Please be patient as the page loads
|
LIRB3_HUMAN
|
||||||
| SwissProt Accessions | O75022, O15471, Q86U49 | Gene names | LILRB3, ILT5, LIR3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily B member 3 precursor (Leukocyte immunoglobulin-like receptor 3) (LIR-3) (Immunoglobulin- like transcript 5) (ILT-5) (Monocyte inhibitory receptor HL9) (CD85a antigen). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
LIRB1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.989732 (rank : 2) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8NHL6, O75024, O75025, Q8NHJ9, Q8NHK0 | Gene names | LILRB1, ILT2, LIR1, MIR7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily B member 1 precursor (Leukocyte immunoglobulin-like receptor 1) (LIR-1) (Immunoglobulin- like transcript 2) (ILT-2) (Monocyte/macrophage immunoglobulin-like receptor 7) (MIR-7) (CD85j antigen). | |||||
|
LIRB2_HUMAN
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.989077 (rank : 3) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8N423, O75017, Q8NHJ7, Q8NHJ8 | Gene names | LILRB2, ILT4, LIR2, MIR10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily B member 2 precursor (Leukocyte immunoglobulin-like receptor 2) (LIR-2) (Immunoglobulin- like transcript 4) (ILT-4) (Monocyte/macrophage immunoglobulin-like receptor 10) (MIR-10) (CD85d antigen). | |||||
|
LIRB3_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 125 | |
| SwissProt Accessions | O75022, O15471, Q86U49 | Gene names | LILRB3, ILT5, LIR3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily B member 3 precursor (Leukocyte immunoglobulin-like receptor 3) (LIR-3) (Immunoglobulin- like transcript 5) (ILT-5) (Monocyte inhibitory receptor HL9) (CD85a antigen). | |||||
|
LIRB5_HUMAN
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.986187 (rank : 6) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O75023, Q8N760 | Gene names | LILRB5, LIR8 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily B member 5 precursor (Leukocyte immunoglobulin-like receptor 8) (LIR-8) (CD85c antigen). | |||||
|
LIRA4_HUMAN
|
||||||
| θ value | 2.17438e-179 (rank : 5) | NC score | 0.986441 (rank : 5) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P59901 | Gene names | LILRA4, ILT7 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily A member 4 precursor (Immunoglobulin-like transcript 7) (ILT-7) (CD85g antigen). | |||||
|
LIRA3_HUMAN
|
||||||
| θ value | 2.67036e-161 (rank : 6) | NC score | 0.982596 (rank : 7) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8N6C8, O15469, O15470, O75016, Q8N151, Q8N154, Q8NHJ1, Q8NHJ2, Q8NHJ3, Q8NHJ4 | Gene names | LILRA3, ILT6, LIR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily A member 3 precursor (Leukocyte immunoglobulin-like receptor 4) (LIR-4) (Immunoglobulin- like transcript 6) (ILT-6) (Monocyte inhibitory receptor HM43/HM31) (CD85e antigen). | |||||
|
LIRA1_HUMAN
|
||||||
| θ value | 3.85598e-160 (rank : 7) | NC score | 0.986914 (rank : 4) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O75019, O75018 | Gene names | LILRA1, LIR6 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily A member 1 precursor (Leukocyte immunoglobulin-like receptor 6) (LIR-6) (CD85i antigen). | |||||
|
LIRA2_HUMAN
|
||||||
| θ value | 6.37065e-155 (rank : 8) | NC score | 0.981094 (rank : 8) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8N149, O75020 | Gene names | LILRA2, ILT1, LIR7 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily A member 2 precursor (Leukocyte immunoglobulin-like receptor 7) (LIR-7) (Immunoglobulin- like transcript 1) (ILT-1) (CD85h antigen). | |||||
|
LIRB4_HUMAN
|
||||||
| θ value | 2.06847e-121 (rank : 9) | NC score | 0.969504 (rank : 9) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8NHJ6, O15468, O75021, Q8N1C7, Q8NHL5 | Gene names | LILRB4, ILT3, LIR5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily B member 4 precursor (Leukocyte immunoglobulin-like receptor 5) (LIR-5) (Immunoglobulin- like transcript 3) (ILT-3) (Monocyte inhibitory receptor HM18) (CD85k antigen). | |||||
|
KI3L2_HUMAN
|
||||||
| θ value | 1.71981e-67 (rank : 10) | NC score | 0.909576 (rank : 20) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P43630, Q13238, Q14947, Q14948, Q92684, Q95367 | Gene names | KIR3DL2, CD158K, NKAT4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Killer cell immunoglobulin-like receptor 3DL2 precursor (MHC class I NK cell receptor) (Natural killer-associated transcript 4) (NKAT-4) (p70 natural killer cell receptor clone CL-5) (CD158k antigen). | |||||
|
KI3L1_HUMAN
|
||||||
| θ value | 2.56992e-63 (rank : 11) | NC score | 0.906339 (rank : 21) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P43629, O43473, Q16541 | Gene names | KIR3DL1, NKAT3, NKB1 | |||
|
Domain Architecture |
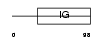
|
|||||
| Description | Killer cell immunoglobulin-like receptor 3DL1 precursor (MHC class I NK cell receptor) (Natural killer-associated transcript 3) (NKAT-3) (p70 natural killer cell receptor clones CL-2/CL-11) (HLA-BW4-specific inhibitory NK cell receptor). | |||||
|
KI3S1_HUMAN
|
||||||
| θ value | 3.35642e-63 (rank : 12) | NC score | 0.912097 (rank : 19) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q14943 | Gene names | KIR3DS1, NKAT10 | |||
|
Domain Architecture |
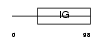
|
|||||
| Description | Killer cell immunoglobulin-like receptor 3DS1 precursor (MHC class I NK cell receptor) (Natural killer-associated transcript 10) (NKAT-10). | |||||
|
KI3L3_HUMAN
|
||||||
| θ value | 1.27544e-62 (rank : 13) | NC score | 0.914212 (rank : 17) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8N743, O95056, Q7Z7N4, Q8NHI2, Q8NHI3, Q9UEI4 | Gene names | KIR3DL3, CD158Z, KIR3DL7, KIRC1 | |||
|
Domain Architecture |
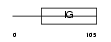
|
|||||
| Description | Killer cell immunoglobulin-like receptor 3DL3 precursor (Killer cell inhibitory receptor 1) (CD158z antigen). | |||||
|
GP49A_MOUSE
|
||||||
| θ value | 7.49467e-55 (rank : 14) | NC score | 0.947482 (rank : 11) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q61450 | Gene names | Gp49a, Gp49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mast cell surface glycoprotein Gp49A precursor. | |||||
|
LIRB4_MOUSE
|
||||||
| θ value | 6.34462e-54 (rank : 15) | NC score | 0.952959 (rank : 10) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q64281, Q64312 | Gene names | Lilrb4, Gp49b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily B member 4 precursor (Mast cell surface glycoprotein Gp49B). | |||||
|
KI2S2_HUMAN
|
||||||
| θ value | 5.55816e-50 (rank : 16) | NC score | 0.892411 (rank : 22) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P43631, Q14955 | Gene names | KIR2DS2, CD158J, NKAT5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Killer cell immunoglobulin-like receptor 2DS2 precursor (MHC class I NK cell receptor) (Natural killer-associated transcript 5) (NKAT-5) (p58 natural killer cell receptor clone CL-49) (p58 NK receptor) (NK receptor 183 ActI) (CD158j antigen). | |||||
|
KI2S4_HUMAN
|
||||||
| θ value | 3.60269e-49 (rank : 17) | NC score | 0.887358 (rank : 25) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P43632 | Gene names | KIR2DS4, CD158I, KKA3, NKAT8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Killer cell immunoglobulin-like receptor 2DS4 precursor (MHC class I NK cell receptor) (Natural killer-associated transcript 8) (NKAT-8) (P58 natural killer cell receptor clone CL-39) (p58 NK receptor) (CL- 17) (CD158i antigen). | |||||
|
KI2S3_HUMAN
|
||||||
| θ value | 2.85459e-46 (rank : 18) | NC score | 0.887458 (rank : 23) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q14952, O00644 | Gene names | KIR2DS3, NKAT7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Killer cell immunoglobulin-like receptor 2DS3 precursor (MHC class I NK cell receptor) (Natural killer-associated transcript 7) (NKAT-7). | |||||
|
KI2L2_HUMAN
|
||||||
| θ value | 6.35935e-46 (rank : 19) | NC score | 0.887034 (rank : 26) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P43627, Q14951 | Gene names | KIR2DL2, NKAT6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Killer cell immunoglobulin-like receptor 2DL2 precursor (MHC class I NK cell receptor) (Natural killer-associated transcript 6) (NKAT-6) (p58 natural killer cell receptor clone CL-43) (p58 NK receptor). | |||||
|
KI2L1_HUMAN
|
||||||
| θ value | 1.85029e-45 (rank : 20) | NC score | 0.887420 (rank : 24) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P43626, O43470 | Gene names | KIR2DL1, CD158A, NKAT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Killer cell immunoglobulin-like receptor 2DL1 precursor (MHC class I NK cell receptor) (Natural killer-associated transcript 1) (NKAT-1) (p58 natural killer cell receptor clones CL-42/47.11) (p58 NK receptor) (p58.1 MHC class-I-specific NK receptor) (CD158a antigen). | |||||
|
KI2L3_HUMAN
|
||||||
| θ value | 8.59492e-43 (rank : 21) | NC score | 0.885573 (rank : 28) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P43628, O43472, P78402, Q14944, Q14945, Q9UM51, Q9UQ70 | Gene names | KIR2DL3, CD158B, KIRCL23, NKAT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Killer cell immunoglobulin-like receptor 2DL3 precursor (MHC class I NK cell receptor) (Natural killer-associated transcript 2) (NKAT-2) (NKAT2a) (NKAT2b) (p58 natural killer cell receptor clone CL-6) (p58 NK receptor) (p58.2 MHC class-I-specific NK receptor) (Killer inhibitory receptor cl 2-3) (KIR-023GB) (CD158b antigen). | |||||
|
KI3L1_MOUSE
|
||||||
| θ value | 1.12253e-42 (rank : 22) | NC score | 0.914205 (rank : 18) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P83555 | Gene names | Kir3dl1 | |||
|
Domain Architecture |
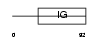
|
|||||
| Description | Killer cell immunoglobulin-like receptor 3DL1 precursor. | |||||
|
KI2S1_HUMAN
|
||||||
| θ value | 1.91475e-42 (rank : 23) | NC score | 0.884436 (rank : 29) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q14954, O43471 | Gene names | KIR2DS1, CD158H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Killer cell immunoglobulin-like receptor 2DS1 precursor (MHC class I NK cell receptor Eb6 ActI) (CD158h antigen). | |||||
|
GPVI_HUMAN
|
||||||
| θ value | 3.26607e-42 (rank : 24) | NC score | 0.936942 (rank : 12) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9HCN6, Q9HCN7, Q9UIF2 | Gene names | GP6, GPVI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Platelet glycoprotein VI precursor. | |||||
|
KI2S5_HUMAN
|
||||||
| θ value | 2.76489e-41 (rank : 25) | NC score | 0.882863 (rank : 30) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q14953 | Gene names | KIR2DS5, CD158G, NKAT9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Killer cell immunoglobulin-like receptor 2DS5 precursor (MHC class I NK cell receptor) (Natural killer-associated transcript 9) (NKAT-9) (CD158g antigen). | |||||
|
NCTR1_HUMAN
|
||||||
| θ value | 3.05695e-40 (rank : 26) | NC score | 0.933240 (rank : 13) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O76036, O76016, O76017, O76018 | Gene names | NCR1, LY94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Natural cytotoxicity triggering receptor 1 precursor (Natural killer cell p46-related protein) (NKp46) (hNKp46) (NK-p46) (NK cell- activating receptor) (Lymphocyte antigen 94 homolog) (CD335 antigen). | |||||
|
NCTR1_MOUSE
|
||||||
| θ value | 2.67802e-36 (rank : 27) | NC score | 0.924874 (rank : 15) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8C567, Q1RLN4, Q80UY6, Q9Z0Q4 | Gene names | Ncr1, Ly94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Natural cytotoxicity triggering receptor 1 precursor (Natural killer cell p46-related protein) (NKp46) (mNKp46) (NK-p46) (Mouse-activating receptor 1) (mAR-1) (Lymphocyte antigen 94). | |||||
|
GPVI_MOUSE
|
||||||
| θ value | 1.32909e-35 (rank : 28) | NC score | 0.931185 (rank : 14) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P0C191 | Gene names | Gp6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Platelet glycoprotein VI precursor (GPVI). | |||||
|
KI2L4_HUMAN
|
||||||
| θ value | 3.27365e-34 (rank : 29) | NC score | 0.885877 (rank : 27) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q99706, O14621, O14622, O14623, O14624, O43534, P78400, P78401, Q99559, Q99560, Q99561, Q99562, Q9UQJ7 | Gene names | KIR2DL4, CD158D, KIR103AS | |||
|
Domain Architecture |
|
|||||
| Description | Killer cell immunoglobulin-like receptor 2DL4 precursor (MHC class I NK cell receptor KIR103AS) (Killer cell inhibitory receptor 103AS) (KIR-103AS) (G9P) (CD158d antigen). | |||||
|
A1BG_HUMAN
|
||||||
| θ value | 5.97985e-28 (rank : 30) | NC score | 0.819793 (rank : 32) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P04217, Q8IYJ6, Q96P39 | Gene names | A1BG | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-1B-glycoprotein precursor (Alpha-1-B glycoprotein). | |||||
|
FCAR_HUMAN
|
||||||
| θ value | 5.06226e-27 (rank : 31) | NC score | 0.918705 (rank : 16) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P24071, Q13603, Q13604, Q15727, Q15728, Q92590 | Gene names | FCAR, CD89 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Immunoglobulin alpha Fc receptor precursor (IgA Fc receptor) (CD89 antigen). | |||||
|
LAIR1_HUMAN
|
||||||
| θ value | 1.80466e-16 (rank : 32) | NC score | 0.794368 (rank : 33) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6GTX8 | Gene names | LAIR1, CD305 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte-associated immunoglobulin-like receptor 1 precursor (LAIR-1) (hLAIR1) (CD305 antigen). | |||||
|
LAIR2_HUMAN
|
||||||
| θ value | 8.95645e-16 (rank : 33) | NC score | 0.835790 (rank : 31) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6ISS4, Q6PEZ4 | Gene names | LAIR2, CD306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte-associated immunoglobulin-like receptor 2 precursor (LAIR-2) (CD306 antigen). | |||||
|
FCGR2_MOUSE
|
||||||
| θ value | 1.29631e-06 (rank : 34) | NC score | 0.227254 (rank : 38) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P08101, P08102, P12316, P97917, Q60938, Q60939, Q60940, Q61170, Q61558 | Gene names | Fcgr2, Fcgr2b, Ly-17 | |||
|
Domain Architecture |
|
|||||
| Description | Low affinity immunoglobulin gamma Fc region receptor II precursor (Fc- gamma RII) (FcRII) (IgG Fc receptor II beta) (Fc gamma receptor IIB) (Fc-gamma-RIIB) (Lymphocyte antigen Ly-17) (CD32 antigen). | |||||
|
FCGR3_MOUSE
|
||||||
| θ value | 4.92598e-06 (rank : 35) | NC score | 0.231275 (rank : 37) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P08508 | Gene names | Fcgr3 | |||
|
Domain Architecture |

|
|||||
| Description | Low affinity immunoglobulin gamma Fc region receptor III precursor (IgG Fc receptor III) (Fc-gamma RIII) (FcRIII). | |||||
|
LAIR1_MOUSE
|
||||||
| θ value | 1.09739e-05 (rank : 36) | NC score | 0.505981 (rank : 34) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BG84, Q80UN5, Q8BHB6, Q8BRR6, Q8C6N8, Q8CEP5 | Gene names | Lair1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte-associated immunoglobulin-like receptor 1 precursor (LAIR-1) (mLAIR1) (CD305 antigen). | |||||
|
CD22_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 37) | NC score | 0.165319 (rank : 40) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P35329, Q9JHL2, Q9JJX9, Q9JJY0, Q9JJY1, Q9R056, Q9R094, Q9WU51 | Gene names | Cd22, Lyb-8 | |||
|
Domain Architecture |

|
|||||
| Description | B-cell receptor CD22 precursor (Leu-14) (B-lymphocyte cell adhesion molecule) (BL-CAM) (Siglec-2). | |||||
|
CD22_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 38) | NC score | 0.135358 (rank : 45) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P20273, O95699, O95701, O95702, O95703, Q01665, Q92872, Q92873, Q9UQA6, Q9UQA7, Q9UQA8, Q9UQA9, Q9UQB0, Q9Y2A6 | Gene names | CD22 | |||
|
Domain Architecture |

|
|||||
| Description | B-cell receptor CD22 precursor (Leu-14) (B-lymphocyte cell adhesion molecule) (BL-CAM) (Siglec-2). | |||||
|
FCRLA_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 39) | NC score | 0.336420 (rank : 36) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q7L513, Q5VXA1, Q5VXA2, Q5VXA3, Q5VXB0, Q8NEW4, Q8WXH3, Q96PC6, Q96PJ0, Q96PJ1, Q96PJ2, Q96PJ4, Q9BR57 | Gene names | FCRLA, FCRL, FCRL1, FCRLM1, FCRX, FREB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fc receptor-like and mucin-like 1 precursor (Fc receptor-like A) (Fc receptor homolog expressed in B-cells) (Fc receptor-related protein X) (FcRX) (Fc receptor-like protein). | |||||
|
FCRLA_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 40) | NC score | 0.409040 (rank : 35) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q920A9, Q3T9Q2, Q8K3U9, Q8VHP5 | Gene names | Fcrla, Fcrl, Fcrl1, Fcrlm1, Fcrx, Freb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fc receptor-like and mucin-like 1 precursor (Fc receptor-like A) (Fc receptor homolog expressed in B-cells) (Fc receptor-related protein X) (FcRX) (Fc receptor-like protein). | |||||
|
PGBM_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 41) | NC score | 0.068229 (rank : 59) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q05793 | Gene names | Hspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
CD276_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 42) | NC score | 0.172898 (rank : 39) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q5ZPR3, Q6P5Y4, Q6UXI2, Q8NBI8, Q8NC34, Q8NCB6, Q9BXR1 | Gene names | CD276, B7H3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CD276 antigen precursor (Costimulatory molecule) (B7 homolog 3) (B7- H3) (4Ig-B7-H3). | |||||
|
PGBM_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 43) | NC score | 0.072442 (rank : 58) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P98160, Q16287, Q9H3V5 | Gene names | HSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
SN_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 44) | NC score | 0.094251 (rank : 52) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q62230, O55216, Q62228, Q62229 | Gene names | Sn, Sa | |||
|
Domain Architecture |

|
|||||
| Description | Sialoadhesin precursor (Sialic acid-binding Ig-like lectin-1) (Siglec- 1) (Sheep erythrocyte receptor) (SER) (CD169 antigen). | |||||
|
VCAM1_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 45) | NC score | 0.097567 (rank : 51) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 320 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P29533 | Gene names | Vcam1, Vcam-1 | |||
|
Domain Architecture |

|
|||||
| Description | Vascular cell adhesion protein 1 precursor (V-CAM 1). | |||||
|
MINT_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 46) | NC score | 0.017129 (rank : 89) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
NTRK1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 47) | NC score | 0.002226 (rank : 130) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1023 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P04629, P08119, Q15655, Q15656, Q5D056, Q5VZS2, Q9UIU7 | Gene names | NTRK1, TRK | |||
|
Domain Architecture |

|
|||||
| Description | High affinity nerve growth factor receptor precursor (EC 2.7.10.1) (Neurotrophic tyrosine kinase receptor type 1) (TRK1 transforming tyrosine kinase protein) (p140-TrkA) (Trk-A). | |||||
|
PTK7_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 48) | NC score | 0.018148 (rank : 87) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1103 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q13308, Q13417 | Gene names | PTK7, CCK4 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase-like 7 precursor (Colon carcinoma kinase 4) (CCK-4). | |||||
|
CCD1A_HUMAN
|
||||||
| θ value | 0.163984 (rank : 49) | NC score | 0.031114 (rank : 70) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6P1N0, Q7Z435, Q86XV0, Q8NF89, Q9H603, Q9NXI1 | Gene names | CC2D1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil and C2 domain-containing protein 1A (Five repressor element under dual repression-binding protein 1) (FRE under dual repression-binding protein 1) (Freud-1) (Putative NF-kappa-B- activating protein 023N). | |||||
|
SFR11_HUMAN
|
||||||
| θ value | 0.163984 (rank : 50) | NC score | 0.023118 (rank : 77) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q05519 | Gene names | SFRS11 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor arginine/serine-rich 11 (Arginine-rich 54 kDa nuclear protein) (p54). | |||||
|
DCC_MOUSE
|
||||||
| θ value | 0.21417 (rank : 51) | NC score | 0.031578 (rank : 69) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P70211 | Gene names | Dcc | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC). | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 52) | NC score | 0.022053 (rank : 78) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
GLU2B_HUMAN
|
||||||
| θ value | 0.279714 (rank : 53) | NC score | 0.019022 (rank : 84) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P14314, Q96BU9, Q96D06, Q9P0W9 | Gene names | PRKCSH, G19P1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glucosidase 2 subunit beta precursor (Glucosidase II subunit beta) (Protein kinase C substrate, 60.1 kDa protein, heavy chain) (PKCSH) (80K-H protein). | |||||
|
FCGR1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 54) | NC score | 0.126111 (rank : 46) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P26151 | Gene names | Fcgr1, Fcg1 | |||
|
Domain Architecture |

|
|||||
| Description | High affinity immunoglobulin gamma Fc receptor I precursor (Fc-gamma RI) (FcRI) (IgG Fc receptor I). | |||||
|
ICAM5_MOUSE
|
||||||
| θ value | 0.365318 (rank : 55) | NC score | 0.060425 (rank : 62) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q60625 | Gene names | Icam5, Tlcn | |||
|
Domain Architecture |

|
|||||
| Description | Intercellular adhesion molecule 5 precursor (ICAM-5) (Telencephalin). | |||||
|
PTPRM_HUMAN
|
||||||
| θ value | 0.365318 (rank : 56) | NC score | 0.014677 (rank : 96) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P28827 | Gene names | PTPRM, PTPRL1 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase mu precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase mu) (R-PTP-mu). | |||||
|
PTPRM_MOUSE
|
||||||
| θ value | 0.365318 (rank : 57) | NC score | 0.014498 (rank : 97) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P28828 | Gene names | Ptprm | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase mu precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase mu) (R-PTP-mu). | |||||
|
CCNK_MOUSE
|
||||||
| θ value | 0.62314 (rank : 58) | NC score | 0.015852 (rank : 93) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
CD008_HUMAN
|
||||||
| θ value | 0.62314 (rank : 59) | NC score | 0.018584 (rank : 85) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P78312, O43607, P78311, P78313, Q9UEG8 | Gene names | C4orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C4orf8 (Protein IT14). | |||||
|
KV2A_MOUSE
|
||||||
| θ value | 0.62314 (rank : 60) | NC score | 0.014245 (rank : 98) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P01626 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig kappa chain V-II region MOPC 167. | |||||
|
TARA_HUMAN
|
||||||
| θ value | 0.62314 (rank : 61) | NC score | 0.012032 (rank : 102) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
PDL2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 62) | NC score | 0.054304 (rank : 64) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9WUL5 | Gene names | Pdcd1lg2, B7dc, Btdc, Cd273, Pdl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Programmed cell death 1 ligand 2 precursor (Programmed death ligand 2) (PD-L2) (PD-1-ligand 2) (PDCD1 ligand 2) (Butyrophilin B7-DC) (B7-DC) (CD273 antigen). | |||||
|
PTPRK_HUMAN
|
||||||
| θ value | 0.813845 (rank : 63) | NC score | 0.019210 (rank : 83) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15262, Q14763 | Gene names | PTPRK, PTPK | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase kappa precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase kappa) (R-PTP-kappa). | |||||
|
PTPRK_MOUSE
|
||||||
| θ value | 0.813845 (rank : 64) | NC score | 0.019411 (rank : 82) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P35822 | Gene names | Ptprk, Ptpk | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase kappa precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase kappa) (R-PTP-kappa). | |||||
|
CD276_MOUSE
|
||||||
| θ value | 1.06291 (rank : 65) | NC score | 0.142331 (rank : 43) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8VE98 | Gene names | Cd276, B7h3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CD276 antigen precursor (Costimulatory molecule) (B7 homolog 3) (B7- H3). | |||||
|
CN155_HUMAN
|
||||||
| θ value | 1.06291 (rank : 66) | NC score | 0.026612 (rank : 73) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
KV2B_MOUSE
|
||||||
| θ value | 1.06291 (rank : 67) | NC score | 0.010557 (rank : 105) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P01627 | Gene names | ||||
|
Domain Architecture |
|
|||||
| Description | Ig kappa chain V-II region VKappa167 precursor. | |||||
|
ADDB_HUMAN
|
||||||
| θ value | 1.38821 (rank : 68) | NC score | 0.008930 (rank : 111) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35612, Q13482, Q16412, Q59G82, Q5U5P4, Q6P0P2, Q6PGQ4, Q7Z688, Q7Z689, Q7Z690, Q7Z691 | Gene names | ADD2, ADDB | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta). | |||||
|
DCC_HUMAN
|
||||||
| θ value | 1.38821 (rank : 69) | NC score | 0.021033 (rank : 80) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 583 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P43146 | Gene names | DCC | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC) (Colorectal cancer suppressor). | |||||
|
DSPP_MOUSE
|
||||||
| θ value | 1.38821 (rank : 70) | NC score | 0.008513 (rank : 113) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
TREX1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 71) | NC score | 0.016111 (rank : 92) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NSU2, Q8TEU2, Q9BPW1, Q9Y4X2 | Gene names | TREX1 | |||
|
Domain Architecture |
|
|||||
| Description | Three prime repair exonuclease 1 (EC 3.1.11.2) (3'-5' exonuclease TREX1) (DNase III). | |||||
|
UBP43_MOUSE
|
||||||
| θ value | 1.38821 (rank : 72) | NC score | 0.010391 (rank : 106) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BUM9, Q8VDP5 | Gene names | Usp43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
ZEP1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 73) | NC score | -0.000168 (rank : 135) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||
|
BASP_HUMAN
|
||||||
| θ value | 1.81305 (rank : 74) | NC score | 0.011459 (rank : 104) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
CB017_MOUSE
|
||||||
| θ value | 1.81305 (rank : 75) | NC score | 0.015437 (rank : 94) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6NS82, Q3TA48, Q8BSD3, Q8CHY1, Q8R0Q3 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C2orf17 homolog. | |||||
|
JPH2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 76) | NC score | 0.023413 (rank : 76) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ET78, Q9ET79 | Gene names | Jph2, Jp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
M3K12_MOUSE
|
||||||
| θ value | 1.81305 (rank : 77) | NC score | -0.002479 (rank : 139) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q60700, P70286, Q3TLL7, Q8C4N7, Q8CBX3, Q8CDL6 | Gene names | Map3k12, Zpk | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 12 (EC 2.7.11.25) (Mixed lineage kinase) (Leucine-zipper protein kinase) (ZPK) (Dual leucine zipper bearing kinase) (DLK) (MAPK-upstream kinase) (MUK). | |||||
|
MYOM1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 78) | NC score | 0.018286 (rank : 86) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q62234 | Gene names | Myom1 | |||
|
Domain Architecture |

|
|||||
| Description | Myomesin-1 (Skelemin). | |||||
|
SN_HUMAN
|
||||||
| θ value | 1.81305 (rank : 79) | NC score | 0.098564 (rank : 50) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9BZZ2, Q96DL4, Q9GZS5, Q9H1H6, Q9H1H7, Q9H7L7 | Gene names | SN | |||
|
Domain Architecture |

|
|||||
| Description | Sialoadhesin precursor (Sialic acid-binding Ig-like lectin-1) (Siglec- 1) (CD169 antigen). | |||||
|
TATD2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 80) | NC score | 0.032837 (rank : 68) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q93075 | Gene names | TATDN2, KIAA0218 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TatD DNase domain-containing deoxyribonuclease 2 (EC 3.1.21.-). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 1.81305 (rank : 81) | NC score | 0.024498 (rank : 74) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
TOB1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 82) | NC score | 0.009780 (rank : 107) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P50616, Q4KMQ0 | Gene names | TOB1, TOB | |||
|
Domain Architecture |

|
|||||
| Description | Tob1 protein (Transducer of erbB-2 1). | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 2.36792 (rank : 83) | NC score | 0.005620 (rank : 126) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
PDL2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 84) | NC score | 0.045902 (rank : 67) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BQ51, Q5T7Z6, Q6JXL8, Q6JXL9 | Gene names | PDCD1LG2, B7DC, CD273, PDCD1L2, PDL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Programmed cell death 1 ligand 2 precursor (Programmed death ligand 2) (PD-L2) (PD-1-ligand 2) (PDCD1 ligand 2) (Butyrophilin B7-DC) (B7-DC) (CD273 antigen). | |||||
|
PELP1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 85) | NC score | 0.011539 (rank : 103) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IZL8, O15450, Q5EGN3, Q6NTE6, Q96FT1, Q9BU60 | Gene names | PELP1, HMX3, MNAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3). | |||||
|
ACINU_HUMAN
|
||||||
| θ value | 3.0926 (rank : 86) | NC score | 0.007215 (rank : 118) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
CDON_HUMAN
|
||||||
| θ value | 3.0926 (rank : 87) | NC score | 0.030698 (rank : 71) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q4KMG0, O14631 | Gene names | CDON, CDO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell adhesion molecule-related/down-regulated by oncogenes precursor. | |||||
|
MDM2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 88) | NC score | 0.009399 (rank : 110) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P23804, Q61040, Q64330 | Gene names | Mdm2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3 Mdm2 (EC 6.3.2.-) (p53-binding protein Mdm2) (Oncoprotein Mdm2) (Double minute 2 protein). | |||||
|
MYOM1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 89) | NC score | 0.016200 (rank : 91) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P52179 | Gene names | MYOM1 | |||
|
Domain Architecture |

|
|||||
| Description | Myomesin-1 (190 kDa titin-associated protein) (190 kDa connectin- associated protein). | |||||
|
NUCL_MOUSE
|
||||||
| θ value | 3.0926 (rank : 90) | NC score | 0.012590 (rank : 99) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P09405, Q548M9, Q61991, Q99K50 | Gene names | Ncl, Nuc | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
41_MOUSE
|
||||||
| θ value | 4.03905 (rank : 91) | NC score | 0.002180 (rank : 131) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48193, Q68FF1, Q6NVF5 | Gene names | Epb41, Epb4.1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein 4.1 (Band 4.1) (P4.1) (4.1R). | |||||
|
CNOT3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 92) | NC score | 0.019969 (rank : 81) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O75175, Q9NZN7, Q9UF76 | Gene names | CNOT3, KIAA0691, NOT3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
HMGN1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 93) | NC score | 0.016300 (rank : 90) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P05114, Q3KQR8 | Gene names | HMGN1, HMG14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nonhistone chromosomal protein HMG-14 (High-mobility group nucleosome- binding domain-containing protein 1). | |||||
|
IPPD_MOUSE
|
||||||
| θ value | 4.03905 (rank : 94) | NC score | 0.012346 (rank : 100) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q60829, Q91XB5 | Gene names | Ppp1r1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dopamine- and cAMP-regulated neuronal phosphoprotein (DARPP-32). | |||||
|
LV0A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 95) | NC score | 0.007082 (rank : 119) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P04211 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig lambda chain V region 4A precursor. | |||||
|
MUC18_MOUSE
|
||||||
| θ value | 4.03905 (rank : 96) | NC score | 0.163444 (rank : 41) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8R2Y2, Q9EPF1, Q9ESS7, Q9JHQ2, Q9JHQ3 | Gene names | Mcam, Muc18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell surface glycoprotein MUC18 precursor (Melanoma-associated antigen MUC18) (Melanoma cell adhesion molecule) (Gicerin). | |||||
|
AHTF1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 97) | NC score | 0.007470 (rank : 115) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CJF7, Q8BVJ5, Q8VD55 | Gene names | Ahctf1, Elys | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-hook-containing transcription factor 1 (Embryonic large molecule derived from yolk sac). | |||||
|
CNTN4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 98) | NC score | 0.021275 (rank : 79) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 449 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8IWV2, Q8IX14, Q8TC35 | Gene names | CNTN4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-4 precursor (Brain-derived immunoglobulin superfamily protein 2) (BIG-2). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 99) | NC score | 0.017551 (rank : 88) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
NBR1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 100) | NC score | 0.007291 (rank : 117) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97432, Q9JIH9 | Gene names | Nbr1, M17s2 | |||
|
Domain Architecture |

|
|||||
| Description | Next to BRCA1 gene 1 protein (Neighbor of BRCA1 gene 1 protein) (Membrane component, chromosome 17, surface marker 2). | |||||
|
TGFR1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 101) | NC score | -0.001284 (rank : 137) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 755 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P36897 | Gene names | TGFBR1 | |||
|
Domain Architecture |

|
|||||
| Description | TGF-beta receptor type-1 precursor (EC 2.7.11.30) (TGF-beta receptor type I) (TGFR-1) (TGF-beta type I receptor) (Serine/threonine-protein kinase receptor R4) (SKR4) (Activin receptor-like kinase 5) (ALK-5). | |||||
|
BASP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | 0.009702 (rank : 108) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
IF39_MOUSE
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | 0.008046 (rank : 114) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8JZQ9, Q922K2 | Gene names | Eif3s9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116). | |||||
|
MALT1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | 0.029570 (rank : 72) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UDY8, Q9NTB7, Q9ULX4 | Gene names | MALT1, MLT | |||
|
Domain Architecture |

|
|||||
| Description | Mucosa-associated lymphoid tissue lymphoma translocation protein 1 (EC 3.4.22.-) (MALT lymphoma-associated translocation) (Paracaspase). | |||||
|
MTFR1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 105) | NC score | 0.008715 (rank : 112) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99MB2, Q9CQZ4 | Gene names | Mtfr1, Kiaa0009 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial fission regulator 1. | |||||
|
NELFE_MOUSE
|
||||||
| θ value | 6.88961 (rank : 106) | NC score | 0.006862 (rank : 122) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P19426, P97485, P97486, Q9CZN3 | Gene names | Rdbp, D17h6s45, Nelfe, Rd | |||
|
Domain Architecture |

|
|||||
| Description | Negative elongation factor E (NELF-E) (RD protein). | |||||
|
NUCKS_HUMAN
|
||||||
| θ value | 6.88961 (rank : 107) | NC score | 0.006455 (rank : 124) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H1E3, Q9H1D6, Q9H723 | Gene names | NUCKS1, NUCKS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (P1). | |||||
|
PDE4D_MOUSE
|
||||||
| θ value | 6.88961 (rank : 108) | NC score | 0.001408 (rank : 134) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q01063, Q6TRH9, Q8C4Q7, Q8CG05 | Gene names | Pde4d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4D (EC 3.1.4.17) (DPDE3). | |||||
|
SRC8_MOUSE
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.001708 (rank : 132) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60598 | Gene names | Cttn, Ems1 | |||
|
Domain Architecture |
|
|||||
| Description | Src substrate cortactin. | |||||
|
TCOF_MOUSE
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.007018 (rank : 120) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
TF7L1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.002788 (rank : 129) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z1J1, O70450, O70573 | Gene names | Tcf7l1, Tcf3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 1 (HMG box transcription factor 3) (TCF-3) (mTCF-3). | |||||
|
TGFR1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 112) | NC score | -0.001424 (rank : 138) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q64729 | Gene names | Tgfbr1 | |||
|
Domain Architecture |

|
|||||
| Description | TGF-beta receptor type-1 precursor (EC 2.7.11.30) (TGF-beta receptor type I) (TGFR-1) (TGF-beta type I receptor) (ESK2). | |||||
|
TNR16_HUMAN
|
||||||
| θ value | 6.88961 (rank : 113) | NC score | 0.006102 (rank : 125) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P08138 | Gene names | NGFR, TNFRSF16 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 16 precursor (Low- affinity nerve growth factor receptor) (NGF receptor) (Gp80-LNGFR) (p75 ICD) (Low affinity neurotrophin receptor p75NTR) (CD271 antigen). | |||||
|
ZMYM3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.012131 (rank : 101) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14202, O15089 | Gene names | ZMYM3, DXS6673E, KIAA0385, ZNF261 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 3 (Zinc finger protein 261). | |||||
|
CNOT3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.014900 (rank : 95) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8K0V4 | Gene names | Cnot3, Not3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
DTX2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.006983 (rank : 121) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86UW9, Q96H69, Q9H890, Q9P200 | Gene names | DTX2, KIAA1528 | |||
|
Domain Architecture |
|
|||||
| Description | Protein deltex-2 (Deltex-2) (Deltex2) (hDTX2). | |||||
|
KV2F_MOUSE
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.003290 (rank : 128) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P01630 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig kappa chain V-II region 7S34.1. | |||||
|
LAP4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | -0.002634 (rank : 140) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14160, Q6P496, Q7Z5D1, Q8WWV8, Q96C69, Q96GG1 | Gene names | SCRIB, CRIB1, KIAA0147, LAP4, SCRB1, VARTUL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog) (hScrib). | |||||
|
MYST3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.001456 (rank : 133) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
NKX26_MOUSE
|
||||||
| θ value | 8.99809 (rank : 120) | NC score | -0.000680 (rank : 136) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P43688 | Gene names | Nkx2-6, Nkx-2.6, Nkx2f | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-2.6 (Homeobox protein NK-2 homolog F). | |||||
|
ROBO4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 121) | NC score | 0.024236 (rank : 75) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8C310, Q9DBW1 | Gene names | Robo4 | |||
|
Domain Architecture |
|
|||||
| Description | Roundabout homolog 4 precursor. | |||||
|
RTN4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 122) | NC score | 0.007452 (rank : 116) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99P72, Q5DTK9, Q7TNB7, Q80W95, Q8BGK7, Q8BGM9, Q8K290, Q8K3G8, Q9CTE3 | Gene names | Rtn4, Kiaa0886, Nogo | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-4 (Neurite outgrowth inhibitor) (Nogo protein). | |||||
|
SMG7_MOUSE
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | 0.004840 (rank : 127) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5RJH6, Q63ZW5, Q6ZQF3 | Gene names | Smg7, Est1c, Kiaa0250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein SMG7 (SMG-7 homolog) (EST1-like protein C). | |||||
|
TCOF_HUMAN
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.009514 (rank : 109) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
TM115_HUMAN
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.006770 (rank : 123) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12893, O14568, Q9UIX3 | Gene names | TMEM115, PL6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 115 (Protein PL6) (Placental protein 6) (PP6). | |||||
|
CEA20_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.066798 (rank : 60) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6UY09 | Gene names | CEACAM20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carcinoembryonic antigen-related cell adhesion molecule 20 precursor. | |||||
|
CEAM5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.050860 (rank : 66) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P06731 | Gene names | CEACAM5, CEA | |||
|
Domain Architecture |

|
|||||
| Description | Carcinoembryonic antigen-related cell adhesion molecule 5 precursor (Carcinoembryonic antigen) (CEA) (Meconium antigen 100) (CD66e antigen). | |||||
|
FCERA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.079650 (rank : 54) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P12319 | Gene names | FCER1A, FCE1A | |||
|
Domain Architecture |
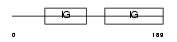
|
|||||
| Description | High affinity immunoglobulin epsilon receptor alpha-subunit precursor (FcERI) (IgE Fc receptor, alpha-subunit) (Fc-epsilon RI-alpha). | |||||
|
FCG2A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.078871 (rank : 55) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P12318 | Gene names | FCGR2A, CD32, FCG2, IGFR2 | |||
|
Domain Architecture |
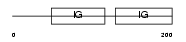
|
|||||
| Description | Low affinity immunoglobulin gamma Fc region receptor II-a precursor (Fc-gamma RII-a) (FcRII-a) (IgG Fc receptor II-a) (Fc-gamma-RIIa) (CD32 antigen) (CDw32). | |||||
|
FCG2B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.111991 (rank : 47) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P31994, O95649, Q53X85, Q5VXA9, Q8NIA1 | Gene names | FCGR2B, CD32, FCG2, IGFR2 | |||
|
Domain Architecture |
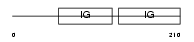
|
|||||
| Description | Low affinity immunoglobulin gamma Fc region receptor II-b precursor (Fc-gamma RII-b) (FcRII-b) (IgG Fc receptor II-b) (Fc-gamma-RIIb) (CD32 antigen) (CDw32). | |||||
|
FCG2C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.109881 (rank : 48) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P31995, O00523, O00524, O00525 | Gene names | FCGR2C, CD32, FCG2, IGFR2 | |||
|
Domain Architecture |
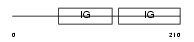
|
|||||
| Description | Low affinity immunoglobulin gamma Fc region receptor II-c precursor (Fc-gamma RII-c) (FcRII-c) (IgG Fc receptor II-c) (Fc-gamma-RIIc) (CD32 antigen) (CDw32). | |||||
|
FCG3A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.063060 (rank : 61) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P08637 | Gene names | FCGR3A, CD16A, FCG3, FCGR3, IGFR3 | |||
|
Domain Architecture |
|
|||||
| Description | Low affinity immunoglobulin gamma Fc region receptor III-A precursor (IgG Fc receptor III-2) (Fc-gamma RIII-alpha) (Fc-gamma RIIIa) (FcRIIIa) (Fc-gamma RIII) (FcRIII) (FcR-10) (CD16a antigen). | |||||
|
FCG3B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.059251 (rank : 63) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O75015 | Gene names | FCGR3B, CD16B, FCG3, FCGR3, IGFR3 | |||
|
Domain Architecture |
|
|||||
| Description | Low affinity immunoglobulin gamma Fc region receptor III-B precursor (IgG Fc receptor III-1) (Fc-gamma RIII-beta) (Fc-gamma RIIIb) (FcRIIIb) (Fc-gamma RIII) (FcRIII) (FcR-10) (CD16b antigen). | |||||
|
FCGR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.136473 (rank : 44) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P12314, P12315, Q92495, Q92663 | Gene names | FCGR1A, FCG1, FCGR1, IGFR1 | |||
|
Domain Architecture |
|
|||||
| Description | High affinity immunoglobulin gamma Fc receptor I precursor (Fc-gamma RI) (FcRI) (IgG Fc receptor I) (CD64 antigen). | |||||
|
FCRL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.101871 (rank : 49) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96LA5, Q6NTA1, Q9BZI4, Q9BZI5, Q9BZI6 | Gene names | FCRL2, FCRH2, IFGP4, IRTA4, SPAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fc receptor-like protein 2 precursor (SH2 domain-containing phosphatase anchor protein 1) (Fc receptor homolog 2) (FcRH2) (Immunoglobulin receptor translocation-associated 4 protein). | |||||
|
FCRL5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.154637 (rank : 42) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96RD9, Q495Q2, Q495Q4, Q5VYK9, Q6UY46 | Gene names | FCRL5, FCRH5, IRTA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fc receptor-like protein 5 precursor (Immunoglobulin receptor translocation-associated gene 2 protein) (BXMAS1) (CD307 antigen). | |||||
|
MUC18_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.072473 (rank : 57) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P43121, O95812, Q59E86, Q6PHR3, Q6ZTR2 | Gene names | MCAM, MUC18 | |||
|
Domain Architecture |

|
|||||
| Description | Cell surface glycoprotein MUC18 precursor (Melanoma-associated antigen MUC18) (Melanoma cell adhesion molecule) (Melanoma-associated antigen A32) (S-endo 1 endothelial-associated antigen) (Cell surface glycoprotein P1H12) (CD146 antigen). | |||||
|
PECA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.090072 (rank : 53) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P16284, Q6LDA9, Q8TBH1, Q96RF5, Q96RF6, Q9NP65, Q9NPB7, Q9NPG9, Q9NQS9, Q9NQT0, Q9NQT1, Q9NQT2 | Gene names | PECAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Platelet endothelial cell adhesion molecule precursor (PECAM-1) (EndoCAM) (GPIIA') (CD31 antigen). | |||||
|
PECA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.051920 (rank : 65) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q08481 | Gene names | Pecam1, Pecam, Pecam-1 | |||
|
Domain Architecture |

|
|||||
| Description | Platelet endothelial cell adhesion molecule precursor (PECAM-1) (CD31 antigen). | |||||
|
VCAM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.074496 (rank : 56) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P19320, Q6NUP8 | Gene names | VCAM1, L1CAM | |||
|
Domain Architecture |

|
|||||
| Description | Vascular cell adhesion protein 1 precursor (V-CAM 1) (CD106 antigen) (INCAM-100). | |||||
|
LIRB3_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 125 | |
| SwissProt Accessions | O75022, O15471, Q86U49 | Gene names | LILRB3, ILT5, LIR3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily B member 3 precursor (Leukocyte immunoglobulin-like receptor 3) (LIR-3) (Immunoglobulin- like transcript 5) (ILT-5) (Monocyte inhibitory receptor HL9) (CD85a antigen). | |||||
|
LIRB1_HUMAN
|
||||||
| NC score | 0.989732 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8NHL6, O75024, O75025, Q8NHJ9, Q8NHK0 | Gene names | LILRB1, ILT2, LIR1, MIR7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily B member 1 precursor (Leukocyte immunoglobulin-like receptor 1) (LIR-1) (Immunoglobulin- like transcript 2) (ILT-2) (Monocyte/macrophage immunoglobulin-like receptor 7) (MIR-7) (CD85j antigen). | |||||
|
LIRB2_HUMAN
|
||||||
| NC score | 0.989077 (rank : 3) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8N423, O75017, Q8NHJ7, Q8NHJ8 | Gene names | LILRB2, ILT4, LIR2, MIR10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily B member 2 precursor (Leukocyte immunoglobulin-like receptor 2) (LIR-2) (Immunoglobulin- like transcript 4) (ILT-4) (Monocyte/macrophage immunoglobulin-like receptor 10) (MIR-10) (CD85d antigen). | |||||
|
LIRA1_HUMAN
|
||||||
| NC score | 0.986914 (rank : 4) | θ value | 3.85598e-160 (rank : 7) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O75019, O75018 | Gene names | LILRA1, LIR6 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily A member 1 precursor (Leukocyte immunoglobulin-like receptor 6) (LIR-6) (CD85i antigen). | |||||
|
LIRA4_HUMAN
|
||||||
| NC score | 0.986441 (rank : 5) | θ value | 2.17438e-179 (rank : 5) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P59901 | Gene names | LILRA4, ILT7 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily A member 4 precursor (Immunoglobulin-like transcript 7) (ILT-7) (CD85g antigen). | |||||
|
LIRB5_HUMAN
|
||||||
| NC score | 0.986187 (rank : 6) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O75023, Q8N760 | Gene names | LILRB5, LIR8 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily B member 5 precursor (Leukocyte immunoglobulin-like receptor 8) (LIR-8) (CD85c antigen). | |||||
|
LIRA3_HUMAN
|
||||||
| NC score | 0.982596 (rank : 7) | θ value | 2.67036e-161 (rank : 6) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8N6C8, O15469, O15470, O75016, Q8N151, Q8N154, Q8NHJ1, Q8NHJ2, Q8NHJ3, Q8NHJ4 | Gene names | LILRA3, ILT6, LIR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily A member 3 precursor (Leukocyte immunoglobulin-like receptor 4) (LIR-4) (Immunoglobulin- like transcript 6) (ILT-6) (Monocyte inhibitory receptor HM43/HM31) (CD85e antigen). | |||||
|
LIRA2_HUMAN
|
||||||
| NC score | 0.981094 (rank : 8) | θ value | 6.37065e-155 (rank : 8) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8N149, O75020 | Gene names | LILRA2, ILT1, LIR7 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily A member 2 precursor (Leukocyte immunoglobulin-like receptor 7) (LIR-7) (Immunoglobulin- like transcript 1) (ILT-1) (CD85h antigen). | |||||
|
LIRB4_HUMAN
|
||||||
| NC score | 0.969504 (rank : 9) | θ value | 2.06847e-121 (rank : 9) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8NHJ6, O15468, O75021, Q8N1C7, Q8NHL5 | Gene names | LILRB4, ILT3, LIR5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily B member 4 precursor (Leukocyte immunoglobulin-like receptor 5) (LIR-5) (Immunoglobulin- like transcript 3) (ILT-3) (Monocyte inhibitory receptor HM18) (CD85k antigen). | |||||
|
LIRB4_MOUSE
|
||||||
| NC score | 0.952959 (rank : 10) | θ value | 6.34462e-54 (rank : 15) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q64281, Q64312 | Gene names | Lilrb4, Gp49b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily B member 4 precursor (Mast cell surface glycoprotein Gp49B). | |||||
|
GP49A_MOUSE
|
||||||
| NC score | 0.947482 (rank : 11) | θ value | 7.49467e-55 (rank : 14) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q61450 | Gene names | Gp49a, Gp49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mast cell surface glycoprotein Gp49A precursor. | |||||
|
GPVI_HUMAN
|
||||||
| NC score | 0.936942 (rank : 12) | θ value | 3.26607e-42 (rank : 24) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9HCN6, Q9HCN7, Q9UIF2 | Gene names | GP6, GPVI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Platelet glycoprotein VI precursor. | |||||
|
NCTR1_HUMAN
|
||||||
| NC score | 0.933240 (rank : 13) | θ value | 3.05695e-40 (rank : 26) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O76036, O76016, O76017, O76018 | Gene names | NCR1, LY94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Natural cytotoxicity triggering receptor 1 precursor (Natural killer cell p46-related protein) (NKp46) (hNKp46) (NK-p46) (NK cell- activating receptor) (Lymphocyte antigen 94 homolog) (CD335 antigen). | |||||
|
GPVI_MOUSE
|
||||||
| NC score | 0.931185 (rank : 14) | θ value | 1.32909e-35 (rank : 28) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P0C191 | Gene names | Gp6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Platelet glycoprotein VI precursor (GPVI). | |||||
|
NCTR1_MOUSE
|
||||||
| NC score | 0.924874 (rank : 15) | θ value | 2.67802e-36 (rank : 27) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8C567, Q1RLN4, Q80UY6, Q9Z0Q4 | Gene names | Ncr1, Ly94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Natural cytotoxicity triggering receptor 1 precursor (Natural killer cell p46-related protein) (NKp46) (mNKp46) (NK-p46) (Mouse-activating receptor 1) (mAR-1) (Lymphocyte antigen 94). | |||||
|
FCAR_HUMAN
|
||||||
| NC score | 0.918705 (rank : 16) | θ value | 5.06226e-27 (rank : 31) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P24071, Q13603, Q13604, Q15727, Q15728, Q92590 | Gene names | FCAR, CD89 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Immunoglobulin alpha Fc receptor precursor (IgA Fc receptor) (CD89 antigen). | |||||
|
KI3L3_HUMAN
|
||||||
| NC score | 0.914212 (rank : 17) | θ value | 1.27544e-62 (rank : 13) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8N743, O95056, Q7Z7N4, Q8NHI2, Q8NHI3, Q9UEI4 | Gene names | KIR3DL3, CD158Z, KIR3DL7, KIRC1 | |||
|
Domain Architecture |

|
|||||
| Description | Killer cell immunoglobulin-like receptor 3DL3 precursor (Killer cell inhibitory receptor 1) (CD158z antigen). | |||||
|
KI3L1_MOUSE
|
||||||
| NC score | 0.914205 (rank : 18) | θ value | 1.12253e-42 (rank : 22) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P83555 | Gene names | Kir3dl1 | |||
|
Domain Architecture |

|
|||||
| Description | Killer cell immunoglobulin-like receptor 3DL1 precursor. | |||||
|
KI3S1_HUMAN
|
||||||
| NC score | 0.912097 (rank : 19) | θ value | 3.35642e-63 (rank : 12) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q14943 | Gene names | KIR3DS1, NKAT10 | |||
|
Domain Architecture |

|
|||||
| Description | Killer cell immunoglobulin-like receptor 3DS1 precursor (MHC class I NK cell receptor) (Natural killer-associated transcript 10) (NKAT-10). | |||||
|
KI3L2_HUMAN
|
||||||
| NC score | 0.909576 (rank : 20) | θ value | 1.71981e-67 (rank : 10) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P43630, Q13238, Q14947, Q14948, Q92684, Q95367 | Gene names | KIR3DL2, CD158K, NKAT4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Killer cell immunoglobulin-like receptor 3DL2 precursor (MHC class I NK cell receptor) (Natural killer-associated transcript 4) (NKAT-4) (p70 natural killer cell receptor clone CL-5) (CD158k antigen). | |||||
|
KI3L1_HUMAN
|
||||||
| NC score | 0.906339 (rank : 21) | θ value | 2.56992e-63 (rank : 11) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P43629, O43473, Q16541 | Gene names | KIR3DL1, NKAT3, NKB1 | |||
|
Domain Architecture |

|
|||||
| Description | Killer cell immunoglobulin-like receptor 3DL1 precursor (MHC class I NK cell receptor) (Natural killer-associated transcript 3) (NKAT-3) (p70 natural killer cell receptor clones CL-2/CL-11) (HLA-BW4-specific inhibitory NK cell receptor). | |||||
|
KI2S2_HUMAN
|
||||||
| NC score | 0.892411 (rank : 22) | θ value | 5.55816e-50 (rank : 16) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P43631, Q14955 | Gene names | KIR2DS2, CD158J, NKAT5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Killer cell immunoglobulin-like receptor 2DS2 precursor (MHC class I NK cell receptor) (Natural killer-associated transcript 5) (NKAT-5) (p58 natural killer cell receptor clone CL-49) (p58 NK receptor) (NK receptor 183 ActI) (CD158j antigen). | |||||
|
KI2S3_HUMAN
|
||||||
| NC score | 0.887458 (rank : 23) | θ value | 2.85459e-46 (rank : 18) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q14952, O00644 | Gene names | KIR2DS3, NKAT7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Killer cell immunoglobulin-like receptor 2DS3 precursor (MHC class I NK cell receptor) (Natural killer-associated transcript 7) (NKAT-7). | |||||
|
KI2L1_HUMAN
|
||||||
| NC score | 0.887420 (rank : 24) | θ value | 1.85029e-45 (rank : 20) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P43626, O43470 | Gene names | KIR2DL1, CD158A, NKAT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Killer cell immunoglobulin-like receptor 2DL1 precursor (MHC class I NK cell receptor) (Natural killer-associated transcript 1) (NKAT-1) (p58 natural killer cell receptor clones CL-42/47.11) (p58 NK receptor) (p58.1 MHC class-I-specific NK receptor) (CD158a antigen). | |||||
|
KI2S4_HUMAN
|
||||||
| NC score | 0.887358 (rank : 25) | θ value | 3.60269e-49 (rank : 17) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P43632 | Gene names | KIR2DS4, CD158I, KKA3, NKAT8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Killer cell immunoglobulin-like receptor 2DS4 precursor (MHC class I NK cell receptor) (Natural killer-associated transcript 8) (NKAT-8) (P58 natural killer cell receptor clone CL-39) (p58 NK receptor) (CL- 17) (CD158i antigen). | |||||
|
KI2L2_HUMAN
|
||||||
| NC score | 0.887034 (rank : 26) | θ value | 6.35935e-46 (rank : 19) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P43627, Q14951 | Gene names | KIR2DL2, NKAT6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Killer cell immunoglobulin-like receptor 2DL2 precursor (MHC class I NK cell receptor) (Natural killer-associated transcript 6) (NKAT-6) (p58 natural killer cell receptor clone CL-43) (p58 NK receptor). | |||||
|
KI2L4_HUMAN
|
||||||
| NC score | 0.885877 (rank : 27) | θ value | 3.27365e-34 (rank : 29) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q99706, O14621, O14622, O14623, O14624, O43534, P78400, P78401, Q99559, Q99560, Q99561, Q99562, Q9UQJ7 | Gene names | KIR2DL4, CD158D, KIR103AS | |||
|
Domain Architecture |

|
|||||
| Description | Killer cell immunoglobulin-like receptor 2DL4 precursor (MHC class I NK cell receptor KIR103AS) (Killer cell inhibitory receptor 103AS) (KIR-103AS) (G9P) (CD158d antigen). | |||||
|
KI2L3_HUMAN
|
||||||
| NC score | 0.885573 (rank : 28) | θ value | 8.59492e-43 (rank : 21) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P43628, O43472, P78402, Q14944, Q14945, Q9UM51, Q9UQ70 | Gene names | KIR2DL3, CD158B, KIRCL23, NKAT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Killer cell immunoglobulin-like receptor 2DL3 precursor (MHC class I NK cell receptor) (Natural killer-associated transcript 2) (NKAT-2) (NKAT2a) (NKAT2b) (p58 natural killer cell receptor clone CL-6) (p58 NK receptor) (p58.2 MHC class-I-specific NK receptor) (Killer inhibitory receptor cl 2-3) (KIR-023GB) (CD158b antigen). | |||||
|
KI2S1_HUMAN
|
||||||
| NC score | 0.884436 (rank : 29) | θ value | 1.91475e-42 (rank : 23) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q14954, O43471 | Gene names | KIR2DS1, CD158H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Killer cell immunoglobulin-like receptor 2DS1 precursor (MHC class I NK cell receptor Eb6 ActI) (CD158h antigen). | |||||
|
KI2S5_HUMAN
|
||||||
| NC score | 0.882863 (rank : 30) | θ value | 2.76489e-41 (rank : 25) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q14953 | Gene names | KIR2DS5, CD158G, NKAT9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Killer cell immunoglobulin-like receptor 2DS5 precursor (MHC class I NK cell receptor) (Natural killer-associated transcript 9) (NKAT-9) (CD158g antigen). | |||||
|
LAIR2_HUMAN
|
||||||
| NC score | 0.835790 (rank : 31) | θ value | 8.95645e-16 (rank : 33) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6ISS4, Q6PEZ4 | Gene names | LAIR2, CD306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte-associated immunoglobulin-like receptor 2 precursor (LAIR-2) (CD306 antigen). | |||||
|
A1BG_HUMAN
|
||||||
| NC score | 0.819793 (rank : 32) | θ value | 5.97985e-28 (rank : 30) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P04217, Q8IYJ6, Q96P39 | Gene names | A1BG | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-1B-glycoprotein precursor (Alpha-1-B glycoprotein). | |||||
|
LAIR1_HUMAN
|
||||||
| NC score | 0.794368 (rank : 33) | θ value | 1.80466e-16 (rank : 32) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6GTX8 | Gene names | LAIR1, CD305 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte-associated immunoglobulin-like receptor 1 precursor (LAIR-1) (hLAIR1) (CD305 antigen). | |||||
|
LAIR1_MOUSE
|
||||||
| NC score | 0.505981 (rank : 34) | θ value | 1.09739e-05 (rank : 36) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BG84, Q80UN5, Q8BHB6, Q8BRR6, Q8C6N8, Q8CEP5 | Gene names | Lair1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte-associated immunoglobulin-like receptor 1 precursor (LAIR-1) (mLAIR1) (CD305 antigen). | |||||
|
FCRLA_MOUSE
|
||||||
| NC score | 0.409040 (rank : 35) | θ value | 0.00134147 (rank : 40) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q920A9, Q3T9Q2, Q8K3U9, Q8VHP5 | Gene names | Fcrla, Fcrl, Fcrl1, Fcrlm1, Fcrx, Freb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fc receptor-like and mucin-like 1 precursor (Fc receptor-like A) (Fc receptor homolog expressed in B-cells) (Fc receptor-related protein X) (FcRX) (Fc receptor-like protein). | |||||
|
FCRLA_HUMAN
|
||||||
| NC score | 0.336420 (rank : 36) | θ value | 0.00134147 (rank : 39) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q7L513, Q5VXA1, Q5VXA2, Q5VXA3, Q5VXB0, Q8NEW4, Q8WXH3, Q96PC6, Q96PJ0, Q96PJ1, Q96PJ2, Q96PJ4, Q9BR57 | Gene names | FCRLA, FCRL, FCRL1, FCRLM1, FCRX, FREB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fc receptor-like and mucin-like 1 precursor (Fc receptor-like A) (Fc receptor homolog expressed in B-cells) (Fc receptor-related protein X) (FcRX) (Fc receptor-like protein). | |||||
|
FCGR3_MOUSE
|
||||||
| NC score | 0.231275 (rank : 37) | θ value | 4.92598e-06 (rank : 35) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P08508 | Gene names | Fcgr3 | |||
|
Domain Architecture |

|
|||||
| Description | Low affinity immunoglobulin gamma Fc region receptor III precursor (IgG Fc receptor III) (Fc-gamma RIII) (FcRIII). | |||||
|
FCGR2_MOUSE
|
||||||
| NC score | 0.227254 (rank : 38) | θ value | 1.29631e-06 (rank : 34) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P08101, P08102, P12316, P97917, Q60938, Q60939, Q60940, Q61170, Q61558 | Gene names | Fcgr2, Fcgr2b, Ly-17 | |||
|
Domain Architecture |

|
|||||
| Description | Low affinity immunoglobulin gamma Fc region receptor II precursor (Fc- gamma RII) (FcRII) (IgG Fc receptor II beta) (Fc gamma receptor IIB) (Fc-gamma-RIIB) (Lymphocyte antigen Ly-17) (CD32 antigen). | |||||
|
CD276_HUMAN
|
||||||
| NC score | 0.172898 (rank : 39) | θ value | 0.00390308 (rank : 42) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q5ZPR3, Q6P5Y4, Q6UXI2, Q8NBI8, Q8NC34, Q8NCB6, Q9BXR1 | Gene names | CD276, B7H3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CD276 antigen precursor (Costimulatory molecule) (B7 homolog 3) (B7- H3) (4Ig-B7-H3). | |||||
|
CD22_MOUSE
|
||||||
| NC score | 0.165319 (rank : 40) | θ value | 4.1701e-05 (rank : 37) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P35329, Q9JHL2, Q9JJX9, Q9JJY0, Q9JJY1, Q9R056, Q9R094, Q9WU51 | Gene names | Cd22, Lyb-8 | |||
|
Domain Architecture |

|
|||||
| Description | B-cell receptor CD22 precursor (Leu-14) (B-lymphocyte cell adhesion molecule) (BL-CAM) (Siglec-2). | |||||
|
MUC18_MOUSE
|
||||||
| NC score | 0.163444 (rank : 41) | θ value | 4.03905 (rank : 96) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8R2Y2, Q9EPF1, Q9ESS7, Q9JHQ2, Q9JHQ3 | Gene names | Mcam, Muc18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell surface glycoprotein MUC18 precursor (Melanoma-associated antigen MUC18) (Melanoma cell adhesion molecule) (Gicerin). | |||||
|
FCRL5_HUMAN
|
||||||
| NC score | 0.154637 (rank : 42) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96RD9, Q495Q2, Q495Q4, Q5VYK9, Q6UY46 | Gene names | FCRL5, FCRH5, IRTA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fc receptor-like protein 5 precursor (Immunoglobulin receptor translocation-associated gene 2 protein) (BXMAS1) (CD307 antigen). | |||||
|
CD276_MOUSE
|
||||||
| NC score | 0.142331 (rank : 43) | θ value | 1.06291 (rank : 65) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8VE98 | Gene names | Cd276, B7h3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CD276 antigen precursor (Costimulatory molecule) (B7 homolog 3) (B7- H3). | |||||
|
FCGR1_HUMAN
|
||||||
| NC score | 0.136473 (rank : 44) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P12314, P12315, Q92495, Q92663 | Gene names | FCGR1A, FCG1, FCGR1, IGFR1 | |||
|
Domain Architecture |

|
|||||
| Description | High affinity immunoglobulin gamma Fc receptor I precursor (Fc-gamma RI) (FcRI) (IgG Fc receptor I) (CD64 antigen). | |||||
|
CD22_HUMAN
|
||||||
| NC score | 0.135358 (rank : 45) | θ value | 0.000786445 (rank : 38) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P20273, O95699, O95701, O95702, O95703, Q01665, Q92872, Q92873, Q9UQA6, Q9UQA7, Q9UQA8, Q9UQA9, Q9UQB0, Q9Y2A6 | Gene names | CD22 | |||
|
Domain Architecture |

|
|||||
| Description | B-cell receptor CD22 precursor (Leu-14) (B-lymphocyte cell adhesion molecule) (BL-CAM) (Siglec-2). | |||||
|
FCGR1_MOUSE
|
||||||
| NC score | 0.126111 (rank : 46) | θ value | 0.365318 (rank : 54) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P26151 | Gene names | Fcgr1, Fcg1 | |||
|
Domain Architecture |

|
|||||
| Description | High affinity immunoglobulin gamma Fc receptor I precursor (Fc-gamma RI) (FcRI) (IgG Fc receptor I). | |||||
|
FCG2B_HUMAN
|
||||||
| NC score | 0.111991 (rank : 47) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P31994, O95649, Q53X85, Q5VXA9, Q8NIA1 | Gene names | FCGR2B, CD32, FCG2, IGFR2 | |||
|
Domain Architecture |

|
|||||
| Description | Low affinity immunoglobulin gamma Fc region receptor II-b precursor (Fc-gamma RII-b) (FcRII-b) (IgG Fc receptor II-b) (Fc-gamma-RIIb) (CD32 antigen) (CDw32). | |||||
|
FCG2C_HUMAN
|
||||||
| NC score | 0.109881 (rank : 48) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P31995, O00523, O00524, O00525 | Gene names | FCGR2C, CD32, FCG2, IGFR2 | |||
|
Domain Architecture |

|
|||||
| Description | Low affinity immunoglobulin gamma Fc region receptor II-c precursor (Fc-gamma RII-c) (FcRII-c) (IgG Fc receptor II-c) (Fc-gamma-RIIc) (CD32 antigen) (CDw32). | |||||
|
FCRL2_HUMAN
|
||||||
| NC score | 0.101871 (rank : 49) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96LA5, Q6NTA1, Q9BZI4, Q9BZI5, Q9BZI6 | Gene names | FCRL2, FCRH2, IFGP4, IRTA4, SPAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fc receptor-like protein 2 precursor (SH2 domain-containing phosphatase anchor protein 1) (Fc receptor homolog 2) (FcRH2) (Immunoglobulin receptor translocation-associated 4 protein). | |||||
|
SN_HUMAN
|
||||||
| NC score | 0.098564 (rank : 50) | θ value | 1.81305 (rank : 79) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9BZZ2, Q96DL4, Q9GZS5, Q9H1H6, Q9H1H7, Q9H7L7 | Gene names | SN | |||
|
Domain Architecture |

|
|||||
| Description | Sialoadhesin precursor (Sialic acid-binding Ig-like lectin-1) (Siglec- 1) (CD169 antigen). | |||||
|
VCAM1_MOUSE
|
||||||
| NC score | 0.097567 (rank : 51) | θ value | 0.0113563 (rank : 45) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 320 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P29533 | Gene names | Vcam1, Vcam-1 | |||
|
Domain Architecture |

|
|||||
| Description | Vascular cell adhesion protein 1 precursor (V-CAM 1). | |||||
|
SN_MOUSE
|
||||||
| NC score | 0.094251 (rank : 52) | θ value | 0.0113563 (rank : 44) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q62230, O55216, Q62228, Q62229 | Gene names | Sn, Sa | |||
|
Domain Architecture |

|
|||||
| Description | Sialoadhesin precursor (Sialic acid-binding Ig-like lectin-1) (Siglec- 1) (Sheep erythrocyte receptor) (SER) (CD169 antigen). | |||||
|
PECA1_HUMAN
|
||||||
| NC score | 0.090072 (rank : 53) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P16284, Q6LDA9, Q8TBH1, Q96RF5, Q96RF6, Q9NP65, Q9NPB7, Q9NPG9, Q9NQS9, Q9NQT0, Q9NQT1, Q9NQT2 | Gene names | PECAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Platelet endothelial cell adhesion molecule precursor (PECAM-1) (EndoCAM) (GPIIA') (CD31 antigen). | |||||
|
FCERA_HUMAN
|
||||||
| NC score | 0.079650 (rank : 54) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P12319 | Gene names | FCER1A, FCE1A | |||
|
Domain Architecture |

|
|||||
| Description | High affinity immunoglobulin epsilon receptor alpha-subunit precursor (FcERI) (IgE Fc receptor, alpha-subunit) (Fc-epsilon RI-alpha). | |||||
|
FCG2A_HUMAN
|
||||||
| NC score | 0.078871 (rank : 55) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P12318 | Gene names | FCGR2A, CD32, FCG2, IGFR2 | |||
|
Domain Architecture |

|
|||||
| Description | Low affinity immunoglobulin gamma Fc region receptor II-a precursor (Fc-gamma RII-a) (FcRII-a) (IgG Fc receptor II-a) (Fc-gamma-RIIa) (CD32 antigen) (CDw32). | |||||
|
VCAM1_HUMAN
|
||||||
| NC score | 0.074496 (rank : 56) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P19320, Q6NUP8 | Gene names | VCAM1, L1CAM | |||
|
Domain Architecture |

|
|||||
| Description | Vascular cell adhesion protein 1 precursor (V-CAM 1) (CD106 antigen) (INCAM-100). | |||||
|
MUC18_HUMAN
|
||||||
| NC score | 0.072473 (rank : 57) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P43121, O95812, Q59E86, Q6PHR3, Q6ZTR2 | Gene names | MCAM, MUC18 | |||
|
Domain Architecture |

|
|||||
| Description | Cell surface glycoprotein MUC18 precursor (Melanoma-associated antigen MUC18) (Melanoma cell adhesion molecule) (Melanoma-associated antigen A32) (S-endo 1 endothelial-associated antigen) (Cell surface glycoprotein P1H12) (CD146 antigen). | |||||
|
PGBM_HUMAN
|
||||||
| NC score | 0.072442 (rank : 58) | θ value | 0.00509761 (rank : 43) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P98160, Q16287, Q9H3V5 | Gene names | HSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
PGBM_MOUSE
|
||||||
| NC score | 0.068229 (rank : 59) | θ value | 0.00134147 (rank : 41) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q05793 | Gene names | Hspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
CEA20_HUMAN
|
||||||
| NC score | 0.066798 (rank : 60) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6UY09 | Gene names | CEACAM20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carcinoembryonic antigen-related cell adhesion molecule 20 precursor. | |||||
|
FCG3A_HUMAN
|
||||||
| NC score | 0.063060 (rank : 61) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P08637 | Gene names | FCGR3A, CD16A, FCG3, FCGR3, IGFR3 | |||
|
Domain Architecture |

|
|||||
| Description | Low affinity immunoglobulin gamma Fc region receptor III-A precursor (IgG Fc receptor III-2) (Fc-gamma RIII-alpha) (Fc-gamma RIIIa) (FcRIIIa) (Fc-gamma RIII) (FcRIII) (FcR-10) (CD16a antigen). | |||||
|
ICAM5_MOUSE
|
||||||
| NC score | 0.060425 (rank : 62) | θ value | 0.365318 (rank : 55) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q60625 | Gene names | Icam5, Tlcn | |||
|
Domain Architecture |

|
|||||
| Description | Intercellular adhesion molecule 5 precursor (ICAM-5) (Telencephalin). | |||||
|
FCG3B_HUMAN
|
||||||
| NC score | 0.059251 (rank : 63) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O75015 | Gene names | FCGR3B, CD16B, FCG3, FCGR3, IGFR3 | |||
|
Domain Architecture |

|
|||||
| Description | Low affinity immunoglobulin gamma Fc region receptor III-B precursor (IgG Fc receptor III-1) (Fc-gamma RIII-beta) (Fc-gamma RIIIb) (FcRIIIb) (Fc-gamma RIII) (FcRIII) (FcR-10) (CD16b antigen). | |||||
|
PDL2_MOUSE
|
||||||
| NC score | 0.054304 (rank : 64) | θ value | 0.813845 (rank : 62) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9WUL5 | Gene names | Pdcd1lg2, B7dc, Btdc, Cd273, Pdl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Programmed cell death 1 ligand 2 precursor (Programmed death ligand 2) (PD-L2) (PD-1-ligand 2) (PDCD1 ligand 2) (Butyrophilin B7-DC) (B7-DC) (CD273 antigen). | |||||
|
PECA1_MOUSE
|
||||||
| NC score | 0.051920 (rank : 65) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q08481 | Gene names | Pecam1, Pecam, Pecam-1 | |||
|
Domain Architecture |

|
|||||
| Description | Platelet endothelial cell adhesion molecule precursor (PECAM-1) (CD31 antigen). | |||||
|
CEAM5_HUMAN
|
||||||
| NC score | 0.050860 (rank : 66) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P06731 | Gene names | CEACAM5, CEA | |||
|
Domain Architecture |

|
|||||
| Description | Carcinoembryonic antigen-related cell adhesion molecule 5 precursor (Carcinoembryonic antigen) (CEA) (Meconium antigen 100) (CD66e antigen). | |||||
|
PDL2_HUMAN
|
||||||
| NC score | 0.045902 (rank : 67) | θ value | 2.36792 (rank : 84) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BQ51, Q5T7Z6, Q6JXL8, Q6JXL9 | Gene names | PDCD1LG2, B7DC, CD273, PDCD1L2, PDL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Programmed cell death 1 ligand 2 precursor (Programmed death ligand 2) (PD-L2) (PD-1-ligand 2) (PDCD1 ligand 2) (Butyrophilin B7-DC) (B7-DC) (CD273 antigen). | |||||
|
TATD2_HUMAN
|
||||||
| NC score | 0.032837 (rank : 68) | θ value | 1.81305 (rank : 80) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q93075 | Gene names | TATDN2, KIAA0218 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TatD DNase domain-containing deoxyribonuclease 2 (EC 3.1.21.-). | |||||
|
DCC_MOUSE
|
||||||
| NC score | 0.031578 (rank : 69) | θ value | 0.21417 (rank : 51) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P70211 | Gene names | Dcc | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC). | |||||
|
CCD1A_HUMAN
|
||||||
| NC score | 0.031114 (rank : 70) | θ value | 0.163984 (rank : 49) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6P1N0, Q7Z435, Q86XV0, Q8NF89, Q9H603, Q9NXI1 | Gene names | CC2D1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil and C2 domain-containing protein 1A (Five repressor element under dual repression-binding protein 1) (FRE under dual repression-binding protein 1) (Freud-1) (Putative NF-kappa-B- activating protein 023N). | |||||
|
CDON_HUMAN
|
||||||
| NC score | 0.030698 (rank : 71) | θ value | 3.0926 (rank : 87) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q4KMG0, O14631 | Gene names | CDON, CDO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell adhesion molecule-related/down-regulated by oncogenes precursor. | |||||
|
MALT1_HUMAN
|
||||||
| NC score | 0.029570 (rank : 72) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UDY8, Q9NTB7, Q9ULX4 | Gene names | MALT1, MLT | |||
|
Domain Architecture |

|
|||||
| Description | Mucosa-associated lymphoid tissue lymphoma translocation protein 1 (EC 3.4.22.-) (MALT lymphoma-associated translocation) (Paracaspase). | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.026612 (rank : 73) | θ value | 1.06291 (rank : 66) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.024498 (rank : 74) | θ value | 1.81305 (rank : 81) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
ROBO4_MOUSE
|
||||||
| NC score | 0.024236 (rank : 75) | θ value | 8.99809 (rank : 121) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8C310, Q9DBW1 | Gene names | Robo4 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 4 precursor. | |||||
|
JPH2_MOUSE
|
||||||
| NC score | 0.023413 (rank : 76) | θ value | 1.81305 (rank : 76) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ET78, Q9ET79 | Gene names | Jph2, Jp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
SFR11_HUMAN
|
||||||
| NC score | 0.023118 (rank : 77) | θ value | 0.163984 (rank : 50) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q05519 | Gene names | SFRS11 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor arginine/serine-rich 11 (Arginine-rich 54 kDa nuclear protein) (p54). | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.022053 (rank : 78) | θ value | 0.21417 (rank : 52) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
CNTN4_HUMAN
|
||||||
| NC score | 0.021275 (rank : 79) | θ value | 5.27518 (rank : 98) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 449 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8IWV2, Q8IX14, Q8TC35 | Gene names | CNTN4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-4 precursor (Brain-derived immunoglobulin superfamily protein 2) (BIG-2). | |||||
|
DCC_HUMAN
|
||||||
| NC score | 0.021033 (rank : 80) | θ value | 1.38821 (rank : 69) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 583 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P43146 | Gene names | DCC | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC) (Colorectal cancer suppressor). | |||||
|
CNOT3_HUMAN
|
||||||
| NC score | 0.019969 (rank : 81) | θ value | 4.03905 (rank : 92) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O75175, Q9NZN7, Q9UF76 | Gene names | CNOT3, KIAA0691, NOT3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
PTPRK_MOUSE
|
||||||
| NC score | 0.019411 (rank : 82) | θ value | 0.813845 (rank : 64) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P35822 | Gene names | Ptprk, Ptpk | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase kappa precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase kappa) (R-PTP-kappa). | |||||
|
PTPRK_HUMAN
|
||||||
| NC score | 0.019210 (rank : 83) | θ value | 0.813845 (rank : 63) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15262, Q14763 | Gene names | PTPRK, PTPK | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase kappa precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase kappa) (R-PTP-kappa). | |||||
|
GLU2B_HUMAN
|
||||||
| NC score | 0.019022 (rank : 84) | θ value | 0.279714 (rank : 53) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P14314, Q96BU9, Q96D06, Q9P0W9 | Gene names | PRKCSH, G19P1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glucosidase 2 subunit beta precursor (Glucosidase II subunit beta) (Protein kinase C substrate, 60.1 kDa protein, heavy chain) (PKCSH) (80K-H protein). | |||||
|
CD008_HUMAN
|
||||||
| NC score | 0.018584 (rank : 85) | θ value | 0.62314 (rank : 59) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P78312, O43607, P78311, P78313, Q9UEG8 | Gene names | C4orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C4orf8 (Protein IT14). | |||||
|
MYOM1_MOUSE
|
||||||
| NC score | 0.018286 (rank : 86) | θ value | 1.81305 (rank : 78) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q62234 | Gene names | Myom1 | |||
|
Domain Architecture |

|
|||||
| Description | Myomesin-1 (Skelemin). | |||||
|
PTK7_HUMAN
|
||||||
| NC score | 0.018148 (rank : 87) | θ value | 0.0563607 (rank : 48) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1103 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q13308, Q13417 | Gene names | PTK7, CCK4 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase-like 7 precursor (Colon carcinoma kinase 4) (CCK-4). | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.017551 (rank : 88) | θ value | 5.27518 (rank : 99) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.017129 (rank : 89) | θ value | 0.0330416 (rank : 46) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
HMGN1_HUMAN
|
||||||
| NC score | 0.016300 (rank : 90) | θ value | 4.03905 (rank : 93) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P05114, Q3KQR8 | Gene names | HMGN1, HMG14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nonhistone chromosomal protein HMG-14 (High-mobility group nucleosome- binding domain-containing protein 1). | |||||
|
MYOM1_HUMAN
|
||||||
| NC score | 0.016200 (rank : 91) | θ value | 3.0926 (rank : 89) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P52179 | Gene names | MYOM1 | |||
|
Domain Architecture |

|
|||||
| Description | Myomesin-1 (190 kDa titin-associated protein) (190 kDa connectin- associated protein). | |||||
|
TREX1_HUMAN
|
||||||
| NC score | 0.016111 (rank : 92) | θ value | 1.38821 (rank : 71) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NSU2, Q8TEU2, Q9BPW1, Q9Y4X2 | Gene names | TREX1 | |||
|
Domain Architecture |
|
|||||
| Description | Three prime repair exonuclease 1 (EC 3.1.11.2) (3'-5' exonuclease TREX1) (DNase III). | |||||
|
CCNK_MOUSE
|
||||||
| NC score | 0.015852 (rank : 93) | θ value | 0.62314 (rank : 58) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
CB017_MOUSE
|
||||||
| NC score | 0.015437 (rank : 94) | θ value | 1.81305 (rank : 75) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6NS82, Q3TA48, Q8BSD3, Q8CHY1, Q8R0Q3 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C2orf17 homolog. | |||||
|
CNOT3_MOUSE
|
||||||
| NC score | 0.014900 (rank : 95) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8K0V4 | Gene names | Cnot3, Not3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
PTPRM_HUMAN
|
||||||
| NC score | 0.014677 (rank : 96) | θ value | 0.365318 (rank : 56) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P28827 | Gene names | PTPRM, PTPRL1 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase mu precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase mu) (R-PTP-mu). | |||||
|
PTPRM_MOUSE
|
||||||
| NC score | 0.014498 (rank : 97) | θ value | 0.365318 (rank : 57) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P28828 | Gene names | Ptprm | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase mu precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase mu) (R-PTP-mu). | |||||
|
KV2A_MOUSE
|
||||||
| NC score | 0.014245 (rank : 98) | θ value | 0.62314 (rank : 60) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P01626 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig kappa chain V-II region MOPC 167. | |||||
|
NUCL_MOUSE
|
||||||
| NC score | 0.012590 (rank : 99) | θ value | 3.0926 (rank : 90) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P09405, Q548M9, Q61991, Q99K50 | Gene names | Ncl, Nuc | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
IPPD_MOUSE
|
||||||
| NC score | 0.012346 (rank : 100) | θ value | 4.03905 (rank : 94) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q60829, Q91XB5 | Gene names | Ppp1r1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dopamine- and cAMP-regulated neuronal phosphoprotein (DARPP-32). | |||||
|
ZMYM3_HUMAN
|
||||||
| NC score | 0.012131 (rank : 101) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14202, O15089 | Gene names | ZMYM3, DXS6673E, KIAA0385, ZNF261 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 3 (Zinc finger protein 261). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.012032 (rank : 102) | θ value | 0.62314 (rank : 61) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
PELP1_HUMAN
|
||||||
| NC score | 0.011539 (rank : 103) | θ value | 2.36792 (rank : 85) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IZL8, O15450, Q5EGN3, Q6NTE6, Q96FT1, Q9BU60 | Gene names | PELP1, HMX3, MNAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3). | |||||
|
BASP_HUMAN
|
||||||
| NC score | 0.011459 (rank : 104) | θ value | 1.81305 (rank : 74) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
KV2B_MOUSE
|
||||||
| NC score | 0.010557 (rank : 105) | θ value | 1.06291 (rank : 67) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P01627 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Ig kappa chain V-II region VKappa167 precursor. | |||||
|
UBP43_MOUSE
|
||||||
| NC score | 0.010391 (rank : 106) | θ value | 1.38821 (rank : 72) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BUM9, Q8VDP5 | Gene names | Usp43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
TOB1_HUMAN
|
||||||
| NC score | 0.009780 (rank : 107) | θ value | 1.81305 (rank : 82) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P50616, Q4KMQ0 | Gene names | TOB1, TOB | |||
|
Domain Architecture |

|
|||||
| Description | Tob1 protein (Transducer of erbB-2 1). | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.009702 (rank : 108) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.009514 (rank : 109) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
MDM2_MOUSE
|
||||||
| NC score | 0.009399 (rank : 110) | θ value | 3.0926 (rank : 88) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P23804, Q61040, Q64330 | Gene names | Mdm2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3 Mdm2 (EC 6.3.2.-) (p53-binding protein Mdm2) (Oncoprotein Mdm2) (Double minute 2 protein). | |||||
|
ADDB_HUMAN
|
||||||
| NC score | 0.008930 (rank : 111) | θ value | 1.38821 (rank : 68) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35612, Q13482, Q16412, Q59G82, Q5U5P4, Q6P0P2, Q6PGQ4, Q7Z688, Q7Z689, Q7Z690, Q7Z691 | Gene names | ADD2, ADDB | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta). | |||||
|
MTFR1_MOUSE
|
||||||
| NC score | 0.008715 (rank : 112) | θ value | 6.88961 (rank : 105) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99MB2, Q9CQZ4 | Gene names | Mtfr1, Kiaa0009 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial fission regulator 1. | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.008513 (rank : 113) | θ value | 1.38821 (rank : 70) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
IF39_MOUSE
|
||||||
| NC score | 0.008046 (rank : 114) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8JZQ9, Q922K2 | Gene names | Eif3s9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116). | |||||
|
AHTF1_MOUSE
|
||||||
| NC score | 0.007470 (rank : 115) | θ value | 5.27518 (rank : 97) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CJF7, Q8BVJ5, Q8VD55 | Gene names | Ahctf1, Elys | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-hook-containing transcription factor 1 (Embryonic large molecule derived from yolk sac). | |||||
|
RTN4_MOUSE
|
||||||
| NC score | 0.007452 (rank : 116) | θ value | 8.99809 (rank : 122) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99P72, Q5DTK9, Q7TNB7, Q80W95, Q8BGK7, Q8BGM9, Q8K290, Q8K3G8, Q9CTE3 | Gene names | Rtn4, Kiaa0886, Nogo | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-4 (Neurite outgrowth inhibitor) (Nogo protein). | |||||
|
NBR1_MOUSE
|
||||||
| NC score | 0.007291 (rank : 117) | θ value | 5.27518 (rank : 100) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97432, Q9JIH9 | Gene names | Nbr1, M17s2 | |||
|
Domain Architecture |

|
|||||
| Description | Next to BRCA1 gene 1 protein (Neighbor of BRCA1 gene 1 protein) (Membrane component, chromosome 17, surface marker 2). | |||||
|
ACINU_HUMAN
|
||||||
| NC score | 0.007215 (rank : 118) | θ value | 3.0926 (rank : 86) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
LV0A_HUMAN
|
||||||
| NC score | 0.007082 (rank : 119) | θ value | 4.03905 (rank : 95) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P04211 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig lambda chain V region 4A precursor. | |||||
|
TCOF_MOUSE
|
||||||
| NC score | 0.007018 (rank : 120) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
DTX2_HUMAN
|
||||||
| NC score | 0.006983 (rank : 121) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86UW9, Q96H69, Q9H890, Q9P200 | Gene names | DTX2, KIAA1528 | |||
|
Domain Architecture |

|
|||||
| Description | Protein deltex-2 (Deltex-2) (Deltex2) (hDTX2). | |||||
|
NELFE_MOUSE
|
||||||
| NC score | 0.006862 (rank : 122) | θ value | 6.88961 (rank : 106) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P19426, P97485, P97486, Q9CZN3 | Gene names | Rdbp, D17h6s45, Nelfe, Rd | |||
|
Domain Architecture |

|
|||||
| Description | Negative elongation factor E (NELF-E) (RD protein). | |||||
|
TM115_HUMAN
|
||||||
| NC score | 0.006770 (rank : 123) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12893, O14568, Q9UIX3 | Gene names | TMEM115, PL6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 115 (Protein PL6) (Placental protein 6) (PP6). | |||||
|
NUCKS_HUMAN
|
||||||
| NC score | 0.006455 (rank : 124) | θ value | 6.88961 (rank : 107) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H1E3, Q9H1D6, Q9H723 | Gene names | NUCKS1, NUCKS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (P1). | |||||
|
TNR16_HUMAN
|
||||||
| NC score | 0.006102 (rank : 125) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P08138 | Gene names | NGFR, TNFRSF16 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 16 precursor (Low- affinity nerve growth factor receptor) (NGF receptor) (Gp80-LNGFR) (p75 ICD) (Low affinity neurotrophin receptor p75NTR) (CD271 antigen). | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.005620 (rank : 126) | θ value | 2.36792 (rank : 83) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
SMG7_MOUSE
|
||||||
| NC score | 0.004840 (rank : 127) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5RJH6, Q63ZW5, Q6ZQF3 | Gene names | Smg7, Est1c, Kiaa0250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein SMG7 (SMG-7 homolog) (EST1-like protein C). | |||||
|
KV2F_MOUSE
|
||||||
| NC score | 0.003290 (rank : 128) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P01630 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig kappa chain V-II region 7S34.1. | |||||
|
TF7L1_MOUSE
|
||||||
| NC score | 0.002788 (rank : 129) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z1J1, O70450, O70573 | Gene names | Tcf7l1, Tcf3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 1 (HMG box transcription factor 3) (TCF-3) (mTCF-3). | |||||
|
NTRK1_HUMAN
|
||||||
| NC score | 0.002226 (rank : 130) | θ value | 0.0563607 (rank : 47) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1023 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P04629, P08119, Q15655, Q15656, Q5D056, Q5VZS2, Q9UIU7 | Gene names | NTRK1, TRK | |||
|
Domain Architecture |

|
|||||
| Description | High affinity nerve growth factor receptor precursor (EC 2.7.10.1) (Neurotrophic tyrosine kinase receptor type 1) (TRK1 transforming tyrosine kinase protein) (p140-TrkA) (Trk-A). | |||||
|
41_MOUSE
|
||||||
| NC score | 0.002180 (rank : 131) | θ value | 4.03905 (rank : 91) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48193, Q68FF1, Q6NVF5 | Gene names | Epb41, Epb4.1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein 4.1 (Band 4.1) (P4.1) (4.1R). | |||||
|
SRC8_MOUSE
|
||||||
| NC score | 0.001708 (rank : 132) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60598 | Gene names | Cttn, Ems1 | |||
|
Domain Architecture |
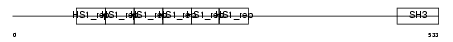
|
|||||
| Description | Src substrate cortactin. | |||||
|
MYST3_MOUSE
|
||||||
| NC score | 0.001456 (rank : 133) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
PDE4D_MOUSE
|
||||||
| NC score | 0.001408 (rank : 134) | θ value | 6.88961 (rank : 108) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q01063, Q6TRH9, Q8C4Q7, Q8CG05 | Gene names | Pde4d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4D (EC 3.1.4.17) (DPDE3). | |||||
|
ZEP1_MOUSE
|
||||||
| NC score | -0.000168 (rank : 135) | θ value | 1.38821 (rank : 73) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||
|
NKX26_MOUSE
|
||||||
| NC score | -0.000680 (rank : 136) | θ value | 8.99809 (rank : 120) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P43688 | Gene names | Nkx2-6, Nkx-2.6, Nkx2f | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-2.6 (Homeobox protein NK-2 homolog F). | |||||
|
TGFR1_HUMAN
|
||||||
| NC score | -0.001284 (rank : 137) | θ value | 5.27518 (rank : 101) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 755 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P36897 | Gene names | TGFBR1 | |||
|
Domain Architecture |

|
|||||
| Description | TGF-beta receptor type-1 precursor (EC 2.7.11.30) (TGF-beta receptor type I) (TGFR-1) (TGF-beta type I receptor) (Serine/threonine-protein kinase receptor R4) (SKR4) (Activin receptor-like kinase 5) (ALK-5). | |||||
|
TGFR1_MOUSE
|
||||||
| NC score | -0.001424 (rank : 138) | θ value | 6.88961 (rank : 112) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q64729 | Gene names | Tgfbr1 | |||
|
Domain Architecture |

|
|||||
| Description | TGF-beta receptor type-1 precursor (EC 2.7.11.30) (TGF-beta receptor type I) (TGFR-1) (TGF-beta type I receptor) (ESK2). | |||||
|
M3K12_MOUSE
|
||||||
| NC score | -0.002479 (rank : 139) | θ value | 1.81305 (rank : 77) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q60700, P70286, Q3TLL7, Q8C4N7, Q8CBX3, Q8CDL6 | Gene names | Map3k12, Zpk | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 12 (EC 2.7.11.25) (Mixed lineage kinase) (Leucine-zipper protein kinase) (ZPK) (Dual leucine zipper bearing kinase) (DLK) (MAPK-upstream kinase) (MUK). | |||||
|
LAP4_HUMAN
|
||||||
| NC score | -0.002634 (rank : 140) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14160, Q6P496, Q7Z5D1, Q8WWV8, Q96C69, Q96GG1 | Gene names | SCRIB, CRIB1, KIAA0147, LAP4, SCRB1, VARTUL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog) (hScrib). | |||||