Please be patient as the page loads
|
SMG5_HUMAN
|
||||||
| SwissProt Accessions | Q9UPR3, Q5QJE7, Q8IXC0, Q8IY09, Q96IJ7 | Gene names | SMG5, EST1B, KIAA1089 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein SMG5 (SMG-5 homolog) (EST1-like protein B) (LPTS-interacting protein) (LPTS-RP1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SMG5_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 180 | |
| SwissProt Accessions | Q9UPR3, Q5QJE7, Q8IXC0, Q8IY09, Q96IJ7 | Gene names | SMG5, EST1B, KIAA1089 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein SMG5 (SMG-5 homolog) (EST1-like protein B) (LPTS-interacting protein) (LPTS-RP1). | |||||
|
SMG5_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.952543 (rank : 2) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6ZPY2, Q497S2, Q8R3S4 | Gene names | Smg5, Est1b, Kiaa1089 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein SMG5 (SMG-5 homolog) (EST1-like protein B). | |||||
|
SMG7_MOUSE
|
||||||
| θ value | 4.44505e-15 (rank : 3) | NC score | 0.469725 (rank : 3) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5RJH6, Q63ZW5, Q6ZQF3 | Gene names | Smg7, Est1c, Kiaa0250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein SMG7 (SMG-7 homolog) (EST1-like protein C). | |||||
|
SMG7_HUMAN
|
||||||
| θ value | 1.29331e-14 (rank : 4) | NC score | 0.467036 (rank : 4) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q92540, Q5T1Q0, Q6PIE0, Q7Z7H9, Q8IXC1, Q8IXC2 | Gene names | SMG7, C1orf16, EST1C, KIAA0250 | |||
|
Domain Architecture |

|
|||||
| Description | Protein SMG7 (SMG-7 homolog) (EST1-like protein C). | |||||
|
EST1A_HUMAN
|
||||||
| θ value | 1.42992e-13 (rank : 5) | NC score | 0.439479 (rank : 5) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86US8, O94837, Q86VH6, Q9UF60 | Gene names | EST1A, C17orf31, KIAA0732 | |||
|
Domain Architecture |

|
|||||
| Description | Telomerase-binding protein EST1A (Ever shorter telomeres 1A) (Telomerase subunit EST1A) (EST1-like protein A) (hSmg5/7a). | |||||
|
EST1A_MOUSE
|
||||||
| θ value | 1.2105e-12 (rank : 6) | NC score | 0.425382 (rank : 6) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P61406, Q5NC64 | Gene names | Est1a, Kiaa0732 | |||
|
Domain Architecture |

|
|||||
| Description | Telomerase-binding protein EST1A (Ever shorter telomeres 1A) (Telomerase subunit EST1A) (EST1-like protein A). | |||||
|
T2FA_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 7) | NC score | 0.151009 (rank : 9) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
DSPP_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 8) | NC score | 0.108795 (rank : 14) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
DSPP_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 9) | NC score | 0.120446 (rank : 12) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
AFF4_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 10) | NC score | 0.106120 (rank : 15) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
GNAS3_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 11) | NC score | 0.125674 (rank : 11) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Z0F1, Q9QXW5, Q9Z0H2 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroendocrine secretory protein 55 (NESP55) [Contains: LHAL tetrapeptide; GPIPIRRH peptide]. | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 12) | NC score | 0.158131 (rank : 7) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 110 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
AFF4_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 13) | NC score | 0.101430 (rank : 17) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
TTBK1_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 14) | NC score | 0.029329 (rank : 120) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1058 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q5TCY1, Q2L6C6, Q8N444, Q96JH2 | Gene names | TTBK1, BDTK, KIAA1855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 1 (EC 2.7.11.1) (Brain-derived tau kinase). | |||||
|
ZN447_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 15) | NC score | 0.022352 (rank : 139) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 899 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8TBC5, Q9BRK7, Q9H9A0 | Gene names | ZNF447 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 447. | |||||
|
CHD2_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 16) | NC score | 0.046745 (rank : 83) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O14647 | Gene names | CHD2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 2 (EC 3.6.1.-) (ATP- dependent helicase CHD2) (CHD-2). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 17) | NC score | 0.154004 (rank : 8) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 117 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
UBP8_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 18) | NC score | 0.049551 (rank : 78) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q80U87, Q80YP2, Q8R0D3, Q9EQU1, Q9WVP5 | Gene names | Usp8, Kiaa0055, Ubpy | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (mUBPy). | |||||
|
ABEC3_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 19) | NC score | 0.063542 (rank : 46) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99J72, Q8C7L0, Q8C8V7 | Gene names | Apobec3 | |||
|
Domain Architecture |
|
|||||
| Description | DNA dC->dU-editing enzyme APOBEC-3 (EC 3.5.4.-) (Apolipoprotein B mRNA-editing complex 3) (Arp3) (CEM15) (CEM-15). | |||||
|
CYLC1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 20) | NC score | 0.080580 (rank : 31) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
RGP1_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 21) | NC score | 0.088000 (rank : 25) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P46061, Q60801 | Gene names | Rangap1, Fug1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
SRCA_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 22) | NC score | 0.059565 (rank : 51) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
AP3B2_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 23) | NC score | 0.066836 (rank : 39) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13367, O14808 | Gene names | AP3B2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (AP-3 complex beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain) (Neuron-specific vesicle coat protein beta-NAP). | |||||
|
FOXD1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 24) | NC score | 0.020644 (rank : 142) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q16676, Q12949 | Gene names | FOXD1, FKHL8, FREAC4 | |||
|
Domain Architecture |
|
|||||
| Description | Forkhead box protein D1 (Forkhead-related protein FKHL8) (Forkhead- related transcription factor 4) (FREAC-4). | |||||
|
PHAR3_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 25) | NC score | 0.068316 (rank : 37) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BYK5, Q8BYS8, Q8C058, Q9DB87 | Gene names | Phactr3, Scapin1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatase and actin regulator 3 (Scapinin) (Scaffold-associated PP1- inhibiting protein). | |||||
|
PTDSR_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 26) | NC score | 0.090523 (rank : 22) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6NYC1, Q86VY0, Q8IUM5, Q9Y4E2 | Gene names | PTDSR, KIAA0585 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein PTDSR (Phosphatidylserine receptor). | |||||
|
NCKX1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 27) | NC score | 0.055945 (rank : 60) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O60721, O43485, O75184 | Gene names | SLC24A1, KIAA0702, NCKX1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium/calcium exchanger 1 (Na(+)/K(+)/Ca(2+)-exchange protein 1) (Retinal rod Na-Ca+K exchanger). | |||||
|
AFF1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 28) | NC score | 0.092619 (rank : 20) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
BAZ2B_HUMAN
|
||||||
| θ value | 0.163984 (rank : 29) | NC score | 0.041900 (rank : 93) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
NSBP1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 30) | NC score | 0.083408 (rank : 30) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
TRI44_HUMAN
|
||||||
| θ value | 0.163984 (rank : 31) | NC score | 0.057849 (rank : 54) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96DX7, Q96QY2, Q9UGK0 | Gene names | TRIM44, DIPB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB). | |||||
|
ABTAP_MOUSE
|
||||||
| θ value | 0.21417 (rank : 32) | NC score | 0.057033 (rank : 57) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q3V1V3 | Gene names | Abtap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
PTDSR_MOUSE
|
||||||
| θ value | 0.21417 (rank : 33) | NC score | 0.092590 (rank : 21) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ERI5, Q80TX1 | Gene names | Ptdsr, Kiaa0585 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein PTDSR (Phosphatidylserine receptor) (Apoptotic cell clearance receptor PtdSerR). | |||||
|
CD19_HUMAN
|
||||||
| θ value | 0.279714 (rank : 34) | NC score | 0.052105 (rank : 72) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P15391, Q96S68, Q9BRD6 | Gene names | CD19 | |||
|
Domain Architecture |
|
|||||
| Description | B-lymphocyte antigen CD19 precursor (Differentiation antigen CD19) (B- lymphocyte surface antigen B4) (Leu-12). | |||||
|
DVL3_MOUSE
|
||||||
| θ value | 0.279714 (rank : 35) | NC score | 0.035163 (rank : 108) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61062 | Gene names | Dvl3 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-3 (Dishevelled-3) (DSH homolog 3). | |||||
|
SALL2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 36) | NC score | 0.017908 (rank : 151) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 836 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9QX96 | Gene names | Sall2, Sal2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 2 (Spalt-like protein 2) (MSal-2). | |||||
|
SET1A_HUMAN
|
||||||
| θ value | 0.279714 (rank : 37) | NC score | 0.059285 (rank : 52) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
ATBF1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 38) | NC score | 0.041514 (rank : 94) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
CTR9_MOUSE
|
||||||
| θ value | 0.365318 (rank : 39) | NC score | 0.065785 (rank : 43) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q62018, Q3UFF5, Q3UY40, Q66JX4, Q7TPS6, Q8BND9, Q8BRD1, Q8C9W7, Q8C9Y3, Q8CHI1 | Gene names | Ctr9, Kiaa0155, Sh2bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1) (Tetratricopeptide repeat-containing, SH2-binding phosphoprotein of 150 kDa) (TPR-containing, SH2-binding phosphoprotein of 150 kDa) (p150TSP). | |||||
|
K1543_HUMAN
|
||||||
| θ value | 0.365318 (rank : 40) | NC score | 0.044305 (rank : 86) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9P1Y5, Q8NDF1 | Gene names | KIAA1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 41) | NC score | 0.132418 (rank : 10) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 108 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
MRP_MOUSE
|
||||||
| θ value | 0.365318 (rank : 42) | NC score | 0.066573 (rank : 41) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P28667, Q3TEZ4, Q91W07 | Gene names | Marcksl1, Mlp, Mrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MARCKS-related protein (MARCKS-like protein 1) (Macrophage myristoylated alanine-rich C kinase substrate) (Mac-MARCKS) (MacMARCKS) (Brain protein F52). | |||||
|
NF2L1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 43) | NC score | 0.040017 (rank : 99) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61985, O70234 | Gene names | Nfe2l1, Nrf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 1 (NF-E2-related factor 1) (NFE2-related factor 1) (Nuclear factor, erythroid derived 2, like 1). | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 44) | NC score | 0.052611 (rank : 70) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
AP3B1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 45) | NC score | 0.063233 (rank : 47) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Z1T1, Q91YR4 | Gene names | Ap3b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
CD2L2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 46) | NC score | 0.014329 (rank : 163) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
DVL3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 47) | NC score | 0.032962 (rank : 115) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92997, O14642, Q13531, Q8N5E9, Q92607 | Gene names | DVL3, KIAA0208 | |||
|
Domain Architecture |
|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-3 (Dishevelled-3) (DSH homolog 3). | |||||
|
RPA1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 48) | NC score | 0.036453 (rank : 103) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95602, Q9UEH0, Q9UFT9 | Gene names | POLR1A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase I largest subunit (EC 2.7.7.6) (RNA polymerase I 194 kDa subunit) (RPA194). | |||||
|
TRI44_MOUSE
|
||||||
| θ value | 0.47712 (rank : 49) | NC score | 0.054186 (rank : 68) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9QXA7, Q80SV9, Q9JHR8 | Gene names | Trim44, Dipb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB) (Protein Mc7). | |||||
|
CDC16_HUMAN
|
||||||
| θ value | 0.62314 (rank : 50) | NC score | 0.086867 (rank : 26) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13042, Q96AE6, Q9Y564 | Gene names | CDC16 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 16 homolog (CDC16Hs) (Anaphase-promoting complex subunit 6) (APC6) (Cyclosome subunit 6). | |||||
|
CYLC2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 51) | NC score | 0.085189 (rank : 27) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
DMP1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 52) | NC score | 0.092993 (rank : 19) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
MYCB2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 53) | NC score | 0.040833 (rank : 96) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7TPH6, Q6PCM8 | Gene names | Mycbp2, Pam, Phr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
PHAR3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 54) | NC score | 0.058309 (rank : 53) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96KR7, Q707P6, Q9H4T4 | Gene names | PHACTR3, C20orf101, SCAPIN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatase and actin regulator 3 (Scapinin) (Scaffold-associated PP1- inhibiting protein). | |||||
|
RNPS1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 55) | NC score | 0.054264 (rank : 66) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q15287, O75308, Q8WY42, Q9NYG3 | Gene names | RNPS1, LDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1 (SR-related protein LDC2). | |||||
|
RNPS1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 56) | NC score | 0.054264 (rank : 67) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q99M28, Q3TMJ1, Q922H8 | Gene names | Rnps1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1. | |||||
|
SFR14_MOUSE
|
||||||
| θ value | 0.62314 (rank : 57) | NC score | 0.037755 (rank : 101) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CH09, Q6PG19, Q80UY8, Q8BY32, Q8CFM0 | Gene names | Sfrs14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative splicing factor, arginine/serine-rich 14 (Arginine/serine- rich-splicing factor 14). | |||||
|
SFRS4_MOUSE
|
||||||
| θ value | 0.62314 (rank : 58) | NC score | 0.044863 (rank : 85) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8VE97, Q9JJC3 | Gene names | Sfrs4 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4. | |||||
|
UBF1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 59) | NC score | 0.051452 (rank : 75) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
CDC16_MOUSE
|
||||||
| θ value | 0.813845 (rank : 60) | NC score | 0.084414 (rank : 29) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8R349, Q9CYX9 | Gene names | Cdc16 | |||
|
Domain Architecture |
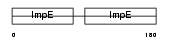
|
|||||
| Description | Cell division cycle protein 16 homolog (Anaphase-promoting complex subunit 6) (APC6) (Cyclosome subunit 6). | |||||
|
CELR2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 61) | NC score | 0.003990 (rank : 192) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HCU4, Q92566 | Gene names | CELSR2, CDHF10, EGFL2, KIAA0279, MEGF3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 2 precursor (Epidermal growth factor-like 2) (Multiple epidermal growth factor-like domains 3) (Flamingo 1). | |||||
|
DAXX_MOUSE
|
||||||
| θ value | 0.813845 (rank : 62) | NC score | 0.066487 (rank : 42) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O35613, Q9QWT8, Q9QWV3 | Gene names | Daxx | |||
|
Domain Architecture |

|
|||||
| Description | Death domain-associated protein 6 (Daxx). | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 63) | NC score | 0.013459 (rank : 169) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
SPT5H_HUMAN
|
||||||
| θ value | 0.813845 (rank : 64) | NC score | 0.054986 (rank : 64) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O00267, O43279, Q59G52, Q99639 | Gene names | SUPT5H, SPT5, SPT5H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (hSPT5) (DRB sensitivity-inducing factor large subunit) (DSIF large subunit) (DSIF p160) (Tat- cotransactivator 1 protein) (Tat-CT1 protein). | |||||
|
ZEP1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 65) | NC score | 0.020075 (rank : 145) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||
|
CBX8_HUMAN
|
||||||
| θ value | 1.06291 (rank : 66) | NC score | 0.043595 (rank : 87) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9HC52, Q96H39, Q9NR07 | Gene names | CBX8, PC3, RC1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 8 (Polycomb 3 homolog) (Pc3) (hPc3) (Rectachrome 1). | |||||
|
EHMT1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 67) | NC score | 0.013378 (rank : 170) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9H9B1, Q8TCN7, Q96F53, Q96JF1, Q96KH4 | Gene names | EHMT1, EUHMTASE1, KIAA1876 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 5 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 5) (H3-K9-HMTase 5) (Euchromatic histone-lysine N-methyltransferase 1) (Eu-HMTase1) (G9a- like protein 1) (GLP1). | |||||
|
GRIN1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 68) | NC score | 0.048142 (rank : 79) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
MYT1L_HUMAN
|
||||||
| θ value | 1.06291 (rank : 69) | NC score | 0.027024 (rank : 126) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UL68, Q6IQ17, Q9UPP6 | Gene names | MYT1L, KIAA1106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (MyT1-L). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 1.06291 (rank : 70) | NC score | 0.034188 (rank : 113) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
AKP8L_HUMAN
|
||||||
| θ value | 1.38821 (rank : 71) | NC score | 0.034487 (rank : 111) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9ULX6, O94792, Q96J58, Q9NRQ0, Q9UGM0 | Gene names | AKAP8L, NAKAP95 | |||
|
Domain Architecture |
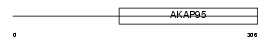
|
|||||
| Description | A-kinase anchor protein 8-like (AKAP8-like protein) (Neighbor of A- kinase-anchoring protein 95) (Neighbor of AKAP95) (Homologous to AKAP95 protein) (HA95) (Helicase A-binding protein 95) (HAP95). | |||||
|
BPTF_HUMAN
|
||||||
| θ value | 1.38821 (rank : 72) | NC score | 0.047571 (rank : 81) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
GLU2B_MOUSE
|
||||||
| θ value | 1.38821 (rank : 73) | NC score | 0.045197 (rank : 84) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O08795, Q921X2 | Gene names | Prkcsh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glucosidase 2 subunit beta precursor (Glucosidase II subunit beta) (Protein kinase C substrate, 60.1 kDa protein, heavy chain) (PKCSH) (80K-H protein). | |||||
|
IRX2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 74) | NC score | 0.017997 (rank : 150) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P81066, O55121, Q9ERN1 | Gene names | Irx2, Irxa2 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-2 (Iroquois homeobox protein 2) (Homeodomain protein IRXA2). | |||||
|
MINK1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 75) | NC score | 0.007127 (rank : 188) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8N4C8, Q9P1X1, Q9P2R8 | Gene names | MINK1, MAP4K6, MINK | |||
|
Domain Architecture |

|
|||||
| Description | Misshapen-like kinase 1 (EC 2.7.11.1) (Mitogen-activated protein kinase kinase kinase kinase 6) (MAPK/ERK kinase kinase kinase 6) (MEK kinase kinase 6) (MEKKK 6) (Misshapen/NIK-related kinase) (GCK family kinase MiNK). | |||||
|
RREB1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 76) | NC score | 0.014057 (rank : 166) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 879 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q92766, O75567 | Gene names | RREB1 | |||
|
Domain Architecture |

|
|||||
| Description | RAS-responsive element-binding protein 1 (RREB-1) (Raf-responsive zinc finger protein LZ321). | |||||
|
ZFHX2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 77) | NC score | 0.020459 (rank : 143) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9C0A1 | Gene names | ZFHX2, KIAA1762 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger homeobox protein 2. | |||||
|
ZN513_HUMAN
|
||||||
| θ value | 1.38821 (rank : 78) | NC score | 0.003424 (rank : 194) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N8E2 | Gene names | ZNF513 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 513. | |||||
|
2B32_HUMAN
|
||||||
| θ value | 1.81305 (rank : 79) | NC score | 0.010923 (rank : 179) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P01913, O78049, P79663, Q29721, Q29809, Q30144, Q9MYA4, Q9MYH3, Q9MYH4, Q9TPB5, Q9TQ21 | Gene names | HLA-DRB3 | |||
|
Domain Architecture |
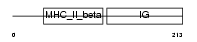
|
|||||
| Description | HLA class II histocompatibility antigen, DRB3-2 beta chain precursor (MHC class I antigen DRB3*2). | |||||
|
BRDT_MOUSE
|
||||||
| θ value | 1.81305 (rank : 80) | NC score | 0.025214 (rank : 132) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q91Y44, Q59HJ4 | Gene names | Brdt, Fsrg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain testis-specific protein (RING3-like protein) (Bromodomain- containing female sterile homeotic-like protein). | |||||
|
CHD1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 81) | NC score | 0.042663 (rank : 91) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
CHD1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 82) | NC score | 0.040268 (rank : 98) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P40201 | Gene names | Chd1, Chd-1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
CT111_HUMAN
|
||||||
| θ value | 1.81305 (rank : 83) | NC score | 0.035052 (rank : 109) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NX31, O95912, Q9NZ84, Q9P0R8 | Gene names | C20orf111 | |||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf111. | |||||
|
FOXD1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 84) | NC score | 0.014492 (rank : 162) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61345 | Gene names | Foxd1, Fkhl8, Freac4, Hfhbf2 | |||
|
Domain Architecture |
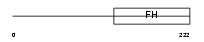
|
|||||
| Description | Forkhead box protein D1 (Forkhead-related protein FKHL8) (Forkhead- related transcription factor 4) (FREAC-4) (Brain factor 2) (HFH-BF-2). | |||||
|
NF2L1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 85) | NC score | 0.034470 (rank : 112) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14494, Q12877, Q96FN6 | Gene names | NFE2L1, HBZ17, NRF1, TCF11 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 1 (NF-E2-related factor 1) (NFE2-related factor 1) (Nuclear factor, erythroid derived 2, like 1) (Transcription factor 11) (Transcription factor HBZ17) (Transcription factor LCR-F1) (Locus control region-factor 1). | |||||
|
PTN21_HUMAN
|
||||||
| θ value | 1.81305 (rank : 86) | NC score | 0.008657 (rank : 185) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16825 | Gene names | PTPN21, PTPD1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 21 (EC 3.1.3.48) (Protein-tyrosine phosphatase D1). | |||||
|
SALL2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 87) | NC score | 0.016366 (rank : 155) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 835 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y467, Q9Y4G1 | Gene names | SALL2, KIAA0360, SAL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 2 (Zinc finger protein SALL2) (HSal2). | |||||
|
SMBT1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 88) | NC score | 0.018700 (rank : 147) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UHJ3, Q96C73, Q9Y4Q9 | Gene names | SFMBT1, RU1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scm-like with four MBT domains protein 1 (Renal ubiquitous protein 1). | |||||
|
SPTA1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 89) | NC score | 0.015818 (rank : 159) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P02549, Q15514, Q6LDY5 | Gene names | SPTA1, SPTA | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 90) | NC score | 0.055845 (rank : 61) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
TCF20_HUMAN
|
||||||
| θ value | 1.81305 (rank : 91) | NC score | 0.042700 (rank : 90) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9UGU0, O14528, Q13078, Q4V353, Q9H4M0 | Gene names | TCF20, KIAA0292, SPBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP) (AR1). | |||||
|
CD2L1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 92) | NC score | 0.013145 (rank : 172) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1463 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P24788, Q3UI03, Q61399, Q7TST4, Q8BP53 | Gene names | Cdc2l1 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1). | |||||
|
CHD9_MOUSE
|
||||||
| θ value | 2.36792 (rank : 93) | NC score | 0.031689 (rank : 117) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
CLUA1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 94) | NC score | 0.037375 (rank : 102) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96AJ1, O75138, Q65ZA3, Q9H8R4, Q9H8T1 | Gene names | CLUAP1, KIAA0643 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Clusterin-associated protein 1. | |||||
|
HTSF1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 95) | NC score | 0.062875 (rank : 48) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BGC0, Q1WWK0, Q9CT41, Q9DAU3 | Gene names | Htatsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 homolog. | |||||
|
IF4B_HUMAN
|
||||||
| θ value | 2.36792 (rank : 96) | NC score | 0.050554 (rank : 76) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P23588 | Gene names | EIF4B | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
NASP_HUMAN
|
||||||
| θ value | 2.36792 (rank : 97) | NC score | 0.079683 (rank : 33) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 2.36792 (rank : 98) | NC score | 0.084821 (rank : 28) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
PHF14_HUMAN
|
||||||
| θ value | 2.36792 (rank : 99) | NC score | 0.036154 (rank : 104) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O94880 | Gene names | PHF14, KIAA0783 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 14. | |||||
|
ZN750_MOUSE
|
||||||
| θ value | 2.36792 (rank : 100) | NC score | 0.059620 (rank : 50) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BH05, Q66JP3, Q8C0L1 | Gene names | Znf750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
ARI3A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 101) | NC score | 0.026712 (rank : 127) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q62431 | Gene names | Arid3a, Dri1, Dril1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 3A (ARID domain- containing protein 3A) (Dead ringer-like protein 1) (B-cell regulator of IgH transcription) (Bright). | |||||
|
ATBF1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 102) | NC score | 0.023643 (rank : 137) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
BRD9_MOUSE
|
||||||
| θ value | 3.0926 (rank : 103) | NC score | 0.025676 (rank : 129) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3UQU0, Q5FWH1, Q811F7 | Gene names | Brd9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 9. | |||||
|
CD2L1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 104) | NC score | 0.012513 (rank : 176) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
CD2L5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 105) | NC score | 0.009249 (rank : 184) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q14004, Q6DKQ9, Q75MH4, Q75MH5, Q96JN4, Q9H4A0, Q9H4A1, Q9UDR4 | Gene names | CDC2L5, CDC2L, CHED, KIAA1791 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle 2-like protein kinase 5 (EC 2.7.11.22) (CDC2- related protein kinase 5) (Cholinesterase-related cell division controller). | |||||
|
CLUA1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 106) | NC score | 0.040949 (rank : 95) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8R3P7, Q6P3Y8 | Gene names | Cluap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Clusterin-associated protein 1. | |||||
|
CPSF1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 107) | NC score | 0.021391 (rank : 141) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EPU4 | Gene names | Cpsf1, Cpsf160 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 160 kDa subunit (CPSF 160 kDa subunit). | |||||
|
DAXX_HUMAN
|
||||||
| θ value | 3.0926 (rank : 108) | NC score | 0.055210 (rank : 63) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UER7, O14747, O15141, O15208, Q9BWI3 | Gene names | DAXX, BING2, DAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Death domain-associated protein 6 (Daxx) (hDaxx) (Fas death domain- associated protein) (ETS1-associated protein 1) (EAP1). | |||||
|
DDX51_HUMAN
|
||||||
| θ value | 3.0926 (rank : 109) | NC score | 0.014812 (rank : 161) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8N8A6, Q5CZ71, Q8IXK5, Q96ED1 | Gene names | DDX51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
FXR2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 110) | NC score | 0.016499 (rank : 154) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P51116, Q8WUM2 | Gene names | FXR2 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 2. | |||||
|
NFM_HUMAN
|
||||||
| θ value | 3.0926 (rank : 111) | NC score | 0.015964 (rank : 158) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
PHF14_MOUSE
|
||||||
| θ value | 3.0926 (rank : 112) | NC score | 0.035406 (rank : 107) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9D4H9, Q810Z6, Q8C284, Q8CGF8, Q9D1Q8 | Gene names | Phf14 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 14. | |||||
|
PIAS4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 113) | NC score | 0.013965 (rank : 167) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JM05, Q8R165 | Gene names | Pias4, Piasg | |||
|
Domain Architecture |

|
|||||
| Description | Protein inhibitor of activated STAT protein 4 (Protein inhibitor of activated STAT protein gamma) (PIAS-gamma) (PIASy). | |||||
|
RUSC2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 114) | NC score | 0.024835 (rank : 133) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8N2Y8, O15080, Q6P1W7 | Gene names | RUSC2, KIAA0375 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 2. | |||||
|
TPR_HUMAN
|
||||||
| θ value | 3.0926 (rank : 115) | NC score | 0.011060 (rank : 178) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
UN13A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 116) | NC score | 0.019557 (rank : 146) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UPW8 | Gene names | UNC13A, KIAA1032 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog A (Munc13-1). | |||||
|
CASC3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 117) | NC score | 0.034187 (rank : 114) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8K3W3, Q8K219, Q99NF0 | Gene names | Casc3, Mln51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC3 (Cancer susceptibility candidate gene 3 protein homolog) (Metastatic lymph node protein 51 homolog) (Protein MLN 51 homolog) (Protein barentsz) (Btz). | |||||
|
CHIC1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 118) | NC score | 0.031460 (rank : 118) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CBW7, O08904, Q8K3S5 | Gene names | Chic1, Brx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich hydrophobic domain 1 protein (Brain X-linked protein). | |||||
|
PCNT_HUMAN
|
||||||
| θ value | 4.03905 (rank : 119) | NC score | 0.017824 (rank : 152) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
PITM2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 120) | NC score | 0.012699 (rank : 174) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BZ72, Q9P271 | Gene names | PITPNM2, KIAA1457, NIR3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 2 (Phosphatidylinositol transfer protein, membrane-associated 2) (Pyk2 N-terminal domain-interacting receptor 3) (NIR-3). | |||||
|
RB3GP_MOUSE
|
||||||
| θ value | 4.03905 (rank : 121) | NC score | 0.020079 (rank : 144) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80UJ7, Q6A0D7, Q8C4Y0, Q8C679, Q8C6A5, Q8K324 | Gene names | Rab3gap1, Kiaa0066, Rab3gap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab3 GTPase-activating protein catalytic subunit (RAB3 GTPase- activating protein 130 kDa subunit) (Rab3-GAP p130) (Rab3-GAP). | |||||
|
ZBT20_HUMAN
|
||||||
| θ value | 4.03905 (rank : 122) | NC score | 0.003710 (rank : 193) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 835 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9HC78 | Gene names | ZBTB20, DPZF, ZNF288 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 20 (Zinc finger protein 288) (Dendritic-derived BTB/POZ zinc finger protein). | |||||
|
ZFY27_MOUSE
|
||||||
| θ value | 4.03905 (rank : 123) | NC score | 0.027898 (rank : 124) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3TXX3, Q8CFP8 | Gene names | Zfyve27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 27. | |||||
|
ZN326_MOUSE
|
||||||
| θ value | 4.03905 (rank : 124) | NC score | 0.029464 (rank : 119) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O88291, Q8BSJ5, Q8K1X9, Q9CYG9 | Gene names | Znf326, Zan75, Zfp326 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 326 (Zinc finger protein-associated with nuclear matrix of 75 kDa). | |||||
|
AN32B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 125) | NC score | 0.023959 (rank : 136) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q92688, O00655, P78458, P78459 | Gene names | ANP32B, APRIL, PHAPI2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member B (PHAPI2 protein) (Silver-stainable protein SSP29) (Acidic protein rich in leucines). | |||||
|
ATAD2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 126) | NC score | 0.029009 (rank : 121) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6PL18, Q658P2, Q68CQ0, Q6PJV6, Q8N890, Q9UHS5 | Gene names | ATAD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
BASP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 127) | NC score | 0.057746 (rank : 55) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
C8AP2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 128) | NC score | 0.035779 (rank : 105) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9WUF3, Q9CSW2 | Gene names | Casp8ap2, Flash | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
CD2L5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 129) | NC score | 0.009292 (rank : 183) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1011 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q69ZA1, Q80V11, Q8BZG1, Q8K0A4 | Gene names | Cdc2l5, Kiaa1791 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle 2-like protein kinase 5 (EC 2.7.11.22) (CDC2- related protein kinase 5). | |||||
|
FMN2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 130) | NC score | 0.018322 (rank : 148) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NZ56, Q59GF6, Q5VU37, Q9NZ55 | Gene names | FMN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-2. | |||||
|
HTSF1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 131) | NC score | 0.043501 (rank : 88) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O43719, Q59G06, Q99730 | Gene names | HTATSF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 (Tat-SF1). | |||||
|
MARCS_MOUSE
|
||||||
| θ value | 5.27518 (rank : 132) | NC score | 0.042968 (rank : 89) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P26645 | Gene names | Marcks, Macs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS). | |||||
|
MYCB2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 133) | NC score | 0.047699 (rank : 80) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O75592, Q5JSX8, Q5VZN6, Q6PIB6, Q9UQ11, Q9Y6E4 | Gene names | MYCBP2, KIAA0916, PAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
ZCPW1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 134) | NC score | 0.024634 (rank : 134) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6IR42 | Gene names | Zcwpw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1 homolog. | |||||
|
AP3B1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 135) | NC score | 0.055215 (rank : 62) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O00203, O00580, Q7Z393, Q9HD66 | Gene names | AP3B1, ADTB3A | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
ARS2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 136) | NC score | 0.032153 (rank : 116) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BXP5, O95808, Q9BWP6, Q9BXP4 | Gene names | ARS2, ASR2 | |||
|
Domain Architecture |

|
|||||
| Description | Arsenite-resistance protein 2. | |||||
|
ECM2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 137) | NC score | 0.003404 (rank : 195) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O94769 | Gene names | ECM2 | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular matrix protein 2 precursor (Matrix glycoprotein SC1/ECM2). | |||||
|
HUWE1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 138) | NC score | 0.012719 (rank : 173) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
LPIN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 139) | NC score | 0.009854 (rank : 182) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14693 | Gene names | LPIN1, KIAA0188 | |||
|
Domain Architecture |
|
|||||
| Description | Lipin-1. | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 140) | NC score | 0.025655 (rank : 130) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MYO3A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 141) | NC score | 0.003042 (rank : 197) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1033 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8NEV4, Q8WX17, Q9NYS8 | Gene names | MYO3A | |||
|
Domain Architecture |

|
|||||
| Description | Myosin IIIA (EC 2.7.11.1). | |||||
|
NCOR1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 142) | NC score | 0.025429 (rank : 131) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
NDF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 143) | NC score | 0.012665 (rank : 175) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13562, O00343, Q13340, Q5U095, Q96TH0, Q99455, Q9UEC8 | Gene names | NEUROD1, NEUROD | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 1 (NeuroD1) (NeuroD). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 6.88961 (rank : 144) | NC score | 0.042574 (rank : 92) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
NKTR_MOUSE
|
||||||
| θ value | 6.88961 (rank : 145) | NC score | 0.028097 (rank : 123) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P30415 | Gene names | Nktr | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
PTMA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 146) | NC score | 0.071081 (rank : 35) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
RERE_MOUSE
|
||||||
| θ value | 6.88961 (rank : 147) | NC score | 0.022284 (rank : 140) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
RFTN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 148) | NC score | 0.016299 (rank : 156) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14699, Q496Y2, Q4QQI7, Q5JB48, Q7Z7P2 | Gene names | RFTN1, KIAA0084 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Raftlin (Raft-linking protein) (Cell migration-inducing gene 2 protein). | |||||
|
SFRS4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 149) | NC score | 0.034774 (rank : 110) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q08170, Q5VXP1, Q9BUA4, Q9UEB5 | Gene names | SFRS4, SRP75 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4 (Pre-mRNA-splicing factor SRP75) (SRP001LB). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 150) | NC score | 0.054428 (rank : 65) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TCF8_MOUSE
|
||||||
| θ value | 6.88961 (rank : 151) | NC score | 0.007616 (rank : 187) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1067 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q64318, Q62519 | Gene names | Tcf8, Zfhx1a, Zfx1a, Zfx1ha | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (Zinc finger homeobox protein 1a) (MEB1) (Delta EF1). | |||||
|
TF3C3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 152) | NC score | 0.026141 (rank : 128) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y5Q9, Q86XJ8, Q8WX84, Q96B44, Q9H5I8, Q9NT97 | Gene names | GTF3C3 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor 3C polypeptide 3 (Transcription factor IIIC-subunit gamma) (TF3C-gamma) (TFIIIC 102 kDa subunit) (TFIIIC102). | |||||
|
YYAP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 153) | NC score | 0.013284 (rank : 171) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H869, Q5VYZ4, Q5VYZ7, Q7L4C3, Q7L5E2, Q8IXA6, Q8TEW5, Q8TF04, Q96HB6, Q9BQ64, Q9NV84 | Gene names | YY1AP1, HCCA2, YY1AP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YY1-associated protein 1 (Hepatocellular carcinoma susceptibility protein) (Hepatocellular carcinoma-associated protein 2). | |||||
|
ZEP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 154) | NC score | 0.016278 (rank : 157) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1036 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P15822, Q14122 | Gene names | HIVEP1, ZNF40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 40 (Human immunodeficiency virus type I enhancer- binding protein 1) (HIV-EP1) (Major histocompatibility complex-binding protein 1) (MBP-1) (Positive regulatory domain II-binding factor 1) (PRDII-BF1). | |||||
|
AGGF1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 155) | NC score | 0.018196 (rank : 149) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7TN31, Q8R2S6, Q9CQR9, Q9CU87, Q9D768 | Gene names | Aggf1, Vg5q | |||
|
Domain Architecture |

|
|||||
| Description | Angiogenic factor with G patch and FHA domains 1 (Angiogenic factor VG5Q) (mVG5Q). | |||||
|
AN32B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 156) | NC score | 0.027818 (rank : 125) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9EST5 | Gene names | Anp32b, Pal31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member B (Proliferation-related acidic leucine-rich protein PAL31). | |||||
|
AP3B2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 157) | NC score | 0.053859 (rank : 69) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
ARI3A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 158) | NC score | 0.028559 (rank : 122) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99856, Q8IZA7 | Gene names | ARID3A, DRIL1, DRX, E2FBP1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 3A (ARID domain- containing protein 3A) (B-cell regulator of IgH transcription) (Bright) (E2F-binding protein 1). | |||||
|
CA026_HUMAN
|
||||||
| θ value | 8.99809 (rank : 159) | NC score | 0.051865 (rank : 73) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5T5J6, Q8NEK9, Q9BZQ7, Q9NXQ0 | Gene names | C1orf26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf26. | |||||
|
CAND2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 160) | NC score | 0.008340 (rank : 186) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6ZQ73, Q6PHT7 | Gene names | Cand2, Kiaa0667, Tip120b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cullin-associated NEDD8-dissociated protein 2 (Cullin-associated and neddylation-dissociated protein 2) (p120 CAND2) (TBP-interacting protein TIP120B) (TBP-interacting protein of 120 kDa B). | |||||
|
CHIC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 161) | NC score | 0.022885 (rank : 138) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5VXU3 | Gene names | CHIC1, BRX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich hydrophobic domain 1 protein (Brain X-linked protein). | |||||
|
DMP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 162) | NC score | 0.095937 (rank : 18) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
DOK7_MOUSE
|
||||||
| θ value | 8.99809 (rank : 163) | NC score | 0.015073 (rank : 160) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q18PE0, Q3TCZ6, Q5FW70, Q8C8U7 | Gene names | Dok7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Dok-7 (Downstream of tyrosine kinase 7). | |||||
|
E41L3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 164) | NC score | 0.013811 (rank : 168) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9WV92, Q9R102 | Gene names | Epb41l3, Dal1, Epb4.1l3 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 3 (4.1B) (Differentially expressed in adenocarcinoma of the lung protein 1) (DAL-1) (DAL1P) (mDAL-1). | |||||
|
IASPP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 165) | NC score | 0.005420 (rank : 189) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8WUF5, Q2PNZ9, Q5DU71, Q5I1X4, Q6P1R7, Q6PKF8, Q9Y290 | Gene names | PPP1R13L, IASPP, NKIP1, PPP1R13BL, RAI | |||
|
Domain Architecture |

|
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
IF4B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 166) | NC score | 0.039410 (rank : 100) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BGD9, Q3TD64 | Gene names | Eif4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
JHD2B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 167) | NC score | 0.010573 (rank : 180) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7LBC6, Q9BVH6, Q9BW93, Q9BZ52, Q9NYF4, Q9UPS0 | Gene names | JMJD1B, C5orf7, JHDM2B, KIAA1082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2B (EC 1.14.11.-) (Jumonji domain-containing protein 1B) (Nuclear protein 5qNCA). | |||||
|
NDF1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 168) | NC score | 0.012482 (rank : 177) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q60867, Q545N9, Q60897 | Gene names | Neurod1, Neurod | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 1 (NeuroD1). | |||||
|
OGT1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 169) | NC score | 0.024596 (rank : 135) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O15294, Q7Z3K0, Q8WWM8, Q96CC1, Q9UG57 | Gene names | OGT | |||
|
Domain Architecture |

|
|||||
| Description | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit (EC 2.4.1.-) (O-GlcNAc transferase p110 subunit). | |||||
|
RBM28_MOUSE
|
||||||
| θ value | 8.99809 (rank : 170) | NC score | 0.014097 (rank : 165) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8CGC6, Q8BG25, Q8VEJ8, Q9CS22, Q9CSE6 | Gene names | Rbm28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 28 (RNA-binding motif protein 28). | |||||
|
SRCH_HUMAN
|
||||||
| θ value | 8.99809 (rank : 171) | NC score | 0.035776 (rank : 106) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P23327, Q504Y6 | Gene names | HRC, HCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcoplasmic reticulum histidine-rich calcium-binding protein precursor. | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 172) | NC score | 0.047270 (rank : 82) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
STK3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 173) | NC score | 0.003130 (rank : 196) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 869 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9JI10, Q60877, Q80UG4, Q8CI58 | Gene names | Stk3, Mess1, Mst2 | |||
|
Domain Architecture |
|
|||||
| Description | Serine/threonine-protein kinase 3 (EC 2.7.11.1) (STE20-like kinase MST2) (MST-2) (Mammalian STE20-like protein kinase 2). | |||||
|
SYP2L_HUMAN
|
||||||
| θ value | 8.99809 (rank : 174) | NC score | 0.010480 (rank : 181) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H987 | Gene names | SYNPO2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin 2-like protein. | |||||
|
TEKT3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 175) | NC score | 0.017413 (rank : 153) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BXF9, Q8N5R5, Q96M48 | Gene names | TEKT3 | |||
|
Domain Architecture |
|
|||||
| Description | Tektin-3. | |||||
|
TLE3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 176) | NC score | 0.005000 (rank : 191) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q04726, Q8IVV6, Q8WVR2, Q9HCM5 | Gene names | TLE3, KIAA1547 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 3 (ESG3). | |||||
|
TRAIP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 177) | NC score | 0.005324 (rank : 190) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BWF2, O00467 | Gene names | TRAIP, TRIP | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
TTC7B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 178) | NC score | 0.014228 (rank : 164) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86TV6, Q86U24, Q86VT3 | Gene names | TTC7B, TTC7L1 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 7B (Tetratricopeptide repeat protein 7-like-1). | |||||
|
UBF1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 179) | NC score | 0.040563 (rank : 97) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P25976 | Gene names | Ubtf, Tcfubf, Ubf-1, Ubf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1). | |||||
|
YTDC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 180) | NC score | 0.057397 (rank : 56) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
ADDB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.051564 (rank : 74) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
AFF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.064837 (rank : 44) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
AFF3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.062390 (rank : 49) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P51826 | Gene names | AFF3, LAF4 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 3 (Protein LAF-4) (Lymphoid nuclear protein related to AF4). | |||||
|
AKA12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.063818 (rank : 45) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
ERIC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.079700 (rank : 32) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
FILA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.056123 (rank : 58) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
GNAS3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.066767 (rank : 40) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95467, O95417 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroendocrine secretory protein 55 (NESP55) [Contains: LHAL tetrapeptide; GPIPIRRH peptide]. | |||||
|
LEO1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.110692 (rank : 13) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 103 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 189) | NC score | 0.088828 (rank : 23) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
NSBP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 190) | NC score | 0.077526 (rank : 34) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 191) | NC score | 0.070570 (rank : 36) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
RBBP6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 192) | NC score | 0.055961 (rank : 59) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
SPRL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 193) | NC score | 0.050428 (rank : 77) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P70663, P97810, Q99L82 | Gene names | Sparcl1, Ecm2, Sc1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (Matrix glycoprotein Sc1) (Extracellular matrix protein 2). | |||||
|
TCAL5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 194) | NC score | 0.088044 (rank : 24) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 195) | NC score | 0.104374 (rank : 16) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
TXND2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 196) | NC score | 0.052298 (rank : 71) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
TXND2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 197) | NC score | 0.068225 (rank : 38) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
SMG5_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 180 | |
| SwissProt Accessions | Q9UPR3, Q5QJE7, Q8IXC0, Q8IY09, Q96IJ7 | Gene names | SMG5, EST1B, KIAA1089 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein SMG5 (SMG-5 homolog) (EST1-like protein B) (LPTS-interacting protein) (LPTS-RP1). | |||||
|
SMG5_MOUSE
|
||||||
| NC score | 0.952543 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6ZPY2, Q497S2, Q8R3S4 | Gene names | Smg5, Est1b, Kiaa1089 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein SMG5 (SMG-5 homolog) (EST1-like protein B). | |||||
|
SMG7_MOUSE
|
||||||
| NC score | 0.469725 (rank : 3) | θ value | 4.44505e-15 (rank : 3) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5RJH6, Q63ZW5, Q6ZQF3 | Gene names | Smg7, Est1c, Kiaa0250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein SMG7 (SMG-7 homolog) (EST1-like protein C). | |||||
|
SMG7_HUMAN
|
||||||
| NC score | 0.467036 (rank : 4) | θ value | 1.29331e-14 (rank : 4) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q92540, Q5T1Q0, Q6PIE0, Q7Z7H9, Q8IXC1, Q8IXC2 | Gene names | SMG7, C1orf16, EST1C, KIAA0250 | |||
|
Domain Architecture |

|
|||||
| Description | Protein SMG7 (SMG-7 homolog) (EST1-like protein C). | |||||
|
EST1A_HUMAN
|
||||||
| NC score | 0.439479 (rank : 5) | θ value | 1.42992e-13 (rank : 5) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86US8, O94837, Q86VH6, Q9UF60 | Gene names | EST1A, C17orf31, KIAA0732 | |||
|
Domain Architecture |

|
|||||
| Description | Telomerase-binding protein EST1A (Ever shorter telomeres 1A) (Telomerase subunit EST1A) (EST1-like protein A) (hSmg5/7a). | |||||
|
EST1A_MOUSE
|
||||||
| NC score | 0.425382 (rank : 6) | θ value | 1.2105e-12 (rank : 6) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P61406, Q5NC64 | Gene names | Est1a, Kiaa0732 | |||
|
Domain Architecture |

|
|||||
| Description | Telomerase-binding protein EST1A (Ever shorter telomeres 1A) (Telomerase subunit EST1A) (EST1-like protein A). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.158131 (rank : 7) | θ value | 0.0148317 (rank : 12) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 110 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.154004 (rank : 8) | θ value | 0.0252991 (rank : 17) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 117 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
T2FA_HUMAN
|
||||||
| NC score | 0.151009 (rank : 9) | θ value | 0.00102713 (rank : 7) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.132418 (rank : 10) | θ value | 0.365318 (rank : 41) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 108 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
GNAS3_MOUSE
|
||||||
| NC score | 0.125674 (rank : 11) | θ value | 0.0148317 (rank : 11) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Z0F1, Q9QXW5, Q9Z0H2 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroendocrine secretory protein 55 (NESP55) [Contains: LHAL tetrapeptide; GPIPIRRH peptide]. | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.120446 (rank : 12) | θ value | 0.00869519 (rank : 9) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.110692 (rank : 13) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 103 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.108795 (rank : 14) | θ value | 0.00509761 (rank : 8) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
AFF4_MOUSE
|
||||||
| NC score | 0.106120 (rank : 15) | θ value | 0.0148317 (rank : 10) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.104374 (rank : 16) | θ value | θ > 10 (rank : 195) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
AFF4_HUMAN
|
||||||
| NC score | 0.101430 (rank : 17) | θ value | 0.0193708 (rank : 13) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
DMP1_HUMAN
|
||||||
| NC score | 0.095937 (rank : 18) | θ value | 8.99809 (rank : 162) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
DMP1_MOUSE
|
||||||
| NC score | 0.092993 (rank : 19) | θ value | 0.62314 (rank : 52) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
AFF1_MOUSE
|
||||||
| NC score | 0.092619 (rank : 20) | θ value | 0.163984 (rank : 28) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
PTDSR_MOUSE
|
||||||
| NC score | 0.092590 (rank : 21) | θ value | 0.21417 (rank : 33) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ERI5, Q80TX1 | Gene names | Ptdsr, Kiaa0585 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein PTDSR (Phosphatidylserine receptor) (Apoptotic cell clearance receptor PtdSerR). | |||||
|
PTDSR_HUMAN
|
||||||
| NC score | 0.090523 (rank : 22) | θ value | 0.0961366 (rank : 26) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6NYC1, Q86VY0, Q8IUM5, Q9Y4E2 | Gene names | PTDSR, KIAA0585 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein PTDSR (Phosphatidylserine receptor). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.088828 (rank : 23) | θ value | θ > 10 (rank : 189) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
TCAL5_HUMAN
|
||||||
| NC score | 0.088044 (rank : 24) | θ value | θ > 10 (rank : 194) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
RGP1_MOUSE
|
||||||
| NC score | 0.088000 (rank : 25) | θ value | 0.0431538 (rank : 21) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P46061, Q60801 | Gene names | Rangap1, Fug1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
CDC16_HUMAN
|
||||||
| NC score | 0.086867 (rank : 26) | θ value | 0.62314 (rank : 50) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13042, Q96AE6, Q9Y564 | Gene names | CDC16 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 16 homolog (CDC16Hs) (Anaphase-promoting complex subunit 6) (APC6) (Cyclosome subunit 6). | |||||
|
CYLC2_HUMAN
|
||||||
| NC score | 0.085189 (rank : 27) | θ value | 0.62314 (rank : 51) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.084821 (rank : 28) | θ value | 2.36792 (rank : 98) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
CDC16_MOUSE
|
||||||
| NC score | 0.084414 (rank : 29) | θ value | 0.813845 (rank : 60) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8R349, Q9CYX9 | Gene names | Cdc16 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 16 homolog (Anaphase-promoting complex subunit 6) (APC6) (Cyclosome subunit 6). | |||||
|
NSBP1_HUMAN
|
||||||
| NC score | 0.083408 (rank : 30) | θ value | 0.163984 (rank : 30) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
CYLC1_HUMAN
|
||||||
| NC score | 0.080580 (rank : 31) | θ value | 0.0431538 (rank : 20) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
ERIC1_HUMAN
|
||||||
| NC score | 0.079700 (rank : 32) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
NASP_HUMAN
|
||||||
| NC score | 0.079683 (rank : 33) | θ value | 2.36792 (rank : 97) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
NSBP1_MOUSE
|
||||||
| NC score | 0.077526 (rank : 34) | θ value | θ > 10 (rank : 190) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
PTMA_MOUSE
|
||||||
| NC score | 0.071081 (rank : 35) | θ value | 6.88961 (rank : 146) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.070570 (rank : 36) | θ value | θ > 10 (rank : 191) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
PHAR3_MOUSE
|
||||||
| NC score | 0.068316 (rank : 37) | θ value | 0.0736092 (rank : 25) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BYK5, Q8BYS8, Q8C058, Q9DB87 | Gene names | Phactr3, Scapin1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatase and actin regulator 3 (Scapinin) (Scaffold-associated PP1- inhibiting protein). | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.068225 (rank : 38) | θ value | θ > 10 (rank : 197) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
AP3B2_HUMAN
|
||||||
| NC score | 0.066836 (rank : 39) | θ value | 0.0736092 (rank : 23) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13367, O14808 | Gene names | AP3B2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (AP-3 complex beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain) (Neuron-specific vesicle coat protein beta-NAP). | |||||
|
GNAS3_HUMAN
|
||||||
| NC score | 0.066767 (rank : 40) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95467, O95417 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroendocrine secretory protein 55 (NESP55) [Contains: LHAL tetrapeptide; GPIPIRRH peptide]. | |||||
|
MRP_MOUSE
|
||||||
| NC score | 0.066573 (rank : 41) | θ value | 0.365318 (rank : 42) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P28667, Q3TEZ4, Q91W07 | Gene names | Marcksl1, Mlp, Mrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MARCKS-related protein (MARCKS-like protein 1) (Macrophage myristoylated alanine-rich C kinase substrate) (Mac-MARCKS) (MacMARCKS) (Brain protein F52). | |||||
|
DAXX_MOUSE
|
||||||
| NC score | 0.066487 (rank : 42) | θ value | 0.813845 (rank : 62) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O35613, Q9QWT8, Q9QWV3 | Gene names | Daxx | |||
|
Domain Architecture |

|
|||||
| Description | Death domain-associated protein 6 (Daxx). | |||||
|
CTR9_MOUSE
|
||||||
| NC score | 0.065785 (rank : 43) | θ value | 0.365318 (rank : 39) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q62018, Q3UFF5, Q3UY40, Q66JX4, Q7TPS6, Q8BND9, Q8BRD1, Q8C9W7, Q8C9Y3, Q8CHI1 | Gene names | Ctr9, Kiaa0155, Sh2bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1) (Tetratricopeptide repeat-containing, SH2-binding phosphoprotein of 150 kDa) (TPR-containing, SH2-binding phosphoprotein of 150 kDa) (p150TSP). | |||||
|
AFF1_HUMAN
|
||||||
| NC score | 0.064837 (rank : 44) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.063818 (rank : 45) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
ABEC3_MOUSE
|
||||||
| NC score | 0.063542 (rank : 46) | θ value | 0.0330416 (rank : 19) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99J72, Q8C7L0, Q8C8V7 | Gene names | Apobec3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA dC->dU-editing enzyme APOBEC-3 (EC 3.5.4.-) (Apolipoprotein B mRNA-editing complex 3) (Arp3) (CEM15) (CEM-15). | |||||
|
AP3B1_MOUSE
|
||||||
| NC score | 0.063233 (rank : 47) | θ value | 0.47712 (rank : 45) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Z1T1, Q91YR4 | Gene names | Ap3b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
HTSF1_MOUSE
|
||||||
| NC score | 0.062875 (rank : 48) | θ value | 2.36792 (rank : 95) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BGC0, Q1WWK0, Q9CT41, Q9DAU3 | Gene names | Htatsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 homolog. | |||||
|
AFF3_HUMAN
|
||||||
| NC score | 0.062390 (rank : 49) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P51826 | Gene names | AFF3, LAF4 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 3 (Protein LAF-4) (Lymphoid nuclear protein related to AF4). | |||||
|
ZN750_MOUSE
|
||||||
| NC score | 0.059620 (rank : 50) | θ value | 2.36792 (rank : 100) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BH05, Q66JP3, Q8C0L1 | Gene names | Znf750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
SRCA_HUMAN
|
||||||
| NC score | 0.059565 (rank : 51) | θ value | 0.0563607 (rank : 22) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.059285 (rank : 52) | θ value | 0.279714 (rank : 37) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
PHAR3_HUMAN
|
||||||
| NC score | 0.058309 (rank : 53) | θ value | 0.62314 (rank : 54) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96KR7, Q707P6, Q9H4T4 | Gene names | PHACTR3, C20orf101, SCAPIN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatase and actin regulator 3 (Scapinin) (Scaffold-associated PP1- inhibiting protein). | |||||
|
TRI44_HUMAN
|
||||||
| NC score | 0.057849 (rank : 54) | θ value | 0.163984 (rank : 31) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96DX7, Q96QY2, Q9UGK0 | Gene names | TRIM44, DIPB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB). | |||||
|
BASP_HUMAN
|
||||||
| NC score | 0.057746 (rank : 55) | θ value | 5.27518 (rank : 127) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
YTDC1_HUMAN
|
||||||
| NC score | 0.057397 (rank : 56) | θ value | 8.99809 (rank : 180) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
ABTAP_MOUSE
|
||||||
| NC score | 0.057033 (rank : 57) | θ value | 0.21417 (rank : 32) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q3V1V3 | Gene names | Abtap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
FILA_HUMAN
|
||||||
| NC score | 0.056123 (rank : 58) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
RBBP6_MOUSE
|
||||||
| NC score | 0.055961 (rank : 59) | θ value | θ > 10 (rank : 192) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
NCKX1_HUMAN
|
||||||
| NC score | 0.055945 (rank : 60) | θ value | 0.125558 (rank : 27) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O60721, O43485, O75184 | Gene names | SLC24A1, KIAA0702, NCKX1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium/calcium exchanger 1 (Na(+)/K(+)/Ca(2+)-exchange protein 1) (Retinal rod Na-Ca+K exchanger). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.055845 (rank : 61) | θ value | 1.81305 (rank : 90) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
AP3B1_HUMAN
|
||||||
| NC score | 0.055215 (rank : 62) | θ value | 6.88961 (rank : 135) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O00203, O00580, Q7Z393, Q9HD66 | Gene names | AP3B1, ADTB3A | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
DAXX_HUMAN
|
||||||
| NC score | 0.055210 (rank : 63) | θ value | 3.0926 (rank : 108) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UER7, O14747, O15141, O15208, Q9BWI3 | Gene names | DAXX, BING2, DAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Death domain-associated protein 6 (Daxx) (hDaxx) (Fas death domain- associated protein) (ETS1-associated protein 1) (EAP1). | |||||
|
SPT5H_HUMAN
|
||||||
| NC score | 0.054986 (rank : 64) | θ value | 0.813845 (rank : 64) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O00267, O43279, Q59G52, Q99639 | Gene names | SUPT5H, SPT5, SPT5H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (hSPT5) (DRB sensitivity-inducing factor large subunit) (DSIF large subunit) (DSIF p160) (Tat- cotransactivator 1 protein) (Tat-CT1 protein). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.054428 (rank : 65) | θ value | 6.88961 (rank : 150) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
RNPS1_HUMAN
|
||||||
| NC score | 0.054264 (rank : 66) | θ value | 0.62314 (rank : 55) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q15287, O75308, Q8WY42, Q9NYG3 | Gene names | RNPS1, LDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1 (SR-related protein LDC2). | |||||
|
RNPS1_MOUSE
|
||||||
| NC score | 0.054264 (rank : 67) | θ value | 0.62314 (rank : 56) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q99M28, Q3TMJ1, Q922H8 | Gene names | Rnps1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1. | |||||
|
TRI44_MOUSE
|
||||||
| NC score | 0.054186 (rank : 68) | θ value | 0.47712 (rank : 49) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9QXA7, Q80SV9, Q9JHR8 | Gene names | Trim44, Dipb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB) (Protein Mc7). | |||||
|
AP3B2_MOUSE
|
||||||
| NC score | 0.053859 (rank : 69) | θ value | 8.99809 (rank : 157) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.052611 (rank : 70) | θ value | 0.365318 (rank : 44) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.052298 (rank : 71) | θ value | θ > 10 (rank : 196) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
CD19_HUMAN
|
||||||
| NC score | 0.052105 (rank : 72) | θ value | 0.279714 (rank : 34) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P15391, Q96S68, Q9BRD6 | Gene names | CD19 | |||
|
Domain Architecture |

|
|||||
| Description | B-lymphocyte antigen CD19 precursor (Differentiation antigen CD19) (B- lymphocyte surface antigen B4) (Leu-12). | |||||
|
CA026_HUMAN
|
||||||
| NC score | 0.051865 (rank : 73) | θ value | 8.99809 (rank : 159) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5T5J6, Q8NEK9, Q9BZQ7, Q9NXQ0 | Gene names | C1orf26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf26. | |||||
|
ADDB_MOUSE
|
||||||
| NC score | 0.051564 (rank : 74) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
UBF1_HUMAN
|
||||||
| NC score | 0.051452 (rank : 75) | θ value | 0.62314 (rank : 59) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
IF4B_HUMAN
|
||||||
| NC score | 0.050554 (rank : 76) | θ value | 2.36792 (rank : 96) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P23588 | Gene names | EIF4B | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
SPRL1_MOUSE
|
||||||
| NC score | 0.050428 (rank : 77) | θ value | θ > 10 (rank : 193) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P70663, P97810, Q99L82 | Gene names | Sparcl1, Ecm2, Sc1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (Matrix glycoprotein Sc1) (Extracellular matrix protein 2). | |||||
|
UBP8_MOUSE
|
||||||
| NC score | 0.049551 (rank : 78) | θ value | 0.0252991 (rank : 18) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q80U87, Q80YP2, Q8R0D3, Q9EQU1, Q9WVP5 | Gene names | Usp8, Kiaa0055, Ubpy | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (mUBPy). | |||||
|
GRIN1_HUMAN
|
||||||
| NC score | 0.048142 (rank : 79) | θ value | 1.06291 (rank : 68) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
MYCB2_HUMAN
|
||||||
| NC score | 0.047699 (rank : 80) | θ value | 5.27518 (rank : 133) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O75592, Q5JSX8, Q5VZN6, Q6PIB6, Q9UQ11, Q9Y6E4 | Gene names | MYCBP2, KIAA0916, PAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
BPTF_HUMAN
|
||||||
| NC score | 0.047571 (rank : 81) | θ value | 1.38821 (rank : 72) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.047270 (rank : 82) | θ value | 8.99809 (rank : 172) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
CHD2_HUMAN
|
||||||
| NC score | 0.046745 (rank : 83) | θ value | 0.0252991 (rank : 16) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O14647 | Gene names | CHD2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 2 (EC 3.6.1.-) (ATP- dependent helicase CHD2) (CHD-2). | |||||
|
GLU2B_MOUSE
|
||||||
| NC score | 0.045197 (rank : 84) | θ value | 1.38821 (rank : 73) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O08795, Q921X2 | Gene names | Prkcsh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glucosidase 2 subunit beta precursor (Glucosidase II subunit beta) (Protein kinase C substrate, 60.1 kDa protein, heavy chain) (PKCSH) (80K-H protein). | |||||
|
SFRS4_MOUSE
|
||||||
| NC score | 0.044863 (rank : 85) | θ value | 0.62314 (rank : 58) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8VE97, Q9JJC3 | Gene names | Sfrs4 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4. | |||||
|
K1543_HUMAN
|
||||||
| NC score | 0.044305 (rank : 86) | θ value | 0.365318 (rank : 40) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9P1Y5, Q8NDF1 | Gene names | KIAA1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
CBX8_HUMAN
|
||||||
| NC score | 0.043595 (rank : 87) | θ value | 1.06291 (rank : 66) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9HC52, Q96H39, Q9NR07 | Gene names | CBX8, PC3, RC1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 8 (Polycomb 3 homolog) (Pc3) (hPc3) (Rectachrome 1). | |||||
|
HTSF1_HUMAN
|
||||||
| NC score | 0.043501 (rank : 88) | θ value | 5.27518 (rank : 131) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O43719, Q59G06, Q99730 | Gene names | HTATSF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 (Tat-SF1). | |||||
|
MARCS_MOUSE
|
||||||
| NC score | 0.042968 (rank : 89) | θ value | 5.27518 (rank : 132) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P26645 | Gene names | Marcks, Macs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS). | |||||
|
TCF20_HUMAN
|
||||||
| NC score | 0.042700 (rank : 90) | θ value | 1.81305 (rank : 91) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9UGU0, O14528, Q13078, Q4V353, Q9H4M0 | Gene names | TCF20, KIAA0292, SPBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP) (AR1). | |||||
|
CHD1_HUMAN
|
||||||
| NC score | 0.042663 (rank : 91) | θ value | 1.81305 (rank : 81) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.042574 (rank : 92) | θ value | 6.88961 (rank : 144) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
BAZ2B_HUMAN
|
||||||
| NC score | 0.041900 (rank : 93) | θ value | 0.163984 (rank : 29) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
ATBF1_MOUSE
|
||||||
| NC score | 0.041514 (rank : 94) | θ value | 0.365318 (rank : 38) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
CLUA1_MOUSE
|
||||||
| NC score | 0.040949 (rank : 95) | θ value | 3.0926 (rank : 106) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8R3P7, Q6P3Y8 | Gene names | Cluap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Clusterin-associated protein 1. | |||||
|
MYCB2_MOUSE
|
||||||
| NC score | 0.040833 (rank : 96) | θ value | 0.62314 (rank : 53) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7TPH6, Q6PCM8 | Gene names | Mycbp2, Pam, Phr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
UBF1_MOUSE
|
||||||
| NC score | 0.040563 (rank : 97) | θ value | 8.99809 (rank : 179) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P25976 | Gene names | Ubtf, Tcfubf, Ubf-1, Ubf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1). | |||||
|
CHD1_MOUSE
|
||||||
| NC score | 0.040268 (rank : 98) | θ value | 1.81305 (rank : 82) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P40201 | Gene names | Chd1, Chd-1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
NF2L1_MOUSE
|
||||||
| NC score | 0.040017 (rank : 99) | θ value | 0.365318 (rank : 43) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61985, O70234 | Gene names | Nfe2l1, Nrf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 1 (NF-E2-related factor 1) (NFE2-related factor 1) (Nuclear factor, erythroid derived 2, like 1). | |||||
|
IF4B_MOUSE
|
||||||
| NC score | 0.039410 (rank : 100) | θ value | 8.99809 (rank : 166) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BGD9, Q3TD64 | Gene names | Eif4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
SFR14_MOUSE
|
||||||
| NC score | 0.037755 (rank : 101) | θ value | 0.62314 (rank : 57) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CH09, Q6PG19, Q80UY8, Q8BY32, Q8CFM0 | Gene names | Sfrs14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative splicing factor, arginine/serine-rich 14 (Arginine/serine- rich-splicing factor 14). | |||||
|
CLUA1_HUMAN
|
||||||
| NC score | 0.037375 (rank : 102) | θ value | 2.36792 (rank : 94) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96AJ1, O75138, Q65ZA3, Q9H8R4, Q9H8T1 | Gene names | CLUAP1, KIAA0643 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Clusterin-associated protein 1. | |||||
|
RPA1_HUMAN
|
||||||
| NC score | 0.036453 (rank : 103) | θ value | 0.47712 (rank : 48) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95602, Q9UEH0, Q9UFT9 | Gene names | POLR1A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase I largest subunit (EC 2.7.7.6) (RNA polymerase I 194 kDa subunit) (RPA194). | |||||
|
PHF14_HUMAN
|
||||||
| NC score | 0.036154 (rank : 104) | θ value | 2.36792 (rank : 99) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O94880 | Gene names | PHF14, KIAA0783 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 14. | |||||
|
C8AP2_MOUSE
|
||||||
| NC score | 0.035779 (rank : 105) | θ value | 5.27518 (rank : 128) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9WUF3, Q9CSW2 | Gene names | Casp8ap2, Flash | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
SRCH_HUMAN
|
||||||
| NC score | 0.035776 (rank : 106) | θ value | 8.99809 (rank : 171) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P23327, Q504Y6 | Gene names | HRC, HCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcoplasmic reticulum histidine-rich calcium-binding protein precursor. | |||||
|
PHF14_MOUSE
|
||||||
| NC score | 0.035406 (rank : 107) | θ value | 3.0926 (rank : 112) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9D4H9, Q810Z6, Q8C284, Q8CGF8, Q9D1Q8 | Gene names | Phf14 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 14. | |||||
|
DVL3_MOUSE
|
||||||
| NC score | 0.035163 (rank : 108) | θ value | 0.279714 (rank : 35) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61062 | Gene names | Dvl3 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-3 (Dishevelled-3) (DSH homolog 3). | |||||
|
CT111_HUMAN
|
||||||
| NC score | 0.035052 (rank : 109) | θ value | 1.81305 (rank : 83) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NX31, O95912, Q9NZ84, Q9P0R8 | Gene names | C20orf111 | |||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf111. | |||||
|
SFRS4_HUMAN
|
||||||
| NC score | 0.034774 (rank : 110) | θ value | 6.88961 (rank : 149) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q08170, Q5VXP1, Q9BUA4, Q9UEB5 | Gene names | SFRS4, SRP75 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4 (Pre-mRNA-splicing factor SRP75) (SRP001LB). | |||||
|
AKP8L_HUMAN
|
||||||
| NC score | 0.034487 (rank : 111) | θ value | 1.38821 (rank : 71) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9ULX6, O94792, Q96J58, Q9NRQ0, Q9UGM0 | Gene names | AKAP8L, NAKAP95 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 8-like (AKAP8-like protein) (Neighbor of A- kinase-anchoring protein 95) (Neighbor of AKAP95) (Homologous to AKAP95 protein) (HA95) (Helicase A-binding protein 95) (HAP95). | |||||
|
NF2L1_HUMAN
|
||||||
| NC score | 0.034470 (rank : 112) | θ value | 1.81305 (rank : 85) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14494, Q12877, Q96FN6 | Gene names | NFE2L1, HBZ17, NRF1, TCF11 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 1 (NF-E2-related factor 1) (NFE2-related factor 1) (Nuclear factor, erythroid derived 2, like 1) (Transcription factor 11) (Transcription factor HBZ17) (Transcription factor LCR-F1) (Locus control region-factor 1). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.034188 (rank : 113) | θ value | 1.06291 (rank : 70) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
CASC3_MOUSE
|
||||||
| NC score | 0.034187 (rank : 114) | θ value | 4.03905 (rank : 117) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8K3W3, Q8K219, Q99NF0 | Gene names | Casc3, Mln51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC3 (Cancer susceptibility candidate gene 3 protein homolog) (Metastatic lymph node protein 51 homolog) (Protein MLN 51 homolog) (Protein barentsz) (Btz). | |||||
|
DVL3_HUMAN
|
||||||
| NC score | 0.032962 (rank : 115) | θ value | 0.47712 (rank : 47) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92997, O14642, Q13531, Q8N5E9, Q92607 | Gene names | DVL3, KIAA0208 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-3 (Dishevelled-3) (DSH homolog 3). | |||||
|
ARS2_HUMAN
|
||||||
| NC score | 0.032153 (rank : 116) | θ value | 6.88961 (rank : 136) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BXP5, O95808, Q9BWP6, Q9BXP4 | Gene names | ARS2, ASR2 | |||
|
Domain Architecture |

|
|||||
| Description | Arsenite-resistance protein 2. | |||||
|
CHD9_MOUSE
|
||||||
| NC score | 0.031689 (rank : 117) | θ value | 2.36792 (rank : 93) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
CHIC1_MOUSE
|
||||||
| NC score | 0.031460 (rank : 118) | θ value | 4.03905 (rank : 118) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CBW7, O08904, Q8K3S5 | Gene names | Chic1, Brx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich hydrophobic domain 1 protein (Brain X-linked protein). | |||||
|
ZN326_MOUSE
|
||||||
| NC score | 0.029464 (rank : 119) | θ value | 4.03905 (rank : 124) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O88291, Q8BSJ5, Q8K1X9, Q9CYG9 | Gene names | Znf326, Zan75, Zfp326 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 326 (Zinc finger protein-associated with nuclear matrix of 75 kDa). | |||||
|
TTBK1_HUMAN
|
||||||
| NC score | 0.029329 (rank : 120) | θ value | 0.0193708 (rank : 14) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1058 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q5TCY1, Q2L6C6, Q8N444, Q96JH2 | Gene names | TTBK1, BDTK, KIAA1855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 1 (EC 2.7.11.1) (Brain-derived tau kinase). | |||||
|
ATAD2_HUMAN
|
||||||
| NC score | 0.029009 (rank : 121) | θ value | 5.27518 (rank : 126) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6PL18, Q658P2, Q68CQ0, Q6PJV6, Q8N890, Q9UHS5 | Gene names | ATAD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
ARI3A_HUMAN
|
||||||
| NC score | 0.028559 (rank : 122) | θ value | 8.99809 (rank : 158) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99856, Q8IZA7 | Gene names | ARID3A, DRIL1, DRX, E2FBP1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 3A (ARID domain- containing protein 3A) (B-cell regulator of IgH transcription) (Bright) (E2F-binding protein 1). | |||||
|
NKTR_MOUSE
|
||||||
| NC score | 0.028097 (rank : 123) | θ value | 6.88961 (rank : 145) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P30415 | Gene names | Nktr | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
ZFY27_MOUSE
|
||||||
| NC score | 0.027898 (rank : 124) | θ value | 4.03905 (rank : 123) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3TXX3, Q8CFP8 | Gene names | Zfyve27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 27. | |||||
|
AN32B_MOUSE
|
||||||
| NC score | 0.027818 (rank : 125) | θ value | 8.99809 (rank : 156) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9EST5 | Gene names | Anp32b, Pal31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member B (Proliferation-related acidic leucine-rich protein PAL31). | |||||
|
MYT1L_HUMAN
|
||||||
| NC score | 0.027024 (rank : 126) | θ value | 1.06291 (rank : 69) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UL68, Q6IQ17, Q9UPP6 | Gene names | MYT1L, KIAA1106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (MyT1-L). | |||||
|
ARI3A_MOUSE
|
||||||
| NC score | 0.026712 (rank : 127) | θ value | 3.0926 (rank : 101) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q62431 | Gene names | Arid3a, Dri1, Dril1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 3A (ARID domain- containing protein 3A) (Dead ringer-like protein 1) (B-cell regulator of IgH transcription) (Bright). | |||||
|
TF3C3_HUMAN
|
||||||
| NC score | 0.026141 (rank : 128) | θ value | 6.88961 (rank : 152) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y5Q9, Q86XJ8, Q8WX84, Q96B44, Q9H5I8, Q9NT97 | Gene names | GTF3C3 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor 3C polypeptide 3 (Transcription factor IIIC-subunit gamma) (TF3C-gamma) (TFIIIC 102 kDa subunit) (TFIIIC102). | |||||
|
BRD9_MOUSE
|
||||||
| NC score | 0.025676 (rank : 129) | θ value | 3.0926 (rank : 103) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3UQU0, Q5FWH1, Q811F7 | Gene names | Brd9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 9. | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.025655 (rank : 130) | θ value | 6.88961 (rank : 140) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
NCOR1_MOUSE
|
||||||
| NC score | 0.025429 (rank : 131) | θ value | 6.88961 (rank : 142) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
BRDT_MOUSE
|
||||||
| NC score | 0.025214 (rank : 132) | θ value | 1.81305 (rank : 80) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q91Y44, Q59HJ4 | Gene names | Brdt, Fsrg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain testis-specific protein (RING3-like protein) (Bromodomain- containing female sterile homeotic-like protein). | |||||
|
RUSC2_HUMAN
|
||||||
| NC score | 0.024835 (rank : 133) | θ value | 3.0926 (rank : 114) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8N2Y8, O15080, Q6P1W7 | Gene names | RUSC2, KIAA0375 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 2. | |||||
|
ZCPW1_MOUSE
|
||||||
| NC score | 0.024634 (rank : 134) | θ value | 5.27518 (rank : 134) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6IR42 | Gene names | Zcwpw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1 homolog. | |||||
|
OGT1_HUMAN
|
||||||
| NC score | 0.024596 (rank : 135) | θ value | 8.99809 (rank : 169) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O15294, Q7Z3K0, Q8WWM8, Q96CC1, Q9UG57 | Gene names | OGT | |||
|
Domain Architecture |

|
|||||
| Description | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit (EC 2.4.1.-) (O-GlcNAc transferase p110 subunit). | |||||
|
AN32B_HUMAN
|
||||||
| NC score | 0.023959 (rank : 136) | θ value | 5.27518 (rank : 125) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q92688, O00655, P78458, P78459 | Gene names | ANP32B, APRIL, PHAPI2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member B (PHAPI2 protein) (Silver-stainable protein SSP29) (Acidic protein rich in leucines). | |||||
|
ATBF1_HUMAN
|
||||||
| NC score | 0.023643 (rank : 137) | θ value | 3.0926 (rank : 102) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
CHIC1_HUMAN
|
||||||
| NC score | 0.022885 (rank : 138) | θ value | 8.99809 (rank : 161) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5VXU3 | Gene names | CHIC1, BRX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich hydrophobic domain 1 protein (Brain X-linked protein). | |||||
|
ZN447_HUMAN
|
||||||
| NC score | 0.022352 (rank : 139) | θ value | 0.0193708 (rank : 15) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 899 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8TBC5, Q9BRK7, Q9H9A0 | Gene names | ZNF447 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 447. | |||||
|
RERE_MOUSE
|
||||||
| NC score | 0.022284 (rank : 140) | θ value | 6.88961 (rank : 147) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
CPSF1_MOUSE
|
||||||
| NC score | 0.021391 (rank : 141) | θ value | 3.0926 (rank : 107) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EPU4 | Gene names | Cpsf1, Cpsf160 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 160 kDa subunit (CPSF 160 kDa subunit). | |||||
|
FOXD1_HUMAN
|
||||||
| NC score | 0.020644 (rank : 142) | θ value | 0.0736092 (rank : 24) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q16676, Q12949 | Gene names | FOXD1, FKHL8, FREAC4 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein D1 (Forkhead-related protein FKHL8) (Forkhead- related transcription factor 4) (FREAC-4). | |||||
|
ZFHX2_HUMAN
|
||||||
| NC score | 0.020459 (rank : 143) | θ value | 1.38821 (rank : 77) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9C0A1 | Gene names | ZFHX2, KIAA1762 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger homeobox protein 2. | |||||
|
RB3GP_MOUSE
|
||||||
| NC score | 0.020079 (rank : 144) | θ value | 4.03905 (rank : 121) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80UJ7, Q6A0D7, Q8C4Y0, Q8C679, Q8C6A5, Q8K324 | Gene names | Rab3gap1, Kiaa0066, Rab3gap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab3 GTPase-activating protein catalytic subunit (RAB3 GTPase- activating protein 130 kDa subunit) (Rab3-GAP p130) (Rab3-GAP). | |||||
|
ZEP1_MOUSE
|
||||||
| NC score | 0.020075 (rank : 145) | θ value | 0.813845 (rank : 65) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||
|
UN13A_HUMAN
|
||||||
| NC score | 0.019557 (rank : 146) | θ value | 3.0926 (rank : 116) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UPW8 | Gene names | UNC13A, KIAA1032 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog A (Munc13-1). | |||||
|
SMBT1_HUMAN
|
||||||
| NC score | 0.018700 (rank : 147) | θ value | 1.81305 (rank : 88) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UHJ3, Q96C73, Q9Y4Q9 | Gene names | SFMBT1, RU1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scm-like with four MBT domains protein 1 (Renal ubiquitous protein 1). | |||||
|
FMN2_HUMAN
|
||||||
| NC score | 0.018322 (rank : 148) | θ value | 5.27518 (rank : 130) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NZ56, Q59GF6, Q5VU37, Q9NZ55 | Gene names | FMN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-2. | |||||
|
AGGF1_MOUSE
|
||||||
| NC score | 0.018196 (rank : 149) | θ value | 8.99809 (rank : 155) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7TN31, Q8R2S6, Q9CQR9, Q9CU87, Q9D768 | Gene names | Aggf1, Vg5q | |||
|
Domain Architecture |

|
|||||
| Description | Angiogenic factor with G patch and FHA domains 1 (Angiogenic factor VG5Q) (mVG5Q). | |||||
|
IRX2_MOUSE
|
||||||
| NC score | 0.017997 (rank : 150) | θ value | 1.38821 (rank : 74) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P81066, O55121, Q9ERN1 | Gene names | Irx2, Irxa2 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-2 (Iroquois homeobox protein 2) (Homeodomain protein IRXA2). | |||||
|
SALL2_MOUSE
|
||||||
| NC score | 0.017908 (rank : 151) | θ value | 0.279714 (rank : 36) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 836 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9QX96 | Gene names | Sall2, Sal2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 2 (Spalt-like protein 2) (MSal-2). | |||||
|
PCNT_HUMAN
|
||||||
| NC score | 0.017824 (rank : 152) | θ value | 4.03905 (rank : 119) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
TEKT3_HUMAN
|
||||||
| NC score | 0.017413 (rank : 153) | θ value | 8.99809 (rank : 175) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BXF9, Q8N5R5, Q96M48 | Gene names | TEKT3 | |||
|
Domain Architecture |

|
|||||
| Description | Tektin-3. | |||||
|
FXR2_HUMAN
|
||||||
| NC score | 0.016499 (rank : 154) | θ value | 3.0926 (rank : 110) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P51116, Q8WUM2 | Gene names | FXR2 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 2. | |||||
|
SALL2_HUMAN
|
||||||
| NC score | 0.016366 (rank : 155) | θ value | 1.81305 (rank : 87) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 835 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y467, Q9Y4G1 | Gene names | SALL2, KIAA0360, SAL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 2 (Zinc finger protein SALL2) (HSal2). | |||||
|
RFTN1_HUMAN
|
||||||
| NC score | 0.016299 (rank : 156) | θ value | 6.88961 (rank : 148) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14699, Q496Y2, Q4QQI7, Q5JB48, Q7Z7P2 | Gene names | RFTN1, KIAA0084 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Raftlin (Raft-linking protein) (Cell migration-inducing gene 2 protein). | |||||
|
ZEP1_HUMAN
|
||||||
| NC score | 0.016278 (rank : 157) | θ value | 6.88961 (rank : 154) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1036 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P15822, Q14122 | Gene names | HIVEP1, ZNF40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 40 (Human immunodeficiency virus type I enhancer- binding protein 1) (HIV-EP1) (Major histocompatibility complex-binding protein 1) (MBP-1) (Positive regulatory domain II-binding factor 1) (PRDII-BF1). | |||||
|
NFM_HUMAN
|
||||||
| NC score | 0.015964 (rank : 158) | θ value | 3.0926 (rank : 111) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
SPTA1_HUMAN
|
||||||
| NC score | 0.015818 (rank : 159) | θ value | 1.81305 (rank : 89) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P02549, Q15514, Q6LDY5 | Gene names | SPTA1, SPTA | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
DOK7_MOUSE
|
||||||
| NC score | 0.015073 (rank : 160) | θ value | 8.99809 (rank : 163) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q18PE0, Q3TCZ6, Q5FW70, Q8C8U7 | Gene names | Dok7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Dok-7 (Downstream of tyrosine kinase 7). | |||||
|
DDX51_HUMAN
|
||||||
| NC score | 0.014812 (rank : 161) | θ value | 3.0926 (rank : 109) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8N8A6, Q5CZ71, Q8IXK5, Q96ED1 | Gene names | DDX51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
FOXD1_MOUSE
|
||||||
| NC score | 0.014492 (rank : 162) | θ value | 1.81305 (rank : 84) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61345 | Gene names | Foxd1, Fkhl8, Freac4, Hfhbf2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein D1 (Forkhead-related protein FKHL8) (Forkhead- related transcription factor 4) (FREAC-4) (Brain factor 2) (HFH-BF-2). | |||||
|
CD2L2_HUMAN
|
||||||
| NC score | 0.014329 (rank : 163) | θ value | 0.47712 (rank : 46) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
TTC7B_HUMAN
|
||||||
| NC score | 0.014228 (rank : 164) | θ value | 8.99809 (rank : 178) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86TV6, Q86U24, Q86VT3 | Gene names | TTC7B, TTC7L1 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 7B (Tetratricopeptide repeat protein 7-like-1). | |||||
|
RBM28_MOUSE
|
||||||
| NC score | 0.014097 (rank : 165) | θ value | 8.99809 (rank : 170) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8CGC6, Q8BG25, Q8VEJ8, Q9CS22, Q9CSE6 | Gene names | Rbm28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 28 (RNA-binding motif protein 28). | |||||
|
RREB1_HUMAN
|
||||||
| NC score | 0.014057 (rank : 166) | θ value | 1.38821 (rank : 76) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 879 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q92766, O75567 | Gene names | RREB1 | |||
|
Domain Architecture |

|
|||||
| Description | RAS-responsive element-binding protein 1 (RREB-1) (Raf-responsive zinc finger protein LZ321). | |||||
|
PIAS4_MOUSE
|
||||||
| NC score | 0.013965 (rank : 167) | θ value | 3.0926 (rank : 113) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JM05, Q8R165 | Gene names | Pias4, Piasg | |||
|
Domain Architecture |

|
|||||
| Description | Protein inhibitor of activated STAT protein 4 (Protein inhibitor of activated STAT protein gamma) (PIAS-gamma) (PIASy). | |||||
|
E41L3_MOUSE
|
||||||
| NC score | 0.013811 (rank : 168) | θ value | 8.99809 (rank : 164) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9WV92, Q9R102 | Gene names | Epb41l3, Dal1, Epb4.1l3 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 3 (4.1B) (Differentially expressed in adenocarcinoma of the lung protein 1) (DAL-1) (DAL1P) (mDAL-1). | |||||
|
MAST4_HUMAN
|
||||||
| NC score | 0.013459 (rank : 169) | θ value | 0.813845 (rank : 63) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
EHMT1_HUMAN
|
||||||
| NC score | 0.013378 (rank : 170) | θ value | 1.06291 (rank : 67) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9H9B1, Q8TCN7, Q96F53, Q96JF1, Q96KH4 | Gene names | EHMT1, EUHMTASE1, KIAA1876 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 5 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 5) (H3-K9-HMTase 5) (Euchromatic histone-lysine N-methyltransferase 1) (Eu-HMTase1) (G9a- like protein 1) (GLP1). | |||||
|
YYAP1_HUMAN
|
||||||
| NC score | 0.013284 (rank : 171) | θ value | 6.88961 (rank : 153) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H869, Q5VYZ4, Q5VYZ7, Q7L4C3, Q7L5E2, Q8IXA6, Q8TEW5, Q8TF04, Q96HB6, Q9BQ64, Q9NV84 | Gene names | YY1AP1, HCCA2, YY1AP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YY1-associated protein 1 (Hepatocellular carcinoma susceptibility protein) (Hepatocellular carcinoma-associated protein 2). | |||||
|
CD2L1_MOUSE
|
||||||
| NC score | 0.013145 (rank : 172) | θ value | 2.36792 (rank : 92) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1463 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P24788, Q3UI03, Q61399, Q7TST4, Q8BP53 | Gene names | Cdc2l1 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1). | |||||
|
HUWE1_MOUSE
|
||||||
| NC score | 0.012719 (rank : 173) | θ value | 6.88961 (rank : 138) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
PITM2_HUMAN
|
||||||
| NC score | 0.012699 (rank : 174) | θ value | 4.03905 (rank : 120) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BZ72, Q9P271 | Gene names | PITPNM2, KIAA1457, NIR3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 2 (Phosphatidylinositol transfer protein, membrane-associated 2) (Pyk2 N-terminal domain-interacting receptor 3) (NIR-3). | |||||
|
NDF1_HUMAN
|
||||||
| NC score | 0.012665 (rank : 175) | θ value | 6.88961 (rank : 143) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13562, O00343, Q13340, Q5U095, Q96TH0, Q99455, Q9UEC8 | Gene names | NEUROD1, NEUROD | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 1 (NeuroD1) (NeuroD). | |||||
|
CD2L1_HUMAN
|
||||||
| NC score | 0.012513 (rank : 176) | θ value | 3.0926 (rank : 104) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
NDF1_MOUSE
|
||||||
| NC score | 0.012482 (rank : 177) | θ value | 8.99809 (rank : 168) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q60867, Q545N9, Q60897 | Gene names | Neurod1, Neurod | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 1 (NeuroD1). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.011060 (rank : 178) | θ value | 3.0926 (rank : 115) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
2B32_HUMAN
|
||||||
| NC score | 0.010923 (rank : 179) | θ value | 1.81305 (rank : 79) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P01913, O78049, P79663, Q29721, Q29809, Q30144, Q9MYA4, Q9MYH3, Q9MYH4, Q9TPB5, Q9TQ21 | Gene names | HLA-DRB3 | |||
|
Domain Architecture |

|
|||||
| Description | HLA class II histocompatibility antigen, DRB3-2 beta chain precursor (MHC class I antigen DRB3*2). | |||||
|
JHD2B_HUMAN
|
||||||
| NC score | 0.010573 (rank : 180) | θ value | 8.99809 (rank : 167) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7LBC6, Q9BVH6, Q9BW93, Q9BZ52, Q9NYF4, Q9UPS0 | Gene names | JMJD1B, C5orf7, JHDM2B, KIAA1082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2B (EC 1.14.11.-) (Jumonji domain-containing protein 1B) (Nuclear protein 5qNCA). | |||||
|
SYP2L_HUMAN
|
||||||
| NC score | 0.010480 (rank : 181) | θ value | 8.99809 (rank : 174) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H987 | Gene names | SYNPO2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin 2-like protein. | |||||
|
LPIN1_HUMAN
|
||||||
| NC score | 0.009854 (rank : 182) | θ value | 6.88961 (rank : 139) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14693 | Gene names | LPIN1, KIAA0188 | |||
|
Domain Architecture |

|
|||||
| Description | Lipin-1. | |||||
|
CD2L5_MOUSE
|
||||||
| NC score | 0.009292 (rank : 183) | θ value | 5.27518 (rank : 129) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1011 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q69ZA1, Q80V11, Q8BZG1, Q8K0A4 | Gene names | Cdc2l5, Kiaa1791 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle 2-like protein kinase 5 (EC 2.7.11.22) (CDC2- related protein kinase 5). | |||||
|
CD2L5_HUMAN
|
||||||
| NC score | 0.009249 (rank : 184) | θ value | 3.0926 (rank : 105) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q14004, Q6DKQ9, Q75MH4, Q75MH5, Q96JN4, Q9H4A0, Q9H4A1, Q9UDR4 | Gene names | CDC2L5, CDC2L, CHED, KIAA1791 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle 2-like protein kinase 5 (EC 2.7.11.22) (CDC2- related protein kinase 5) (Cholinesterase-related cell division controller). | |||||
|
PTN21_HUMAN
|
||||||
| NC score | 0.008657 (rank : 185) | θ value | 1.81305 (rank : 86) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16825 | Gene names | PTPN21, PTPD1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 21 (EC 3.1.3.48) (Protein-tyrosine phosphatase D1). | |||||
|
CAND2_MOUSE
|
||||||
| NC score | 0.008340 (rank : 186) | θ value | 8.99809 (rank : 160) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6ZQ73, Q6PHT7 | Gene names | Cand2, Kiaa0667, Tip120b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cullin-associated NEDD8-dissociated protein 2 (Cullin-associated and neddylation-dissociated protein 2) (p120 CAND2) (TBP-interacting protein TIP120B) (TBP-interacting protein of 120 kDa B). | |||||
|
TCF8_MOUSE
|
||||||
| NC score | 0.007616 (rank : 187) | θ value | 6.88961 (rank : 151) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1067 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q64318, Q62519 | Gene names | Tcf8, Zfhx1a, Zfx1a, Zfx1ha | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (Zinc finger homeobox protein 1a) (MEB1) (Delta EF1). | |||||
|
MINK1_HUMAN
|
||||||
| NC score | 0.007127 (rank : 188) | θ value | 1.38821 (rank : 75) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8N4C8, Q9P1X1, Q9P2R8 | Gene names | MINK1, MAP4K6, MINK | |||
|
Domain Architecture |

|
|||||
| Description | Misshapen-like kinase 1 (EC 2.7.11.1) (Mitogen-activated protein kinase kinase kinase kinase 6) (MAPK/ERK kinase kinase kinase 6) (MEK kinase kinase 6) (MEKKK 6) (Misshapen/NIK-related kinase) (GCK family kinase MiNK). | |||||
|
IASPP_HUMAN
|
||||||
| NC score | 0.005420 (rank : 189) | θ value | 8.99809 (rank : 165) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8WUF5, Q2PNZ9, Q5DU71, Q5I1X4, Q6P1R7, Q6PKF8, Q9Y290 | Gene names | PPP1R13L, IASPP, NKIP1, PPP1R13BL, RAI | |||
|
Domain Architecture |

|
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
TRAIP_HUMAN
|
||||||
| NC score | 0.005324 (rank : 190) | θ value | 8.99809 (rank : 177) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BWF2, O00467 | Gene names | TRAIP, TRIP | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
TLE3_HUMAN
|
||||||
| NC score | 0.005000 (rank : 191) | θ value | 8.99809 (rank : 176) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q04726, Q8IVV6, Q8WVR2, Q9HCM5 | Gene names | TLE3, KIAA1547 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 3 (ESG3). | |||||
|
CELR2_HUMAN
|
||||||
| NC score | 0.003990 (rank : 192) | θ value | 0.813845 (rank : 61) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HCU4, Q92566 | Gene names | CELSR2, CDHF10, EGFL2, KIAA0279, MEGF3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 2 precursor (Epidermal growth factor-like 2) (Multiple epidermal growth factor-like domains 3) (Flamingo 1). | |||||
|
ZBT20_HUMAN
|
||||||
| NC score | 0.003710 (rank : 193) | θ value | 4.03905 (rank : 122) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 835 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9HC78 | Gene names | ZBTB20, DPZF, ZNF288 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 20 (Zinc finger protein 288) (Dendritic-derived BTB/POZ zinc finger protein). | |||||
|
ZN513_HUMAN
|
||||||
| NC score | 0.003424 (rank : 194) | θ value | 1.38821 (rank : 78) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N8E2 | Gene names | ZNF513 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 513. | |||||
|
ECM2_HUMAN
|
||||||
| NC score | 0.003404 (rank : 195) | θ value | 6.88961 (rank : 137) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O94769 | Gene names | ECM2 | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular matrix protein 2 precursor (Matrix glycoprotein SC1/ECM2). | |||||
|
STK3_MOUSE
|
||||||
| NC score | 0.003130 (rank : 196) | θ value | 8.99809 (rank : 173) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 869 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9JI10, Q60877, Q80UG4, Q8CI58 | Gene names | Stk3, Mess1, Mst2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 3 (EC 2.7.11.1) (STE20-like kinase MST2) (MST-2) (Mammalian STE20-like protein kinase 2). | |||||
|
MYO3A_HUMAN
|
||||||
| NC score | 0.003042 (rank : 197) | θ value | 6.88961 (rank : 141) | |||
| Query Neighborhood Hits | 180 | Target Neighborhood Hits | 1033 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8NEV4, Q8WX17, Q9NYS8 | Gene names | MYO3A | |||
|
Domain Architecture |

|
|||||
| Description | Myosin IIIA (EC 2.7.11.1). | |||||