Please be patient as the page loads
|
ADDB_HUMAN
|
||||||
| SwissProt Accessions | P35612, Q13482, Q16412, Q59G82, Q5U5P4, Q6P0P2, Q6PGQ4, Q7Z688, Q7Z689, Q7Z690, Q7Z691 | Gene names | ADD2, ADDB | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ADDB_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | P35612, Q13482, Q16412, Q59G82, Q5U5P4, Q6P0P2, Q6PGQ4, Q7Z688, Q7Z689, Q7Z690, Q7Z691 | Gene names | ADD2, ADDB | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta). | |||||
|
ADDB_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.877296 (rank : 6) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
ADDG_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.951190 (rank : 3) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UEY8, O43243, Q92773, Q9UEY7 | Gene names | ADD3, ADDL | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-adducin (Adducin-like protein 70). | |||||
|
ADDG_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.955089 (rank : 2) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QYB5, Q9D2M5, Q9QYB6 | Gene names | Add3, Addl | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-adducin (Adducin-like protein 70). | |||||
|
ADDA_HUMAN
|
||||||
| θ value | 7.25522e-167 (rank : 5) | NC score | 0.937028 (rank : 4) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P35611, Q13734, Q14729, Q16156, Q9UJB6 | Gene names | ADD1, ADDA | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-adducin (Erythrocyte adducin subunit alpha). | |||||
|
ADDA_MOUSE
|
||||||
| θ value | 3.04821e-165 (rank : 6) | NC score | 0.926090 (rank : 5) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9QYC0, Q9JLE3 | Gene names | Add1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-adducin (Erythrocyte adducin subunit alpha). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 3.77169e-06 (rank : 7) | NC score | 0.113834 (rank : 10) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
YTDC1_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 8) | NC score | 0.101903 (rank : 11) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
TXND2_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 9) | NC score | 0.089756 (rank : 15) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 10) | NC score | 0.068623 (rank : 33) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 11) | NC score | 0.049461 (rank : 90) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
CD2L1_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 12) | NC score | 0.026351 (rank : 134) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
TP53B_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 13) | NC score | 0.090252 (rank : 14) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
GP158_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 14) | NC score | 0.065932 (rank : 36) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5T848, Q6QR81, Q9ULT3 | Gene names | GPR158, KIAA1136 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 158 precursor. | |||||
|
CF010_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 15) | NC score | 0.114855 (rank : 9) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q5SRN2, Q5SPI9, Q5SPJ0, Q5SPK9, Q5SPL0, Q5SRN3, Q5TG25, Q5TG26, Q8N4B6 | Gene names | C6orf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf10. | |||||
|
KTN1_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 16) | NC score | 0.041591 (rank : 101) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1324 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q61595, Q8BG49, Q8BHF4, Q8BHM8, Q8C9Y5, Q8CG51, Q8CG52, Q8CG53, Q8CG54, Q8CG55, Q8CG56, Q8CG57, Q8CG58, Q8CG59, Q8CG60, Q8CG61, Q8CG62, Q8CG63 | Gene names | Ktn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin. | |||||
|
LRRF1_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 17) | NC score | 0.079389 (rank : 22) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q32MZ4, O75766, O75799, Q32MZ5, Q53T49, Q6PKG2 | Gene names | LRRFIP1, GCF2, TRIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (TAR RNA-interacting protein) (GC-binding factor 2). | |||||
|
MYLK_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 18) | NC score | 0.014930 (rank : 168) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q15746, O95796, O95797, O95798, O95799, Q14844, Q16794, Q5MY99, Q5MYA0, Q7Z4J0, Q9C0L5, Q9UBG5, Q9UIT9 | Gene names | MYLK, MLCK | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light chain kinase, smooth muscle (EC 2.7.11.18) (MLCK) (Telokin) (Kinase-related protein) (KRP). | |||||
|
SELPL_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 19) | NC score | 0.066339 (rank : 35) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
SNUT3_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 20) | NC score | 0.166648 (rank : 7) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K194, Q8BRA8, Q9CSV0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 3. | |||||
|
LIMA1_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 21) | NC score | 0.060241 (rank : 52) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9ERG0, Q9ERG1 | Gene names | Lima1, D15Ertd366e, Eplin | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain and actin-binding protein 1 (Epithelial protein lost in neoplasm) (mEPLIN). | |||||
|
AN30A_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 22) | NC score | 0.037342 (rank : 107) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1119 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9BXX3, Q5W025 | Gene names | ANKRD30A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 30A (NY-BR-1 antigen). | |||||
|
MUC1_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 23) | NC score | 0.069544 (rank : 32) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
PALM_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 24) | NC score | 0.058990 (rank : 60) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Z0P4, Q9Z0P3 | Gene names | Palm | |||
|
Domain Architecture |

|
|||||
| Description | Paralemmin. | |||||
|
CJ047_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 25) | NC score | 0.060603 (rank : 50) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8C5R2, Q8R017, Q8R2Z9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf47 homolog. | |||||
|
TP53B_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 26) | NC score | 0.084723 (rank : 18) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q12888, Q2M1Z7, Q4LE46, Q5FWZ3, Q7Z3U4 | Gene names | TP53BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
EMSY_HUMAN
|
||||||
| θ value | 0.125558 (rank : 27) | NC score | 0.056207 (rank : 65) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7Z589, Q4G109, Q8NBU6, Q8TE50, Q9H8I9, Q9NRH0 | Gene names | EMSY, C11orf30 | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
SNUT3_HUMAN
|
||||||
| θ value | 0.125558 (rank : 28) | NC score | 0.137294 (rank : 8) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8WVK2, Q15410 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 3 (U4/U6.U5 tri-snRNP-associated 27 kDa protein) (27K) (Nucleic acid-binding protein RY-1). | |||||
|
BCL6_HUMAN
|
||||||
| θ value | 0.21417 (rank : 29) | NC score | 0.003799 (rank : 190) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 922 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P41182 | Gene names | BCL6, BCL5, LAZ3, ZBTB27, ZNF51 | |||
|
Domain Architecture |
|
|||||
| Description | B-cell lymphoma 6 protein (BCL-6) (Zinc finger protein 51) (LAZ-3 protein) (BCL-5) (Zinc finger and BTB domain-containing protein 27). | |||||
|
NALP5_MOUSE
|
||||||
| θ value | 0.21417 (rank : 30) | NC score | 0.030458 (rank : 119) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
RBBP6_MOUSE
|
||||||
| θ value | 0.21417 (rank : 31) | NC score | 0.099325 (rank : 12) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
SLK_MOUSE
|
||||||
| θ value | 0.21417 (rank : 32) | NC score | 0.019336 (rank : 156) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1798 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O54988, Q80U65, Q8CAU2, Q8CDW2, Q9WU41 | Gene names | Slk, Kiaa0204, Stk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (mSLK) (Serine/threonine-protein kinase 2) (STE20-related kinase SMAK) (Etk4). | |||||
|
TRI44_HUMAN
|
||||||
| θ value | 0.21417 (rank : 33) | NC score | 0.055776 (rank : 66) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96DX7, Q96QY2, Q9UGK0 | Gene names | TRIM44, DIPB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB). | |||||
|
NOC3L_HUMAN
|
||||||
| θ value | 0.279714 (rank : 34) | NC score | 0.057874 (rank : 62) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8WTT2, Q9H5M6, Q9H9D8 | Gene names | NOC3L, AD24, C10orf117, FAD24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar complex protein 3 homolog (NOC3 protein homolog) (NOC3-like protein) (Nucleolar complex-associated protein 3-like protein) (Factor for adipocyte differentiation 24). | |||||
|
ZCPW1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 35) | NC score | 0.040109 (rank : 105) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H0M4, Q8NA98, Q9BUD0, Q9NWF7 | Gene names | ZCWPW1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1. | |||||
|
SRBS1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 36) | NC score | 0.024114 (rank : 145) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q62417, Q80TF8, Q8BZI3, Q8K3Y2, Q921F8, Q9Z0Z8, Q9Z0Z9 | Gene names | Sorbs1, Kiaa1296, Sh3d5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorbin and SH3 domain-containing protein 1 (Ponsin) (c-Cbl-associated protein) (CAP) (SH3 domain protein 5) (SH3P12). | |||||
|
HRH1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 37) | NC score | 0.004224 (rank : 188) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P70174, Q91V75, Q91XN0, Q91XN1, Q91XN2, Q91XN3 | Gene names | Hrh1, Bphs | |||
|
Domain Architecture |
|
|||||
| Description | Histamine H1 receptor. | |||||
|
KMHN1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 38) | NC score | 0.033599 (rank : 113) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 950 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8TBZ0, Q86YI9, Q8N7W0 | Gene names | KMHN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1. | |||||
|
LUZP1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 39) | NC score | 0.037032 (rank : 108) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8R4U7, Q3UV18, Q7TS71, Q8BQW1, Q99NG3, Q9CSL6 | Gene names | Luzp1, Luzp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1 (Leucine zipper motif-containing protein). | |||||
|
MY18A_HUMAN
|
||||||
| θ value | 0.47712 (rank : 40) | NC score | 0.030190 (rank : 120) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
SLK_HUMAN
|
||||||
| θ value | 0.47712 (rank : 41) | NC score | 0.018012 (rank : 159) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1775 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9H2G2, O00211, Q6P1Z4, Q86WU7, Q86WW1, Q92603, Q9NQL0, Q9NQL1 | Gene names | SLK, KIAA0204, STK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (hSLK) (Serine/threonine-protein kinase 2) (CTCL tumor antigen se20-9). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 0.47712 (rank : 42) | NC score | 0.026072 (rank : 135) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
BCAS1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 43) | NC score | 0.044615 (rank : 98) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q80YN3, Q9CVA1 | Gene names | Bcas1, Nabc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast carcinoma amplified sequence 1 homolog (Novel amplified in breast cancer 1 homolog). | |||||
|
G3BP2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 44) | NC score | 0.057895 (rank : 61) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UN86, O60606, O75149, Q9UPA1 | Gene names | G3BP2, KIAA0660 | |||
|
Domain Architecture |

|
|||||
| Description | Ras-GTPase-activating protein-binding protein 2 (GAP SH3-domain- binding protein 2) (G3BP-2). | |||||
|
MYH11_MOUSE
|
||||||
| θ value | 0.813845 (rank : 45) | NC score | 0.028045 (rank : 125) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | 0.813845 (rank : 46) | NC score | 0.073050 (rank : 30) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
SIA7A_HUMAN
|
||||||
| θ value | 0.813845 (rank : 47) | NC score | 0.025573 (rank : 136) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NSC7, Q6UW90, Q9NSC6 | Gene names | ST6GALNAC1, SIAT7A | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 (EC 2.4.99.3) (GalNAc alpha-2,6-sialyltransferase I) (ST6GalNAc I) (Sialyltransferase 7A). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 48) | NC score | 0.034695 (rank : 111) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
CAC1C_HUMAN
|
||||||
| θ value | 1.06291 (rank : 49) | NC score | 0.013852 (rank : 170) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13936, Q13917, Q13918, Q13919, Q13920, Q13921, Q13922, Q13923, Q13924, Q13925, Q13926, Q13927, Q13928, Q13929, Q13930, Q13932, Q13933, Q14743, Q14744, Q15877, Q99025, Q99241, Q99875 | Gene names | CACNA1C, CACH2, CACN2, CACNL1A1, CCHL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle). | |||||
|
MYH11_HUMAN
|
||||||
| θ value | 1.06291 (rank : 50) | NC score | 0.026966 (rank : 130) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MYH9_MOUSE
|
||||||
| θ value | 1.06291 (rank : 51) | NC score | 0.026401 (rank : 133) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MYST3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 52) | NC score | 0.050744 (rank : 83) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 1.06291 (rank : 53) | NC score | 0.056484 (rank : 64) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
NOC3L_MOUSE
|
||||||
| θ value | 1.06291 (rank : 54) | NC score | 0.041422 (rank : 102) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8VI84, Q9JKM3 | Gene names | Noc3l, Ad24, Fad24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar complex protein 3 homolog (NOC3 protein homolog) (NOC3-like protein) (Nucleolar complex-associated protein 3-like protein) (Factor for adipocyte differentiation 24). | |||||
|
TTBK1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 55) | NC score | 0.027022 (rank : 129) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1058 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5TCY1, Q2L6C6, Q8N444, Q96JH2 | Gene names | TTBK1, BDTK, KIAA1855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 1 (EC 2.7.11.1) (Brain-derived tau kinase). | |||||
|
ZEP1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 56) | NC score | 0.014456 (rank : 169) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1036 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P15822, Q14122 | Gene names | HIVEP1, ZNF40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 40 (Human immunodeficiency virus type I enhancer- binding protein 1) (HIV-EP1) (Major histocompatibility complex-binding protein 1) (MBP-1) (Positive regulatory domain II-binding factor 1) (PRDII-BF1). | |||||
|
COPA_HUMAN
|
||||||
| θ value | 1.38821 (rank : 57) | NC score | 0.008904 (rank : 181) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P53621 | Gene names | COPA | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit alpha (Alpha-coat protein) (Alpha-COP) (HEPCOP) (HEP- COP) [Contains: Xenin (Xenopsin-related peptide); Proxenin]. | |||||
|
GCC2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 58) | NC score | 0.031286 (rank : 118) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
LIRB3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 59) | NC score | 0.008930 (rank : 180) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75022, O15471, Q86U49 | Gene names | LILRB3, ILT5, LIR3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily B member 3 precursor (Leukocyte immunoglobulin-like receptor 3) (LIR-3) (Immunoglobulin- like transcript 5) (ILT-5) (Monocyte inhibitory receptor HL9) (CD85a antigen). | |||||
|
MAP4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 60) | NC score | 0.049113 (rank : 91) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
RBP24_HUMAN
|
||||||
| θ value | 1.38821 (rank : 61) | NC score | 0.031289 (rank : 117) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
SART3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 62) | NC score | 0.020618 (rank : 155) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9JLI8, Q6ZQI2, Q8BPK9, Q8C3B7, Q8CFU9 | Gene names | Sart3, Kiaa0156 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Squamous cell carcinoma antigen recognized by T-cells 3 (SART-3) (mSART-3) (Tumor-rejection antigen SART3). | |||||
|
VEZA_HUMAN
|
||||||
| θ value | 1.38821 (rank : 63) | NC score | 0.048819 (rank : 93) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9HBM0, Q9H2F4, Q9H2U5, Q9NT70, Q9NVW0, Q9UF91 | Gene names | VEZT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vezatin. | |||||
|
WAPL_MOUSE
|
||||||
| θ value | 1.38821 (rank : 64) | NC score | 0.032400 (rank : 116) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q65Z40, Q6ZQE9 | Gene names | Wapal, Kiaa0261, Wapl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wings apart-like protein homolog (Dioxin-inducible factor 2) (DIF-2). | |||||
|
ABCF1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 65) | NC score | 0.027724 (rank : 128) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8NE71, O14897, Q69YP6 | Gene names | ABCF1, ABC50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family F member 1 (ATP-binding cassette 50) (TNF-alpha-stimulated ABC protein). | |||||
|
BRCA2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 66) | NC score | 0.024414 (rank : 143) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P97929, O35922, P97383 | Gene names | Brca2, Fancd1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 2 susceptibility protein homolog (Fanconi anemia group D1 protein homolog). | |||||
|
DMP1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 67) | NC score | 0.075712 (rank : 26) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
EVI1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 68) | NC score | 0.001382 (rank : 193) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 826 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P14404 | Gene names | Evi1, Evi-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ecotropic virus integration 1 site protein. | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 1.81305 (rank : 69) | NC score | 0.077906 (rank : 23) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
MYRIP_HUMAN
|
||||||
| θ value | 1.81305 (rank : 70) | NC score | 0.023915 (rank : 146) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8NFW9, Q8IUF5, Q9Y3V4 | Gene names | MYRIP, SLAC2C | |||
|
Domain Architecture |

|
|||||
| Description | Rab effector MyRIP (Myosin-VIIa- and Rab-interacting protein) (Exophilin-8) (Slp homolog lacking C2 domains c). | |||||
|
UBP1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 71) | NC score | 0.024538 (rank : 142) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O94782, Q9UFR0, Q9UNJ3 | Gene names | USP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 1 (EC 3.1.2.15) (Ubiquitin thioesterase 1) (Ubiquitin-specific-processing protease 1) (Deubiquitinating enzyme 1) (hUBP). | |||||
|
CTDP1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 72) | NC score | 0.045483 (rank : 97) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y5B0, Q7Z644, Q96BZ1, Q9Y6F5 | Gene names | CTDP1, FCP1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II subunit A C-terminal domain phosphatase (EC 3.1.3.16) (TFIIF-associating CTD phosphatase). | |||||
|
DDX46_HUMAN
|
||||||
| θ value | 2.36792 (rank : 73) | NC score | 0.014991 (rank : 167) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q7L014, O94894, Q96EI0, Q9Y658 | Gene names | DDX46, KIAA0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46) (PRP5 homolog). | |||||
|
HELZ_HUMAN
|
||||||
| θ value | 2.36792 (rank : 74) | NC score | 0.022023 (rank : 153) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P42694 | Gene names | HELZ, KIAA0054 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable helicase with zinc-finger domain (EC 3.6.1.-). | |||||
|
KI21A_MOUSE
|
||||||
| θ value | 2.36792 (rank : 75) | NC score | 0.018205 (rank : 158) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9QXL2, Q6P5H1, Q6ZPJ8, Q8BWZ9, Q8BXF1 | Gene names | Kif21a, Kiaa1708 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A. | |||||
|
PEPL_HUMAN
|
||||||
| θ value | 2.36792 (rank : 76) | NC score | 0.025406 (rank : 138) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1141 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O60437, O60314, O60454 | Gene names | PPL, KIAA0568 | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin (195 kDa cornified envelope precursor protein) (190 kDa paraneoplastic pemphigus antigen). | |||||
|
TRHY_HUMAN
|
||||||
| θ value | 2.36792 (rank : 77) | NC score | 0.032572 (rank : 115) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
TRI44_MOUSE
|
||||||
| θ value | 2.36792 (rank : 78) | NC score | 0.046755 (rank : 95) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9QXA7, Q80SV9, Q9JHR8 | Gene names | Trim44, Dipb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB) (Protein Mc7). | |||||
|
CCAR1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 79) | NC score | 0.040382 (rank : 104) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8CH18, Q6AXC9, Q6PAR2, Q80XE4, Q8BJY0, Q8BVN2, Q8CGG1, Q9CSR5 | Gene names | Ccar1, Carp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle and apoptosis regulator protein 1 (Cell cycle and apoptosis regulatory protein 1) (CARP-1). | |||||
|
CE290_HUMAN
|
||||||
| θ value | 3.0926 (rank : 80) | NC score | 0.030151 (rank : 121) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
CEP35_HUMAN
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.048934 (rank : 92) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q5VT06, O75068, Q8TDK3, Q8WY20 | Gene names | CEP350, CAP350, GM133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein 350 (Centrosome-associated protein of 350 kDa). | |||||
|
INVO_MOUSE
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | 0.039445 (rank : 106) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P48997 | Gene names | Ivl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Involucrin. | |||||
|
MDC1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 83) | NC score | 0.063123 (rank : 44) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 3.0926 (rank : 84) | NC score | 0.035893 (rank : 109) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
PP4R1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 85) | NC score | 0.017177 (rank : 161) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K2V1, Q8R0R8 | Gene names | Ppp4r1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 4 regulatory subunit 1. | |||||
|
TCF8_HUMAN
|
||||||
| θ value | 3.0926 (rank : 86) | NC score | 0.009710 (rank : 178) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P37275, Q12924, Q13800 | Gene names | TCF8, AREB6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (NIL-2-A zinc finger protein) (Negative regulator of IL2). | |||||
|
TIP60_HUMAN
|
||||||
| θ value | 3.0926 (rank : 87) | NC score | 0.027975 (rank : 126) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q92993, O95624, Q13430, Q6GSE8, Q9BWK7 | Gene names | HTATIP, TIP60 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase HTATIP (EC 2.3.1.48) (EC 2.3.1.-) (60 kDa Tat interactive protein) (Tip60) (HIV-1 Tat interactive protein) (cPLA(2)-interacting protein). | |||||
|
TIP60_MOUSE
|
||||||
| θ value | 3.0926 (rank : 88) | NC score | 0.028591 (rank : 123) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8CHK4, Q8CGZ3, Q8CGZ4, Q8VIH0 | Gene names | Htatip, Tip60 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase HTATIP (EC 2.3.1.48) (EC 2.3.1.-) (60 kDa Tat interactive protein) (Tip60). | |||||
|
BPAEA_MOUSE
|
||||||
| θ value | 4.03905 (rank : 89) | NC score | 0.023146 (rank : 149) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
CE110_MOUSE
|
||||||
| θ value | 4.03905 (rank : 90) | NC score | 0.016946 (rank : 162) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7TSH4, Q6A072 | Gene names | Cep110, Cp110, Kiaa0419 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 110 kDa (Cep110 protein). | |||||
|
CGRE1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 91) | NC score | 0.027880 (rank : 127) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8R1U2, Q9D1K1 | Gene names | Cgref1, Cgr11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
CPLX4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 92) | NC score | 0.023454 (rank : 148) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z7G2 | Gene names | CPLX4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complexin-4 (Complexin IV) (CPX IV). | |||||
|
CSPG2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 93) | NC score | 0.006504 (rank : 184) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q62059, Q62058, Q9CUU0 | Gene names | Cspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M). | |||||
|
CYLC1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 94) | NC score | 0.074307 (rank : 28) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
EVL_MOUSE
|
||||||
| θ value | 4.03905 (rank : 95) | NC score | 0.017323 (rank : 160) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P70429, Q9ERU8 | Gene names | Evl | |||
|
Domain Architecture |

|
|||||
| Description | Ena/VASP-like protein (Ena/vasodilator-stimulated phosphoprotein- like). | |||||
|
NEB1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 96) | NC score | 0.026803 (rank : 131) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9ULJ8, O76059, Q9NXT2 | Gene names | PPP1R9A, KIAA1222 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-1 (Neurabin-I) (Neural tissue-specific F-actin-binding protein I) (Protein phosphatase 1 regulatory subunit 9A). | |||||
|
SYTL2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 97) | NC score | 0.021852 (rank : 154) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9HCH5, Q2YDA7, Q8ND34, Q96BJ2, Q9H768, Q9NXM1 | Gene names | SYTL2, KIAA1597, SLP2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
TBC10_MOUSE
|
||||||
| θ value | 4.03905 (rank : 98) | NC score | 0.016722 (rank : 163) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P58802 | Gene names | Tbc1d10a, Epi64, Tbc1d10 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 10A (EBP50-PDX interactor of 64 kDa) (EPI64 protein). | |||||
|
TRPS1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 99) | NC score | 0.012865 (rank : 171) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q925H1 | Gene names | Trps1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger transcription factor Trps1. | |||||
|
CAMKV_HUMAN
|
||||||
| θ value | 5.27518 (rank : 100) | NC score | 0.007159 (rank : 183) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8NCB2, Q6FIB8, Q8NBS8, Q8NC85, Q8NDU4, Q8WTT8, Q9BQC9, Q9H0Q5 | Gene names | CAMKV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CaM kinase-like vesicle-associated protein. | |||||
|
CCD45_MOUSE
|
||||||
| θ value | 5.27518 (rank : 101) | NC score | 0.024571 (rank : 141) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BVV7 | Gene names | Ccdc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 45. | |||||
|
CPLX4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 102) | NC score | 0.022474 (rank : 150) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80WM3, Q8C8Y0, Q91WE5 | Gene names | Cplx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complexin-4 (Complexin IV) (CPX IV). | |||||
|
ERCC5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 103) | NC score | 0.022384 (rank : 151) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P35689, Q61528, Q64248 | Gene names | Ercc5, Ercc-5, Xpg | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-G cells homolog (Xeroderma pigmentosum group G-complementing protein homolog) (DNA excision repair protein ERCC-5). | |||||
|
GP179_HUMAN
|
||||||
| θ value | 5.27518 (rank : 104) | NC score | 0.040574 (rank : 103) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6PRD1 | Gene names | GPR179, GPR158L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 179 precursor (Probable G-protein coupled receptor 158-like 1). | |||||
|
ICB1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 105) | NC score | 0.015038 (rank : 166) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91YX0 | Gene names | Icb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Induced by contact to basement membrane 1 protein homolog (ICB-1 protein). | |||||
|
LATS1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 106) | NC score | 0.005808 (rank : 186) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O95835, Q6PKD0 | Gene names | LATS1, WARTS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS1 (EC 2.7.11.1) (Large tumor suppressor homolog 1) (WARTS protein kinase) (h-warts). | |||||
|
NF2L2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.011775 (rank : 174) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q60795 | Gene names | Nfe2l2, Nrf-2, Nrf2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 2 (NF-E2-related factor 2) (NFE2-related factor 2) (Nuclear factor, erythroid derived 2, like 2). | |||||
|
PB1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 108) | NC score | 0.012739 (rank : 172) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86U86, Q9H2T3, Q9H2T4, Q9H2T5, Q9H301, Q9H314 | Gene names | PB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein polybromo-1 (hPB1) (Polybromo-1D) (BRG1-associated factor 180) (BAF180). | |||||
|
PLK3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 109) | NC score | 0.003974 (rank : 189) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 863 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q60806, Q60822, Q9R009 | Gene names | Plk3, Cnk, Fnk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PLK3 (EC 2.7.11.21) (Polo-like kinase 3) (PLK-3) (Cytokine-inducible serine/threonine-protein kinase) (FGF- inducible kinase). | |||||
|
REST_MOUSE
|
||||||
| θ value | 5.27518 (rank : 110) | NC score | 0.024578 (rank : 140) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
SEPT9_HUMAN
|
||||||
| θ value | 5.27518 (rank : 111) | NC score | 0.015992 (rank : 164) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
SETBP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 112) | NC score | 0.023677 (rank : 147) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y6X0, Q9UEF3 | Gene names | SETBP1, KIAA0437 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET-binding protein (SEB). | |||||
|
SIA7A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 113) | NC score | 0.028161 (rank : 124) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QZ39, Q9JJP5 | Gene names | St6galnac1, Siat7a | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 (EC 2.4.99.3) (GalNAc alpha-2,6-sialyltransferase I) (ST6GalNAc I) (Sialyltransferase 7A). | |||||
|
ZBT40_HUMAN
|
||||||
| θ value | 5.27518 (rank : 114) | NC score | 0.000392 (rank : 194) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NUA8, O75066, Q5TFU5, Q8N1R1 | Gene names | ZBTB40, KIAA0478 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 40. | |||||
|
ANLN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.022267 (rank : 152) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9NQW6, Q5CZ78, Q6NSK5, Q9H8Y4, Q9NVN9, Q9NVP0 | Gene names | ANLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding protein anillin. | |||||
|
CCD19_HUMAN
|
||||||
| θ value | 6.88961 (rank : 116) | NC score | 0.035304 (rank : 110) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UL16 | Gene names | CCDC19, NESG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 19 (Nasopharyngeal epithelium- specific protein 1). | |||||
|
DEN4C_HUMAN
|
||||||
| θ value | 6.88961 (rank : 117) | NC score | 0.009756 (rank : 177) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5VZ89, Q6AI48, Q6ZUB3, Q8NCY7, Q9H6N4, Q9NUT1, Q9NWA5 | Gene names | DENND4C, C9orf55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 4C. | |||||
|
GLU2B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 118) | NC score | 0.074506 (rank : 27) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P14314, Q96BU9, Q96D06, Q9P0W9 | Gene names | PRKCSH, G19P1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glucosidase 2 subunit beta precursor (Glucosidase II subunit beta) (Protein kinase C substrate, 60.1 kDa protein, heavy chain) (PKCSH) (80K-H protein). | |||||
|
LGR6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 119) | NC score | 0.001392 (rank : 192) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HBX8, Q5T509, Q5T512, Q6UY15, Q86VU0, Q96K69, Q9BYD7 | Gene names | LGR6 | |||
|
Domain Architecture |
|
|||||
| Description | Leucine-rich repeat-containing G-protein coupled receptor 6 (VTS20631). | |||||
|
LMAN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 120) | NC score | 0.008999 (rank : 179) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49257, Q12895, Q8N5I7, Q9UQG1, Q9UQG2, Q9UQG3, Q9UQG4, Q9UQG5, Q9UQG6, Q9UQG7, Q9UQG8, Q9UQG9, Q9UQH0, Q9UQH1, Q9UQH2 | Gene names | LMAN1, ERGIC53, F5F8D | |||
|
Domain Architecture |
|
|||||
| Description | ERGIC-53 protein precursor (ER-Golgi intermediate compartment 53 kDa protein) (Lectin, mannose-binding 1) (Gp58) (Intracellular mannose- specific lectin MR60). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 121) | NC score | 0.047278 (rank : 94) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MYH1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 122) | NC score | 0.025527 (rank : 137) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
MYH4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 123) | NC score | 0.024355 (rank : 144) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
PPIG_HUMAN
|
||||||
| θ value | 6.88961 (rank : 124) | NC score | 0.034061 (rank : 112) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
REST_HUMAN
|
||||||
| θ value | 6.88961 (rank : 125) | NC score | 0.026579 (rank : 132) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
SART3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 126) | NC score | 0.015253 (rank : 165) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 301 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15020, Q58F06, Q8IUS1, Q96J95 | Gene names | SART3, KIAA0156, TIP110 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Squamous cell carcinoma antigen recognized by T-cells 3 (SART-3) (hSART-3) (Tat-interacting protein of 110 kDa) (Tip110). | |||||
|
ZN174_HUMAN
|
||||||
| θ value | 6.88961 (rank : 127) | NC score | -0.000196 (rank : 195) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15697, Q9BQ34 | Gene names | ZNF174, ZSCAN8 | |||
|
Domain Architecture |
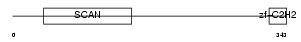
|
|||||
| Description | Zinc finger protein 174 (AW-1) (Zinc finger and SCAN domain-containing protein 8). | |||||
|
DDX46_MOUSE
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.010356 (rank : 176) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q569Z5, Q6ZQ42, Q8R0R6 | Gene names | Ddx46, Kiaa0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46). | |||||
|
G3BP2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | 0.043088 (rank : 99) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P97379, Q9R1B8 | Gene names | G3bp2 | |||
|
Domain Architecture |

|
|||||
| Description | Ras-GTPase-activating protein-binding protein 2 (GAP SH3-domain- binding protein 2) (G3BP-2). | |||||
|
IL4RA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | 0.011720 (rank : 175) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P24394, Q96P01, Q9H181, Q9H182, Q9H183, Q9H184, Q9H185, Q9H186, Q9H187, Q9H188 | Gene names | IL4R, 582J2.1, IL4RA | |||
|
Domain Architecture |
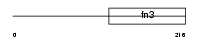
|
|||||
| Description | Interleukin-4 receptor alpha chain precursor (IL-4R-alpha) (CD124 antigen) [Contains: Soluble interleukin-4 receptor alpha chain (sIL4Ralpha/prot) (IL-4-binding protein) (IL4-BP)]. | |||||
|
LPIN2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.012638 (rank : 173) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q92539 | Gene names | LPIN2, KIAA0249 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipin-2. | |||||
|
MAP2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | 0.028792 (rank : 122) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P20357 | Gene names | Map2, Mtap2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2). | |||||
|
MYH8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | 0.024764 (rank : 139) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
MYST3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 134) | NC score | 0.046485 (rank : 96) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
NFM_HUMAN
|
||||||
| θ value | 8.99809 (rank : 135) | NC score | 0.041677 (rank : 100) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
PCDLK_HUMAN
|
||||||
| θ value | 8.99809 (rank : 136) | NC score | 0.002112 (rank : 191) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BYE9, Q9NXP8 | Gene names | PCLKC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin LKC precursor (PC-LKC). | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 137) | NC score | 0.059474 (rank : 57) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
SIPA1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 138) | NC score | 0.006405 (rank : 185) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P46062, P70204 | Gene names | Sipa1, Spa-1, Spa1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal-induced proliferation-associated protein 1 (Sipa-1) (GTPase- activating protein Spa-1). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 139) | NC score | 0.033182 (rank : 114) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TBX2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 140) | NC score | 0.007581 (rank : 182) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13207, Q16424, Q7Z647 | Gene names | TBX2 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX2 (T-box protein 2). | |||||
|
TOP2B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 141) | NC score | 0.018629 (rank : 157) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q64511, Q7TQG4 | Gene names | Top2b | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 2-beta (EC 5.99.1.3) (DNA topoisomerase II, beta isozyme). | |||||
|
WWP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 142) | NC score | 0.005623 (rank : 187) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O00308, Q96CZ2, Q9BWN6 | Gene names | WWP2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2) (Atrophin-1-interacting protein 2) (AIP2). | |||||
|
ABTAP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.050599 (rank : 85) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q3V1V3 | Gene names | Abtap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
ACINU_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.050281 (rank : 87) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
ACINU_MOUSE
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.050120 (rank : 89) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
AFF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.060222 (rank : 53) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
AKA12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.062758 (rank : 45) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
BASP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.081580 (rank : 21) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
BASP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.067854 (rank : 34) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
CN155_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.051418 (rank : 81) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
DEK_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.064751 (rank : 40) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q7TNV0, Q80VC5, Q8BZV6 | Gene names | Dek | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
DMP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.062266 (rank : 46) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
DNMT1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.050153 (rank : 88) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P26358, Q9UHG5, Q9ULA2, Q9UMZ6 | Gene names | DNMT1, AIM, DNMT | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 1 (EC 2.1.1.37) (Dnmt1) (DNA methyltransferase HsaI) (DNA MTase HsaI) (MCMT) (M.HsaI). | |||||
|
ENL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.051112 (rank : 82) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q03111, Q14768 | Gene names | MLLT1, ENL, LTG19, YEATS1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ENL (YEATS domain-containing protein 1). | |||||
|
GLU2B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.051867 (rank : 79) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O08795, Q921X2 | Gene names | Prkcsh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glucosidase 2 subunit beta precursor (Glucosidase II subunit beta) (Protein kinase C substrate, 60.1 kDa protein, heavy chain) (PKCSH) (80K-H protein). | |||||
|
ICAL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.052766 (rank : 76) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P51125, Q9EQV4, Q9EQV5, Q9QXQ3, Q9QXQ4, Q9R0N1 | Gene names | Cast | |||
|
Domain Architecture |

|
|||||
| Description | Calpastatin (Calpain inhibitor). | |||||
|
IF2P_HUMAN
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.053699 (rank : 74) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.055659 (rank : 67) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.065908 (rank : 37) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
LAD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.059611 (rank : 56) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P57016 | Gene names | Lad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (Linear IgA disease autoantigen). | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.064191 (rank : 41) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
MARCS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.065187 (rank : 39) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
MINT_MOUSE
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.050577 (rank : 86) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.050722 (rank : 84) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
NASP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.060409 (rank : 51) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
NEUM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.076280 (rank : 25) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P17677 | Gene names | GAP43 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (PP46) (Neural phosphoprotein B-50). | |||||
|
NEUM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.087780 (rank : 16) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P06837 | Gene names | Gap43, Basp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (Calmodulin-binding protein P-57). | |||||
|
NFH_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.064138 (rank : 42) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.098027 (rank : 13) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
NSBP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.054100 (rank : 71) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
NUCKS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.086482 (rank : 17) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H1E3, Q9H1D6, Q9H723 | Gene names | NUCKS1, NUCKS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (P1). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.055422 (rank : 68) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PININ_MOUSE
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.054565 (rank : 70) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
PRG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.054012 (rank : 72) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRG4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.061366 (rank : 48) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
R51A1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.053870 (rank : 73) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8C551, O55219, Q8BP36, Q99L94, Q9D0J0 | Gene names | Rad51ap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RAD51-associated protein 1 (RAD51-interacting protein) (RAB22). | |||||
|
RL1D1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.055151 (rank : 69) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O76021, Q6PL22, Q8IWS7, Q8WUZ1, Q9HDA9, Q9Y3Z9 | Gene names | RSL1D1, CATX11, CSIG, PBK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal L1 domain-containing protein 1 (Cellular senescence- inhibited gene protein) (Protein PBK1) (CATX-11). | |||||
|
SFR12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.052641 (rank : 77) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.052368 (rank : 78) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.061299 (rank : 49) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.059884 (rank : 54) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.059256 (rank : 59) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
T2FA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.059358 (rank : 58) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
TAF3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.057059 (rank : 63) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q5HZG4, Q3U490, Q3UWX2, Q8BIU8, Q99JH4 | Gene names | Taf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
TCOF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.063294 (rank : 43) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
TCOF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.065709 (rank : 38) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
TGON1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.059615 (rank : 55) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q62313 | Gene names | Tgoln1, Ttgn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 1 precursor (TGN38A). | |||||
|
TGON2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.061407 (rank : 47) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q62314 | Gene names | Tgoln2, Ttgn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (TGN38B). | |||||
|
TRDN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 189) | NC score | 0.072218 (rank : 31) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
TXLNB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 190) | NC score | 0.051737 (rank : 80) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8N3L3, Q76L25, Q86T52, Q8N3S2 | Gene names | TXLNB, C6orf198, MDP77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77) (hMDP77). | |||||
|
TXND2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 191) | NC score | 0.073653 (rank : 29) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
VCX1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 192) | NC score | 0.077627 (rank : 24) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H320, Q9P0H3 | Gene names | VCX, VCX1, VCX10R, VCXB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 1 (VCX-B1 protein) (Variably charged protein X-B1) (Variable charge protein on X with ten repeats) (VCX- 10r). | |||||
|
VCX3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 193) | NC score | 0.081715 (rank : 20) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NNX9, Q9P0H4 | Gene names | VCX3A, VCX3, VCX8R, VCXA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 3 (VCX-A protein) (Variably charged protein X-A) (Variable charge protein on X with eight repeats) (VCX- 8r). | |||||
|
VCXC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 194) | NC score | 0.083591 (rank : 19) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H321 | Gene names | VCXC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | VCX-C protein (Variably charged protein X-C). | |||||
|
ZCPW1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 195) | NC score | 0.053131 (rank : 75) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6IR42 | Gene names | Zcwpw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1 homolog. | |||||
|
ADDB_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | P35612, Q13482, Q16412, Q59G82, Q5U5P4, Q6P0P2, Q6PGQ4, Q7Z688, Q7Z689, Q7Z690, Q7Z691 | Gene names | ADD2, ADDB | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta). | |||||
|
ADDG_MOUSE
|
||||||
| NC score | 0.955089 (rank : 2) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QYB5, Q9D2M5, Q9QYB6 | Gene names | Add3, Addl | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-adducin (Adducin-like protein 70). | |||||
|
ADDG_HUMAN
|
||||||
| NC score | 0.951190 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UEY8, O43243, Q92773, Q9UEY7 | Gene names | ADD3, ADDL | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-adducin (Adducin-like protein 70). | |||||
|
ADDA_HUMAN
|
||||||
| NC score | 0.937028 (rank : 4) | θ value | 7.25522e-167 (rank : 5) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P35611, Q13734, Q14729, Q16156, Q9UJB6 | Gene names | ADD1, ADDA | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-adducin (Erythrocyte adducin subunit alpha). | |||||
|
ADDA_MOUSE
|
||||||
| NC score | 0.926090 (rank : 5) | θ value | 3.04821e-165 (rank : 6) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9QYC0, Q9JLE3 | Gene names | Add1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-adducin (Erythrocyte adducin subunit alpha). | |||||
|
ADDB_MOUSE
|
||||||
| NC score | 0.877296 (rank : 6) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
SNUT3_MOUSE
|
||||||
| NC score | 0.166648 (rank : 7) | θ value | 0.0148317 (rank : 20) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K194, Q8BRA8, Q9CSV0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 3. | |||||
|
SNUT3_HUMAN
|
||||||
| NC score | 0.137294 (rank : 8) | θ value | 0.125558 (rank : 28) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8WVK2, Q15410 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 3 (U4/U6.U5 tri-snRNP-associated 27 kDa protein) (27K) (Nucleic acid-binding protein RY-1). | |||||
|
CF010_HUMAN
|
||||||
| NC score | 0.114855 (rank : 9) | θ value | 0.00665767 (rank : 15) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q5SRN2, Q5SPI9, Q5SPJ0, Q5SPK9, Q5SPL0, Q5SRN3, Q5TG25, Q5TG26, Q8N4B6 | Gene names | C6orf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf10. | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.113834 (rank : 10) | θ value | 3.77169e-06 (rank : 7) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
YTDC1_HUMAN
|
||||||
| NC score | 0.101903 (rank : 11) | θ value | 0.00035302 (rank : 8) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
RBBP6_MOUSE
|
||||||
| NC score | 0.099325 (rank : 12) | θ value | 0.21417 (rank : 31) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.098027 (rank : 13) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
TP53B_MOUSE
|
||||||
| NC score | 0.090252 (rank : 14) | θ value | 0.00298849 (rank : 13) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.089756 (rank : 15) | θ value | 0.000461057 (rank : 9) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
NEUM_MOUSE
|
||||||
| NC score | 0.087780 (rank : 16) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P06837 | Gene names | Gap43, Basp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (Calmodulin-binding protein P-57). | |||||
|
NUCKS_HUMAN
|
||||||
| NC score | 0.086482 (rank : 17) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H1E3, Q9H1D6, Q9H723 | Gene names | NUCKS1, NUCKS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (P1). | |||||
|
TP53B_HUMAN
|
||||||
| NC score | 0.084723 (rank : 18) | θ value | 0.0736092 (rank : 26) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q12888, Q2M1Z7, Q4LE46, Q5FWZ3, Q7Z3U4 | Gene names | TP53BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
VCXC_HUMAN
|
||||||
| NC score | 0.083591 (rank : 19) | θ value | θ > 10 (rank : 194) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H321 | Gene names | VCXC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | VCX-C protein (Variably charged protein X-C). | |||||
|
VCX3_HUMAN
|
||||||
| NC score | 0.081715 (rank : 20) | θ value | θ > 10 (rank : 193) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NNX9, Q9P0H4 | Gene names | VCX3A, VCX3, VCX8R, VCXA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 3 (VCX-A protein) (Variably charged protein X-A) (Variable charge protein on X with eight repeats) (VCX- 8r). | |||||
|
BASP_HUMAN
|
||||||
| NC score | 0.081580 (rank : 21) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
LRRF1_HUMAN
|
||||||
| NC score | 0.079389 (rank : 22) | θ value | 0.00869519 (rank : 17) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q32MZ4, O75766, O75799, Q32MZ5, Q53T49, Q6PKG2 | Gene names | LRRFIP1, GCF2, TRIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (TAR RNA-interacting protein) (GC-binding factor 2). | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.077906 (rank : 23) | θ value | 1.81305 (rank : 69) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
VCX1_HUMAN
|
||||||
| NC score | 0.077627 (rank : 24) | θ value | θ > 10 (rank : 192) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H320, Q9P0H3 | Gene names | VCX, VCX1, VCX10R, VCXB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 1 (VCX-B1 protein) (Variably charged protein X-B1) (Variable charge protein on X with ten repeats) (VCX- 10r). | |||||
|
NEUM_HUMAN
|
||||||
| NC score | 0.076280 (rank : 25) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P17677 | Gene names | GAP43 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (PP46) (Neural phosphoprotein B-50). | |||||
|
DMP1_MOUSE
|
||||||
| NC score | 0.075712 (rank : 26) | θ value | 1.81305 (rank : 67) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
GLU2B_HUMAN
|
||||||
| NC score | 0.074506 (rank : 27) | θ value | 6.88961 (rank : 118) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P14314, Q96BU9, Q96D06, Q9P0W9 | Gene names | PRKCSH, G19P1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glucosidase 2 subunit beta precursor (Glucosidase II subunit beta) (Protein kinase C substrate, 60.1 kDa protein, heavy chain) (PKCSH) (80K-H protein). | |||||
|
CYLC1_HUMAN
|
||||||
| NC score | 0.074307 (rank : 28) | θ value | 4.03905 (rank : 94) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.073653 (rank : 29) | θ value | θ > 10 (rank : 191) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.073050 (rank : 30) | θ value | 0.813845 (rank : 46) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
TRDN_HUMAN
|
||||||
| NC score | 0.072218 (rank : 31) | θ value | θ > 10 (rank : 189) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
MUC1_MOUSE
|
||||||
| NC score | 0.069544 (rank : 32) | θ value | 0.0330416 (rank : 23) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.068623 (rank : 33) | θ value | 0.000602161 (rank : 10) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.067854 (rank : 34) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
SELPL_MOUSE
|
||||||
| NC score | 0.066339 (rank : 35) | θ value | 0.0148317 (rank : 19) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
GP158_HUMAN
|
||||||
| NC score | 0.065932 (rank : 36) | θ value | 0.00390308 (rank : 14) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5T848, Q6QR81, Q9ULT3 | Gene names | GPR158, KIAA1136 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 158 precursor. | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.065908 (rank : 37) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
TCOF_MOUSE
|
||||||
| NC score | 0.065709 (rank : 38) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
MARCS_HUMAN
|
||||||
| NC score | 0.065187 (rank : 39) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
DEK_MOUSE
|
||||||
| NC score | 0.064751 (rank : 40) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q7TNV0, Q80VC5, Q8BZV6 | Gene names | Dek | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.064191 (rank : 41) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.064138 (rank : 42) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.063294 (rank : 43) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.063123 (rank : 44) | θ value | 3.0926 (rank : 83) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.062758 (rank : 45) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
DMP1_HUMAN
|
||||||
| NC score | 0.062266 (rank : 46) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
TGON2_MOUSE
|
||||||
| NC score | 0.061407 (rank : 47) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q62314 | Gene names | Tgoln2, Ttgn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (TGN38B). | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.061366 (rank : 48) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.061299 (rank : 49) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
CJ047_MOUSE
|
||||||
| NC score | 0.060603 (rank : 50) | θ value | 0.0736092 (rank : 25) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8C5R2, Q8R017, Q8R2Z9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf47 homolog. | |||||
|
NASP_HUMAN
|
||||||
| NC score | 0.060409 (rank : 51) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
LIMA1_MOUSE
|
||||||
| NC score | 0.060241 (rank : 52) | θ value | 0.0252991 (rank : 21) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9ERG0, Q9ERG1 | Gene names | Lima1, D15Ertd366e, Eplin | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain and actin-binding protein 1 (Epithelial protein lost in neoplasm) (mEPLIN). | |||||
|
AFF1_HUMAN
|
||||||
| NC score | 0.060222 (rank : 53) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.059884 (rank : 54) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
TGON1_MOUSE
|
||||||
| NC score | 0.059615 (rank : 55) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q62313 | Gene names | Tgoln1, Ttgn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 1 precursor (TGN38A). | |||||
|
LAD1_MOUSE
|
||||||
| NC score | 0.059611 (rank : 56) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P57016 | Gene names | Lad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (Linear IgA disease autoantigen). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.059474 (rank : 57) | θ value | 8.99809 (rank : 137) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
T2FA_HUMAN
|
||||||
| NC score | 0.059358 (rank : 58) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.059256 (rank : 59) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
PALM_MOUSE
|
||||||
| NC score | 0.058990 (rank : 60) | θ value | 0.0431538 (rank : 24) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Z0P4, Q9Z0P3 | Gene names | Palm | |||
|
Domain Architecture |

|
|||||
| Description | Paralemmin. | |||||
|
G3BP2_HUMAN
|
||||||
| NC score | 0.057895 (rank : 61) | θ value | 0.62314 (rank : 44) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UN86, O60606, O75149, Q9UPA1 | Gene names | G3BP2, KIAA0660 | |||
|
Domain Architecture |

|
|||||
| Description | Ras-GTPase-activating protein-binding protein 2 (GAP SH3-domain- binding protein 2) (G3BP-2). | |||||
|
NOC3L_HUMAN
|
||||||
| NC score | 0.057874 (rank : 62) | θ value | 0.279714 (rank : 34) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8WTT2, Q9H5M6, Q9H9D8 | Gene names | NOC3L, AD24, C10orf117, FAD24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar complex protein 3 homolog (NOC3 protein homolog) (NOC3-like protein) (Nucleolar complex-associated protein 3-like protein) (Factor for adipocyte differentiation 24). | |||||
|
TAF3_MOUSE
|
||||||
| NC score | 0.057059 (rank : 63) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q5HZG4, Q3U490, Q3UWX2, Q8BIU8, Q99JH4 | Gene names | Taf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.056484 (rank : 64) | θ value | 1.06291 (rank : 53) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
EMSY_HUMAN
|
||||||
| NC score | 0.056207 (rank : 65) | θ value | 0.125558 (rank : 27) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7Z589, Q4G109, Q8NBU6, Q8TE50, Q9H8I9, Q9NRH0 | Gene names | EMSY, C11orf30 | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
TRI44_HUMAN
|
||||||
| NC score | 0.055776 (rank : 66) | θ value | 0.21417 (rank : 33) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96DX7, Q96QY2, Q9UGK0 | Gene names | TRIM44, DIPB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.055659 (rank : 67) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.055422 (rank : 68) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
RL1D1_HUMAN
|
||||||
| NC score | 0.055151 (rank : 69) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O76021, Q6PL22, Q8IWS7, Q8WUZ1, Q9HDA9, Q9Y3Z9 | Gene names | RSL1D1, CATX11, CSIG, PBK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal L1 domain-containing protein 1 (Cellular senescence- inhibited gene protein) (Protein PBK1) (CATX-11). | |||||
|
PININ_MOUSE
|
||||||
| NC score | 0.054565 (rank : 70) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
NSBP1_HUMAN
|
||||||
| NC score | 0.054100 (rank : 71) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.054012 (rank : 72) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
R51A1_MOUSE
|
||||||
| NC score | 0.053870 (rank : 73) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8C551, O55219, Q8BP36, Q99L94, Q9D0J0 | Gene names | Rad51ap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RAD51-associated protein 1 (RAD51-interacting protein) (RAB22). | |||||
|
IF2P_HUMAN
|
||||||
| NC score | 0.053699 (rank : 74) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
ZCPW1_MOUSE
|
||||||
| NC score | 0.053131 (rank : 75) | θ value | θ > 10 (rank : 195) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6IR42 | Gene names | Zcwpw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1 homolog. | |||||
|
ICAL_MOUSE
|
||||||
| NC score | 0.052766 (rank : 76) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P51125, Q9EQV4, Q9EQV5, Q9QXQ3, Q9QXQ4, Q9R0N1 | Gene names | Cast | |||
|
Domain Architecture |

|
|||||
| Description | Calpastatin (Calpain inhibitor). | |||||
|
SFR12_HUMAN
|
||||||
| NC score | 0.052641 (rank : 77) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.052368 (rank : 78) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
GLU2B_MOUSE
|
||||||
| NC score | 0.051867 (rank : 79) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O08795, Q921X2 | Gene names | Prkcsh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glucosidase 2 subunit beta precursor (Glucosidase II subunit beta) (Protein kinase C substrate, 60.1 kDa protein, heavy chain) (PKCSH) (80K-H protein). | |||||
|
TXLNB_HUMAN
|
||||||
| NC score | 0.051737 (rank : 80) | θ value | θ > 10 (rank : 190) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8N3L3, Q76L25, Q86T52, Q8N3S2 | Gene names | TXLNB, C6orf198, MDP77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77) (hMDP77). | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.051418 (rank : 81) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
ENL_HUMAN
|
||||||
| NC score | 0.051112 (rank : 82) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q03111, Q14768 | Gene names | MLLT1, ENL, LTG19, YEATS1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ENL (YEATS domain-containing protein 1). | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.050744 (rank : 83) | θ value | 1.06291 (rank : 52) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.050722 (rank : 84) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
ABTAP_MOUSE
|
||||||
| NC score | 0.050599 (rank : 85) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q3V1V3 | Gene names | Abtap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.050577 (rank : 86) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
ACINU_HUMAN
|
||||||
| NC score | 0.050281 (rank : 87) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
DNMT1_HUMAN
|
||||||
| NC score | 0.050153 (rank : 88) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P26358, Q9UHG5, Q9ULA2, Q9UMZ6 | Gene names | DNMT1, AIM, DNMT | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 1 (EC 2.1.1.37) (Dnmt1) (DNA methyltransferase HsaI) (DNA MTase HsaI) (MCMT) (M.HsaI). | |||||
|
ACINU_MOUSE
|
||||||
| NC score | 0.050120 (rank : 89) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.049461 (rank : 90) | θ value | 0.00175202 (rank : 11) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
MAP4_MOUSE
|
||||||
| NC score | 0.049113 (rank : 91) | θ value | 1.38821 (rank : 60) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
CEP35_HUMAN
|
||||||
| NC score | 0.048934 (rank : 92) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q5VT06, O75068, Q8TDK3, Q8WY20 | Gene names | CEP350, CAP350, GM133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein 350 (Centrosome-associated protein of 350 kDa). | |||||
|
VEZA_HUMAN
|
||||||
| NC score | 0.048819 (rank : 93) | θ value | 1.38821 (rank : 63) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9HBM0, Q9H2F4, Q9H2U5, Q9NT70, Q9NVW0, Q9UF91 | Gene names | VEZT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vezatin. | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.047278 (rank : 94) | θ value | 6.88961 (rank : 121) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
TRI44_MOUSE
|
||||||
| NC score | 0.046755 (rank : 95) | θ value | 2.36792 (rank : 78) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9QXA7, Q80SV9, Q9JHR8 | Gene names | Trim44, Dipb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB) (Protein Mc7). | |||||
|
MYST3_MOUSE
|
||||||
| NC score | 0.046485 (rank : 96) | θ value | 8.99809 (rank : 134) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
CTDP1_HUMAN
|
||||||
| NC score | 0.045483 (rank : 97) | θ value | 2.36792 (rank : 72) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y5B0, Q7Z644, Q96BZ1, Q9Y6F5 | Gene names | CTDP1, FCP1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II subunit A C-terminal domain phosphatase (EC 3.1.3.16) (TFIIF-associating CTD phosphatase). | |||||
|
BCAS1_MOUSE
|
||||||
| NC score | 0.044615 (rank : 98) | θ value | 0.62314 (rank : 43) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q80YN3, Q9CVA1 | Gene names | Bcas1, Nabc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast carcinoma amplified sequence 1 homolog (Novel amplified in breast cancer 1 homolog). | |||||
|
G3BP2_MOUSE
|
||||||
| NC score | 0.043088 (rank : 99) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P97379, Q9R1B8 | Gene names | G3bp2 | |||
|
Domain Architecture |

|
|||||
| Description | Ras-GTPase-activating protein-binding protein 2 (GAP SH3-domain- binding protein 2) (G3BP-2). | |||||
|
NFM_HUMAN
|
||||||
| NC score | 0.041677 (rank : 100) | θ value | 8.99809 (rank : 135) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
KTN1_MOUSE
|
||||||
| NC score | 0.041591 (rank : 101) | θ value | 0.00665767 (rank : 16) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1324 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q61595, Q8BG49, Q8BHF4, Q8BHM8, Q8C9Y5, Q8CG51, Q8CG52, Q8CG53, Q8CG54, Q8CG55, Q8CG56, Q8CG57, Q8CG58, Q8CG59, Q8CG60, Q8CG61, Q8CG62, Q8CG63 | Gene names | Ktn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin. | |||||
|
NOC3L_MOUSE
|
||||||
| NC score | 0.041422 (rank : 102) | θ value | 1.06291 (rank : 54) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8VI84, Q9JKM3 | Gene names | Noc3l, Ad24, Fad24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar complex protein 3 homolog (NOC3 protein homolog) (NOC3-like protein) (Nucleolar complex-associated protein 3-like protein) (Factor for adipocyte differentiation 24). | |||||
|
GP179_HUMAN
|
||||||
| NC score | 0.040574 (rank : 103) | θ value | 5.27518 (rank : 104) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6PRD1 | Gene names | GPR179, GPR158L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 179 precursor (Probable G-protein coupled receptor 158-like 1). | |||||
|
CCAR1_MOUSE
|
||||||
| NC score | 0.040382 (rank : 104) | θ value | 3.0926 (rank : 79) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8CH18, Q6AXC9, Q6PAR2, Q80XE4, Q8BJY0, Q8BVN2, Q8CGG1, Q9CSR5 | Gene names | Ccar1, Carp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle and apoptosis regulator protein 1 (Cell cycle and apoptosis regulatory protein 1) (CARP-1). | |||||
|
ZCPW1_HUMAN
|
||||||
| NC score | 0.040109 (rank : 105) | θ value | 0.279714 (rank : 35) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H0M4, Q8NA98, Q9BUD0, Q9NWF7 | Gene names | ZCWPW1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1. | |||||
|
INVO_MOUSE
|
||||||
| NC score | 0.039445 (rank : 106) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P48997 | Gene names | Ivl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Involucrin. | |||||
|
AN30A_HUMAN
|
||||||
| NC score | 0.037342 (rank : 107) | θ value | 0.0330416 (rank : 22) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1119 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9BXX3, Q5W025 | Gene names | ANKRD30A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 30A (NY-BR-1 antigen). | |||||
|
LUZP1_MOUSE
|
||||||
| NC score | 0.037032 (rank : 108) | θ value | 0.47712 (rank : 39) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8R4U7, Q3UV18, Q7TS71, Q8BQW1, Q99NG3, Q9CSL6 | Gene names | Luzp1, Luzp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1 (Leucine zipper motif-containing protein). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.035893 (rank : 109) | θ value | 3.0926 (rank : 84) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
CCD19_HUMAN
|
||||||
| NC score | 0.035304 (rank : 110) | θ value | 6.88961 (rank : 116) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UL16 | Gene names | CCDC19, NESG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 19 (Nasopharyngeal epithelium- specific protein 1). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.034695 (rank : 111) | θ value | 1.06291 (rank : 48) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
PPIG_HUMAN
|
||||||
| NC score | 0.034061 (rank : 112) | θ value | 6.88961 (rank : 124) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
KMHN1_HUMAN
|
||||||
| NC score | 0.033599 (rank : 113) | θ value | 0.47712 (rank : 38) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 950 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8TBZ0, Q86YI9, Q8N7W0 | Gene names | KMHN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1. | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.033182 (rank : 114) | θ value | 8.99809 (rank : 139) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TRHY_HUMAN
|
||||||
| NC score | 0.032572 (rank : 115) | θ value | 2.36792 (rank : 77) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
WAPL_MOUSE
|
||||||
| NC score | 0.032400 (rank : 116) | θ value | 1.38821 (rank : 64) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q65Z40, Q6ZQE9 | Gene names | Wapal, Kiaa0261, Wapl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wings apart-like protein homolog (Dioxin-inducible factor 2) (DIF-2). | |||||
|
RBP24_HUMAN
|
||||||
| NC score | 0.031289 (rank : 117) | θ value | 1.38821 (rank : 61) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
GCC2_HUMAN
|
||||||
| NC score | 0.031286 (rank : 118) | θ value | 1.38821 (rank : 58) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
NALP5_MOUSE
|
||||||
| NC score | 0.030458 (rank : 119) | θ value | 0.21417 (rank : 30) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
MY18A_HUMAN
|
||||||
| NC score | 0.030190 (rank : 120) | θ value | 0.47712 (rank : 40) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
CE290_HUMAN
|
||||||
| NC score | 0.030151 (rank : 121) | θ value | 3.0926 (rank : 80) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
MAP2_MOUSE
|
||||||
| NC score | 0.028792 (rank : 122) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P20357 | Gene names | Map2, Mtap2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2). | |||||
|
TIP60_MOUSE
|
||||||
| NC score | 0.028591 (rank : 123) | θ value | 3.0926 (rank : 88) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8CHK4, Q8CGZ3, Q8CGZ4, Q8VIH0 | Gene names | Htatip, Tip60 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase HTATIP (EC 2.3.1.48) (EC 2.3.1.-) (60 kDa Tat interactive protein) (Tip60). | |||||
|
SIA7A_MOUSE
|
||||||
| NC score | 0.028161 (rank : 124) | θ value | 5.27518 (rank : 113) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QZ39, Q9JJP5 | Gene names | St6galnac1, Siat7a | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 (EC 2.4.99.3) (GalNAc alpha-2,6-sialyltransferase I) (ST6GalNAc I) (Sialyltransferase 7A). | |||||
|
MYH11_MOUSE
|
||||||
| NC score | 0.028045 (rank : 125) | θ value | 0.813845 (rank : 45) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
TIP60_HUMAN
|
||||||
| NC score | 0.027975 (rank : 126) | θ value | 3.0926 (rank : 87) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q92993, O95624, Q13430, Q6GSE8, Q9BWK7 | Gene names | HTATIP, TIP60 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase HTATIP (EC 2.3.1.48) (EC 2.3.1.-) (60 kDa Tat interactive protein) (Tip60) (HIV-1 Tat interactive protein) (cPLA(2)-interacting protein). | |||||
|
CGRE1_MOUSE
|
||||||
| NC score | 0.027880 (rank : 127) | θ value | 4.03905 (rank : 91) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8R1U2, Q9D1K1 | Gene names | Cgref1, Cgr11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
ABCF1_HUMAN
|
||||||
| NC score | 0.027724 (rank : 128) | θ value | 1.81305 (rank : 65) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8NE71, O14897, Q69YP6 | Gene names | ABCF1, ABC50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family F member 1 (ATP-binding cassette 50) (TNF-alpha-stimulated ABC protein). | |||||
|
TTBK1_HUMAN
|
||||||
| NC score | 0.027022 (rank : 129) | θ value | 1.06291 (rank : 55) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1058 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5TCY1, Q2L6C6, Q8N444, Q96JH2 | Gene names | TTBK1, BDTK, KIAA1855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 1 (EC 2.7.11.1) (Brain-derived tau kinase). | |||||
|
MYH11_HUMAN
|
||||||
| NC score | 0.026966 (rank : 130) | θ value | 1.06291 (rank : 50) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
NEB1_HUMAN
|
||||||
| NC score | 0.026803 (rank : 131) | θ value | 4.03905 (rank : 96) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9ULJ8, O76059, Q9NXT2 | Gene names | PPP1R9A, KIAA1222 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-1 (Neurabin-I) (Neural tissue-specific F-actin-binding protein I) (Protein phosphatase 1 regulatory subunit 9A). | |||||
|
REST_HUMAN
|
||||||
| NC score | 0.026579 (rank : 132) | θ value | 6.88961 (rank : 125) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
MYH9_MOUSE
|
||||||
| NC score | 0.026401 (rank : 133) | θ value | 1.06291 (rank : 51) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
CD2L1_HUMAN
|
||||||
| NC score | 0.026351 (rank : 134) | θ value | 0.00228821 (rank : 12) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.026072 (rank : 135) | θ value | 0.47712 (rank : 42) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
SIA7A_HUMAN
|
||||||
| NC score | 0.025573 (rank : 136) | θ value | 0.813845 (rank : 47) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NSC7, Q6UW90, Q9NSC6 | Gene names | ST6GALNAC1, SIAT7A | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 (EC 2.4.99.3) (GalNAc alpha-2,6-sialyltransferase I) (ST6GalNAc I) (Sialyltransferase 7A). | |||||
|
MYH1_HUMAN
|
||||||
| NC score | 0.025527 (rank : 137) | θ value | 6.88961 (rank : 122) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
PEPL_HUMAN
|
||||||
| NC score | 0.025406 (rank : 138) | θ value | 2.36792 (rank : 76) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1141 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O60437, O60314, O60454 | Gene names | PPL, KIAA0568 | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin (195 kDa cornified envelope precursor protein) (190 kDa paraneoplastic pemphigus antigen). | |||||
|
MYH8_HUMAN
|
||||||
| NC score | 0.024764 (rank : 139) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
REST_MOUSE
|
||||||
| NC score | 0.024578 (rank : 140) | θ value | 5.27518 (rank : 110) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
CCD45_MOUSE
|
||||||
| NC score | 0.024571 (rank : 141) | θ value | 5.27518 (rank : 101) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BVV7 | Gene names | Ccdc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 45. | |||||
|
UBP1_HUMAN
|
||||||
| NC score | 0.024538 (rank : 142) | θ value | 1.81305 (rank : 71) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O94782, Q9UFR0, Q9UNJ3 | Gene names | USP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 1 (EC 3.1.2.15) (Ubiquitin thioesterase 1) (Ubiquitin-specific-processing protease 1) (Deubiquitinating enzyme 1) (hUBP). | |||||
|
BRCA2_MOUSE
|
||||||
| NC score | 0.024414 (rank : 143) | θ value | 1.81305 (rank : 66) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P97929, O35922, P97383 | Gene names | Brca2, Fancd1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 2 susceptibility protein homolog (Fanconi anemia group D1 protein homolog). | |||||
|
MYH4_MOUSE
|
||||||
| NC score | 0.024355 (rank : 144) | θ value | 6.88961 (rank : 123) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
SRBS1_MOUSE
|
||||||
| NC score | 0.024114 (rank : 145) | θ value | 0.365318 (rank : 36) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q62417, Q80TF8, Q8BZI3, Q8K3Y2, Q921F8, Q9Z0Z8, Q9Z0Z9 | Gene names | Sorbs1, Kiaa1296, Sh3d5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorbin and SH3 domain-containing protein 1 (Ponsin) (c-Cbl-associated protein) (CAP) (SH3 domain protein 5) (SH3P12). | |||||
|
MYRIP_HUMAN
|
||||||
| NC score | 0.023915 (rank : 146) | θ value | 1.81305 (rank : 70) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8NFW9, Q8IUF5, Q9Y3V4 | Gene names | MYRIP, SLAC2C | |||
|
Domain Architecture |

|
|||||
| Description | Rab effector MyRIP (Myosin-VIIa- and Rab-interacting protein) (Exophilin-8) (Slp homolog lacking C2 domains c). | |||||
|
SETBP_HUMAN
|
||||||
| NC score | 0.023677 (rank : 147) | θ value | 5.27518 (rank : 112) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y6X0, Q9UEF3 | Gene names | SETBP1, KIAA0437 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET-binding protein (SEB). | |||||
|
CPLX4_HUMAN
|
||||||
| NC score | 0.023454 (rank : 148) | θ value | 4.03905 (rank : 92) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z7G2 | Gene names | CPLX4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complexin-4 (Complexin IV) (CPX IV). | |||||
|
BPAEA_MOUSE
|
||||||
| NC score | 0.023146 (rank : 149) | θ value | 4.03905 (rank : 89) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
CPLX4_MOUSE
|
||||||
| NC score | 0.022474 (rank : 150) | θ value | 5.27518 (rank : 102) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80WM3, Q8C8Y0, Q91WE5 | Gene names | Cplx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complexin-4 (Complexin IV) (CPX IV). | |||||
|
ERCC5_MOUSE
|
||||||
| NC score | 0.022384 (rank : 151) | θ value | 5.27518 (rank : 103) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P35689, Q61528, Q64248 | Gene names | Ercc5, Ercc-5, Xpg | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-G cells homolog (Xeroderma pigmentosum group G-complementing protein homolog) (DNA excision repair protein ERCC-5). | |||||
|
ANLN_HUMAN
|
||||||
| NC score | 0.022267 (rank : 152) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9NQW6, Q5CZ78, Q6NSK5, Q9H8Y4, Q9NVN9, Q9NVP0 | Gene names | ANLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding protein anillin. | |||||
|
HELZ_HUMAN
|
||||||
| NC score | 0.022023 (rank : 153) | θ value | 2.36792 (rank : 74) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P42694 | Gene names | HELZ, KIAA0054 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable helicase with zinc-finger domain (EC 3.6.1.-). | |||||
|
SYTL2_HUMAN
|
||||||
| NC score | 0.021852 (rank : 154) | θ value | 4.03905 (rank : 97) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9HCH5, Q2YDA7, Q8ND34, Q96BJ2, Q9H768, Q9NXM1 | Gene names | SYTL2, KIAA1597, SLP2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
SART3_MOUSE
|
||||||
| NC score | 0.020618 (rank : 155) | θ value | 1.38821 (rank : 62) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9JLI8, Q6ZQI2, Q8BPK9, Q8C3B7, Q8CFU9 | Gene names | Sart3, Kiaa0156 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Squamous cell carcinoma antigen recognized by T-cells 3 (SART-3) (mSART-3) (Tumor-rejection antigen SART3). | |||||
|
SLK_MOUSE
|
||||||
| NC score | 0.019336 (rank : 156) | θ value | 0.21417 (rank : 32) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1798 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O54988, Q80U65, Q8CAU2, Q8CDW2, Q9WU41 | Gene names | Slk, Kiaa0204, Stk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (mSLK) (Serine/threonine-protein kinase 2) (STE20-related kinase SMAK) (Etk4). | |||||
|
TOP2B_MOUSE
|
||||||
| NC score | 0.018629 (rank : 157) | θ value | 8.99809 (rank : 141) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q64511, Q7TQG4 | Gene names | Top2b | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 2-beta (EC 5.99.1.3) (DNA topoisomerase II, beta isozyme). | |||||
|
KI21A_MOUSE
|
||||||
| NC score | 0.018205 (rank : 158) | θ value | 2.36792 (rank : 75) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9QXL2, Q6P5H1, Q6ZPJ8, Q8BWZ9, Q8BXF1 | Gene names | Kif21a, Kiaa1708 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A. | |||||
|
SLK_HUMAN
|
||||||
| NC score | 0.018012 (rank : 159) | θ value | 0.47712 (rank : 41) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1775 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9H2G2, O00211, Q6P1Z4, Q86WU7, Q86WW1, Q92603, Q9NQL0, Q9NQL1 | Gene names | SLK, KIAA0204, STK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (hSLK) (Serine/threonine-protein kinase 2) (CTCL tumor antigen se20-9). | |||||
|
EVL_MOUSE
|
||||||
| NC score | 0.017323 (rank : 160) | θ value | 4.03905 (rank : 95) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P70429, Q9ERU8 | Gene names | Evl | |||
|
Domain Architecture |

|
|||||
| Description | Ena/VASP-like protein (Ena/vasodilator-stimulated phosphoprotein- like). | |||||
|
PP4R1_MOUSE
|
||||||
| NC score | 0.017177 (rank : 161) | θ value | 3.0926 (rank : 85) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K2V1, Q8R0R8 | Gene names | Ppp4r1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 4 regulatory subunit 1. | |||||
|
CE110_MOUSE
|
||||||
| NC score | 0.016946 (rank : 162) | θ value | 4.03905 (rank : 90) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7TSH4, Q6A072 | Gene names | Cep110, Cp110, Kiaa0419 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 110 kDa (Cep110 protein). | |||||
|
TBC10_MOUSE
|
||||||
| NC score | 0.016722 (rank : 163) | θ value | 4.03905 (rank : 98) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P58802 | Gene names | Tbc1d10a, Epi64, Tbc1d10 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 10A (EBP50-PDX interactor of 64 kDa) (EPI64 protein). | |||||
|
SEPT9_HUMAN
|
||||||
| NC score | 0.015992 (rank : 164) | θ value | 5.27518 (rank : 111) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
SART3_HUMAN
|
||||||
| NC score | 0.015253 (rank : 165) | θ value | 6.88961 (rank : 126) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 301 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15020, Q58F06, Q8IUS1, Q96J95 | Gene names | SART3, KIAA0156, TIP110 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Squamous cell carcinoma antigen recognized by T-cells 3 (SART-3) (hSART-3) (Tat-interacting protein of 110 kDa) (Tip110). | |||||
|
ICB1_MOUSE
|
||||||
| NC score | 0.015038 (rank : 166) | θ value | 5.27518 (rank : 105) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91YX0 | Gene names | Icb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Induced by contact to basement membrane 1 protein homolog (ICB-1 protein). | |||||
|
DDX46_HUMAN
|
||||||
| NC score | 0.014991 (rank : 167) | θ value | 2.36792 (rank : 73) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q7L014, O94894, Q96EI0, Q9Y658 | Gene names | DDX46, KIAA0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46) (PRP5 homolog). | |||||
|
MYLK_HUMAN
|
||||||
| NC score | 0.014930 (rank : 168) | θ value | 0.00869519 (rank : 18) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q15746, O95796, O95797, O95798, O95799, Q14844, Q16794, Q5MY99, Q5MYA0, Q7Z4J0, Q9C0L5, Q9UBG5, Q9UIT9 | Gene names | MYLK, MLCK | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light chain kinase, smooth muscle (EC 2.7.11.18) (MLCK) (Telokin) (Kinase-related protein) (KRP). | |||||
|
ZEP1_HUMAN
|
||||||
| NC score | 0.014456 (rank : 169) | θ value | 1.06291 (rank : 56) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1036 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P15822, Q14122 | Gene names | HIVEP1, ZNF40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 40 (Human immunodeficiency virus type I enhancer- binding protein 1) (HIV-EP1) (Major histocompatibility complex-binding protein 1) (MBP-1) (Positive regulatory domain II-binding factor 1) (PRDII-BF1). | |||||
|
CAC1C_HUMAN
|
||||||
| NC score | 0.013852 (rank : 170) | θ value | 1.06291 (rank : 49) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13936, Q13917, Q13918, Q13919, Q13920, Q13921, Q13922, Q13923, Q13924, Q13925, Q13926, Q13927, Q13928, Q13929, Q13930, Q13932, Q13933, Q14743, Q14744, Q15877, Q99025, Q99241, Q99875 | Gene names | CACNA1C, CACH2, CACN2, CACNL1A1, CCHL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle). | |||||
|
TRPS1_MOUSE
|
||||||
| NC score | 0.012865 (rank : 171) | θ value | 4.03905 (rank : 99) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q925H1 | Gene names | Trps1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger transcription factor Trps1. | |||||
|
PB1_HUMAN
|
||||||
| NC score | 0.012739 (rank : 172) | θ value | 5.27518 (rank : 108) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86U86, Q9H2T3, Q9H2T4, Q9H2T5, Q9H301, Q9H314 | Gene names | PB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein polybromo-1 (hPB1) (Polybromo-1D) (BRG1-associated factor 180) (BAF180). | |||||
|
LPIN2_HUMAN
|
||||||
| NC score | 0.012638 (rank : 173) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q92539 | Gene names | LPIN2, KIAA0249 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipin-2. | |||||
|
NF2L2_MOUSE
|
||||||
| NC score | 0.011775 (rank : 174) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q60795 | Gene names | Nfe2l2, Nrf-2, Nrf2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 2 (NF-E2-related factor 2) (NFE2-related factor 2) (Nuclear factor, erythroid derived 2, like 2). | |||||
|
IL4RA_HUMAN
|
||||||
| NC score | 0.011720 (rank : 175) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P24394, Q96P01, Q9H181, Q9H182, Q9H183, Q9H184, Q9H185, Q9H186, Q9H187, Q9H188 | Gene names | IL4R, 582J2.1, IL4RA | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-4 receptor alpha chain precursor (IL-4R-alpha) (CD124 antigen) [Contains: Soluble interleukin-4 receptor alpha chain (sIL4Ralpha/prot) (IL-4-binding protein) (IL4-BP)]. | |||||
|
DDX46_MOUSE
|
||||||
| NC score | 0.010356 (rank : 176) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q569Z5, Q6ZQ42, Q8R0R6 | Gene names | Ddx46, Kiaa0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46). | |||||
|
DEN4C_HUMAN
|
||||||
| NC score | 0.009756 (rank : 177) | θ value | 6.88961 (rank : 117) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5VZ89, Q6AI48, Q6ZUB3, Q8NCY7, Q9H6N4, Q9NUT1, Q9NWA5 | Gene names | DENND4C, C9orf55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 4C. | |||||
|
TCF8_HUMAN
|
||||||
| NC score | 0.009710 (rank : 178) | θ value | 3.0926 (rank : 86) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P37275, Q12924, Q13800 | Gene names | TCF8, AREB6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (NIL-2-A zinc finger protein) (Negative regulator of IL2). | |||||
|
LMAN1_HUMAN
|
||||||
| NC score | 0.008999 (rank : 179) | θ value | 6.88961 (rank : 120) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49257, Q12895, Q8N5I7, Q9UQG1, Q9UQG2, Q9UQG3, Q9UQG4, Q9UQG5, Q9UQG6, Q9UQG7, Q9UQG8, Q9UQG9, Q9UQH0, Q9UQH1, Q9UQH2 | Gene names | LMAN1, ERGIC53, F5F8D | |||
|
Domain Architecture |

|
|||||
| Description | ERGIC-53 protein precursor (ER-Golgi intermediate compartment 53 kDa protein) (Lectin, mannose-binding 1) (Gp58) (Intracellular mannose- specific lectin MR60). | |||||
|
LIRB3_HUMAN
|
||||||
| NC score | 0.008930 (rank : 180) | θ value | 1.38821 (rank : 59) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75022, O15471, Q86U49 | Gene names | LILRB3, ILT5, LIR3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily B member 3 precursor (Leukocyte immunoglobulin-like receptor 3) (LIR-3) (Immunoglobulin- like transcript 5) (ILT-5) (Monocyte inhibitory receptor HL9) (CD85a antigen). | |||||
|
COPA_HUMAN
|
||||||
| NC score | 0.008904 (rank : 181) | θ value | 1.38821 (rank : 57) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P53621 | Gene names | COPA | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit alpha (Alpha-coat protein) (Alpha-COP) (HEPCOP) (HEP- COP) [Contains: Xenin (Xenopsin-related peptide); Proxenin]. | |||||
|
TBX2_HUMAN
|
||||||
| NC score | 0.007581 (rank : 182) | θ value | 8.99809 (rank : 140) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13207, Q16424, Q7Z647 | Gene names | TBX2 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX2 (T-box protein 2). | |||||
|
CAMKV_HUMAN
|
||||||
| NC score | 0.007159 (rank : 183) | θ value | 5.27518 (rank : 100) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8NCB2, Q6FIB8, Q8NBS8, Q8NC85, Q8NDU4, Q8WTT8, Q9BQC9, Q9H0Q5 | Gene names | CAMKV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CaM kinase-like vesicle-associated protein. | |||||
|
CSPG2_MOUSE
|
||||||
| NC score | 0.006504 (rank : 184) | θ value | 4.03905 (rank : 93) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q62059, Q62058, Q9CUU0 | Gene names | Cspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M). | |||||
|
SIPA1_MOUSE
|
||||||
| NC score | 0.006405 (rank : 185) | θ value | 8.99809 (rank : 138) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P46062, P70204 | Gene names | Sipa1, Spa-1, Spa1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal-induced proliferation-associated protein 1 (Sipa-1) (GTPase- activating protein Spa-1). | |||||
|
LATS1_HUMAN
|
||||||
| NC score | 0.005808 (rank : 186) | θ value | 5.27518 (rank : 106) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O95835, Q6PKD0 | Gene names | LATS1, WARTS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS1 (EC 2.7.11.1) (Large tumor suppressor homolog 1) (WARTS protein kinase) (h-warts). | |||||
|
WWP2_HUMAN
|
||||||
| NC score | 0.005623 (rank : 187) | θ value | 8.99809 (rank : 142) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O00308, Q96CZ2, Q9BWN6 | Gene names | WWP2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2) (Atrophin-1-interacting protein 2) (AIP2). | |||||
|
HRH1_MOUSE
|
||||||
| NC score | 0.004224 (rank : 188) | θ value | 0.47712 (rank : 37) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P70174, Q91V75, Q91XN0, Q91XN1, Q91XN2, Q91XN3 | Gene names | Hrh1, Bphs | |||
|
Domain Architecture |

|
|||||
| Description | Histamine H1 receptor. | |||||
|
PLK3_MOUSE
|
||||||
| NC score | 0.003974 (rank : 189) | θ value | 5.27518 (rank : 109) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 863 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q60806, Q60822, Q9R009 | Gene names | Plk3, Cnk, Fnk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PLK3 (EC 2.7.11.21) (Polo-like kinase 3) (PLK-3) (Cytokine-inducible serine/threonine-protein kinase) (FGF- inducible kinase). | |||||
|
BCL6_HUMAN
|
||||||
| NC score | 0.003799 (rank : 190) | θ value | 0.21417 (rank : 29) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 922 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P41182 | Gene names | BCL6, BCL5, LAZ3, ZBTB27, ZNF51 | |||
|
Domain Architecture |

|
|||||
| Description | B-cell lymphoma 6 protein (BCL-6) (Zinc finger protein 51) (LAZ-3 protein) (BCL-5) (Zinc finger and BTB domain-containing protein 27). | |||||
|
PCDLK_HUMAN
|
||||||
| NC score | 0.002112 (rank : 191) | θ value | 8.99809 (rank : 136) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BYE9, Q9NXP8 | Gene names | PCLKC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin LKC precursor (PC-LKC). | |||||
|
LGR6_HUMAN
|
||||||
| NC score | 0.001392 (rank : 192) | θ value | 6.88961 (rank : 119) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HBX8, Q5T509, Q5T512, Q6UY15, Q86VU0, Q96K69, Q9BYD7 | Gene names | LGR6 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeat-containing G-protein coupled receptor 6 (VTS20631). | |||||
|
EVI1_MOUSE
|
||||||
| NC score | 0.001382 (rank : 193) | θ value | 1.81305 (rank : 68) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 826 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P14404 | Gene names | Evi1, Evi-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ecotropic virus integration 1 site protein. | |||||
|
ZBT40_HUMAN
|
||||||
| NC score | 0.000392 (rank : 194) | θ value | 5.27518 (rank : 114) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NUA8, O75066, Q5TFU5, Q8N1R1 | Gene names | ZBTB40, KIAA0478 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 40. | |||||
|
ZN174_HUMAN
|
||||||
| NC score | -0.000196 (rank : 195) | θ value | 6.88961 (rank : 127) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15697, Q9BQ34 | Gene names | ZNF174, ZSCAN8 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 174 (AW-1) (Zinc finger and SCAN domain-containing protein 8). | |||||