Please be patient as the page loads
|
FXR2_HUMAN
|
||||||
| SwissProt Accessions | P51116, Q8WUM2 | Gene names | FXR2 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 2. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
FXR1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.979665 (rank : 4) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P51114, Q7Z450, Q8N6R8 | Gene names | FXR1 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 1 (hFXR1P). | |||||
|
FXR1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.982393 (rank : 3) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61584, Q8VCU4, Q9R1E2, Q9R1E3, Q9R1E4, Q9R1E5, Q9WUA7, Q9WUA8, Q9WUA9 | Gene names | Fxr1, Fxr1h | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 1 (mFxr1p). | |||||
|
FXR2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P51116, Q8WUM2 | Gene names | FXR2 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 2. | |||||
|
FXR2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.990533 (rank : 2) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9WVR4, Q9WVR5 | Gene names | Fxr2, Fxr2h | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 2. | |||||
|
FMR1_MOUSE
|
||||||
| θ value | 8.93083e-141 (rank : 5) | NC score | 0.962699 (rank : 6) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P35922 | Gene names | Fmr1, Fmr-1 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation protein 1 homolog (Protein FMR-1) (FMRP) (mFmr1p). | |||||
|
FMR1_HUMAN
|
||||||
| θ value | 4.58675e-137 (rank : 6) | NC score | 0.962941 (rank : 5) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q06787, Q16578, Q99054 | Gene names | FMR1 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation 1 protein (Protein FMR-1) (FMRP). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 7) | NC score | 0.078209 (rank : 28) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 8) | NC score | 0.090137 (rank : 25) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
TDRKH_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 9) | NC score | 0.238928 (rank : 7) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y2W6, Q8N582, Q9NYV5 | Gene names | TDRKH, TDRD2 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor and KH domain-containing protein. | |||||
|
TDRKH_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 10) | NC score | 0.236366 (rank : 8) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q80VL1 | Gene names | Tdrkh | |||
|
Domain Architecture |

|
|||||
| Description | Tudor and KH domain-containing protein. | |||||
|
FUBP3_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 11) | NC score | 0.177857 (rank : 9) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96I24, Q92946, Q9BVB6 | Gene names | FUBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 3 (FUSE-binding protein 3). | |||||
|
HNRPK_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 12) | NC score | 0.171333 (rank : 10) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P61978, Q07244, Q15671, Q60577, Q922Y7, Q96J62 | Gene names | HNRPK, HNRNPK | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein K (hnRNP K) (Transformation up-regulated nuclear protein) (TUNP). | |||||
|
HNRPK_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 13) | NC score | 0.171333 (rank : 11) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P61979, Q07244, Q15671, Q60577, Q8BGQ8, Q922Y7, Q96J62 | Gene names | Hnrpk, Hnrnpk | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein K. | |||||
|
RPTN_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 14) | NC score | 0.043702 (rank : 37) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P97347 | Gene names | Rptn | |||
|
Domain Architecture |

|
|||||
| Description | Repetin. | |||||
|
RRBP1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 15) | NC score | 0.025018 (rank : 46) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
ELOA1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 16) | NC score | 0.030761 (rank : 42) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14241, Q8IXH1 | Gene names | TCEB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor B polypeptide 3 (RNA polymerase II transcription factor SIII subunit A1) (SIII p110) (Elongin-A) (EloA) (Elongin 110 kDa subunit). | |||||
|
FUBP1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 17) | NC score | 0.165770 (rank : 14) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96AE4, Q12828 | Gene names | FUBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 1 (FUSE-binding protein 1) (FBP) (DNA helicase V) (HDH V). | |||||
|
FUBP1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 18) | NC score | 0.166222 (rank : 12) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q91WJ8, Q8C0Y8 | Gene names | Fubp1, D3Ertd330e | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 1 (FUSE-binding protein 1) (FBP). | |||||
|
FUBP2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 19) | NC score | 0.166149 (rank : 13) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q92945, O00301, Q9UNT5, Q9UQH5 | Gene names | KHSRP, FUBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 2 (FUSE-binding protein 2) (KH type-splicing regulatory protein) (KSRP) (p75). | |||||
|
LAP2B_HUMAN
|
||||||
| θ value | 0.21417 (rank : 20) | NC score | 0.048554 (rank : 35) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P42167, Q14861 | Gene names | TMPO, LAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamina-associated polypeptide 2, isoforms beta/gamma (Thymopoietin, isoforms beta/gamma) (TP beta/gamma) (Thymopoietin-related peptide isoforms beta/gamma) (TPRP isoforms beta/gamma) [Contains: Thymopoietin (TP) (Splenin); Thymopentin (TP5)]. | |||||
|
PCBP3_HUMAN
|
||||||
| θ value | 0.21417 (rank : 21) | NC score | 0.118428 (rank : 18) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P57721, Q8N9K6, Q96EP6 | Gene names | PCBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 3 (Alpha-CP3). | |||||
|
PCBP3_MOUSE
|
||||||
| θ value | 0.21417 (rank : 22) | NC score | 0.118495 (rank : 17) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P57722, Q8BSB0, Q8C544 | Gene names | Pcbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 3 (Alpha-CP3). | |||||
|
REXO1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 23) | NC score | 0.033419 (rank : 41) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7TT28, Q3UMP1, Q69ZR0, Q6NSQ6, Q6PI95, Q9DA29 | Gene names | Rexo1, Kiaa1138, Tceb3bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 1 homolog (EC 3.1.-.-) (Transcription elongation factor B polypeptide 3-binding protein 1). | |||||
|
ZN326_HUMAN
|
||||||
| θ value | 0.279714 (rank : 24) | NC score | 0.042985 (rank : 38) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5BKZ1, Q5VW93, Q5VW94, Q5VW96, Q5VW97, Q6NSA2, Q7Z638, Q7Z6C2 | Gene names | ZNF326 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 326. | |||||
|
FBX34_MOUSE
|
||||||
| θ value | 0.47712 (rank : 25) | NC score | 0.027411 (rank : 45) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80XI1, Q9D290 | Gene names | Fbxo34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 34. | |||||
|
MYH6_HUMAN
|
||||||
| θ value | 0.47712 (rank : 26) | NC score | 0.013331 (rank : 62) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 27) | NC score | 0.023844 (rank : 48) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
TNK1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 28) | NC score | 0.006758 (rank : 73) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 815 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13470, O95364, Q8IYI4 | Gene names | TNK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Non-receptor tyrosine-protein kinase TNK1 (EC 2.7.10.2) (CD38 negative kinase 1). | |||||
|
MYH6_MOUSE
|
||||||
| θ value | 0.62314 (rank : 29) | NC score | 0.013008 (rank : 63) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
PCBP2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 30) | NC score | 0.104613 (rank : 19) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q15366, Q6PKG5 | Gene names | PCBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 2 (Alpha-CP2) (hnRNP-E2). | |||||
|
PCBP2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 31) | NC score | 0.104066 (rank : 20) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q61990, Q61383, Q62042 | Gene names | Pcbp2, Cbp, Hnrnpx, Hnrpx | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 2 (Alpha-CP2) (Putative heterogeneous nuclear ribonucleoprotein X) (hnRNP X) (CTBP) (CBP). | |||||
|
ZN326_MOUSE
|
||||||
| θ value | 0.62314 (rank : 32) | NC score | 0.039873 (rank : 40) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88291, Q8BSJ5, Q8K1X9, Q9CYG9 | Gene names | Znf326, Zan75, Zfp326 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 326 (Zinc finger protein-associated with nuclear matrix of 75 kDa). | |||||
|
PNPT1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 33) | NC score | 0.066529 (rank : 33) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8TCS8, Q53SU0, Q68CN1, Q7Z7D1, Q8IWX1, Q96T05, Q9BRU3, Q9BVX0 | Gene names | PNPT1, PNPASE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyribonucleotide nucleotidyltransferase 1, mitochondrial precursor (EC 2.7.7.8) (PNPase 1) (Polynucleotide phosphorylase-like protein) (PNPase old-35) (3'-5' RNA exonuclease OLD35). | |||||
|
SRBS1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 34) | NC score | 0.021804 (rank : 50) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q62417, Q80TF8, Q8BZI3, Q8K3Y2, Q921F8, Q9Z0Z8, Q9Z0Z9 | Gene names | Sorbs1, Kiaa1296, Sh3d5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorbin and SH3 domain-containing protein 1 (Ponsin) (c-Cbl-associated protein) (CAP) (SH3 domain protein 5) (SH3P12). | |||||
|
MLF2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 35) | NC score | 0.028655 (rank : 43) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99KX1 | Gene names | Mlf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myeloid leukemia factor 2 (Myelodysplasia-myeloid leukemia factor 2). | |||||
|
NOVA1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 36) | NC score | 0.148339 (rank : 15) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P51513 | Gene names | NOVA1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein Nova-1 (Neuro-oncological ventral antigen 1) (Onconeural ventral antigen 1) (Paraneoplastic Ri antigen) (Ventral neuron-specific protein 1). | |||||
|
PCBP1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 37) | NC score | 0.100720 (rank : 21) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q15365, Q13157, Q14975 | Gene names | PCBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 1 (Alpha-CP1) (hnRNP-E1) (Nucleic acid- binding protein SUB2.3). | |||||
|
PCBP1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 38) | NC score | 0.100720 (rank : 22) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P60335 | Gene names | Pcbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 1 (Alpha-CP1) (hnRNP-E1). | |||||
|
MYH7_HUMAN
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.012071 (rank : 64) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
RTN1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.018481 (rank : 54) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8K0T0, Q8K4S4 | Gene names | Rtn1, Nsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulon-1 (Neuroendocrine-specific protein). | |||||
|
CHD6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.011243 (rank : 65) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8TD26, Q5JYQ0, Q5TGZ9, Q5TH00, Q5TH01, Q8IZR2, Q8WTY0, Q9H4H6, Q9H6D4, Q9NTT7, Q9P2L1 | Gene names | CHD6, CHD5, KIAA1335, RIGB | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 6 (EC 3.6.1.-) (ATP- dependent helicase CHD6) (CHD-6) (Radiation-induced gene B protein). | |||||
|
PNPT1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.047362 (rank : 36) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K1R3, Q3UEP9, Q810U7, Q812B3, Q8R2U3, Q9DC52 | Gene names | Pnpt1, Pnpase | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyribonucleotide nucleotidyltransferase 1, mitochondrial precursor (EC 2.7.7.8) (PNPase 1) (Polynucleotide phosphorylase-like protein) (PNPase old-35) (3'-5' RNA exonuclease OLD35). | |||||
|
VGFR2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.001528 (rank : 78) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1089 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35918 | Gene names | Kdr, Flk-1, Flk1 | |||
|
Domain Architecture |

|
|||||
| Description | Vascular endothelial growth factor receptor 2 precursor (EC 2.7.10.1) (VEGFR-2) (Protein-tyrosine kinase receptor flk-1) (Fetal liver kinase 1) (Kinase NYK) (CD309 antigen). | |||||
|
AL2S4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.028348 (rank : 44) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q3V0J1, Q3TIS2 | Gene names | Als2cr4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 4 protein homolog. | |||||
|
NOVA2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.146331 (rank : 16) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UNW9, O43267, Q9UEA1 | Gene names | NOVA2, ANOVA, NOVA3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein Nova-2 (Neuro-oncological ventral antigen 2) (Astrocytic NOVA1-like RNA-binding protein). | |||||
|
SMG5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.016499 (rank : 56) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UPR3, Q5QJE7, Q8IXC0, Q8IY09, Q96IJ7 | Gene names | SMG5, EST1B, KIAA1089 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein SMG5 (SMG-5 homolog) (EST1-like protein B) (LPTS-interacting protein) (LPTS-RP1). | |||||
|
UBP53_MOUSE
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.014335 (rank : 60) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P15975, Q8CB37, Q8R251 | Gene names | Usp53, Phxr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 53 (Inactive ubiquitin- specific peptidase 53) (Per-hexamer repeat protein 3). | |||||
|
VSX1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.008024 (rank : 69) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91V10 | Gene names | Vsx1, Rinx | |||
|
Domain Architecture |

|
|||||
| Description | Visual system homeobox 1 (Transcription factor VSX1) (Retinal inner nuclear layer homeobox protein) (Homeodomain protein RINX). | |||||
|
CAC1G_HUMAN
|
||||||
| θ value | 4.03905 (rank : 49) | NC score | 0.006111 (rank : 75) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
HMMR_MOUSE
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.010390 (rank : 66) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q00547 | Gene names | Hmmr, Ihabp, Rhamm | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility). | |||||
|
TEP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.009658 (rank : 67) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97499 | Gene names | Tep1, Tp1 | |||
|
Domain Architecture |

|
|||||
| Description | Telomerase protein component 1 (Telomerase-associated protein 1) (Telomerase protein 1) (p240) (p80 homolog). | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.053246 (rank : 34) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
AKAP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.074695 (rank : 30) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q92667, Q13320, Q9BW14 | Gene names | AKAP1, AKAP149 | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 1, mitochondrial precursor (Protein kinase A- anchoring protein 1) (PRKA1) (A-kinase anchor protein 149 kDa) (AKAP 149) (Dual specificity A-kinase-anchoring protein 1) (D-AKAP-1) (Spermatid A-kinase anchor protein 84) (S-AKAP84). | |||||
|
AKAP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.071490 (rank : 32) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O08715, O08714, P97488 | Gene names | Akap1, Akap | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 1, mitochondrial precursor (Protein kinase A- anchoring protein 1) (PRKA1) (Dual specificity A-kinase-anchoring protein 1) (D-AKAP-1) (Spermatid A-kinase anchor protein) (S-AKAP). | |||||
|
CSPG3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.007126 (rank : 70) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P55066, Q6P1E3, Q8C4F8 | Gene names | Cspg3, Ncan | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
IF2B2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.098083 (rank : 23) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Y6M1 | Gene names | IGF2BP2, IMP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin-like growth factor 2 mRNA-binding protein 2 (IGF-II mRNA- binding protein 2) (IMP-2) (Hepatocellular carcinoma autoantigen p62). | |||||
|
IF2B2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.096485 (rank : 24) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5SF07, Q3TCU4, Q7TQF9 | Gene names | Igf2bp2, Imp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin-like growth factor 2 mRNA-binding protein 2 (IGF-II mRNA- binding protein 2) (IMP-2). | |||||
|
RAD_MOUSE
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.003329 (rank : 77) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O88667 | Gene names | Rrad | |||
|
Domain Architecture |

|
|||||
| Description | GTP-binding protein RAD. | |||||
|
SEM6A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.006991 (rank : 71) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H2E6, Q9P2H9 | Gene names | SEMA6A, KIAA1368 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6A precursor (Semaphorin VIA) (Sema VIA) (Semaphorin 6A-1) (SEMA6A-1). | |||||
|
SEM6A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.006931 (rank : 72) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35464, Q6P5A8, Q6PCN9, Q9EQ71 | Gene names | Sema6a, Semaq | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6A precursor (Semaphorin VIA) (Sema VIA) (Semaphorin 6A-1) (SEMA6A-1) (Semaphorin Q) (Sema Q). | |||||
|
CAH9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.006370 (rank : 74) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q16790 | Gene names | CA9, G250, MN | |||
|
Domain Architecture |

|
|||||
| Description | Carbonic anhydrase 9 precursor (EC 4.2.1.1) (Carbonic anhydrase IX) (Carbonate dehydratase IX) (CA-IX) (CAIX) (Membrane antigen MN) (P54/58N) (Renal cell carcinoma-associated antigen G250) (RCC- associated antigen G250) (pMW1). | |||||
|
CT043_MOUSE
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.020415 (rank : 52) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99K95, Q543D3, Q80X88, Q9CV27 | Gene names | ||||
|
Domain Architecture |
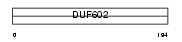
|
|||||
| Description | Uncharacterized protein C20orf43 homolog. | |||||
|
JADE2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.005188 (rank : 76) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
MARK2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | -0.001017 (rank : 81) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1056 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7KZI7, Q15449, Q15524, Q5XGA3, Q68A18, Q96HB3, Q96RG0 | Gene names | MARK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MARK2 (EC 2.7.11.1) (MAP/microtubule affinity-regulating kinase 2) (ELKL motif kinase) (EMK1) (PAR1 homolog). | |||||
|
MINT_MOUSE
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.022771 (rank : 49) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
RBM15_HUMAN
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.021514 (rank : 51) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96T37, Q5D058, Q96PE4, Q96SC5, Q96SC6, Q96SC9, Q96SD0, Q96T38, Q9BRA5, Q9H6R8, Q9H9Y0 | Gene names | RBM15, OTT | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein 15 (RNA-binding motif protein 15) (One- twenty two protein). | |||||
|
TTC16_HUMAN
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.015722 (rank : 59) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NEE8, Q5JU66, Q96M72 | Gene names | TTC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
WNK2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.000426 (rank : 79) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
BLNK_MOUSE
|
||||||
| θ value | 8.99809 (rank : 69) | NC score | 0.008249 (rank : 68) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QUN3, O88504 | Gene names | Blnk, Bash, Ly57, Slp65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell linker protein (Cytoplasmic adapter protein) (B-cell adapter containing SH2 domain protein) (B-cell adapter containing Src homology 2 domain protein) (Src homology 2 domain-containing leukocyte protein of 65 kDa) (Slp-65) (Lymphocyte antigen 57). | |||||
|
CAMP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.013960 (rank : 61) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49913, Q71SN9 | Gene names | CAMP, CAP18, FALL39 | |||
|
Domain Architecture |
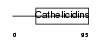
|
|||||
| Description | Cathelicidin antimicrobial peptide precursor (18 kDa cationic antimicrobial protein) (CAP-18) (hCAP-18) [Contains: Antibacterial protein FALL-39 (FALL-39 peptide antibiotic); Antibacterial protein LL-37]. | |||||
|
CPSF5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.015997 (rank : 57) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43809, Q6IB85, Q6NE84 | Gene names | NUDT21, CFIM25, CPSF25, CPSF5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 5 (Cleavage and polyadenylation specificity factor 25 kDa subunit) (CPSF 25 kDa subunit) (Pre-mRNA cleavage factor Im 25 kDa subunit) (Nucleoside diphosphate-linked moiety X motif 21) (Nudix motif 21). | |||||
|
CPSF5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.015996 (rank : 58) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CQF3, Q3UJK1 | Gene names | Nudt21, Cpsf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 5 (Nucleoside diphosphate-linked moiety X motif 21) (Nudix motif 21). | |||||
|
FMN1B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.017950 (rank : 55) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q05859 | Gene names | Fmn, Ld | |||
|
Domain Architecture |

|
|||||
| Description | Formin-1 isoform IV (Limb deformity protein). | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.000051 (rank : 80) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
MLF2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.020254 (rank : 53) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15773 | Gene names | MLF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myeloid leukemia factor 2 (Myelodysplasia-myeloid leukemia factor 2). | |||||
|
RKHD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.042977 (rank : 39) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q86XN8 | Gene names | RKHD1, KIAA2031 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger and KH domain-containing protein 1. | |||||
|
S39A6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.024200 (rank : 47) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8C145, Q7TPP9, Q7TQE0, Q8R518 | Gene names | Slc39a6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc transporter SLC39A6 precursor (Solute carrier family 39 member 6) (Endoplasmic reticulum membrane-linked protein) (Ermelin). | |||||
|
PCBP4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.087296 (rank : 27) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P57723, Q96AH7 | Gene names | PCBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 4 (Alpha-CP4). | |||||
|
PCBP4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.087888 (rank : 26) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P57724, Q91VR3 | Gene names | Pcbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 4 (Alpha-CP4). | |||||
|
VIGLN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.074491 (rank : 31) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q00341, Q9UCY3 | Gene names | HDLBP, HBP, VGL | |||
|
Domain Architecture |

|
|||||
| Description | Vigilin (High density lipoprotein-binding protein) (HDL-binding protein). | |||||
|
VIGLN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.076949 (rank : 29) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8VDJ3 | Gene names | Hdlbp | |||
|
Domain Architecture |

|
|||||
| Description | Vigilin (High density lipoprotein-binding protein) (HDL-binding protein). | |||||
|
FXR2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P51116, Q8WUM2 | Gene names | FXR2 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 2. | |||||
|
FXR2_MOUSE
|
||||||
| NC score | 0.990533 (rank : 2) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9WVR4, Q9WVR5 | Gene names | Fxr2, Fxr2h | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 2. | |||||
|
FXR1_MOUSE
|
||||||
| NC score | 0.982393 (rank : 3) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61584, Q8VCU4, Q9R1E2, Q9R1E3, Q9R1E4, Q9R1E5, Q9WUA7, Q9WUA8, Q9WUA9 | Gene names | Fxr1, Fxr1h | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 1 (mFxr1p). | |||||
|
FXR1_HUMAN
|
||||||
| NC score | 0.979665 (rank : 4) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P51114, Q7Z450, Q8N6R8 | Gene names | FXR1 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 1 (hFXR1P). | |||||
|
FMR1_HUMAN
|
||||||
| NC score | 0.962941 (rank : 5) | θ value | 4.58675e-137 (rank : 6) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q06787, Q16578, Q99054 | Gene names | FMR1 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation 1 protein (Protein FMR-1) (FMRP). | |||||
|
FMR1_MOUSE
|
||||||
| NC score | 0.962699 (rank : 6) | θ value | 8.93083e-141 (rank : 5) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P35922 | Gene names | Fmr1, Fmr-1 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation protein 1 homolog (Protein FMR-1) (FMRP) (mFmr1p). | |||||
|
TDRKH_HUMAN
|
||||||
| NC score | 0.238928 (rank : 7) | θ value | 0.00102713 (rank : 9) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y2W6, Q8N582, Q9NYV5 | Gene names | TDRKH, TDRD2 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor and KH domain-containing protein. | |||||
|
TDRKH_MOUSE
|
||||||
| NC score | 0.236366 (rank : 8) | θ value | 0.00102713 (rank : 10) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q80VL1 | Gene names | Tdrkh | |||
|
Domain Architecture |

|
|||||
| Description | Tudor and KH domain-containing protein. | |||||
|
FUBP3_HUMAN
|
||||||
| NC score | 0.177857 (rank : 9) | θ value | 0.0193708 (rank : 11) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96I24, Q92946, Q9BVB6 | Gene names | FUBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 3 (FUSE-binding protein 3). | |||||
|
HNRPK_HUMAN
|
||||||
| NC score | 0.171333 (rank : 10) | θ value | 0.0330416 (rank : 12) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P61978, Q07244, Q15671, Q60577, Q922Y7, Q96J62 | Gene names | HNRPK, HNRNPK | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein K (hnRNP K) (Transformation up-regulated nuclear protein) (TUNP). | |||||
|
HNRPK_MOUSE
|
||||||
| NC score | 0.171333 (rank : 11) | θ value | 0.0330416 (rank : 13) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P61979, Q07244, Q15671, Q60577, Q8BGQ8, Q922Y7, Q96J62 | Gene names | Hnrpk, Hnrnpk | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein K. | |||||
|
FUBP1_MOUSE
|
||||||
| NC score | 0.166222 (rank : 12) | θ value | 0.21417 (rank : 18) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q91WJ8, Q8C0Y8 | Gene names | Fubp1, D3Ertd330e | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 1 (FUSE-binding protein 1) (FBP). | |||||
|
FUBP2_HUMAN
|
||||||
| NC score | 0.166149 (rank : 13) | θ value | 0.21417 (rank : 19) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q92945, O00301, Q9UNT5, Q9UQH5 | Gene names | KHSRP, FUBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 2 (FUSE-binding protein 2) (KH type-splicing regulatory protein) (KSRP) (p75). | |||||
|
FUBP1_HUMAN
|
||||||
| NC score | 0.165770 (rank : 14) | θ value | 0.21417 (rank : 17) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96AE4, Q12828 | Gene names | FUBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 1 (FUSE-binding protein 1) (FBP) (DNA helicase V) (HDH V). | |||||
|
NOVA1_HUMAN
|
||||||
| NC score | 0.148339 (rank : 15) | θ value | 1.38821 (rank : 36) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P51513 | Gene names | NOVA1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein Nova-1 (Neuro-oncological ventral antigen 1) (Onconeural ventral antigen 1) (Paraneoplastic Ri antigen) (Ventral neuron-specific protein 1). | |||||
|
NOVA2_HUMAN
|
||||||
| NC score | 0.146331 (rank : 16) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UNW9, O43267, Q9UEA1 | Gene names | NOVA2, ANOVA, NOVA3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein Nova-2 (Neuro-oncological ventral antigen 2) (Astrocytic NOVA1-like RNA-binding protein). | |||||
|
PCBP3_MOUSE
|
||||||
| NC score | 0.118495 (rank : 17) | θ value | 0.21417 (rank : 22) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P57722, Q8BSB0, Q8C544 | Gene names | Pcbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 3 (Alpha-CP3). | |||||
|
PCBP3_HUMAN
|
||||||
| NC score | 0.118428 (rank : 18) | θ value | 0.21417 (rank : 21) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P57721, Q8N9K6, Q96EP6 | Gene names | PCBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 3 (Alpha-CP3). | |||||
|
PCBP2_HUMAN
|
||||||
| NC score | 0.104613 (rank : 19) | θ value | 0.62314 (rank : 30) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q15366, Q6PKG5 | Gene names | PCBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 2 (Alpha-CP2) (hnRNP-E2). | |||||
|
PCBP2_MOUSE
|
||||||
| NC score | 0.104066 (rank : 20) | θ value | 0.62314 (rank : 31) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q61990, Q61383, Q62042 | Gene names | Pcbp2, Cbp, Hnrnpx, Hnrpx | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 2 (Alpha-CP2) (Putative heterogeneous nuclear ribonucleoprotein X) (hnRNP X) (CTBP) (CBP). | |||||
|
PCBP1_HUMAN
|
||||||
| NC score | 0.100720 (rank : 21) | θ value | 1.38821 (rank : 37) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q15365, Q13157, Q14975 | Gene names | PCBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 1 (Alpha-CP1) (hnRNP-E1) (Nucleic acid- binding protein SUB2.3). | |||||
|
PCBP1_MOUSE
|
||||||
| NC score | 0.100720 (rank : 22) | θ value | 1.38821 (rank : 38) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P60335 | Gene names | Pcbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 1 (Alpha-CP1) (hnRNP-E1). | |||||
|
IF2B2_HUMAN
|
||||||
| NC score | 0.098083 (rank : 23) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Y6M1 | Gene names | IGF2BP2, IMP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin-like growth factor 2 mRNA-binding protein 2 (IGF-II mRNA- binding protein 2) (IMP-2) (Hepatocellular carcinoma autoantigen p62). | |||||
|
IF2B2_MOUSE
|
||||||
| NC score | 0.096485 (rank : 24) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5SF07, Q3TCU4, Q7TQF9 | Gene names | Igf2bp2, Imp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin-like growth factor 2 mRNA-binding protein 2 (IGF-II mRNA- binding protein 2) (IMP-2). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.090137 (rank : 25) | θ value | 0.00035302 (rank : 8) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
PCBP4_MOUSE
|
||||||
| NC score | 0.087888 (rank : 26) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P57724, Q91VR3 | Gene names | Pcbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 4 (Alpha-CP4). | |||||
|
PCBP4_HUMAN
|
||||||
| NC score | 0.087296 (rank : 27) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P57723, Q96AH7 | Gene names | PCBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 4 (Alpha-CP4). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.078209 (rank : 28) | θ value | 1.87187e-05 (rank : 7) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
VIGLN_MOUSE
|
||||||
| NC score | 0.076949 (rank : 29) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8VDJ3 | Gene names | Hdlbp | |||
|
Domain Architecture |

|
|||||
| Description | Vigilin (High density lipoprotein-binding protein) (HDL-binding protein). | |||||
|
AKAP1_HUMAN
|
||||||
| NC score | 0.074695 (rank : 30) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q92667, Q13320, Q9BW14 | Gene names | AKAP1, AKAP149 | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 1, mitochondrial precursor (Protein kinase A- anchoring protein 1) (PRKA1) (A-kinase anchor protein 149 kDa) (AKAP 149) (Dual specificity A-kinase-anchoring protein 1) (D-AKAP-1) (Spermatid A-kinase anchor protein 84) (S-AKAP84). | |||||
|
VIGLN_HUMAN
|
||||||
| NC score | 0.074491 (rank : 31) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q00341, Q9UCY3 | Gene names | HDLBP, HBP, VGL | |||
|
Domain Architecture |

|
|||||
| Description | Vigilin (High density lipoprotein-binding protein) (HDL-binding protein). | |||||
|
AKAP1_MOUSE
|
||||||
| NC score | 0.071490 (rank : 32) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O08715, O08714, P97488 | Gene names | Akap1, Akap | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 1, mitochondrial precursor (Protein kinase A- anchoring protein 1) (PRKA1) (Dual specificity A-kinase-anchoring protein 1) (D-AKAP-1) (Spermatid A-kinase anchor protein) (S-AKAP). | |||||
|
PNPT1_HUMAN
|
||||||
| NC score | 0.066529 (rank : 33) | θ value | 0.813845 (rank : 33) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8TCS8, Q53SU0, Q68CN1, Q7Z7D1, Q8IWX1, Q96T05, Q9BRU3, Q9BVX0 | Gene names | PNPT1, PNPASE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyribonucleotide nucleotidyltransferase 1, mitochondrial precursor (EC 2.7.7.8) (PNPase 1) (Polynucleotide phosphorylase-like protein) (PNPase old-35) (3'-5' RNA exonuclease OLD35). | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.053246 (rank : 34) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
LAP2B_HUMAN
|
||||||
| NC score | 0.048554 (rank : 35) | θ value | 0.21417 (rank : 20) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P42167, Q14861 | Gene names | TMPO, LAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamina-associated polypeptide 2, isoforms beta/gamma (Thymopoietin, isoforms beta/gamma) (TP beta/gamma) (Thymopoietin-related peptide isoforms beta/gamma) (TPRP isoforms beta/gamma) [Contains: Thymopoietin (TP) (Splenin); Thymopentin (TP5)]. | |||||
|
PNPT1_MOUSE
|
||||||
| NC score | 0.047362 (rank : 36) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K1R3, Q3UEP9, Q810U7, Q812B3, Q8R2U3, Q9DC52 | Gene names | Pnpt1, Pnpase | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyribonucleotide nucleotidyltransferase 1, mitochondrial precursor (EC 2.7.7.8) (PNPase 1) (Polynucleotide phosphorylase-like protein) (PNPase old-35) (3'-5' RNA exonuclease OLD35). | |||||
|
RPTN_MOUSE
|
||||||
| NC score | 0.043702 (rank : 37) | θ value | 0.0563607 (rank : 14) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P97347 | Gene names | Rptn | |||
|
Domain Architecture |

|
|||||
| Description | Repetin. | |||||
|
ZN326_HUMAN
|
||||||
| NC score | 0.042985 (rank : 38) | θ value | 0.279714 (rank : 24) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5BKZ1, Q5VW93, Q5VW94, Q5VW96, Q5VW97, Q6NSA2, Q7Z638, Q7Z6C2 | Gene names | ZNF326 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 326. | |||||
|
RKHD1_HUMAN
|
||||||
| NC score | 0.042977 (rank : 39) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q86XN8 | Gene names | RKHD1, KIAA2031 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger and KH domain-containing protein 1. | |||||
|
ZN326_MOUSE
|
||||||
| NC score | 0.039873 (rank : 40) | θ value | 0.62314 (rank : 32) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88291, Q8BSJ5, Q8K1X9, Q9CYG9 | Gene names | Znf326, Zan75, Zfp326 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 326 (Zinc finger protein-associated with nuclear matrix of 75 kDa). | |||||
|
REXO1_MOUSE
|
||||||
| NC score | 0.033419 (rank : 41) | θ value | 0.21417 (rank : 23) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7TT28, Q3UMP1, Q69ZR0, Q6NSQ6, Q6PI95, Q9DA29 | Gene names | Rexo1, Kiaa1138, Tceb3bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 1 homolog (EC 3.1.-.-) (Transcription elongation factor B polypeptide 3-binding protein 1). | |||||
|
ELOA1_HUMAN
|
||||||
| NC score | 0.030761 (rank : 42) | θ value | 0.163984 (rank : 16) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14241, Q8IXH1 | Gene names | TCEB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor B polypeptide 3 (RNA polymerase II transcription factor SIII subunit A1) (SIII p110) (Elongin-A) (EloA) (Elongin 110 kDa subunit). | |||||
|
MLF2_MOUSE
|
||||||
| NC score | 0.028655 (rank : 43) | θ value | 1.38821 (rank : 35) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99KX1 | Gene names | Mlf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myeloid leukemia factor 2 (Myelodysplasia-myeloid leukemia factor 2). | |||||
|
AL2S4_MOUSE
|
||||||
| NC score | 0.028348 (rank : 44) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q3V0J1, Q3TIS2 | Gene names | Als2cr4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 4 protein homolog. | |||||
|
FBX34_MOUSE
|
||||||
| NC score | 0.027411 (rank : 45) | θ value | 0.47712 (rank : 25) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80XI1, Q9D290 | Gene names | Fbxo34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 34. | |||||
|
RRBP1_MOUSE
|
||||||
| NC score | 0.025018 (rank : 46) | θ value | 0.0961366 (rank : 15) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
S39A6_MOUSE
|
||||||
| NC score | 0.024200 (rank : 47) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8C145, Q7TPP9, Q7TQE0, Q8R518 | Gene names | Slc39a6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc transporter SLC39A6 precursor (Solute carrier family 39 member 6) (Endoplasmic reticulum membrane-linked protein) (Ermelin). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.023844 (rank : 48) | θ value | 0.47712 (rank : 27) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.022771 (rank : 49) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
SRBS1_MOUSE
|
||||||
| NC score | 0.021804 (rank : 50) | θ value | 1.06291 (rank : 34) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q62417, Q80TF8, Q8BZI3, Q8K3Y2, Q921F8, Q9Z0Z8, Q9Z0Z9 | Gene names | Sorbs1, Kiaa1296, Sh3d5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorbin and SH3 domain-containing protein 1 (Ponsin) (c-Cbl-associated protein) (CAP) (SH3 domain protein 5) (SH3P12). | |||||
|
RBM15_HUMAN
|
||||||
| NC score | 0.021514 (rank : 51) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96T37, Q5D058, Q96PE4, Q96SC5, Q96SC6, Q96SC9, Q96SD0, Q96T38, Q9BRA5, Q9H6R8, Q9H9Y0 | Gene names | RBM15, OTT | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein 15 (RNA-binding motif protein 15) (One- twenty two protein). | |||||
|
CT043_MOUSE
|
||||||
| NC score | 0.020415 (rank : 52) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99K95, Q543D3, Q80X88, Q9CV27 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf43 homolog. | |||||
|
MLF2_HUMAN
|
||||||
| NC score | 0.020254 (rank : 53) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15773 | Gene names | MLF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myeloid leukemia factor 2 (Myelodysplasia-myeloid leukemia factor 2). | |||||
|
RTN1_MOUSE
|
||||||
| NC score | 0.018481 (rank : 54) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8K0T0, Q8K4S4 | Gene names | Rtn1, Nsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulon-1 (Neuroendocrine-specific protein). | |||||
|
FMN1B_MOUSE
|
||||||
| NC score | 0.017950 (rank : 55) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q05859 | Gene names | Fmn, Ld | |||
|
Domain Architecture |

|
|||||
| Description | Formin-1 isoform IV (Limb deformity protein). | |||||
|
SMG5_HUMAN
|
||||||
| NC score | 0.016499 (rank : 56) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UPR3, Q5QJE7, Q8IXC0, Q8IY09, Q96IJ7 | Gene names | SMG5, EST1B, KIAA1089 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein SMG5 (SMG-5 homolog) (EST1-like protein B) (LPTS-interacting protein) (LPTS-RP1). | |||||
|
CPSF5_HUMAN
|
||||||
| NC score | 0.015997 (rank : 57) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43809, Q6IB85, Q6NE84 | Gene names | NUDT21, CFIM25, CPSF25, CPSF5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 5 (Cleavage and polyadenylation specificity factor 25 kDa subunit) (CPSF 25 kDa subunit) (Pre-mRNA cleavage factor Im 25 kDa subunit) (Nucleoside diphosphate-linked moiety X motif 21) (Nudix motif 21). | |||||
|
CPSF5_MOUSE
|
||||||
| NC score | 0.015996 (rank : 58) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CQF3, Q3UJK1 | Gene names | Nudt21, Cpsf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 5 (Nucleoside diphosphate-linked moiety X motif 21) (Nudix motif 21). | |||||
|
TTC16_HUMAN
|
||||||
| NC score | 0.015722 (rank : 59) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NEE8, Q5JU66, Q96M72 | Gene names | TTC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
UBP53_MOUSE
|
||||||
| NC score | 0.014335 (rank : 60) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P15975, Q8CB37, Q8R251 | Gene names | Usp53, Phxr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 53 (Inactive ubiquitin- specific peptidase 53) (Per-hexamer repeat protein 3). | |||||
|
CAMP_HUMAN
|
||||||
| NC score | 0.013960 (rank : 61) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49913, Q71SN9 | Gene names | CAMP, CAP18, FALL39 | |||
|
Domain Architecture |

|
|||||
| Description | Cathelicidin antimicrobial peptide precursor (18 kDa cationic antimicrobial protein) (CAP-18) (hCAP-18) [Contains: Antibacterial protein FALL-39 (FALL-39 peptide antibiotic); Antibacterial protein LL-37]. | |||||
|
MYH6_HUMAN
|
||||||
| NC score | 0.013331 (rank : 62) | θ value | 0.47712 (rank : 26) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYH6_MOUSE
|
||||||
| NC score | 0.013008 (rank : 63) | θ value | 0.62314 (rank : 29) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYH7_HUMAN
|
||||||
| NC score | 0.012071 (rank : 64) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
CHD6_HUMAN
|
||||||
| NC score | 0.011243 (rank : 65) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8TD26, Q5JYQ0, Q5TGZ9, Q5TH00, Q5TH01, Q8IZR2, Q8WTY0, Q9H4H6, Q9H6D4, Q9NTT7, Q9P2L1 | Gene names | CHD6, CHD5, KIAA1335, RIGB | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 6 (EC 3.6.1.-) (ATP- dependent helicase CHD6) (CHD-6) (Radiation-induced gene B protein). | |||||
|
HMMR_MOUSE
|
||||||
| NC score | 0.010390 (rank : 66) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q00547 | Gene names | Hmmr, Ihabp, Rhamm | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility). | |||||
|
TEP1_MOUSE
|
||||||
| NC score | 0.009658 (rank : 67) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97499 | Gene names | Tep1, Tp1 | |||
|
Domain Architecture |

|
|||||
| Description | Telomerase protein component 1 (Telomerase-associated protein 1) (Telomerase protein 1) (p240) (p80 homolog). | |||||
|
BLNK_MOUSE
|
||||||
| NC score | 0.008249 (rank : 68) | θ value | 8.99809 (rank : 69) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QUN3, O88504 | Gene names | Blnk, Bash, Ly57, Slp65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell linker protein (Cytoplasmic adapter protein) (B-cell adapter containing SH2 domain protein) (B-cell adapter containing Src homology 2 domain protein) (Src homology 2 domain-containing leukocyte protein of 65 kDa) (Slp-65) (Lymphocyte antigen 57). | |||||
|
VSX1_MOUSE
|
||||||
| NC score | 0.008024 (rank : 69) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91V10 | Gene names | Vsx1, Rinx | |||
|
Domain Architecture |

|
|||||
| Description | Visual system homeobox 1 (Transcription factor VSX1) (Retinal inner nuclear layer homeobox protein) (Homeodomain protein RINX). | |||||
|
CSPG3_MOUSE
|
||||||
| NC score | 0.007126 (rank : 70) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P55066, Q6P1E3, Q8C4F8 | Gene names | Cspg3, Ncan | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
SEM6A_HUMAN
|
||||||
| NC score | 0.006991 (rank : 71) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H2E6, Q9P2H9 | Gene names | SEMA6A, KIAA1368 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6A precursor (Semaphorin VIA) (Sema VIA) (Semaphorin 6A-1) (SEMA6A-1). | |||||
|
SEM6A_MOUSE
|
||||||
| NC score | 0.006931 (rank : 72) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35464, Q6P5A8, Q6PCN9, Q9EQ71 | Gene names | Sema6a, Semaq | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6A precursor (Semaphorin VIA) (Sema VIA) (Semaphorin 6A-1) (SEMA6A-1) (Semaphorin Q) (Sema Q). | |||||
|
TNK1_HUMAN
|
||||||
| NC score | 0.006758 (rank : 73) | θ value | 0.47712 (rank : 28) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 815 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13470, O95364, Q8IYI4 | Gene names | TNK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Non-receptor tyrosine-protein kinase TNK1 (EC 2.7.10.2) (CD38 negative kinase 1). | |||||
|
CAH9_HUMAN
|
||||||
| NC score | 0.006370 (rank : 74) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q16790 | Gene names | CA9, G250, MN | |||
|
Domain Architecture |

|
|||||
| Description | Carbonic anhydrase 9 precursor (EC 4.2.1.1) (Carbonic anhydrase IX) (Carbonate dehydratase IX) (CA-IX) (CAIX) (Membrane antigen MN) (P54/58N) (Renal cell carcinoma-associated antigen G250) (RCC- associated antigen G250) (pMW1). | |||||
|
CAC1G_HUMAN
|
||||||
| NC score | 0.006111 (rank : 75) | θ value | 4.03905 (rank : 49) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
JADE2_MOUSE
|
||||||
| NC score | 0.005188 (rank : 76) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
RAD_MOUSE
|
||||||
| NC score | 0.003329 (rank : 77) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O88667 | Gene names | Rrad | |||
|
Domain Architecture |

|
|||||
| Description | GTP-binding protein RAD. | |||||
|
VGFR2_MOUSE
|
||||||
| NC score | 0.001528 (rank : 78) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1089 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35918 | Gene names | Kdr, Flk-1, Flk1 | |||
|
Domain Architecture |

|
|||||
| Description | Vascular endothelial growth factor receptor 2 precursor (EC 2.7.10.1) (VEGFR-2) (Protein-tyrosine kinase receptor flk-1) (Fetal liver kinase 1) (Kinase NYK) (CD309 antigen). | |||||
|
WNK2_HUMAN
|
||||||
| NC score | 0.000426 (rank : 79) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
MAST4_HUMAN
|
||||||
| NC score | 0.000051 (rank : 80) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
MARK2_HUMAN
|
||||||
| NC score | -0.001017 (rank : 81) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1056 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7KZI7, Q15449, Q15524, Q5XGA3, Q68A18, Q96HB3, Q96RG0 | Gene names | MARK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MARK2 (EC 2.7.11.1) (MAP/microtubule affinity-regulating kinase 2) (ELKL motif kinase) (EMK1) (PAR1 homolog). | |||||