Please be patient as the page loads
|
K0329_HUMAN
|
||||||
| SwissProt Accessions | O15040, Q9UEG6 | Gene names | KIAA0329, KIAA0297 | |||
|
Domain Architecture |

|
|||||
| Description | Protein KIAA0329/KIAA0297. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
K0329_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | O15040, Q9UEG6 | Gene names | KIAA0329, KIAA0297 | |||
|
Domain Architecture |

|
|||||
| Description | Protein KIAA0329/KIAA0297. | |||||
|
HPS5_MOUSE
|
||||||
| θ value | 1.16704e-23 (rank : 2) | NC score | 0.545197 (rank : 2) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P59438 | Gene names | Hps5, Ru2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hermansky-Pudlak syndrome 5 protein homolog (Ruby-eye protein 2) (Ru2). | |||||
|
HPS5_HUMAN
|
||||||
| θ value | 5.79196e-23 (rank : 3) | NC score | 0.542882 (rank : 3) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UPZ3, O95942, Q8N4U0 | Gene names | HPS5, AIBP63, KIAA1017 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hermansky-Pudlak syndrome 5 protein (Alpha-integrin-binding protein 63) (Ruby-eye protein 2 homolog) (Ru2). | |||||
|
NIBA_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 4) | NC score | 0.134708 (rank : 4) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BZQ8, Q5TEM8, Q8TEI5, Q9H593, Q9H9Y8, Q9HCB9 | Gene names | C1orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Niban protein. | |||||
|
EPN3_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 5) | NC score | 0.096799 (rank : 5) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q91W69, Q9CV55 | Gene names | Epn3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
PI5PA_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 6) | NC score | 0.061293 (rank : 19) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
TCOF_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 7) | NC score | 0.090260 (rank : 8) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
TCF20_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 8) | NC score | 0.077951 (rank : 13) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
EPN3_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 9) | NC score | 0.090024 (rank : 9) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H201, Q9BVN6, Q9NWK2 | Gene names | EPN3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 10) | NC score | 0.065913 (rank : 17) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
AHI1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 11) | NC score | 0.054026 (rank : 28) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 537 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8N157, Q504T3, Q5TCP9, Q6P098, Q6PIT6, Q8NDX0, Q9H0H2 | Gene names | AHI1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Jouberin (Abelson helper integration site 1 protein homolog) (AHI-1). | |||||
|
NSD1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 12) | NC score | 0.047194 (rank : 37) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
DDB2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 13) | NC score | 0.093452 (rank : 6) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q92466, Q76E54, Q76E55, Q76E56, Q76E57 | Gene names | DDB2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA damage-binding protein 2 (Damage-specific DNA-binding protein 2) (DDB p48 subunit) (DDBb) (UV-damaged DNA-binding protein 2) (UV-DDB 2). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 14) | NC score | 0.047675 (rank : 36) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
GT2D1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 15) | NC score | 0.065048 (rank : 18) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JI57, Q8VHD5, Q8VI58, Q9EQE7, Q9ESZ6, Q9ESZ7 | Gene names | Gtf2ird1, Ben | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor II-I repeat domain-containing protein 1 (GTF2I repeat domain-containing protein 1) (Binding factor for early enhancer). | |||||
|
IF140_HUMAN
|
||||||
| θ value | 0.279714 (rank : 16) | NC score | 0.090460 (rank : 7) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96RY7, O60332 | Gene names | IFT140, KIAA0590, WDTC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 140 homolog (WD and tetratricopeptide repeats protein 2). | |||||
|
GON4L_HUMAN
|
||||||
| θ value | 0.365318 (rank : 17) | NC score | 0.050016 (rank : 32) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
WDR26_HUMAN
|
||||||
| θ value | 0.365318 (rank : 18) | NC score | 0.069458 (rank : 16) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9H7D7, Q86UY4 | Gene names | WDR26 | |||
|
Domain Architecture |
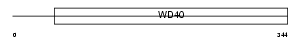
|
|||||
| Description | WD repeat protein 26. | |||||
|
WDR26_MOUSE
|
||||||
| θ value | 0.365318 (rank : 19) | NC score | 0.070048 (rank : 15) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8C6G8, Q8C4F3, Q8VDZ6 | Gene names | Wdr26 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 26. | |||||
|
DDB2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 20) | NC score | 0.087410 (rank : 10) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99J79 | Gene names | Ddb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA damage-binding protein 2 (Damage-specific DNA-binding protein 2). | |||||
|
WDR19_MOUSE
|
||||||
| θ value | 0.47712 (rank : 21) | NC score | 0.083968 (rank : 11) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q3UGF1, Q6ZPK8, Q80VQ6, Q8C794, Q8K3R5 | Gene names | Wdr19, Kiaa1638 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 19. | |||||
|
A16L1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 22) | NC score | 0.051251 (rank : 30) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 776 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8C0J2, Q6KAT7, Q80U97, Q80U98, Q80U99, Q80Y53, Q9DB63 | Gene names | Atg16l1, Apg16l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autophagy-related protein 16-1 (APG16-like 1). | |||||
|
BRWD2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 23) | NC score | 0.071732 (rank : 14) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BZH6, Q5VWA1 | Gene names | BRWD2, WDR11 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain and WD repeat domain-containing protein 2 (WD repeat protein 11). | |||||
|
SM1L2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 24) | NC score | 0.018453 (rank : 67) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1019 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NDV3, Q5TIC3, Q9Y3G5 | Gene names | SMC1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 2 protein (SMC1beta protein). | |||||
|
WDR16_HUMAN
|
||||||
| θ value | 0.62314 (rank : 25) | NC score | 0.059344 (rank : 20) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8N1V2, Q5DX23, Q8TC73, Q8TCI3 | Gene names | WDR16, WDRPUH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 16 (WD40-repeat protein up-regulated in HCC). | |||||
|
WDR16_MOUSE
|
||||||
| θ value | 0.62314 (rank : 26) | NC score | 0.059084 (rank : 21) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5F201, Q5F200, Q9D432, Q9DA68 | Gene names | WDR16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 16. | |||||
|
WDR19_HUMAN
|
||||||
| θ value | 0.62314 (rank : 27) | NC score | 0.082609 (rank : 12) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8NEZ3, Q8N5B4, Q9H5S0, Q9HCD4 | Gene names | WDR19, KIAA1638 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 19. | |||||
|
A16L1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 28) | NC score | 0.051529 (rank : 29) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 664 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q676U5, Q6IPN1, Q6UXW4, Q6ZVZ5, Q8NCY2, Q96JV5, Q9H619 | Gene names | ATG16L1, APG16L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autophagy-related protein 16-1 (APG16-like 1). | |||||
|
OXR1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 29) | NC score | 0.040198 (rank : 40) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8N573, Q3LIB5, Q6ZVK9, Q8N8V0, Q9H266, Q9NWC7 | Gene names | OXR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidation resistance protein 1. | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 30) | NC score | 0.040381 (rank : 39) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
NLE1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 31) | NC score | 0.048736 (rank : 33) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9NVX2, O60868, Q9BU54 | Gene names | NLE1 | |||
|
Domain Architecture |
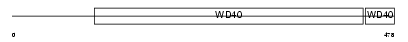
|
|||||
| Description | Notchless homolog 1. | |||||
|
NLE1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 32) | NC score | 0.048438 (rank : 34) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8VEJ4 | Gene names | Nle1 | |||
|
Domain Architecture |
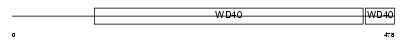
|
|||||
| Description | Notchless homolog 1. | |||||
|
PWP2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 33) | NC score | 0.054031 (rank : 27) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q15269, Q96A77 | Gene names | PWP2H, PWP2 | |||
|
Domain Architecture |

|
|||||
| Description | Periodic tryptophan protein 2 homolog. | |||||
|
PWP2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 34) | NC score | 0.055173 (rank : 25) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BU03, Q8BTR0 | Gene names | Pwp2h | |||
|
Domain Architecture |

|
|||||
| Description | Periodic tryptophan protein 2 homolog. | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 35) | NC score | 0.038195 (rank : 42) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | 1.06291 (rank : 36) | NC score | 0.029713 (rank : 52) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
IRS2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 37) | NC score | 0.035880 (rank : 45) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P81122 | Gene names | Irs2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin receptor substrate 2 (IRS-2) (4PS). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 38) | NC score | 0.017707 (rank : 68) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
REV1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.033623 (rank : 47) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q920Q2, Q9QXV2 | Gene names | Rev1l, Rev1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein REV1 (EC 2.7.7.-) (Rev1-like terminal deoxycytidyl transferase). | |||||
|
KPCD1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.006071 (rank : 77) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 901 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15139 | Gene names | PRKD1, PKD, PKD1, PRKCM | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase D1 (EC 2.7.11.13) (nPKC-D1) (Protein kinase D) (Protein kinase C mu type) (nPKC-mu). | |||||
|
KPCD1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.006126 (rank : 76) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 891 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q62101 | Gene names | Prkd1, Pkcm, Pkd, Prkcm | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase D1 (EC 2.7.11.13) (nPKC-D1) (Protein kinase D) (Protein kinase C mu type) (nPKC-mu). | |||||
|
TCAL5_HUMAN
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.027987 (rank : 56) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
TOIP2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.036426 (rank : 43) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BYU6, Q6NY10, Q8BJP0 | Gene names | Tor1aip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Torsin-1A-interacting protein 2. | |||||
|
ZN409_HUMAN
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.018863 (rank : 66) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UPU6 | Gene names | ZNF409, KIAA1056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 409. | |||||
|
ZNF18_MOUSE
|
||||||
| θ value | 2.36792 (rank : 45) | NC score | 0.002834 (rank : 82) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 743 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q810A1, Q9D972 | Gene names | Znf18, Zfp535 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 18 (Zinc finger protein 535). | |||||
|
GT2D1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.050137 (rank : 31) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UHL9, O95444, Q75MX7, Q86UM3, Q8WVC4, Q9UHK8, Q9UI91 | Gene names | GTF2IRD1, CREAM1, GTF3, MUSTRD1, RBAP2, WBSCR11, WBSCR12 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor II-I repeat domain-containing protein 1 (GTF2I repeat domain-containing protein 1) (Muscle TFII-I repeat domain-containing protein 1) (General transcription factor III) (Slow- muscle-fiber enhancer-binding protein) (USE B1-binding protein) (MusTRD1/BEN) (Williams-Beuren syndrome chromosome region 11 protein). | |||||
|
HSPB8_MOUSE
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.028444 (rank : 55) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JK92 | Gene names | Hspb8, Cryac, Hsp22 | |||
|
Domain Architecture |
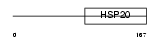
|
|||||
| Description | Heat-shock protein beta-8 (HspB8) (Alpha crystallin C chain) (Small stress protein-like protein HSP22). | |||||
|
M3K12_MOUSE
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.005026 (rank : 78) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q60700, P70286, Q3TLL7, Q8C4N7, Q8CBX3, Q8CDL6 | Gene names | Map3k12, Zpk | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 12 (EC 2.7.11.25) (Mixed lineage kinase) (Leucine-zipper protein kinase) (ZPK) (Dual leucine zipper bearing kinase) (DLK) (MAPK-upstream kinase) (MUK). | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.058796 (rank : 22) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
SON_MOUSE
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.023302 (rank : 63) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
WDR22_HUMAN
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.056906 (rank : 23) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96JK2, O60559, Q8N3V3, Q8N3V5 | Gene names | WDR22, BCRG2, KIAA1824 | |||
|
Domain Architecture |
|
|||||
| Description | WD repeat protein 22 (Breakpoint cluster region protein 2) (BCRP2). | |||||
|
WDR22_MOUSE
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.056327 (rank : 24) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q80T85, Q80VT3, Q80ZW6, Q8BIP8, Q8BVK5, Q8K0Q6 | Gene names | Wdr22, Kiaa1824 | |||
|
Domain Architecture |
|
|||||
| Description | WD repeat protein 22. | |||||
|
WDR60_MOUSE
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | 0.040126 (rank : 41) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8C761, Q3UNM4 | Gene names | Wdr60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 60. | |||||
|
GEMI5_MOUSE
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.043468 (rank : 38) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 424 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BX17 | Gene names | Gemin5 | |||
|
Domain Architecture |

|
|||||
| Description | Gem-associated protein 5 (Gemin5). | |||||
|
MAP4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.028766 (rank : 54) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
TCGAP_MOUSE
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.027537 (rank : 58) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80YF9, Q6P7W6 | Gene names | Snx26, Tcgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
CIR1A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.033908 (rank : 46) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8R2N2, Q62306, Q69Z48, Q6NVG9, Q8VEL5 | Gene names | Cirh1a, Kiaa1988, Tex292 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cirhin (Testis-expressed gene 292 protein). | |||||
|
EP300_HUMAN
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.024312 (rank : 62) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
GFPT1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.025501 (rank : 60) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q06210, Q9BXF8 | Gene names | GFPT1, GFAT, GFPT | |||
|
Domain Architecture |

|
|||||
| Description | Glucosamine--fructose-6-phosphate aminotransferase [isomerizing] 1 (EC 2.6.1.16) (Hexosephosphate aminotransferase 1) (D-fructose-6- phosphate amidotransferase 1) (GFAT 1) (GFAT1). | |||||
|
GFPT1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.025509 (rank : 59) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P47856, Q91XG9, Q99LP7, Q99MJ4 | Gene names | Gfpt1, Gfpt | |||
|
Domain Architecture |

|
|||||
| Description | Glucosamine--fructose-6-phosphate aminotransferase [isomerizing] 1 (EC 2.6.1.16) (Hexosephosphate aminotransferase 1) (D-fructose-6- phosphate amidotransferase 1) (GFAT 1) (GFAT1). | |||||
|
GLIS1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.002708 (rank : 83) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NBF1, Q8N9V9 | Gene names | GLIS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLIS1 (GLI-similar 1). | |||||
|
M3K12_HUMAN
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.004595 (rank : 80) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q12852, Q86VQ5, Q8WY25 | Gene names | MAP3K12, ZPK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 12 (EC 2.7.11.25) (Mixed lineage kinase) (Leucine-zipper protein kinase) (ZPK) (Dual leucine zipper bearing kinase) (DLK) (MAPK-upstream kinase) (MUK). | |||||
|
MYCB2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.029643 (rank : 53) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7TPH6, Q6PCM8 | Gene names | Mycbp2, Pam, Phr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
RFWD2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.031702 (rank : 50) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8NHY2, Q6H103, Q9H6L7 | Gene names | RFWD2, COP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger and WD repeat domain protein 2 (EC 6.3.2.-) (Ubiquitin- protein ligase COP1) (Constitutive photomorphogenesis protein 1 homolog) (hCOP1). | |||||
|
RFWD2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.031550 (rank : 51) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9R1A8 | Gene names | Rfwd2, Cop1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger and WD repeat domain protein 2 (EC 6.3.2.-) (Ubiquitin- protein ligase COP1) (Constitutive photomorphogenesis protein 1 homolog) (mCOP1). | |||||
|
SYBU_HUMAN
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.020932 (rank : 65) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NX95, Q5R1T1, Q5R1T2, Q5R1T3, Q5Y2M6, Q8ND49, Q8TCR6, Q9P256 | Gene names | SYBU, GOLSYN, KIAA1472 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntabulin (Syntaxin-1-binding protein) (Golgi-localized syntaphilin- related protein). | |||||
|
TCF20_HUMAN
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.054386 (rank : 26) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UGU0, O14528, Q13078, Q4V353, Q9H4M0 | Gene names | TCF20, KIAA0292, SPBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP) (AR1). | |||||
|
WIPI2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.024387 (rank : 61) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80W47 | Gene names | Wipi2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat domain phosphoinositide-interacting protein 2 (WIPI-2). | |||||
|
AKA12_HUMAN
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.036049 (rank : 44) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
ELK1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.010432 (rank : 74) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P19419, O75606, O95058, Q969X8, Q9UJM4 | Gene names | ELK1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-1. | |||||
|
FBX40_HUMAN
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.016904 (rank : 69) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UH90, Q9ULM5 | Gene names | FBXO40, FBX40, KIAA1195 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 40 (Muscle disease-related protein). | |||||
|
IKAR_HUMAN
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.002198 (rank : 85) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13422, O00598, Q8WVA3 | Gene names | IKZF1, IK1, IKAROS, LYF1, ZNFN1A1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-binding protein Ikaros (IKAROS family zinc finger protein 1) (Lymphoid transcription factor LyF-1). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.033569 (rank : 48) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
MYCB2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.027682 (rank : 57) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75592, Q5JSX8, Q5VZN6, Q6PIB6, Q9UQ11, Q9Y6E4 | Gene names | MYCBP2, KIAA0916, PAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
NEST_HUMAN
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.013619 (rank : 71) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
WDR43_HUMAN
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.047994 (rank : 35) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q15061, Q15395, Q92577 | Gene names | WDR43, KIAA0007 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 43. | |||||
|
WDR60_HUMAN
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.032761 (rank : 49) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WVS4, Q9NW58 | Gene names | WDR60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 60. | |||||
|
CDS2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.012843 (rank : 73) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95674, Q5TDY2, Q5TDY3, Q5TDY4, Q5TDY5, Q9BYK5, Q9NTT2 | Gene names | CDS2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidate cytidylyltransferase 2 (EC 2.7.7.41) (CDP-diglyceride synthetase 2) (CDP-diglyceride pyrophosphorylase 2) (CDP- diacylglycerol synthase 2) (CDS 2) (CTP:phosphatidate cytidylyltransferase 2) (CDP-DAG synthase 2) (CDP-DG synthetase 2). | |||||
|
LRP1B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.003192 (rank : 81) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 606 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JI18, Q8BZD3, Q8BZM7 | Gene names | Lrp1b, Lrpdit | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1B precursor (Low- density lipoprotein receptor-related protein-deleted in tumor) (LRP- DIT). | |||||
|
PCP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.013377 (rank : 72) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P42785 | Gene names | PRCP, PCP | |||
|
Domain Architecture |
|
|||||
| Description | Lysosomal Pro-X carboxypeptidase precursor (EC 3.4.16.2) (Prolylcarboxypeptidase) (PRCP) (Proline carboxypeptidase) (Angiotensinase C) (Lysosomal carboxypeptidase C). | |||||
|
PEBP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.016555 (rank : 70) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P30086 | Gene names | PEBP1, PBP, PEBP | |||
|
Domain Architecture |
|
|||||
| Description | Phosphatidylethanolamine-binding protein 1 (PEBP-1) (Prostatic-binding protein) (HCNPpp) (Neuropolypeptide h3) (Raf kinase inhibitor protein) (RKIP) [Contains: Hippocampal cholinergic neurostimulating peptide (HCNP)]. | |||||
|
SALL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.002691 (rank : 84) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NSC2, Q99881, Q9NSC3, Q9P1R0 | Gene names | SALL1, SAL1 | |||
|
Domain Architecture |

|
|||||
| Description | Sal-like protein 1 (Zinc finger protein SALL1) (Spalt-like transcription factor 1) (HSal1). | |||||
|
SYT6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.004690 (rank : 79) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R0N8 | Gene names | Syt6 | |||
|
Domain Architecture |
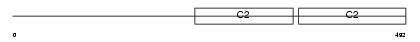
|
|||||
| Description | Synaptotagmin-6 (Synaptotagmin VI) (SytVI). | |||||
|
UHRF2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.009927 (rank : 75) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TMI3, Q8BG56, Q8BJP6, Q8BY30, Q8K1I5 | Gene names | Uhrf2, Nirf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 2 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 2) (Np95-like ring finger protein) (Nuclear zinc finger protein Np97) (NIRF). | |||||
|
WIPI2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.021284 (rank : 64) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y4P8, Q5MNZ8, Q6FI96, Q75L50, Q96IE4, Q9Y364 | Gene names | WIPI2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat domain phosphoinositide-interacting protein 2 (WIPI-2) (WIPI49-like protein 2). | |||||
|
K0329_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | O15040, Q9UEG6 | Gene names | KIAA0329, KIAA0297 | |||
|
Domain Architecture |

|
|||||
| Description | Protein KIAA0329/KIAA0297. | |||||
|
HPS5_MOUSE
|
||||||
| NC score | 0.545197 (rank : 2) | θ value | 1.16704e-23 (rank : 2) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P59438 | Gene names | Hps5, Ru2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hermansky-Pudlak syndrome 5 protein homolog (Ruby-eye protein 2) (Ru2). | |||||
|
HPS5_HUMAN
|
||||||
| NC score | 0.542882 (rank : 3) | θ value | 5.79196e-23 (rank : 3) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UPZ3, O95942, Q8N4U0 | Gene names | HPS5, AIBP63, KIAA1017 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hermansky-Pudlak syndrome 5 protein (Alpha-integrin-binding protein 63) (Ruby-eye protein 2 homolog) (Ru2). | |||||
|
NIBA_HUMAN
|
||||||
| NC score | 0.134708 (rank : 4) | θ value | 0.00035302 (rank : 4) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BZQ8, Q5TEM8, Q8TEI5, Q9H593, Q9H9Y8, Q9HCB9 | Gene names | C1orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Niban protein. | |||||
|
EPN3_MOUSE
|
||||||
| NC score | 0.096799 (rank : 5) | θ value | 0.00509761 (rank : 5) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q91W69, Q9CV55 | Gene names | Epn3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
DDB2_HUMAN
|
||||||
| NC score | 0.093452 (rank : 6) | θ value | 0.163984 (rank : 13) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q92466, Q76E54, Q76E55, Q76E56, Q76E57 | Gene names | DDB2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA damage-binding protein 2 (Damage-specific DNA-binding protein 2) (DDB p48 subunit) (DDBb) (UV-damaged DNA-binding protein 2) (UV-DDB 2). | |||||
|
IF140_HUMAN
|
||||||
| NC score | 0.090460 (rank : 7) | θ value | 0.279714 (rank : 16) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96RY7, O60332 | Gene names | IFT140, KIAA0590, WDTC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 140 homolog (WD and tetratricopeptide repeats protein 2). | |||||
|
TCOF_MOUSE
|
||||||
| NC score | 0.090260 (rank : 8) | θ value | 0.0148317 (rank : 7) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
EPN3_HUMAN
|
||||||
| NC score | 0.090024 (rank : 9) | θ value | 0.0431538 (rank : 9) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H201, Q9BVN6, Q9NWK2 | Gene names | EPN3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
DDB2_MOUSE
|
||||||
| NC score | 0.087410 (rank : 10) | θ value | 0.47712 (rank : 20) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99J79 | Gene names | Ddb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA damage-binding protein 2 (Damage-specific DNA-binding protein 2). | |||||
|
WDR19_MOUSE
|
||||||
| NC score | 0.083968 (rank : 11) | θ value | 0.47712 (rank : 21) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q3UGF1, Q6ZPK8, Q80VQ6, Q8C794, Q8K3R5 | Gene names | Wdr19, Kiaa1638 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 19. | |||||
|
WDR19_HUMAN
|
||||||
| NC score | 0.082609 (rank : 12) | θ value | 0.62314 (rank : 27) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8NEZ3, Q8N5B4, Q9H5S0, Q9HCD4 | Gene names | WDR19, KIAA1638 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 19. | |||||
|
TCF20_MOUSE
|
||||||
| NC score | 0.077951 (rank : 13) | θ value | 0.0330416 (rank : 8) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
BRWD2_HUMAN
|
||||||
| NC score | 0.071732 (rank : 14) | θ value | 0.62314 (rank : 23) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BZH6, Q5VWA1 | Gene names | BRWD2, WDR11 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain and WD repeat domain-containing protein 2 (WD repeat protein 11). | |||||
|
WDR26_MOUSE
|
||||||
| NC score | 0.070048 (rank : 15) | θ value | 0.365318 (rank : 19) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8C6G8, Q8C4F3, Q8VDZ6 | Gene names | Wdr26 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 26. | |||||
|
WDR26_HUMAN
|
||||||
| NC score | 0.069458 (rank : 16) | θ value | 0.365318 (rank : 18) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9H7D7, Q86UY4 | Gene names | WDR26 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 26. | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.065913 (rank : 17) | θ value | 0.0961366 (rank : 10) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
GT2D1_MOUSE
|
||||||
| NC score | 0.065048 (rank : 18) | θ value | 0.279714 (rank : 15) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JI57, Q8VHD5, Q8VI58, Q9EQE7, Q9ESZ6, Q9ESZ7 | Gene names | Gtf2ird1, Ben | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor II-I repeat domain-containing protein 1 (GTF2I repeat domain-containing protein 1) (Binding factor for early enhancer). | |||||
|
PI5PA_HUMAN
|
||||||
| NC score | 0.061293 (rank : 19) | θ value | 0.0113563 (rank : 6) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
WDR16_HUMAN
|
||||||
| NC score | 0.059344 (rank : 20) | θ value | 0.62314 (rank : 25) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8N1V2, Q5DX23, Q8TC73, Q8TCI3 | Gene names | WDR16, WDRPUH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 16 (WD40-repeat protein up-regulated in HCC). | |||||
|
WDR16_MOUSE
|
||||||
| NC score | 0.059084 (rank : 21) | θ value | 0.62314 (rank : 26) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5F201, Q5F200, Q9D432, Q9DA68 | Gene names | WDR16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 16. | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.058796 (rank : 22) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
WDR22_HUMAN
|
||||||
| NC score | 0.056906 (rank : 23) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96JK2, O60559, Q8N3V3, Q8N3V5 | Gene names | WDR22, BCRG2, KIAA1824 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 22 (Breakpoint cluster region protein 2) (BCRP2). | |||||
|
WDR22_MOUSE
|
||||||
| NC score | 0.056327 (rank : 24) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q80T85, Q80VT3, Q80ZW6, Q8BIP8, Q8BVK5, Q8K0Q6 | Gene names | Wdr22, Kiaa1824 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 22. | |||||
|
PWP2_MOUSE
|
||||||
| NC score | 0.055173 (rank : 25) | θ value | 1.06291 (rank : 34) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BU03, Q8BTR0 | Gene names | Pwp2h | |||
|
Domain Architecture |

|
|||||
| Description | Periodic tryptophan protein 2 homolog. | |||||
|
TCF20_HUMAN
|
||||||
| NC score | 0.054386 (rank : 26) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UGU0, O14528, Q13078, Q4V353, Q9H4M0 | Gene names | TCF20, KIAA0292, SPBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP) (AR1). | |||||
|
PWP2_HUMAN
|
||||||
| NC score | 0.054031 (rank : 27) | θ value | 1.06291 (rank : 33) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q15269, Q96A77 | Gene names | PWP2H, PWP2 | |||
|
Domain Architecture |

|
|||||
| Description | Periodic tryptophan protein 2 homolog. | |||||
|
AHI1_HUMAN
|
||||||
| NC score | 0.054026 (rank : 28) | θ value | 0.125558 (rank : 11) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 537 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8N157, Q504T3, Q5TCP9, Q6P098, Q6PIT6, Q8NDX0, Q9H0H2 | Gene names | AHI1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Jouberin (Abelson helper integration site 1 protein homolog) (AHI-1). | |||||
|
A16L1_HUMAN
|
||||||
| NC score | 0.051529 (rank : 29) | θ value | 0.813845 (rank : 28) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 664 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q676U5, Q6IPN1, Q6UXW4, Q6ZVZ5, Q8NCY2, Q96JV5, Q9H619 | Gene names | ATG16L1, APG16L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autophagy-related protein 16-1 (APG16-like 1). | |||||
|
A16L1_MOUSE
|
||||||
| NC score | 0.051251 (rank : 30) | θ value | 0.62314 (rank : 22) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 776 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8C0J2, Q6KAT7, Q80U97, Q80U98, Q80U99, Q80Y53, Q9DB63 | Gene names | Atg16l1, Apg16l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autophagy-related protein 16-1 (APG16-like 1). | |||||
|
GT2D1_HUMAN
|
||||||
| NC score | 0.050137 (rank : 31) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UHL9, O95444, Q75MX7, Q86UM3, Q8WVC4, Q9UHK8, Q9UI91 | Gene names | GTF2IRD1, CREAM1, GTF3, MUSTRD1, RBAP2, WBSCR11, WBSCR12 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor II-I repeat domain-containing protein 1 (GTF2I repeat domain-containing protein 1) (Muscle TFII-I repeat domain-containing protein 1) (General transcription factor III) (Slow- muscle-fiber enhancer-binding protein) (USE B1-binding protein) (MusTRD1/BEN) (Williams-Beuren syndrome chromosome region 11 protein). | |||||
|
GON4L_HUMAN
|
||||||
| NC score | 0.050016 (rank : 32) | θ value | 0.365318 (rank : 17) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
NLE1_HUMAN
|
||||||
| NC score | 0.048736 (rank : 33) | θ value | 1.06291 (rank : 31) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9NVX2, O60868, Q9BU54 | Gene names | NLE1 | |||
|
Domain Architecture |

|
|||||
| Description | Notchless homolog 1. | |||||
|
NLE1_MOUSE
|
||||||
| NC score | 0.048438 (rank : 34) | θ value | 1.06291 (rank : 32) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8VEJ4 | Gene names | Nle1 | |||
|
Domain Architecture |

|
|||||
| Description | Notchless homolog 1. | |||||
|
WDR43_HUMAN
|
||||||
| NC score | 0.047994 (rank : 35) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q15061, Q15395, Q92577 | Gene names | WDR43, KIAA0007 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 43. | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.047675 (rank : 36) | θ value | 0.21417 (rank : 14) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
NSD1_MOUSE
|
||||||
| NC score | 0.047194 (rank : 37) | θ value | 0.125558 (rank : 12) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
GEMI5_MOUSE
|
||||||
| NC score | 0.043468 (rank : 38) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 424 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BX17 | Gene names | Gemin5 | |||
|
Domain Architecture |

|
|||||
| Description | Gem-associated protein 5 (Gemin5). | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.040381 (rank : 39) | θ value | 1.06291 (rank : 30) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
OXR1_HUMAN
|
||||||
| NC score | 0.040198 (rank : 40) | θ value | 0.813845 (rank : 29) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8N573, Q3LIB5, Q6ZVK9, Q8N8V0, Q9H266, Q9NWC7 | Gene names | OXR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidation resistance protein 1. | |||||
|
WDR60_MOUSE
|
||||||
| NC score | 0.040126 (rank : 41) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8C761, Q3UNM4 | Gene names | Wdr60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 60. | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.038195 (rank : 42) | θ value | 1.06291 (rank : 35) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
TOIP2_MOUSE
|
||||||
| NC score | 0.036426 (rank : 43) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BYU6, Q6NY10, Q8BJP0 | Gene names | Tor1aip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Torsin-1A-interacting protein 2. | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.036049 (rank : 44) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
IRS2_MOUSE
|
||||||
| NC score | 0.035880 (rank : 45) | θ value | 1.38821 (rank : 37) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P81122 | Gene names | Irs2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin receptor substrate 2 (IRS-2) (4PS). | |||||
|
CIR1A_MOUSE
|
||||||
| NC score | 0.033908 (rank : 46) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8R2N2, Q62306, Q69Z48, Q6NVG9, Q8VEL5 | Gene names | Cirh1a, Kiaa1988, Tex292 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cirhin (Testis-expressed gene 292 protein). | |||||
|
REV1_MOUSE
|
||||||
| NC score | 0.033623 (rank : 47) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q920Q2, Q9QXV2 | Gene names | Rev1l, Rev1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein REV1 (EC 2.7.7.-) (Rev1-like terminal deoxycytidyl transferase). | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.033569 (rank : 48) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
WDR60_HUMAN
|
||||||
| NC score | 0.032761 (rank : 49) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WVS4, Q9NW58 | Gene names | WDR60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 60. | |||||
|
RFWD2_HUMAN
|
||||||
| NC score | 0.031702 (rank : 50) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8NHY2, Q6H103, Q9H6L7 | Gene names | RFWD2, COP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger and WD repeat domain protein 2 (EC 6.3.2.-) (Ubiquitin- protein ligase COP1) (Constitutive photomorphogenesis protein 1 homolog) (hCOP1). | |||||
|
RFWD2_MOUSE
|
||||||
| NC score | 0.031550 (rank : 51) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9R1A8 | Gene names | Rfwd2, Cop1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger and WD repeat domain protein 2 (EC 6.3.2.-) (Ubiquitin- protein ligase COP1) (Constitutive photomorphogenesis protein 1 homolog) (mCOP1). | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.029713 (rank : 52) | θ value | 1.06291 (rank : 36) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
MYCB2_MOUSE
|
||||||
| NC score | 0.029643 (rank : 53) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7TPH6, Q6PCM8 | Gene names | Mycbp2, Pam, Phr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
MAP4_MOUSE
|
||||||
| NC score | 0.028766 (rank : 54) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
HSPB8_MOUSE
|
||||||
| NC score | 0.028444 (rank : 55) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JK92 | Gene names | Hspb8, Cryac, Hsp22 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein beta-8 (HspB8) (Alpha crystallin C chain) (Small stress protein-like protein HSP22). | |||||
|
TCAL5_HUMAN
|
||||||
| NC score | 0.027987 (rank : 56) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
MYCB2_HUMAN
|
||||||
| NC score | 0.027682 (rank : 57) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75592, Q5JSX8, Q5VZN6, Q6PIB6, Q9UQ11, Q9Y6E4 | Gene names | MYCBP2, KIAA0916, PAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
TCGAP_MOUSE
|
||||||
| NC score | 0.027537 (rank : 58) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80YF9, Q6P7W6 | Gene names | Snx26, Tcgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
GFPT1_MOUSE
|
||||||
| NC score | 0.025509 (rank : 59) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P47856, Q91XG9, Q99LP7, Q99MJ4 | Gene names | Gfpt1, Gfpt | |||
|
Domain Architecture |

|
|||||
| Description | Glucosamine--fructose-6-phosphate aminotransferase [isomerizing] 1 (EC 2.6.1.16) (Hexosephosphate aminotransferase 1) (D-fructose-6- phosphate amidotransferase 1) (GFAT 1) (GFAT1). | |||||
|
GFPT1_HUMAN
|
||||||
| NC score | 0.025501 (rank : 60) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q06210, Q9BXF8 | Gene names | GFPT1, GFAT, GFPT | |||
|
Domain Architecture |

|
|||||
| Description | Glucosamine--fructose-6-phosphate aminotransferase [isomerizing] 1 (EC 2.6.1.16) (Hexosephosphate aminotransferase 1) (D-fructose-6- phosphate amidotransferase 1) (GFAT 1) (GFAT1). | |||||
|
WIPI2_MOUSE
|
||||||
| NC score | 0.024387 (rank : 61) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80W47 | Gene names | Wipi2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat domain phosphoinositide-interacting protein 2 (WIPI-2). | |||||
|
EP300_HUMAN
|
||||||
| NC score | 0.024312 (rank : 62) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.023302 (rank : 63) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
WIPI2_HUMAN
|
||||||
| NC score | 0.021284 (rank : 64) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y4P8, Q5MNZ8, Q6FI96, Q75L50, Q96IE4, Q9Y364 | Gene names | WIPI2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat domain phosphoinositide-interacting protein 2 (WIPI-2) (WIPI49-like protein 2). | |||||
|
SYBU_HUMAN
|
||||||
| NC score | 0.020932 (rank : 65) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NX95, Q5R1T1, Q5R1T2, Q5R1T3, Q5Y2M6, Q8ND49, Q8TCR6, Q9P256 | Gene names | SYBU, GOLSYN, KIAA1472 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntabulin (Syntaxin-1-binding protein) (Golgi-localized syntaphilin- related protein). | |||||
|
ZN409_HUMAN
|
||||||
| NC score | 0.018863 (rank : 66) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UPU6 | Gene names | ZNF409, KIAA1056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 409. | |||||
|
SM1L2_HUMAN
|
||||||
| NC score | 0.018453 (rank : 67) | θ value | 0.62314 (rank : 24) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1019 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NDV3, Q5TIC3, Q9Y3G5 | Gene names | SMC1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 2 protein (SMC1beta protein). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.017707 (rank : 68) | θ value | 1.81305 (rank : 38) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
FBX40_HUMAN
|
||||||
| NC score | 0.016904 (rank : 69) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UH90, Q9ULM5 | Gene names | FBXO40, FBX40, KIAA1195 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 40 (Muscle disease-related protein). | |||||
|
PEBP1_HUMAN
|
||||||
| NC score | 0.016555 (rank : 70) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P30086 | Gene names | PEBP1, PBP, PEBP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylethanolamine-binding protein 1 (PEBP-1) (Prostatic-binding protein) (HCNPpp) (Neuropolypeptide h3) (Raf kinase inhibitor protein) (RKIP) [Contains: Hippocampal cholinergic neurostimulating peptide (HCNP)]. | |||||
|
NEST_HUMAN
|
||||||
| NC score | 0.013619 (rank : 71) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
PCP_HUMAN
|
||||||
| NC score | 0.013377 (rank : 72) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P42785 | Gene names | PRCP, PCP | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal Pro-X carboxypeptidase precursor (EC 3.4.16.2) (Prolylcarboxypeptidase) (PRCP) (Proline carboxypeptidase) (Angiotensinase C) (Lysosomal carboxypeptidase C). | |||||
|
CDS2_HUMAN
|
||||||
| NC score | 0.012843 (rank : 73) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95674, Q5TDY2, Q5TDY3, Q5TDY4, Q5TDY5, Q9BYK5, Q9NTT2 | Gene names | CDS2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidate cytidylyltransferase 2 (EC 2.7.7.41) (CDP-diglyceride synthetase 2) (CDP-diglyceride pyrophosphorylase 2) (CDP- diacylglycerol synthase 2) (CDS 2) (CTP:phosphatidate cytidylyltransferase 2) (CDP-DAG synthase 2) (CDP-DG synthetase 2). | |||||
|
ELK1_HUMAN
|
||||||
| NC score | 0.010432 (rank : 74) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P19419, O75606, O95058, Q969X8, Q9UJM4 | Gene names | ELK1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-1. | |||||
|
UHRF2_MOUSE
|
||||||
| NC score | 0.009927 (rank : 75) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TMI3, Q8BG56, Q8BJP6, Q8BY30, Q8K1I5 | Gene names | Uhrf2, Nirf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 2 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 2) (Np95-like ring finger protein) (Nuclear zinc finger protein Np97) (NIRF). | |||||
|
KPCD1_MOUSE
|
||||||
| NC score | 0.006126 (rank : 76) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 891 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q62101 | Gene names | Prkd1, Pkcm, Pkd, Prkcm | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase D1 (EC 2.7.11.13) (nPKC-D1) (Protein kinase D) (Protein kinase C mu type) (nPKC-mu). | |||||
|
KPCD1_HUMAN
|
||||||
| NC score | 0.006071 (rank : 77) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 901 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15139 | Gene names | PRKD1, PKD, PKD1, PRKCM | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase D1 (EC 2.7.11.13) (nPKC-D1) (Protein kinase D) (Protein kinase C mu type) (nPKC-mu). | |||||
|
M3K12_MOUSE
|
||||||
| NC score | 0.005026 (rank : 78) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q60700, P70286, Q3TLL7, Q8C4N7, Q8CBX3, Q8CDL6 | Gene names | Map3k12, Zpk | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 12 (EC 2.7.11.25) (Mixed lineage kinase) (Leucine-zipper protein kinase) (ZPK) (Dual leucine zipper bearing kinase) (DLK) (MAPK-upstream kinase) (MUK). | |||||
|
SYT6_MOUSE
|
||||||
| NC score | 0.004690 (rank : 79) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R0N8 | Gene names | Syt6 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-6 (Synaptotagmin VI) (SytVI). | |||||
|
M3K12_HUMAN
|
||||||
| NC score | 0.004595 (rank : 80) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q12852, Q86VQ5, Q8WY25 | Gene names | MAP3K12, ZPK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 12 (EC 2.7.11.25) (Mixed lineage kinase) (Leucine-zipper protein kinase) (ZPK) (Dual leucine zipper bearing kinase) (DLK) (MAPK-upstream kinase) (MUK). | |||||
|
LRP1B_MOUSE
|
||||||
| NC score | 0.003192 (rank : 81) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 606 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JI18, Q8BZD3, Q8BZM7 | Gene names | Lrp1b, Lrpdit | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1B precursor (Low- density lipoprotein receptor-related protein-deleted in tumor) (LRP- DIT). | |||||
|
ZNF18_MOUSE
|
||||||
| NC score | 0.002834 (rank : 82) | θ value | 2.36792 (rank : 45) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 743 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q810A1, Q9D972 | Gene names | Znf18, Zfp535 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 18 (Zinc finger protein 535). | |||||
|
GLIS1_HUMAN
|
||||||
| NC score | 0.002708 (rank : 83) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NBF1, Q8N9V9 | Gene names | GLIS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLIS1 (GLI-similar 1). | |||||
|
SALL1_HUMAN
|
||||||
| NC score | 0.002691 (rank : 84) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NSC2, Q99881, Q9NSC3, Q9P1R0 | Gene names | SALL1, SAL1 | |||
|
Domain Architecture |

|
|||||
| Description | Sal-like protein 1 (Zinc finger protein SALL1) (Spalt-like transcription factor 1) (HSal1). | |||||
|
IKAR_HUMAN
|
||||||
| NC score | 0.002198 (rank : 85) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13422, O00598, Q8WVA3 | Gene names | IKZF1, IK1, IKAROS, LYF1, ZNFN1A1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-binding protein Ikaros (IKAROS family zinc finger protein 1) (Lymphoid transcription factor LyF-1). | |||||