Please be patient as the page loads
|
SMG7_MOUSE
|
||||||
| SwissProt Accessions | Q5RJH6, Q63ZW5, Q6ZQF3 | Gene names | Smg7, Est1c, Kiaa0250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein SMG7 (SMG-7 homolog) (EST1-like protein C). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SMG7_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.984958 (rank : 2) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q92540, Q5T1Q0, Q6PIE0, Q7Z7H9, Q8IXC1, Q8IXC2 | Gene names | SMG7, C1orf16, EST1C, KIAA0250 | |||
|
Domain Architecture |

|
|||||
| Description | Protein SMG7 (SMG-7 homolog) (EST1-like protein C). | |||||
|
SMG7_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 113 | |
| SwissProt Accessions | Q5RJH6, Q63ZW5, Q6ZQF3 | Gene names | Smg7, Est1c, Kiaa0250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein SMG7 (SMG-7 homolog) (EST1-like protein C). | |||||
|
EST1A_HUMAN
|
||||||
| θ value | 9.24701e-21 (rank : 3) | NC score | 0.531325 (rank : 3) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86US8, O94837, Q86VH6, Q9UF60 | Gene names | EST1A, C17orf31, KIAA0732 | |||
|
Domain Architecture |

|
|||||
| Description | Telomerase-binding protein EST1A (Ever shorter telomeres 1A) (Telomerase subunit EST1A) (EST1-like protein A) (hSmg5/7a). | |||||
|
EST1A_MOUSE
|
||||||
| θ value | 3.51386e-20 (rank : 4) | NC score | 0.519478 (rank : 4) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P61406, Q5NC64 | Gene names | Est1a, Kiaa0732 | |||
|
Domain Architecture |

|
|||||
| Description | Telomerase-binding protein EST1A (Ever shorter telomeres 1A) (Telomerase subunit EST1A) (EST1-like protein A). | |||||
|
SMG5_HUMAN
|
||||||
| θ value | 4.44505e-15 (rank : 5) | NC score | 0.469725 (rank : 6) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UPR3, Q5QJE7, Q8IXC0, Q8IY09, Q96IJ7 | Gene names | SMG5, EST1B, KIAA1089 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein SMG5 (SMG-5 homolog) (EST1-like protein B) (LPTS-interacting protein) (LPTS-RP1). | |||||
|
SMG5_MOUSE
|
||||||
| θ value | 9.90251e-15 (rank : 6) | NC score | 0.491005 (rank : 5) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6ZPY2, Q497S2, Q8R3S4 | Gene names | Smg5, Est1b, Kiaa1089 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein SMG5 (SMG-5 homolog) (EST1-like protein B). | |||||
|
K1802_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 7) | NC score | 0.053227 (rank : 35) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 8) | NC score | 0.071069 (rank : 16) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 9) | NC score | 0.084640 (rank : 12) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
BSN_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 10) | NC score | 0.072587 (rank : 15) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CCNK_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 11) | NC score | 0.082580 (rank : 13) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O75909, Q96B63, Q9NNY9 | Gene names | CCNK, CPR4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-K. | |||||
|
ZIM10_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 12) | NC score | 0.086229 (rank : 11) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6P1E1, Q6PDK9, Q6PF85, Q8BW47 | Gene names | Rai17, Zimp10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
GP152_HUMAN
|
||||||
| θ value | 0.163984 (rank : 13) | NC score | 0.047585 (rank : 48) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8TDT2, Q86SM0 | Gene names | GPR152, PGR5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152 (G-protein coupled receptor PGR5). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 14) | NC score | 0.053919 (rank : 32) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 0.21417 (rank : 15) | NC score | 0.039472 (rank : 60) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
TPO_MOUSE
|
||||||
| θ value | 0.21417 (rank : 16) | NC score | 0.093383 (rank : 7) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P40226 | Gene names | Thpo | |||
|
Domain Architecture |

|
|||||
| Description | Thrombopoietin precursor (Megakaryocyte colony-stimulating factor) (Myeloproliferative leukemia virus oncogene ligand) (C-mpl ligand) (ML) (Megakaryocyte growth and development factor) (MGDF). | |||||
|
CDC16_HUMAN
|
||||||
| θ value | 0.47712 (rank : 17) | NC score | 0.092726 (rank : 9) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13042, Q96AE6, Q9Y564 | Gene names | CDC16 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 16 homolog (CDC16Hs) (Anaphase-promoting complex subunit 6) (APC6) (Cyclosome subunit 6). | |||||
|
CDC16_MOUSE
|
||||||
| θ value | 0.47712 (rank : 18) | NC score | 0.092150 (rank : 10) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R349, Q9CYX9 | Gene names | Cdc16 | |||
|
Domain Architecture |
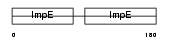
|
|||||
| Description | Cell division cycle protein 16 homolog (Anaphase-promoting complex subunit 6) (APC6) (Cyclosome subunit 6). | |||||
|
CIZ1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 19) | NC score | 0.092811 (rank : 8) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9ULV3, Q9NYM8, Q9UHK4, Q9Y3F9, Q9Y3G0 | Gene names | CIZ1, LSFR1, NP94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cip1-interacting zinc finger protein (Nuclear protein NP94). | |||||
|
EP400_MOUSE
|
||||||
| θ value | 0.47712 (rank : 20) | NC score | 0.052133 (rank : 37) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
EYA4_HUMAN
|
||||||
| θ value | 0.47712 (rank : 21) | NC score | 0.045963 (rank : 51) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95677, O95464, O95679, Q8IW39, Q9NTR7 | Gene names | EYA4 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 4 (EC 3.1.3.48). | |||||
|
PAIP1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 22) | NC score | 0.054268 (rank : 31) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H074, O60455, Q96B61, Q9BS63 | Gene names | PAIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein-interacting protein 1 (Poly(A)-binding protein-interacting protein 1) (PABP-interacting protein 1) (PAIP-1). | |||||
|
SON_HUMAN
|
||||||
| θ value | 0.47712 (rank : 23) | NC score | 0.045086 (rank : 56) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
GON4L_MOUSE
|
||||||
| θ value | 0.62314 (rank : 24) | NC score | 0.051208 (rank : 42) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9DB00, Q80TB4, Q91YI9 | Gene names | Gon4l, Gon4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
OGT1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 25) | NC score | 0.047286 (rank : 49) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O15294, Q7Z3K0, Q8WWM8, Q96CC1, Q9UG57 | Gene names | OGT | |||
|
Domain Architecture |

|
|||||
| Description | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit (EC 2.4.1.-) (O-GlcNAc transferase p110 subunit). | |||||
|
BRD4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 26) | NC score | 0.053871 (rank : 33) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
DAZ2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 27) | NC score | 0.054956 (rank : 30) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13117, Q96P41, Q9NR91 | Gene names | DAZ2 | |||
|
Domain Architecture |
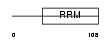
|
|||||
| Description | Deleted in azoospermia protein 2. | |||||
|
NEK4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 28) | NC score | 0.005878 (rank : 110) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 883 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P51957 | Gene names | NEK4, STK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek4 (EC 2.7.11.1) (NimA-related protein kinase 4) (Serine/threonine-protein kinase 2) (Serine/threonine-protein kinase NRK2). | |||||
|
ARI1A_HUMAN
|
||||||
| θ value | 1.06291 (rank : 29) | NC score | 0.062427 (rank : 22) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 1.06291 (rank : 30) | NC score | 0.072859 (rank : 14) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
ZIM10_HUMAN
|
||||||
| θ value | 1.06291 (rank : 31) | NC score | 0.067324 (rank : 19) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
CAF1A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 32) | NC score | 0.028679 (rank : 74) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9QWF0 | Gene names | Chaf1a, Caip150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
DAZ3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 33) | NC score | 0.051685 (rank : 41) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NR90 | Gene names | DAZ3 | |||
|
Domain Architecture |
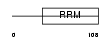
|
|||||
| Description | Deleted in azoospermia protein 3. | |||||
|
EMSY_HUMAN
|
||||||
| θ value | 1.38821 (rank : 34) | NC score | 0.060173 (rank : 23) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q7Z589, Q4G109, Q8NBU6, Q8TE50, Q9H8I9, Q9NRH0 | Gene names | EMSY, C11orf30 | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
LARP4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 35) | NC score | 0.055543 (rank : 28) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BWW4, Q3UR69 | Gene names | Larp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | La-related protein 4 (La ribonucleoprotein domain family member 4). | |||||
|
LPP_HUMAN
|
||||||
| θ value | 1.38821 (rank : 36) | NC score | 0.027829 (rank : 76) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 37) | NC score | 0.058773 (rank : 24) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
NRX2A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 38) | NC score | 0.030236 (rank : 69) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
NRX2B_HUMAN
|
||||||
| θ value | 1.38821 (rank : 39) | NC score | 0.040875 (rank : 57) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
RUSC2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 40) | NC score | 0.050149 (rank : 44) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8N2Y8, O15080, Q6P1W7 | Gene names | RUSC2, KIAA0375 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 2. | |||||
|
RUSC2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 41) | NC score | 0.045837 (rank : 52) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q80U22, Q8R1D6 | Gene names | Rusc2, Kiaa0375 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 2. | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.013840 (rank : 97) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 1.81305 (rank : 43) | NC score | 0.051791 (rank : 39) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
NFAT5_HUMAN
|
||||||
| θ value | 1.81305 (rank : 44) | NC score | 0.036628 (rank : 65) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O94916, O95693, Q9UN18 | Gene names | NFAT5, KIAA0827, TONEBP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells 5 (T-cell transcription factor NFAT5) (NF-AT5) (Tonicity-responsive enhancer-binding protein) (TonE- binding protein) (TonEBP). | |||||
|
PCQAP_HUMAN
|
||||||
| θ value | 1.81305 (rank : 45) | NC score | 0.070983 (rank : 17) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96RN5, O15413, Q6IC31, Q8NF16, Q96CT0, Q96IH7, Q9P1T3 | Gene names | PCQAP, ARC105, CTG7A, TIG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (TPA-inducible gene 1 protein) (TIG-1) (Activator-recruited cofactor 105 kDa component) (ARC105) (CTG repeat protein 7a). | |||||
|
PHC2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 46) | NC score | 0.034247 (rank : 66) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8IXK0, Q5T0C1, Q6NUJ6, Q6ZQR1, Q8N306, Q8TAG8, Q96BL4, Q9Y4Y7 | Gene names | PHC2, EDR2, PH2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 2 (hPH2) (Early development regulatory protein 2). | |||||
|
RASF1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 47) | NC score | 0.029636 (rank : 71) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NS23, O14571, O60539, O60710, Q9HB04, Q9HB18, Q9NS22, Q9UND4, Q9UND5 | Gene names | RASSF1, RDA32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras association domain-containing protein 1. | |||||
|
SC24C_HUMAN
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.046741 (rank : 50) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
SCAM2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 49) | NC score | 0.037159 (rank : 64) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15127, Q9BQE8 | Gene names | SCAMP2 | |||
|
Domain Architecture |

|
|||||
| Description | Secretory carrier-associated membrane protein 2 (Secretory carrier membrane protein 2). | |||||
|
THA10_HUMAN
|
||||||
| θ value | 1.81305 (rank : 50) | NC score | 0.045616 (rank : 53) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9P2Z0 | Gene names | THAP10 | |||
|
Domain Architecture |
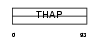
|
|||||
| Description | THAP domain-containing protein 10. | |||||
|
AMELX_HUMAN
|
||||||
| θ value | 2.36792 (rank : 51) | NC score | 0.069163 (rank : 18) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q99217, Q96NW6, Q9UCA7 | Gene names | AMELX, AMG, AMGX | |||
|
Domain Architecture |

|
|||||
| Description | Amelogenin, X isoform precursor. | |||||
|
CBP_MOUSE
|
||||||
| θ value | 2.36792 (rank : 52) | NC score | 0.052010 (rank : 38) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
K1718_HUMAN
|
||||||
| θ value | 2.36792 (rank : 53) | NC score | 0.019557 (rank : 84) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6ZMT4, Q6MZL8, Q9C0E5 | Gene names | KIAA1718 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1718. | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 2.36792 (rank : 54) | NC score | 0.053279 (rank : 34) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PGBM_HUMAN
|
||||||
| θ value | 2.36792 (rank : 55) | NC score | 0.011119 (rank : 104) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P98160, Q16287, Q9H3V5 | Gene names | HSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
CBP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | 0.051753 (rank : 40) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
HRBL_HUMAN
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.024450 (rank : 78) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O95081, O75429, Q96AB9, Q96GL4 | Gene names | HRBL, RABR | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.065167 (rank : 21) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
NCOR2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.040628 (rank : 59) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
PCNT_MOUSE
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | 0.015464 (rank : 93) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1207 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P48725 | Gene names | Pcnt, Pcnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pericentrin. | |||||
|
PO6F2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 61) | NC score | 0.029362 (rank : 72) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P78424, P78425, Q75ME8, Q86UM6, Q9UDS7 | Gene names | POU6F2, RPF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 2 (Retina-derived POU-domain factor 1) (RPF-1). | |||||
|
PRG4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 62) | NC score | 0.048044 (rank : 47) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
ZC3H6_MOUSE
|
||||||
| θ value | 3.0926 (rank : 63) | NC score | 0.022707 (rank : 80) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BYK8, Q80UK3, Q8C1H1, Q9D604 | Gene names | Zc3h6, Zc3hdc6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
AAK1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.020179 (rank : 83) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q3UHJ0, Q3TY53, Q6ZPZ6, Q80XP6 | Gene names | Aak1, Kiaa1048 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP2-associated protein kinase 1 (EC 2.7.11.1) (Adaptor-associated kinase 1). | |||||
|
CBX2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.022677 (rank : 81) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14781, Q9BTB1 | Gene names | CBX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromobox protein homolog 2. | |||||
|
CT077_HUMAN
|
||||||
| θ value | 4.03905 (rank : 66) | NC score | 0.021641 (rank : 82) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NQG5 | Gene names | C20orf77 | |||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf77. | |||||
|
DAZ1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.045097 (rank : 55) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NQZ3, Q9NQZ4 | Gene names | DAZ1, DAZ | |||
|
Domain Architecture |
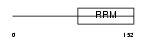
|
|||||
| Description | Deleted in azoospermia protein 1. | |||||
|
DAZ4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.045222 (rank : 54) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86SG3, Q9NR88, Q9NR89 | Gene names | DAZ4 | |||
|
Domain Architecture |

|
|||||
| Description | Deleted in azoospermia protein 4. | |||||
|
NALDL_MOUSE
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.012878 (rank : 99) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7M758 | Gene names | Naaladl1, Naaladasel | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylated-alpha-linked acidic dipeptidase-like protein (EC 3.4.17.21) (NAALADase L). | |||||
|
NFM_HUMAN
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.011620 (rank : 101) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
NGEF_MOUSE
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.017440 (rank : 90) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CHT1, Q8R204, Q923H2, Q9JHT9 | Gene names | Ngef | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ephexin-1 (Eph-interacting exchange protein) (Neuronal guanine nucleotide exchange factor). | |||||
|
ZAN_MOUSE
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.015189 (rank : 94) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
AMELX_MOUSE
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.037457 (rank : 62) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P63277, P45559, P70592, Q61293 | Gene names | Amelx, Amel | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amelogenin, X isoform precursor (Leucine-rich amelogenin peptide) (LRAP). | |||||
|
CASL_HUMAN
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.019413 (rank : 87) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14511 | Gene names | NEDD9, CASL | |||
|
Domain Architecture |
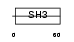
|
|||||
| Description | Enhancer of filamentation 1 (HEF1) (CRK-associated substrate-related protein) (CAS-L) (CasL) (p105) (Protein NEDD9) (NY-REN-12 antigen). | |||||
|
CASL_MOUSE
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.019517 (rank : 85) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O35177, Q8BJL8, Q8BK90, Q8BL52, Q8BM94, Q8BMI9, Q99KE7 | Gene names | Nedd9, Casl | |||
|
Domain Architecture |
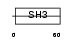
|
|||||
| Description | Enhancer of filamentation 1 (MEF1) (CRK-associated substrate-related protein) (CAS-L) (p105) (Protein NEDD9) (Neural precursor cell expressed developmentally down-regulated protein 9). | |||||
|
EDA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.029198 (rank : 73) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O54693, O35705, Q9QWJ8, Q9QZ01, Q9QZ02 | Gene names | Eda, Ed1, Ta | |||
|
Domain Architecture |

|
|||||
| Description | Ectodysplasin-A (EDA protein homolog) (Tabby protein) [Contains: Ectodysplasin-A, membrane form; Ectodysplasin-A, secreted form]. | |||||
|
HS105_HUMAN
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.008707 (rank : 105) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q92598, O95739, Q5TBM6, Q5TBM8, Q9UPC4 | Gene names | HSPH1, HSP105, HSP110, KIAA0201 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein 105 kDa (Heat shock 110 kDa protein) (Antigen NY- CO-25). | |||||
|
ITSN1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.011644 (rank : 100) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
MAML3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.056530 (rank : 25) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96JK9 | Gene names | MAML3, KIAA1816 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 3 (Mam-3). | |||||
|
R3HD2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.066755 (rank : 20) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q80TM6, Q80YB1, Q8BLS5, Q9CW50 | Gene names | R3hdm2, Kiaa1002 | |||
|
Domain Architecture |

|
|||||
| Description | R3H domain-containing protein 2. | |||||
|
YLPM1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.048894 (rank : 45) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
AFAD_HUMAN
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.013980 (rank : 96) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P55196, O75087, O75088, O75089, Q5TIG6, Q9NSN7, Q9NU92 | Gene names | MLLT4, AF6 | |||
|
Domain Architecture |

|
|||||
| Description | Afadin (Protein AF-6). | |||||
|
ALMS1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.027575 (rank : 77) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8TCU4, Q53S05, Q580Q8, Q86VP9, Q9Y4G4 | Gene names | ALMS1, KIAA0328 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alstrom syndrome protein 1. | |||||
|
DOT1L_HUMAN
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.030277 (rank : 68) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
RUNX2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.019492 (rank : 86) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13950, O14614, O14615, O95181 | Gene names | RUNX2, AML3, CBFA1, OSF2, PEBP2A | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 2 (Core-binding factor, alpha 1 subunit) (CBF-alpha 1) (Acute myeloid leukemia 3 protein) (Oncogene AML-3) (Polyomavirus enhancer-binding protein 2 alpha A subunit) (PEBP2-alpha A) (PEA2-alpha A) (SL3-3 enhancer factor 1 alpha A subunit) (SL3/AKV core-binding factor alpha A subunit) (Osteoblast- specific transcription factor 2) (OSF-2). | |||||
|
RUNX2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.018474 (rank : 89) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q08775, O35183, Q08776, Q9JLN0, Q9QUQ6, Q9QY29, Q9R0U4, Q9Z2J7 | Gene names | Runx2, Aml3, Cbfa1, Osf2, Pebp2a | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 2 (Core-binding factor, alpha 1 subunit) (CBF-alpha 1) (Acute myeloid leukemia 3 protein) (Oncogene AML-3) (Polyomavirus enhancer-binding protein 2 alpha A subunit) (PEBP2-alpha A) (PEA2-alpha A) (SL3-3 enhancer factor 1 alpha A subunit) (SL3/AKV core-binding factor alpha A subunit) (Osteoblast- specific transcription factor 2) (OSF-2). | |||||
|
SPR1B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.028589 (rank : 75) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P22528, P22529, P22530, Q5T524 | Gene names | SPRR1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin B (Small proline-rich protein IB) (SPR-IB) (14.9 kDa pancornulin). | |||||
|
TBX21_HUMAN
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.011269 (rank : 103) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UL17 | Gene names | TBX21, TBET, TBLYM | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX21 (T-box protein 21) (Transcription factor TBLYM) (T-cell-specific T-box transcription factor T-bet). | |||||
|
TXLNA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.011367 (rank : 102) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 982 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P40222, Q66K62, Q86T54, Q86T85, Q86T86, Q86Y86, Q86YW3, Q8N2Y3 | Gene names | TXLNA, TXLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
APLP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.016287 (rank : 91) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q03157, Q8VC38 | Gene names | Aplp1 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 1 precursor (APLP) (APLP-1) [Contains: C30]. | |||||
|
BPTF_HUMAN
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.033818 (rank : 67) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
CCD45_MOUSE
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.024303 (rank : 79) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BVV7 | Gene names | Ccdc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 45. | |||||
|
DDX4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.002842 (rank : 114) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NQI0, Q86VX0, Q9NT92, Q9NYB1 | Gene names | DDX4, VASA | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX4 (EC 3.6.1.-) (DEAD box protein 4) (VASA homolog). | |||||
|
DMD_HUMAN
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.006920 (rank : 108) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
DMD_MOUSE
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.007260 (rank : 107) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
E41LB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.004769 (rank : 112) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H329, Q9H328, Q9P2V3 | Gene names | EPB41L4B, EHM2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 4B (Protein EHM2) (FERM-containing protein CG1). | |||||
|
FOXM1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.018700 (rank : 88) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q08050, O43258, O43259, O43260, Q4ZGG7, Q9BRL2 | Gene names | FOXM1, FKHL16, HFH11, MPP2, WIN | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein M1 (Forkhead-related protein FKHL16) (Hepatocyte nuclear factor 3 forkhead homolog 11) (HNF-3/fork-head homolog 11) (HFH-11) (Winged helix factor from INS-1 cells) (M-phase phosphoprotein 2) (MPM-2 reactive phosphoprotein 2) (Transcription factor Trident). | |||||
|
FOXO4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.006417 (rank : 109) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P98177, O43821, Q13720, Q3KPF1, Q8TDK9 | Gene names | MLLT7, AFX, AFX1, FOXO4 | |||
|
Domain Architecture |

|
|||||
| Description | Fork head domain transcription factor AFX1 (Forkhead box protein O4). | |||||
|
GLIS3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.000598 (rank : 116) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 861 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6XP49, Q8BI60 | Gene names | Glis3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLIS3 (GLI-similar 3). | |||||
|
GPV_MOUSE
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.001740 (rank : 115) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O08742 | Gene names | Gp5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Platelet glycoprotein V precursor (GPV) (CD42D antigen). | |||||
|
HXD3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.003385 (rank : 113) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P09027, Q3UUD3 | Gene names | Hoxd3, Hox-4.1, Hoxd-3 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D3 (Hox-4.1) (Homeobox protein MH-19). | |||||
|
LIRB3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.004840 (rank : 111) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75022, O15471, Q86U49 | Gene names | LILRB3, ILT5, LIR3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily B member 3 precursor (Leukocyte immunoglobulin-like receptor 3) (LIR-3) (Immunoglobulin- like transcript 5) (ILT-5) (Monocyte inhibitory receptor HL9) (CD85a antigen). | |||||
|
MAML1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.030140 (rank : 70) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q92585, Q9NZ12 | Gene names | MAML1, KIAA0200 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 1 (Mam-1). | |||||
|
MAP4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.038691 (rank : 61) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
MCPH1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.014686 (rank : 95) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TT79, Q8BNI9, Q8C9I7 | Gene names | Mcph1 | |||
|
Domain Architecture |

|
|||||
| Description | Microcephalin. | |||||
|
MTMR7_MOUSE
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.013490 (rank : 98) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z2C9, Q8C4J6 | Gene names | Mtmr7 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 7 (EC 3.1.3.-). | |||||
|
NFH_HUMAN
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.015909 (rank : 92) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
NU214_HUMAN
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.037347 (rank : 63) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.048497 (rank : 46) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
PORIM_MOUSE
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.055547 (rank : 27) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q91Z22, Q3T9F6, Q3TLL2, Q3U1R6, Q8CEX4 | Gene names | Tmem123 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Porimin precursor (Transmembrane protein 123). | |||||
|
TAU_MOUSE
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.040833 (rank : 58) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.007913 (rank : 106) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
ZN710_HUMAN
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | -0.000025 (rank : 117) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 796 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8N1W2, Q6ZMK9, Q8NDU0 | Gene names | ZNF710 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 710. | |||||
|
BSN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.055653 (rank : 26) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
CA026_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.055102 (rank : 29) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBQ9, Q6PAN2, Q7TMQ6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf26 homolog. | |||||
|
PRP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.051076 (rank : 43) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.052185 (rank : 36) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
SMG7_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 113 | |
| SwissProt Accessions | Q5RJH6, Q63ZW5, Q6ZQF3 | Gene names | Smg7, Est1c, Kiaa0250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein SMG7 (SMG-7 homolog) (EST1-like protein C). | |||||
|
SMG7_HUMAN
|
||||||
| NC score | 0.984958 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q92540, Q5T1Q0, Q6PIE0, Q7Z7H9, Q8IXC1, Q8IXC2 | Gene names | SMG7, C1orf16, EST1C, KIAA0250 | |||
|
Domain Architecture |

|
|||||
| Description | Protein SMG7 (SMG-7 homolog) (EST1-like protein C). | |||||
|
EST1A_HUMAN
|
||||||
| NC score | 0.531325 (rank : 3) | θ value | 9.24701e-21 (rank : 3) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86US8, O94837, Q86VH6, Q9UF60 | Gene names | EST1A, C17orf31, KIAA0732 | |||
|
Domain Architecture |

|
|||||
| Description | Telomerase-binding protein EST1A (Ever shorter telomeres 1A) (Telomerase subunit EST1A) (EST1-like protein A) (hSmg5/7a). | |||||
|
EST1A_MOUSE
|
||||||
| NC score | 0.519478 (rank : 4) | θ value | 3.51386e-20 (rank : 4) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P61406, Q5NC64 | Gene names | Est1a, Kiaa0732 | |||
|
Domain Architecture |

|
|||||
| Description | Telomerase-binding protein EST1A (Ever shorter telomeres 1A) (Telomerase subunit EST1A) (EST1-like protein A). | |||||
|
SMG5_MOUSE
|
||||||
| NC score | 0.491005 (rank : 5) | θ value | 9.90251e-15 (rank : 6) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6ZPY2, Q497S2, Q8R3S4 | Gene names | Smg5, Est1b, Kiaa1089 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein SMG5 (SMG-5 homolog) (EST1-like protein B). | |||||
|
SMG5_HUMAN
|
||||||
| NC score | 0.469725 (rank : 6) | θ value | 4.44505e-15 (rank : 5) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UPR3, Q5QJE7, Q8IXC0, Q8IY09, Q96IJ7 | Gene names | SMG5, EST1B, KIAA1089 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein SMG5 (SMG-5 homolog) (EST1-like protein B) (LPTS-interacting protein) (LPTS-RP1). | |||||
|
TPO_MOUSE
|
||||||
| NC score | 0.093383 (rank : 7) | θ value | 0.21417 (rank : 16) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P40226 | Gene names | Thpo | |||
|
Domain Architecture |

|
|||||
| Description | Thrombopoietin precursor (Megakaryocyte colony-stimulating factor) (Myeloproliferative leukemia virus oncogene ligand) (C-mpl ligand) (ML) (Megakaryocyte growth and development factor) (MGDF). | |||||
|
CIZ1_HUMAN
|
||||||
| NC score | 0.092811 (rank : 8) | θ value | 0.47712 (rank : 19) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9ULV3, Q9NYM8, Q9UHK4, Q9Y3F9, Q9Y3G0 | Gene names | CIZ1, LSFR1, NP94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cip1-interacting zinc finger protein (Nuclear protein NP94). | |||||
|
CDC16_HUMAN
|
||||||
| NC score | 0.092726 (rank : 9) | θ value | 0.47712 (rank : 17) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13042, Q96AE6, Q9Y564 | Gene names | CDC16 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 16 homolog (CDC16Hs) (Anaphase-promoting complex subunit 6) (APC6) (Cyclosome subunit 6). | |||||
|
CDC16_MOUSE
|
||||||
| NC score | 0.092150 (rank : 10) | θ value | 0.47712 (rank : 18) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R349, Q9CYX9 | Gene names | Cdc16 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 16 homolog (Anaphase-promoting complex subunit 6) (APC6) (Cyclosome subunit 6). | |||||
|
ZIM10_MOUSE
|
||||||
| NC score | 0.086229 (rank : 11) | θ value | 0.0563607 (rank : 12) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6P1E1, Q6PDK9, Q6PF85, Q8BW47 | Gene names | Rai17, Zimp10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.084640 (rank : 12) | θ value | 0.0252991 (rank : 9) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
CCNK_HUMAN
|
||||||
| NC score | 0.082580 (rank : 13) | θ value | 0.0563607 (rank : 11) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O75909, Q96B63, Q9NNY9 | Gene names | CCNK, CPR4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-K. | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.072859 (rank : 14) | θ value | 1.06291 (rank : 30) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.072587 (rank : 15) | θ value | 0.0330416 (rank : 10) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.071069 (rank : 16) | θ value | 0.00869519 (rank : 8) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
PCQAP_HUMAN
|
||||||
| NC score | 0.070983 (rank : 17) | θ value | 1.81305 (rank : 45) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96RN5, O15413, Q6IC31, Q8NF16, Q96CT0, Q96IH7, Q9P1T3 | Gene names | PCQAP, ARC105, CTG7A, TIG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (TPA-inducible gene 1 protein) (TIG-1) (Activator-recruited cofactor 105 kDa component) (ARC105) (CTG repeat protein 7a). | |||||
|
AMELX_HUMAN
|
||||||
| NC score | 0.069163 (rank : 18) | θ value | 2.36792 (rank : 51) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q99217, Q96NW6, Q9UCA7 | Gene names | AMELX, AMG, AMGX | |||
|
Domain Architecture |

|
|||||
| Description | Amelogenin, X isoform precursor. | |||||
|
ZIM10_HUMAN
|
||||||
| NC score | 0.067324 (rank : 19) | θ value | 1.06291 (rank : 31) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
R3HD2_MOUSE
|
||||||
| NC score | 0.066755 (rank : 20) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q80TM6, Q80YB1, Q8BLS5, Q9CW50 | Gene names | R3hdm2, Kiaa1002 | |||
|
Domain Architecture |

|
|||||
| Description | R3H domain-containing protein 2. | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.065167 (rank : 21) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
ARI1A_HUMAN
|
||||||
| NC score | 0.062427 (rank : 22) | θ value | 1.06291 (rank : 29) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
EMSY_HUMAN
|
||||||
| NC score | 0.060173 (rank : 23) | θ value | 1.38821 (rank : 34) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q7Z589, Q4G109, Q8NBU6, Q8TE50, Q9H8I9, Q9NRH0 | Gene names | EMSY, C11orf30 | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.058773 (rank : 24) | θ value | 1.38821 (rank : 37) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MAML3_HUMAN
|
||||||
| NC score | 0.056530 (rank : 25) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96JK9 | Gene names | MAML3, KIAA1816 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 3 (Mam-3). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.055653 (rank : 26) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
PORIM_MOUSE
|
||||||
| NC score | 0.055547 (rank : 27) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q91Z22, Q3T9F6, Q3TLL2, Q3U1R6, Q8CEX4 | Gene names | Tmem123 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Porimin precursor (Transmembrane protein 123). | |||||
|
LARP4_MOUSE
|
||||||
| NC score | 0.055543 (rank : 28) | θ value | 1.38821 (rank : 35) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BWW4, Q3UR69 | Gene names | Larp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | La-related protein 4 (La ribonucleoprotein domain family member 4). | |||||
|
CA026_MOUSE
|
||||||
| NC score | 0.055102 (rank : 29) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBQ9, Q6PAN2, Q7TMQ6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf26 homolog. | |||||
|
DAZ2_HUMAN
|
||||||
| NC score | 0.054956 (rank : 30) | θ value | 0.813845 (rank : 27) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13117, Q96P41, Q9NR91 | Gene names | DAZ2 | |||
|
Domain Architecture |

|
|||||
| Description | Deleted in azoospermia protein 2. | |||||
|
PAIP1_HUMAN
|
||||||
| NC score | 0.054268 (rank : 31) | θ value | 0.47712 (rank : 22) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H074, O60455, Q96B61, Q9BS63 | Gene names | PAIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein-interacting protein 1 (Poly(A)-binding protein-interacting protein 1) (PABP-interacting protein 1) (PAIP-1). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.053919 (rank : 32) | θ value | 0.163984 (rank : 14) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
BRD4_HUMAN
|
||||||
| NC score | 0.053871 (rank : 33) | θ value | 0.813845 (rank : 26) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.053279 (rank : 34) | θ value | 2.36792 (rank : 54) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.053227 (rank : 35) | θ value | 0.00390308 (rank : 7) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.052185 (rank : 36) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
EP400_MOUSE
|
||||||
| NC score | 0.052133 (rank : 37) | θ value | 0.47712 (rank : 20) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.052010 (rank : 38) | θ value | 2.36792 (rank : 52) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.051791 (rank : 39) | θ value | 1.81305 (rank : 43) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.051753 (rank : 40) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
DAZ3_HUMAN
|
||||||
| NC score | 0.051685 (rank : 41) | θ value | 1.38821 (rank : 33) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NR90 | Gene names | DAZ3 | |||
|
Domain Architecture |

|
|||||
| Description | Deleted in azoospermia protein 3. | |||||
|
GON4L_MOUSE
|
||||||
| NC score | 0.051208 (rank : 42) | θ value | 0.62314 (rank : 24) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9DB00, Q80TB4, Q91YI9 | Gene names | Gon4l, Gon4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.051076 (rank : 43) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
RUSC2_HUMAN
|
||||||
| NC score | 0.050149 (rank : 44) | θ value | 1.38821 (rank : 40) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8N2Y8, O15080, Q6P1W7 | Gene names | RUSC2, KIAA0375 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 2. | |||||
|
YLPM1_MOUSE
|
||||||
| NC score | 0.048894 (rank : 45) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.048497 (rank : 46) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.048044 (rank : 47) | θ value | 3.0926 (rank : 62) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
GP152_HUMAN
|
||||||
| NC score | 0.047585 (rank : 48) | θ value | 0.163984 (rank : 13) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8TDT2, Q86SM0 | Gene names | GPR152, PGR5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152 (G-protein coupled receptor PGR5). | |||||
|
OGT1_HUMAN
|
||||||
| NC score | 0.047286 (rank : 49) | θ value | 0.62314 (rank : 25) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O15294, Q7Z3K0, Q8WWM8, Q96CC1, Q9UG57 | Gene names | OGT | |||
|
Domain Architecture |

|
|||||
| Description | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit (EC 2.4.1.-) (O-GlcNAc transferase p110 subunit). | |||||
|
SC24C_HUMAN
|
||||||
| NC score | 0.046741 (rank : 50) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
EYA4_HUMAN
|
||||||
| NC score | 0.045963 (rank : 51) | θ value | 0.47712 (rank : 21) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95677, O95464, O95679, Q8IW39, Q9NTR7 | Gene names | EYA4 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 4 (EC 3.1.3.48). | |||||
|
RUSC2_MOUSE
|
||||||
| NC score | 0.045837 (rank : 52) | θ value | 1.38821 (rank : 41) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q80U22, Q8R1D6 | Gene names | Rusc2, Kiaa0375 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 2. | |||||
|
THA10_HUMAN
|
||||||
| NC score | 0.045616 (rank : 53) | θ value | 1.81305 (rank : 50) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9P2Z0 | Gene names | THAP10 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 10. | |||||
|
DAZ4_HUMAN
|
||||||
| NC score | 0.045222 (rank : 54) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86SG3, Q9NR88, Q9NR89 | Gene names | DAZ4 | |||
|
Domain Architecture |

|
|||||
| Description | Deleted in azoospermia protein 4. | |||||
|
DAZ1_HUMAN
|
||||||
| NC score | 0.045097 (rank : 55) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NQZ3, Q9NQZ4 | Gene names | DAZ1, DAZ | |||
|
Domain Architecture |

|
|||||
| Description | Deleted in azoospermia protein 1. | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.045086 (rank : 56) | θ value | 0.47712 (rank : 23) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
NRX2B_HUMAN
|
||||||
| NC score | 0.040875 (rank : 57) | θ value | 1.38821 (rank : 39) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
TAU_MOUSE
|
||||||
| NC score | 0.040833 (rank : 58) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
NCOR2_HUMAN
|
||||||
| NC score | 0.040628 (rank : 59) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.039472 (rank : 60) | θ value | 0.21417 (rank : 15) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
MAP4_HUMAN
|
||||||
| NC score | 0.038691 (rank : 61) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
AMELX_MOUSE
|
||||||
| NC score | 0.037457 (rank : 62) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P63277, P45559, P70592, Q61293 | Gene names | Amelx, Amel | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amelogenin, X isoform precursor (Leucine-rich amelogenin peptide) (LRAP). | |||||
|
NU214_HUMAN
|
||||||
| NC score | 0.037347 (rank : 63) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
SCAM2_HUMAN
|
||||||
| NC score | 0.037159 (rank : 64) | θ value | 1.81305 (rank : 49) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15127, Q9BQE8 | Gene names | SCAMP2 | |||
|
Domain Architecture |

|
|||||
| Description | Secretory carrier-associated membrane protein 2 (Secretory carrier membrane protein 2). | |||||
|
NFAT5_HUMAN
|
||||||
| NC score | 0.036628 (rank : 65) | θ value | 1.81305 (rank : 44) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O94916, O95693, Q9UN18 | Gene names | NFAT5, KIAA0827, TONEBP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells 5 (T-cell transcription factor NFAT5) (NF-AT5) (Tonicity-responsive enhancer-binding protein) (TonE- binding protein) (TonEBP). | |||||
|
PHC2_HUMAN
|
||||||
| NC score | 0.034247 (rank : 66) | θ value | 1.81305 (rank : 46) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8IXK0, Q5T0C1, Q6NUJ6, Q6ZQR1, Q8N306, Q8TAG8, Q96BL4, Q9Y4Y7 | Gene names | PHC2, EDR2, PH2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 2 (hPH2) (Early development regulatory protein 2). | |||||
|
BPTF_HUMAN
|
||||||
| NC score | 0.033818 (rank : 67) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
DOT1L_HUMAN
|
||||||
| NC score | 0.030277 (rank : 68) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
NRX2A_HUMAN
|
||||||
| NC score | 0.030236 (rank : 69) | θ value | 1.38821 (rank : 38) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
MAML1_HUMAN
|
||||||
| NC score | 0.030140 (rank : 70) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q92585, Q9NZ12 | Gene names | MAML1, KIAA0200 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 1 (Mam-1). | |||||
|
RASF1_HUMAN
|
||||||
| NC score | 0.029636 (rank : 71) | θ value | 1.81305 (rank : 47) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NS23, O14571, O60539, O60710, Q9HB04, Q9HB18, Q9NS22, Q9UND4, Q9UND5 | Gene names | RASSF1, RDA32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras association domain-containing protein 1. | |||||
|
PO6F2_HUMAN
|
||||||
| NC score | 0.029362 (rank : 72) | θ value | 3.0926 (rank : 61) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P78424, P78425, Q75ME8, Q86UM6, Q9UDS7 | Gene names | POU6F2, RPF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 2 (Retina-derived POU-domain factor 1) (RPF-1). | |||||
|
EDA_MOUSE
|
||||||
| NC score | 0.029198 (rank : 73) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O54693, O35705, Q9QWJ8, Q9QZ01, Q9QZ02 | Gene names | Eda, Ed1, Ta | |||
|
Domain Architecture |

|
|||||
| Description | Ectodysplasin-A (EDA protein homolog) (Tabby protein) [Contains: Ectodysplasin-A, membrane form; Ectodysplasin-A, secreted form]. | |||||
|
CAF1A_MOUSE
|
||||||
| NC score | 0.028679 (rank : 74) | θ value | 1.38821 (rank : 32) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9QWF0 | Gene names | Chaf1a, Caip150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
SPR1B_HUMAN
|
||||||
| NC score | 0.028589 (rank : 75) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P22528, P22529, P22530, Q5T524 | Gene names | SPRR1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin B (Small proline-rich protein IB) (SPR-IB) (14.9 kDa pancornulin). | |||||
|
LPP_HUMAN
|
||||||
| NC score | 0.027829 (rank : 76) | θ value | 1.38821 (rank : 36) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
ALMS1_HUMAN
|
||||||
| NC score | 0.027575 (rank : 77) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8TCU4, Q53S05, Q580Q8, Q86VP9, Q9Y4G4 | Gene names | ALMS1, KIAA0328 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alstrom syndrome protein 1. | |||||
|
HRBL_HUMAN
|
||||||
| NC score | 0.024450 (rank : 78) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O95081, O75429, Q96AB9, Q96GL4 | Gene names | HRBL, RABR | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
CCD45_MOUSE
|
||||||
| NC score | 0.024303 (rank : 79) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BVV7 | Gene names | Ccdc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 45. | |||||
|
ZC3H6_MOUSE
|
||||||
| NC score | 0.022707 (rank : 80) | θ value | 3.0926 (rank : 63) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BYK8, Q80UK3, Q8C1H1, Q9D604 | Gene names | Zc3h6, Zc3hdc6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
CBX2_HUMAN
|
||||||
| NC score | 0.022677 (rank : 81) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14781, Q9BTB1 | Gene names | CBX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromobox protein homolog 2. | |||||
|
CT077_HUMAN
|
||||||
| NC score | 0.021641 (rank : 82) | θ value | 4.03905 (rank : 66) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NQG5 | Gene names | C20orf77 | |||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf77. | |||||
|
AAK1_MOUSE
|
||||||
| NC score | 0.020179 (rank : 83) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q3UHJ0, Q3TY53, Q6ZPZ6, Q80XP6 | Gene names | Aak1, Kiaa1048 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP2-associated protein kinase 1 (EC 2.7.11.1) (Adaptor-associated kinase 1). | |||||
|
K1718_HUMAN
|
||||||
| NC score | 0.019557 (rank : 84) | θ value | 2.36792 (rank : 53) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6ZMT4, Q6MZL8, Q9C0E5 | Gene names | KIAA1718 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1718. | |||||
|
CASL_MOUSE
|
||||||
| NC score | 0.019517 (rank : 85) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O35177, Q8BJL8, Q8BK90, Q8BL52, Q8BM94, Q8BMI9, Q99KE7 | Gene names | Nedd9, Casl | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of filamentation 1 (MEF1) (CRK-associated substrate-related protein) (CAS-L) (p105) (Protein NEDD9) (Neural precursor cell expressed developmentally down-regulated protein 9). | |||||
|
RUNX2_HUMAN
|
||||||
| NC score | 0.019492 (rank : 86) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13950, O14614, O14615, O95181 | Gene names | RUNX2, AML3, CBFA1, OSF2, PEBP2A | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 2 (Core-binding factor, alpha 1 subunit) (CBF-alpha 1) (Acute myeloid leukemia 3 protein) (Oncogene AML-3) (Polyomavirus enhancer-binding protein 2 alpha A subunit) (PEBP2-alpha A) (PEA2-alpha A) (SL3-3 enhancer factor 1 alpha A subunit) (SL3/AKV core-binding factor alpha A subunit) (Osteoblast- specific transcription factor 2) (OSF-2). | |||||
|
CASL_HUMAN
|
||||||
| NC score | 0.019413 (rank : 87) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14511 | Gene names | NEDD9, CASL | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of filamentation 1 (HEF1) (CRK-associated substrate-related protein) (CAS-L) (CasL) (p105) (Protein NEDD9) (NY-REN-12 antigen). | |||||
|
FOXM1_HUMAN
|
||||||
| NC score | 0.018700 (rank : 88) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q08050, O43258, O43259, O43260, Q4ZGG7, Q9BRL2 | Gene names | FOXM1, FKHL16, HFH11, MPP2, WIN | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein M1 (Forkhead-related protein FKHL16) (Hepatocyte nuclear factor 3 forkhead homolog 11) (HNF-3/fork-head homolog 11) (HFH-11) (Winged helix factor from INS-1 cells) (M-phase phosphoprotein 2) (MPM-2 reactive phosphoprotein 2) (Transcription factor Trident). | |||||
|
RUNX2_MOUSE
|
||||||
| NC score | 0.018474 (rank : 89) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q08775, O35183, Q08776, Q9JLN0, Q9QUQ6, Q9QY29, Q9R0U4, Q9Z2J7 | Gene names | Runx2, Aml3, Cbfa1, Osf2, Pebp2a | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 2 (Core-binding factor, alpha 1 subunit) (CBF-alpha 1) (Acute myeloid leukemia 3 protein) (Oncogene AML-3) (Polyomavirus enhancer-binding protein 2 alpha A subunit) (PEBP2-alpha A) (PEA2-alpha A) (SL3-3 enhancer factor 1 alpha A subunit) (SL3/AKV core-binding factor alpha A subunit) (Osteoblast- specific transcription factor 2) (OSF-2). | |||||
|
NGEF_MOUSE
|
||||||
| NC score | 0.017440 (rank : 90) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CHT1, Q8R204, Q923H2, Q9JHT9 | Gene names | Ngef | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ephexin-1 (Eph-interacting exchange protein) (Neuronal guanine nucleotide exchange factor). | |||||
|
APLP1_MOUSE
|
||||||
| NC score | 0.016287 (rank : 91) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q03157, Q8VC38 | Gene names | Aplp1 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 1 precursor (APLP) (APLP-1) [Contains: C30]. | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.015909 (rank : 92) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
PCNT_MOUSE
|
||||||
| NC score | 0.015464 (rank : 93) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1207 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P48725 | Gene names | Pcnt, Pcnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pericentrin. | |||||
|
ZAN_MOUSE
|
||||||
| NC score | 0.015189 (rank : 94) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
MCPH1_MOUSE
|
||||||
| NC score | 0.014686 (rank : 95) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TT79, Q8BNI9, Q8C9I7 | Gene names | Mcph1 | |||
|
Domain Architecture |

|
|||||
| Description | Microcephalin. | |||||
|
AFAD_HUMAN
|
||||||
| NC score | 0.013980 (rank : 96) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P55196, O75087, O75088, O75089, Q5TIG6, Q9NSN7, Q9NU92 | Gene names | MLLT4, AF6 | |||
|
Domain Architecture |

|
|||||
| Description | Afadin (Protein AF-6). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.013840 (rank : 97) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
MTMR7_MOUSE
|
||||||
| NC score | 0.013490 (rank : 98) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z2C9, Q8C4J6 | Gene names | Mtmr7 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 7 (EC 3.1.3.-). | |||||
|
NALDL_MOUSE
|
||||||
| NC score | 0.012878 (rank : 99) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7M758 | Gene names | Naaladl1, Naaladasel | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylated-alpha-linked acidic dipeptidase-like protein (EC 3.4.17.21) (NAALADase L). | |||||
|
ITSN1_HUMAN
|
||||||
| NC score | 0.011644 (rank : 100) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
NFM_HUMAN
|
||||||
| NC score | 0.011620 (rank : 101) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
TXLNA_HUMAN
|
||||||
| NC score | 0.011367 (rank : 102) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 982 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P40222, Q66K62, Q86T54, Q86T85, Q86T86, Q86Y86, Q86YW3, Q8N2Y3 | Gene names | TXLNA, TXLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
TBX21_HUMAN
|
||||||
| NC score | 0.011269 (rank : 103) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UL17 | Gene names | TBX21, TBET, TBLYM | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX21 (T-box protein 21) (Transcription factor TBLYM) (T-cell-specific T-box transcription factor T-bet). | |||||
|
PGBM_HUMAN
|
||||||
| NC score | 0.011119 (rank : 104) | θ value | 2.36792 (rank : 55) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P98160, Q16287, Q9H3V5 | Gene names | HSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
HS105_HUMAN
|
||||||
| NC score | 0.008707 (rank : 105) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q92598, O95739, Q5TBM6, Q5TBM8, Q9UPC4 | Gene names | HSPH1, HSP105, HSP110, KIAA0201 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein 105 kDa (Heat shock 110 kDa protein) (Antigen NY- CO-25). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.007913 (rank : 106) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
DMD_MOUSE
|
||||||
| NC score | 0.007260 (rank : 107) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
DMD_HUMAN
|
||||||
| NC score | 0.006920 (rank : 108) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
FOXO4_HUMAN
|
||||||
| NC score | 0.006417 (rank : 109) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P98177, O43821, Q13720, Q3KPF1, Q8TDK9 | Gene names | MLLT7, AFX, AFX1, FOXO4 | |||
|
Domain Architecture |

|
|||||
| Description | Fork head domain transcription factor AFX1 (Forkhead box protein O4). | |||||
|
NEK4_HUMAN
|
||||||
| NC score | 0.005878 (rank : 110) | θ value | 0.813845 (rank : 28) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 883 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P51957 | Gene names | NEK4, STK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek4 (EC 2.7.11.1) (NimA-related protein kinase 4) (Serine/threonine-protein kinase 2) (Serine/threonine-protein kinase NRK2). | |||||
|
LIRB3_HUMAN
|
||||||
| NC score | 0.004840 (rank : 111) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75022, O15471, Q86U49 | Gene names | LILRB3, ILT5, LIR3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily B member 3 precursor (Leukocyte immunoglobulin-like receptor 3) (LIR-3) (Immunoglobulin- like transcript 5) (ILT-5) (Monocyte inhibitory receptor HL9) (CD85a antigen). | |||||
|
E41LB_HUMAN
|
||||||
| NC score | 0.004769 (rank : 112) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H329, Q9H328, Q9P2V3 | Gene names | EPB41L4B, EHM2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 4B (Protein EHM2) (FERM-containing protein CG1). | |||||
|
HXD3_MOUSE
|
||||||
| NC score | 0.003385 (rank : 113) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P09027, Q3UUD3 | Gene names | Hoxd3, Hox-4.1, Hoxd-3 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D3 (Hox-4.1) (Homeobox protein MH-19). | |||||
|
DDX4_HUMAN
|
||||||
| NC score | 0.002842 (rank : 114) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NQI0, Q86VX0, Q9NT92, Q9NYB1 | Gene names | DDX4, VASA | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX4 (EC 3.6.1.-) (DEAD box protein 4) (VASA homolog). | |||||
|
GPV_MOUSE
|
||||||
| NC score | 0.001740 (rank : 115) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O08742 | Gene names | Gp5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Platelet glycoprotein V precursor (GPV) (CD42D antigen). | |||||
|
GLIS3_MOUSE
|
||||||
| NC score | 0.000598 (rank : 116) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 861 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6XP49, Q8BI60 | Gene names | Glis3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLIS3 (GLI-similar 3). | |||||
|
ZN710_HUMAN
|
||||||
| NC score | -0.000025 (rank : 117) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 796 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8N1W2, Q6ZMK9, Q8NDU0 | Gene names | ZNF710 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 710. | |||||