Please be patient as the page loads
|
ELK1_HUMAN
|
||||||
| SwissProt Accessions | P19419, O75606, O95058, Q969X8, Q9UJM4 | Gene names | ELK1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-1. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ELK1_HUMAN
|
||||||
| θ value | 1.04523e-173 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 112 | |
| SwissProt Accessions | P19419, O75606, O95058, Q969X8, Q9UJM4 | Gene names | ELK1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-1. | |||||
|
ELK1_MOUSE
|
||||||
| θ value | 2.85304e-163 (rank : 2) | NC score | 0.990370 (rank : 2) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P41969 | Gene names | Elk1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-1. | |||||
|
ELK4_HUMAN
|
||||||
| θ value | 7.25919e-50 (rank : 3) | NC score | 0.944914 (rank : 6) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P28324, P28323 | Gene names | ELK4, SAP1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
|
ELK3_HUMAN
|
||||||
| θ value | 1.2411e-41 (rank : 4) | NC score | 0.955805 (rank : 3) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P41970, Q6FG57, Q6GU29, Q9UD17 | Gene names | ELK3, NET, SAP2 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-3 (ETS-related protein NET) (ETS- related protein ERP) (SRF accessory protein 2) (SAP-2). | |||||
|
ELK3_MOUSE
|
||||||
| θ value | 1.79215e-40 (rank : 5) | NC score | 0.953017 (rank : 4) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P41971, P97747, Q62346 | Gene names | Elk3, Erp, Net | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-3 (ETS-related protein NET) (ETS- related protein ERP). | |||||
|
ELK4_MOUSE
|
||||||
| θ value | 5.21438e-40 (rank : 6) | NC score | 0.952547 (rank : 5) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P41158 | Gene names | Elk4, Sap1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
|
ETV4_HUMAN
|
||||||
| θ value | 4.28545e-26 (rank : 7) | NC score | 0.874555 (rank : 28) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P43268, Q96AW9 | Gene names | ETV4, E1AF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 4 (Adenovirus E1A enhancer-binding protein) (E1A-F). | |||||
|
ETV4_MOUSE
|
||||||
| θ value | 4.28545e-26 (rank : 8) | NC score | 0.875152 (rank : 27) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P28322 | Gene names | Etv4, Pea-3, Pea3 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 4 (Polyomavirus enhancer activator 3) (Protein PEA3). | |||||
|
ETV1_HUMAN
|
||||||
| θ value | 7.30988e-26 (rank : 9) | NC score | 0.877646 (rank : 26) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P50549, O75849, Q9UQ71, Q9Y636 | Gene names | ETV1, ER81 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 1 (ER81 protein). | |||||
|
ETV1_MOUSE
|
||||||
| θ value | 7.30988e-26 (rank : 10) | NC score | 0.878575 (rank : 22) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P41164 | Gene names | Etv1, Er81, Etsrp81 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 1 (ER81 protein). | |||||
|
ETV5_HUMAN
|
||||||
| θ value | 1.62847e-25 (rank : 11) | NC score | 0.873017 (rank : 29) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P41161 | Gene names | ETV5, ERM | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 5 (Ets-related protein ERM). | |||||
|
ETV5_MOUSE
|
||||||
| θ value | 1.62847e-25 (rank : 12) | NC score | 0.852485 (rank : 36) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9CXC9, Q3TG49, Q8C0F3, Q9JHB1 | Gene names | Etv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 5. | |||||
|
ETS2_HUMAN
|
||||||
| θ value | 1.37858e-24 (rank : 13) | NC score | 0.898834 (rank : 9) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P15036 | Gene names | ETS2 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-2 protein. | |||||
|
ETS2_MOUSE
|
||||||
| θ value | 1.37858e-24 (rank : 14) | NC score | 0.897381 (rank : 12) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P15037 | Gene names | Ets2 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-2 protein. | |||||
|
ERG_MOUSE
|
||||||
| θ value | 3.07116e-24 (rank : 15) | NC score | 0.881722 (rank : 20) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P81270, Q8C5L4, Q920K7, Q920K8, Q920K9 | Gene names | Erg, Erg-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ERG. | |||||
|
ETS1_HUMAN
|
||||||
| θ value | 5.23862e-24 (rank : 16) | NC score | 0.897692 (rank : 10) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P14921, Q14278, Q16080 | Gene names | ETS1 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-1 protein (p54). | |||||
|
ETS1_MOUSE
|
||||||
| θ value | 5.23862e-24 (rank : 17) | NC score | 0.897683 (rank : 11) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P27577, Q61403 | Gene names | Ets1, Ets-1 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-1 protein (p54). | |||||
|
ETV3_MOUSE
|
||||||
| θ value | 5.23862e-24 (rank : 18) | NC score | 0.878072 (rank : 25) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8R4Z4, Q9QZW1 | Gene names | Etv3, Mets, Pe1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
ETV3_HUMAN
|
||||||
| θ value | 6.84181e-24 (rank : 19) | NC score | 0.853636 (rank : 34) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P41162, Q8TAC8, Q9BX30 | Gene names | ETV3, METS, PE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
ERG_HUMAN
|
||||||
| θ value | 1.16704e-23 (rank : 20) | NC score | 0.882059 (rank : 19) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P11308, Q16113 | Gene names | ERG | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ERG (Transforming protein ERG). | |||||
|
FLI1_HUMAN
|
||||||
| θ value | 1.16704e-23 (rank : 21) | NC score | 0.887316 (rank : 16) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q01543, Q14319, Q92480, Q9UE07 | Gene names | FLI1 | |||
|
Domain Architecture |

|
|||||
| Description | Friend leukemia integration 1 transcription factor (Fli-1 proto- oncogene) (ERGB transcription factor). | |||||
|
FLI1_MOUSE
|
||||||
| θ value | 1.16704e-23 (rank : 22) | NC score | 0.888505 (rank : 15) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P26323 | Gene names | Fli1, Fli-1 | |||
|
Domain Architecture |

|
|||||
| Description | Friend leukemia integration 1 transcription factor (Retroviral integration site protein Fli-1). | |||||
|
ELF1_HUMAN
|
||||||
| θ value | 4.43474e-23 (rank : 23) | NC score | 0.861675 (rank : 31) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P32519, Q9UDE1 | Gene names | ELF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
ERF_HUMAN
|
||||||
| θ value | 2.87452e-22 (rank : 24) | NC score | 0.878249 (rank : 24) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P50548, Q9UPI7 | Gene names | ERF | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing transcription factor ERF (Ets2 repressor factor). | |||||
|
ERF_MOUSE
|
||||||
| θ value | 2.87452e-22 (rank : 25) | NC score | 0.878415 (rank : 23) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P70459 | Gene names | Erf | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing transcription factor ERF. | |||||
|
ELF1_MOUSE
|
||||||
| θ value | 8.36355e-22 (rank : 26) | NC score | 0.853137 (rank : 35) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q60775 | Gene names | Elf1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
GABPA_HUMAN
|
||||||
| θ value | 7.0802e-21 (rank : 27) | NC score | 0.895862 (rank : 14) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q06546, Q12939 | Gene names | GABPA, E4TF1A | |||
|
Domain Architecture |

|
|||||
| Description | GA-binding protein alpha chain (GABP-subunit alpha) (Transcription factor E4TF1-60) (Nuclear respiratory factor 2 subunit alpha). | |||||
|
GABPA_MOUSE
|
||||||
| θ value | 7.0802e-21 (rank : 28) | NC score | 0.897327 (rank : 13) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q00422 | Gene names | Gabpa, E4tf1a | |||
|
Domain Architecture |

|
|||||
| Description | GA-binding protein alpha chain (GABP-subunit alpha). | |||||
|
ELF4_MOUSE
|
||||||
| θ value | 9.24701e-21 (rank : 29) | NC score | 0.837753 (rank : 41) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Z2U4 | Gene names | Elf4, Mef | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-4 (E74-like factor 4) (Myeloid Elf-1-like factor). | |||||
|
ETV2_HUMAN
|
||||||
| θ value | 1.2077e-20 (rank : 30) | NC score | 0.911012 (rank : 7) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O00321, Q9UEA0 | Gene names | ETV2, ER71, ETSRP71 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 2 (Ets-related protein 71). | |||||
|
ELF4_HUMAN
|
||||||
| θ value | 2.06002e-20 (rank : 31) | NC score | 0.860030 (rank : 33) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q99607, O60435 | Gene names | ELF4, ELFR, MEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-4 (E74-like factor 4) (Myeloid Elf-1-like factor). | |||||
|
ELF2_MOUSE
|
||||||
| θ value | 5.99374e-20 (rank : 32) | NC score | 0.862393 (rank : 30) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9JHC9, Q6NST2, Q8BTX8, Q9JHC7, Q9JHC8, Q9JHD0 | Gene names | Elf2, Nerf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-2 (E74-like factor 2) (New ETS- related factor). | |||||
|
ELF2_HUMAN
|
||||||
| θ value | 1.33526e-19 (rank : 33) | NC score | 0.860795 (rank : 32) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q15723, Q15724, Q15725, Q6P1K5 | Gene names | ELF2, NERF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-2 (E74-like factor 2) (New ETS- related factor). | |||||
|
ETV2_MOUSE
|
||||||
| θ value | 8.65492e-19 (rank : 34) | NC score | 0.907173 (rank : 8) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P41163 | Gene names | Etv2, Er71, Etsrp71 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 2 (Ets-related protein 71). | |||||
|
SPDEF_HUMAN
|
||||||
| θ value | 7.32683e-18 (rank : 35) | NC score | 0.883237 (rank : 18) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O95238 | Gene names | SPDEF, PDEF, PSE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAM pointed domain-containing Ets transcription factor (Prostate- derived Ets factor) (Prostate epithelium-specific Ets transcription factor) (Prostate-specific Ets). | |||||
|
SPDEF_MOUSE
|
||||||
| θ value | 9.56915e-18 (rank : 36) | NC score | 0.881562 (rank : 21) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9WTP3 | Gene names | Spdef, Pdef, Pse | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAM pointed domain-containing Ets transcription factor (Prostate- derived Ets factor) (Prostate epithelium-specific Ets transcription factor) (Prostate-specific Ets). | |||||
|
ETV6_HUMAN
|
||||||
| θ value | 1.63225e-17 (rank : 37) | NC score | 0.848087 (rank : 38) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P41212, Q9UMF6 | Gene names | ETV6, TEL, TEL1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV6 (ETS-related protein Tel1) (Tel) (ETS translocation variant 6). | |||||
|
ETV6_MOUSE
|
||||||
| θ value | 1.63225e-17 (rank : 38) | NC score | 0.851656 (rank : 37) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P97360 | Gene names | Etv6, Tel, Tel1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV6 (ETS-related protein Tel1) (Tel) (ETS translocation variant 6). | |||||
|
ETV7_HUMAN
|
||||||
| θ value | 1.80466e-16 (rank : 39) | NC score | 0.886683 (rank : 17) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9Y603, Q9NZ65, Q9NZ66, Q9NZ68, Q9NZR8, Q9UNJ7, Q9Y5K4, Q9Y604 | Gene names | ETV7, TEL2, TELB, TREF | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV7 (Transcription factor Tel-2) (ETS-related protein Tel2) (Tel-related Ets factor). | |||||
|
ELF5_HUMAN
|
||||||
| θ value | 4.16044e-13 (rank : 40) | NC score | 0.840244 (rank : 40) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UKW6, O95175, Q8N2K9, Q96QY3, Q9UKW5 | Gene names | ELF5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-5 (E74-like factor 5) (Epithelium-specific Ets transcription factor 2) (ESE-2) (Epithelium- restricted ESE-1-related Ets factor). | |||||
|
ELF5_MOUSE
|
||||||
| θ value | 4.16044e-13 (rank : 41) | NC score | 0.841507 (rank : 39) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8VDK3, Q921H5, Q9Z2K6 | Gene names | Elf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-5 (E74-like factor 5). | |||||
|
SPI1_HUMAN
|
||||||
| θ value | 4.45536e-07 (rank : 42) | NC score | 0.662749 (rank : 43) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P17947 | Gene names | SPI1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor PU.1 (31 kDa transforming protein). | |||||
|
SPI1_MOUSE
|
||||||
| θ value | 4.45536e-07 (rank : 43) | NC score | 0.664141 (rank : 42) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P17433, Q99L57 | Gene names | Spi1, Sfpi-1, Sfpi1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor PU.1 (31 kDa transforming protein) (SFFV proviral integration 1 protein). | |||||
|
SPIB_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 44) | NC score | 0.650539 (rank : 44) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q01892, Q15359 | Gene names | SPIB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Spi-B. | |||||
|
SPIB_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 45) | NC score | 0.636618 (rank : 45) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O35906, O35907, O35909, O55199 | Gene names | Spib | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-B. | |||||
|
SPIC_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 46) | NC score | 0.614177 (rank : 46) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8N5J4 | Gene names | SPIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-C. | |||||
|
SPIC_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 47) | NC score | 0.581869 (rank : 47) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6P3D7, Q9Z0Y0 | Gene names | Spic | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-C (Pu.1-related factor) (Prf). | |||||
|
TXND2_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 48) | NC score | 0.036067 (rank : 50) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 49) | NC score | 0.044855 (rank : 49) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
PHC2_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 50) | NC score | 0.033351 (rank : 51) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8IXK0, Q5T0C1, Q6NUJ6, Q6ZQR1, Q8N306, Q8TAG8, Q96BL4, Q9Y4Y7 | Gene names | PHC2, EDR2, PH2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 2 (hPH2) (Early development regulatory protein 2). | |||||
|
GON4L_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 51) | NC score | 0.031900 (rank : 52) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9DB00, Q80TB4, Q91YI9 | Gene names | Gon4l, Gon4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 52) | NC score | 0.023344 (rank : 57) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
APXL_HUMAN
|
||||||
| θ value | 0.125558 (rank : 53) | NC score | 0.030978 (rank : 53) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13796 | Gene names | APXL | |||
|
Domain Architecture |

|
|||||
| Description | Apical-like protein (Protein APXL). | |||||
|
SSBP4_HUMAN
|
||||||
| θ value | 0.125558 (rank : 54) | NC score | 0.029982 (rank : 54) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 0.21417 (rank : 55) | NC score | 0.018214 (rank : 65) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
TAF10_HUMAN
|
||||||
| θ value | 0.279714 (rank : 56) | NC score | 0.045815 (rank : 48) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q12962, O00703, Q13175 | Gene names | TAF10, TAF2H, TAFII30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 10 (Transcription initiation factor TFIID 30 kDa subunit) (TAF(II)30) (TAFII-30) (TAFII30) (STAF28). | |||||
|
GHR_HUMAN
|
||||||
| θ value | 0.62314 (rank : 57) | NC score | 0.027504 (rank : 55) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P10912, Q9HCX2 | Gene names | GHR | |||
|
Domain Architecture |
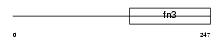
|
|||||
| Description | Growth hormone receptor precursor (GH receptor) (Somatotropin receptor) [Contains: Growth hormone-binding protein (GH-binding protein) (GHBP) (Serum-binding protein)]. | |||||
|
LIRB5_HUMAN
|
||||||
| θ value | 0.813845 (rank : 58) | NC score | 0.007657 (rank : 91) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75023, Q8N760 | Gene names | LILRB5, LIR8 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily B member 5 precursor (Leukocyte immunoglobulin-like receptor 8) (LIR-8) (CD85c antigen). | |||||
|
SC24C_HUMAN
|
||||||
| θ value | 0.813845 (rank : 59) | NC score | 0.020591 (rank : 61) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
DOCK6_HUMAN
|
||||||
| θ value | 1.38821 (rank : 60) | NC score | 0.011006 (rank : 77) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96HP0, Q9P2F2 | Gene names | DOCK6, KIAA1395 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 6. | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 1.38821 (rank : 61) | NC score | 0.019179 (rank : 62) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
FIGN_HUMAN
|
||||||
| θ value | 1.81305 (rank : 62) | NC score | 0.010706 (rank : 80) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5HY92, Q9H6M5, Q9NVZ9 | Gene names | FIGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
GLI2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 63) | NC score | -0.002219 (rank : 108) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 971 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P10070, O60252, O60253, O60254, O60255, Q15590, Q15591 | Gene names | GLI2, THP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI2 (Tax helper protein). | |||||
|
LIMD1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 64) | NC score | 0.010226 (rank : 86) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9QXD8, Q8C8G4, Q9CW55 | Gene names | Limd1 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain-containing protein 1. | |||||
|
NEST_HUMAN
|
||||||
| θ value | 2.36792 (rank : 65) | NC score | 0.003441 (rank : 103) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
PO121_MOUSE
|
||||||
| θ value | 2.36792 (rank : 66) | NC score | 0.022831 (rank : 58) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
ATF6B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 67) | NC score | 0.012601 (rank : 74) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99941, Q13269, Q14343, Q14345, Q99635, Q99637, Q9H3V9, Q9H3W1, Q9NPL0 | Gene names | CREBL1, G13 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-6 beta (Activating transcription factor 6 beta) (ATF6-beta) (cAMP-responsive element- binding protein-like 1) (cAMP response element-binding protein-related protein) (Creb-rp) (Protein G13). | |||||
|
COAA1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 68) | NC score | 0.008843 (rank : 89) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q03692 | Gene names | COL10A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(X) chain precursor. | |||||
|
FOXK2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 69) | NC score | 0.007290 (rank : 92) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q01167, Q13622, Q13623, Q13624 | Gene names | FOXK2, ILF, ILF1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein K2 (Interleukin enhancer-binding factor 1) (Cellular transcription factor ILF-1). | |||||
|
MYCD_MOUSE
|
||||||
| θ value | 3.0926 (rank : 70) | NC score | 0.011288 (rank : 76) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8VIM5, Q8C3W6, Q8VIL4 | Gene names | Myocd, Bsac2, Mycd, Srfcp | |||
|
Domain Architecture |

|
|||||
| Description | Myocardin (SRF cofactor protein) (Basic SAP coiled-coil transcription activator 2). | |||||
|
PRGR_MOUSE
|
||||||
| θ value | 3.0926 (rank : 71) | NC score | 0.004438 (rank : 101) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q00175 | Gene names | Pgr, Nr3c3, Pr | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone receptor (PR). | |||||
|
SC24A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 72) | NC score | 0.021820 (rank : 60) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q3U2P1, Q3TQ05, Q3TRG7, Q8BIS0 | Gene names | Sec24a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein transport protein Sec24A (SEC24-related protein A). | |||||
|
SIGIR_MOUSE
|
||||||
| θ value | 3.0926 (rank : 73) | NC score | 0.014764 (rank : 71) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JLZ8, Q52L48 | Gene names | Sigirr, Tir8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Single Ig IL-1-related receptor (Single Ig IL-1R-related molecule) (Single immunoglobulin domain-containing IL1R-related protein) (Toll/interleukin-1 receptor 8) (TIR8). | |||||
|
CO1A1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 74) | NC score | 0.007289 (rank : 93) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P02452, P78441, Q13896, Q13902, Q13903, Q14037, Q14992, Q15176, Q15201, Q16050, Q7KZ30, Q7KZ34, Q8IVI5, Q9UML6, Q9UMM7 | Gene names | COL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(I) chain precursor. | |||||
|
FIGN_MOUSE
|
||||||
| θ value | 4.03905 (rank : 75) | NC score | 0.009409 (rank : 87) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ERZ6, Q3TPB0, Q3UP57, Q6PCM0 | Gene names | Fign | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
GMIP_MOUSE
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | 0.005206 (rank : 99) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PGG2, Q6P9S3 | Gene names | Gmip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GEM-interacting protein (GMIP). | |||||
|
GT2D1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.010241 (rank : 85) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UHL9, O95444, Q75MX7, Q86UM3, Q8WVC4, Q9UHK8, Q9UI91 | Gene names | GTF2IRD1, CREAM1, GTF3, MUSTRD1, RBAP2, WBSCR11, WBSCR12 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor II-I repeat domain-containing protein 1 (GTF2I repeat domain-containing protein 1) (Muscle TFII-I repeat domain-containing protein 1) (General transcription factor III) (Slow- muscle-fiber enhancer-binding protein) (USE B1-binding protein) (MusTRD1/BEN) (Williams-Beuren syndrome chromosome region 11 protein). | |||||
|
K0460_MOUSE
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | 0.022560 (rank : 59) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6NXI6, Q3U3L8, Q6ZQA7 | Gene names | Kiaa0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
RECQ5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.008010 (rank : 90) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O94762, Q9P1W7, Q9UNC8 | Gene names | RECQL5, RECQ5 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q5 (EC 3.6.1.-) (RecQ protein-like 5) (RecQ5). | |||||
|
SFPQ_MOUSE
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.017541 (rank : 67) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
ZSWM5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 81) | NC score | 0.010846 (rank : 78) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P217, Q5SXQ9 | Gene names | ZSWIM5, KIAA1511 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger SWIM domain-containing protein 5. | |||||
|
ATF7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.015540 (rank : 70) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P17544, Q13814 | Gene names | ATF7, ATFA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
GNAS1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.005597 (rank : 97) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6R0H7, Q6R0H4, Q6R0H5, Q6R2J5, Q9JJX0, Q9Z1N8 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | 0.018602 (rank : 64) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
PHLPP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.002767 (rank : 104) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CHE4, Q6PCN0, Q8QZU8, Q8R5E5, Q99KL6 | Gene names | Phlpp, Kiaa0606, Plekhe1, Scop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein). | |||||
|
RBM12_MOUSE
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.019029 (rank : 63) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8R4X3, Q8K373, Q8R302, Q9CS80 | Gene names | Rbm12 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN). | |||||
|
RIMB1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.000554 (rank : 106) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
YBOX1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.010765 (rank : 79) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P67809, P16990, P16991, Q14972, Q15325, Q5FVF0 | Gene names | YBX1, NSEP1, YB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclease sensitive element-binding protein 1 (Y-box-binding protein 1) (Y-box transcription factor) (YB-1) (CCAAT-binding transcription factor I subunit A) (CBF-A) (Enhancer factor I subunit A) (EFI-A) (DNA-binding protein B) (DBPB). | |||||
|
ZFP95_HUMAN
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | -0.005232 (rank : 111) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y2L8 | Gene names | ZFP95, KIAA1015 | |||
|
Domain Architecture |
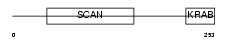
|
|||||
| Description | Zinc finger protein 95 homolog (Zfp-95). | |||||
|
ABL2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | -0.004536 (rank : 110) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1045 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P42684, Q5T0X6, Q6NZY6 | Gene names | ABL2, ABLL, ARG | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase ABL2 (EC 2.7.10.2) (Abelson murine leukemia viral oncogene homolog 2) (Tyrosine kinase ARG). | |||||
|
ARI1B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.014153 (rank : 72) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
CO2A1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.005408 (rank : 98) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 483 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P28481 | Gene names | Col2a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(II) chain precursor [Contains: Chondrocalcin]. | |||||
|
DOCK4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.005901 (rank : 96) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8N1I0, O14584, O94824, Q8NB45 | Gene names | DOCK4, KIAA0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
EDA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.011969 (rank : 75) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92838, O75910, Q5JUM7, Q9UP77, Q9Y6L0, Q9Y6L1, Q9Y6L2, Q9Y6L3, Q9Y6L4 | Gene names | EDA, ED1 | |||
|
Domain Architecture |
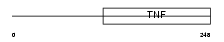
|
|||||
| Description | Ectodysplasin-A (Ectodermal dysplasia protein) (EDA protein) [Contains: Ectodysplasin-A, membrane form; Ectodysplasin-A, secreted form]. | |||||
|
EDA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.015918 (rank : 68) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O54693, O35705, Q9QWJ8, Q9QZ01, Q9QZ02 | Gene names | Eda, Ed1, Ta | |||
|
Domain Architecture |

|
|||||
| Description | Ectodysplasin-A (EDA protein homolog) (Tabby protein) [Contains: Ectodysplasin-A, membrane form; Ectodysplasin-A, secreted form]. | |||||
|
EP400_MOUSE
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.006354 (rank : 95) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
IASPP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.007182 (rank : 94) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
JADE2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.010501 (rank : 81) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
K0329_HUMAN
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.010432 (rank : 82) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15040, Q9UEG6 | Gene names | KIAA0329, KIAA0297 | |||
|
Domain Architecture |

|
|||||
| Description | Protein KIAA0329/KIAA0297. | |||||
|
NOS1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.003481 (rank : 102) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P29475 | Gene names | NOS1 | |||
|
Domain Architecture |

|
|||||
| Description | Nitric-oxide synthase, brain (EC 1.14.13.39) (NOS type I) (Neuronal NOS) (N-NOS) (nNOS) (Constitutive NOS) (NC-NOS) (bNOS). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.018192 (rank : 66) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
TCOF_MOUSE
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | 0.023843 (rank : 56) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
ULK1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | -0.005309 (rank : 112) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1214 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O70405, Q6PGB2 | Gene names | Ulk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1) (Serine/threonine-protein kinase Unc51.1). | |||||
|
VGLL2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | 0.010363 (rank : 84) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N8G2 | Gene names | VGLL2, VITO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription cofactor vestigial-like protein 2 (Vgl-2) (Protein VITO1). | |||||
|
CAC1G_HUMAN
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.002590 (rank : 105) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
CECR2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.004740 (rank : 100) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BXF3, Q658Z4, Q96P58, Q9C0C3 | Gene names | CECR2, KIAA1740 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 2. | |||||
|
CSPG3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | -0.000309 (rank : 107) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P55066, Q6P1E3, Q8C4F8 | Gene names | Cspg3, Ncan | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
FOXN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.010399 (rank : 83) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O15353, O15352 | Gene names | FOXN1, WHN | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein N1 (Transcription factor winged-helix nude). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.015874 (rank : 69) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
PRR3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.014140 (rank : 73) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P79522, Q5RJB5 | Gene names | PRR3, CAT56 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 3 (MHC class I region proline-rich protein CAT56). | |||||
|
YBOX1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.009273 (rank : 88) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P62960, P22568, P27817, P43482 | Gene names | Ybx1, Msy-1, Msy1, Nsep1, Yb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclease sensitive element-binding protein 1 (Y-box-binding protein 1) (Y-box transcription factor) (YB-1) (CCAAT-binding transcription factor I subunit A) (CBF-A) (Enhancer factor I subunit A) (EFI-A) (DNA-binding protein B) (DBPB). | |||||
|
ZN342_HUMAN
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | -0.003899 (rank : 109) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 705 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WUU4 | Gene names | ZNF342 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 342. | |||||
|
ELK1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.04523e-173 (rank : 1) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 112 | |
| SwissProt Accessions | P19419, O75606, O95058, Q969X8, Q9UJM4 | Gene names | ELK1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-1. | |||||
|
ELK1_MOUSE
|
||||||
| NC score | 0.990370 (rank : 2) | θ value | 2.85304e-163 (rank : 2) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P41969 | Gene names | Elk1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-1. | |||||
|
ELK3_HUMAN
|
||||||
| NC score | 0.955805 (rank : 3) | θ value | 1.2411e-41 (rank : 4) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P41970, Q6FG57, Q6GU29, Q9UD17 | Gene names | ELK3, NET, SAP2 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-3 (ETS-related protein NET) (ETS- related protein ERP) (SRF accessory protein 2) (SAP-2). | |||||
|
ELK3_MOUSE
|
||||||
| NC score | 0.953017 (rank : 4) | θ value | 1.79215e-40 (rank : 5) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P41971, P97747, Q62346 | Gene names | Elk3, Erp, Net | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-3 (ETS-related protein NET) (ETS- related protein ERP). | |||||
|
ELK4_MOUSE
|
||||||
| NC score | 0.952547 (rank : 5) | θ value | 5.21438e-40 (rank : 6) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P41158 | Gene names | Elk4, Sap1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
|
ELK4_HUMAN
|
||||||
| NC score | 0.944914 (rank : 6) | θ value | 7.25919e-50 (rank : 3) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P28324, P28323 | Gene names | ELK4, SAP1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
|
ETV2_HUMAN
|
||||||
| NC score | 0.911012 (rank : 7) | θ value | 1.2077e-20 (rank : 30) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O00321, Q9UEA0 | Gene names | ETV2, ER71, ETSRP71 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 2 (Ets-related protein 71). | |||||
|
ETV2_MOUSE
|
||||||
| NC score | 0.907173 (rank : 8) | θ value | 8.65492e-19 (rank : 34) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P41163 | Gene names | Etv2, Er71, Etsrp71 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 2 (Ets-related protein 71). | |||||
|
ETS2_HUMAN
|
||||||
| NC score | 0.898834 (rank : 9) | θ value | 1.37858e-24 (rank : 13) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P15036 | Gene names | ETS2 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-2 protein. | |||||
|
ETS1_HUMAN
|
||||||
| NC score | 0.897692 (rank : 10) | θ value | 5.23862e-24 (rank : 16) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P14921, Q14278, Q16080 | Gene names | ETS1 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-1 protein (p54). | |||||
|
ETS1_MOUSE
|
||||||
| NC score | 0.897683 (rank : 11) | θ value | 5.23862e-24 (rank : 17) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P27577, Q61403 | Gene names | Ets1, Ets-1 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-1 protein (p54). | |||||
|
ETS2_MOUSE
|
||||||
| NC score | 0.897381 (rank : 12) | θ value | 1.37858e-24 (rank : 14) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P15037 | Gene names | Ets2 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-2 protein. | |||||
|
GABPA_MOUSE
|
||||||
| NC score | 0.897327 (rank : 13) | θ value | 7.0802e-21 (rank : 28) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q00422 | Gene names | Gabpa, E4tf1a | |||
|
Domain Architecture |

|
|||||
| Description | GA-binding protein alpha chain (GABP-subunit alpha). | |||||
|
GABPA_HUMAN
|
||||||
| NC score | 0.895862 (rank : 14) | θ value | 7.0802e-21 (rank : 27) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q06546, Q12939 | Gene names | GABPA, E4TF1A | |||
|
Domain Architecture |

|
|||||
| Description | GA-binding protein alpha chain (GABP-subunit alpha) (Transcription factor E4TF1-60) (Nuclear respiratory factor 2 subunit alpha). | |||||
|
FLI1_MOUSE
|
||||||
| NC score | 0.888505 (rank : 15) | θ value | 1.16704e-23 (rank : 22) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P26323 | Gene names | Fli1, Fli-1 | |||
|
Domain Architecture |

|
|||||
| Description | Friend leukemia integration 1 transcription factor (Retroviral integration site protein Fli-1). | |||||
|
FLI1_HUMAN
|
||||||
| NC score | 0.887316 (rank : 16) | θ value | 1.16704e-23 (rank : 21) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q01543, Q14319, Q92480, Q9UE07 | Gene names | FLI1 | |||
|
Domain Architecture |

|
|||||
| Description | Friend leukemia integration 1 transcription factor (Fli-1 proto- oncogene) (ERGB transcription factor). | |||||
|
ETV7_HUMAN
|
||||||
| NC score | 0.886683 (rank : 17) | θ value | 1.80466e-16 (rank : 39) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9Y603, Q9NZ65, Q9NZ66, Q9NZ68, Q9NZR8, Q9UNJ7, Q9Y5K4, Q9Y604 | Gene names | ETV7, TEL2, TELB, TREF | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV7 (Transcription factor Tel-2) (ETS-related protein Tel2) (Tel-related Ets factor). | |||||
|
SPDEF_HUMAN
|
||||||
| NC score | 0.883237 (rank : 18) | θ value | 7.32683e-18 (rank : 35) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O95238 | Gene names | SPDEF, PDEF, PSE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAM pointed domain-containing Ets transcription factor (Prostate- derived Ets factor) (Prostate epithelium-specific Ets transcription factor) (Prostate-specific Ets). | |||||
|
ERG_HUMAN
|
||||||
| NC score | 0.882059 (rank : 19) | θ value | 1.16704e-23 (rank : 20) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P11308, Q16113 | Gene names | ERG | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ERG (Transforming protein ERG). | |||||
|
ERG_MOUSE
|
||||||
| NC score | 0.881722 (rank : 20) | θ value | 3.07116e-24 (rank : 15) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P81270, Q8C5L4, Q920K7, Q920K8, Q920K9 | Gene names | Erg, Erg-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ERG. | |||||
|
SPDEF_MOUSE
|
||||||
| NC score | 0.881562 (rank : 21) | θ value | 9.56915e-18 (rank : 36) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9WTP3 | Gene names | Spdef, Pdef, Pse | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAM pointed domain-containing Ets transcription factor (Prostate- derived Ets factor) (Prostate epithelium-specific Ets transcription factor) (Prostate-specific Ets). | |||||
|
ETV1_MOUSE
|
||||||
| NC score | 0.878575 (rank : 22) | θ value | 7.30988e-26 (rank : 10) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P41164 | Gene names | Etv1, Er81, Etsrp81 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 1 (ER81 protein). | |||||
|
ERF_MOUSE
|
||||||
| NC score | 0.878415 (rank : 23) | θ value | 2.87452e-22 (rank : 25) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P70459 | Gene names | Erf | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing transcription factor ERF. | |||||
|
ERF_HUMAN
|
||||||
| NC score | 0.878249 (rank : 24) | θ value | 2.87452e-22 (rank : 24) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P50548, Q9UPI7 | Gene names | ERF | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing transcription factor ERF (Ets2 repressor factor). | |||||
|
ETV3_MOUSE
|
||||||
| NC score | 0.878072 (rank : 25) | θ value | 5.23862e-24 (rank : 18) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8R4Z4, Q9QZW1 | Gene names | Etv3, Mets, Pe1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
ETV1_HUMAN
|
||||||
| NC score | 0.877646 (rank : 26) | θ value | 7.30988e-26 (rank : 9) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P50549, O75849, Q9UQ71, Q9Y636 | Gene names | ETV1, ER81 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 1 (ER81 protein). | |||||
|
ETV4_MOUSE
|
||||||
| NC score | 0.875152 (rank : 27) | θ value | 4.28545e-26 (rank : 8) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P28322 | Gene names | Etv4, Pea-3, Pea3 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 4 (Polyomavirus enhancer activator 3) (Protein PEA3). | |||||
|
ETV4_HUMAN
|
||||||
| NC score | 0.874555 (rank : 28) | θ value | 4.28545e-26 (rank : 7) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P43268, Q96AW9 | Gene names | ETV4, E1AF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 4 (Adenovirus E1A enhancer-binding protein) (E1A-F). | |||||
|
ETV5_HUMAN
|
||||||
| NC score | 0.873017 (rank : 29) | θ value | 1.62847e-25 (rank : 11) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P41161 | Gene names | ETV5, ERM | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 5 (Ets-related protein ERM). | |||||
|
ELF2_MOUSE
|
||||||
| NC score | 0.862393 (rank : 30) | θ value | 5.99374e-20 (rank : 32) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9JHC9, Q6NST2, Q8BTX8, Q9JHC7, Q9JHC8, Q9JHD0 | Gene names | Elf2, Nerf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-2 (E74-like factor 2) (New ETS- related factor). | |||||
|
ELF1_HUMAN
|
||||||
| NC score | 0.861675 (rank : 31) | θ value | 4.43474e-23 (rank : 23) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P32519, Q9UDE1 | Gene names | ELF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
ELF2_HUMAN
|
||||||
| NC score | 0.860795 (rank : 32) | θ value | 1.33526e-19 (rank : 33) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q15723, Q15724, Q15725, Q6P1K5 | Gene names | ELF2, NERF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-2 (E74-like factor 2) (New ETS- related factor). | |||||
|
ELF4_HUMAN
|
||||||
| NC score | 0.860030 (rank : 33) | θ value | 2.06002e-20 (rank : 31) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q99607, O60435 | Gene names | ELF4, ELFR, MEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-4 (E74-like factor 4) (Myeloid Elf-1-like factor). | |||||
|
ETV3_HUMAN
|
||||||
| NC score | 0.853636 (rank : 34) | θ value | 6.84181e-24 (rank : 19) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P41162, Q8TAC8, Q9BX30 | Gene names | ETV3, METS, PE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
ELF1_MOUSE
|
||||||
| NC score | 0.853137 (rank : 35) | θ value | 8.36355e-22 (rank : 26) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q60775 | Gene names | Elf1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
ETV5_MOUSE
|
||||||
| NC score | 0.852485 (rank : 36) | θ value | 1.62847e-25 (rank : 12) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9CXC9, Q3TG49, Q8C0F3, Q9JHB1 | Gene names | Etv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 5. | |||||
|
ETV6_MOUSE
|
||||||
| NC score | 0.851656 (rank : 37) | θ value | 1.63225e-17 (rank : 38) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P97360 | Gene names | Etv6, Tel, Tel1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV6 (ETS-related protein Tel1) (Tel) (ETS translocation variant 6). | |||||
|
ETV6_HUMAN
|
||||||
| NC score | 0.848087 (rank : 38) | θ value | 1.63225e-17 (rank : 37) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P41212, Q9UMF6 | Gene names | ETV6, TEL, TEL1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV6 (ETS-related protein Tel1) (Tel) (ETS translocation variant 6). | |||||
|
ELF5_MOUSE
|
||||||
| NC score | 0.841507 (rank : 39) | θ value | 4.16044e-13 (rank : 41) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8VDK3, Q921H5, Q9Z2K6 | Gene names | Elf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-5 (E74-like factor 5). | |||||
|
ELF5_HUMAN
|
||||||
| NC score | 0.840244 (rank : 40) | θ value | 4.16044e-13 (rank : 40) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UKW6, O95175, Q8N2K9, Q96QY3, Q9UKW5 | Gene names | ELF5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-5 (E74-like factor 5) (Epithelium-specific Ets transcription factor 2) (ESE-2) (Epithelium- restricted ESE-1-related Ets factor). | |||||
|
ELF4_MOUSE
|
||||||
| NC score | 0.837753 (rank : 41) | θ value | 9.24701e-21 (rank : 29) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Z2U4 | Gene names | Elf4, Mef | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-4 (E74-like factor 4) (Myeloid Elf-1-like factor). | |||||
|
SPI1_MOUSE
|
||||||
| NC score | 0.664141 (rank : 42) | θ value | 4.45536e-07 (rank : 43) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P17433, Q99L57 | Gene names | Spi1, Sfpi-1, Sfpi1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor PU.1 (31 kDa transforming protein) (SFFV proviral integration 1 protein). | |||||
|
SPI1_HUMAN
|
||||||
| NC score | 0.662749 (rank : 43) | θ value | 4.45536e-07 (rank : 42) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P17947 | Gene names | SPI1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor PU.1 (31 kDa transforming protein). | |||||
|
SPIB_HUMAN
|
||||||
| NC score | 0.650539 (rank : 44) | θ value | 1.09739e-05 (rank : 44) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q01892, Q15359 | Gene names | SPIB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Spi-B. | |||||
|
SPIB_MOUSE
|
||||||
| NC score | 0.636618 (rank : 45) | θ value | 1.43324e-05 (rank : 45) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O35906, O35907, O35909, O55199 | Gene names | Spib | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-B. | |||||
|
SPIC_HUMAN
|
||||||
| NC score | 0.614177 (rank : 46) | θ value | 7.1131e-05 (rank : 46) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8N5J4 | Gene names | SPIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-C. | |||||
|
SPIC_MOUSE
|
||||||
| NC score | 0.581869 (rank : 47) | θ value | 0.000602161 (rank : 47) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6P3D7, Q9Z0Y0 | Gene names | Spic | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-C (Pu.1-related factor) (Prf). | |||||
|
TAF10_HUMAN
|
||||||
| NC score | 0.045815 (rank : 48) | θ value | 0.279714 (rank : 56) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q12962, O00703, Q13175 | Gene names | TAF10, TAF2H, TAFII30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 10 (Transcription initiation factor TFIID 30 kDa subunit) (TAF(II)30) (TAFII-30) (TAFII30) (STAF28). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.044855 (rank : 49) | θ value | 0.0193708 (rank : 49) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.036067 (rank : 50) | θ value | 0.000786445 (rank : 48) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
PHC2_HUMAN
|
||||||
| NC score | 0.033351 (rank : 51) | θ value | 0.0330416 (rank : 50) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8IXK0, Q5T0C1, Q6NUJ6, Q6ZQR1, Q8N306, Q8TAG8, Q96BL4, Q9Y4Y7 | Gene names | PHC2, EDR2, PH2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 2 (hPH2) (Early development regulatory protein 2). | |||||
|
GON4L_MOUSE
|
||||||
| NC score | 0.031900 (rank : 52) | θ value | 0.0736092 (rank : 51) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9DB00, Q80TB4, Q91YI9 | Gene names | Gon4l, Gon4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
APXL_HUMAN
|
||||||
| NC score | 0.030978 (rank : 53) | θ value | 0.125558 (rank : 53) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13796 | Gene names | APXL | |||
|
Domain Architecture |

|
|||||
| Description | Apical-like protein (Protein APXL). | |||||
|
SSBP4_HUMAN
|
||||||
| NC score | 0.029982 (rank : 54) | θ value | 0.125558 (rank : 54) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
GHR_HUMAN
|
||||||
| NC score | 0.027504 (rank : 55) | θ value | 0.62314 (rank : 57) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P10912, Q9HCX2 | Gene names | GHR | |||
|
Domain Architecture |

|
|||||
| Description | Growth hormone receptor precursor (GH receptor) (Somatotropin receptor) [Contains: Growth hormone-binding protein (GH-binding protein) (GHBP) (Serum-binding protein)]. | |||||
|
TCOF_MOUSE
|
||||||
| NC score | 0.023843 (rank : 56) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.023344 (rank : 57) | θ value | 0.0961366 (rank : 52) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
PO121_MOUSE
|
||||||
| NC score | 0.022831 (rank : 58) | θ value | 2.36792 (rank : 66) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
K0460_MOUSE
|
||||||
| NC score | 0.022560 (rank : 59) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6NXI6, Q3U3L8, Q6ZQA7 | Gene names | Kiaa0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
SC24A_MOUSE
|
||||||
| NC score | 0.021820 (rank : 60) | θ value | 3.0926 (rank : 72) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q3U2P1, Q3TQ05, Q3TRG7, Q8BIS0 | Gene names | Sec24a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein transport protein Sec24A (SEC24-related protein A). | |||||
|
SC24C_HUMAN
|
||||||
| NC score | 0.020591 (rank : 61) | θ value | 0.813845 (rank : 59) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.019179 (rank : 62) | θ value | 1.38821 (rank : 61) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
RBM12_MOUSE
|
||||||
| NC score | 0.019029 (rank : 63) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8R4X3, Q8K373, Q8R302, Q9CS80 | Gene names | Rbm12 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.018602 (rank : 64) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.018214 (rank : 65) | θ value | 0.21417 (rank : 55) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.018192 (rank : 66) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
SFPQ_MOUSE
|
||||||
| NC score | 0.017541 (rank : 67) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
EDA_MOUSE
|
||||||
| NC score | 0.015918 (rank : 68) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O54693, O35705, Q9QWJ8, Q9QZ01, Q9QZ02 | Gene names | Eda, Ed1, Ta | |||
|
Domain Architecture |

|
|||||
| Description | Ectodysplasin-A (EDA protein homolog) (Tabby protein) [Contains: Ectodysplasin-A, membrane form; Ectodysplasin-A, secreted form]. | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.015874 (rank : 69) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
ATF7_HUMAN
|
||||||
| NC score | 0.015540 (rank : 70) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P17544, Q13814 | Gene names | ATF7, ATFA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
SIGIR_MOUSE
|
||||||
| NC score | 0.014764 (rank : 71) | θ value | 3.0926 (rank : 73) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JLZ8, Q52L48 | Gene names | Sigirr, Tir8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Single Ig IL-1-related receptor (Single Ig IL-1R-related molecule) (Single immunoglobulin domain-containing IL1R-related protein) (Toll/interleukin-1 receptor 8) (TIR8). | |||||
|
ARI1B_HUMAN
|
||||||
| NC score | 0.014153 (rank : 72) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
PRR3_HUMAN
|
||||||
| NC score | 0.014140 (rank : 73) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P79522, Q5RJB5 | Gene names | PRR3, CAT56 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 3 (MHC class I region proline-rich protein CAT56). | |||||
|
ATF6B_HUMAN
|
||||||
| NC score | 0.012601 (rank : 74) | θ value | 3.0926 (rank : 67) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99941, Q13269, Q14343, Q14345, Q99635, Q99637, Q9H3V9, Q9H3W1, Q9NPL0 | Gene names | CREBL1, G13 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-6 beta (Activating transcription factor 6 beta) (ATF6-beta) (cAMP-responsive element- binding protein-like 1) (cAMP response element-binding protein-related protein) (Creb-rp) (Protein G13). | |||||
|
EDA_HUMAN
|
||||||
| NC score | 0.011969 (rank : 75) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92838, O75910, Q5JUM7, Q9UP77, Q9Y6L0, Q9Y6L1, Q9Y6L2, Q9Y6L3, Q9Y6L4 | Gene names | EDA, ED1 | |||
|
Domain Architecture |

|
|||||
| Description | Ectodysplasin-A (Ectodermal dysplasia protein) (EDA protein) [Contains: Ectodysplasin-A, membrane form; Ectodysplasin-A, secreted form]. | |||||
|
MYCD_MOUSE
|
||||||
| NC score | 0.011288 (rank : 76) | θ value | 3.0926 (rank : 70) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8VIM5, Q8C3W6, Q8VIL4 | Gene names | Myocd, Bsac2, Mycd, Srfcp | |||
|
Domain Architecture |

|
|||||
| Description | Myocardin (SRF cofactor protein) (Basic SAP coiled-coil transcription activator 2). | |||||
|
DOCK6_HUMAN
|
||||||
| NC score | 0.011006 (rank : 77) | θ value | 1.38821 (rank : 60) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96HP0, Q9P2F2 | Gene names | DOCK6, KIAA1395 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 6. | |||||
|
ZSWM5_HUMAN
|
||||||
| NC score | 0.010846 (rank : 78) | θ value | 4.03905 (rank : 81) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P217, Q5SXQ9 | Gene names | ZSWIM5, KIAA1511 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger SWIM domain-containing protein 5. | |||||
|
YBOX1_HUMAN
|
||||||
| NC score | 0.010765 (rank : 79) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P67809, P16990, P16991, Q14972, Q15325, Q5FVF0 | Gene names | YBX1, NSEP1, YB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclease sensitive element-binding protein 1 (Y-box-binding protein 1) (Y-box transcription factor) (YB-1) (CCAAT-binding transcription factor I subunit A) (CBF-A) (Enhancer factor I subunit A) (EFI-A) (DNA-binding protein B) (DBPB). | |||||
|
FIGN_HUMAN
|
||||||
| NC score | 0.010706 (rank : 80) | θ value | 1.81305 (rank : 62) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5HY92, Q9H6M5, Q9NVZ9 | Gene names | FIGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
JADE2_MOUSE
|
||||||
| NC score | 0.010501 (rank : 81) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
K0329_HUMAN
|
||||||
| NC score | 0.010432 (rank : 82) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15040, Q9UEG6 | Gene names | KIAA0329, KIAA0297 | |||
|
Domain Architecture |

|
|||||
| Description | Protein KIAA0329/KIAA0297. | |||||
|
FOXN1_HUMAN
|
||||||
| NC score | 0.010399 (rank : 83) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O15353, O15352 | Gene names | FOXN1, WHN | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein N1 (Transcription factor winged-helix nude). | |||||
|
VGLL2_HUMAN
|
||||||
| NC score | 0.010363 (rank : 84) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N8G2 | Gene names | VGLL2, VITO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription cofactor vestigial-like protein 2 (Vgl-2) (Protein VITO1). | |||||
|
GT2D1_HUMAN
|
||||||
| NC score | 0.010241 (rank : 85) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UHL9, O95444, Q75MX7, Q86UM3, Q8WVC4, Q9UHK8, Q9UI91 | Gene names | GTF2IRD1, CREAM1, GTF3, MUSTRD1, RBAP2, WBSCR11, WBSCR12 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor II-I repeat domain-containing protein 1 (GTF2I repeat domain-containing protein 1) (Muscle TFII-I repeat domain-containing protein 1) (General transcription factor III) (Slow- muscle-fiber enhancer-binding protein) (USE B1-binding protein) (MusTRD1/BEN) (Williams-Beuren syndrome chromosome region 11 protein). | |||||
|
LIMD1_MOUSE
|
||||||
| NC score | 0.010226 (rank : 86) | θ value | 2.36792 (rank : 64) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9QXD8, Q8C8G4, Q9CW55 | Gene names | Limd1 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain-containing protein 1. | |||||
|
FIGN_MOUSE
|
||||||
| NC score | 0.009409 (rank : 87) | θ value | 4.03905 (rank : 75) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ERZ6, Q3TPB0, Q3UP57, Q6PCM0 | Gene names | Fign | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
YBOX1_MOUSE
|
||||||
| NC score | 0.009273 (rank : 88) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P62960, P22568, P27817, P43482 | Gene names | Ybx1, Msy-1, Msy1, Nsep1, Yb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclease sensitive element-binding protein 1 (Y-box-binding protein 1) (Y-box transcription factor) (YB-1) (CCAAT-binding transcription factor I subunit A) (CBF-A) (Enhancer factor I subunit A) (EFI-A) (DNA-binding protein B) (DBPB). | |||||
|
COAA1_HUMAN
|
||||||
| NC score | 0.008843 (rank : 89) | θ value | 3.0926 (rank : 68) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q03692 | Gene names | COL10A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(X) chain precursor. | |||||
|
RECQ5_HUMAN
|
||||||
| NC score | 0.008010 (rank : 90) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O94762, Q9P1W7, Q9UNC8 | Gene names | RECQL5, RECQ5 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q5 (EC 3.6.1.-) (RecQ protein-like 5) (RecQ5). | |||||
|
LIRB5_HUMAN
|
||||||
| NC score | 0.007657 (rank : 91) | θ value | 0.813845 (rank : 58) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75023, Q8N760 | Gene names | LILRB5, LIR8 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily B member 5 precursor (Leukocyte immunoglobulin-like receptor 8) (LIR-8) (CD85c antigen). | |||||
|
FOXK2_HUMAN
|
||||||
| NC score | 0.007290 (rank : 92) | θ value | 3.0926 (rank : 69) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q01167, Q13622, Q13623, Q13624 | Gene names | FOXK2, ILF, ILF1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein K2 (Interleukin enhancer-binding factor 1) (Cellular transcription factor ILF-1). | |||||
|
CO1A1_HUMAN
|
||||||
| NC score | 0.007289 (rank : 93) | θ value | 4.03905 (rank : 74) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P02452, P78441, Q13896, Q13902, Q13903, Q14037, Q14992, Q15176, Q15201, Q16050, Q7KZ30, Q7KZ34, Q8IVI5, Q9UML6, Q9UMM7 | Gene names | COL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(I) chain precursor. | |||||
|
IASPP_MOUSE
|
||||||
| NC score | 0.007182 (rank : 94) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
EP400_MOUSE
|
||||||
| NC score | 0.006354 (rank : 95) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
DOCK4_HUMAN
|
||||||
| NC score | 0.005901 (rank : 96) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8N1I0, O14584, O94824, Q8NB45 | Gene names | DOCK4, KIAA0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
GNAS1_MOUSE
|
||||||
| NC score | 0.005597 (rank : 97) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6R0H7, Q6R0H4, Q6R0H5, Q6R2J5, Q9JJX0, Q9Z1N8 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
CO2A1_MOUSE
|
||||||
| NC score | 0.005408 (rank : 98) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 483 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P28481 | Gene names | Col2a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(II) chain precursor [Contains: Chondrocalcin]. | |||||
|
GMIP_MOUSE
|
||||||
| NC score | 0.005206 (rank : 99) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PGG2, Q6P9S3 | Gene names | Gmip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GEM-interacting protein (GMIP). | |||||
|
CECR2_HUMAN
|
||||||
| NC score | 0.004740 (rank : 100) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BXF3, Q658Z4, Q96P58, Q9C0C3 | Gene names | CECR2, KIAA1740 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 2. | |||||
|
PRGR_MOUSE
|
||||||
| NC score | 0.004438 (rank : 101) | θ value | 3.0926 (rank : 71) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q00175 | Gene names | Pgr, Nr3c3, Pr | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone receptor (PR). | |||||
|
NOS1_HUMAN
|
||||||
| NC score | 0.003481 (rank : 102) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P29475 | Gene names | NOS1 | |||
|
Domain Architecture |

|
|||||
| Description | Nitric-oxide synthase, brain (EC 1.14.13.39) (NOS type I) (Neuronal NOS) (N-NOS) (nNOS) (Constitutive NOS) (NC-NOS) (bNOS). | |||||
|
NEST_HUMAN
|
||||||
| NC score | 0.003441 (rank : 103) | θ value | 2.36792 (rank : 65) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
PHLPP_MOUSE
|
||||||
| NC score | 0.002767 (rank : 104) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CHE4, Q6PCN0, Q8QZU8, Q8R5E5, Q99KL6 | Gene names | Phlpp, Kiaa0606, Plekhe1, Scop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein). | |||||
|
CAC1G_HUMAN
|
||||||
| NC score | 0.002590 (rank : 105) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
RIMB1_MOUSE
|
||||||
| NC score | 0.000554 (rank : 106) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
CSPG3_MOUSE
|
||||||
| NC score | -0.000309 (rank : 107) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P55066, Q6P1E3, Q8C4F8 | Gene names | Cspg3, Ncan | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
GLI2_HUMAN
|
||||||
| NC score | -0.002219 (rank : 108) | θ value | 1.81305 (rank : 63) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 971 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P10070, O60252, O60253, O60254, O60255, Q15590, Q15591 | Gene names | GLI2, THP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI2 (Tax helper protein). | |||||
|
ZN342_HUMAN
|
||||||
| NC score | -0.003899 (rank : 109) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 705 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WUU4 | Gene names | ZNF342 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 342. | |||||
|
ABL2_HUMAN
|
||||||
| NC score | -0.004536 (rank : 110) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1045 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P42684, Q5T0X6, Q6NZY6 | Gene names | ABL2, ABLL, ARG | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase ABL2 (EC 2.7.10.2) (Abelson murine leukemia viral oncogene homolog 2) (Tyrosine kinase ARG). | |||||
|
ZFP95_HUMAN
|
||||||
| NC score | -0.005232 (rank : 111) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y2L8 | Gene names | ZFP95, KIAA1015 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 95 homolog (Zfp-95). | |||||
|
ULK1_MOUSE
|
||||||
| NC score | -0.005309 (rank : 112) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1214 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O70405, Q6PGB2 | Gene names | Ulk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1) (Serine/threonine-protein kinase Unc51.1). | |||||