Please be patient as the page loads
|
NF2L2_MOUSE
|
||||||
| SwissProt Accessions | Q60795 | Gene names | Nfe2l2, Nrf-2, Nrf2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 2 (NF-E2-related factor 2) (NFE2-related factor 2) (Nuclear factor, erythroid derived 2, like 2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NF2L2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.958956 (rank : 2) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q16236, Q96F71 | Gene names | NFE2L2, NRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 2 (NF-E2-related factor 2) (NFE2-related factor 2) (Nuclear factor, erythroid derived 2, like 2) (HEBP1). | |||||
|
NF2L2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 183 | |
| SwissProt Accessions | Q60795 | Gene names | Nfe2l2, Nrf-2, Nrf2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 2 (NF-E2-related factor 2) (NFE2-related factor 2) (Nuclear factor, erythroid derived 2, like 2). | |||||
|
NF2L1_HUMAN
|
||||||
| θ value | 1.49966e-79 (rank : 3) | NC score | 0.889582 (rank : 4) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q14494, Q12877, Q96FN6 | Gene names | NFE2L1, HBZ17, NRF1, TCF11 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 1 (NF-E2-related factor 1) (NFE2-related factor 1) (Nuclear factor, erythroid derived 2, like 1) (Transcription factor 11) (Transcription factor HBZ17) (Transcription factor LCR-F1) (Locus control region-factor 1). | |||||
|
NF2L1_MOUSE
|
||||||
| θ value | 1.7756e-72 (rank : 4) | NC score | 0.874182 (rank : 5) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q61985, O70234 | Gene names | Nfe2l1, Nrf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 1 (NF-E2-related factor 1) (NFE2-related factor 1) (Nuclear factor, erythroid derived 2, like 1). | |||||
|
NFE2_HUMAN
|
||||||
| θ value | 7.25919e-50 (rank : 5) | NC score | 0.892722 (rank : 3) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q16621, Q07720, Q6ICV9 | Gene names | NFE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor NF-E2 45 kDa subunit (Nuclear factor, erythroid- derived 2 45 kDa subunit) (p45 NF-E2) (Leucine zipper protein NF-E2). | |||||
|
NF2L3_HUMAN
|
||||||
| θ value | 5.95217e-44 (rank : 6) | NC score | 0.843105 (rank : 7) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Y4A8, Q6NUS0, Q7Z498, Q86UJ4, Q86VR5, Q9UQA4 | Gene names | NFE2L3, NRF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor erythroid 2-related factor 3 (NF-E2-related factor 3) (NFE2-related factor 3) (Nuclear factor, erythroid derived 2, like 3). | |||||
|
NF2L3_MOUSE
|
||||||
| θ value | 2.11701e-41 (rank : 7) | NC score | 0.848652 (rank : 6) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9WTM4 | Gene names | Nfe2l3, Nrf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor erythroid 2-related factor 3 (NF-E2-related factor 3) (NFE2-related factor 3) (Nuclear factor, erythroid derived 2, like 3). | |||||
|
BACH2_HUMAN
|
||||||
| θ value | 1.52774e-15 (rank : 8) | NC score | 0.372608 (rank : 11) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9BYV9, Q59H70, Q5T793, Q9NTS5 | Gene names | BACH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription regulator protein BACH2 (BTB and CNC homolog 2). | |||||
|
BACH2_MOUSE
|
||||||
| θ value | 1.99529e-15 (rank : 9) | NC score | 0.374770 (rank : 10) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P97303 | Gene names | Bach2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription regulator protein BACH2 (BTB and CNC homolog 2). | |||||
|
BACH1_MOUSE
|
||||||
| θ value | 6.41864e-14 (rank : 10) | NC score | 0.377384 (rank : 9) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P97302 | Gene names | Bach1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription regulator protein BACH1 (BTB and CNC homolog 1). | |||||
|
BACH1_HUMAN
|
||||||
| θ value | 8.38298e-14 (rank : 11) | NC score | 0.378055 (rank : 8) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O14867, O43285 | Gene names | BACH1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription regulator protein BACH1 (BTB and CNC homolog 1) (HA2303). | |||||
|
JUNB_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 12) | NC score | 0.349460 (rank : 14) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P17275, Q96GH3 | Gene names | JUNB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-B. | |||||
|
JUNB_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 13) | NC score | 0.343358 (rank : 17) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P09450, Q8C2G9 | Gene names | Junb, Jun-b | |||
|
Domain Architecture |
|
|||||
| Description | Transcription factor jun-B. | |||||
|
JUND_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 14) | NC score | 0.354781 (rank : 12) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P15066 | Gene names | Jund, Jun-d, Jund1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-D. | |||||
|
FA44A_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 15) | NC score | 0.073896 (rank : 56) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8NFC6, Q6P0M8 | Gene names | FAM44A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM44A. | |||||
|
XBP1_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 16) | NC score | 0.238241 (rank : 18) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P17861, Q8WYK6, Q969P1, Q96BD7 | Gene names | XBP1, TREB5, XBP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X box-binding protein 1 (XBP-1) (Tax-responsive element-binding protein 5). | |||||
|
JUN_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 17) | NC score | 0.346061 (rank : 16) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P05412, Q96G93 | Gene names | JUN | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-1 (Activator protein 1) (AP1) (Proto-oncogene c-jun) (V-jun avian sarcoma virus 17 oncogene homolog) (p39). | |||||
|
JUN_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 18) | NC score | 0.346724 (rank : 15) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P05627 | Gene names | Jun | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-1 (Activator protein 1) (AP1) (Proto-oncogene c-jun) (V-jun avian sarcoma virus 17 oncogene homolog) (Jun A) (AH119). | |||||
|
JUND_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 19) | NC score | 0.353757 (rank : 13) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P17535 | Gene names | JUND | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-D. | |||||
|
XBP1_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 20) | NC score | 0.221892 (rank : 19) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O35426, Q8VHM0, Q922G5, Q9ESS3 | Gene names | Xbp1, Treb5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X box-binding protein 1 (XBP-1) (Tax-responsive element-binding protein 5 homolog). | |||||
|
SALL3_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 21) | NC score | 0.015166 (rank : 161) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 1066 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BXA9, Q9UGH1 | Gene names | SALL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 3 (Zinc finger protein SALL3) (hSALL3). | |||||
|
MAFG_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 22) | NC score | 0.199120 (rank : 27) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O15525 | Gene names | MAFG | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafG (V-maf musculoaponeurotic fibrosarcoma oncogene homolog G) (hMAF). | |||||
|
TRIM6_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 23) | NC score | 0.021120 (rank : 133) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9C030, Q86WZ8, Q9HCR1 | Gene names | TRIM6, RNF89 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 6 (RING finger protein 89). | |||||
|
MAFG_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 24) | NC score | 0.198686 (rank : 28) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O54790 | Gene names | Mafg | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafG (V-maf musculoaponeurotic fibrosarcoma oncogene homolog G). | |||||
|
NCOR1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 25) | NC score | 0.059667 (rank : 61) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
WDR60_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 26) | NC score | 0.046172 (rank : 73) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8C761, Q3UNM4 | Gene names | Wdr60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 60. | |||||
|
DBP_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 27) | NC score | 0.062929 (rank : 60) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q10586 | Gene names | DBP | |||
|
Domain Architecture |

|
|||||
| Description | D site-binding protein (Albumin D box-binding protein) (Albumin D- element-binding protein) (TAXREB302). | |||||
|
MAFK_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 28) | NC score | 0.209920 (rank : 25) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O60675 | Gene names | MAFK | |||
|
Domain Architecture |
|
|||||
| Description | Transcription factor MafK (Erythroid transcription factor NF-E2 p18 subunit). | |||||
|
MAF_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 29) | NC score | 0.211772 (rank : 23) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O75444, Q9UP93 | Gene names | MAF | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Maf (Proto-oncogene c-maf). | |||||
|
MAFK_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 30) | NC score | 0.206765 (rank : 26) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q61827, Q60600 | Gene names | Mafk, Nfe2u | |||
|
Domain Architecture |
|
|||||
| Description | Transcription factor MafK (Erythroid transcription factor NF-E2 p18 subunit). | |||||
|
ZN291_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 31) | NC score | 0.044708 (rank : 74) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 647 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9BY12 | Gene names | ZNF291 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 291. | |||||
|
BAZ2A_MOUSE
|
||||||
| θ value | 0.125558 (rank : 32) | NC score | 0.037832 (rank : 83) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
FA21C_HUMAN
|
||||||
| θ value | 0.125558 (rank : 33) | NC score | 0.063215 (rank : 59) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y4E1, Q5SQU4, Q5SQU5, Q7L521, Q9UG79 | Gene names | FAM21C, KIAA0592 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM21C. | |||||
|
FOSB_HUMAN
|
||||||
| θ value | 0.125558 (rank : 34) | NC score | 0.167396 (rank : 36) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P53539 | Gene names | FOSB, G0S3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein fosB (G0/G1 switch regulatory protein 3). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 35) | NC score | 0.053455 (rank : 66) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
MAP2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 36) | NC score | 0.050451 (rank : 68) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P11137, Q99975, Q99976 | Gene names | MAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2) (MAP-2). | |||||
|
S23IP_HUMAN
|
||||||
| θ value | 0.125558 (rank : 37) | NC score | 0.040184 (rank : 80) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y6Y8, Q8IXH5, Q9BUK5 | Gene names | SEC23IP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SEC23-interacting protein (p125). | |||||
|
CEP35_HUMAN
|
||||||
| θ value | 0.163984 (rank : 38) | NC score | 0.049390 (rank : 70) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q5VT06, O75068, Q8TDK3, Q8WY20 | Gene names | CEP350, CAP350, GM133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein 350 (Centrosome-associated protein of 350 kDa). | |||||
|
DSPP_HUMAN
|
||||||
| θ value | 0.163984 (rank : 39) | NC score | 0.059375 (rank : 63) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
MAF_MOUSE
|
||||||
| θ value | 0.163984 (rank : 40) | NC score | 0.210531 (rank : 24) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P54843 | Gene names | Maf, Maf2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Maf (Proto-oncogene c-maf). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 41) | NC score | 0.030721 (rank : 100) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
DBP_MOUSE
|
||||||
| θ value | 0.21417 (rank : 42) | NC score | 0.059558 (rank : 62) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q60925, Q8VCX3 | Gene names | Dbp | |||
|
Domain Architecture |

|
|||||
| Description | D site-binding protein (Albumin D box-binding protein) (Albumin D- element-binding protein). | |||||
|
FOSL2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 43) | NC score | 0.167604 (rank : 35) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P15408 | Gene names | FOSL2, FRA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fos-related antigen 2. | |||||
|
ICAL_MOUSE
|
||||||
| θ value | 0.21417 (rank : 44) | NC score | 0.050741 (rank : 67) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P51125, Q9EQV4, Q9EQV5, Q9QXQ3, Q9QXQ4, Q9R0N1 | Gene names | Cast | |||
|
Domain Architecture |

|
|||||
| Description | Calpastatin (Calpain inhibitor). | |||||
|
MBD5_HUMAN
|
||||||
| θ value | 0.21417 (rank : 45) | NC score | 0.054356 (rank : 65) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9P267, Q9NUV6 | Gene names | MBD5, KIAA1461 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 5 (Methyl-CpG-binding protein MBD5). | |||||
|
FOSB_MOUSE
|
||||||
| θ value | 0.279714 (rank : 46) | NC score | 0.165562 (rank : 37) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P13346 | Gene names | Fosb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein fosB. | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 47) | NC score | 0.033478 (rank : 96) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
TMF1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 48) | NC score | 0.029891 (rank : 105) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P82094 | Gene names | TMF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA element modulatory factor (TMF). | |||||
|
BRD4_MOUSE
|
||||||
| θ value | 0.365318 (rank : 49) | NC score | 0.030190 (rank : 102) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
FOSL2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 50) | NC score | 0.164244 (rank : 38) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P47930 | Gene names | Fosl2, Fra-2, Fra2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fos-related antigen 2. | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 51) | NC score | 0.027840 (rank : 111) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
CREB3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 52) | NC score | 0.182030 (rank : 32) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q61817, Q99M21, Q9CVK9 | Gene names | Creb3, Lzip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-responsive element-binding protein 3 (Transcription factor LZIP). | |||||
|
DSPP_MOUSE
|
||||||
| θ value | 0.47712 (rank : 53) | NC score | 0.058183 (rank : 64) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
HSF1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 54) | NC score | 0.039883 (rank : 81) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q00613 | Gene names | HSF1, HSTF1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
MAFB_HUMAN
|
||||||
| θ value | 0.47712 (rank : 55) | NC score | 0.214795 (rank : 21) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Y5Q3, Q9H1F1 | Gene names | MAFB, KRML | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafB (V-maf musculoaponeurotic fibrosarcoma oncogene homolog B). | |||||
|
NR1D1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 56) | NC score | 0.012110 (rank : 169) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P20393, Q15304 | Gene names | NR1D1, EAR1, HREV, THRAL | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR1D1 (V-erbA-related protein EAR-1) (Rev- erbA-alpha). | |||||
|
PPRB_HUMAN
|
||||||
| θ value | 0.47712 (rank : 57) | NC score | 0.043442 (rank : 75) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q15648, O43810, O75447, Q9HD39 | Gene names | PPARBP, ARC205, DRIP205, TRAP220, TRIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2) (p53 regulatory protein RB18A) (Vitamin D receptor-interacting protein complex component DRIP205) (Activator- recruited cofactor 205 kDa component) (ARC205). | |||||
|
ATN1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 58) | NC score | 0.047531 (rank : 71) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
CENPU_MOUSE
|
||||||
| θ value | 0.62314 (rank : 59) | NC score | 0.040552 (rank : 79) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8C4M7, Q6UNA2, Q9D9U1 | Gene names | Mlf1ip, Cenpu | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein U (CENP-U) (MLF1-interacting protein). | |||||
|
CK5P2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 60) | NC score | 0.035362 (rank : 91) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
FOSL1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 61) | NC score | 0.172834 (rank : 33) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P15407 | Gene names | FOSL1, FRA1 | |||
|
Domain Architecture |

|
|||||
| Description | Fos-related antigen 1 (FRA-1). | |||||
|
FOSL1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 62) | NC score | 0.171972 (rank : 34) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P48755, O35285 | Gene names | Fosl1, Fra1 | |||
|
Domain Architecture |

|
|||||
| Description | Fos-related antigen 1 (FRA-1). | |||||
|
INCE_HUMAN
|
||||||
| θ value | 0.62314 (rank : 63) | NC score | 0.029394 (rank : 109) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
MAFB_MOUSE
|
||||||
| θ value | 0.62314 (rank : 64) | NC score | 0.215089 (rank : 20) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P54841 | Gene names | Mafb, Krml, Maf1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafB (V-maf musculoaponeurotic fibrosarcoma oncogene homolog B) (Transcription factor MAF1) (Segmentation protein KR) (Kreisler). | |||||
|
NCOR1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 65) | NC score | 0.047224 (rank : 72) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
TP53B_HUMAN
|
||||||
| θ value | 0.62314 (rank : 66) | NC score | 0.040671 (rank : 78) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q12888, Q2M1Z7, Q4LE46, Q5FWZ3, Q7Z3U4 | Gene names | TP53BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
ATF3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 67) | NC score | 0.185382 (rank : 30) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P18847 | Gene names | ATF3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-3 (Activating transcription factor 3). | |||||
|
ATF3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 68) | NC score | 0.190502 (rank : 29) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q60765 | Gene names | Atf3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-3 (Activating transcription factor 3) (Transcription factor LRG-21). | |||||
|
ATF6A_HUMAN
|
||||||
| θ value | 0.813845 (rank : 69) | NC score | 0.121291 (rank : 44) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P18850, O15139, Q5VW62, Q6IPB5, Q9UEC9 | Gene names | ATF6 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-6 alpha (Activating transcription factor 6 alpha) (ATF6-alpha). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 0.813845 (rank : 70) | NC score | 0.041817 (rank : 76) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
AP3B2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 71) | NC score | 0.028206 (rank : 110) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
BPTF_HUMAN
|
||||||
| θ value | 1.06291 (rank : 72) | NC score | 0.036880 (rank : 86) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
EP15R_HUMAN
|
||||||
| θ value | 1.06291 (rank : 73) | NC score | 0.031274 (rank : 97) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 1083 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9UBC2 | Gene names | EPS15L1, EPS15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R). | |||||
|
EP15R_MOUSE
|
||||||
| θ value | 1.06291 (rank : 74) | NC score | 0.036394 (rank : 89) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q60902, Q8CB60, Q8CB70, Q91WH8 | Gene names | Eps15l1, Eps15-rs, Eps15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R) (Epidermal growth factor receptor pathway substrate 15-related sequence) (Eps15-rs). | |||||
|
FOS_HUMAN
|
||||||
| θ value | 1.06291 (rank : 75) | NC score | 0.161048 (rank : 40) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P01100, P18849 | Gene names | FOS, G0S7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene protein c-fos (Cellular oncogene fos) (G0/G1 switch regulatory protein 7). | |||||
|
FOS_MOUSE
|
||||||
| θ value | 1.06291 (rank : 76) | NC score | 0.162557 (rank : 39) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P01101 | Gene names | Fos | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene protein c-fos (Cellular oncogene fos). | |||||
|
MYEF2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 77) | NC score | 0.017197 (rank : 153) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P2K5, Q6NUM5, Q7L388, Q7Z4B7, Q9H922, Q9NUQ1 | Gene names | MYEF2, KIAA1341 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin expression factor 2 (MyEF-2) (MST156). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 1.06291 (rank : 78) | NC score | 0.037113 (rank : 85) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
RERE_MOUSE
|
||||||
| θ value | 1.06291 (rank : 79) | NC score | 0.037803 (rank : 84) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
SAPS3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 80) | NC score | 0.019540 (rank : 142) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5H9R7, Q3B7I1, Q3I4Y0, Q3KR35, Q68CR3, Q7L4R8, Q8N3B2, Q96MB2, Q9H2K5, Q9H2K6, Q9HCL4, Q9NUY3 | Gene names | SAPS3, C11orf23, KIAA1558, PP6R3, SAPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAPS domain family member 3 (Sporulation-induced transcript 4- associated protein SAPL) (Protein phosphatase 6 regulatory subunit 3). | |||||
|
ASPP1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 81) | NC score | 0.021176 (rank : 132) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 956 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q96KQ4, O94870 | Gene names | PPP1R13B, ASPP1, KIAA0771 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
CAF1A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 82) | NC score | 0.035204 (rank : 92) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q13111, Q7Z7K3, Q9UJY8 | Gene names | CHAF1A, CAF, CAF1P150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
CLSPN_MOUSE
|
||||||
| θ value | 1.38821 (rank : 83) | NC score | 0.031069 (rank : 98) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q80YR7, Q69GM2 | Gene names | Clspn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin. | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 1.38821 (rank : 84) | NC score | 0.036447 (rank : 88) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
CX039_MOUSE
|
||||||
| θ value | 1.81305 (rank : 85) | NC score | 0.036474 (rank : 87) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K2D0, Q3U3R2 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein CXorf39 homolog. | |||||
|
G3BP_MOUSE
|
||||||
| θ value | 1.81305 (rank : 86) | NC score | 0.016084 (rank : 158) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97855 | Gene names | G3bp, G3bp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ras-GTPase-activating protein-binding protein 1 (EC 3.6.1.-) (ATP- dependent DNA helicase VIII) (GAP SH3-domain-binding protein 1) (G3BP- 1) (HDH-VIII). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 87) | NC score | 0.029604 (rank : 108) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
HNF1A_MOUSE
|
||||||
| θ value | 1.81305 (rank : 88) | NC score | 0.030317 (rank : 101) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P22361 | Gene names | Tcf1, Hnf-1, Hnf-1a, Hnf1a | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 1-alpha (HNF-1A) (Liver-specific transcription factor LF-B1) (LFB1). | |||||
|
HSF1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 89) | NC score | 0.033726 (rank : 94) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P38532, O70462 | Gene names | Hsf1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 90) | NC score | 0.049396 (rank : 69) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
MAFF_HUMAN
|
||||||
| θ value | 1.81305 (rank : 91) | NC score | 0.142257 (rank : 41) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9ULX9, Q9Y525 | Gene names | MAFF | |||
|
Domain Architecture |
|
|||||
| Description | Transcription factor MafF (V-maf musculoaponeurotic fibrosarcoma oncogene homolog F) (U-Maf). | |||||
|
TMPSD_MOUSE
|
||||||
| θ value | 1.81305 (rank : 92) | NC score | 0.004335 (rank : 184) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5U405, Q8CFE0, Q91VQ8 | Gene names | Tmprss13, Msp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 13 (EC 3.4.21.-) (Mosaic serine protease) (Membrane-type mosaic serine protease). | |||||
|
VITRN_HUMAN
|
||||||
| θ value | 1.81305 (rank : 93) | NC score | 0.012125 (rank : 168) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6UXI7, Q6P7T3, Q96DT1 | Gene names | VIT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vitrin precursor. | |||||
|
BSN_HUMAN
|
||||||
| θ value | 2.36792 (rank : 94) | NC score | 0.040715 (rank : 77) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
CCNK_MOUSE
|
||||||
| θ value | 2.36792 (rank : 95) | NC score | 0.026076 (rank : 118) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
CREM_HUMAN
|
||||||
| θ value | 2.36792 (rank : 96) | NC score | 0.089041 (rank : 49) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q03060, Q16114, Q16116, Q9NZB9 | Gene names | CREM | |||
|
Domain Architecture |
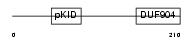
|
|||||
| Description | cAMP-responsive element modulator. | |||||
|
CREM_MOUSE
|
||||||
| θ value | 2.36792 (rank : 97) | NC score | 0.090045 (rank : 48) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P27699, P27698 | Gene names | Crem | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-responsive element modulator. | |||||
|
EDD1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 98) | NC score | 0.014282 (rank : 164) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95071, O94970, Q9NPL3 | Gene names | EDD1, EDD, HYD, KIAA0896 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog) (hHYD) (Progestin-induced protein). | |||||
|
EPAS1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 99) | NC score | 0.017720 (rank : 151) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97481, O08787, O55046 | Gene names | Epas1, Hif2a | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Hypoxia- inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (MHLF) (HIF-related factor) (HRF). | |||||
|
ERBB4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 100) | NC score | -0.001351 (rank : 188) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 910 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15303 | Gene names | ERBB4, HER4 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor tyrosine-protein kinase erbB-4 precursor (EC 2.7.10.1) (p180erbB4) (Tyrosine kinase-type cell surface receptor HER4). | |||||
|
MAFF_MOUSE
|
||||||
| θ value | 2.36792 (rank : 101) | NC score | 0.141879 (rank : 42) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O54791 | Gene names | Maff | |||
|
Domain Architecture |
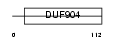
|
|||||
| Description | Transcription factor MafF (V-maf musculoaponeurotic fibrosarcoma oncogene homolog F). | |||||
|
RADI_MOUSE
|
||||||
| θ value | 2.36792 (rank : 102) | NC score | 0.018484 (rank : 147) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 773 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P26043, Q9QW27 | Gene names | Rdx | |||
|
Domain Architecture |

|
|||||
| Description | Radixin (ESP10). | |||||
|
SKIL_HUMAN
|
||||||
| θ value | 2.36792 (rank : 103) | NC score | 0.023077 (rank : 122) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P12757, O00464, P12756, Q07501 | Gene names | SKIL, SNO | |||
|
Domain Architecture |
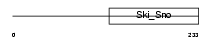
|
|||||
| Description | Ski-like protein (Ski-related protein) (Ski-related oncogene). | |||||
|
CKAP4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 104) | NC score | 0.030023 (rank : 103) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BMK4, Q8BTK8, Q8R3F2 | Gene names | Ckap4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 4 (63 kDa membrane protein) (p63). | |||||
|
CMTA2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 105) | NC score | 0.016548 (rank : 156) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O94983, Q7Z6M8, Q8N3V0, Q8NDG4, Q96G17 | Gene names | CAMTA2, KIAA0909 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-binding transcription activator 2. | |||||
|
CX039_HUMAN
|
||||||
| θ value | 3.0926 (rank : 106) | NC score | 0.033668 (rank : 95) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PEV8, Q8WVP6, Q96AV3 | Gene names | CXorf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein CXorf39. | |||||
|
GGA3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 107) | NC score | 0.013538 (rank : 165) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BMI3, Q80U71 | Gene names | Gga3, Kiaa0154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
IL3RB_HUMAN
|
||||||
| θ value | 3.0926 (rank : 108) | NC score | 0.019483 (rank : 144) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P32927 | Gene names | CSF2RB, IL3RB, IL5RB | |||
|
Domain Architecture |

|
|||||
| Description | Cytokine receptor common beta chain precursor (GM-CSF/IL-3/IL-5 receptor common beta-chain) (CD131 antigen) (CDw131). | |||||
|
K1683_HUMAN
|
||||||
| θ value | 3.0926 (rank : 109) | NC score | 0.033741 (rank : 93) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H0B3, Q8N4G8, Q96M14, Q9C0I0 | Gene names | KIAA1683 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1683. | |||||
|
MYH11_MOUSE
|
||||||
| θ value | 3.0926 (rank : 110) | NC score | 0.021503 (rank : 129) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
RABE1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 111) | NC score | 0.038361 (rank : 82) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O35551, Q99L08, Q9CRP3, Q9EQF9, Q9JI94 | Gene names | Rabep1, Rab5ep, Rabpt5, Rabpt5a | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha). | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 112) | NC score | 0.029638 (rank : 107) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
ZN645_HUMAN
|
||||||
| θ value | 3.0926 (rank : 113) | NC score | 0.029933 (rank : 104) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8N7E2, Q6DJY9 | Gene names | ZNF645 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 645. | |||||
|
AMPH_MOUSE
|
||||||
| θ value | 4.03905 (rank : 114) | NC score | 0.026869 (rank : 113) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q7TQF7, Q8R1C4 | Gene names | Amph, Amph1 | |||
|
Domain Architecture |

|
|||||
| Description | Amphiphysin. | |||||
|
BRWD1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 115) | NC score | 0.009321 (rank : 175) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NSI6, O43721, Q5R2V0, Q5R2V1, Q8TCV3, Q96QG9, Q96QH0, Q9NUK1 | Gene names | BRWD1, C21orf107, WDR9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain and WD repeat domain-containing protein 1 (WD repeat protein 9). | |||||
|
DDX50_HUMAN
|
||||||
| θ value | 4.03905 (rank : 116) | NC score | 0.004253 (rank : 185) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BQ39, Q8WV76, Q9BWI8 | Gene names | DDX50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
GCC2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 117) | NC score | 0.023057 (rank : 123) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
MAK_MOUSE
|
||||||
| θ value | 4.03905 (rank : 118) | NC score | -0.002781 (rank : 190) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 873 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q04859 | Gene names | Mak, Rck | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase MAK (EC 2.7.11.22) (Male germ cell- associated kinase) (Protein kinase RCK). | |||||
|
MOES_MOUSE
|
||||||
| θ value | 4.03905 (rank : 119) | NC score | 0.018233 (rank : 148) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 923 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P26041, Q3UL28, Q8BSN4 | Gene names | Msn | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
MYH9_MOUSE
|
||||||
| θ value | 4.03905 (rank : 120) | NC score | 0.023300 (rank : 121) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
NCOA5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 121) | NC score | 0.018853 (rank : 145) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9HCD5, Q6HA99, Q9H1F2, Q9H2T2, Q9H4Y9 | Gene names | NCOA5, KIAA1637 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 5 (NCoA-5) (Coactivator independent of AF-2) (CIA). | |||||
|
NOP56_MOUSE
|
||||||
| θ value | 4.03905 (rank : 122) | NC score | 0.016426 (rank : 157) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D6Z1, Q3UD45, Q8BVL1, Q8VDT2, Q99LT8, Q9CT15 | Gene names | Nol5a, Nop56 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar protein Nop56 (Nucleolar protein 5A). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 4.03905 (rank : 123) | NC score | 0.029732 (rank : 106) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
OSBP2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 124) | NC score | 0.011294 (rank : 171) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q969R2, O60396, Q9BY96, Q9BZF0 | Gene names | OSBP2, KIAA1664, ORP4, OSBPL4 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein 2 (Oxysterol-binding protein-related protein 4) (OSBP-related protein 4) (ORP-4). | |||||
|
RBP24_HUMAN
|
||||||
| θ value | 4.03905 (rank : 125) | NC score | 0.023036 (rank : 124) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
SRCA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 126) | NC score | 0.019527 (rank : 143) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
TRAIP_MOUSE
|
||||||
| θ value | 4.03905 (rank : 127) | NC score | 0.026395 (rank : 116) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8VIG6, O08854, Q922M8, Q9CPP4 | Gene names | Traip, Trip | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
TRI29_HUMAN
|
||||||
| θ value | 4.03905 (rank : 128) | NC score | 0.020530 (rank : 137) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14134, Q96AA9, Q9BZY7 | Gene names | TRIM29, ATDC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 29 (Ataxia-telangiectasia group D- associated protein). | |||||
|
UBP37_HUMAN
|
||||||
| θ value | 4.03905 (rank : 129) | NC score | 0.010142 (rank : 173) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q86T82, Q9HCH8 | Gene names | USP37, KIAA1594 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 37 (EC 3.1.2.15) (Ubiquitin thioesterase 37) (Ubiquitin-specific-processing protease 37) (Deubiquitinating enzyme 37). | |||||
|
XE7_HUMAN
|
||||||
| θ value | 4.03905 (rank : 130) | NC score | 0.026447 (rank : 115) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 738 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q02040, Q02832 | Gene names | CXYorf3, DXYS155E, XE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-lymphocyte antigen precursor (B-lymphocyte surface antigen) (721P) (Protein XE7). | |||||
|
ADDB_HUMAN
|
||||||
| θ value | 5.27518 (rank : 131) | NC score | 0.011775 (rank : 170) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P35612, Q13482, Q16412, Q59G82, Q5U5P4, Q6P0P2, Q6PGQ4, Q7Z688, Q7Z689, Q7Z690, Q7Z691 | Gene names | ADD2, ADDB | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta). | |||||
|
AIM1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 132) | NC score | 0.017764 (rank : 149) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y4K1, O00296, Q5VWJ2 | Gene names | AIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Absent in melanoma 1 protein. | |||||
|
ALX4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 133) | NC score | 0.001882 (rank : 187) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H161, Q96JN7, Q9H198, Q9HAY9 | Gene names | ALX4, KIAA1788 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein aristaless-like 4. | |||||
|
ATF1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 134) | NC score | 0.080679 (rank : 53) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P18846, P25168 | Gene names | ATF1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-1 (Activating transcription factor 1) (TREB36 protein). | |||||
|
ATF1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 135) | NC score | 0.077807 (rank : 55) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P81269 | Gene names | Atf1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-1 (Activating transcription factor 1) (TCR-ATF1). | |||||
|
BATF_MOUSE
|
||||||
| θ value | 5.27518 (rank : 136) | NC score | 0.212590 (rank : 22) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O35284 | Gene names | Batf | |||
|
Domain Architecture |
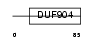
|
|||||
| Description | ATF-like basic leucine zipper transcription factor B-ATF. | |||||
|
CASP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 137) | NC score | 0.021465 (rank : 130) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 1152 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P70403, Q7TQJ4, Q91WN6 | Gene names | Cutl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
DACH2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 138) | NC score | 0.016775 (rank : 155) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q925Q8, Q8BMA0 | Gene names | Dach2 | |||
|
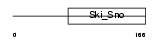
Domain Architecture |
|
|||||
| Description | Dachshund homolog 2 (Dach2). | |||||
|
FARP2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 139) | NC score | 0.006497 (rank : 181) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91VS8, Q3TAP2, Q69ZZ0 | Gene names | Farp2, Kiaa0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
HNF1A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 140) | NC score | 0.026613 (rank : 114) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P20823, Q99861 | Gene names | TCF1, HNF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 1-alpha (HNF-1A) (Liver-specific transcription factor LF-B1) (LFB1) (Transcription factor 1) (TCF-1). | |||||
|
IF35_HUMAN
|
||||||
| θ value | 5.27518 (rank : 141) | NC score | 0.019571 (rank : 141) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O00303, Q8N978 | Gene names | EIF3S5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 5 (eIF-3 epsilon) (eIF3 p47 subunit) (eIF3f). | |||||
|
KIF17_MOUSE
|
||||||
| θ value | 5.27518 (rank : 142) | NC score | 0.006053 (rank : 182) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q99PW8 | Gene names | Kif17 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF17 (MmKIF17). | |||||
|
NFAC4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 143) | NC score | 0.012432 (rank : 167) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14934 | Gene names | NFATC4, NFAT3 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 4 (NF-ATc4) (NFATc4) (T cell transcription factor NFAT3) (NF-AT3). | |||||
|
NFH_HUMAN
|
||||||
| θ value | 5.27518 (rank : 144) | NC score | 0.017327 (rank : 152) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
PHC3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 145) | NC score | 0.015282 (rank : 160) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8NDX5, Q5HYF0, Q6NSG2, Q8NFT7, Q8NFZ1, Q8TBM2, Q9H971, Q9H9I4 | Gene names | PHC3, EDR3, PH3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 3 (hPH3) (Homolog of polyhomeotic 3) (Early development regulatory protein 3). | |||||
|
PSL1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 146) | NC score | 0.011173 (rank : 172) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3TD49, Q3TLQ8, Q3UG60, Q80VZ6 | Gene names | Sppl2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b). | |||||
|
RABE1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 147) | NC score | 0.036301 (rank : 90) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q15276, O95369, Q8IVX3 | Gene names | RABEP1, RABPT5, RABPT5A | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha) (Rabaptin-4) (NY-REN-17 antigen). | |||||
|
TACC1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 148) | NC score | 0.022664 (rank : 125) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6Y685, Q6Y686 | Gene names | Tacc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transforming acidic coiled-coil-containing protein 1. | |||||
|
UBL7_MOUSE
|
||||||
| θ value | 5.27518 (rank : 149) | NC score | 0.015470 (rank : 159) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91W67, Q9D7P5 | Gene names | Ubl7 | |||
|
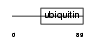
Domain Architecture |
|
|||||
| Description | Ubiquitin-like protein 7. | |||||
|
AKAP8_MOUSE
|
||||||
| θ value | 6.88961 (rank : 150) | NC score | 0.018652 (rank : 146) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9DBR0, Q9R0L8 | Gene names | Akap8, Akap95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 8 (A-kinase anchor protein 95 kDa) (AKAP 95). | |||||
|
AMPH_HUMAN
|
||||||
| θ value | 6.88961 (rank : 151) | NC score | 0.025695 (rank : 119) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P49418, O43538, Q75MJ8, Q75MK5, Q75MM3, Q8N4G0 | Gene names | AMPH, AMPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Amphiphysin. | |||||
|
ASPX_MOUSE
|
||||||
| θ value | 6.88961 (rank : 152) | NC score | 0.020758 (rank : 135) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P50289, Q9DAM6 | Gene names | Acrv1 | |||
|
Domain Architecture |

|
|||||
| Description | Acrosomal protein SP-10 precursor (Acrosomal vesicle protein 1) (MSA- 63). | |||||
|
BASP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 153) | NC score | 0.031051 (rank : 99) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
BATF_HUMAN
|
||||||
| θ value | 6.88961 (rank : 154) | NC score | 0.183611 (rank : 31) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q16520 | Gene names | BATF | |||
|
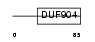
Domain Architecture |
|
|||||
| Description | ATF-like basic leucine zipper transcriptional factor B-ATF (SF-HT- activated gene 2) (SFA-2). | |||||
|
CASP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 155) | NC score | 0.021304 (rank : 131) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q13948, Q53GU9, Q8TBS3 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CL026_HUMAN
|
||||||
| θ value | 6.88961 (rank : 156) | NC score | 0.013075 (rank : 166) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N6Q8, Q9H5Y3 | Gene names | C12orf26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C12orf26. | |||||
|
CUTL1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 157) | NC score | 0.022282 (rank : 126) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
KCNN3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 158) | NC score | 0.006570 (rank : 180) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P58391 | Gene names | Kcnn3, Sk3 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 3 (SK3). | |||||
|
KNTC2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 159) | NC score | 0.026174 (rank : 117) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9D0F1, Q3TQT6, Q3UWM5, Q99P70 | Gene names | Kntc2, Hec1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Hec1 (Kinetochore-associated protein 2). | |||||
|
LIPB2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 160) | NC score | 0.014760 (rank : 162) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 599 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8ND30, O75337, Q8WW26 | Gene names | PPFIBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 2) (PTPRF-interacting protein-binding protein 2). | |||||
|
MYH11_HUMAN
|
||||||
| θ value | 6.88961 (rank : 161) | NC score | 0.021105 (rank : 134) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MYH6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 162) | NC score | 0.019989 (rank : 139) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYH6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 163) | NC score | 0.019617 (rank : 140) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYH9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 164) | NC score | 0.020752 (rank : 136) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
RREB1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 165) | NC score | 0.003189 (rank : 186) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 879 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q92766, O75567 | Gene names | RREB1 | |||
|
Domain Architecture |

|
|||||
| Description | RAS-responsive element-binding protein 1 (RREB-1) (Raf-responsive zinc finger protein LZ321). | |||||
|
TRAIP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 166) | NC score | 0.025362 (rank : 120) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9BWF2, O00467 | Gene names | TRAIP, TRIP | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
TTC15_MOUSE
|
||||||
| θ value | 6.88961 (rank : 167) | NC score | 0.017744 (rank : 150) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8K2L8, Q8BX27 | Gene names | Ttc15 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 15 (TPR repeat protein 15). | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 168) | NC score | 0.021818 (rank : 128) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
CCD41_MOUSE
|
||||||
| θ value | 8.99809 (rank : 169) | NC score | 0.027194 (rank : 112) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9D5R3, Q3U7X7, Q3UX57, Q80VF0 | Gene names | Ccdc41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41. | |||||
|
CREB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 170) | NC score | 0.071237 (rank : 58) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P16220, P21934, Q9UMA7 | Gene names | CREB1 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP response element-binding protein (CREB). | |||||
|
CREB1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 171) | NC score | 0.071243 (rank : 57) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q01147 | Gene names | Creb1, Creb-1 | |||
|
Domain Architecture |
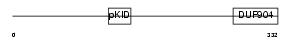
|
|||||
| Description | cAMP response element-binding protein (CREB). | |||||
|
DHX29_HUMAN
|
||||||
| θ value | 8.99809 (rank : 172) | NC score | 0.008711 (rank : 176) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7Z478, O75549, Q63HN0, Q63HN3, Q8IWW2, Q8N3A1, Q9UMH2 | Gene names | DHX29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX29 (EC 3.6.1.-) (DEAH box protein 29) (Nucleic acid helicase DDXx). | |||||
|
FA5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 173) | NC score | 0.007481 (rank : 177) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P12259, Q14285, Q6UPU6 | Gene names | F5 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor V precursor (Activated protein C cofactor) [Contains: Coagulation factor V heavy chain; Coagulation factor V light chain]. | |||||
|
IMA4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 174) | NC score | 0.006972 (rank : 178) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00629, O00190 | Gene names | KPNA4, QIP1 | |||
|
Domain Architecture |
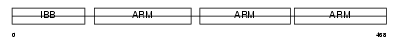
|
|||||
| Description | Importin alpha-4 subunit (Karyopherin alpha-4 subunit) (Qip1 protein). | |||||
|
IMA4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 175) | NC score | 0.006812 (rank : 179) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35343 | Gene names | Kpna4 | |||
|
Domain Architecture |
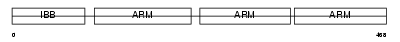
|
|||||
| Description | Importin alpha-4 subunit (Karyopherin alpha-4 subunit) (Importin alpha Q1). | |||||
|
K0310_HUMAN
|
||||||
| θ value | 8.99809 (rank : 176) | NC score | 0.020201 (rank : 138) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O15027, Q5SXP0, Q5SXP1, Q8N347, Q96HP1 | Gene names | KIAA0310 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0310. | |||||
|
MARCS_HUMAN
|
||||||
| θ value | 8.99809 (rank : 177) | NC score | 0.022034 (rank : 127) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
NRDC_MOUSE
|
||||||
| θ value | 8.99809 (rank : 178) | NC score | 0.009844 (rank : 174) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BHG1 | Gene names | Nrd1 | |||
|
Domain Architecture |

|
|||||
| Description | Nardilysin precursor (EC 3.4.24.61) (N-arginine dibasic convertase) (NRD convertase) (NRD-C). | |||||
|
NRL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 179) | NC score | 0.126318 (rank : 43) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P54846 | Gene names | Nrl | |||
|
Domain Architecture |
|
|||||
| Description | Neural retina-specific leucine zipper protein (NRL). | |||||
|
PKN2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 180) | NC score | -0.002323 (rank : 189) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 892 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q16513, Q9H1W4 | Gene names | PKN2, PRK2, PRKCL2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase N2 (EC 2.7.11.13) (Protein kinase C- like 2) (Protein-kinase C-related kinase 2). | |||||
|
RIN3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 181) | NC score | 0.014671 (rank : 163) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P59729 | Gene names | Rin3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras and Rab interactor 3 (Ras interaction/interference protein 3). | |||||
|
SYT12_MOUSE
|
||||||
| θ value | 8.99809 (rank : 182) | NC score | 0.004942 (rank : 183) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q920N7 | Gene names | Syt12 | |||
|
Domain Architecture |
|
|||||
| Description | Synaptotagmin-12 (Synaptotagmin XII) (SytXII). | |||||
|
TRI29_MOUSE
|
||||||
| θ value | 8.99809 (rank : 183) | NC score | 0.017094 (rank : 154) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8R2Q0, Q8CEE2, Q922Y3, Q99PN5, Q9CSC9 | Gene names | Trim29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 29. | |||||
|
ATF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.081774 (rank : 52) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P15336, Q13000 | Gene names | ATF2, CREB2, CREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (HB16). | |||||
|
ATF2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.081961 (rank : 51) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P16951, Q64089, Q64090, Q64091 | Gene names | Atf2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (MXBP protein). | |||||
|
ATF7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.086922 (rank : 50) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P17544, Q13814 | Gene names | ATF7, ATFA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
ATF7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.080553 (rank : 54) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8R0S1 | Gene names | Atf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
CREB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.090990 (rank : 47) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O43889, O14671, O14919, Q96GK8, Q9H2W3, Q9UE77 | Gene names | CREB3, LZIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-responsive element-binding protein 3 (Luman protein) (Transcription factor LZIP-alpha). | |||||
|
CREB5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 189) | NC score | 0.109390 (rank : 46) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q02930, Q05886, Q06246 | Gene names | CREB5, CREBPA | |||
|
Domain Architecture |

|
|||||
| Description | cAMP response element-binding protein 5 (CRE-BPa). | |||||
|
NRL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 190) | NC score | 0.113854 (rank : 45) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P54845 | Gene names | NRL | |||
|
Domain Architecture |
|
|||||
| Description | Neural retina-specific leucine zipper protein (NRL) (D14S46E). | |||||
|
NF2L2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 183 | |
| SwissProt Accessions | Q60795 | Gene names | Nfe2l2, Nrf-2, Nrf2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 2 (NF-E2-related factor 2) (NFE2-related factor 2) (Nuclear factor, erythroid derived 2, like 2). | |||||
|
NF2L2_HUMAN
|
||||||
| NC score | 0.958956 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q16236, Q96F71 | Gene names | NFE2L2, NRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 2 (NF-E2-related factor 2) (NFE2-related factor 2) (Nuclear factor, erythroid derived 2, like 2) (HEBP1). | |||||
|
NFE2_HUMAN
|
||||||
| NC score | 0.892722 (rank : 3) | θ value | 7.25919e-50 (rank : 5) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q16621, Q07720, Q6ICV9 | Gene names | NFE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor NF-E2 45 kDa subunit (Nuclear factor, erythroid- derived 2 45 kDa subunit) (p45 NF-E2) (Leucine zipper protein NF-E2). | |||||
|
NF2L1_HUMAN
|
||||||
| NC score | 0.889582 (rank : 4) | θ value | 1.49966e-79 (rank : 3) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q14494, Q12877, Q96FN6 | Gene names | NFE2L1, HBZ17, NRF1, TCF11 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 1 (NF-E2-related factor 1) (NFE2-related factor 1) (Nuclear factor, erythroid derived 2, like 1) (Transcription factor 11) (Transcription factor HBZ17) (Transcription factor LCR-F1) (Locus control region-factor 1). | |||||
|
NF2L1_MOUSE
|
||||||
| NC score | 0.874182 (rank : 5) | θ value | 1.7756e-72 (rank : 4) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q61985, O70234 | Gene names | Nfe2l1, Nrf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 1 (NF-E2-related factor 1) (NFE2-related factor 1) (Nuclear factor, erythroid derived 2, like 1). | |||||
|
NF2L3_MOUSE
|
||||||
| NC score | 0.848652 (rank : 6) | θ value | 2.11701e-41 (rank : 7) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9WTM4 | Gene names | Nfe2l3, Nrf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor erythroid 2-related factor 3 (NF-E2-related factor 3) (NFE2-related factor 3) (Nuclear factor, erythroid derived 2, like 3). | |||||
|
NF2L3_HUMAN
|
||||||
| NC score | 0.843105 (rank : 7) | θ value | 5.95217e-44 (rank : 6) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Y4A8, Q6NUS0, Q7Z498, Q86UJ4, Q86VR5, Q9UQA4 | Gene names | NFE2L3, NRF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor erythroid 2-related factor 3 (NF-E2-related factor 3) (NFE2-related factor 3) (Nuclear factor, erythroid derived 2, like 3). | |||||
|
BACH1_HUMAN
|
||||||
| NC score | 0.378055 (rank : 8) | θ value | 8.38298e-14 (rank : 11) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O14867, O43285 | Gene names | BACH1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription regulator protein BACH1 (BTB and CNC homolog 1) (HA2303). | |||||
|
BACH1_MOUSE
|
||||||
| NC score | 0.377384 (rank : 9) | θ value | 6.41864e-14 (rank : 10) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P97302 | Gene names | Bach1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription regulator protein BACH1 (BTB and CNC homolog 1). | |||||
|
BACH2_MOUSE
|
||||||
| NC score | 0.374770 (rank : 10) | θ value | 1.99529e-15 (rank : 9) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P97303 | Gene names | Bach2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription regulator protein BACH2 (BTB and CNC homolog 2). | |||||
|
BACH2_HUMAN
|
||||||
| NC score | 0.372608 (rank : 11) | θ value | 1.52774e-15 (rank : 8) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9BYV9, Q59H70, Q5T793, Q9NTS5 | Gene names | BACH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription regulator protein BACH2 (BTB and CNC homolog 2). | |||||
|
JUND_MOUSE
|
||||||
| NC score | 0.354781 (rank : 12) | θ value | 0.000461057 (rank : 14) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P15066 | Gene names | Jund, Jun-d, Jund1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-D. | |||||
|
JUND_HUMAN
|
||||||
| NC score | 0.353757 (rank : 13) | θ value | 0.00665767 (rank : 19) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P17535 | Gene names | JUND | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-D. | |||||
|
JUNB_HUMAN
|
||||||
| NC score | 0.349460 (rank : 14) | θ value | 7.1131e-05 (rank : 12) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P17275, Q96GH3 | Gene names | JUNB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-B. | |||||
|
JUN_MOUSE
|
||||||
| NC score | 0.346724 (rank : 15) | θ value | 0.00390308 (rank : 18) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P05627 | Gene names | Jun | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-1 (Activator protein 1) (AP1) (Proto-oncogene c-jun) (V-jun avian sarcoma virus 17 oncogene homolog) (Jun A) (AH119). | |||||
|
JUN_HUMAN
|
||||||
| NC score | 0.346061 (rank : 16) | θ value | 0.00390308 (rank : 17) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P05412, Q96G93 | Gene names | JUN | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-1 (Activator protein 1) (AP1) (Proto-oncogene c-jun) (V-jun avian sarcoma virus 17 oncogene homolog) (p39). | |||||
|
JUNB_MOUSE
|
||||||
| NC score | 0.343358 (rank : 17) | θ value | 0.000270298 (rank : 13) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P09450, Q8C2G9 | Gene names | Junb, Jun-b | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-B. | |||||
|
XBP1_HUMAN
|
||||||
| NC score | 0.238241 (rank : 18) | θ value | 0.00228821 (rank : 16) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P17861, Q8WYK6, Q969P1, Q96BD7 | Gene names | XBP1, TREB5, XBP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X box-binding protein 1 (XBP-1) (Tax-responsive element-binding protein 5). | |||||
|
XBP1_MOUSE
|
||||||
| NC score | 0.221892 (rank : 19) | θ value | 0.0148317 (rank : 20) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O35426, Q8VHM0, Q922G5, Q9ESS3 | Gene names | Xbp1, Treb5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X box-binding protein 1 (XBP-1) (Tax-responsive element-binding protein 5 homolog). | |||||
|
MAFB_MOUSE
|
||||||
| NC score | 0.215089 (rank : 20) | θ value | 0.62314 (rank : 64) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P54841 | Gene names | Mafb, Krml, Maf1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafB (V-maf musculoaponeurotic fibrosarcoma oncogene homolog B) (Transcription factor MAF1) (Segmentation protein KR) (Kreisler). | |||||
|
MAFB_HUMAN
|
||||||
| NC score | 0.214795 (rank : 21) | θ value | 0.47712 (rank : 55) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Y5Q3, Q9H1F1 | Gene names | MAFB, KRML | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafB (V-maf musculoaponeurotic fibrosarcoma oncogene homolog B). | |||||
|
BATF_MOUSE
|
||||||
| NC score | 0.212590 (rank : 22) | θ value | 5.27518 (rank : 136) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O35284 | Gene names | Batf | |||
|
Domain Architecture |

|
|||||
| Description | ATF-like basic leucine zipper transcription factor B-ATF. | |||||
|
MAF_HUMAN
|
||||||
| NC score | 0.211772 (rank : 23) | θ value | 0.0736092 (rank : 29) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O75444, Q9UP93 | Gene names | MAF | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Maf (Proto-oncogene c-maf). | |||||
|
MAF_MOUSE
|
||||||
| NC score | 0.210531 (rank : 24) | θ value | 0.163984 (rank : 40) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P54843 | Gene names | Maf, Maf2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Maf (Proto-oncogene c-maf). | |||||
|
MAFK_HUMAN
|
||||||
| NC score | 0.209920 (rank : 25) | θ value | 0.0736092 (rank : 28) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O60675 | Gene names | MAFK | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafK (Erythroid transcription factor NF-E2 p18 subunit). | |||||
|
MAFK_MOUSE
|
||||||
| NC score | 0.206765 (rank : 26) | θ value | 0.0961366 (rank : 30) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q61827, Q60600 | Gene names | Mafk, Nfe2u | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafK (Erythroid transcription factor NF-E2 p18 subunit). | |||||
|
MAFG_HUMAN
|
||||||
| NC score | 0.199120 (rank : 27) | θ value | 0.0431538 (rank : 22) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O15525 | Gene names | MAFG | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafG (V-maf musculoaponeurotic fibrosarcoma oncogene homolog G) (hMAF). | |||||
|
MAFG_MOUSE
|
||||||
| NC score | 0.198686 (rank : 28) | θ value | 0.0563607 (rank : 24) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O54790 | Gene names | Mafg | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafG (V-maf musculoaponeurotic fibrosarcoma oncogene homolog G). | |||||
|
ATF3_MOUSE
|
||||||
| NC score | 0.190502 (rank : 29) | θ value | 0.813845 (rank : 68) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q60765 | Gene names | Atf3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-3 (Activating transcription factor 3) (Transcription factor LRG-21). | |||||
|
ATF3_HUMAN
|
||||||
| NC score | 0.185382 (rank : 30) | θ value | 0.813845 (rank : 67) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P18847 | Gene names | ATF3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-3 (Activating transcription factor 3). | |||||
|
BATF_HUMAN
|
||||||
| NC score | 0.183611 (rank : 31) | θ value | 6.88961 (rank : 154) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q16520 | Gene names | BATF | |||
|
Domain Architecture |

|
|||||
| Description | ATF-like basic leucine zipper transcriptional factor B-ATF (SF-HT- activated gene 2) (SFA-2). | |||||
|
CREB3_MOUSE
|
||||||
| NC score | 0.182030 (rank : 32) | θ value | 0.47712 (rank : 52) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q61817, Q99M21, Q9CVK9 | Gene names | Creb3, Lzip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-responsive element-binding protein 3 (Transcription factor LZIP). | |||||
|
FOSL1_HUMAN
|
||||||
| NC score | 0.172834 (rank : 33) | θ value | 0.62314 (rank : 61) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P15407 | Gene names | FOSL1, FRA1 | |||
|
Domain Architecture |

|
|||||
| Description | Fos-related antigen 1 (FRA-1). | |||||
|
FOSL1_MOUSE
|
||||||
| NC score | 0.171972 (rank : 34) | θ value | 0.62314 (rank : 62) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P48755, O35285 | Gene names | Fosl1, Fra1 | |||
|
Domain Architecture |

|
|||||
| Description | Fos-related antigen 1 (FRA-1). | |||||
|
FOSL2_HUMAN
|
||||||
| NC score | 0.167604 (rank : 35) | θ value | 0.21417 (rank : 43) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P15408 | Gene names | FOSL2, FRA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fos-related antigen 2. | |||||
|
FOSB_HUMAN
|
||||||
| NC score | 0.167396 (rank : 36) | θ value | 0.125558 (rank : 34) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P53539 | Gene names | FOSB, G0S3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein fosB (G0/G1 switch regulatory protein 3). | |||||
|
FOSB_MOUSE
|
||||||
| NC score | 0.165562 (rank : 37) | θ value | 0.279714 (rank : 46) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P13346 | Gene names | Fosb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein fosB. | |||||
|
FOSL2_MOUSE
|
||||||
| NC score | 0.164244 (rank : 38) | θ value | 0.365318 (rank : 50) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P47930 | Gene names | Fosl2, Fra-2, Fra2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fos-related antigen 2. | |||||
|
FOS_MOUSE
|
||||||
| NC score | 0.162557 (rank : 39) | θ value | 1.06291 (rank : 76) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P01101 | Gene names | Fos | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene protein c-fos (Cellular oncogene fos). | |||||
|
FOS_HUMAN
|
||||||
| NC score | 0.161048 (rank : 40) | θ value | 1.06291 (rank : 75) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P01100, P18849 | Gene names | FOS, G0S7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene protein c-fos (Cellular oncogene fos) (G0/G1 switch regulatory protein 7). | |||||
|
MAFF_HUMAN
|
||||||
| NC score | 0.142257 (rank : 41) | θ value | 1.81305 (rank : 91) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9ULX9, Q9Y525 | Gene names | MAFF | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafF (V-maf musculoaponeurotic fibrosarcoma oncogene homolog F) (U-Maf). | |||||
|
MAFF_MOUSE
|
||||||
| NC score | 0.141879 (rank : 42) | θ value | 2.36792 (rank : 101) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O54791 | Gene names | Maff | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafF (V-maf musculoaponeurotic fibrosarcoma oncogene homolog F). | |||||
|
NRL_MOUSE
|
||||||
| NC score | 0.126318 (rank : 43) | θ value | 8.99809 (rank : 179) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P54846 | Gene names | Nrl | |||
|
Domain Architecture |

|
|||||
| Description | Neural retina-specific leucine zipper protein (NRL). | |||||
|
ATF6A_HUMAN
|
||||||
| NC score | 0.121291 (rank : 44) | θ value | 0.813845 (rank : 69) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P18850, O15139, Q5VW62, Q6IPB5, Q9UEC9 | Gene names | ATF6 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-6 alpha (Activating transcription factor 6 alpha) (ATF6-alpha). | |||||
|
NRL_HUMAN
|
||||||
| NC score | 0.113854 (rank : 45) | θ value | θ > 10 (rank : 190) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P54845 | Gene names | NRL | |||
|
Domain Architecture |

|
|||||
| Description | Neural retina-specific leucine zipper protein (NRL) (D14S46E). | |||||
|
CREB5_HUMAN
|
||||||
| NC score | 0.109390 (rank : 46) | θ value | θ > 10 (rank : 189) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q02930, Q05886, Q06246 | Gene names | CREB5, CREBPA | |||
|
Domain Architecture |

|
|||||
| Description | cAMP response element-binding protein 5 (CRE-BPa). | |||||
|
CREB3_HUMAN
|
||||||
| NC score | 0.090990 (rank : 47) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O43889, O14671, O14919, Q96GK8, Q9H2W3, Q9UE77 | Gene names | CREB3, LZIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-responsive element-binding protein 3 (Luman protein) (Transcription factor LZIP-alpha). | |||||
|
CREM_MOUSE
|
||||||
| NC score | 0.090045 (rank : 48) | θ value | 2.36792 (rank : 97) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P27699, P27698 | Gene names | Crem | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-responsive element modulator. | |||||
|
CREM_HUMAN
|
||||||
| NC score | 0.089041 (rank : 49) | θ value | 2.36792 (rank : 96) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q03060, Q16114, Q16116, Q9NZB9 | Gene names | CREM | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-responsive element modulator. | |||||
|
ATF7_HUMAN
|
||||||
| NC score | 0.086922 (rank : 50) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P17544, Q13814 | Gene names | ATF7, ATFA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
ATF2_MOUSE
|
||||||
| NC score | 0.081961 (rank : 51) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P16951, Q64089, Q64090, Q64091 | Gene names | Atf2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (MXBP protein). | |||||
|
ATF2_HUMAN
|
||||||
| NC score | 0.081774 (rank : 52) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P15336, Q13000 | Gene names | ATF2, CREB2, CREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (HB16). | |||||
|
ATF1_HUMAN
|
||||||
| NC score | 0.080679 (rank : 53) | θ value | 5.27518 (rank : 134) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P18846, P25168 | Gene names | ATF1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-1 (Activating transcription factor 1) (TREB36 protein). | |||||
|
ATF7_MOUSE
|
||||||
| NC score | 0.080553 (rank : 54) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8R0S1 | Gene names | Atf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
ATF1_MOUSE
|
||||||
| NC score | 0.077807 (rank : 55) | θ value | 5.27518 (rank : 135) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P81269 | Gene names | Atf1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-1 (Activating transcription factor 1) (TCR-ATF1). | |||||
|
FA44A_HUMAN
|
||||||
| NC score | 0.073896 (rank : 56) | θ value | 0.00228821 (rank : 15) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8NFC6, Q6P0M8 | Gene names | FAM44A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM44A. | |||||
|
CREB1_MOUSE
|
||||||
| NC score | 0.071243 (rank : 57) | θ value | 8.99809 (rank : 171) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q01147 | Gene names | Creb1, Creb-1 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP response element-binding protein (CREB). | |||||
|
CREB1_HUMAN
|
||||||
| NC score | 0.071237 (rank : 58) | θ value | 8.99809 (rank : 170) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P16220, P21934, Q9UMA7 | Gene names | CREB1 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP response element-binding protein (CREB). | |||||
|
FA21C_HUMAN
|
||||||
| NC score | 0.063215 (rank : 59) | θ value | 0.125558 (rank : 33) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y4E1, Q5SQU4, Q5SQU5, Q7L521, Q9UG79 | Gene names | FAM21C, KIAA0592 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM21C. | |||||
|
DBP_HUMAN
|
||||||
| NC score | 0.062929 (rank : 60) | θ value | 0.0736092 (rank : 27) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q10586 | Gene names | DBP | |||
|
Domain Architecture |

|
|||||
| Description | D site-binding protein (Albumin D box-binding protein) (Albumin D- element-binding protein) (TAXREB302). | |||||
|
NCOR1_MOUSE
|
||||||
| NC score | 0.059667 (rank : 61) | θ value | 0.0563607 (rank : 25) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
DBP_MOUSE
|
||||||
| NC score | 0.059558 (rank : 62) | θ value | 0.21417 (rank : 42) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q60925, Q8VCX3 | Gene names | Dbp | |||
|
Domain Architecture |

|
|||||
| Description | D site-binding protein (Albumin D box-binding protein) (Albumin D- element-binding protein). | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.059375 (rank : 63) | θ value | 0.163984 (rank : 39) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.058183 (rank : 64) | θ value | 0.47712 (rank : 53) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
MBD5_HUMAN
|
||||||
| NC score | 0.054356 (rank : 65) | θ value | 0.21417 (rank : 45) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9P267, Q9NUV6 | Gene names | MBD5, KIAA1461 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 5 (Methyl-CpG-binding protein MBD5). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.053455 (rank : 66) | θ value | 0.125558 (rank : 35) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
ICAL_MOUSE
|
||||||
| NC score | 0.050741 (rank : 67) | θ value | 0.21417 (rank : 44) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P51125, Q9EQV4, Q9EQV5, Q9QXQ3, Q9QXQ4, Q9R0N1 | Gene names | Cast | |||
|
Domain Architecture |

|
|||||
| Description | Calpastatin (Calpain inhibitor). | |||||
|
MAP2_HUMAN
|
||||||
| NC score | 0.050451 (rank : 68) | θ value | 0.125558 (rank : 36) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P11137, Q99975, Q99976 | Gene names | MAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2) (MAP-2). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.049396 (rank : 69) | θ value | 1.81305 (rank : 90) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
CEP35_HUMAN
|
||||||
| NC score | 0.049390 (rank : 70) | θ value | 0.163984 (rank : 38) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q5VT06, O75068, Q8TDK3, Q8WY20 | Gene names | CEP350, CAP350, GM133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein 350 (Centrosome-associated protein of 350 kDa). | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.047531 (rank : 71) | θ value | 0.62314 (rank : 58) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
NCOR1_HUMAN
|
||||||
| NC score | 0.047224 (rank : 72) | θ value | 0.62314 (rank : 65) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
WDR60_MOUSE
|
||||||
| NC score | 0.046172 (rank : 73) | θ value | 0.0563607 (rank : 26) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8C761, Q3UNM4 | Gene names | Wdr60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 60. | |||||
|
ZN291_HUMAN
|
||||||
| NC score | 0.044708 (rank : 74) | θ value | 0.0961366 (rank : 31) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 647 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9BY12 | Gene names | ZNF291 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 291. | |||||
|
PPRB_HUMAN
|
||||||
| NC score | 0.043442 (rank : 75) | θ value | 0.47712 (rank : 57) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q15648, O43810, O75447, Q9HD39 | Gene names | PPARBP, ARC205, DRIP205, TRAP220, TRIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2) (p53 regulatory protein RB18A) (Vitamin D receptor-interacting protein complex component DRIP205) (Activator- recruited cofactor 205 kDa component) (ARC205). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.041817 (rank : 76) | θ value | 0.813845 (rank : 70) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.040715 (rank : 77) | θ value | 2.36792 (rank : 94) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
TP53B_HUMAN
|
||||||
| NC score | 0.040671 (rank : 78) | θ value | 0.62314 (rank : 66) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q12888, Q2M1Z7, Q4LE46, Q5FWZ3, Q7Z3U4 | Gene names | TP53BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
CENPU_MOUSE
|
||||||
| NC score | 0.040552 (rank : 79) | θ value | 0.62314 (rank : 59) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8C4M7, Q6UNA2, Q9D9U1 | Gene names | Mlf1ip, Cenpu | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein U (CENP-U) (MLF1-interacting protein). | |||||
|
S23IP_HUMAN
|
||||||
| NC score | 0.040184 (rank : 80) | θ value | 0.125558 (rank : 37) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y6Y8, Q8IXH5, Q9BUK5 | Gene names | SEC23IP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SEC23-interacting protein (p125). | |||||
|
HSF1_HUMAN
|
||||||
| NC score | 0.039883 (rank : 81) | θ value | 0.47712 (rank : 54) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q00613 | Gene names | HSF1, HSTF1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
RABE1_MOUSE
|
||||||
| NC score | 0.038361 (rank : 82) | θ value | 3.0926 (rank : 111) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O35551, Q99L08, Q9CRP3, Q9EQF9, Q9JI94 | Gene names | Rabep1, Rab5ep, Rabpt5, Rabpt5a | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha). | |||||
|
BAZ2A_MOUSE
|
||||||
| NC score | 0.037832 (rank : 83) | θ value | 0.125558 (rank : 32) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
RERE_MOUSE
|
||||||
| NC score | 0.037803 (rank : 84) | θ value | 1.06291 (rank : 79) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.037113 (rank : 85) | θ value | 1.06291 (rank : 78) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
BPTF_HUMAN
|
||||||
| NC score | 0.036880 (rank : 86) | θ value | 1.06291 (rank : 72) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
CX039_MOUSE
|
||||||
| NC score | 0.036474 (rank : 87) | θ value | 1.81305 (rank : 85) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K2D0, Q3U3R2 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein CXorf39 homolog. | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.036447 (rank : 88) | θ value | 1.38821 (rank : 84) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
EP15R_MOUSE
|
||||||
| NC score | 0.036394 (rank : 89) | θ value | 1.06291 (rank : 74) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q60902, Q8CB60, Q8CB70, Q91WH8 | Gene names | Eps15l1, Eps15-rs, Eps15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R) (Epidermal growth factor receptor pathway substrate 15-related sequence) (Eps15-rs). | |||||
|
RABE1_HUMAN
|
||||||
| NC score | 0.036301 (rank : 90) | θ value | 5.27518 (rank : 147) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q15276, O95369, Q8IVX3 | Gene names | RABEP1, RABPT5, RABPT5A | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha) (Rabaptin-4) (NY-REN-17 antigen). | |||||
|
CK5P2_HUMAN
|
||||||
| NC score | 0.035362 (rank : 91) | θ value | 0.62314 (rank : 60) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
CAF1A_HUMAN
|
||||||
| NC score | 0.035204 (rank : 92) | θ value | 1.38821 (rank : 82) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q13111, Q7Z7K3, Q9UJY8 | Gene names | CHAF1A, CAF, CAF1P150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
K1683_HUMAN
|
||||||
| NC score | 0.033741 (rank : 93) | θ value | 3.0926 (rank : 109) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H0B3, Q8N4G8, Q96M14, Q9C0I0 | Gene names | KIAA1683 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1683. | |||||
|
HSF1_MOUSE
|
||||||
| NC score | 0.033726 (rank : 94) | θ value | 1.81305 (rank : 89) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P38532, O70462 | Gene names | Hsf1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
CX039_HUMAN
|
||||||
| NC score | 0.033668 (rank : 95) | θ value | 3.0926 (rank : 106) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PEV8, Q8WVP6, Q96AV3 | Gene names | CXorf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein CXorf39. | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.033478 (rank : 96) | θ value | 0.279714 (rank : 47) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
EP15R_HUMAN
|
||||||
| NC score | 0.031274 (rank : 97) | θ value | 1.06291 (rank : 73) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 1083 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9UBC2 | Gene names | EPS15L1, EPS15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R). | |||||
|
CLSPN_MOUSE
|
||||||
| NC score | 0.031069 (rank : 98) | θ value | 1.38821 (rank : 83) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q80YR7, Q69GM2 | Gene names | Clspn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin. | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.031051 (rank : 99) | θ value | 6.88961 (rank : 153) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.030721 (rank : 100) | θ value | 0.163984 (rank : 41) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
HNF1A_MOUSE
|
||||||
| NC score | 0.030317 (rank : 101) | θ value | 1.81305 (rank : 88) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P22361 | Gene names | Tcf1, Hnf-1, Hnf-1a, Hnf1a | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 1-alpha (HNF-1A) (Liver-specific transcription factor LF-B1) (LFB1). | |||||
|
BRD4_MOUSE
|
||||||
| NC score | 0.030190 (rank : 102) | θ value | 0.365318 (rank : 49) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
CKAP4_MOUSE
|
||||||
| NC score | 0.030023 (rank : 103) | θ value | 3.0926 (rank : 104) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BMK4, Q8BTK8, Q8R3F2 | Gene names | Ckap4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 4 (63 kDa membrane protein) (p63). | |||||
|
ZN645_HUMAN
|
||||||
| NC score | 0.029933 (rank : 104) | θ value | 3.0926 (rank : 113) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8N7E2, Q6DJY9 | Gene names | ZNF645 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 645. | |||||
|
TMF1_HUMAN
|
||||||
| NC score | 0.029891 (rank : 105) | θ value | 0.279714 (rank : 48) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P82094 | Gene names | TMF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA element modulatory factor (TMF). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.029732 (rank : 106) | θ value | 4.03905 (rank : 123) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.029638 (rank : 107) | θ value | 3.0926 (rank : 112) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.029604 (rank : 108) | θ value | 1.81305 (rank : 87) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
INCE_HUMAN
|
||||||
| NC score | 0.029394 (rank : 109) | θ value | 0.62314 (rank : 63) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
AP3B2_MOUSE
|
||||||
| NC score | 0.028206 (rank : 110) | θ value | 1.06291 (rank : 71) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.027840 (rank : 111) | θ value | 0.47712 (rank : 51) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
CCD41_MOUSE
|
||||||
| NC score | 0.027194 (rank : 112) | θ value | 8.99809 (rank : 169) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9D5R3, Q3U7X7, Q3UX57, Q80VF0 | Gene names | Ccdc41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41. | |||||
|
AMPH_MOUSE
|
||||||
| NC score | 0.026869 (rank : 113) | θ value | 4.03905 (rank : 114) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q7TQF7, Q8R1C4 | Gene names | Amph, Amph1 | |||
|
Domain Architecture |

|
|||||
| Description | Amphiphysin. | |||||
|
HNF1A_HUMAN
|
||||||
| NC score | 0.026613 (rank : 114) | θ value | 5.27518 (rank : 140) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P20823, Q99861 | Gene names | TCF1, HNF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 1-alpha (HNF-1A) (Liver-specific transcription factor LF-B1) (LFB1) (Transcription factor 1) (TCF-1). | |||||
|
XE7_HUMAN
|
||||||
| NC score | 0.026447 (rank : 115) | θ value | 4.03905 (rank : 130) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 738 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q02040, Q02832 | Gene names | CXYorf3, DXYS155E, XE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-lymphocyte antigen precursor (B-lymphocyte surface antigen) (721P) (Protein XE7). | |||||
|
TRAIP_MOUSE
|
||||||
| NC score | 0.026395 (rank : 116) | θ value | 4.03905 (rank : 127) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8VIG6, O08854, Q922M8, Q9CPP4 | Gene names | Traip, Trip | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
KNTC2_MOUSE
|
||||||
| NC score | 0.026174 (rank : 117) | θ value | 6.88961 (rank : 159) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9D0F1, Q3TQT6, Q3UWM5, Q99P70 | Gene names | Kntc2, Hec1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Hec1 (Kinetochore-associated protein 2). | |||||
|
CCNK_MOUSE
|
||||||
| NC score | 0.026076 (rank : 118) | θ value | 2.36792 (rank : 95) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
AMPH_HUMAN
|
||||||
| NC score | 0.025695 (rank : 119) | θ value | 6.88961 (rank : 151) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P49418, O43538, Q75MJ8, Q75MK5, Q75MM3, Q8N4G0 | Gene names | AMPH, AMPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Amphiphysin. | |||||
|
TRAIP_HUMAN
|
||||||
| NC score | 0.025362 (rank : 120) | θ value | 6.88961 (rank : 166) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9BWF2, O00467 | Gene names | TRAIP, TRIP | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
MYH9_MOUSE
|
||||||
| NC score | 0.023300 (rank : 121) | θ value | 4.03905 (rank : 120) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
SKIL_HUMAN
|
||||||
| NC score | 0.023077 (rank : 122) | θ value | 2.36792 (rank : 103) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P12757, O00464, P12756, Q07501 | Gene names | SKIL, SNO | |||
|
Domain Architecture |

|
|||||
| Description | Ski-like protein (Ski-related protein) (Ski-related oncogene). | |||||
|
GCC2_HUMAN
|
||||||
| NC score | 0.023057 (rank : 123) | θ value | 4.03905 (rank : 117) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
RBP24_HUMAN
|
||||||
| NC score | 0.023036 (rank : 124) | θ value | 4.03905 (rank : 125) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
TACC1_MOUSE
|
||||||
| NC score | 0.022664 (rank : 125) | θ value | 5.27518 (rank : 148) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6Y685, Q6Y686 | Gene names | Tacc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transforming acidic coiled-coil-containing protein 1. | |||||
|
CUTL1_HUMAN
|
||||||
| NC score | 0.022282 (rank : 126) | θ value | 6.88961 (rank : 157) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
MARCS_HUMAN
|
||||||
| NC score | 0.022034 (rank : 127) | θ value | 8.99809 (rank : 177) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.021818 (rank : 128) | θ value | 8.99809 (rank : 168) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
MYH11_MOUSE
|
||||||
| NC score | 0.021503 (rank : 129) | θ value | 3.0926 (rank : 110) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
CASP_MOUSE
|
||||||
| NC score | 0.021465 (rank : 130) | θ value | 5.27518 (rank : 137) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 1152 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P70403, Q7TQJ4, Q91WN6 | Gene names | Cutl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CASP_HUMAN
|
||||||
| NC score | 0.021304 (rank : 131) | θ value | 6.88961 (rank : 155) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q13948, Q53GU9, Q8TBS3 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
ASPP1_HUMAN
|
||||||
| NC score | 0.021176 (rank : 132) | θ value | 1.38821 (rank : 81) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 956 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q96KQ4, O94870 | Gene names | PPP1R13B, ASPP1, KIAA0771 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
TRIM6_HUMAN
|
||||||
| NC score | 0.021120 (rank : 133) | θ value | 0.0431538 (rank : 23) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9C030, Q86WZ8, Q9HCR1 | Gene names | TRIM6, RNF89 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 6 (RING finger protein 89). | |||||
|
MYH11_HUMAN
|
||||||
| NC score | 0.021105 (rank : 134) | θ value | 6.88961 (rank : 161) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
ASPX_MOUSE
|
||||||
| NC score | 0.020758 (rank : 135) | θ value | 6.88961 (rank : 152) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P50289, Q9DAM6 | Gene names | Acrv1 | |||
|
Domain Architecture |

|
|||||
| Description | Acrosomal protein SP-10 precursor (Acrosomal vesicle protein 1) (MSA- 63). | |||||
|
MYH9_HUMAN
|
||||||
| NC score | 0.020752 (rank : 136) | θ value | 6.88961 (rank : 164) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
TRI29_HUMAN
|
||||||
| NC score | 0.020530 (rank : 137) | θ value | 4.03905 (rank : 128) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14134, Q96AA9, Q9BZY7 | Gene names | TRIM29, ATDC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 29 (Ataxia-telangiectasia group D- associated protein). | |||||
|
K0310_HUMAN
|
||||||
| NC score | 0.020201 (rank : 138) | θ value | 8.99809 (rank : 176) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O15027, Q5SXP0, Q5SXP1, Q8N347, Q96HP1 | Gene names | KIAA0310 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0310. | |||||
|
MYH6_HUMAN
|
||||||
| NC score | 0.019989 (rank : 139) | θ value | 6.88961 (rank : 162) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYH6_MOUSE
|
||||||
| NC score | 0.019617 (rank : 140) | θ value | 6.88961 (rank : 163) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
IF35_HUMAN
|
||||||
| NC score | 0.019571 (rank : 141) | θ value | 5.27518 (rank : 141) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O00303, Q8N978 | Gene names | EIF3S5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 5 (eIF-3 epsilon) (eIF3 p47 subunit) (eIF3f). | |||||
|
SAPS3_HUMAN
|
||||||
| NC score | 0.019540 (rank : 142) | θ value | 1.06291 (rank : 80) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5H9R7, Q3B7I1, Q3I4Y0, Q3KR35, Q68CR3, Q7L4R8, Q8N3B2, Q96MB2, Q9H2K5, Q9H2K6, Q9HCL4, Q9NUY3 | Gene names | SAPS3, C11orf23, KIAA1558, PP6R3, SAPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAPS domain family member 3 (Sporulation-induced transcript 4- associated protein SAPL) (Protein phosphatase 6 regulatory subunit 3). | |||||
|
SRCA_HUMAN
|
||||||
| NC score | 0.019527 (rank : 143) | θ value | 4.03905 (rank : 126) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
IL3RB_HUMAN
|
||||||
| NC score | 0.019483 (rank : 144) | θ value | 3.0926 (rank : 108) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P32927 | Gene names | CSF2RB, IL3RB, IL5RB | |||
|
Domain Architecture |

|
|||||
| Description | Cytokine receptor common beta chain precursor (GM-CSF/IL-3/IL-5 receptor common beta-chain) (CD131 antigen) (CDw131). | |||||
|
NCOA5_HUMAN
|
||||||
| NC score | 0.018853 (rank : 145) | θ value | 4.03905 (rank : 121) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9HCD5, Q6HA99, Q9H1F2, Q9H2T2, Q9H4Y9 | Gene names | NCOA5, KIAA1637 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 5 (NCoA-5) (Coactivator independent of AF-2) (CIA). | |||||
|
AKAP8_MOUSE
|
||||||
| NC score | 0.018652 (rank : 146) | θ value | 6.88961 (rank : 150) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9DBR0, Q9R0L8 | Gene names | Akap8, Akap95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 8 (A-kinase anchor protein 95 kDa) (AKAP 95). | |||||
|
RADI_MOUSE
|
||||||
| NC score | 0.018484 (rank : 147) | θ value | 2.36792 (rank : 102) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 773 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P26043, Q9QW27 | Gene names | Rdx | |||
|
Domain Architecture |

|
|||||
| Description | Radixin (ESP10). | |||||
|
MOES_MOUSE
|
||||||
| NC score | 0.018233 (rank : 148) | θ value | 4.03905 (rank : 119) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 923 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P26041, Q3UL28, Q8BSN4 | Gene names | Msn | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
AIM1_HUMAN
|
||||||
| NC score | 0.017764 (rank : 149) | θ value | 5.27518 (rank : 132) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y4K1, O00296, Q5VWJ2 | Gene names | AIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Absent in melanoma 1 protein. | |||||
|
TTC15_MOUSE
|
||||||
| NC score | 0.017744 (rank : 150) | θ value | 6.88961 (rank : 167) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8K2L8, Q8BX27 | Gene names | Ttc15 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 15 (TPR repeat protein 15). | |||||
|
EPAS1_MOUSE
|
||||||
| NC score | 0.017720 (rank : 151) | θ value | 2.36792 (rank : 99) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97481, O08787, O55046 | Gene names | Epas1, Hif2a | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Hypoxia- inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (MHLF) (HIF-related factor) (HRF). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.017327 (rank : 152) | θ value | 5.27518 (rank : 144) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
MYEF2_HUMAN
|
||||||
| NC score | 0.017197 (rank : 153) | θ value | 1.06291 (rank : 77) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P2K5, Q6NUM5, Q7L388, Q7Z4B7, Q9H922, Q9NUQ1 | Gene names | MYEF2, KIAA1341 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin expression factor 2 (MyEF-2) (MST156). | |||||
|
TRI29_MOUSE
|
||||||
| NC score | 0.017094 (rank : 154) | θ value | 8.99809 (rank : 183) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8R2Q0, Q8CEE2, Q922Y3, Q99PN5, Q9CSC9 | Gene names | Trim29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 29. | |||||
|
DACH2_MOUSE
|
||||||
| NC score | 0.016775 (rank : 155) | θ value | 5.27518 (rank : 138) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q925Q8, Q8BMA0 | Gene names | Dach2 | |||
|
Domain Architecture |

|
|||||
| Description | Dachshund homolog 2 (Dach2). | |||||
|
CMTA2_HUMAN
|
||||||
| NC score | 0.016548 (rank : 156) | θ value | 3.0926 (rank : 105) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O94983, Q7Z6M8, Q8N3V0, Q8NDG4, Q96G17 | Gene names | CAMTA2, KIAA0909 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-binding transcription activator 2. | |||||
|
NOP56_MOUSE
|
||||||
| NC score | 0.016426 (rank : 157) | θ value | 4.03905 (rank : 122) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D6Z1, Q3UD45, Q8BVL1, Q8VDT2, Q99LT8, Q9CT15 | Gene names | Nol5a, Nop56 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar protein Nop56 (Nucleolar protein 5A). | |||||
|
G3BP_MOUSE
|
||||||
| NC score | 0.016084 (rank : 158) | θ value | 1.81305 (rank : 86) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97855 | Gene names | G3bp, G3bp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ras-GTPase-activating protein-binding protein 1 (EC 3.6.1.-) (ATP- dependent DNA helicase VIII) (GAP SH3-domain-binding protein 1) (G3BP- 1) (HDH-VIII). | |||||
|
UBL7_MOUSE
|
||||||
| NC score | 0.015470 (rank : 159) | θ value | 5.27518 (rank : 149) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91W67, Q9D7P5 | Gene names | Ubl7 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like protein 7. | |||||
|
PHC3_HUMAN
|
||||||
| NC score | 0.015282 (rank : 160) | θ value | 5.27518 (rank : 145) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8NDX5, Q5HYF0, Q6NSG2, Q8NFT7, Q8NFZ1, Q8TBM2, Q9H971, Q9H9I4 | Gene names | PHC3, EDR3, PH3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 3 (hPH3) (Homolog of polyhomeotic 3) (Early development regulatory protein 3). | |||||
|
SALL3_HUMAN
|
||||||
| NC score | 0.015166 (rank : 161) | θ value | 0.0252991 (rank : 21) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 1066 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BXA9, Q9UGH1 | Gene names | SALL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 3 (Zinc finger protein SALL3) (hSALL3). | |||||
|
LIPB2_HUMAN
|
||||||
| NC score | 0.014760 (rank : 162) | θ value | 6.88961 (rank : 160) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 599 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8ND30, O75337, Q8WW26 | Gene names | PPFIBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 2) (PTPRF-interacting protein-binding protein 2). | |||||
|
RIN3_MOUSE
|
||||||
| NC score | 0.014671 (rank : 163) | θ value | 8.99809 (rank : 181) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P59729 | Gene names | Rin3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras and Rab interactor 3 (Ras interaction/interference protein 3). | |||||
|
EDD1_HUMAN
|
||||||
| NC score | 0.014282 (rank : 164) | θ value | 2.36792 (rank : 98) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95071, O94970, Q9NPL3 | Gene names | EDD1, EDD, HYD, KIAA0896 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog) (hHYD) (Progestin-induced protein). | |||||
|
GGA3_MOUSE
|
||||||
| NC score | 0.013538 (rank : 165) | θ value | 3.0926 (rank : 107) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BMI3, Q80U71 | Gene names | Gga3, Kiaa0154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
CL026_HUMAN
|
||||||
| NC score | 0.013075 (rank : 166) | θ value | 6.88961 (rank : 156) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N6Q8, Q9H5Y3 | Gene names | C12orf26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C12orf26. | |||||
|
NFAC4_HUMAN
|
||||||
| NC score | 0.012432 (rank : 167) | θ value | 5.27518 (rank : 143) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14934 | Gene names | NFATC4, NFAT3 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 4 (NF-ATc4) (NFATc4) (T cell transcription factor NFAT3) (NF-AT3). | |||||
|
VITRN_HUMAN
|
||||||
| NC score | 0.012125 (rank : 168) | θ value | 1.81305 (rank : 93) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6UXI7, Q6P7T3, Q96DT1 | Gene names | VIT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vitrin precursor. | |||||
|
NR1D1_HUMAN
|
||||||
| NC score | 0.012110 (rank : 169) | θ value | 0.47712 (rank : 56) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P20393, Q15304 | Gene names | NR1D1, EAR1, HREV, THRAL | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR1D1 (V-erbA-related protein EAR-1) (Rev- erbA-alpha). | |||||
|
ADDB_HUMAN
|
||||||
| NC score | 0.011775 (rank : 170) | θ value | 5.27518 (rank : 131) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P35612, Q13482, Q16412, Q59G82, Q5U5P4, Q6P0P2, Q6PGQ4, Q7Z688, Q7Z689, Q7Z690, Q7Z691 | Gene names | ADD2, ADDB | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta). | |||||
|
OSBP2_HUMAN
|
||||||
| NC score | 0.011294 (rank : 171) | θ value | 4.03905 (rank : 124) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q969R2, O60396, Q9BY96, Q9BZF0 | Gene names | OSBP2, KIAA1664, ORP4, OSBPL4 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein 2 (Oxysterol-binding protein-related protein 4) (OSBP-related protein 4) (ORP-4). | |||||
|
PSL1_MOUSE
|
||||||
| NC score | 0.011173 (rank : 172) | θ value | 5.27518 (rank : 146) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3TD49, Q3TLQ8, Q3UG60, Q80VZ6 | Gene names | Sppl2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b). | |||||
|
UBP37_HUMAN
|
||||||
| NC score | 0.010142 (rank : 173) | θ value | 4.03905 (rank : 129) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q86T82, Q9HCH8 | Gene names | USP37, KIAA1594 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 37 (EC 3.1.2.15) (Ubiquitin thioesterase 37) (Ubiquitin-specific-processing protease 37) (Deubiquitinating enzyme 37). | |||||
|
NRDC_MOUSE
|
||||||
| NC score | 0.009844 (rank : 174) | θ value | 8.99809 (rank : 178) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BHG1 | Gene names | Nrd1 | |||
|
Domain Architecture |

|
|||||
| Description | Nardilysin precursor (EC 3.4.24.61) (N-arginine dibasic convertase) (NRD convertase) (NRD-C). | |||||
|
BRWD1_HUMAN
|
||||||
| NC score | 0.009321 (rank : 175) | θ value | 4.03905 (rank : 115) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NSI6, O43721, Q5R2V0, Q5R2V1, Q8TCV3, Q96QG9, Q96QH0, Q9NUK1 | Gene names | BRWD1, C21orf107, WDR9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain and WD repeat domain-containing protein 1 (WD repeat protein 9). | |||||
|
DHX29_HUMAN
|
||||||
| NC score | 0.008711 (rank : 176) | θ value | 8.99809 (rank : 172) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7Z478, O75549, Q63HN0, Q63HN3, Q8IWW2, Q8N3A1, Q9UMH2 | Gene names | DHX29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX29 (EC 3.6.1.-) (DEAH box protein 29) (Nucleic acid helicase DDXx). | |||||
|
FA5_HUMAN
|
||||||
| NC score | 0.007481 (rank : 177) | θ value | 8.99809 (rank : 173) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P12259, Q14285, Q6UPU6 | Gene names | F5 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor V precursor (Activated protein C cofactor) [Contains: Coagulation factor V heavy chain; Coagulation factor V light chain]. | |||||
|
IMA4_HUMAN
|
||||||
| NC score | 0.006972 (rank : 178) | θ value | 8.99809 (rank : 174) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00629, O00190 | Gene names | KPNA4, QIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-4 subunit (Karyopherin alpha-4 subunit) (Qip1 protein). | |||||
|
IMA4_MOUSE
|
||||||
| NC score | 0.006812 (rank : 179) | θ value | 8.99809 (rank : 175) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35343 | Gene names | Kpna4 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-4 subunit (Karyopherin alpha-4 subunit) (Importin alpha Q1). | |||||
|
KCNN3_MOUSE
|
||||||
| NC score | 0.006570 (rank : 180) | θ value | 6.88961 (rank : 158) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P58391 | Gene names | Kcnn3, Sk3 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 3 (SK3). | |||||
|
FARP2_MOUSE
|
||||||
| NC score | 0.006497 (rank : 181) | θ value | 5.27518 (rank : 139) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91VS8, Q3TAP2, Q69ZZ0 | Gene names | Farp2, Kiaa0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
KIF17_MOUSE
|
||||||
| NC score | 0.006053 (rank : 182) | θ value | 5.27518 (rank : 142) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q99PW8 | Gene names | Kif17 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF17 (MmKIF17). | |||||
|
SYT12_MOUSE
|
||||||
| NC score | 0.004942 (rank : 183) | θ value | 8.99809 (rank : 182) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q920N7 | Gene names | Syt12 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-12 (Synaptotagmin XII) (SytXII). | |||||
|
TMPSD_MOUSE
|
||||||
| NC score | 0.004335 (rank : 184) | θ value | 1.81305 (rank : 92) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5U405, Q8CFE0, Q91VQ8 | Gene names | Tmprss13, Msp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 13 (EC 3.4.21.-) (Mosaic serine protease) (Membrane-type mosaic serine protease). | |||||
|
DDX50_HUMAN
|
||||||
| NC score | 0.004253 (rank : 185) | θ value | 4.03905 (rank : 116) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BQ39, Q8WV76, Q9BWI8 | Gene names | DDX50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
RREB1_HUMAN
|
||||||
| NC score | 0.003189 (rank : 186) | θ value | 6.88961 (rank : 165) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 879 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q92766, O75567 | Gene names | RREB1 | |||
|
Domain Architecture |

|
|||||
| Description | RAS-responsive element-binding protein 1 (RREB-1) (Raf-responsive zinc finger protein LZ321). | |||||
|
ALX4_HUMAN
|
||||||
| NC score | 0.001882 (rank : 187) | θ value | 5.27518 (rank : 133) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H161, Q96JN7, Q9H198, Q9HAY9 | Gene names | ALX4, KIAA1788 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein aristaless-like 4. | |||||
|
ERBB4_HUMAN
|
||||||
| NC score | -0.001351 (rank : 188) | θ value | 2.36792 (rank : 100) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 910 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15303 | Gene names | ERBB4, HER4 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor tyrosine-protein kinase erbB-4 precursor (EC 2.7.10.1) (p180erbB4) (Tyrosine kinase-type cell surface receptor HER4). | |||||
|
PKN2_HUMAN
|
||||||
| NC score | -0.002323 (rank : 189) | θ value | 8.99809 (rank : 180) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 892 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q16513, Q9H1W4 | Gene names | PKN2, PRK2, PRKCL2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase N2 (EC 2.7.11.13) (Protein kinase C- like 2) (Protein-kinase C-related kinase 2). | |||||
|
MAK_MOUSE
|
||||||
| NC score | -0.002781 (rank : 190) | θ value | 4.03905 (rank : 118) | |||
| Query Neighborhood Hits | 183 | Target Neighborhood Hits | 873 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q04859 | Gene names | Mak, Rck | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase MAK (EC 2.7.11.22) (Male germ cell- associated kinase) (Protein kinase RCK). | |||||