Please be patient as the page loads
|
TOIP2_MOUSE
|
||||||
| SwissProt Accessions | Q8BYU6, Q6NY10, Q8BJP0 | Gene names | Tor1aip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Torsin-1A-interacting protein 2. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TOIP2_MOUSE
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 115 | |
| SwissProt Accessions | Q8BYU6, Q6NY10, Q8BJP0 | Gene names | Tor1aip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Torsin-1A-interacting protein 2. | |||||
|
TOIP2_HUMAN
|
||||||
| θ value | 6.80647e-157 (rank : 2) | NC score | 0.926659 (rank : 2) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8NFQ8 | Gene names | TOR1AIP2, LULL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Torsin-1A-interacting protein 2 (Lumenal domain-like LAP1). | |||||
|
TOIP1_HUMAN
|
||||||
| θ value | 1.83746e-69 (rank : 3) | NC score | 0.859428 (rank : 3) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5JTV8, Q5JTV6, Q8IZ65, Q9H8Y6, Q9HAJ1, Q9NV52, Q9Y3X5 | Gene names | TOR1AIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Torsin-1A-interacting protein 1. | |||||
|
TOIP1_MOUSE
|
||||||
| θ value | 1.04338e-64 (rank : 4) | NC score | 0.856538 (rank : 4) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q921T2, Q3U7A4 | Gene names | Tor1aip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Torsin-1A-interacting protein 1 (Lamina-associated polypeptide 1B). | |||||
|
DSPP_HUMAN
|
||||||
| θ value | 2.88788e-06 (rank : 5) | NC score | 0.182342 (rank : 6) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
DSPP_MOUSE
|
||||||
| θ value | 3.77169e-06 (rank : 6) | NC score | 0.195740 (rank : 5) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 7) | NC score | 0.115254 (rank : 13) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 8) | NC score | 0.022719 (rank : 95) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 9) | NC score | 0.079954 (rank : 25) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
MDN1_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 10) | NC score | 0.138152 (rank : 9) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
GNAS1_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 11) | NC score | 0.038205 (rank : 75) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6R0H7, Q6R0H4, Q6R0H5, Q6R2J5, Q9JJX0, Q9Z1N8 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
SPRL1_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 12) | NC score | 0.102552 (rank : 17) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P70663, P97810, Q99L82 | Gene names | Sparcl1, Ecm2, Sc1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (Matrix glycoprotein Sc1) (Extracellular matrix protein 2). | |||||
|
CGRE1_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 13) | NC score | 0.111125 (rank : 14) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q99674 | Gene names | CGREF1, CGR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
TTC15_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 14) | NC score | 0.089341 (rank : 23) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8WVT3, Q8WVW1, Q9Y395 | Gene names | TTC15 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 15 (TPR repeat protein 15). | |||||
|
AMPH_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 15) | NC score | 0.069517 (rank : 31) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7TQF7, Q8R1C4 | Gene names | Amph, Amph1 | |||
|
Domain Architecture |

|
|||||
| Description | Amphiphysin. | |||||
|
FILA_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 16) | NC score | 0.117378 (rank : 12) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
SRCA_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 17) | NC score | 0.095909 (rank : 20) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q7TQ48, Q8BS43 | Gene names | Srl, Sar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
NSBP1_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 18) | NC score | 0.125790 (rank : 11) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 19) | NC score | 0.137076 (rank : 10) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
DMP1_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 20) | NC score | 0.160238 (rank : 7) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
NFM_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 21) | NC score | 0.028441 (rank : 88) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
NCKX1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 22) | NC score | 0.072564 (rank : 28) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O60721, O43485, O75184 | Gene names | SLC24A1, KIAA0702, NCKX1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium/calcium exchanger 1 (Na(+)/K(+)/Ca(2+)-exchange protein 1) (Retinal rod Na-Ca+K exchanger). | |||||
|
DNM3A_HUMAN
|
||||||
| θ value | 0.163984 (rank : 23) | NC score | 0.055370 (rank : 51) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y6K1, Q8WXU9 | Gene names | DNMT3A | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 3A (EC 2.1.1.37) (Dnmt3a) (DNA methyltransferase HsaIIIA) (DNA MTase HsaIIIA) (M.HsaIIIA). | |||||
|
FBLN2_MOUSE
|
||||||
| θ value | 0.163984 (rank : 24) | NC score | 0.015600 (rank : 112) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P37889, Q9WUI2 | Gene names | Fbln2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-2 precursor. | |||||
|
SRCA_HUMAN
|
||||||
| θ value | 0.163984 (rank : 25) | NC score | 0.110200 (rank : 15) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
DNM3A_MOUSE
|
||||||
| θ value | 0.279714 (rank : 26) | NC score | 0.052529 (rank : 57) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O88508, Q3UH24, Q8CJ60, Q922J0, Q9CSE1 | Gene names | Dnmt3a | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 3A (EC 2.1.1.37) (Dnmt3a) (DNA methyltransferase MmuIIIA) (DNA MTase MmuIIIA) (M.MmuIIIA). | |||||
|
CAC1B_MOUSE
|
||||||
| θ value | 0.365318 (rank : 27) | NC score | 0.025765 (rank : 93) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
DDX21_MOUSE
|
||||||
| θ value | 0.365318 (rank : 28) | NC score | 0.021399 (rank : 100) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JIK5, Q9WV45 | Gene names | Ddx21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DMP1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 29) | NC score | 0.157932 (rank : 8) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
DOK7_MOUSE
|
||||||
| θ value | 0.365318 (rank : 30) | NC score | 0.047896 (rank : 65) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q18PE0, Q3TCZ6, Q5FW70, Q8C8U7 | Gene names | Dok7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Dok-7 (Downstream of tyrosine kinase 7). | |||||
|
ZBT17_HUMAN
|
||||||
| θ value | 0.365318 (rank : 31) | NC score | 0.005833 (rank : 130) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 925 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13105, Q15932, Q5JYB2, Q9NUC9 | Gene names | ZBTB17, MIZ1, ZNF151 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 17 (Zinc finger protein 151) (Myc-interacting zinc finger protein) (Miz-1 protein). | |||||
|
CLIC6_HUMAN
|
||||||
| θ value | 0.47712 (rank : 32) | NC score | 0.072419 (rank : 29) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96NY7, Q8IX31 | Gene names | CLIC6, CLIC1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
IF4G1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 33) | NC score | 0.038809 (rank : 74) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
PGCA_MOUSE
|
||||||
| θ value | 0.47712 (rank : 34) | NC score | 0.037580 (rank : 76) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
BAZ2B_HUMAN
|
||||||
| θ value | 0.62314 (rank : 35) | NC score | 0.048680 (rank : 62) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
LPIN3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 36) | NC score | 0.029341 (rank : 85) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BQK8, Q5TDB9, Q9NPY8, Q9UJE5 | Gene names | LPIN3, LIPN3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipin-3 (Lipin 3-like). | |||||
|
ZBT17_MOUSE
|
||||||
| θ value | 0.62314 (rank : 37) | NC score | 0.006257 (rank : 128) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q60821, Q60699 | Gene names | Zbtb17, Zfp100, Znf151 | |||
|
Domain Architecture |
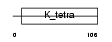
|
|||||
| Description | Zinc finger and BTB domain-containing protein 17 (Zinc finger protein 151) (Zinc finger protein 100) (Zfp-100) (Polyomavirus late initiator promoter-binding protein) (LP-1) (Zinc finger protein Z13). | |||||
|
CAC1H_MOUSE
|
||||||
| θ value | 0.813845 (rank : 38) | NC score | 0.028880 (rank : 87) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88427, Q80TJ2, Q9JKU5 | Gene names | Cacna1h, Kiaa1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2). | |||||
|
CHD9_HUMAN
|
||||||
| θ value | 0.813845 (rank : 39) | NC score | 0.020722 (rank : 104) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
NC6IP_MOUSE
|
||||||
| θ value | 0.813845 (rank : 40) | NC score | 0.069957 (rank : 30) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q923W1, Q6DI60, Q6PEA7, Q8R0W9 | Gene names | Ncoa6ip, Pimt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA methyltransferase NCOA6IP (EC 2.1.1.-) (Nuclear receptor coactivator 6-interacting protein) (PRIP-interacting protein with methyltransferase motif) (PIPMT) (PIMT). | |||||
|
PDXL2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 41) | NC score | 0.049057 (rank : 61) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8CAE9, Q8CFW3 | Gene names | Podxl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Podocalyxin-like protein 2 precursor (Endoglycan). | |||||
|
PININ_HUMAN
|
||||||
| θ value | 0.813845 (rank : 42) | NC score | 0.043745 (rank : 69) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9H307, O60899, Q53EM7, Q6P5X4, Q7KYL1, Q99738, Q9UHZ9, Q9UQR9 | Gene names | PNN, DRS, MEMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin (140 kDa nuclear and cell adhesion-related phosphoprotein) (Domain-rich serine protein) (DRS-protein) (DRSP) (Melanoma metastasis clone A protein) (Desmosome-associated protein) (SR-like protein) (Nuclear protein SDK3). | |||||
|
SRCH_HUMAN
|
||||||
| θ value | 0.813845 (rank : 43) | NC score | 0.089787 (rank : 22) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P23327, Q504Y6 | Gene names | HRC, HCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcoplasmic reticulum histidine-rich calcium-binding protein precursor. | |||||
|
X3CL1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 44) | NC score | 0.042153 (rank : 71) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O35188, O35933 | Gene names | Cx3cl1, Cx3c, Fkn, Scyd1 | |||
|
Domain Architecture |
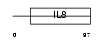
|
|||||
| Description | Fractalkine precursor (CX3CL1) (Neurotactin) (CX3C membrane-anchored chemokine) (Small inducible cytokine D1). | |||||
|
CBP_MOUSE
|
||||||
| θ value | 1.06291 (rank : 45) | NC score | 0.038823 (rank : 73) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CLIC6_MOUSE
|
||||||
| θ value | 1.06291 (rank : 46) | NC score | 0.041158 (rank : 72) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BHB9 | Gene names | Clic6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
EDD1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 47) | NC score | 0.027227 (rank : 90) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80TP3, Q698K9, Q6PEQ8, Q6PFQ9, Q80VL4, Q810V6, Q9CXE9 | Gene names | Edd1, Edd, Kiaa0896 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog). | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 1.06291 (rank : 48) | NC score | 0.073336 (rank : 27) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
ATOH1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 49) | NC score | 0.022405 (rank : 97) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P48985 | Gene names | Atoh1, Ath1 | |||
|
Domain Architecture |

|
|||||
| Description | Atonal protein homolog 1 (Helix-loop-helix protein mATH-1) (MATH1). | |||||
|
GTF2I_MOUSE
|
||||||
| θ value | 1.38821 (rank : 50) | NC score | 0.030196 (rank : 84) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9ESZ8, O54700, O55030, O55031, Q8VHD1, Q8VHD2, Q8VHD3, Q8VHD4, Q9CSB5, Q9D9K9 | Gene names | Gtf2i, Bap135, Diws1t | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor II-I (GTFII-I) (TFII-I) (Bruton tyrosine kinase-associated protein 135) (BTK-associated protein 135) (BAP-135). | |||||
|
HORN_HUMAN
|
||||||
| θ value | 1.38821 (rank : 51) | NC score | 0.063177 (rank : 38) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q86YZ3 | Gene names | HRNR, S100A18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hornerin. | |||||
|
LRRF1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 52) | NC score | 0.090006 (rank : 21) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q3UZ39, O70323, Q3TS94 | Gene names | Lrrfip1, Flap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (FLI-LRR-associated protein 1) (Flap-1) (H186 FLAP). | |||||
|
PAK4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 53) | NC score | 0.007272 (rank : 127) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1098 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BTW9, Q6ZPX0, Q80Z97, Q9CS71 | Gene names | Pak4, Kiaa1142 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PAK 4 (EC 2.7.11.1) (p21-activated kinase 4) (PAK-4). | |||||
|
CAC1H_HUMAN
|
||||||
| θ value | 1.81305 (rank : 54) | NC score | 0.026737 (rank : 92) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
CMGA_MOUSE
|
||||||
| θ value | 1.81305 (rank : 55) | NC score | 0.051619 (rank : 58) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P26339 | Gene names | Chga | |||
|
Domain Architecture |

|
|||||
| Description | Chromogranin A precursor (CgA) [Contains: Pancreastatin; Beta-granin; WE-14]. | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 56) | NC score | 0.109439 (rank : 16) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
NPAS3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 57) | NC score | 0.018445 (rank : 108) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9QZQ0, Q9EQP4 | Gene names | Npas3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
PERQ1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 58) | NC score | 0.048525 (rank : 63) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99MR1 | Gene names | Perq1 | |||
|
Domain Architecture |

|
|||||
| Description | PERQ amino acid rich with GYF domain protein 1. | |||||
|
SAM14_HUMAN
|
||||||
| θ value | 1.81305 (rank : 59) | NC score | 0.034362 (rank : 79) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IZD0, Q8N2X0 | Gene names | SAMD14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sterile alpha motif domain-containing protein 14. | |||||
|
VCX1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 60) | NC score | 0.046496 (rank : 67) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H320, Q9P0H3 | Gene names | VCX, VCX1, VCX10R, VCXB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 1 (VCX-B1 protein) (Variably charged protein X-B1) (Variable charge protein on X with ten repeats) (VCX- 10r). | |||||
|
ZN165_HUMAN
|
||||||
| θ value | 1.81305 (rank : 61) | NC score | 0.002099 (rank : 138) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 734 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P49910 | Gene names | ZNF165, ZPF165, ZSCAN7 | |||
|
Domain Architecture |
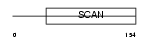
|
|||||
| Description | Zinc finger protein 165 (LD65) (Zinc finger and SCAN domain-containing protein 7). | |||||
|
ATX7_MOUSE
|
||||||
| θ value | 2.36792 (rank : 62) | NC score | 0.024864 (rank : 94) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R4I1, Q8BL17 | Gene names | Atxn7, Sca7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein homolog). | |||||
|
CYLC2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 63) | NC score | 0.055292 (rank : 52) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
K0329_HUMAN
|
||||||
| θ value | 2.36792 (rank : 64) | NC score | 0.036426 (rank : 77) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O15040, Q9UEG6 | Gene names | KIAA0329, KIAA0297 | |||
|
Domain Architecture |

|
|||||
| Description | Protein KIAA0329/KIAA0297. | |||||
|
MINT_HUMAN
|
||||||
| θ value | 2.36792 (rank : 65) | NC score | 0.046982 (rank : 66) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
SAFB2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 66) | NC score | 0.026901 (rank : 91) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q14151, Q8TB13 | Gene names | SAFB2, KIAA0138 | |||
|
Domain Architecture |

|
|||||
| Description | Scaffold attachment factor B2. | |||||
|
CN155_HUMAN
|
||||||
| θ value | 3.0926 (rank : 67) | NC score | 0.048036 (rank : 64) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 68) | NC score | 0.058474 (rank : 47) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
PZRN3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 69) | NC score | 0.016196 (rank : 110) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UPQ7, Q8N2N7, Q96CC2, Q9NSQ2 | Gene names | PDZRN3, KIAA1095, LNX3, SEMCAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing RING finger protein 3 (Ligand of Numb-protein X 3) (Semaphorin cytoplasmic domain-associated protein 3) (SEMACAP3 protein). | |||||
|
SOX10_HUMAN
|
||||||
| θ value | 3.0926 (rank : 70) | NC score | 0.011057 (rank : 122) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P56693 | Gene names | SOX10 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-10. | |||||
|
TRPS1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 71) | NC score | 0.022033 (rank : 98) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q925H1 | Gene names | Trps1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger transcription factor Trps1. | |||||
|
CBP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.034347 (rank : 80) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
FUS_HUMAN
|
||||||
| θ value | 4.03905 (rank : 73) | NC score | 0.021336 (rank : 101) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35637 | Gene names | FUS, TLS | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein FUS (Oncogene FUS) (Oncogene TLS) (Translocated in liposarcoma protein) (POMp75) (75 kDa DNA-pairing protein). | |||||
|
HIRP3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 74) | NC score | 0.051072 (rank : 60) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BW71, O75707, O75708 | Gene names | HIRIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIRA-interacting protein 3. | |||||
|
KCT2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 75) | NC score | 0.033812 (rank : 81) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NC54, Q9NRG2 | Gene names | KCT2, C5orf15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratinocytes-associated transmembrane protein 2 precursor. | |||||
|
MAP4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | 0.028887 (rank : 86) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
SH3BG_MOUSE
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.059876 (rank : 43) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9WUZ7 | Gene names | Sh3bgr | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding glutamic acid-rich protein (SH3BGR protein). | |||||
|
APC_MOUSE
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.019682 (rank : 105) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61315, Q62044 | Gene names | Apc | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC) (mAPC). | |||||
|
ATBF1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.011142 (rank : 121) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
BPA1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.008284 (rank : 126) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
CO1A2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.013563 (rank : 115) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q01149, Q8CGA5 | Gene names | Col1a2, Cola2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(I) chain precursor. | |||||
|
ERIC1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.095930 (rank : 19) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
GPTC8_HUMAN
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.043032 (rank : 70) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
HNRLL_MOUSE
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | 0.013474 (rank : 117) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q921F4, Q8BIP6, Q99J40 | Gene names | Hnrpll | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein L-like. | |||||
|
HNRPU_HUMAN
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.021539 (rank : 99) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q00839, O75507, Q96HY9 | Gene names | HNRPU, SAFA, U21.1 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein U (hnRNP U) (Scaffold attachment factor A) (SAF-A) (p120) (pp120). | |||||
|
HXA5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.006011 (rank : 129) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P20719, O43367, Q96CY6 | Gene names | HOXA5, HOX1C | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A5 (Hox-1C). | |||||
|
IF4G1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.031288 (rank : 82) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q04637, O43177, O95066, Q5HYG0, Q6ZN21, Q8N102 | Gene names | EIF4G1, EIF4G, EIF4GI | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1) (p220). | |||||
|
MAVS_MOUSE
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.020996 (rank : 102) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VCF0 | Gene names | Mavs, Ips1, Visa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial antiviral signaling protein (Interferon-beta promoter stimulator protein 1) (IPS-1) (Virus-induced signaling adapter) (CARD adapter inducing interferon-beta) (Cardif). | |||||
|
MINT_MOUSE
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.045456 (rank : 68) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MRP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.030741 (rank : 83) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P28667, Q3TEZ4, Q91W07 | Gene names | Marcksl1, Mlp, Mrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MARCKS-related protein (MARCKS-like protein 1) (Macrophage myristoylated alanine-rich C kinase substrate) (Mac-MARCKS) (MacMARCKS) (Brain protein F52). | |||||
|
NEK11_HUMAN
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.004353 (rank : 133) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1043 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8NG66, Q5JPC0, Q8NG65, Q8TBY1, Q9H5F4 | Gene names | NEK11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase Nek11 (EC 2.7.11.1) (NimA-related protein kinase 11) (Never in mitosis A-related kinase 11). | |||||
|
PACN2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.011413 (rank : 120) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UNF0, O95921, Q96HV9, Q9H0D3, Q9NPN1, Q9Y4V2 | Gene names | PACSIN2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C and casein kinase substrate in neurons protein 2. | |||||
|
PDZD2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.034699 (rank : 78) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
PLK3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.004176 (rank : 134) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 865 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H4B4, Q15767, Q96CV1 | Gene names | PLK3, CNK, FNK, PRK | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PLK3 (EC 2.7.11.21) (Polo-like kinase 3) (PLK-3) (Cytokine-inducible serine/threonine-protein kinase) (FGF- inducible kinase) (Proliferation-related kinase). | |||||
|
RNF12_MOUSE
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.020881 (rank : 103) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9WTV7 | Gene names | Rnf12, Rlim | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 12 (LIM domain-interacting RING finger protein) (RING finger LIM domain-binding protein) (R-LIM). | |||||
|
SAC3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 96) | NC score | 0.015770 (rank : 111) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91WF7, Q6A092 | Gene names | Sac3, Kiaa0274 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAC domain-containing protein 3. | |||||
|
SLK_MOUSE
|
||||||
| θ value | 5.27518 (rank : 97) | NC score | 0.008407 (rank : 125) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1798 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O54988, Q80U65, Q8CAU2, Q8CDW2, Q9WU41 | Gene names | Slk, Kiaa0204, Stk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (mSLK) (Serine/threonine-protein kinase 2) (STE20-related kinase SMAK) (Etk4). | |||||
|
CD99_MOUSE
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.067918 (rank : 35) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VCN6, Q4L299, Q4L2A4, Q8BLN1 | Gene names | Cd99, Pilrl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CD99 antigen precursor (Paired immunoglobin-like type 2 receptor- ligand) (PILR-L). | |||||
|
CHFR_MOUSE
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.012499 (rank : 118) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q810L3, Q8BJZ9, Q8BWH4 | Gene names | Chfr | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase CHFR (EC 6.3.2.-) (Checkpoint with forkhead and RING finger domains protein). | |||||
|
CLAP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.013847 (rank : 114) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z460, O75118, Q8N5B8, Q9BQT5 | Gene names | CLASP1, KIAA0622, MAST1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CLIP-associating protein 1 (Cytoplasmic linker-associated protein 1) (Multiple asters homolog 1). | |||||
|
HUWE1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.017360 (rank : 109) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
HXA5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | 0.005611 (rank : 132) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P09021, Q91VQ5 | Gene names | Hoxa5, Hox-1.3, Hoxa-5 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A5 (Hox-1.3) (M2). | |||||
|
IMMT_HUMAN
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | 0.013501 (rank : 116) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 494 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q16891, Q14539, Q15092, Q68D41, Q69HW5, Q6IBL0, Q7Z3X1, Q8TAJ5, Q9P0V2 | Gene names | IMMT, HMP, PIG4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial inner membrane protein (Mitofilin) (p87/89) (Proliferation-inducing gene 4 protein). | |||||
|
K0427_MOUSE
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | 0.019396 (rank : 106) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6PEE2 | Gene names | Gm672 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0427 homolog. | |||||
|
AKAP8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.027299 (rank : 89) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O43823 | Gene names | AKAP8, AKAP95 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 8 (A-kinase anchor protein 95 kDa) (AKAP 95). | |||||
|
ATS19_MOUSE
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.002889 (rank : 137) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P59509 | Gene names | Adamts19 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-19 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 19) (ADAM-TS 19) (ADAM-TS19). | |||||
|
CO1A2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.013988 (rank : 113) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P08123, P02464, Q9UEB6, Q9UPH0 | Gene names | COL1A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(I) chain precursor. | |||||
|
CSPG5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.019339 (rank : 107) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95196, Q71M39, Q71M40 | Gene names | CSPG5, CALEB, NGC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate proteoglycan 5 precursor (Neuroglycan C) (Acidic leucine-rich EGF-like domain-containing brain protein). | |||||
|
FOXC1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.004104 (rank : 135) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61572, O88409, Q61582 | Gene names | Foxc1, Fkh1, Fkhl7, Freac3, Mf1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein C1 (Forkhead-related protein FKHL7) (Forkhead- related transcription factor 3) (FREAC-3) (Transcription factor FKH-1) (Mesoderm/mesenchyme forkhead 1) (MF-1). | |||||
|
HNRPC_MOUSE
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.009442 (rank : 124) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z204, Q3TLB5, Q501Q3, Q8C2G5, Q99KE2, Q9CQT3, Q9CY83 | Gene names | Hnrpc, Hnrnpc | |||
|
Domain Architecture |
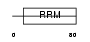
|
|||||
| Description | Heterogeneous nuclear ribonucleoproteins C1/C2 (hnRNP C1 / hnRNP C2). | |||||
|
MSH6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.010641 (rank : 123) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P54276, O54710 | Gene names | Msh6, Gtmbp | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein MSH6 (MutS-alpha 160 kDa subunit) (G/T mismatch-binding protein) (GTBP) (GTMBP) (p160). | |||||
|
MYST4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.022629 (rank : 96) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
PLK3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.003769 (rank : 136) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 863 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q60806, Q60822, Q9R009 | Gene names | Plk3, Cnk, Fnk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PLK3 (EC 2.7.11.21) (Polo-like kinase 3) (PLK-3) (Cytokine-inducible serine/threonine-protein kinase) (FGF- inducible kinase). | |||||
|
RLF_HUMAN
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.005658 (rank : 131) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13129, Q9NU60 | Gene names | RLF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein Rlf (Rearranged L-myc fusion gene protein) (Zn-15- related protein). | |||||
|
SMCA4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.012394 (rank : 119) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P51532, O95052, Q9HBD3 | Gene names | SMARCA4, BRG1, SNF2B, SNF2L4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L4 (EC 3.6.1.-) (ATP- dependent helicase SMARCA4) (SNF2-beta) (BRG-1 protein) (Mitotic growth and transcription activator) (Brahma protein homolog 1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4). | |||||
|
AKA12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.069399 (rank : 32) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
CGRE1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.065589 (rank : 37) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8R1U2, Q9D1K1 | Gene names | Cgref1, Cgr11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.084999 (rank : 24) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.078364 (rank : 26) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
LRRF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.054023 (rank : 55) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q32MZ4, O75766, O75799, Q32MZ5, Q53T49, Q6PKG2 | Gene names | LRRFIP1, GCF2, TRIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (TAR RNA-interacting protein) (GC-binding factor 2). | |||||
|
NASP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.058835 (rank : 46) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.069312 (rank : 33) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.060795 (rank : 40) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
NSBP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.056126 (rank : 49) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
OXR1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.058903 (rank : 45) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q4KMM3, Q5FWW1, Q99L06, Q99MK1, Q99MP4 | Gene names | Oxr1, C7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidation resistance protein 1 (Protein C7). | |||||
|
PTMA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.054923 (rank : 53) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
PTMA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.060310 (rank : 41) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
RP1L1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.051165 (rank : 59) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8CGM2, Q8K0I4 | Gene names | Rp1l1, Rp1hl1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein (Retinitis pigmentosa 1-like protein 1). | |||||
|
RPTN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.056307 (rank : 48) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6XPR3 | Gene names | RPTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Repetin. | |||||
|
RPTN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.067106 (rank : 36) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P97347 | Gene names | Rptn | |||
|
Domain Architecture |

|
|||||
| Description | Repetin. | |||||
|
RRBP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.054024 (rank : 54) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
SG16_MOUSE
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.055954 (rank : 50) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P02815, Q62263 | Gene names | Spt1, Spt-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 16.5 kDa submandibular gland glycoprotein precursor (Salivary protein 1). | |||||
|
TCAL5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.062100 (rank : 39) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.098957 (rank : 18) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
TP53B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.060300 (rank : 42) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
TTC15_MOUSE
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.053347 (rank : 56) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K2L8, Q8BX27 | Gene names | Ttc15 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 15 (TPR repeat protein 15). | |||||
|
TXND2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.059841 (rank : 44) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
TXND2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.069182 (rank : 34) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
TOIP2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 115 | |
| SwissProt Accessions | Q8BYU6, Q6NY10, Q8BJP0 | Gene names | Tor1aip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Torsin-1A-interacting protein 2. | |||||
|
TOIP2_HUMAN
|
||||||
| NC score | 0.926659 (rank : 2) | θ value | 6.80647e-157 (rank : 2) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8NFQ8 | Gene names | TOR1AIP2, LULL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Torsin-1A-interacting protein 2 (Lumenal domain-like LAP1). | |||||
|
TOIP1_HUMAN
|
||||||
| NC score | 0.859428 (rank : 3) | θ value | 1.83746e-69 (rank : 3) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5JTV8, Q5JTV6, Q8IZ65, Q9H8Y6, Q9HAJ1, Q9NV52, Q9Y3X5 | Gene names | TOR1AIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Torsin-1A-interacting protein 1. | |||||
|
TOIP1_MOUSE
|
||||||
| NC score | 0.856538 (rank : 4) | θ value | 1.04338e-64 (rank : 4) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q921T2, Q3U7A4 | Gene names | Tor1aip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Torsin-1A-interacting protein 1 (Lamina-associated polypeptide 1B). | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.195740 (rank : 5) | θ value | 3.77169e-06 (rank : 6) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.182342 (rank : 6) | θ value | 2.88788e-06 (rank : 5) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
DMP1_MOUSE
|
||||||
| NC score | 0.160238 (rank : 7) | θ value | 0.0431538 (rank : 20) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
DMP1_HUMAN
|
||||||
| NC score | 0.157932 (rank : 8) | θ value | 0.365318 (rank : 29) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
MDN1_HUMAN
|
||||||
| NC score | 0.138152 (rank : 9) | θ value | 0.00228821 (rank : 10) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.137076 (rank : 10) | θ value | 0.0330416 (rank : 19) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
NSBP1_MOUSE
|
||||||
| NC score | 0.125790 (rank : 11) | θ value | 0.0252991 (rank : 18) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
FILA_HUMAN
|
||||||
| NC score | 0.117378 (rank : 12) | θ value | 0.0193708 (rank : 16) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.115254 (rank : 13) | θ value | 1.87187e-05 (rank : 7) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
CGRE1_HUMAN
|
||||||
| NC score | 0.111125 (rank : 14) | θ value | 0.00509761 (rank : 13) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q99674 | Gene names | CGREF1, CGR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
SRCA_HUMAN
|
||||||
| NC score | 0.110200 (rank : 15) | θ value | 0.163984 (rank : 25) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.109439 (rank : 16) | θ value | 1.81305 (rank : 56) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
SPRL1_MOUSE
|
||||||
| NC score | 0.102552 (rank : 17) | θ value | 0.00390308 (rank : 12) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P70663, P97810, Q99L82 | Gene names | Sparcl1, Ecm2, Sc1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (Matrix glycoprotein Sc1) (Extracellular matrix protein 2). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.098957 (rank : 18) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
ERIC1_HUMAN
|
||||||
| NC score | 0.095930 (rank : 19) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
SRCA_MOUSE
|
||||||
| NC score | 0.095909 (rank : 20) | θ value | 0.0193708 (rank : 17) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q7TQ48, Q8BS43 | Gene names | Srl, Sar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
LRRF1_MOUSE
|
||||||
| NC score | 0.090006 (rank : 21) | θ value | 1.38821 (rank : 52) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q3UZ39, O70323, Q3TS94 | Gene names | Lrrfip1, Flap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (FLI-LRR-associated protein 1) (Flap-1) (H186 FLAP). | |||||
|
SRCH_HUMAN
|
||||||
| NC score | 0.089787 (rank : 22) | θ value | 0.813845 (rank : 43) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P23327, Q504Y6 | Gene names | HRC, HCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcoplasmic reticulum histidine-rich calcium-binding protein precursor. | |||||
|
TTC15_HUMAN
|
||||||
| NC score | 0.089341 (rank : 23) | θ value | 0.00509761 (rank : 14) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8WVT3, Q8WVW1, Q9Y395 | Gene names | TTC15 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 15 (TPR repeat protein 15). | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.084999 (rank : 24) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.079954 (rank : 25) | θ value | 0.00175202 (rank : 9) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.078364 (rank : 26) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.073336 (rank : 27) | θ value | 1.06291 (rank : 48) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
NCKX1_HUMAN
|
||||||
| NC score | 0.072564 (rank : 28) | θ value | 0.0961366 (rank : 22) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O60721, O43485, O75184 | Gene names | SLC24A1, KIAA0702, NCKX1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium/calcium exchanger 1 (Na(+)/K(+)/Ca(2+)-exchange protein 1) (Retinal rod Na-Ca+K exchanger). | |||||
|
CLIC6_HUMAN
|
||||||
| NC score | 0.072419 (rank : 29) | θ value | 0.47712 (rank : 32) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96NY7, Q8IX31 | Gene names | CLIC6, CLIC1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
NC6IP_MOUSE
|
||||||
| NC score | 0.069957 (rank : 30) | θ value | 0.813845 (rank : 40) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q923W1, Q6DI60, Q6PEA7, Q8R0W9 | Gene names | Ncoa6ip, Pimt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA methyltransferase NCOA6IP (EC 2.1.1.-) (Nuclear receptor coactivator 6-interacting protein) (PRIP-interacting protein with methyltransferase motif) (PIPMT) (PIMT). | |||||
|
AMPH_MOUSE
|
||||||
| NC score | 0.069517 (rank : 31) | θ value | 0.00665767 (rank : 15) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7TQF7, Q8R1C4 | Gene names | Amph, Amph1 | |||
|
Domain Architecture |

|
|||||
| Description | Amphiphysin. | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.069399 (rank : 32) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.069312 (rank : 33) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.069182 (rank : 34) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
CD99_MOUSE
|
||||||
| NC score | 0.067918 (rank : 35) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VCN6, Q4L299, Q4L2A4, Q8BLN1 | Gene names | Cd99, Pilrl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CD99 antigen precursor (Paired immunoglobin-like type 2 receptor- ligand) (PILR-L). | |||||
|
RPTN_MOUSE
|
||||||
| NC score | 0.067106 (rank : 36) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P97347 | Gene names | Rptn | |||
|
Domain Architecture |

|
|||||
| Description | Repetin. | |||||
|
CGRE1_MOUSE
|
||||||
| NC score | 0.065589 (rank : 37) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8R1U2, Q9D1K1 | Gene names | Cgref1, Cgr11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
HORN_HUMAN
|
||||||
| NC score | 0.063177 (rank : 38) | θ value | 1.38821 (rank : 51) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q86YZ3 | Gene names | HRNR, S100A18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hornerin. | |||||
|
TCAL5_HUMAN
|
||||||
| NC score | 0.062100 (rank : 39) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.060795 (rank : 40) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
PTMA_MOUSE
|
||||||
| NC score | 0.060310 (rank : 41) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
TP53B_MOUSE
|
||||||
| NC score | 0.060300 (rank : 42) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
SH3BG_MOUSE
|
||||||
| NC score | 0.059876 (rank : 43) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9WUZ7 | Gene names | Sh3bgr | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding glutamic acid-rich protein (SH3BGR protein). | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.059841 (rank : 44) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
OXR1_MOUSE
|
||||||
| NC score | 0.058903 (rank : 45) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q4KMM3, Q5FWW1, Q99L06, Q99MK1, Q99MP4 | Gene names | Oxr1, C7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidation resistance protein 1 (Protein C7). | |||||
|
NASP_HUMAN
|
||||||
| NC score | 0.058835 (rank : 46) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.058474 (rank : 47) | θ value | 3.0926 (rank : 68) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
RPTN_HUMAN
|
||||||
| NC score | 0.056307 (rank : 48) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6XPR3 | Gene names | RPTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Repetin. | |||||
|
NSBP1_HUMAN
|
||||||
| NC score | 0.056126 (rank : 49) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
SG16_MOUSE
|
||||||
| NC score | 0.055954 (rank : 50) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P02815, Q62263 | Gene names | Spt1, Spt-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 16.5 kDa submandibular gland glycoprotein precursor (Salivary protein 1). | |||||
|
DNM3A_HUMAN
|
||||||
| NC score | 0.055370 (rank : 51) | θ value | 0.163984 (rank : 23) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y6K1, Q8WXU9 | Gene names | DNMT3A | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 3A (EC 2.1.1.37) (Dnmt3a) (DNA methyltransferase HsaIIIA) (DNA MTase HsaIIIA) (M.HsaIIIA). | |||||
|
CYLC2_HUMAN
|
||||||
| NC score | 0.055292 (rank : 52) | θ value | 2.36792 (rank : 63) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
PTMA_HUMAN
|
||||||
| NC score | 0.054923 (rank : 53) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
RRBP1_MOUSE
|
||||||
| NC score | 0.054024 (rank : 54) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
LRRF1_HUMAN
|
||||||
| NC score | 0.054023 (rank : 55) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q32MZ4, O75766, O75799, Q32MZ5, Q53T49, Q6PKG2 | Gene names | LRRFIP1, GCF2, TRIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (TAR RNA-interacting protein) (GC-binding factor 2). | |||||
|
TTC15_MOUSE
|
||||||
| NC score | 0.053347 (rank : 56) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K2L8, Q8BX27 | Gene names | Ttc15 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 15 (TPR repeat protein 15). | |||||
|
DNM3A_MOUSE
|
||||||
| NC score | 0.052529 (rank : 57) | θ value | 0.279714 (rank : 26) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O88508, Q3UH24, Q8CJ60, Q922J0, Q9CSE1 | Gene names | Dnmt3a | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 3A (EC 2.1.1.37) (Dnmt3a) (DNA methyltransferase MmuIIIA) (DNA MTase MmuIIIA) (M.MmuIIIA). | |||||
|
CMGA_MOUSE
|
||||||
| NC score | 0.051619 (rank : 58) | θ value | 1.81305 (rank : 55) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P26339 | Gene names | Chga | |||
|
Domain Architecture |

|
|||||
| Description | Chromogranin A precursor (CgA) [Contains: Pancreastatin; Beta-granin; WE-14]. | |||||
|
RP1L1_MOUSE
|
||||||
| NC score | 0.051165 (rank : 59) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8CGM2, Q8K0I4 | Gene names | Rp1l1, Rp1hl1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein (Retinitis pigmentosa 1-like protein 1). | |||||
|
HIRP3_HUMAN
|
||||||
| NC score | 0.051072 (rank : 60) | θ value | 4.03905 (rank : 74) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BW71, O75707, O75708 | Gene names | HIRIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIRA-interacting protein 3. | |||||
|
PDXL2_MOUSE
|
||||||
| NC score | 0.049057 (rank : 61) | θ value | 0.813845 (rank : 41) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8CAE9, Q8CFW3 | Gene names | Podxl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Podocalyxin-like protein 2 precursor (Endoglycan). | |||||
|
BAZ2B_HUMAN
|
||||||
| NC score | 0.048680 (rank : 62) | θ value | 0.62314 (rank : 35) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
PERQ1_MOUSE
|
||||||
| NC score | 0.048525 (rank : 63) | θ value | 1.81305 (rank : 58) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99MR1 | Gene names | Perq1 | |||
|
Domain Architecture |

|
|||||
| Description | PERQ amino acid rich with GYF domain protein 1. | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.048036 (rank : 64) | θ value | 3.0926 (rank : 67) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
DOK7_MOUSE
|
||||||
| NC score | 0.047896 (rank : 65) | θ value | 0.365318 (rank : 30) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q18PE0, Q3TCZ6, Q5FW70, Q8C8U7 | Gene names | Dok7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Dok-7 (Downstream of tyrosine kinase 7). | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.046982 (rank : 66) | θ value | 2.36792 (rank : 65) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
VCX1_HUMAN
|
||||||
| NC score | 0.046496 (rank : 67) | θ value | 1.81305 (rank : 60) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H320, Q9P0H3 | Gene names | VCX, VCX1, VCX10R, VCXB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 1 (VCX-B1 protein) (Variably charged protein X-B1) (Variable charge protein on X with ten repeats) (VCX- 10r). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.045456 (rank : 68) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
PININ_HUMAN
|
||||||
| NC score | 0.043745 (rank : 69) | θ value | 0.813845 (rank : 42) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9H307, O60899, Q53EM7, Q6P5X4, Q7KYL1, Q99738, Q9UHZ9, Q9UQR9 | Gene names | PNN, DRS, MEMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin (140 kDa nuclear and cell adhesion-related phosphoprotein) (Domain-rich serine protein) (DRS-protein) (DRSP) (Melanoma metastasis clone A protein) (Desmosome-associated protein) (SR-like protein) (Nuclear protein SDK3). | |||||
|
GPTC8_HUMAN
|
||||||
| NC score | 0.043032 (rank : 70) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
X3CL1_MOUSE
|
||||||
| NC score | 0.042153 (rank : 71) | θ value | 0.813845 (rank : 44) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O35188, O35933 | Gene names | Cx3cl1, Cx3c, Fkn, Scyd1 | |||
|
Domain Architecture |

|
|||||
| Description | Fractalkine precursor (CX3CL1) (Neurotactin) (CX3C membrane-anchored chemokine) (Small inducible cytokine D1). | |||||
|
CLIC6_MOUSE
|
||||||
| NC score | 0.041158 (rank : 72) | θ value | 1.06291 (rank : 46) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BHB9 | Gene names | Clic6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.038823 (rank : 73) | θ value | 1.06291 (rank : 45) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
IF4G1_MOUSE
|
||||||
| NC score | 0.038809 (rank : 74) | θ value | 0.47712 (rank : 33) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
GNAS1_MOUSE
|
||||||
| NC score | 0.038205 (rank : 75) | θ value | 0.00390308 (rank : 11) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6R0H7, Q6R0H4, Q6R0H5, Q6R2J5, Q9JJX0, Q9Z1N8 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
PGCA_MOUSE
|
||||||
| NC score | 0.037580 (rank : 76) | θ value | 0.47712 (rank : 34) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
K0329_HUMAN
|
||||||
| NC score | 0.036426 (rank : 77) | θ value | 2.36792 (rank : 64) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O15040, Q9UEG6 | Gene names | KIAA0329, KIAA0297 | |||
|
Domain Architecture |

|
|||||
| Description | Protein KIAA0329/KIAA0297. | |||||
|
PDZD2_HUMAN
|
||||||
| NC score | 0.034699 (rank : 78) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
SAM14_HUMAN
|
||||||
| NC score | 0.034362 (rank : 79) | θ value | 1.81305 (rank : 59) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IZD0, Q8N2X0 | Gene names | SAMD14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sterile alpha motif domain-containing protein 14. | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.034347 (rank : 80) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
KCT2_HUMAN
|
||||||
| NC score | 0.033812 (rank : 81) | θ value | 4.03905 (rank : 75) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NC54, Q9NRG2 | Gene names | KCT2, C5orf15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratinocytes-associated transmembrane protein 2 precursor. | |||||
|
IF4G1_HUMAN
|
||||||
| NC score | 0.031288 (rank : 82) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q04637, O43177, O95066, Q5HYG0, Q6ZN21, Q8N102 | Gene names | EIF4G1, EIF4G, EIF4GI | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1) (p220). | |||||
|
MRP_MOUSE
|
||||||
| NC score | 0.030741 (rank : 83) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P28667, Q3TEZ4, Q91W07 | Gene names | Marcksl1, Mlp, Mrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MARCKS-related protein (MARCKS-like protein 1) (Macrophage myristoylated alanine-rich C kinase substrate) (Mac-MARCKS) (MacMARCKS) (Brain protein F52). | |||||
|
GTF2I_MOUSE
|
||||||
| NC score | 0.030196 (rank : 84) | θ value | 1.38821 (rank : 50) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9ESZ8, O54700, O55030, O55031, Q8VHD1, Q8VHD2, Q8VHD3, Q8VHD4, Q9CSB5, Q9D9K9 | Gene names | Gtf2i, Bap135, Diws1t | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor II-I (GTFII-I) (TFII-I) (Bruton tyrosine kinase-associated protein 135) (BTK-associated protein 135) (BAP-135). | |||||
|
LPIN3_HUMAN
|
||||||
| NC score | 0.029341 (rank : 85) | θ value | 0.62314 (rank : 36) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BQK8, Q5TDB9, Q9NPY8, Q9UJE5 | Gene names | LPIN3, LIPN3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipin-3 (Lipin 3-like). | |||||
|
MAP4_MOUSE
|
||||||
| NC score | 0.028887 (rank : 86) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
CAC1H_MOUSE
|
||||||
| NC score | 0.028880 (rank : 87) | θ value | 0.813845 (rank : 38) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88427, Q80TJ2, Q9JKU5 | Gene names | Cacna1h, Kiaa1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2). | |||||
|
NFM_MOUSE
|
||||||
| NC score | 0.028441 (rank : 88) | θ value | 0.0431538 (rank : 21) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
AKAP8_HUMAN
|
||||||
| NC score | 0.027299 (rank : 89) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O43823 | Gene names | AKAP8, AKAP95 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 8 (A-kinase anchor protein 95 kDa) (AKAP 95). | |||||
|
EDD1_MOUSE
|
||||||
| NC score | 0.027227 (rank : 90) | θ value | 1.06291 (rank : 47) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80TP3, Q698K9, Q6PEQ8, Q6PFQ9, Q80VL4, Q810V6, Q9CXE9 | Gene names | Edd1, Edd, Kiaa0896 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog). | |||||
|
SAFB2_HUMAN
|
||||||
| NC score | 0.026901 (rank : 91) | θ value | 2.36792 (rank : 66) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q14151, Q8TB13 | Gene names | SAFB2, KIAA0138 | |||
|
Domain Architecture |

|
|||||
| Description | Scaffold attachment factor B2. | |||||
|
CAC1H_HUMAN
|
||||||
| NC score | 0.026737 (rank : 92) | θ value | 1.81305 (rank : 54) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
CAC1B_MOUSE
|
||||||
| NC score | 0.025765 (rank : 93) | θ value | 0.365318 (rank : 27) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
ATX7_MOUSE
|
||||||
| NC score | 0.024864 (rank : 94) | θ value | 2.36792 (rank : 62) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R4I1, Q8BL17 | Gene names | Atxn7, Sca7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein homolog). | |||||
|
MAST4_HUMAN
|
||||||
| NC score | 0.022719 (rank : 95) | θ value | 0.000270298 (rank : 8) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
MYST4_HUMAN
|
||||||
| NC score | 0.022629 (rank : 96) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
ATOH1_MOUSE
|
||||||
| NC score | 0.022405 (rank : 97) | θ value | 1.38821 (rank : 49) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P48985 | Gene names | Atoh1, Ath1 | |||
|
Domain Architecture |

|
|||||
| Description | Atonal protein homolog 1 (Helix-loop-helix protein mATH-1) (MATH1). | |||||
|
TRPS1_MOUSE
|
||||||
| NC score | 0.022033 (rank : 98) | θ value | 3.0926 (rank : 71) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q925H1 | Gene names | Trps1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger transcription factor Trps1. | |||||
|
HNRPU_HUMAN
|
||||||
| NC score | 0.021539 (rank : 99) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q00839, O75507, Q96HY9 | Gene names | HNRPU, SAFA, U21.1 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein U (hnRNP U) (Scaffold attachment factor A) (SAF-A) (p120) (pp120). | |||||
|
DDX21_MOUSE
|
||||||
| NC score | 0.021399 (rank : 100) | θ value | 0.365318 (rank : 28) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JIK5, Q9WV45 | Gene names | Ddx21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
FUS_HUMAN
|
||||||
| NC score | 0.021336 (rank : 101) | θ value | 4.03905 (rank : 73) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35637 | Gene names | FUS, TLS | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein FUS (Oncogene FUS) (Oncogene TLS) (Translocated in liposarcoma protein) (POMp75) (75 kDa DNA-pairing protein). | |||||
|
MAVS_MOUSE
|
||||||
| NC score | 0.020996 (rank : 102) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VCF0 | Gene names | Mavs, Ips1, Visa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial antiviral signaling protein (Interferon-beta promoter stimulator protein 1) (IPS-1) (Virus-induced signaling adapter) (CARD adapter inducing interferon-beta) (Cardif). | |||||
|
RNF12_MOUSE
|
||||||
| NC score | 0.020881 (rank : 103) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9WTV7 | Gene names | Rnf12, Rlim | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 12 (LIM domain-interacting RING finger protein) (RING finger LIM domain-binding protein) (R-LIM). | |||||
|
CHD9_HUMAN
|
||||||
| NC score | 0.020722 (rank : 104) | θ value | 0.813845 (rank : 39) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
APC_MOUSE
|
||||||
| NC score | 0.019682 (rank : 105) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61315, Q62044 | Gene names | Apc | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC) (mAPC). | |||||
|
K0427_MOUSE
|
||||||
| NC score | 0.019396 (rank : 106) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6PEE2 | Gene names | Gm672 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0427 homolog. | |||||
|
CSPG5_HUMAN
|
||||||
| NC score | 0.019339 (rank : 107) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95196, Q71M39, Q71M40 | Gene names | CSPG5, CALEB, NGC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate proteoglycan 5 precursor (Neuroglycan C) (Acidic leucine-rich EGF-like domain-containing brain protein). | |||||
|
NPAS3_MOUSE
|
||||||
| NC score | 0.018445 (rank : 108) | θ value | 1.81305 (rank : 57) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9QZQ0, Q9EQP4 | Gene names | Npas3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
HUWE1_HUMAN
|
||||||
| NC score | 0.017360 (rank : 109) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
PZRN3_HUMAN
|
||||||
| NC score | 0.016196 (rank : 110) | θ value | 3.0926 (rank : 69) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UPQ7, Q8N2N7, Q96CC2, Q9NSQ2 | Gene names | PDZRN3, KIAA1095, LNX3, SEMCAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing RING finger protein 3 (Ligand of Numb-protein X 3) (Semaphorin cytoplasmic domain-associated protein 3) (SEMACAP3 protein). | |||||
|
SAC3_MOUSE
|
||||||
| NC score | 0.015770 (rank : 111) | θ value | 5.27518 (rank : 96) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91WF7, Q6A092 | Gene names | Sac3, Kiaa0274 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAC domain-containing protein 3. | |||||
|
FBLN2_MOUSE
|
||||||
| NC score | 0.015600 (rank : 112) | θ value | 0.163984 (rank : 24) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P37889, Q9WUI2 | Gene names | Fbln2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-2 precursor. | |||||
|
CO1A2_HUMAN
|
||||||
| NC score | 0.013988 (rank : 113) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P08123, P02464, Q9UEB6, Q9UPH0 | Gene names | COL1A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(I) chain precursor. | |||||
|
CLAP1_HUMAN
|
||||||
| NC score | 0.013847 (rank : 114) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z460, O75118, Q8N5B8, Q9BQT5 | Gene names | CLASP1, KIAA0622, MAST1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CLIP-associating protein 1 (Cytoplasmic linker-associated protein 1) (Multiple asters homolog 1). | |||||
|
CO1A2_MOUSE
|
||||||
| NC score | 0.013563 (rank : 115) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q01149, Q8CGA5 | Gene names | Col1a2, Cola2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(I) chain precursor. | |||||
|
IMMT_HUMAN
|
||||||
| NC score | 0.013501 (rank : 116) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 494 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q16891, Q14539, Q15092, Q68D41, Q69HW5, Q6IBL0, Q7Z3X1, Q8TAJ5, Q9P0V2 | Gene names | IMMT, HMP, PIG4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial inner membrane protein (Mitofilin) (p87/89) (Proliferation-inducing gene 4 protein). | |||||
|
HNRLL_MOUSE
|
||||||
| NC score | 0.013474 (rank : 117) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q921F4, Q8BIP6, Q99J40 | Gene names | Hnrpll | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein L-like. | |||||
|
CHFR_MOUSE
|
||||||
| NC score | 0.012499 (rank : 118) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q810L3, Q8BJZ9, Q8BWH4 | Gene names | Chfr | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase CHFR (EC 6.3.2.-) (Checkpoint with forkhead and RING finger domains protein). | |||||
|
SMCA4_HUMAN
|
||||||
| NC score | 0.012394 (rank : 119) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P51532, O95052, Q9HBD3 | Gene names | SMARCA4, BRG1, SNF2B, SNF2L4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L4 (EC 3.6.1.-) (ATP- dependent helicase SMARCA4) (SNF2-beta) (BRG-1 protein) (Mitotic growth and transcription activator) (Brahma protein homolog 1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4). | |||||
|
PACN2_HUMAN
|
||||||
| NC score | 0.011413 (rank : 120) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UNF0, O95921, Q96HV9, Q9H0D3, Q9NPN1, Q9Y4V2 | Gene names | PACSIN2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C and casein kinase substrate in neurons protein 2. | |||||
|
ATBF1_HUMAN
|
||||||
| NC score | 0.011142 (rank : 121) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
SOX10_HUMAN
|
||||||
| NC score | 0.011057 (rank : 122) | θ value | 3.0926 (rank : 70) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P56693 | Gene names | SOX10 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-10. | |||||
|
MSH6_MOUSE
|
||||||
| NC score | 0.010641 (rank : 123) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P54276, O54710 | Gene names | Msh6, Gtmbp | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein MSH6 (MutS-alpha 160 kDa subunit) (G/T mismatch-binding protein) (GTBP) (GTMBP) (p160). | |||||
|
HNRPC_MOUSE
|
||||||
| NC score | 0.009442 (rank : 124) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z204, Q3TLB5, Q501Q3, Q8C2G5, Q99KE2, Q9CQT3, Q9CY83 | Gene names | Hnrpc, Hnrnpc | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoproteins C1/C2 (hnRNP C1 / hnRNP C2). | |||||
|
SLK_MOUSE
|
||||||
| NC score | 0.008407 (rank : 125) | θ value | 5.27518 (rank : 97) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1798 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O54988, Q80U65, Q8CAU2, Q8CDW2, Q9WU41 | Gene names | Slk, Kiaa0204, Stk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (mSLK) (Serine/threonine-protein kinase 2) (STE20-related kinase SMAK) (Etk4). | |||||
|
BPA1_MOUSE
|
||||||
| NC score | 0.008284 (rank : 126) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
PAK4_MOUSE
|
||||||
| NC score | 0.007272 (rank : 127) | θ value | 1.38821 (rank : 53) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1098 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BTW9, Q6ZPX0, Q80Z97, Q9CS71 | Gene names | Pak4, Kiaa1142 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PAK 4 (EC 2.7.11.1) (p21-activated kinase 4) (PAK-4). | |||||
|
ZBT17_MOUSE
|
||||||
| NC score | 0.006257 (rank : 128) | θ value | 0.62314 (rank : 37) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q60821, Q60699 | Gene names | Zbtb17, Zfp100, Znf151 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 17 (Zinc finger protein 151) (Zinc finger protein 100) (Zfp-100) (Polyomavirus late initiator promoter-binding protein) (LP-1) (Zinc finger protein Z13). | |||||
|
HXA5_HUMAN
|
||||||
| NC score | 0.006011 (rank : 129) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P20719, O43367, Q96CY6 | Gene names | HOXA5, HOX1C | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A5 (Hox-1C). | |||||
|
ZBT17_HUMAN
|
||||||
| NC score | 0.005833 (rank : 130) | θ value | 0.365318 (rank : 31) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 925 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13105, Q15932, Q5JYB2, Q9NUC9 | Gene names | ZBTB17, MIZ1, ZNF151 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 17 (Zinc finger protein 151) (Myc-interacting zinc finger protein) (Miz-1 protein). | |||||
|
RLF_HUMAN
|
||||||
| NC score | 0.005658 (rank : 131) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13129, Q9NU60 | Gene names | RLF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein Rlf (Rearranged L-myc fusion gene protein) (Zn-15- related protein). | |||||
|
HXA5_MOUSE
|
||||||
| NC score | 0.005611 (rank : 132) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P09021, Q91VQ5 | Gene names | Hoxa5, Hox-1.3, Hoxa-5 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A5 (Hox-1.3) (M2). | |||||
|
NEK11_HUMAN
|
||||||
| NC score | 0.004353 (rank : 133) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1043 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8NG66, Q5JPC0, Q8NG65, Q8TBY1, Q9H5F4 | Gene names | NEK11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase Nek11 (EC 2.7.11.1) (NimA-related protein kinase 11) (Never in mitosis A-related kinase 11). | |||||
|
PLK3_HUMAN
|
||||||
| NC score | 0.004176 (rank : 134) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 865 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H4B4, Q15767, Q96CV1 | Gene names | PLK3, CNK, FNK, PRK | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PLK3 (EC 2.7.11.21) (Polo-like kinase 3) (PLK-3) (Cytokine-inducible serine/threonine-protein kinase) (FGF- inducible kinase) (Proliferation-related kinase). | |||||
|
FOXC1_MOUSE
|
||||||
| NC score | 0.004104 (rank : 135) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61572, O88409, Q61582 | Gene names | Foxc1, Fkh1, Fkhl7, Freac3, Mf1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein C1 (Forkhead-related protein FKHL7) (Forkhead- related transcription factor 3) (FREAC-3) (Transcription factor FKH-1) (Mesoderm/mesenchyme forkhead 1) (MF-1). | |||||
|
PLK3_MOUSE
|
||||||
| NC score | 0.003769 (rank : 136) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 863 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q60806, Q60822, Q9R009 | Gene names | Plk3, Cnk, Fnk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PLK3 (EC 2.7.11.21) (Polo-like kinase 3) (PLK-3) (Cytokine-inducible serine/threonine-protein kinase) (FGF- inducible kinase). | |||||
|
ATS19_MOUSE
|
||||||
| NC score | 0.002889 (rank : 137) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P59509 | Gene names | Adamts19 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-19 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 19) (ADAM-TS 19) (ADAM-TS19). | |||||
|
ZN165_HUMAN
|
||||||
| NC score | 0.002099 (rank : 138) | θ value | 1.81305 (rank : 61) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 734 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P49910 | Gene names | ZNF165, ZPF165, ZSCAN7 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 165 (LD65) (Zinc finger and SCAN domain-containing protein 7). | |||||