Please be patient as the page loads
|
SMG5_MOUSE
|
||||||
| SwissProt Accessions | Q6ZPY2, Q497S2, Q8R3S4 | Gene names | Smg5, Est1b, Kiaa1089 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein SMG5 (SMG-5 homolog) (EST1-like protein B). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SMG5_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.952543 (rank : 2) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UPR3, Q5QJE7, Q8IXC0, Q8IY09, Q96IJ7 | Gene names | SMG5, EST1B, KIAA1089 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein SMG5 (SMG-5 homolog) (EST1-like protein B) (LPTS-interacting protein) (LPTS-RP1). | |||||
|
SMG5_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q6ZPY2, Q497S2, Q8R3S4 | Gene names | Smg5, Est1b, Kiaa1089 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein SMG5 (SMG-5 homolog) (EST1-like protein B). | |||||
|
EST1A_HUMAN
|
||||||
| θ value | 9.90251e-15 (rank : 3) | NC score | 0.464552 (rank : 5) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86US8, O94837, Q86VH6, Q9UF60 | Gene names | EST1A, C17orf31, KIAA0732 | |||
|
Domain Architecture |

|
|||||
| Description | Telomerase-binding protein EST1A (Ever shorter telomeres 1A) (Telomerase subunit EST1A) (EST1-like protein A) (hSmg5/7a). | |||||
|
SMG7_MOUSE
|
||||||
| θ value | 9.90251e-15 (rank : 4) | NC score | 0.491005 (rank : 3) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5RJH6, Q63ZW5, Q6ZQF3 | Gene names | Smg7, Est1c, Kiaa0250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein SMG7 (SMG-7 homolog) (EST1-like protein C). | |||||
|
SMG7_HUMAN
|
||||||
| θ value | 1.29331e-14 (rank : 5) | NC score | 0.488559 (rank : 4) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92540, Q5T1Q0, Q6PIE0, Q7Z7H9, Q8IXC1, Q8IXC2 | Gene names | SMG7, C1orf16, EST1C, KIAA0250 | |||
|
Domain Architecture |

|
|||||
| Description | Protein SMG7 (SMG-7 homolog) (EST1-like protein C). | |||||
|
EST1A_MOUSE
|
||||||
| θ value | 8.38298e-14 (rank : 6) | NC score | 0.450613 (rank : 6) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P61406, Q5NC64 | Gene names | Est1a, Kiaa0732 | |||
|
Domain Architecture |

|
|||||
| Description | Telomerase-binding protein EST1A (Ever shorter telomeres 1A) (Telomerase subunit EST1A) (EST1-like protein A). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 7) | NC score | 0.083673 (rank : 12) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
ATBF1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 8) | NC score | 0.032566 (rank : 36) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 9) | NC score | 0.078884 (rank : 13) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
ADDB_MOUSE
|
||||||
| θ value | 0.163984 (rank : 10) | NC score | 0.039972 (rank : 30) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
RGP1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 11) | NC score | 0.090035 (rank : 9) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P46061, Q60801 | Gene names | Rangap1, Fug1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
UBP8_MOUSE
|
||||||
| θ value | 0.365318 (rank : 12) | NC score | 0.038999 (rank : 31) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q80U87, Q80YP2, Q8R0D3, Q9EQU1, Q9WVP5 | Gene names | Usp8, Kiaa0055, Ubpy | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (mUBPy). | |||||
|
CDC16_HUMAN
|
||||||
| θ value | 0.62314 (rank : 13) | NC score | 0.086780 (rank : 10) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13042, Q96AE6, Q9Y564 | Gene names | CDC16 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 16 homolog (CDC16Hs) (Anaphase-promoting complex subunit 6) (APC6) (Cyclosome subunit 6). | |||||
|
SPTA1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 14) | NC score | 0.021446 (rank : 50) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P02549, Q15514, Q6LDY5 | Gene names | SPTA1, SPTA | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 15) | NC score | 0.037582 (rank : 32) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
ATAD2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 16) | NC score | 0.024759 (rank : 47) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6PL18, Q658P2, Q68CQ0, Q6PJV6, Q8N890, Q9UHS5 | Gene names | ATAD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
CDC16_MOUSE
|
||||||
| θ value | 0.813845 (rank : 17) | NC score | 0.084249 (rank : 11) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R349, Q9CYX9 | Gene names | Cdc16 | |||
|
Domain Architecture |
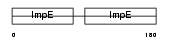
|
|||||
| Description | Cell division cycle protein 16 homolog (Anaphase-promoting complex subunit 6) (APC6) (Cyclosome subunit 6). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 18) | NC score | 0.050348 (rank : 25) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | 1.06291 (rank : 19) | NC score | 0.033071 (rank : 35) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
ZN750_MOUSE
|
||||||
| θ value | 1.38821 (rank : 20) | NC score | 0.045406 (rank : 28) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BH05, Q66JP3, Q8C0L1 | Gene names | Znf750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
2B32_HUMAN
|
||||||
| θ value | 1.81305 (rank : 21) | NC score | 0.012685 (rank : 59) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P01913, O78049, P79663, Q29721, Q29809, Q30144, Q9MYA4, Q9MYH3, Q9MYH4, Q9TPB5, Q9TQ21 | Gene names | HLA-DRB3 | |||
|
Domain Architecture |
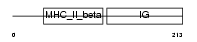
|
|||||
| Description | HLA class II histocompatibility antigen, DRB3-2 beta chain precursor (MHC class I antigen DRB3*2). | |||||
|
TCAL5_HUMAN
|
||||||
| θ value | 1.81305 (rank : 22) | NC score | 0.026225 (rank : 45) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
ABEC3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 23) | NC score | 0.051521 (rank : 23) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99J72, Q8C7L0, Q8C8V7 | Gene names | Apobec3 | |||
|
Domain Architecture |
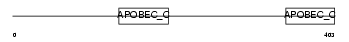
|
|||||
| Description | DNA dC->dU-editing enzyme APOBEC-3 (EC 3.5.4.-) (Apolipoprotein B mRNA-editing complex 3) (Arp3) (CEM15) (CEM-15). | |||||
|
AFF1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 24) | NC score | 0.057155 (rank : 18) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 2.36792 (rank : 25) | NC score | 0.017614 (rank : 54) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
PCNT_HUMAN
|
||||||
| θ value | 2.36792 (rank : 26) | NC score | 0.017752 (rank : 53) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
CBX8_HUMAN
|
||||||
| θ value | 3.0926 (rank : 27) | NC score | 0.036622 (rank : 34) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9HC52, Q96H39, Q9NR07 | Gene names | CBX8, PC3, RC1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 8 (Polycomb 3 homolog) (Pc3) (hPc3) (Rectachrome 1). | |||||
|
PEG3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 28) | NC score | 0.006541 (rank : 62) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1101 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q3URU2, O54978, Q3TQ69, Q5EBP7, Q61138, Q6GQS0, Q80U47, Q8R5N0, Q9QX53 | Gene names | Peg3, Kiaa0287, Pw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paternally expressed gene 3 protein (ASF-1). | |||||
|
PTDSR_MOUSE
|
||||||
| θ value | 3.0926 (rank : 29) | NC score | 0.062438 (rank : 15) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ERI5, Q80TX1 | Gene names | Ptdsr, Kiaa0585 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein PTDSR (Phosphatidylserine receptor) (Apoptotic cell clearance receptor PtdSerR). | |||||
|
T2FA_HUMAN
|
||||||
| θ value | 3.0926 (rank : 30) | NC score | 0.119119 (rank : 7) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
B4GN3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 31) | NC score | 0.015781 (rank : 56) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6L9W6, Q6ZNC1, Q8N7T6 | Gene names | B4GALNT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 2 (EC 2.4.1.244) (NGalNAc-T2) (Beta- 1,4-N-acetylgalactosaminyltransferase III) (Beta4GalNAc-T3) (Beta4GalNAcT3). | |||||
|
BIN3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 32) | NC score | 0.027279 (rank : 40) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JI08 | Gene names | Bin3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bridging integrator 3. | |||||
|
GLU2B_MOUSE
|
||||||
| θ value | 4.03905 (rank : 33) | NC score | 0.043746 (rank : 29) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O08795, Q921X2 | Gene names | Prkcsh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glucosidase 2 subunit beta precursor (Glucosidase II subunit beta) (Protein kinase C substrate, 60.1 kDa protein, heavy chain) (PKCSH) (80K-H protein). | |||||
|
GNAS3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 34) | NC score | 0.048000 (rank : 26) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95467, O95417 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroendocrine secretory protein 55 (NESP55) [Contains: LHAL tetrapeptide; GPIPIRRH peptide]. | |||||
|
GRIN1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 35) | NC score | 0.031256 (rank : 38) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
SYNE1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 36) | NC score | 0.009408 (rank : 61) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
TEKT3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 37) | NC score | 0.026886 (rank : 43) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BXF9, Q8N5R5, Q96M48 | Gene names | TEKT3 | |||
|
Domain Architecture |
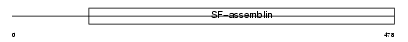
|
|||||
| Description | Tektin-3. | |||||
|
TXND2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 38) | NC score | 0.027024 (rank : 41) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
AAKG2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 39) | NC score | 0.013173 (rank : 58) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UGJ0, Q53Y07, Q6NUI0, Q75MP4, Q9NUZ9, Q9UDN8, Q9ULX8 | Gene names | PRKAG2 | |||
|
Domain Architecture |

|
|||||
| Description | 5'-AMP-activated protein kinase subunit gamma-2 (AMPK gamma-2 chain) (AMPK gamma2) (H91620p). | |||||
|
FMN2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 40) | NC score | 0.020387 (rank : 51) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NZ56, Q59GF6, Q5VU37, Q9NZ55 | Gene names | FMN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-2. | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 41) | NC score | 0.024677 (rank : 48) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
AFF3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 42) | NC score | 0.026252 (rank : 44) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P51826 | Gene names | AFF3, LAF4 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 3 (Protein LAF-4) (Lymphoid nuclear protein related to AF4). | |||||
|
CSKI1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 43) | NC score | 0.006292 (rank : 63) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
DMP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 44) | NC score | 0.046182 (rank : 27) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
EPIPL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 45) | NC score | 0.010763 (rank : 60) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R0W0 | Gene names | Eppk1 | |||
|
Domain Architecture |

|
|||||
| Description | Epiplakin. | |||||
|
MYH14_MOUSE
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.003933 (rank : 64) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
TEKT3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.020354 (rank : 52) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6X6Z7 | Gene names | Tekt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tektin-3. | |||||
|
ZFY27_MOUSE
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | 0.026997 (rank : 42) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3TXX3, Q8CFP8 | Gene names | Zfyve27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 27. | |||||
|
ANK1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | 0.003223 (rank : 65) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P16157, O43400, Q13768, Q59FP2, Q8N604, Q99407 | Gene names | ANK1, ANK | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-1 (Erythrocyte ankyrin) (Ankyrin-R). | |||||
|
BIN3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.023184 (rank : 49) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NQY0, Q9BVG2, Q9NVY9 | Gene names | BIN3 | |||
|
Domain Architecture |
|
|||||
| Description | Bridging integrator 3. | |||||
|
BPTF_HUMAN
|
||||||
| θ value | 8.99809 (rank : 51) | NC score | 0.025455 (rank : 46) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
CENPT_MOUSE
|
||||||
| θ value | 8.99809 (rank : 52) | NC score | 0.016056 (rank : 55) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3TJM4, Q3TPY6, Q3UPD2, Q8BTH0, Q8BTP2, Q8R5E9 | Gene names | Cenpt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein T (CENP-T). | |||||
|
CLUA1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | 0.028160 (rank : 39) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8R3P7, Q6P3Y8 | Gene names | Cluap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Clusterin-associated protein 1. | |||||
|
CN044_MOUSE
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.014085 (rank : 57) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CB59, Q8BQX8, Q8BSL9, Q8CBZ3 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf44 homolog. | |||||
|
PTDSR_HUMAN
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.060046 (rank : 17) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6NYC1, Q86VY0, Q8IUM5, Q9Y4E2 | Gene names | PTDSR, KIAA0585 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein PTDSR (Phosphatidylserine receptor). | |||||
|
RGP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.037112 (rank : 33) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P46060 | Gene names | RANGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
SEC62_MOUSE
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.031820 (rank : 37) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BU14, Q6NX81 | Gene names | Tloc1, Sec62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Translocation protein SEC62 (Translocation protein 1) (TP-1). | |||||
|
ZNF43_HUMAN
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | -0.000488 (rank : 66) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P17038, P28160, Q96DG1 | Gene names | ZNF43, KOX27, ZNF39 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 43 (Zinc protein HTF6) (Zinc finger protein KOX27). | |||||
|
CYLC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.056487 (rank : 19) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
CYLC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.053637 (rank : 22) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
DMP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.050370 (rank : 24) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
DSPP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.054355 (rank : 21) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
DSPP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.056467 (rank : 20) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
GNAS3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.103743 (rank : 8) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Z0F1, Q9QXW5, Q9Z0H2 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroendocrine secretory protein 55 (NESP55) [Contains: LHAL tetrapeptide; GPIPIRRH peptide]. | |||||
|
NSBP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.067394 (rank : 14) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
PTMA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.061153 (rank : 16) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
SMG5_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q6ZPY2, Q497S2, Q8R3S4 | Gene names | Smg5, Est1b, Kiaa1089 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein SMG5 (SMG-5 homolog) (EST1-like protein B). | |||||
|
SMG5_HUMAN
|
||||||
| NC score | 0.952543 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UPR3, Q5QJE7, Q8IXC0, Q8IY09, Q96IJ7 | Gene names | SMG5, EST1B, KIAA1089 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein SMG5 (SMG-5 homolog) (EST1-like protein B) (LPTS-interacting protein) (LPTS-RP1). | |||||
|
SMG7_MOUSE
|
||||||
| NC score | 0.491005 (rank : 3) | θ value | 9.90251e-15 (rank : 4) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5RJH6, Q63ZW5, Q6ZQF3 | Gene names | Smg7, Est1c, Kiaa0250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein SMG7 (SMG-7 homolog) (EST1-like protein C). | |||||
|
SMG7_HUMAN
|
||||||
| NC score | 0.488559 (rank : 4) | θ value | 1.29331e-14 (rank : 5) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92540, Q5T1Q0, Q6PIE0, Q7Z7H9, Q8IXC1, Q8IXC2 | Gene names | SMG7, C1orf16, EST1C, KIAA0250 | |||
|
Domain Architecture |

|
|||||
| Description | Protein SMG7 (SMG-7 homolog) (EST1-like protein C). | |||||
|
EST1A_HUMAN
|
||||||
| NC score | 0.464552 (rank : 5) | θ value | 9.90251e-15 (rank : 3) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86US8, O94837, Q86VH6, Q9UF60 | Gene names | EST1A, C17orf31, KIAA0732 | |||
|
Domain Architecture |

|
|||||
| Description | Telomerase-binding protein EST1A (Ever shorter telomeres 1A) (Telomerase subunit EST1A) (EST1-like protein A) (hSmg5/7a). | |||||
|
EST1A_MOUSE
|
||||||
| NC score | 0.450613 (rank : 6) | θ value | 8.38298e-14 (rank : 6) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P61406, Q5NC64 | Gene names | Est1a, Kiaa0732 | |||
|
Domain Architecture |

|
|||||
| Description | Telomerase-binding protein EST1A (Ever shorter telomeres 1A) (Telomerase subunit EST1A) (EST1-like protein A). | |||||
|
T2FA_HUMAN
|
||||||
| NC score | 0.119119 (rank : 7) | θ value | 3.0926 (rank : 30) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
GNAS3_MOUSE
|
||||||
| NC score | 0.103743 (rank : 8) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Z0F1, Q9QXW5, Q9Z0H2 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroendocrine secretory protein 55 (NESP55) [Contains: LHAL tetrapeptide; GPIPIRRH peptide]. | |||||
|
RGP1_MOUSE
|
||||||
| NC score | 0.090035 (rank : 9) | θ value | 0.365318 (rank : 11) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P46061, Q60801 | Gene names | Rangap1, Fug1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
CDC16_HUMAN
|
||||||
| NC score | 0.086780 (rank : 10) | θ value | 0.62314 (rank : 13) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13042, Q96AE6, Q9Y564 | Gene names | CDC16 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 16 homolog (CDC16Hs) (Anaphase-promoting complex subunit 6) (APC6) (Cyclosome subunit 6). | |||||
|
CDC16_MOUSE
|
||||||
| NC score | 0.084249 (rank : 11) | θ value | 0.813845 (rank : 17) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R349, Q9CYX9 | Gene names | Cdc16 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 16 homolog (Anaphase-promoting complex subunit 6) (APC6) (Cyclosome subunit 6). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.083673 (rank : 12) | θ value | 0.0113563 (rank : 7) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.078884 (rank : 13) | θ value | 0.125558 (rank : 9) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
NSBP1_HUMAN
|
||||||
| NC score | 0.067394 (rank : 14) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
PTDSR_MOUSE
|
||||||
| NC score | 0.062438 (rank : 15) | θ value | 3.0926 (rank : 29) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ERI5, Q80TX1 | Gene names | Ptdsr, Kiaa0585 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein PTDSR (Phosphatidylserine receptor) (Apoptotic cell clearance receptor PtdSerR). | |||||
|
PTMA_MOUSE
|
||||||
| NC score | 0.061153 (rank : 16) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
PTDSR_HUMAN
|
||||||
| NC score | 0.060046 (rank : 17) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6NYC1, Q86VY0, Q8IUM5, Q9Y4E2 | Gene names | PTDSR, KIAA0585 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein PTDSR (Phosphatidylserine receptor). | |||||
|
AFF1_MOUSE
|
||||||
| NC score | 0.057155 (rank : 18) | θ value | 2.36792 (rank : 24) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
CYLC1_HUMAN
|
||||||
| NC score | 0.056487 (rank : 19) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.056467 (rank : 20) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.054355 (rank : 21) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
CYLC2_HUMAN
|
||||||
| NC score | 0.053637 (rank : 22) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
ABEC3_MOUSE
|
||||||
| NC score | 0.051521 (rank : 23) | θ value | 2.36792 (rank : 23) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99J72, Q8C7L0, Q8C8V7 | Gene names | Apobec3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA dC->dU-editing enzyme APOBEC-3 (EC 3.5.4.-) (Apolipoprotein B mRNA-editing complex 3) (Arp3) (CEM15) (CEM-15). | |||||
|
DMP1_MOUSE
|
||||||
| NC score | 0.050370 (rank : 24) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.050348 (rank : 25) | θ value | 0.813845 (rank : 18) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
GNAS3_HUMAN
|
||||||
| NC score | 0.048000 (rank : 26) | θ value | 4.03905 (rank : 34) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95467, O95417 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroendocrine secretory protein 55 (NESP55) [Contains: LHAL tetrapeptide; GPIPIRRH peptide]. | |||||
|
DMP1_HUMAN
|
||||||
| NC score | 0.046182 (rank : 27) | θ value | 6.88961 (rank : 44) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
ZN750_MOUSE
|
||||||
| NC score | 0.045406 (rank : 28) | θ value | 1.38821 (rank : 20) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BH05, Q66JP3, Q8C0L1 | Gene names | Znf750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
GLU2B_MOUSE
|
||||||
| NC score | 0.043746 (rank : 29) | θ value | 4.03905 (rank : 33) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O08795, Q921X2 | Gene names | Prkcsh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glucosidase 2 subunit beta precursor (Glucosidase II subunit beta) (Protein kinase C substrate, 60.1 kDa protein, heavy chain) (PKCSH) (80K-H protein). | |||||
|
ADDB_MOUSE
|
||||||
| NC score | 0.039972 (rank : 30) | θ value | 0.163984 (rank : 10) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
UBP8_MOUSE
|
||||||
| NC score | 0.038999 (rank : 31) | θ value | 0.365318 (rank : 12) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q80U87, Q80YP2, Q8R0D3, Q9EQU1, Q9WVP5 | Gene names | Usp8, Kiaa0055, Ubpy | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (mUBPy). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.037582 (rank : 32) | θ value | 0.62314 (rank : 15) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
RGP1_HUMAN
|
||||||
| NC score | 0.037112 (rank : 33) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P46060 | Gene names | RANGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
CBX8_HUMAN
|
||||||
| NC score | 0.036622 (rank : 34) | θ value | 3.0926 (rank : 27) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9HC52, Q96H39, Q9NR07 | Gene names | CBX8, PC3, RC1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 8 (Polycomb 3 homolog) (Pc3) (hPc3) (Rectachrome 1). | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.033071 (rank : 35) | θ value | 1.06291 (rank : 19) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
ATBF1_MOUSE
|
||||||
| NC score | 0.032566 (rank : 36) | θ value | 0.0736092 (rank : 8) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
SEC62_MOUSE
|
||||||
| NC score | 0.031820 (rank : 37) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BU14, Q6NX81 | Gene names | Tloc1, Sec62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Translocation protein SEC62 (Translocation protein 1) (TP-1). | |||||
|
GRIN1_HUMAN
|
||||||
| NC score | 0.031256 (rank : 38) | θ value | 4.03905 (rank : 35) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
CLUA1_MOUSE
|
||||||
| NC score | 0.028160 (rank : 39) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8R3P7, Q6P3Y8 | Gene names | Cluap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Clusterin-associated protein 1. | |||||
|
BIN3_MOUSE
|
||||||
| NC score | 0.027279 (rank : 40) | θ value | 4.03905 (rank : 32) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JI08 | Gene names | Bin3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bridging integrator 3. | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.027024 (rank : 41) | θ value | 4.03905 (rank : 38) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
ZFY27_MOUSE
|
||||||
| NC score | 0.026997 (rank : 42) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3TXX3, Q8CFP8 | Gene names | Zfyve27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 27. | |||||
|
TEKT3_HUMAN
|
||||||
| NC score | 0.026886 (rank : 43) | θ value | 4.03905 (rank : 37) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BXF9, Q8N5R5, Q96M48 | Gene names | TEKT3 | |||
|
Domain Architecture |

|
|||||
| Description | Tektin-3. | |||||
|
AFF3_HUMAN
|
||||||
| NC score | 0.026252 (rank : 44) | θ value | 6.88961 (rank : 42) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P51826 | Gene names | AFF3, LAF4 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 3 (Protein LAF-4) (Lymphoid nuclear protein related to AF4). | |||||
|
TCAL5_HUMAN
|
||||||
| NC score | 0.026225 (rank : 45) | θ value | 1.81305 (rank : 22) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
BPTF_HUMAN
|
||||||
| NC score | 0.025455 (rank : 46) | θ value | 8.99809 (rank : 51) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
ATAD2_HUMAN
|
||||||
| NC score | 0.024759 (rank : 47) | θ value | 0.813845 (rank : 16) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6PL18, Q658P2, Q68CQ0, Q6PJV6, Q8N890, Q9UHS5 | Gene names | ATAD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.024677 (rank : 48) | θ value | 5.27518 (rank : 41) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
BIN3_HUMAN
|
||||||
| NC score | 0.023184 (rank : 49) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NQY0, Q9BVG2, Q9NVY9 | Gene names | BIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Bridging integrator 3. | |||||
|
SPTA1_HUMAN
|
||||||
| NC score | 0.021446 (rank : 50) | θ value | 0.62314 (rank : 14) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P02549, Q15514, Q6LDY5 | Gene names | SPTA1, SPTA | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
FMN2_HUMAN
|
||||||
| NC score | 0.020387 (rank : 51) | θ value | 5.27518 (rank : 40) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NZ56, Q59GF6, Q5VU37, Q9NZ55 | Gene names | FMN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-2. | |||||
|
TEKT3_MOUSE
|
||||||
| NC score | 0.020354 (rank : 52) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6X6Z7 | Gene names | Tekt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tektin-3. | |||||
|
PCNT_HUMAN
|
||||||
| NC score | 0.017752 (rank : 53) | θ value | 2.36792 (rank : 26) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.017614 (rank : 54) | θ value | 2.36792 (rank : 25) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
CENPT_MOUSE
|
||||||
| NC score | 0.016056 (rank : 55) | θ value | 8.99809 (rank : 52) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3TJM4, Q3TPY6, Q3UPD2, Q8BTH0, Q8BTP2, Q8R5E9 | Gene names | Cenpt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein T (CENP-T). | |||||
|
B4GN3_HUMAN
|
||||||
| NC score | 0.015781 (rank : 56) | θ value | 4.03905 (rank : 31) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6L9W6, Q6ZNC1, Q8N7T6 | Gene names | B4GALNT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 2 (EC 2.4.1.244) (NGalNAc-T2) (Beta- 1,4-N-acetylgalactosaminyltransferase III) (Beta4GalNAc-T3) (Beta4GalNAcT3). | |||||
|
CN044_MOUSE
|
||||||
| NC score | 0.014085 (rank : 57) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CB59, Q8BQX8, Q8BSL9, Q8CBZ3 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf44 homolog. | |||||
|
AAKG2_HUMAN
|
||||||
| NC score | 0.013173 (rank : 58) | θ value | 5.27518 (rank : 39) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UGJ0, Q53Y07, Q6NUI0, Q75MP4, Q9NUZ9, Q9UDN8, Q9ULX8 | Gene names | PRKAG2 | |||
|
Domain Architecture |

|
|||||
| Description | 5'-AMP-activated protein kinase subunit gamma-2 (AMPK gamma-2 chain) (AMPK gamma2) (H91620p). | |||||
|
2B32_HUMAN
|
||||||
| NC score | 0.012685 (rank : 59) | θ value | 1.81305 (rank : 21) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P01913, O78049, P79663, Q29721, Q29809, Q30144, Q9MYA4, Q9MYH3, Q9MYH4, Q9TPB5, Q9TQ21 | Gene names | HLA-DRB3 | |||
|
Domain Architecture |

|
|||||
| Description | HLA class II histocompatibility antigen, DRB3-2 beta chain precursor (MHC class I antigen DRB3*2). | |||||
|
EPIPL_MOUSE
|
||||||
| NC score | 0.010763 (rank : 60) | θ value | 6.88961 (rank : 45) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R0W0 | Gene names | Eppk1 | |||
|
Domain Architecture |

|
|||||
| Description | Epiplakin. | |||||
|
SYNE1_HUMAN
|
||||||
| NC score | 0.009408 (rank : 61) | θ value | 4.03905 (rank : 36) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
PEG3_MOUSE
|
||||||
| NC score | 0.006541 (rank : 62) | θ value | 3.0926 (rank : 28) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1101 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q3URU2, O54978, Q3TQ69, Q5EBP7, Q61138, Q6GQS0, Q80U47, Q8R5N0, Q9QX53 | Gene names | Peg3, Kiaa0287, Pw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paternally expressed gene 3 protein (ASF-1). | |||||
|
CSKI1_MOUSE
|
||||||
| NC score | 0.006292 (rank : 63) | θ value | 6.88961 (rank : 43) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
MYH14_MOUSE
|
||||||
| NC score | 0.003933 (rank : 64) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
ANK1_HUMAN
|
||||||
| NC score | 0.003223 (rank : 65) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P16157, O43400, Q13768, Q59FP2, Q8N604, Q99407 | Gene names | ANK1, ANK | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-1 (Erythrocyte ankyrin) (Ankyrin-R). | |||||
|
ZNF43_HUMAN
|
||||||
| NC score | -0.000488 (rank : 66) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P17038, P28160, Q96DG1 | Gene names | ZNF43, KOX27, ZNF39 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 43 (Zinc protein HTF6) (Zinc finger protein KOX27). | |||||