rank : 4) | NC score | 0.743217 (rank : 3) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9ULX6, O94792, Q96J58, Q9NRQ0, Q9UGM0 | Gene names | AKAP8L, NAKAP95 | |||
|
Domain Architecture |
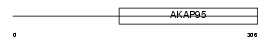
|
|||||
| Description | A-kinase anchor protein 8-like (AKAP8-like protein) (Neighbor of A- kinase-anchoring protein 95) (Neighbor of AKAP95) (Homologous to AKAP95 protein) (HA95) (Helicase A-binding protein 95) (HAP95). | |||||
|
AKAP8_HUMAN
|
||||||
| θ value | 3.17079e-29 (rank : 5) | NC score | 0.716624 (rank : 6) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O43823 | Gene names | AKAP8, AKAP95 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 8 (A-kinase anchor protein 95 kDa) (AKAP 95). | |||||
|
AKAP8_MOUSE
|
||||||
| θ value | 1.33217e-27 (rank : 6) | NC score | 0.730776 (rank : 5) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9DBR0, Q9R0L8 | Gene names | Akap8, Akap95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 8 (A-kinase anchor protein 95 kDa) (AKAP 95). | |||||
|
ROA1_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 7) | NC score | 0.130998 (rank : 11) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P09651, Q6PJZ7 | Gene names | HNRPA1 | |||
|
Domain Architecture |
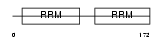
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein A1 (Helix-destabilizing protein) (Single-strand RNA-binding protein) (hnRNP core protein A1). | |||||
|
FUS_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 8) | NC score | 0.182069 (rank : 7) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P56959 | Gene names | Fus | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein FUS (Pigpen protein). | |||||
|
HNRPG_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 9) | NC score | 0.127286 (rank : 12) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P38159, Q5JQ67, Q969R3 | Gene names | RBMX, HNRPG, RBMXP1 | |||
|
Domain Architecture |
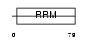
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein G (hnRNP G) (RNA-binding motif protein, X chromosome) (Glycoprotein p43). | |||||
|
HNRPG_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 10) | NC score | 0.132718 (rank : 10) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O35479 | Gene names | Rbmx, Hnrnpg, Hnrpg, Rbmxp1, Rbmxrt | |||
|
Domain Architecture |
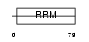
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein G (hnRNP G) (RNA-binding motif protein, X chromosome). | |||||
|
FUS_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 11) | NC score | 0.171418 (rank : 9) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P35637 | Gene names | FUS, TLS | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein FUS (Oncogene FUS) (Oncogene TLS) (Translocated in liposarcoma protein) (POMp75) (75 kDa DNA-pairing protein). | |||||
|
RBP56_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 12) | NC score | 0.174438 (rank : 8) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q92804, Q92751 | Gene names | TAF15, RBP56, TAF2N | |||
|
Domain Architecture |

|
|||||
| Description | TATA-binding protein-associated factor 2N (RNA-binding protein 56) (TAFII68) (TAF(II)68). | |||||
|
ROA3_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 13) | NC score | 0.117050 (rank : 16) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P51991, Q6URK5 | Gene names | HNRPA3 | |||
|
Domain Architecture |
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein A3 (hnRNP A3). | |||||
|
ROA3_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 14) | NC score | 0.121662 (rank : 14) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BG05, Q8BHF8 | Gene names | Hnrpa3, Hnrnpa3 | |||
|
Domain Architecture |
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein A3 (hnRNP A3). | |||||
|
FILA_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 15) | NC score | 0.094473 (rank : 23) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
ILF3_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 16) | NC score | 0.109355 (rank : 21) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z1X4, Q80VD5, Q812A1, Q8BP80, Q8C2H8, Q8K588 | Gene names | Ilf3 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin enhancer-binding factor 3. | |||||
|
K1C10_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 17) | NC score | 0.044187 (rank : 68) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 466 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P02535, P08731, Q8BUX3, Q8BV09, Q8BVU3, Q9CXH6, Q9JKB4 | Gene names | Krt10, Krt1-10 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 10 (Cytokeratin-10) (CK-10) (Keratin-10) (K10) (56 kDa cytokeratin) (Keratin, type I cytoskeletal 59 kDa). | |||||
|
HNRPR_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 18) | NC score | 0.080672 (rank : 29) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O43390 | Gene names | HNRPR | |||
|
Domain Architecture |
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein R (hnRNP R). | |||||
|
UACA_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 19) | NC score | 0.033793 (rank : 81) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8CGB3, Q69ZG3, Q7TN77, Q8BJC8 | Gene names | Uaca, Kiaa1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein (Nucling) (Nuclear membrane-binding protein). | |||||
|
MY18A_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 20) | NC score | 0.036846 (rank : 76) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
TCAL5_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 21) | NC score | 0.064058 (rank : 37) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
ILF3_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 22) | NC score | 0.112675 (rank : 18) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q12906, O43409, Q6P1X1, Q86XY7, Q99544, Q99545, Q9BZH4, Q9BZH5, Q9NQ95, Q9NQ96, Q9NQ97, Q9NQ98, Q9NQ99, Q9NQA0, Q9NQA1, Q9NQA2, Q9NRN2, Q9NRN3, Q9NRN4, Q9UMZ9, Q9UN00, Q9UN84, Q9UNA2 | Gene names | ILF3, DRBF, MPHOSPH4, NF90 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin enhancer-binding factor 3 (Nuclear factor of activated T- cells 90 kDa) (NF-AT-90) (Double-stranded RNA-binding protein 76) (DRBP76) (Translational control protein 80) (TCP80) (Nuclear factor associated with dsRNA) (NFAR) (M-phase phosphoprotein 4) (MPP4). | |||||
|
RBY1A_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 23) | NC score | 0.119736 (rank : 15) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q15414, Q15377, Q8NHR0 | Gene names | RBMY1A1, RBM1, YRRM1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding motif protein, Y chromosome, family 1 member A1 (RNA- binding motif protein 1). | |||||
|
FLIP1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 24) | NC score | 0.037424 (rank : 74) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
K1C9_HUMAN
|
||||||
| θ value | 0.125558 (rank : 25) | NC score | 0.044228 (rank : 67) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 526 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P35527, O00109, Q14665 | Gene names | KRT9 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 9 (Cytokeratin-9) (CK-9) (Keratin-9) (K9). | |||||
|
K2C1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 26) | NC score | 0.037616 (rank : 73) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P04264, Q14720, Q6GSJ0, Q9H298 | Gene names | KRT1, KRTA | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 1 (Cytokeratin-1) (CK-1) (Keratin-1) (K1) (67 kDa cytokeratin) (Hair alpha protein). | |||||
|
UACA_HUMAN
|
||||||
| θ value | 0.125558 (rank : 27) | NC score | 0.035754 (rank : 80) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1622 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9BZF9, Q8N3B8, Q9HCL1, Q9NWC6 | Gene names | UACA, KIAA1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein. | |||||
|
CGBP1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 28) | NC score | 0.122198 (rank : 13) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BHG9, Q8K2K1 | Gene names | Cggbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CGG triplet repeat-binding protein 1 (CGG-binding protein 1) (p20- CGGBP DNA-binding protein) (20 kDa CGG-binding protein). | |||||
|
ROA0_HUMAN
|
||||||
| θ value | 0.21417 (rank : 29) | NC score | 0.113678 (rank : 17) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13151 | Gene names | HNRPA0 | |||
|
Domain Architecture |
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein A0 (hnRNP A0). | |||||
|
FXR2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 30) | NC score | 0.042985 (rank : 69) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P51116, Q8WUM2 | Gene names | FXR2 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 2. | |||||
|
ROA2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 31) | NC score | 0.109859 (rank : 19) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P22626, P22627 | Gene names | HNRPA2B1 | |||
|
Domain Architecture |
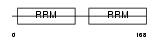
|
|||||
| Description | Heterogeneous nuclear ribonucleoproteins A2/B1 (hnRNP A2 / hnRNP B1). | |||||
|
CIRBP_MOUSE
|
||||||
| θ value | 0.365318 (rank : 32) | NC score | 0.089409 (rank : 26) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P60824, O09069, O09148, Q3V1A6, Q61413 | Gene names | Cirbp, Cirp | |||
|
Domain Architecture |
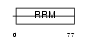
|
|||||
| Description | Cold-inducible RNA-binding protein (Glycine-rich RNA-binding protein CIRP) (A18 hnRNP). | |||||
|
FLIP1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 33) | NC score | 0.039310 (rank : 71) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1176 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9CS72 | Gene names | Filip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
K2C1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 34) | NC score | 0.036554 (rank : 77) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P04104 | Gene names | Krt1, Krt2-1 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 1 (Cytokeratin-1) (CK-1) (Keratin-1) (K1) (67 kDa cytokeratin). | |||||
|
LORI_MOUSE
|
||||||
| θ value | 0.365318 (rank : 35) | NC score | 0.109666 (rank : 20) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P18165, Q8R0I9 | Gene names | Lor | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Loricrin. | |||||
|
NCOAT_HUMAN
|
||||||
| θ value | 0.47712 (rank : 36) | NC score | 0.055802 (rank : 53) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60502, O75166, Q86WV0, Q8IV98, Q9BVA5, Q9HAR0 | Gene names | MGEA5, HEXC, KIAA0679, MEA5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional protein NCOAT (Nuclear cytoplasmic O-GlcNAcase and acetyltransferase) (Meningioma-expressed antigen 5) [Includes: Beta- hexosaminidase (EC 3.2.1.52) (N-acetyl-beta-glucosaminidase) (Beta-N- acetylhexosaminidase) (Hexosaminidase C) (N-acetyl-beta-D- glucosaminidase) (O-GlcNAcase); Histone acetyltransferase (EC 2.3.1.48) (HAT)]. | |||||
|
SYCP1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 37) | NC score | 0.029969 (rank : 87) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
ARNT_MOUSE
|
||||||
| θ value | 0.62314 (rank : 38) | NC score | 0.025901 (rank : 94) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
DHX9_MOUSE
|
||||||
| θ value | 0.62314 (rank : 39) | NC score | 0.025193 (rank : 97) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O70133, O35931, O54703, Q5FWY1, Q6R5F7, Q9CSA2 | Gene names | Dhx9, Ddx9 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase A (EC 3.6.1.-) (Nuclear DNA helicase II) (NDH II) (DEAH box protein 9) (mHEL-5). | |||||
|
MY18A_MOUSE
|
||||||
| θ value | 0.62314 (rank : 40) | NC score | 0.030469 (rank : 86) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
CGRE1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 41) | NC score | 0.069815 (rank : 30) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8R1U2, Q9D1K1 | Gene names | Cgref1, Cgr11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
FYB_HUMAN
|
||||||
| θ value | 0.813845 (rank : 42) | NC score | 0.036504 (rank : 78) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O15117, O00359 | Gene names | FYB, SLAP130 | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
FA98A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 43) | NC score | 0.058120 (rank : 47) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NCA5 | Gene names | FAM98A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM98A. | |||||
|
K1C10_HUMAN
|
||||||
| θ value | 1.38821 (rank : 44) | NC score | 0.037319 (rank : 75) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 526 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P13645, Q14664, Q8N175 | Gene names | KRT10 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 10 (Cytokeratin-10) (CK-10) (Keratin-10) (K10). | |||||
|
MDN1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 45) | NC score | 0.044756 (rank : 66) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
CGBP1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 46) | NC score | 0.093695 (rank : 24) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UFW8, O15183 | Gene names | CGGBP1, CGGBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CGG triplet repeat-binding protein 1 (CGG-binding protein 1) (p20- CGGBP DNA-binding protein) (20 kDa CGG-binding protein). | |||||
|
HORN_HUMAN
|
||||||
| θ value | 1.81305 (rank : 47) | NC score | 0.056314 (rank : 50) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86YZ3 | Gene names | HRNR, S100A18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hornerin. | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.051948 (rank : 61) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
NCOAT_MOUSE
|
||||||
| θ value | 1.81305 (rank : 49) | NC score | 0.050191 (rank : 63) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EQQ9, Q3ULY7, Q6ZQ71, Q8BK05, Q8BTT2, Q8CFX2, Q9CSJ4, Q9CUR7 | Gene names | Mgea5, Hexc, Kiaa0679 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional protein NCOAT (Nuclear cytoplasmic O-GlcNAcase and acetyltransferase) (Meningioma-expressed antigen 5) [Includes: Beta- hexosaminidase (EC 3.2.1.52) (N-acetyl-beta-glucosaminidase) (Beta-N- acetylhexosaminidase) (Hexosaminidase C) (N-acetyl-beta-D- glucosaminidase) (O-GlcNAcase); Histone acetyltransferase (EC 2.3.1.48) (HAT)]. | |||||
|
ROA2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 50) | NC score | 0.104141 (rank : 22) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O88569 | Gene names | Hnrpa2b1 | |||
|
Domain Architecture |
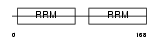
|
|||||
| Description | Heterogeneous nuclear ribonucleoproteins A2/B1 (hnRNP A2 / hnRNP B1). | |||||
|
ARI5B_HUMAN
|
||||||
| θ value | 2.36792 (rank : 51) | NC score | 0.035880 (rank : 79) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14865, Q7Z3M4, Q8N421, Q9H786 | Gene names | ARID5B, DESRT, MRF2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (Mrf1-like) (Modulator recognition factor 2) (MRF-2). | |||||
|
BRD8_MOUSE
|
||||||
| θ value | 2.36792 (rank : 52) | NC score | 0.028840 (rank : 91) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
CO5A3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 53) | NC score | 0.010033 (rank : 111) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P25940, Q9NZQ6 | Gene names | COL5A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(V) chain precursor. | |||||
|
EWS_MOUSE
|
||||||
| θ value | 2.36792 (rank : 54) | NC score | 0.090026 (rank : 25) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q61545 | Gene names | Ewsr1, Ews, Ewsh | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein EWS. | |||||
|
KS6A3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 55) | NC score | 0.000093 (rank : 118) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 860 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P18654, Q03140, Q8K3J8 | Gene names | Rps6ka3, Rps6ka-rs1, Rsk2 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosomal protein S6 kinase alpha-3 (EC 2.7.11.1) (S6K-alpha 3) (90 kDa ribosomal protein S6 kinase 3) (p90-RSK 3) (Ribosomal S6 kinase 2) (RSK-2) (pp90RSK2) (MAP kinase-activated protein kinase 1b) (MAPKAPK1B). | |||||
|
NKTR_MOUSE
|
||||||
| θ value | 2.36792 (rank : 56) | NC score | 0.029672 (rank : 89) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P30415 | Gene names | Nktr | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
PPIG_HUMAN
|
||||||
| θ value | 2.36792 (rank : 57) | NC score | 0.025672 (rank : 95) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
RNH1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 58) | NC score | 0.030863 (rank : 85) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60930, O60523, O60857, Q57Z93, Q5U0C1, Q6FHD4 | Gene names | RNASEH1, RNH1 | |||
|
Domain Architecture |
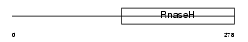
|
|||||
| Description | Ribonuclease H1 (EC 3.1.26.4) (RNase H1) (Ribonuclease H type II). | |||||
|
HM20A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.032005 (rank : 84) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NP66, Q53G31, Q9NSF6 | Gene names | HMG20A, HMGX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein 20A (HMG box-containing protein 20A) (HMG domain-containing protein HMGX1). | |||||
|
HM20A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | 0.032009 (rank : 83) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9DC33, Q3LSF9, Q6NV87, Q8BSK1, Q8C3C1, Q8CAA0, Q9CYG2 | Gene names | Hmg20a, Ibraf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein 20A (HMG box-containing protein 20A) (iBRAF) (Inhibitor of BRAF35) (HMG domain protein HMGX1). | |||||
|
PDCL2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 61) | NC score | 0.038145 (rank : 72) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q78Y63 | Gene names | Pdcl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosducin-like protein 2. | |||||
|
S20A1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.021075 (rank : 98) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8WUM9, Q08344, Q6DHX8, Q9UQ82 | Gene names | SLC20A1, GLVR1, PIT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium-dependent phosphate transporter 1 (Solute carrier family 20 member 1) (Phosphate transporter 1) (PiT-1) (Gibbon ape leukemia virus receptor 1) (GLVR-1) (Leukemia virus receptor 1 homolog). | |||||
|
SPC24_HUMAN
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.029869 (rank : 88) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NBT2 | Gene names | SPBC24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Spc24 (hSpc24). | |||||
|
WDFY3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.017934 (rank : 102) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IZQ1, Q4W5K5, Q6P0Q5, Q8N1T2, Q96BS7, Q96N85, Q9Y2J7 | Gene names | WDFY3, KIAA0993 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat and FYVE domain-containing protein 3 (Autophagy-linked FYVE protein) (Alfy). | |||||
|
ZBT24_MOUSE
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | -0.001161 (rank : 119) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 894 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80X44, Q3UIB7, Q7TPC4, Q8CJC9 | Gene names | Zbtb24, Bif1, Bsg1, Znf450 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and BTB domain-containing protein 24 (Zinc finger protein 450) (Bone morphogenetic protein-induced factor 1) (Brain-specific protein 1). | |||||
|
EWS_HUMAN
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.087236 (rank : 27) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q01844, Q92635 | Gene names | EWSR1, EWS | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein EWS (EWS oncogene) (Ewing sarcoma breakpoint region 1 protein). | |||||
|
HORN_MOUSE
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.040763 (rank : 70) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8VHD8 | Gene names | Hrnr | |||
|
Domain Architecture |

|
|||||
| Description | Hornerin. | |||||
|
MELK_MOUSE
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.001776 (rank : 116) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 860 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61846, Q61804, Q6ZQH6 | Gene names | Melk, Kiaa0175, Pk38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Maternal embryonic leucine zipper kinase (EC 2.7.11.1) (Protein kinase PK38) (mPK38). | |||||
|
OSBL5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.014661 (rank : 107) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ER64, Q8R510, Q99NF5 | Gene names | Osbpl5, Obph1, Osbp2 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 5 (OSBP-related protein 5) (ORP-5) (Oxysterol-binding protein homolog 1). | |||||
|
PDCL2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.032576 (rank : 82) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N4E4 | Gene names | PDCL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosducin-like protein 2. | |||||
|
YTDC1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.027905 (rank : 93) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
ZN409_HUMAN
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.018225 (rank : 101) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UPU6 | Gene names | ZNF409, KIAA1056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 409. | |||||
|
ATBF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.011418 (rank : 109) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
ATBF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.015796 (rank : 105) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
CH60_MOUSE
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.020151 (rank : 100) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P63038, P19226, P19227, P97602 | Gene names | Hspd1, Hsp60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 60 kDa heat shock protein, mitochondrial precursor (Hsp60) (60 kDa chaperonin) (CPN60) (Heat shock protein 60) (HSP-60) (Mitochondrial matrix protein P1) (HSP-65). | |||||
|
CHD2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.009617 (rank : 112) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O14647 | Gene names | CHD2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 2 (EC 3.6.1.-) (ATP- dependent helicase CHD2) (CHD-2). | |||||
|
K1C16_HUMAN
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.028985 (rank : 90) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P08779, P30654, Q16402, Q9UBG8 | Gene names | KRT16, KRT16A | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 16 (Cytokeratin-16) (CK-16) (Keratin-16) (K16). | |||||
|
MELK_HUMAN
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.001026 (rank : 117) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 847 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14680, Q7L3C3 | Gene names | MELK, KIAA0175 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Maternal embryonic leucine zipper kinase (EC 2.7.11.1) (hMELK) (Protein kinase PK38) (hPK38). | |||||
|
PZRN3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.007905 (rank : 115) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UPQ7, Q8N2N7, Q96CC2, Q9NSQ2 | Gene names | PDZRN3, KIAA1095, LNX3, SEMCAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing RING finger protein 3 (Ligand of Numb-protein X 3) (Semaphorin cytoplasmic domain-associated protein 3) (SEMACAP3 protein). | |||||
|
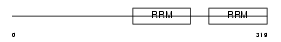
TRIPC_HUMAN
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.009478 (rank : 113) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14669, Q15644 | Gene names | TRIP12, KIAA0045 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 12 (TRIP12). | |||||
|
VASP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.010996 (rank : 110) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P50552, Q6PIZ1, Q93035 | Gene names | VASP | |||
|
Domain Architecture |

|
|||||
| Description | Vasodilator-stimulated phosphoprotein (VASP). | |||||
|
WDFY3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.015715 (rank : 106) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6VNB8, Q8C8H7 | Gene names | Wdfy3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat and FYVE domain-containing protein 3 (Beach domain, WD repeat and FYVE domain-containing protein 1) (BWF1). | |||||
|
CDSN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.020317 (rank : 99) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TPC1 | Gene names | Cdsn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Corneodesmosin precursor. | |||||
|
CH60_HUMAN
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.016612 (rank : 103) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P10809 | Gene names | HSPD1, HSP60 | |||
|
Domain Architecture |

|
|||||
| Description | 60 kDa heat shock protein, mitochondrial precursor (Hsp60) (60 kDa chaperonin) (CPN60) (Heat shock protein 60) (HSP-60) (Mitochondrial matrix protein P1) (P60 lymphocyte protein) (HuCHA60). | |||||
|
K22E_HUMAN
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.027972 (rank : 92) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 515 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P35908 | Gene names | KRT2A, KRT2E | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 2 epidermal (Cytokeratin-2e) (K2e) (CK 2e). | |||||
|
LIPA2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.016559 (rank : 104) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1282 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BSS9, Q8BN73 | Gene names | Ppfia2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
NSBP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.053042 (rank : 60) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
OSBL8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.025423 (rank : 96) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BZF1, Q8WXP8, Q9P277 | Gene names | OSBPL8, KIAA1451, ORP8, OSBP10 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 8 (OSBP-related protein 8) (ORP-8). | |||||
|
PEG3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.009208 (rank : 114) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1101 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q3URU2, O54978, Q3TQ69, Q5EBP7, Q61138, Q6GQS0, Q80U47, Q8R5N0, Q9QX53 | Gene names | Peg3, Kiaa0287, Pw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paternally expressed gene 3 protein (ASF-1). | |||||
|
SSXT_MOUSE
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.012781 (rank : 108) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q62280 | Gene names | Ss18, Ssxt, Syt | |||
|
Domain Architecture |
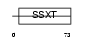
|
|||||
| Description | SSXT protein (SYT protein) (Synovial sarcoma-associated Ss18-alpha). | |||||
|
CGRE1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.068968 (rank : 31) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99674 | Gene names | CGREF1, CGR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
CIRBP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.065965 (rank : 32) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q14011 | Gene names | CIRBP, A18HNRNP, CIRP | |||
|
Domain Architecture |
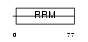
|
|||||
| Description | Cold-inducible RNA-binding protein (Glycine-rich RNA-binding protein CIRP) (A18 hnRNP). | |||||
|
DAZP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.063857 (rank : 38) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96EP5, Q96MJ3, Q9NRR9 | Gene names | DAZAP1 | |||
|
Domain Architecture |
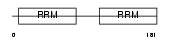
|
|||||
| Description | DAZ-associated protein 1 (Deleted in azoospermia-associated protein 1). | |||||
|
DAZP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.063495 (rank : 39) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JII5 | Gene names | Dazap1 | |||
|
Domain Architecture |
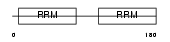
|
|||||
| Description | DAZ-associated protein 1 (Deleted in azoospermia-associated protein 1). | |||||
|
ERIC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.055977 (rank : 51) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
HNRPD_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.065503 (rank : 34) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14103, Q01858, Q14100, Q14101, Q14102 | Gene names | HNRPD, AUF1 | |||
|
Domain Architecture |
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein D0 (hnRNP D0) (AU-rich element RNA-binding protein 1). | |||||
|
HNRPD_MOUSE
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.065353 (rank : 35) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q60668, Q60667, Q80ZJ0, Q91X94 | Gene names | Hnrpd, Auf1 | |||
|
Domain Architecture |
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein D0 (hnRNP D0) (AU-rich element RNA-binding protein 1). | |||||
|
MSI1H_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.059761 (rank : 44) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O43347, Q96PU0, Q96PU1, Q96PU2, Q96PU3 | Gene names | MSI1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein Musashi homolog 1 (Musashi-1). | |||||
|
MSI1H_MOUSE
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.059753 (rank : 45) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61474, Q8BNC7 | Gene names | Msi1, Msi1h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein Musashi homolog 1 (Musashi-1). | |||||
|
MSI2H_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.061071 (rank : 41) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96DH6, Q7Z6M7, Q8N9T4 | Gene names | MSI2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein Musashi homolog 2 (Musashi-2). | |||||
|
MSI2H_MOUSE
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.060377 (rank : 43) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q920Q6, Q8BQ90, Q920Q7 | Gene names | Msi2, Msi2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein Musashi homolog 2 (Musashi-2). | |||||
|
NUCL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.058599 (rank : 46) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P19338, Q53SK1, Q8NB06 | Gene names | NCL | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
NUCL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.054101 (rank : 54) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P09405, Q548M9, Q61991, Q99K50 | Gene names | Ncl, Nuc | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
RBM23_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.050015 (rank : 65) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q86U06, Q8ND16, Q8TB88, Q8WY40, Q9BUJ1, Q9NVV7 | Gene names | RBM23, RNPC4 | |||
|
Domain Architecture |
|
|||||
| Description | Probable RNA-binding protein 23 (RNA-binding motif protein 23) (RNA- binding region-containing protein 4) (Splicing factor SF2). | |||||
|
RBM3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.065551 (rank : 33) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P98179 | Gene names | RBM3, RNPL | |||
|
Domain Architecture |
|
|||||
| Description | Putative RNA-binding protein 3 (RNA-binding motif protein 3) (RNPL). | |||||
|
RBM3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.065028 (rank : 36) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O89086 | Gene names | Rbm3 | |||
|
Domain Architecture |
|
|||||
| Description | Putative RNA-binding protein 3 (RNA-binding motif protein 3). | |||||
|
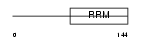
RBM8A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.053235 (rank : 56) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y5S9, Q6FHD1, Q9GZX8, Q9NZI4 | Gene names | RBM8A, RBM8 | |||
|
Domain Architecture |
|
|||||
| Description | RNA-binding protein 8A (RNA-binding motif protein 8A) (Ribonucleoprotein RBM8A) (RNA-binding protein Y14) (Binder of OVCA1- 1) (BOV-1). | |||||
|
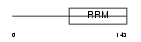
RBM8A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.053235 (rank : 57) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9CWZ3, Q9D6U5 | Gene names | Rbm8a, Rbm8 | |||
|
Domain Architecture |
|
|||||
| Description | RNA-binding protein 8A (RNA-binding motif protein 8A) (Ribonucleoprotein RBM8A). | |||||
|
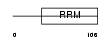
RBMX2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.050155 (rank : 64) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y388, Q5JY82, Q9Y3I8 | Gene names | RBMX2 | |||
|
Domain Architecture |
|
|||||
| Description | RNA-binding motif protein, X-linked 2. | |||||
|
RBMX2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.053457 (rank : 55) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8R0F5, Q3TI42 | Gene names | Rbmx2 | |||
|
Domain Architecture |
|
|||||
| Description | RNA-binding motif protein, X-linked 2. | |||||
|
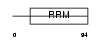
RNPC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.056488 (rank : 49) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9H0Z9, Q15350, Q15351, Q9BYK3, Q9BYK4 | Gene names | RNPC1, SEB4 | |||
|
Domain Architecture |
|
|||||
| Description | RNA-binding region-containing protein 1 (HSRNASEB) (ssDNA-binding protein SEB4) (CLL-associated antigen KW-5). | |||||
|
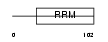
RNPC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.055946 (rank : 52) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q62176, Q922Z2 | Gene names | Rnpc1, Seb4, Seb4l | |||
|
Domain Architecture |
|
|||||
| Description | RNA-binding region-containing protein 1 (ssDNA-binding protein SEB4). | |||||
|
RNPC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.053165 (rank : 59) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14498, Q14499 | Gene names | RNPC2, HCC1 | |||
|
Domain Architecture |
|
|||||
| Description | RNA-binding region-containing protein 2 (Hepatocellular carcinoma protein 1) (Splicing factor HCC1). | |||||
|
RNPC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.053209 (rank : 58) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8VH51, Q99KV0 | Gene names | Rnpc2, Caper | |||
|
Domain Architecture |
|
|||||
| Description | RNA-binding region-containing protein 2 (Coactivator of activating protein 1 and estrogen receptors) (Coactivator of AP-1 and ERs) (Transcription coactivator CAPER). | |||||
|
ROA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.084668 (rank : 28) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P49312, P97312, Q3V269 | Gene names | Hnrpa1, Fli-2, Tis | |||
|
Domain Architecture |
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein A1 (Helix-destabilizing protein) (Single-strand-binding protein) (hnRNP core protein A1) (HDP- 1) (Topoisomerase-inhibitor suppressed). | |||||
|
ROAA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.063104 (rank : 40) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99729, Q04150, Q8N7U3, Q9BQ99 | Gene names | HNRPAB, ABBP1 | |||
|
Domain Architecture |
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein A/B (hnRNP A/B) (APOBEC-1- binding protein 1) (ABBP-1). | |||||
|
ROAA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.060937 (rank : 42) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99020, Q8BPE0 | Gene names | Hnrpab, Cbf-a, Cgbfa | |||
|
Domain Architecture |
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein A/B (hnRNP A/B) (CArG-binding factor-A) (CBF-A). | |||||
|
RPTN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.057109 (rank : 48) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6XPR3 | Gene names | RPTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Repetin. | |||||
|
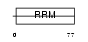
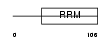
RRBP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.051159 (rank : 62) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
ZN326_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q5BKZ1, Q5VW93, Q5VW94, Q5VW96, Q5VW97, Q6NSA2, Q7Z638, Q7Z6C2 | Gene names | ZNF326 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 326. | |||||
|
ZN326_MOUSE
|
||||||
| NC score | 0.972274 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O88291, Q8BSJ5, Q8K1X9, Q9CYG9 | Gene names | Znf326, Zan75, Zfp326 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 326 (Zinc finger protein-associated with nuclear matrix of 75 kDa). | |||||
|
AKP8L_HUMAN
|
||||||
| NC score | 0.743217 (rank : 3) | θ value | 1.08979e-29 (rank : 4) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9ULX6, O94792, Q96J58, Q9NRQ0, Q9UGM0 | Gene names | AKAP8L, NAKAP95 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 8-like (AKAP8-like protein) (Neighbor of A- kinase-anchoring protein 95) (Neighbor of AKAP95) (Homologous to AKAP95 protein) (HA95) (Helicase A-binding protein 95) (HAP95). | |||||
|
AKP8L_MOUSE
|
||||||
| NC score | 0.741762 (rank : 4) | θ value | 4.42448e-31 (rank : 3) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9R0L7 | Gene names | Akap8l, Nakap95 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 8-like (AKAP8-like protein) (Neighbor of A- kinase-anchoring protein 95) (Neighbor of AKAP95). | |||||
|
AKAP8_MOUSE
|
||||||
| NC score | 0.730776 (rank : 5) | θ value | 1.33217e-27 (rank : 6) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9DBR0, Q9R0L8 | Gene names | Akap8, Akap95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 8 (A-kinase anchor protein 95 kDa) (AKAP 95). | |||||
|
AKAP8_HUMAN
|
||||||
| NC score | 0.716624 (rank : 6) | θ value | 3.17079e-29 (rank : 5) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O43823 | Gene names | AKAP8, AKAP95 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 8 (A-kinase anchor protein 95 kDa) (AKAP 95). | |||||
|
FUS_MOUSE
|
||||||
| NC score | 0.182069 (rank : 7) | θ value | 0.00228821 (rank : 8) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P56959 | Gene names | Fus | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein FUS (Pigpen protein). | |||||
|
RBP56_HUMAN
|
||||||
| NC score | 0.174438 (rank : 8) | θ value | 0.0113563 (rank : 12) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q92804, Q92751 | Gene names | TAF15, RBP56, TAF2N | |||
|
Domain Architecture |

|
|||||
| Description | TATA-binding protein-associated factor 2N (RNA-binding protein 56) (TAFII68) (TAF(II)68). | |||||
|
FUS_HUMAN
|
||||||
| NC score | 0.171418 (rank : 9) | θ value | 0.00665767 (rank : 11) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P35637 | Gene names | FUS, TLS | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein FUS (Oncogene FUS) (Oncogene TLS) (Translocated in liposarcoma protein) (POMp75) (75 kDa DNA-pairing protein). | |||||
|
HNRPG_MOUSE
|
||||||
| NC score | 0.132718 (rank : 10) | θ value | 0.00298849 (rank : 10) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O35479 | Gene names | Rbmx, Hnrnpg, Hnrpg, Rbmxp1, Rbmxrt | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein G (hnRNP G) (RNA-binding motif protein, X chromosome). | |||||
|
ROA1_HUMAN
|
||||||
| NC score | 0.130998 (rank : 11) | θ value | 0.000121331 (rank : 7) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P09651, Q6PJZ7 | Gene names | HNRPA1 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein A1 (Helix-destabilizing protein) (Single-strand RNA-binding protein) (hnRNP core protein A1). | |||||
|
HNRPG_HUMAN
|
||||||
| NC score | 0.127286 (rank : 12) | θ value | 0.00298849 (rank : 9) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P38159, Q5JQ67, Q969R3 | Gene names | RBMX, HNRPG, RBMXP1 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein G (hnRNP G) (RNA-binding motif protein, X chromosome) (Glycoprotein p43). | |||||
|
CGBP1_MOUSE
|
||||||
| NC score | 0.122198 (rank : 13) | θ value | 0.21417 (rank : 28) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BHG9, Q8K2K1 | Gene names | Cggbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CGG triplet repeat-binding protein 1 (CGG-binding protein 1) (p20- CGGBP DNA-binding protein) (20 kDa CGG-binding protein). | |||||
|
ROA3_MOUSE
|
||||||
| NC score | 0.121662 (rank : 14) | θ value | 0.0252991 (rank : 14) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BG05, Q8BHF8 | Gene names | Hnrpa3, Hnrnpa3 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein A3 (hnRNP A3). | |||||
|
RBY1A_HUMAN
|
||||||
| NC score | 0.119736 (rank : 15) | θ value | 0.0736092 (rank : 23) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q15414, Q15377, Q8NHR0 | Gene names | RBMY1A1, RBM1, YRRM1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding motif protein, Y chromosome, family 1 member A1 (RNA- binding motif protein 1). | |||||
|
ROA3_HUMAN
|
||||||
| NC score | 0.117050 (rank : 16) | θ value | 0.0113563 (rank : 13) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P51991, Q6URK5 | Gene names | HNRPA3 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein A3 (hnRNP A3). | |||||
|
ROA0_HUMAN
|
||||||
| NC score | 0.113678 (rank : 17) | θ value | 0.21417 (rank : 29) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13151 | Gene names | HNRPA0 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein A0 (hnRNP A0). | |||||
|
ILF3_HUMAN
|
||||||
| NC score | 0.112675 (rank : 18) | θ value | 0.0736092 (rank : 22) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q12906, O43409, Q6P1X1, Q86XY7, Q99544, Q99545, Q9BZH4, Q9BZH5, Q9NQ95, Q9NQ96, Q9NQ97, Q9NQ98, Q9NQ99, Q9NQA0, Q9NQA1, Q9NQA2, Q9NRN2, Q9NRN3, Q9NRN4, Q9UMZ9, Q9UN00, Q9UN84, Q9UNA2 | Gene names | ILF3, DRBF, MPHOSPH4, NF90 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin enhancer-binding factor 3 (Nuclear factor of activated T- cells 90 kDa) (NF-AT-90) (Double-stranded RNA-binding protein 76) (DRBP76) (Translational control protein 80) (TCP80) (Nuclear factor associated with dsRNA) (NFAR) (M-phase phosphoprotein 4) (MPP4). | |||||
|
ROA2_HUMAN
|
||||||
| NC score | 0.109859 (rank : 19) | θ value | 0.279714 (rank : 31) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P22626, P22627 | Gene names | HNRPA2B1 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoproteins A2/B1 (hnRNP A2 / hnRNP B1). | |||||
|
LORI_MOUSE
|
||||||
| NC score | 0.109666 (rank : 20) | θ value | 0.365318 (rank : 35) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P18165, Q8R0I9 | Gene names | Lor | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Loricrin. | |||||
|
ILF3_MOUSE
|
||||||
| NC score | 0.109355 (rank : 21) | θ value | 0.0330416 (rank : 16) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z1X4, Q80VD5, Q812A1, Q8BP80, Q8C2H8, Q8K588 | Gene names | Ilf3 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin enhancer-binding factor 3. | |||||
|
ROA2_MOUSE
|
||||||
| NC score | 0.104141 (rank : 22) | θ value | 1.81305 (rank : 50) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O88569 | Gene names | Hnrpa2b1 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoproteins A2/B1 (hnRNP A2 / hnRNP B1). | |||||
|
FILA_HUMAN
|
||||||
| NC score | 0.094473 (rank : 23) | θ value | 0.0330416 (rank : 15) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
CGBP1_HUMAN
|
||||||
| NC score | 0.093695 (rank : 24) | θ value | 1.81305 (rank : 46) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UFW8, O15183 | Gene names | CGGBP1, CGGBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CGG triplet repeat-binding protein 1 (CGG-binding protein 1) (p20- CGGBP DNA-binding protein) (20 kDa CGG-binding protein). | |||||
|
EWS_MOUSE
|
||||||
| NC score | 0.090026 (rank : 25) | θ value | 2.36792 (rank : 54) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q61545 | Gene names | Ewsr1, Ews, Ewsh | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein EWS. | |||||
|
CIRBP_MOUSE
|
||||||
| NC score | 0.089409 (rank : 26) | θ value | 0.365318 (rank : 32) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P60824, O09069, O09148, Q3V1A6, Q61413 | Gene names | Cirbp, Cirp | |||
|
Domain Architecture |

|
|||||
| Description | Cold-inducible RNA-binding protein (Glycine-rich RNA-binding protein CIRP) (A18 hnRNP). | |||||
|
EWS_HUMAN
|
||||||
| NC score | 0.087236 (rank : 27) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q01844, Q92635 | Gene names | EWSR1, EWS | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein EWS (EWS oncogene) (Ewing sarcoma breakpoint region 1 protein). | |||||
|
ROA1_MOUSE
|
||||||
| NC score | 0.084668 (rank : 28) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P49312, P97312, Q3V269 | Gene names | Hnrpa1, Fli-2, Tis | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein A1 (Helix-destabilizing protein) (Single-strand-binding protein) (hnRNP core protein A1) (HDP- 1) (Topoisomerase-inhibitor suppressed). | |||||
|
HNRPR_HUMAN
|
||||||
| NC score | 0.080672 (rank : 29) | θ value | 0.0431538 (rank : 18) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O43390 | Gene names | HNRPR | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein R (hnRNP R). | |||||
|
CGRE1_MOUSE
|
||||||
| NC score | 0.069815 (rank : 30) | θ value | 0.813845 (rank : 41) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8R1U2, Q9D1K1 | Gene names | Cgref1, Cgr11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
CGRE1_HUMAN
|
||||||
| NC score | 0.068968 (rank : 31) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99674 | Gene names | CGREF1, CGR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
CIRBP_HUMAN
|
||||||
| NC score | 0.065965 (rank : 32) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q14011 | Gene names | CIRBP, A18HNRNP, CIRP | |||
|
Domain Architecture |

|
|||||
| Description | Cold-inducible RNA-binding protein (Glycine-rich RNA-binding protein CIRP) (A18 hnRNP). | |||||
|
RBM3_HUMAN
|
||||||
| NC score | 0.065551 (rank : 33) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P98179 | Gene names | RBM3, RNPL | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein 3 (RNA-binding motif protein 3) (RNPL). | |||||
|
HNRPD_HUMAN
|
||||||
| NC score | 0.065503 (rank : 34) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14103, Q01858, Q14100, Q14101, Q14102 | Gene names | HNRPD, AUF1 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein D0 (hnRNP D0) (AU-rich element RNA-binding protein 1). | |||||
|
HNRPD_MOUSE
|
||||||
| NC score | 0.065353 (rank : 35) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q60668, Q60667, Q80ZJ0, Q91X94 | Gene names | Hnrpd, Auf1 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein D0 (hnRNP D0) (AU-rich element RNA-binding protein 1). | |||||
|
RBM3_MOUSE
|
||||||
| NC score | 0.065028 (rank : 36) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O89086 | Gene names | Rbm3 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein 3 (RNA-binding motif protein 3). | |||||
|
TCAL5_HUMAN
|
||||||
| NC score | 0.064058 (rank : 37) | θ value | 0.0563607 (rank : 21) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
DAZP1_HUMAN
|
||||||
| NC score | 0.063857 (rank : 38) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96EP5, Q96MJ3, Q9NRR9 | Gene names | DAZAP1 | |||
|
Domain Architecture |

|
|||||
| Description | DAZ-associated protein 1 (Deleted in azoospermia-associated protein 1). | |||||
|
DAZP1_MOUSE
|
||||||
| NC score | 0.063495 (rank : 39) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JII5 | Gene names | Dazap1 | |||
|
Domain Architecture |

|
|||||
| Description | DAZ-associated protein 1 (Deleted in azoospermia-associated protein 1). | |||||
|
ROAA_HUMAN
|
||||||
| NC score | 0.063104 (rank : 40) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99729, Q04150, Q8N7U3, Q9BQ99 | Gene names | HNRPAB, ABBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein A/B (hnRNP A/B) (APOBEC-1- binding protein 1) (ABBP-1). | |||||
|
MSI2H_HUMAN
|
||||||
| NC score | 0.061071 (rank : 41) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96DH6, Q7Z6M7, Q8N9T4 | Gene names | MSI2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein Musashi homolog 2 (Musashi-2). | |||||
|
ROAA_MOUSE
|
||||||
| NC score | 0.060937 (rank : 42) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99020, Q8BPE0 | Gene names | Hnrpab, Cbf-a, Cgbfa | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein A/B (hnRNP A/B) (CArG-binding factor-A) (CBF-A). | |||||
|
MSI2H_MOUSE
|
||||||
| NC score | 0.060377 (rank : 43) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q920Q6, Q8BQ90, Q920Q7 | Gene names | Msi2, Msi2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein Musashi homolog 2 (Musashi-2). | |||||
|
MSI1H_HUMAN
|
||||||
| NC score | 0.059761 (rank : 44) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O43347, Q96PU0, Q96PU1, Q96PU2, Q96PU3 | Gene names | MSI1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein Musashi homolog 1 (Musashi-1). | |||||
|
MSI1H_MOUSE
|
||||||
| NC score | 0.059753 (rank : 45) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61474, Q8BNC7 | Gene names | Msi1, Msi1h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein Musashi homolog 1 (Musashi-1). | |||||
|
NUCL_HUMAN
|
||||||
| NC score | 0.058599 (rank : 46) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P19338, Q53SK1, Q8NB06 | Gene names | NCL | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
FA98A_HUMAN
|
||||||
| NC score | 0.058120 (rank : 47) | θ value | 1.38821 (rank : 43) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NCA5 | Gene names | FAM98A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM98A. | |||||
|
RPTN_HUMAN
|
||||||
| NC score | 0.057109 (rank : 48) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6XPR3 | Gene names | RPTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Repetin. | |||||
|
RNPC1_HUMAN
|
||||||
| NC score | 0.056488 (rank : 49) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9H0Z9, Q15350, Q15351, Q9BYK3, Q9BYK4 | Gene names | RNPC1, SEB4 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding region-containing protein 1 (HSRNASEB) (ssDNA-binding protein SEB4) (CLL-associated antigen KW-5). | |||||
|
HORN_HUMAN
|
||||||
| NC score | 0.056314 (rank : 50) | θ value | 1.81305 (rank : 47) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86YZ3 | Gene names | HRNR, S100A18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hornerin. | |||||
|
ERIC1_HUMAN
|
||||||
| NC score | 0.055977 (rank : 51) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
RNPC1_MOUSE
|
||||||
| NC score | 0.055946 (rank : 52) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q62176, Q922Z2 | Gene names | Rnpc1, Seb4, Seb4l | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding region-containing protein 1 (ssDNA-binding protein SEB4). | |||||
|
NCOAT_HUMAN
|
||||||
| NC score | 0.055802 (rank : 53) | θ value | 0.47712 (rank : 36) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60502, O75166, Q86WV0, Q8IV98, Q9BVA5, Q9HAR0 | Gene names | MGEA5, HEXC, KIAA0679, MEA5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional protein NCOAT (Nuclear cytoplasmic O-GlcNAcase and acetyltransferase) (Meningioma-expressed antigen 5) [Includes: Beta- hexosaminidase (EC 3.2.1.52) (N-acetyl-beta-glucosaminidase) (Beta-N- acetylhexosaminidase) (Hexosaminidase C) (N-acetyl-beta-D- glucosaminidase) (O-GlcNAcase); Histone acetyltransferase (EC 2.3.1.48) (HAT)]. | |||||
|
NUCL_MOUSE
|
||||||
| NC score | 0.054101 (rank : 54) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P09405, Q548M9, Q61991, Q99K50 | Gene names | Ncl, Nuc | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
RBMX2_MOUSE
|
||||||
| NC score | 0.053457 (rank : 55) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8R0F5, Q3TI42 | Gene names | Rbmx2 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding motif protein, X-linked 2. | |||||
|
RBM8A_HUMAN
|
||||||
| NC score | 0.053235 (rank : 56) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y5S9, Q6FHD1, Q9GZX8, Q9NZI4 | Gene names | RBM8A, RBM8 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 8A (RNA-binding motif protein 8A) (Ribonucleoprotein RBM8A) (RNA-binding protein Y14) (Binder of OVCA1- 1) (BOV-1). | |||||
|
RBM8A_MOUSE
|
||||||
| NC score | 0.053235 (rank : 57) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9CWZ3, Q9D6U5 | Gene names | Rbm8a, Rbm8 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 8A (RNA-binding motif protein 8A) (Ribonucleoprotein RBM8A). | |||||
|
RNPC2_MOUSE
|
||||||
| NC score | 0.053209 (rank : 58) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8VH51, Q99KV0 | Gene names | Rnpc2, Caper | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding region-containing protein 2 (Coactivator of activating protein 1 and estrogen receptors) (Coactivator of AP-1 and ERs) (Transcription coactivator CAPER). | |||||
|
RNPC2_HUMAN
|
||||||
| NC score | 0.053165 (rank : 59) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14498, Q14499 | Gene names | RNPC2, HCC1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding region-containing protein 2 (Hepatocellular carcinoma protein 1) (Splicing factor HCC1). | |||||
|
NSBP1_MOUSE
|
||||||
| NC score | 0.053042 (rank : 60) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.051948 (rank : 61) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
RRBP1_MOUSE
|
||||||
| NC score | 0.051159 (rank : 62) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
NCOAT_MOUSE
|
||||||
| NC score | 0.050191 (rank : 63) | θ value | 1.81305 (rank : 49) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EQQ9, Q3ULY7, Q6ZQ71, Q8BK05, Q8BTT2, Q8CFX2, Q9CSJ4, Q9CUR7 | Gene names | Mgea5, Hexc, Kiaa0679 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional protein NCOAT (Nuclear cytoplasmic O-GlcNAcase and acetyltransferase) (Meningioma-expressed antigen 5) [Includes: Beta- hexosaminidase (EC 3.2.1.52) (N-acetyl-beta-glucosaminidase) (Beta-N- acetylhexosaminidase) (Hexosaminidase C) (N-acetyl-beta-D- glucosaminidase) (O-GlcNAcase); Histone acetyltransferase (EC 2.3.1.48) (HAT)]. | |||||
|
RBMX2_HUMAN
|
||||||
| NC score | 0.050155 (rank : 64) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y388, Q5JY82, Q9Y3I8 | Gene names | RBMX2 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding motif protein, X-linked 2. | |||||
|
RBM23_HUMAN
|
||||||
| NC score | 0.050015 (rank : 65) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q86U06, Q8ND16, Q8TB88, Q8WY40, Q9BUJ1, Q9NVV7 | Gene names | RBM23, RNPC4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable RNA-binding protein 23 (RNA-binding motif protein 23) (RNA- binding region-containing protein 4) (Splicing factor SF2). | |||||
|
MDN1_HUMAN
|
||||||
| NC score | 0.044756 (rank : 66) | θ value | 1.38821 (rank : 45) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
K1C9_HUMAN
|
||||||
| NC score | 0.044228 (rank : 67) | θ value | 0.125558 (rank : 25) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 526 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P35527, O00109, Q14665 | Gene names | KRT9 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 9 (Cytokeratin-9) (CK-9) (Keratin-9) (K9). | |||||
|
K1C10_MOUSE
|
||||||
| NC score | 0.044187 (rank : 68) | θ value | 0.0330416 (rank : 17) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 466 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P02535, P08731, Q8BUX3, Q8BV09, Q8BVU3, Q9CXH6, Q9JKB4 | Gene names | Krt10, Krt1-10 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 10 (Cytokeratin-10) (CK-10) (Keratin-10) (K10) (56 kDa cytokeratin) (Keratin, type I cytoskeletal 59 kDa). | |||||
|
FXR2_HUMAN
|
||||||
| NC score | 0.042985 (rank : 69) | θ value | 0.279714 (rank : 30) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P51116, Q8WUM2 | Gene names | FXR2 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 2. | |||||
|
HORN_MOUSE
|
||||||
| NC score | 0.040763 (rank : 70) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8VHD8 | Gene names | Hrnr | |||
|
Domain Architecture |

|
|||||
| Description | Hornerin. | |||||
|
FLIP1_MOUSE
|
||||||
| NC score | 0.039310 (rank : 71) | θ value | 0.365318 (rank : 33) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1176 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9CS72 | Gene names | Filip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
PDCL2_MOUSE
|
||||||
| NC score | 0.038145 (rank : 72) | θ value | 3.0926 (rank : 61) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q78Y63 | Gene names | Pdcl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosducin-like protein 2. | |||||
|
K2C1_HUMAN
|
||||||
| NC score | 0.037616 (rank : 73) | θ value | 0.125558 (rank : 26) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P04264, Q14720, Q6GSJ0, Q9H298 | Gene names | KRT1, KRTA | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 1 (Cytokeratin-1) (CK-1) (Keratin-1) (K1) (67 kDa cytokeratin) (Hair alpha protein). | |||||
|
FLIP1_HUMAN
|
||||||
| NC score | 0.037424 (rank : 74) | θ value | 0.125558 (rank : 24) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
K1C10_HUMAN
|
||||||
| NC score | 0.037319 (rank : 75) | θ value | 1.38821 (rank : 44) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 526 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P13645, Q14664, Q8N175 | Gene names | KRT10 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 10 (Cytokeratin-10) (CK-10) (Keratin-10) (K10). | |||||
|
MY18A_HUMAN
|
||||||
| NC score | 0.036846 (rank : 76) | θ value | 0.0563607 (rank : 20) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
K2C1_MOUSE
|
||||||
| NC score | 0.036554 (rank : 77) | θ value | 0.365318 (rank : 34) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P04104 | Gene names | Krt1, Krt2-1 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 1 (Cytokeratin-1) (CK-1) (Keratin-1) (K1) (67 kDa cytokeratin). | |||||
|
FYB_HUMAN
|
||||||
| NC score | 0.036504 (rank : 78) | θ value | 0.813845 (rank : 42) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O15117, O00359 | Gene names | FYB, SLAP130 | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
ARI5B_HUMAN
|
||||||
| NC score | 0.035880 (rank : 79) | θ value | 2.36792 (rank : 51) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14865, Q7Z3M4, Q8N421, Q9H786 | Gene names | ARID5B, DESRT, MRF2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (Mrf1-like) (Modulator recognition factor 2) (MRF-2). | |||||
|
UACA_HUMAN
|
||||||
| NC score | 0.035754 (rank : 80) | θ value | 0.125558 (rank : 27) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1622 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9BZF9, Q8N3B8, Q9HCL1, Q9NWC6 | Gene names | UACA, KIAA1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein. | |||||
|
UACA_MOUSE
|
||||||
| NC score | 0.033793 (rank : 81) | θ value | 0.0431538 (rank : 19) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8CGB3, Q69ZG3, Q7TN77, Q8BJC8 | Gene names | Uaca, Kiaa1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein (Nucling) (Nuclear membrane-binding protein). | |||||
|
PDCL2_HUMAN
|
||||||
| NC score | 0.032576 (rank : 82) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N4E4 | Gene names | PDCL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosducin-like protein 2. | |||||
|
HM20A_MOUSE
|
||||||
| NC score | 0.032009 (rank : 83) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9DC33, Q3LSF9, Q6NV87, Q8BSK1, Q8C3C1, Q8CAA0, Q9CYG2 | Gene names | Hmg20a, Ibraf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein 20A (HMG box-containing protein 20A) (iBRAF) (Inhibitor of BRAF35) (HMG domain protein HMGX1). | |||||
|
HM20A_HUMAN
|
||||||
| NC score | 0.032005 (rank : 84) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NP66, Q53G31, Q9NSF6 | Gene names | HMG20A, HMGX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein 20A (HMG box-containing protein 20A) (HMG domain-containing protein HMGX1). | |||||
|
RNH1_HUMAN
|
||||||
| NC score | 0.030863 (rank : 85) | θ value | 2.36792 (rank : 58) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60930, O60523, O60857, Q57Z93, Q5U0C1, Q6FHD4 | Gene names | RNASEH1, RNH1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribonuclease H1 (EC 3.1.26.4) (RNase H1) (Ribonuclease H type II). | |||||
|
MY18A_MOUSE
|
||||||
| NC score | 0.030469 (rank : 86) | θ value | 0.62314 (rank : 40) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
SYCP1_MOUSE
|
||||||
| NC score | 0.029969 (rank : 87) | θ value | 0.47712 (rank : 37) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
SPC24_HUMAN
|
||||||
| NC score | 0.029869 (rank : 88) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NBT2 | Gene names | SPBC24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Spc24 (hSpc24). | |||||
|
NKTR_MOUSE
|
||||||
| NC score | 0.029672 (rank : 89) | θ value | 2.36792 (rank : 56) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P30415 | Gene names | Nktr | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
K1C16_HUMAN
|
||||||
| NC score | 0.028985 (rank : 90) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P08779, P30654, Q16402, Q9UBG8 | Gene names | KRT16, KRT16A | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 16 (Cytokeratin-16) (CK-16) (Keratin-16) (K16). | |||||
|
BRD8_MOUSE
|
||||||
| NC score | 0.028840 (rank : 91) | θ value | 2.36792 (rank : 52) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
K22E_HUMAN
|
||||||
| NC score | 0.027972 (rank : 92) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 515 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P35908 | Gene names | KRT2A, KRT2E | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 2 epidermal (Cytokeratin-2e) (K2e) (CK 2e). | |||||
|
YTDC1_HUMAN
|
||||||
| NC score | 0.027905 (rank : 93) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
ARNT_MOUSE
|
||||||
| NC score | 0.025901 (rank : 94) | θ value | 0.62314 (rank : 38) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
PPIG_HUMAN
|
||||||
| NC score | 0.025672 (rank : 95) | θ value | 2.36792 (rank : 57) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
OSBL8_HUMAN
|
||||||
| NC score | 0.025423 (rank : 96) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BZF1, Q8WXP8, Q9P277 | Gene names | OSBPL8, KIAA1451, ORP8, OSBP10 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 8 (OSBP-related protein 8) (ORP-8). | |||||
|
DHX9_MOUSE
|
||||||
| NC score | 0.025193 (rank : 97) | θ value | 0.62314 (rank : 39) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O70133, O35931, O54703, Q5FWY1, Q6R5F7, Q9CSA2 | Gene names | Dhx9, Ddx9 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase A (EC 3.6.1.-) (Nuclear DNA helicase II) (NDH II) (DEAH box protein 9) (mHEL-5). | |||||
|
S20A1_HUMAN
|
||||||
| NC score | 0.021075 (rank : 98) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8WUM9, Q08344, Q6DHX8, Q9UQ82 | Gene names | SLC20A1, GLVR1, PIT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium-dependent phosphate transporter 1 (Solute carrier family 20 member 1) (Phosphate transporter 1) (PiT-1) (Gibbon ape leukemia virus receptor 1) (GLVR-1) (Leukemia virus receptor 1 homolog). | |||||
|
CDSN_MOUSE
|
||||||
| NC score | 0.020317 (rank : 99) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TPC1 | Gene names | Cdsn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Corneodesmosin precursor. | |||||
|
CH60_MOUSE
|
||||||
| NC score | 0.020151 (rank : 100) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P63038, P19226, P19227, P97602 | Gene names | Hspd1, Hsp60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 60 kDa heat shock protein, mitochondrial precursor (Hsp60) (60 kDa chaperonin) (CPN60) (Heat shock protein 60) (HSP-60) (Mitochondrial matrix protein P1) (HSP-65). | |||||
|
ZN409_HUMAN
|
||||||
| NC score | 0.018225 (rank : 101) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UPU6 | Gene names | ZNF409, KIAA1056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 409. | |||||
|
WDFY3_HUMAN
|
||||||
| NC score | 0.017934 (rank : 102) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IZQ1, Q4W5K5, Q6P0Q5, Q8N1T2, Q96BS7, Q96N85, Q9Y2J7 | Gene names | WDFY3, KIAA0993 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat and FYVE domain-containing protein 3 (Autophagy-linked FYVE protein) (Alfy). | |||||
|
CH60_HUMAN
|
||||||
| NC score | 0.016612 (rank : 103) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P10809 | Gene names | HSPD1, HSP60 | |||
|
Domain Architecture |

|
|||||
| Description | 60 kDa heat shock protein, mitochondrial precursor (Hsp60) (60 kDa chaperonin) (CPN60) (Heat shock protein 60) (HSP-60) (Mitochondrial matrix protein P1) (P60 lymphocyte protein) (HuCHA60). | |||||
|
LIPA2_MOUSE
|
||||||
| NC score | 0.016559 (rank : 104) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1282 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BSS9, Q8BN73 | Gene names | Ppfia2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
ATBF1_MOUSE
|
||||||
| NC score | 0.015796 (rank : 105) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
WDFY3_MOUSE
|
||||||
| NC score | 0.015715 (rank : 106) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6VNB8, Q8C8H7 | Gene names | Wdfy3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat and FYVE domain-containing protein 3 (Beach domain, WD repeat and FYVE domain-containing protein 1) (BWF1). | |||||
|
OSBL5_MOUSE
|
||||||
| NC score | 0.014661 (rank : 107) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ER64, Q8R510, Q99NF5 | Gene names | Osbpl5, Obph1, Osbp2 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 5 (OSBP-related protein 5) (ORP-5) (Oxysterol-binding protein homolog 1). | |||||
|
SSXT_MOUSE
|
||||||
| NC score | 0.012781 (rank : 108) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q62280 | Gene names | Ss18, Ssxt, Syt | |||
|
Domain Architecture |

|
|||||
| Description | SSXT protein (SYT protein) (Synovial sarcoma-associated Ss18-alpha). | |||||
|
ATBF1_HUMAN
|
||||||
| NC score | 0.011418 (rank : 109) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
VASP_HUMAN
|
||||||
| NC score | 0.010996 (rank : 110) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P50552, Q6PIZ1, Q93035 | Gene names | VASP | |||
|
Domain Architecture |

|
|||||
| Description | Vasodilator-stimulated phosphoprotein (VASP). | |||||
|
CO5A3_HUMAN
|
||||||
| NC score | 0.010033 (rank : 111) | θ value | 2.36792 (rank : 53) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P25940, Q9NZQ6 | Gene names | COL5A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(V) chain precursor. | |||||
|
CHD2_HUMAN
|
||||||
| NC score | 0.009617 (rank : 112) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O14647 | Gene names | CHD2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 2 (EC 3.6.1.-) (ATP- dependent helicase CHD2) (CHD-2). | |||||
|
TRIPC_HUMAN
|
||||||
| NC score | 0.009478 (rank : 113) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14669, Q15644 | Gene names | TRIP12, KIAA0045 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 12 (TRIP12). | |||||
|
PEG3_MOUSE
|
||||||
| NC score | 0.009208 (rank : 114) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1101 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q3URU2, O54978, Q3TQ69, Q5EBP7, Q61138, Q6GQS0, Q80U47, Q8R5N0, Q9QX53 | Gene names | Peg3, Kiaa0287, Pw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paternally expressed gene 3 protein (ASF-1). | |||||
|
PZRN3_HUMAN
|
||||||
| NC score | 0.007905 (rank : 115) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UPQ7, Q8N2N7, Q96CC2, Q9NSQ2 | Gene names | PDZRN3, KIAA1095, LNX3, SEMCAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing RING finger protein 3 (Ligand of Numb-protein X 3) (Semaphorin cytoplasmic domain-associated protein 3) (SEMACAP3 protein). | |||||
|
MELK_MOUSE
|
||||||
| NC score | 0.001776 (rank : 116) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 860 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61846, Q61804, Q6ZQH6 | Gene names | Melk, Kiaa0175, Pk38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Maternal embryonic leucine zipper kinase (EC 2.7.11.1) (Protein kinase PK38) (mPK38). | |||||
|
MELK_HUMAN
|
||||||
| NC score | 0.001026 (rank : 117) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 847 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14680, Q7L3C3 | Gene names | MELK, KIAA0175 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Maternal embryonic leucine zipper kinase (EC 2.7.11.1) (hMELK) (Protein kinase PK38) (hPK38). | |||||
|
KS6A3_MOUSE
|
||||||
| NC score | 0.000093 (rank : 118) | θ value | 2.36792 (rank : 55) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 860 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P18654, Q03140, Q8K3J8 | Gene names | Rps6ka3, Rps6ka-rs1, Rsk2 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosomal protein S6 kinase alpha-3 (EC 2.7.11.1) (S6K-alpha 3) (90 kDa ribosomal protein S6 kinase 3) (p90-RSK 3) (Ribosomal S6 kinase 2) (RSK-2) (pp90RSK2) (MAP kinase-activated protein kinase 1b) (MAPKAPK1B). | |||||
|
ZBT24_MOUSE
|
||||||
| NC score | -0.001161 (rank : 119) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 894 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80X44, Q3UIB7, Q7TPC4, Q8CJC9 | Gene names | Zbtb24, Bif1, Bsg1, Znf450 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and BTB domain-containing protein 24 (Zinc finger protein 450) (Bone morphogenetic protein-induced factor 1) (Brain-specific protein 1). | |||||