Please be patient as the page loads
|
TTC15_MOUSE
|
||||||
| SwissProt Accessions | Q8K2L8, Q8BX27 | Gene names | Ttc15 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 15 (TPR repeat protein 15). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TTC15_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.920736 (rank : 2) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8WVT3, Q8WVW1, Q9Y395 | Gene names | TTC15 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 15 (TPR repeat protein 15). | |||||
|
TTC15_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 113 | |
| SwissProt Accessions | Q8K2L8, Q8BX27 | Gene names | Ttc15 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 15 (TPR repeat protein 15). | |||||
|
SYNJ1_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 3) | NC score | 0.076952 (rank : 21) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
MAP4_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 4) | NC score | 0.084265 (rank : 17) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
SGTB_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 5) | NC score | 0.133719 (rank : 4) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96EQ0 | Gene names | SGTB, SGT2 | |||
|
Domain Architecture |
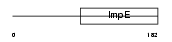
|
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein B (Small glutamine-rich protein with tetratricopeptide repeats 2). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 6) | NC score | 0.071697 (rank : 24) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
DNJC7_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 7) | NC score | 0.098261 (rank : 12) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9QYI3, Q3TKR1, Q8BPG3, Q8CIL2, Q9CT29, Q9D026 | Gene names | Dnajc7, Ttc2 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 7 (Tetratricopeptide repeat protein 2) (TPR repeat protein 2) (MDj11). | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 8) | NC score | 0.071015 (rank : 26) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
DNJC7_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 9) | NC score | 0.097932 (rank : 13) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99615 | Gene names | DNAJC7, TPR2, TTC2 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 7 (Tetratricopeptide repeat protein 2) (TPR repeat protein 2). | |||||
|
IFT88_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 10) | NC score | 0.118565 (rank : 11) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13099 | Gene names | IFT88, TG737, TTC10 | |||
|
Domain Architecture |

|
|||||
| Description | Intraflagellar transport 88 homolog (Tetratricopeptide repeat protein 10) (TPR repeat protein 10) (Recessive polycystic kidney disease protein Tg737 homolog). | |||||
|
SGTB_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 11) | NC score | 0.128069 (rank : 8) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8VD33 | Gene names | Sgtb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein B. | |||||
|
DMP1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 12) | NC score | 0.081911 (rank : 19) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
OGT1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 13) | NC score | 0.129426 (rank : 7) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O15294, Q7Z3K0, Q8WWM8, Q96CC1, Q9UG57 | Gene names | OGT | |||
|
Domain Architecture |

|
|||||
| Description | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit (EC 2.4.1.-) (O-GlcNAc transferase p110 subunit). | |||||
|
CTR9_HUMAN
|
||||||
| θ value | 0.163984 (rank : 14) | NC score | 0.122719 (rank : 9) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6PD62, Q15015 | Gene names | CTR9, KIAA0155, SH2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1). | |||||
|
CTR9_MOUSE
|
||||||
| θ value | 0.163984 (rank : 15) | NC score | 0.121107 (rank : 10) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q62018, Q3UFF5, Q3UY40, Q66JX4, Q7TPS6, Q8BND9, Q8BRD1, Q8C9W7, Q8C9Y3, Q8CHI1 | Gene names | Ctr9, Kiaa0155, Sh2bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1) (Tetratricopeptide repeat-containing, SH2-binding phosphoprotein of 150 kDa) (TPR-containing, SH2-binding phosphoprotein of 150 kDa) (p150TSP). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 16) | NC score | 0.059364 (rank : 36) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
TTC13_HUMAN
|
||||||
| θ value | 0.163984 (rank : 17) | NC score | 0.136092 (rank : 3) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8NBP0, Q8IVP8, Q8NBI0, Q8ND20 | Gene names | TTC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 13 (TPR repeat protein 13). | |||||
|
TTC8_HUMAN
|
||||||
| θ value | 0.163984 (rank : 18) | NC score | 0.131538 (rank : 5) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8TAM2, Q86SY0, Q86TV9, Q86U26, Q8NDH9, Q96DG8 | Gene names | TTC8, BBS8 | |||
|
Domain Architecture |
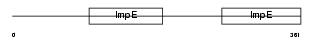
|
|||||
| Description | Tetratricopeptide repeat protein 8 (TPR repeat protein 8) (Bardet- Biedl syndrome 8 protein). | |||||
|
DSPP_HUMAN
|
||||||
| θ value | 0.21417 (rank : 19) | NC score | 0.073452 (rank : 22) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
COCA1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 20) | NC score | 0.027225 (rank : 80) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q60847, P70322 | Gene names | Col12a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XII) chain precursor. | |||||
|
SPAG1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 21) | NC score | 0.079433 (rank : 20) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q07617, Q7Z5G1 | Gene names | SPAG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 1 (Infertility-related sperm protein Spag-1) (HSD-3.8). | |||||
|
TIF1A_MOUSE
|
||||||
| θ value | 0.279714 (rank : 22) | NC score | 0.037169 (rank : 65) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q64127, Q64126 | Gene names | Trim24, Tif1, Tif1a | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24). | |||||
|
TTC8_MOUSE
|
||||||
| θ value | 0.279714 (rank : 23) | NC score | 0.131454 (rank : 6) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8VD72, Q9DCP7 | Gene names | Ttc8, Bbs8 | |||
|
Domain Architecture |
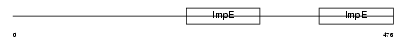
|
|||||
| Description | Tetratricopeptide repeat protein 8 (TPR repeat protein 8) (Bardet- Biedl syndrome 8 protein homolog). | |||||
|
ATCAY_HUMAN
|
||||||
| θ value | 0.365318 (rank : 24) | NC score | 0.037012 (rank : 66) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86WG3, Q8NAQ2, Q8TAQ3, Q96HC6, Q96JF5 | Gene names | ATCAY, KIAA1872 | |||
|
Domain Architecture |

|
|||||
| Description | Caytaxin (Ataxia Cayman type protein) (BNIP-H). | |||||
|
MYST4_MOUSE
|
||||||
| θ value | 0.365318 (rank : 25) | NC score | 0.044239 (rank : 54) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
CT077_HUMAN
|
||||||
| θ value | 0.47712 (rank : 26) | NC score | 0.045140 (rank : 53) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NQG5 | Gene names | C20orf77 | |||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf77. | |||||
|
CYLC2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 27) | NC score | 0.055042 (rank : 42) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
RTN4_MOUSE
|
||||||
| θ value | 0.47712 (rank : 28) | NC score | 0.041052 (rank : 60) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99P72, Q5DTK9, Q7TNB7, Q80W95, Q8BGK7, Q8BGM9, Q8K290, Q8K3G8, Q9CTE3 | Gene names | Rtn4, Kiaa0886, Nogo | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-4 (Neurite outgrowth inhibitor) (Nogo protein). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 29) | NC score | 0.060339 (rank : 35) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
TARA_MOUSE
|
||||||
| θ value | 0.47712 (rank : 30) | NC score | 0.069860 (rank : 28) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
DSPP_MOUSE
|
||||||
| θ value | 0.62314 (rank : 31) | NC score | 0.071081 (rank : 25) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
MUC2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 32) | NC score | 0.037304 (rank : 64) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
AF10_MOUSE
|
||||||
| θ value | 0.813845 (rank : 33) | NC score | 0.026936 (rank : 83) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O54826 | Gene names | Mllt10, Af10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-10. | |||||
|
CD2L2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 34) | NC score | 0.006335 (rank : 123) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 35) | NC score | 0.069969 (rank : 27) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
SOX30_HUMAN
|
||||||
| θ value | 0.813845 (rank : 36) | NC score | 0.029676 (rank : 73) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O94993, O94995, Q8IYX6 | Gene names | SOX30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
ARI1B_HUMAN
|
||||||
| θ value | 1.06291 (rank : 37) | NC score | 0.035242 (rank : 68) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
EP15R_HUMAN
|
||||||
| θ value | 1.06291 (rank : 38) | NC score | 0.025048 (rank : 87) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1083 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UBC2 | Gene names | EPS15L1, EPS15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R). | |||||
|
NBR1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 39) | NC score | 0.040014 (rank : 62) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P97432, Q9JIH9 | Gene names | Nbr1, M17s2 | |||
|
Domain Architecture |

|
|||||
| Description | Next to BRCA1 gene 1 protein (Neighbor of BRCA1 gene 1 protein) (Membrane component, chromosome 17, surface marker 2). | |||||
|
TXLNB_HUMAN
|
||||||
| θ value | 1.06291 (rank : 40) | NC score | 0.024348 (rank : 90) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8N3L3, Q76L25, Q86T52, Q8N3S2 | Gene names | TXLNB, C6orf198, MDP77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77) (hMDP77). | |||||
|
AFF4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 41) | NC score | 0.045883 (rank : 52) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
CENPB_MOUSE
|
||||||
| θ value | 1.38821 (rank : 42) | NC score | 0.042086 (rank : 57) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P27790 | Gene names | Cenpb, Cenp-b | |||
|
Domain Architecture |

|
|||||
| Description | Major centromere autoantigen B (Centromere protein B) (CENP-B). | |||||
|
EP15R_MOUSE
|
||||||
| θ value | 1.38821 (rank : 43) | NC score | 0.026257 (rank : 85) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q60902, Q8CB60, Q8CB70, Q91WH8 | Gene names | Eps15l1, Eps15-rs, Eps15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R) (Epidermal growth factor receptor pathway substrate 15-related sequence) (Eps15-rs). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 44) | NC score | 0.056675 (rank : 40) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
SEPT9_MOUSE
|
||||||
| θ value | 1.38821 (rank : 45) | NC score | 0.022299 (rank : 94) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80UG5, Q3URP2, Q80TM7, Q9QYX9 | Gene names | Sept9, Kiaa0991, Sint1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (SL3-3 integration site 1 protein). | |||||
|
ATRIP_MOUSE
|
||||||
| θ value | 1.81305 (rank : 46) | NC score | 0.041060 (rank : 59) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BMG1 | Gene names | Atrip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATR-interacting protein (ATM and Rad3-related-interacting protein). | |||||
|
BI2L1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 47) | NC score | 0.029570 (rank : 75) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DBJ3 | Gene names | Baiap2l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-specific angiogenesis inhibitor 1-associated protein 2-like protein 1 (BAI1-associated protein 2-like protein 1). | |||||
|
CELR1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.006111 (rank : 124) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35161 | Gene names | Celsr1 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 1 precursor. | |||||
|
JIP4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 49) | NC score | 0.034557 (rank : 70) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 585 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O60271, O60905, Q3KQU8, Q3MKM7, Q86WC7, Q86WC8, Q8IZX7, Q96II0, Q9H811 | Gene names | SPAG9, HSS, KIAA0516, MAPK8IP4, SYD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 4 (JNK-interacting protein 4) (JIP-4) (JNK-associated leucine-zipper protein) (JLP) (Sperm-associated antigen 9) (Mitogen-activated protein kinase 8- interacting protein 4) (Human lung cancer protein 6) (HLC-6) (Proliferation-inducing protein 6) (Sperm-specific protein) (Sperm surface protein) (Protein highly expressed in testis) (PHET) (Sunday driver 1). | |||||
|
STIP1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 50) | NC score | 0.094079 (rank : 15) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P31948 | Gene names | STIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-induced-phosphoprotein 1 (STI1) (Hsc70/Hsp90-organizing protein) (Hop) (Transformation-sensitive protein IEF SSP 3521) (NY- REN-11 antigen). | |||||
|
STIP1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 51) | NC score | 0.096602 (rank : 14) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q60864, Q3TT16, Q8BPH3, Q99L66 | Gene names | Stip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stress-induced-phosphoprotein 1 (STI1) (Hsc70/Hsp90-organizing protein) (Hop) (mSTI1). | |||||
|
TTC28_MOUSE
|
||||||
| θ value | 1.81305 (rank : 52) | NC score | 0.052418 (rank : 46) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q80XJ3, Q6P9J6, Q80TL5, Q8BV10, Q8C0F2 | Gene names | Ttc28, Kiaa1043 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
ASPX_MOUSE
|
||||||
| θ value | 2.36792 (rank : 53) | NC score | 0.041289 (rank : 58) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P50289, Q9DAM6 | Gene names | Acrv1 | |||
|
Domain Architecture |

|
|||||
| Description | Acrosomal protein SP-10 precursor (Acrosomal vesicle protein 1) (MSA- 63). | |||||
|
CDC16_HUMAN
|
||||||
| θ value | 2.36792 (rank : 54) | NC score | 0.085726 (rank : 16) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13042, Q96AE6, Q9Y564 | Gene names | CDC16 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 16 homolog (CDC16Hs) (Anaphase-promoting complex subunit 6) (APC6) (Cyclosome subunit 6). | |||||
|
CDC16_MOUSE
|
||||||
| θ value | 2.36792 (rank : 55) | NC score | 0.084169 (rank : 18) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8R349, Q9CYX9 | Gene names | Cdc16 | |||
|
Domain Architecture |
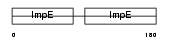
|
|||||
| Description | Cell division cycle protein 16 homolog (Anaphase-promoting complex subunit 6) (APC6) (Cyclosome subunit 6). | |||||
|
DAB2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 56) | NC score | 0.032122 (rank : 71) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P98078, Q3U3K1, Q91W56, Q923E1 | Gene names | Dab2, Doc2 | |||
|
Domain Architecture |
|
|||||
| Description | Disabled homolog 2 (DOC-2) (Mitogen-responsive phosphoprotein). | |||||
|
NOTC2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 57) | NC score | 0.008432 (rank : 121) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q04721, Q99734, Q9H240 | Gene names | NOTCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (hN2) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
SYNJ1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 58) | NC score | 0.056885 (rank : 39) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O43426, O43425, O94984 | Gene names | SYNJ1, KIAA0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 2.36792 (rank : 59) | NC score | 0.009841 (rank : 118) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
YAP1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 60) | NC score | 0.028675 (rank : 76) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P46937 | Gene names | YAP1, YAP65 | |||
|
Domain Architecture |

|
|||||
| Description | 65 kDa Yes-associated protein (YAP65). | |||||
|
JIP4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 61) | NC score | 0.034740 (rank : 69) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q58A65, Q3UH77, Q3UHF0, Q58VQ4, Q5NC70, Q5NC78, Q6A057, Q6PAS3, Q8CJC2 | Gene names | Spag9, Jip4, Jsap2, Kiaa0516, Mapk8ip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 4 (JNK-interacting protein 4) (JIP-4) (JNK-associated leucine-zipper protein) (JLP) (Sperm-associated antigen 9) (Mitogen-activated protein kinase 8- interacting protein 4) (JNK/SAPK-associated protein 2) (JSAP2). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 62) | NC score | 0.068526 (rank : 29) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MYO9B_MOUSE
|
||||||
| θ value | 3.0926 (rank : 63) | NC score | 0.006925 (rank : 122) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QY06, Q9QY07, Q9QY08, Q9QY09 | Gene names | Myo9b, Myr5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
NBEAL_HUMAN
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.019595 (rank : 98) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZS30, Q6Y876, Q6ZQY5, Q96Q30 | Gene names | NBEAL1, ALS2CR17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurobeachin-like 1 (Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 17 protein). | |||||
|
PTMS_HUMAN
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.058865 (rank : 37) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P20962 | Gene names | PTMS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
SRBS1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.023444 (rank : 91) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q62417, Q80TF8, Q8BZI3, Q8K3Y2, Q921F8, Q9Z0Z8, Q9Z0Z9 | Gene names | Sorbs1, Kiaa1296, Sh3d5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorbin and SH3 domain-containing protein 1 (Ponsin) (c-Cbl-associated protein) (CAP) (SH3 domain protein 5) (SH3P12). | |||||
|
SRCA_HUMAN
|
||||||
| θ value | 3.0926 (rank : 67) | NC score | 0.025090 (rank : 86) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
AN30A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.013119 (rank : 114) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1119 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BXX3, Q5W025 | Gene names | ANKRD30A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 30A (NY-BR-1 antigen). | |||||
|
LEUK_HUMAN
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.044227 (rank : 55) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P16150 | Gene names | SPN, CD43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukosialin precursor (Leukocyte sialoglycoprotein) (Sialophorin) (Galactoglycoprotein) (GALGP) (CD43 antigen). | |||||
|
MYST3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.039361 (rank : 63) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
PRGC2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.024873 (rank : 88) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86YN6, Q86YN3, Q86YN4, Q86YN5, Q8N1N9, Q8TDE4, Q8TDE5 | Gene names | PPARGC1B, PERC, PGC1, PGC1B, PPARGC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-beta (PPAR gamma coactivator-1beta) (PGC-1-related estrogen receptor alpha coactivator) (PPARGC-1-beta) (PGC-1-beta). | |||||
|
REPS1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.027167 (rank : 81) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O54916, Q8C9J9, Q99LR8 | Gene names | Reps1 | |||
|
Domain Architecture |

|
|||||
| Description | RalBP1-associated Eps domain-containing protein 1 (RalBP1-interacting protein 1). | |||||
|
RSF1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 73) | NC score | 0.036983 (rank : 67) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
TIF1A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 74) | NC score | 0.030121 (rank : 72) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O15164, O95854 | Gene names | TRIM24, RNF82, TIF1, TIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24) (RING finger protein 82). | |||||
|
UN45B_MOUSE
|
||||||
| θ value | 4.03905 (rank : 75) | NC score | 0.042832 (rank : 56) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CGY6, Q5XG72, Q8BHC5, Q8BWK3 | Gene names | Unc45b, Cmya4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog B (UNC-45B). | |||||
|
ZNF23_HUMAN
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | -0.000819 (rank : 126) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P17027, Q96IT3, Q9UG42 | Gene names | ZNF23, KOX16, ZNF359 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 23 (Zinc finger protein 359) (Zinc finger protein KOX16). | |||||
|
AFF4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.046914 (rank : 51) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
BSN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.024463 (rank : 89) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
COCA1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.022383 (rank : 93) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99715, Q99716 | Gene names | COL12A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XII) chain precursor. | |||||
|
CUTL2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.014344 (rank : 112) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 771 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P70298 | Gene names | Cutl2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
ELOA3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.026655 (rank : 84) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8NG57 | Gene names | TCEB3C, TCEB3L2 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II transcription factor SIII subunit A3 (Elongin-A3) (EloA3) (Transcription elongation factor B polypeptide 3C). | |||||
|
MYST3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.040159 (rank : 61) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
OTOAN_MOUSE
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.018957 (rank : 101) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K561 | Gene names | Otoa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Otoancorin precursor. | |||||
|
SYP2L_MOUSE
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | 0.018219 (rank : 104) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BWB1 | Gene names | Synpo2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin 2-like protein. | |||||
|
TAF11_HUMAN
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.027094 (rank : 82) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15544, Q9UHS0 | Gene names | TAF11, TAF2I | |||
|
Domain Architecture |
|
|||||
| Description | Transcription initiation factor TFIID subunit 11 (Transcription initiation factor TFIID 28 kDa subunit) (TAF(II)28) (TAFII-28) (TAFII28) (TFIID subunit p30-beta). | |||||
|
TEX14_HUMAN
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.012212 (rank : 116) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1103 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8IWB6, Q7RTP3, Q8ND97, Q9BXT9 | Gene names | TEX14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed protein 14 (Testis-expressed sequence 14). | |||||
|
ZBT16_HUMAN
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | -0.000057 (rank : 125) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1010 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q05516, Q8TAL4 | Gene names | ZBTB16, PLZF, ZNF145 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 16 (Zinc finger protein PLZF) (Promyelocytic leukemia zinc finger protein) (Zinc finger protein 145). | |||||
|
ZN750_MOUSE
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.055897 (rank : 41) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BH05, Q66JP3, Q8C0L1 | Gene names | Znf750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
BACH2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.008668 (rank : 120) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97303 | Gene names | Bach2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription regulator protein BACH2 (BTB and CNC homolog 2). | |||||
|
CIC_MOUSE
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.018789 (rank : 102) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
FYCO1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.009143 (rank : 119) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8VDC1, Q3V385, Q7TMT0, Q8BJN2 | Gene names | Fyco1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1. | |||||
|
NF2L2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.017744 (rank : 106) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q60795 | Gene names | Nfe2l2, Nrf-2, Nrf2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 2 (NF-E2-related factor 2) (NFE2-related factor 2) (Nuclear factor, erythroid derived 2, like 2). | |||||
|
NU153_HUMAN
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.027703 (rank : 78) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P49790 | Gene names | NUP153 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup153 (Nucleoporin Nup153) (153 kDa nucleoporin). | |||||
|
PAG1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.019590 (rank : 99) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NWQ8, Q2M1Z9, Q5BKU4, Q9NYK0 | Gene names | PAG1, CBP, PAG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphoprotein associated with glycosphingolipid-enriched microdomains 1 (Transmembrane adapter protein PAG) (Csk-binding protein) (Transmembrane phosphoprotein Cbp). | |||||
|
SNX1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.010580 (rank : 117) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WV80, Q9EQZ9 | Gene names | Snx1 | |||
|
Domain Architecture |
|
|||||
| Description | Sorting nexin-1. | |||||
|
SPAG1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.064492 (rank : 32) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q80ZX8, Q8CCK7, Q9QZJ3, Q9QZJ4 | Gene names | Spag1, Tpis | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 1 (Infertility-related sperm protein Spag-1) (TPR-containing protein involved in spermatogenesis) (TPIS). | |||||
|
SYN1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.020056 (rank : 97) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O88935, Q62279, Q8QZT8 | Gene names | Syn1, Syn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I). | |||||
|
TENR_MOUSE
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.016215 (rank : 108) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BYI9, O88717 | Gene names | Tnr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tenascin-R precursor (TN-R) (Restrictin) (Janusin) (Neural recognition molecule J1-160/180). | |||||
|
TRI66_HUMAN
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.027783 (rank : 77) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O15016, Q9BQQ4 | Gene names | TRIM66, KIAA0298 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
ZCH11_HUMAN
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.018482 (rank : 103) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5TAX3, Q12764, Q5TAX4, Q86XZ3 | Gene names | ZCCHC11, KIAA0191 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 11. | |||||
|
ZF106_MOUSE
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.018047 (rank : 105) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O88466, O55185, O88465, O88467, Q792M1, Q792P4, Q8CDZ8, Q8R3I4, Q9ESU3 | Gene names | Zfp106, H3a, Sh3bp3, Sirm, Znf474 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 106 (Zfp-106) (Zinc finger protein 474) (H3a minor histocompatibility antigen) (Son of insulin receptor mutant). | |||||
|
AP3B2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.015114 (rank : 111) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13367, O14808 | Gene names | AP3B2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (AP-3 complex beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain) (Neuron-specific vesicle coat protein beta-NAP). | |||||
|
CLSPN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.019258 (rank : 100) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80YR7, Q69GM2 | Gene names | Clspn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin. | |||||
|
EP400_MOUSE
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.012981 (rank : 115) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.029665 (rank : 74) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MUC13_MOUSE
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.027444 (rank : 79) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
NUPL2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.017154 (rank : 107) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CIC2, Q8BJX3, Q8BVP7, Q924S0 | Gene names | Nupl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin-like 2 (NLP-1). | |||||
|
PP1RA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.015651 (rank : 109) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96QC0, O00405 | Gene names | PPP1R10, CAT53, FB19, PNUTS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 1 regulatory subunit 10 (Phosphatase 1 nuclear targeting subunit) (MHC class I region proline- rich protein CAT53) (FB19 protein) (PP1-binding protein of 114 kDa) (p99). | |||||
|
SYN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.022485 (rank : 92) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P17600, O75825, Q5H9A9 | Gene names | SYN1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I) (Brain protein 4.1). | |||||
|
TENA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.013692 (rank : 113) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 631 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P24821, Q14583, Q15567 | Gene names | TNC, HXB | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin precursor (TN) (Tenascin-C) (TN-C) (Hexabrachion) (Cytotactin) (Neuronectin) (GMEM) (JI) (Myotendinous antigen) (Glioma- associated-extracellular matrix antigen) (GP 150-225). | |||||
|
TENR_HUMAN
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.015503 (rank : 110) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 526 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92752, Q15568, Q5R3G0 | Gene names | TNR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tenascin-R precursor (TN-R) (Restrictin) (Janusin). | |||||
|
VPS52_HUMAN
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.020397 (rank : 96) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N1B4, Q53GR4, Q5JPA0, Q5SQW1, Q8IUN6, Q9NPT5 | Gene names | VPS52, SACM2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associated protein 52 (SAC2 suppressor of actin mutations 2-like protein). | |||||
|
VPS52_MOUSE
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.020464 (rank : 95) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C754, Q6P4N7, Q8BQ15, Q9QWT6, Q9QWV2 | Gene names | Vps52 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associated protein 52. | |||||
|
BBS4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.058131 (rank : 38) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96RK4, Q8NHU9, Q96H45 | Gene names | BBS4 | |||
|
Domain Architecture |
|
|||||
| Description | Bardet-Biedl syndrome 4 protein. | |||||
|
BBS4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.054735 (rank : 43) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8C1Z7, Q3UYF0, Q8CA57 | Gene names | Bbs4 | |||
|
Domain Architecture |
|
|||||
| Description | Bardet-Biedl syndrome 4 protein homolog. | |||||
|
CDC27_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.065245 (rank : 31) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P30260, Q16349, Q96F35 | Gene names | CDC27 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 27 homolog (CDC27Hs) (H-NUC). | |||||
|
DMP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.052598 (rank : 45) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
IFT88_MOUSE
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.072905 (rank : 23) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q61371 | Gene names | Ift88, Tg737, Tg737Rpw, TgN737Rpw, Ttc10 | |||
|
Domain Architecture |

|
|||||
| Description | Intraflagellar transport 88 homolog (Tetratricopeptide repeat protein 10) (TPR repeat protein 10) (Recessive polycystic kidney disease protein Tg737) (TgN(Imorpk)737Rpw). | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.051041 (rank : 49) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
MAP4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.050887 (rank : 50) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
SGTA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.065264 (rank : 30) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O43765, Q9BTZ9 | Gene names | SGTA, SGT | |||
|
Domain Architecture |
|
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein A (Vpu-binding protein) (UBP). | |||||
|
SGTA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.063160 (rank : 33) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BJU0, Q8BGA6, Q99L52 | Gene names | Sgta, Sgt | |||
|
Domain Architecture |
|
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein A. | |||||
|
TOIP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.053347 (rank : 44) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BYU6, Q6NY10, Q8BJP0 | Gene names | Tor1aip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Torsin-1A-interacting protein 2. | |||||
|
TTC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.051423 (rank : 47) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99614, Q9BVT3 | Gene names | TTC1, TPR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
TTC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.051222 (rank : 48) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q91Z38, Q3TLM0 | Gene names | Ttc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
TTC6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.062705 (rank : 34) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q86TZ1, Q96CE6 | Gene names | TTC6 | |||
|
Domain Architecture |
|
|||||
| Description | Tetratricopeptide repeat protein 6 (TPR repeat protein 6). | |||||
|
TTC15_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 113 | |
| SwissProt Accessions | Q8K2L8, Q8BX27 | Gene names | Ttc15 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 15 (TPR repeat protein 15). | |||||
|
TTC15_HUMAN
|
||||||
| NC score | 0.920736 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8WVT3, Q8WVW1, Q9Y395 | Gene names | TTC15 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 15 (TPR repeat protein 15). | |||||
|
TTC13_HUMAN
|
||||||
| NC score | 0.136092 (rank : 3) | θ value | 0.163984 (rank : 17) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8NBP0, Q8IVP8, Q8NBI0, Q8ND20 | Gene names | TTC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 13 (TPR repeat protein 13). | |||||
|
SGTB_HUMAN
|
||||||
| NC score | 0.133719 (rank : 4) | θ value | 0.0252991 (rank : 5) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96EQ0 | Gene names | SGTB, SGT2 | |||
|
Domain Architecture |

|
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein B (Small glutamine-rich protein with tetratricopeptide repeats 2). | |||||
|
TTC8_HUMAN
|
||||||
| NC score | 0.131538 (rank : 5) | θ value | 0.163984 (rank : 18) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8TAM2, Q86SY0, Q86TV9, Q86U26, Q8NDH9, Q96DG8 | Gene names | TTC8, BBS8 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 8 (TPR repeat protein 8) (Bardet- Biedl syndrome 8 protein). | |||||
|
TTC8_MOUSE
|
||||||
| NC score | 0.131454 (rank : 6) | θ value | 0.279714 (rank : 23) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8VD72, Q9DCP7 | Gene names | Ttc8, Bbs8 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 8 (TPR repeat protein 8) (Bardet- Biedl syndrome 8 protein homolog). | |||||
|
OGT1_HUMAN
|
||||||
| NC score | 0.129426 (rank : 7) | θ value | 0.0961366 (rank : 13) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O15294, Q7Z3K0, Q8WWM8, Q96CC1, Q9UG57 | Gene names | OGT | |||
|
Domain Architecture |

|
|||||
| Description | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit (EC 2.4.1.-) (O-GlcNAc transferase p110 subunit). | |||||
|
SGTB_MOUSE
|
||||||
| NC score | 0.128069 (rank : 8) | θ value | 0.0563607 (rank : 11) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8VD33 | Gene names | Sgtb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein B. | |||||
|
CTR9_HUMAN
|
||||||
| NC score | 0.122719 (rank : 9) | θ value | 0.163984 (rank : 14) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6PD62, Q15015 | Gene names | CTR9, KIAA0155, SH2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1). | |||||
|
CTR9_MOUSE
|
||||||
| NC score | 0.121107 (rank : 10) | θ value | 0.163984 (rank : 15) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q62018, Q3UFF5, Q3UY40, Q66JX4, Q7TPS6, Q8BND9, Q8BRD1, Q8C9W7, Q8C9Y3, Q8CHI1 | Gene names | Ctr9, Kiaa0155, Sh2bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1) (Tetratricopeptide repeat-containing, SH2-binding phosphoprotein of 150 kDa) (TPR-containing, SH2-binding phosphoprotein of 150 kDa) (p150TSP). | |||||
|
IFT88_HUMAN
|
||||||
| NC score | 0.118565 (rank : 11) | θ value | 0.0563607 (rank : 10) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13099 | Gene names | IFT88, TG737, TTC10 | |||
|
Domain Architecture |

|
|||||
| Description | Intraflagellar transport 88 homolog (Tetratricopeptide repeat protein 10) (TPR repeat protein 10) (Recessive polycystic kidney disease protein Tg737 homolog). | |||||
|
DNJC7_MOUSE
|
||||||
| NC score | 0.098261 (rank : 12) | θ value | 0.0431538 (rank : 7) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9QYI3, Q3TKR1, Q8BPG3, Q8CIL2, Q9CT29, Q9D026 | Gene names | Dnajc7, Ttc2 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 7 (Tetratricopeptide repeat protein 2) (TPR repeat protein 2) (MDj11). | |||||
|
DNJC7_HUMAN
|
||||||
| NC score | 0.097932 (rank : 13) | θ value | 0.0563607 (rank : 9) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99615 | Gene names | DNAJC7, TPR2, TTC2 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 7 (Tetratricopeptide repeat protein 2) (TPR repeat protein 2). | |||||
|
STIP1_MOUSE
|
||||||
| NC score | 0.096602 (rank : 14) | θ value | 1.81305 (rank : 51) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q60864, Q3TT16, Q8BPH3, Q99L66 | Gene names | Stip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stress-induced-phosphoprotein 1 (STI1) (Hsc70/Hsp90-organizing protein) (Hop) (mSTI1). | |||||
|
STIP1_HUMAN
|
||||||
| NC score | 0.094079 (rank : 15) | θ value | 1.81305 (rank : 50) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P31948 | Gene names | STIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-induced-phosphoprotein 1 (STI1) (Hsc70/Hsp90-organizing protein) (Hop) (Transformation-sensitive protein IEF SSP 3521) (NY- REN-11 antigen). | |||||
|
CDC16_HUMAN
|
||||||
| NC score | 0.085726 (rank : 16) | θ value | 2.36792 (rank : 54) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13042, Q96AE6, Q9Y564 | Gene names | CDC16 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 16 homolog (CDC16Hs) (Anaphase-promoting complex subunit 6) (APC6) (Cyclosome subunit 6). | |||||
|
MAP4_HUMAN
|
||||||
| NC score | 0.084265 (rank : 17) | θ value | 0.00298849 (rank : 4) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
CDC16_MOUSE
|
||||||
| NC score | 0.084169 (rank : 18) | θ value | 2.36792 (rank : 55) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8R349, Q9CYX9 | Gene names | Cdc16 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 16 homolog (Anaphase-promoting complex subunit 6) (APC6) (Cyclosome subunit 6). | |||||
|
DMP1_MOUSE
|
||||||
| NC score | 0.081911 (rank : 19) | θ value | 0.0961366 (rank : 12) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
SPAG1_HUMAN
|
||||||
| NC score | 0.079433 (rank : 20) | θ value | 0.279714 (rank : 21) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q07617, Q7Z5G1 | Gene names | SPAG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 1 (Infertility-related sperm protein Spag-1) (HSD-3.8). | |||||
|
SYNJ1_MOUSE
|
||||||
| NC score | 0.076952 (rank : 21) | θ value | 0.00134147 (rank : 3) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.073452 (rank : 22) | θ value | 0.21417 (rank : 19) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
IFT88_MOUSE
|
||||||
| NC score | 0.072905 (rank : 23) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q61371 | Gene names | Ift88, Tg737, Tg737Rpw, TgN737Rpw, Ttc10 | |||
|
Domain Architecture |

|
|||||
| Description | Intraflagellar transport 88 homolog (Tetratricopeptide repeat protein 10) (TPR repeat protein 10) (Recessive polycystic kidney disease protein Tg737) (TgN(Imorpk)737Rpw). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.071697 (rank : 24) | θ value | 0.0330416 (rank : 6) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.071081 (rank : 25) | θ value | 0.62314 (rank : 31) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.071015 (rank : 26) | θ value | 0.0431538 (rank : 8) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.069969 (rank : 27) | θ value | 0.813845 (rank : 35) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.069860 (rank : 28) | θ value | 0.47712 (rank : 30) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.068526 (rank : 29) | θ value | 3.0926 (rank : 62) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
SGTA_HUMAN
|
||||||
| NC score | 0.065264 (rank : 30) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O43765, Q9BTZ9 | Gene names | SGTA, SGT | |||
|
Domain Architecture |

|
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein A (Vpu-binding protein) (UBP). | |||||
|
CDC27_HUMAN
|
||||||
| NC score | 0.065245 (rank : 31) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P30260, Q16349, Q96F35 | Gene names | CDC27 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 27 homolog (CDC27Hs) (H-NUC). | |||||
|
SPAG1_MOUSE
|
||||||
| NC score | 0.064492 (rank : 32) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q80ZX8, Q8CCK7, Q9QZJ3, Q9QZJ4 | Gene names | Spag1, Tpis | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 1 (Infertility-related sperm protein Spag-1) (TPR-containing protein involved in spermatogenesis) (TPIS). | |||||
|
SGTA_MOUSE
|
||||||
| NC score | 0.063160 (rank : 33) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BJU0, Q8BGA6, Q99L52 | Gene names | Sgta, Sgt | |||
|
Domain Architecture |

|
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein A. | |||||
|
TTC6_HUMAN
|
||||||
| NC score | 0.062705 (rank : 34) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q86TZ1, Q96CE6 | Gene names | TTC6 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 6 (TPR repeat protein 6). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.060339 (rank : 35) | θ value | 0.47712 (rank : 29) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.059364 (rank : 36) | θ value | 0.163984 (rank : 16) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
PTMS_HUMAN
|
||||||
| NC score | 0.058865 (rank : 37) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P20962 | Gene names | PTMS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
BBS4_HUMAN
|
||||||
| NC score | 0.058131 (rank : 38) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96RK4, Q8NHU9, Q96H45 | Gene names | BBS4 | |||
|
Domain Architecture |

|
|||||
| Description | Bardet-Biedl syndrome 4 protein. | |||||
|
SYNJ1_HUMAN
|
||||||
| NC score | 0.056885 (rank : 39) | θ value | 2.36792 (rank : 58) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O43426, O43425, O94984 | Gene names | SYNJ1, KIAA0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.056675 (rank : 40) | θ value | 1.38821 (rank : 44) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
ZN750_MOUSE
|
||||||
| NC score | 0.055897 (rank : 41) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BH05, Q66JP3, Q8C0L1 | Gene names | Znf750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
CYLC2_HUMAN
|
||||||
| NC score | 0.055042 (rank : 42) | θ value | 0.47712 (rank : 27) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
BBS4_MOUSE
|
||||||
| NC score | 0.054735 (rank : 43) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8C1Z7, Q3UYF0, Q8CA57 | Gene names | Bbs4 | |||
|
Domain Architecture |

|
|||||
| Description | Bardet-Biedl syndrome 4 protein homolog. | |||||
|
TOIP2_MOUSE
|
||||||
| NC score | 0.053347 (rank : 44) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BYU6, Q6NY10, Q8BJP0 | Gene names | Tor1aip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Torsin-1A-interacting protein 2. | |||||
|
DMP1_HUMAN
|
||||||
| NC score | 0.052598 (rank : 45) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
TTC28_MOUSE
|
||||||
| NC score | 0.052418 (rank : 46) | θ value | 1.81305 (rank : 52) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q80XJ3, Q6P9J6, Q80TL5, Q8BV10, Q8C0F2 | Gene names | Ttc28, Kiaa1043 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
TTC1_HUMAN
|
||||||
| NC score | 0.051423 (rank : 47) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99614, Q9BVT3 | Gene names | TTC1, TPR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
TTC1_MOUSE
|
||||||
| NC score | 0.051222 (rank : 48) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q91Z38, Q3TLM0 | Gene names | Ttc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.051041 (rank : 49) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
MAP4_MOUSE
|
||||||
| NC score | 0.050887 (rank : 50) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
AFF4_MOUSE
|
||||||
| NC score | 0.046914 (rank : 51) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
AFF4_HUMAN
|
||||||
| NC score | 0.045883 (rank : 52) | θ value | 1.38821 (rank : 41) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
CT077_HUMAN
|
||||||
| NC score | 0.045140 (rank : 53) | θ value | 0.47712 (rank : 26) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NQG5 | Gene names | C20orf77 | |||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf77. | |||||
|
MYST4_MOUSE
|
||||||
| NC score | 0.044239 (rank : 54) | θ value | 0.365318 (rank : 25) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
LEUK_HUMAN
|
||||||
| NC score | 0.044227 (rank : 55) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P16150 | Gene names | SPN, CD43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukosialin precursor (Leukocyte sialoglycoprotein) (Sialophorin) (Galactoglycoprotein) (GALGP) (CD43 antigen). | |||||
|
UN45B_MOUSE
|
||||||
| NC score | 0.042832 (rank : 56) | θ value | 4.03905 (rank : 75) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CGY6, Q5XG72, Q8BHC5, Q8BWK3 | Gene names | Unc45b, Cmya4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog B (UNC-45B). | |||||
|
CENPB_MOUSE
|
||||||
| NC score | 0.042086 (rank : 57) | θ value | 1.38821 (rank : 42) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P27790 | Gene names | Cenpb, Cenp-b | |||
|
Domain Architecture |

|
|||||
| Description | Major centromere autoantigen B (Centromere protein B) (CENP-B). | |||||
|
ASPX_MOUSE
|
||||||
| NC score | 0.041289 (rank : 58) | θ value | 2.36792 (rank : 53) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P50289, Q9DAM6 | Gene names | Acrv1 | |||
|
Domain Architecture |

|
|||||
| Description | Acrosomal protein SP-10 precursor (Acrosomal vesicle protein 1) (MSA- 63). | |||||
|
ATRIP_MOUSE
|
||||||
| NC score | 0.041060 (rank : 59) | θ value | 1.81305 (rank : 46) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BMG1 | Gene names | Atrip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATR-interacting protein (ATM and Rad3-related-interacting protein). | |||||
|
RTN4_MOUSE
|
||||||
| NC score | 0.041052 (rank : 60) | θ value | 0.47712 (rank : 28) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99P72, Q5DTK9, Q7TNB7, Q80W95, Q8BGK7, Q8BGM9, Q8K290, Q8K3G8, Q9CTE3 | Gene names | Rtn4, Kiaa0886, Nogo | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-4 (Neurite outgrowth inhibitor) (Nogo protein). | |||||
|
MYST3_MOUSE
|
||||||
| NC score | 0.040159 (rank : 61) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
NBR1_MOUSE
|
||||||
| NC score | 0.040014 (rank : 62) | θ value | 1.06291 (rank : 39) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P97432, Q9JIH9 | Gene names | Nbr1, M17s2 | |||
|
Domain Architecture |

|
|||||
| Description | Next to BRCA1 gene 1 protein (Neighbor of BRCA1 gene 1 protein) (Membrane component, chromosome 17, surface marker 2). | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.039361 (rank : 63) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.037304 (rank : 64) | θ value | 0.62314 (rank : 32) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
TIF1A_MOUSE
|
||||||
| NC score | 0.037169 (rank : 65) | θ value | 0.279714 (rank : 22) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q64127, Q64126 | Gene names | Trim24, Tif1, Tif1a | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24). | |||||
|
ATCAY_HUMAN
|
||||||
| NC score | 0.037012 (rank : 66) | θ value | 0.365318 (rank : 24) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86WG3, Q8NAQ2, Q8TAQ3, Q96HC6, Q96JF5 | Gene names | ATCAY, KIAA1872 | |||
|
Domain Architecture |

|
|||||
| Description | Caytaxin (Ataxia Cayman type protein) (BNIP-H). | |||||
|
RSF1_HUMAN
|
||||||
| NC score | 0.036983 (rank : 67) | θ value | 4.03905 (rank : 73) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
ARI1B_HUMAN
|
||||||
| NC score | 0.035242 (rank : 68) | θ value | 1.06291 (rank : 37) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
JIP4_MOUSE
|
||||||
| NC score | 0.034740 (rank : 69) | θ value | 3.0926 (rank : 61) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q58A65, Q3UH77, Q3UHF0, Q58VQ4, Q5NC70, Q5NC78, Q6A057, Q6PAS3, Q8CJC2 | Gene names | Spag9, Jip4, Jsap2, Kiaa0516, Mapk8ip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 4 (JNK-interacting protein 4) (JIP-4) (JNK-associated leucine-zipper protein) (JLP) (Sperm-associated antigen 9) (Mitogen-activated protein kinase 8- interacting protein 4) (JNK/SAPK-associated protein 2) (JSAP2). | |||||
|
JIP4_HUMAN
|
||||||
| NC score | 0.034557 (rank : 70) | θ value | 1.81305 (rank : 49) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 585 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O60271, O60905, Q3KQU8, Q3MKM7, Q86WC7, Q86WC8, Q8IZX7, Q96II0, Q9H811 | Gene names | SPAG9, HSS, KIAA0516, MAPK8IP4, SYD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 4 (JNK-interacting protein 4) (JIP-4) (JNK-associated leucine-zipper protein) (JLP) (Sperm-associated antigen 9) (Mitogen-activated protein kinase 8- interacting protein 4) (Human lung cancer protein 6) (HLC-6) (Proliferation-inducing protein 6) (Sperm-specific protein) (Sperm surface protein) (Protein highly expressed in testis) (PHET) (Sunday driver 1). | |||||
|
DAB2_MOUSE
|
||||||
| NC score | 0.032122 (rank : 71) | θ value | 2.36792 (rank : 56) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P98078, Q3U3K1, Q91W56, Q923E1 | Gene names | Dab2, Doc2 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 2 (DOC-2) (Mitogen-responsive phosphoprotein). | |||||
|
TIF1A_HUMAN
|
||||||
| NC score | 0.030121 (rank : 72) | θ value | 4.03905 (rank : 74) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O15164, O95854 | Gene names | TRIM24, RNF82, TIF1, TIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24) (RING finger protein 82). | |||||
|
SOX30_HUMAN
|
||||||
| NC score | 0.029676 (rank : 73) | θ value | 0.813845 (rank : 36) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O94993, O94995, Q8IYX6 | Gene names | SOX30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.029665 (rank : 74) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
BI2L1_MOUSE
|
||||||
| NC score | 0.029570 (rank : 75) | θ value | 1.81305 (rank : 47) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DBJ3 | Gene names | Baiap2l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-specific angiogenesis inhibitor 1-associated protein 2-like protein 1 (BAI1-associated protein 2-like protein 1). | |||||
|
YAP1_HUMAN
|
||||||
| NC score | 0.028675 (rank : 76) | θ value | 2.36792 (rank : 60) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P46937 | Gene names | YAP1, YAP65 | |||
|
Domain Architecture |

|
|||||
| Description | 65 kDa Yes-associated protein (YAP65). | |||||
|
TRI66_HUMAN
|
||||||
| NC score | 0.027783 (rank : 77) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O15016, Q9BQQ4 | Gene names | TRIM66, KIAA0298 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
NU153_HUMAN
|
||||||
| NC score | 0.027703 (rank : 78) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P49790 | Gene names | NUP153 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup153 (Nucleoporin Nup153) (153 kDa nucleoporin). | |||||
|
MUC13_MOUSE
|
||||||
| NC score | 0.027444 (rank : 79) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
COCA1_MOUSE
|
||||||
| NC score | 0.027225 (rank : 80) | θ value | 0.279714 (rank : 20) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q60847, P70322 | Gene names | Col12a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XII) chain precursor. | |||||
|
REPS1_MOUSE
|
||||||
| NC score | 0.027167 (rank : 81) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O54916, Q8C9J9, Q99LR8 | Gene names | Reps1 | |||
|
Domain Architecture |

|
|||||
| Description | RalBP1-associated Eps domain-containing protein 1 (RalBP1-interacting protein 1). | |||||
|
TAF11_HUMAN
|
||||||
| NC score | 0.027094 (rank : 82) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15544, Q9UHS0 | Gene names | TAF11, TAF2I | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 11 (Transcription initiation factor TFIID 28 kDa subunit) (TAF(II)28) (TAFII-28) (TAFII28) (TFIID subunit p30-beta). | |||||
|
AF10_MOUSE
|
||||||
| NC score | 0.026936 (rank : 83) | θ value | 0.813845 (rank : 33) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O54826 | Gene names | Mllt10, Af10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-10. | |||||
|
ELOA3_HUMAN
|
||||||
| NC score | 0.026655 (rank : 84) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8NG57 | Gene names | TCEB3C, TCEB3L2 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II transcription factor SIII subunit A3 (Elongin-A3) (EloA3) (Transcription elongation factor B polypeptide 3C). | |||||
|
EP15R_MOUSE
|
||||||
| NC score | 0.026257 (rank : 85) | θ value | 1.38821 (rank : 43) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q60902, Q8CB60, Q8CB70, Q91WH8 | Gene names | Eps15l1, Eps15-rs, Eps15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R) (Epidermal growth factor receptor pathway substrate 15-related sequence) (Eps15-rs). | |||||
|
SRCA_HUMAN
|
||||||
| NC score | 0.025090 (rank : 86) | θ value | 3.0926 (rank : 67) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
EP15R_HUMAN
|
||||||
| NC score | 0.025048 (rank : 87) | θ value | 1.06291 (rank : 38) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1083 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UBC2 | Gene names | EPS15L1, EPS15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R). | |||||
|
PRGC2_HUMAN
|
||||||
| NC score | 0.024873 (rank : 88) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86YN6, Q86YN3, Q86YN4, Q86YN5, Q8N1N9, Q8TDE4, Q8TDE5 | Gene names | PPARGC1B, PERC, PGC1, PGC1B, PPARGC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-beta (PPAR gamma coactivator-1beta) (PGC-1-related estrogen receptor alpha coactivator) (PPARGC-1-beta) (PGC-1-beta). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.024463 (rank : 89) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
TXLNB_HUMAN
|
||||||
| NC score | 0.024348 (rank : 90) | θ value | 1.06291 (rank : 40) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8N3L3, Q76L25, Q86T52, Q8N3S2 | Gene names | TXLNB, C6orf198, MDP77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77) (hMDP77). | |||||
|
SRBS1_MOUSE
|
||||||
| NC score | 0.023444 (rank : 91) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q62417, Q80TF8, Q8BZI3, Q8K3Y2, Q921F8, Q9Z0Z8, Q9Z0Z9 | Gene names | Sorbs1, Kiaa1296, Sh3d5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorbin and SH3 domain-containing protein 1 (Ponsin) (c-Cbl-associated protein) (CAP) (SH3 domain protein 5) (SH3P12). | |||||
|
SYN1_HUMAN
|
||||||
| NC score | 0.022485 (rank : 92) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P17600, O75825, Q5H9A9 | Gene names | SYN1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I) (Brain protein 4.1). | |||||
|
COCA1_HUMAN
|
||||||
| NC score | 0.022383 (rank : 93) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99715, Q99716 | Gene names | COL12A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XII) chain precursor. | |||||
|
SEPT9_MOUSE
|
||||||
| NC score | 0.022299 (rank : 94) | θ value | 1.38821 (rank : 45) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80UG5, Q3URP2, Q80TM7, Q9QYX9 | Gene names | Sept9, Kiaa0991, Sint1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (SL3-3 integration site 1 protein). | |||||
|
VPS52_MOUSE
|
||||||
| NC score | 0.020464 (rank : 95) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C754, Q6P4N7, Q8BQ15, Q9QWT6, Q9QWV2 | Gene names | Vps52 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associated protein 52. | |||||
|
VPS52_HUMAN
|
||||||
| NC score | 0.020397 (rank : 96) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N1B4, Q53GR4, Q5JPA0, Q5SQW1, Q8IUN6, Q9NPT5 | Gene names | VPS52, SACM2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associated protein 52 (SAC2 suppressor of actin mutations 2-like protein). | |||||
|
SYN1_MOUSE
|
||||||
| NC score | 0.020056 (rank : 97) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O88935, Q62279, Q8QZT8 | Gene names | Syn1, Syn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I). | |||||
|
NBEAL_HUMAN
|
||||||
| NC score | 0.019595 (rank : 98) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZS30, Q6Y876, Q6ZQY5, Q96Q30 | Gene names | NBEAL1, ALS2CR17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurobeachin-like 1 (Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 17 protein). | |||||
|
PAG1_HUMAN
|
||||||
| NC score | 0.019590 (rank : 99) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NWQ8, Q2M1Z9, Q5BKU4, Q9NYK0 | Gene names | PAG1, CBP, PAG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphoprotein associated with glycosphingolipid-enriched microdomains 1 (Transmembrane adapter protein PAG) (Csk-binding protein) (Transmembrane phosphoprotein Cbp). | |||||
|
CLSPN_MOUSE
|
||||||
| NC score | 0.019258 (rank : 100) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80YR7, Q69GM2 | Gene names | Clspn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin. | |||||
|
OTOAN_MOUSE
|
||||||
| NC score | 0.018957 (rank : 101) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K561 | Gene names | Otoa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Otoancorin precursor. | |||||
|
CIC_MOUSE
|
||||||
| NC score | 0.018789 (rank : 102) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
ZCH11_HUMAN
|
||||||
| NC score | 0.018482 (rank : 103) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5TAX3, Q12764, Q5TAX4, Q86XZ3 | Gene names | ZCCHC11, KIAA0191 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 11. | |||||
|
SYP2L_MOUSE
|
||||||
| NC score | 0.018219 (rank : 104) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BWB1 | Gene names | Synpo2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin 2-like protein. | |||||
|
ZF106_MOUSE
|
||||||
| NC score | 0.018047 (rank : 105) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O88466, O55185, O88465, O88467, Q792M1, Q792P4, Q8CDZ8, Q8R3I4, Q9ESU3 | Gene names | Zfp106, H3a, Sh3bp3, Sirm, Znf474 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 106 (Zfp-106) (Zinc finger protein 474) (H3a minor histocompatibility antigen) (Son of insulin receptor mutant). | |||||
|
NF2L2_MOUSE
|
||||||
| NC score | 0.017744 (rank : 106) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q60795 | Gene names | Nfe2l2, Nrf-2, Nrf2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 2 (NF-E2-related factor 2) (NFE2-related factor 2) (Nuclear factor, erythroid derived 2, like 2). | |||||
|
NUPL2_MOUSE
|
||||||
| NC score | 0.017154 (rank : 107) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CIC2, Q8BJX3, Q8BVP7, Q924S0 | Gene names | Nupl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin-like 2 (NLP-1). | |||||
|
TENR_MOUSE
|
||||||
| NC score | 0.016215 (rank : 108) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BYI9, O88717 | Gene names | Tnr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tenascin-R precursor (TN-R) (Restrictin) (Janusin) (Neural recognition molecule J1-160/180). | |||||
|
PP1RA_HUMAN
|
||||||
| NC score | 0.015651 (rank : 109) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96QC0, O00405 | Gene names | PPP1R10, CAT53, FB19, PNUTS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 1 regulatory subunit 10 (Phosphatase 1 nuclear targeting subunit) (MHC class I region proline- rich protein CAT53) (FB19 protein) (PP1-binding protein of 114 kDa) (p99). | |||||
|
TENR_HUMAN
|
||||||
| NC score | 0.015503 (rank : 110) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 526 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92752, Q15568, Q5R3G0 | Gene names | TNR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tenascin-R precursor (TN-R) (Restrictin) (Janusin). | |||||
|
AP3B2_HUMAN
|
||||||
| NC score | 0.015114 (rank : 111) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13367, O14808 | Gene names | AP3B2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (AP-3 complex beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain) (Neuron-specific vesicle coat protein beta-NAP). | |||||
|
CUTL2_MOUSE
|
||||||
| NC score | 0.014344 (rank : 112) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 771 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P70298 | Gene names | Cutl2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
TENA_HUMAN
|
||||||
| NC score | 0.013692 (rank : 113) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 631 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P24821, Q14583, Q15567 | Gene names | TNC, HXB | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin precursor (TN) (Tenascin-C) (TN-C) (Hexabrachion) (Cytotactin) (Neuronectin) (GMEM) (JI) (Myotendinous antigen) (Glioma- associated-extracellular matrix antigen) (GP 150-225). | |||||
|
AN30A_HUMAN
|
||||||
| NC score | 0.013119 (rank : 114) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1119 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BXX3, Q5W025 | Gene names | ANKRD30A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 30A (NY-BR-1 antigen). | |||||
|
EP400_MOUSE
|
||||||
| NC score | 0.012981 (rank : 115) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
TEX14_HUMAN
|
||||||
| NC score | 0.012212 (rank : 116) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1103 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8IWB6, Q7RTP3, Q8ND97, Q9BXT9 | Gene names | TEX14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed protein 14 (Testis-expressed sequence 14). | |||||
|
SNX1_MOUSE
|
||||||
| NC score | 0.010580 (rank : 117) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WV80, Q9EQZ9 | Gene names | Snx1 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-1. | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.009841 (rank : 118) | θ value | 2.36792 (rank : 59) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
FYCO1_MOUSE
|
||||||
| NC score | 0.009143 (rank : 119) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8VDC1, Q3V385, Q7TMT0, Q8BJN2 | Gene names | Fyco1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1. | |||||
|
BACH2_MOUSE
|
||||||
| NC score | 0.008668 (rank : 120) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97303 | Gene names | Bach2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription regulator protein BACH2 (BTB and CNC homolog 2). | |||||
|
NOTC2_HUMAN
|
||||||
| NC score | 0.008432 (rank : 121) | θ value | 2.36792 (rank : 57) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q04721, Q99734, Q9H240 | Gene names | NOTCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (hN2) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
MYO9B_MOUSE
|
||||||
| NC score | 0.006925 (rank : 122) | θ value | 3.0926 (rank : 63) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QY06, Q9QY07, Q9QY08, Q9QY09 | Gene names | Myo9b, Myr5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
CD2L2_HUMAN
|
||||||
| NC score | 0.006335 (rank : 123) | θ value | 0.813845 (rank : 34) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
CELR1_MOUSE
|
||||||
| NC score | 0.006111 (rank : 124) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35161 | Gene names | Celsr1 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 1 precursor. | |||||
|
ZBT16_HUMAN
|
||||||
| NC score | -0.000057 (rank : 125) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1010 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q05516, Q8TAL4 | Gene names | ZBTB16, PLZF, ZNF145 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 16 (Zinc finger protein PLZF) (Promyelocytic leukemia zinc finger protein) (Zinc finger protein 145). | |||||
|
ZNF23_HUMAN
|
||||||
| NC score | -0.000819 (rank : 126) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P17027, Q96IT3, Q9UG42 | Gene names | ZNF23, KOX16, ZNF359 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 23 (Zinc finger protein 359) (Zinc finger protein KOX16). | |||||