Please be patient as the page loads
|
ETV5_HUMAN
|
||||||
| SwissProt Accessions | P41161 | Gene names | ETV5, ERM | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 5 (Ets-related protein ERM). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ETV5_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 181 | |
| SwissProt Accessions | P41161 | Gene names | ETV5, ERM | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 5 (Ets-related protein ERM). | |||||
|
ETV5_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.972469 (rank : 4) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 125 | |
| SwissProt Accessions | Q9CXC9, Q3TG49, Q8C0F3, Q9JHB1 | Gene names | Etv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 5. | |||||
|
ETV1_HUMAN
|
||||||
| θ value | 1.19589e-169 (rank : 3) | NC score | 0.980633 (rank : 3) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P50549, O75849, Q9UQ71, Q9Y636 | Gene names | ETV1, ER81 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 1 (ER81 protein). | |||||
|
ETV1_MOUSE
|
||||||
| θ value | 2.94559e-168 (rank : 4) | NC score | 0.981219 (rank : 2) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P41164 | Gene names | Etv1, Er81, Etsrp81 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 1 (ER81 protein). | |||||
|
ETV4_HUMAN
|
||||||
| θ value | 1.75107e-120 (rank : 5) | NC score | 0.968104 (rank : 5) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P43268, Q96AW9 | Gene names | ETV4, E1AF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 4 (Adenovirus E1A enhancer-binding protein) (E1A-F). | |||||
|
ETV4_MOUSE
|
||||||
| θ value | 4.03687e-117 (rank : 6) | NC score | 0.966686 (rank : 6) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P28322 | Gene names | Etv4, Pea-3, Pea3 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 4 (Polyomavirus enhancer activator 3) (Protein PEA3). | |||||
|
ETS1_HUMAN
|
||||||
| θ value | 1.08979e-29 (rank : 7) | NC score | 0.870027 (rank : 13) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P14921, Q14278, Q16080 | Gene names | ETS1 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-1 protein (p54). | |||||
|
ETS1_MOUSE
|
||||||
| θ value | 1.08979e-29 (rank : 8) | NC score | 0.870085 (rank : 12) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P27577, Q61403 | Gene names | Ets1, Ets-1 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-1 protein (p54). | |||||
|
ETS2_HUMAN
|
||||||
| θ value | 5.40856e-29 (rank : 9) | NC score | 0.867608 (rank : 16) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P15036 | Gene names | ETS2 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-2 protein. | |||||
|
ETS2_MOUSE
|
||||||
| θ value | 5.40856e-29 (rank : 10) | NC score | 0.866953 (rank : 17) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P15037 | Gene names | Ets2 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-2 protein. | |||||
|
ELK4_MOUSE
|
||||||
| θ value | 8.63488e-27 (rank : 11) | NC score | 0.871634 (rank : 9) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P41158 | Gene names | Elk4, Sap1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
|
FLI1_HUMAN
|
||||||
| θ value | 1.92365e-26 (rank : 12) | NC score | 0.844066 (rank : 22) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q01543, Q14319, Q92480, Q9UE07 | Gene names | FLI1 | |||
|
Domain Architecture |

|
|||||
| Description | Friend leukemia integration 1 transcription factor (Fli-1 proto- oncogene) (ERGB transcription factor). | |||||
|
FLI1_MOUSE
|
||||||
| θ value | 2.51237e-26 (rank : 13) | NC score | 0.845457 (rank : 21) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P26323 | Gene names | Fli1, Fli-1 | |||
|
Domain Architecture |

|
|||||
| Description | Friend leukemia integration 1 transcription factor (Retroviral integration site protein Fli-1). | |||||
|
ELK4_HUMAN
|
||||||
| θ value | 5.59698e-26 (rank : 14) | NC score | 0.858141 (rank : 18) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P28324, P28323 | Gene names | ELK4, SAP1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
|
ERG_MOUSE
|
||||||
| θ value | 7.30988e-26 (rank : 15) | NC score | 0.842520 (rank : 23) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P81270, Q8C5L4, Q920K7, Q920K8, Q920K9 | Gene names | Erg, Erg-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ERG. | |||||
|
ERG_HUMAN
|
||||||
| θ value | 1.24688e-25 (rank : 16) | NC score | 0.841862 (rank : 24) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P11308, Q16113 | Gene names | ERG | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ERG (Transforming protein ERG). | |||||
|
ELK1_HUMAN
|
||||||
| θ value | 1.62847e-25 (rank : 17) | NC score | 0.873017 (rank : 8) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P19419, O75606, O95058, Q969X8, Q9UJM4 | Gene names | ELK1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-1. | |||||
|
ELK1_MOUSE
|
||||||
| θ value | 1.62847e-25 (rank : 18) | NC score | 0.870566 (rank : 11) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P41969 | Gene names | Elk1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-1. | |||||
|
ELK3_MOUSE
|
||||||
| θ value | 8.08199e-25 (rank : 19) | NC score | 0.869422 (rank : 14) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P41971, P97747, Q62346 | Gene names | Elk3, Erp, Net | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-3 (ETS-related protein NET) (ETS- related protein ERP). | |||||
|
ELK3_HUMAN
|
||||||
| θ value | 1.05554e-24 (rank : 20) | NC score | 0.871632 (rank : 10) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P41970, Q6FG57, Q6GU29, Q9UD17 | Gene names | ELK3, NET, SAP2 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-3 (ETS-related protein NET) (ETS- related protein ERP) (SRF accessory protein 2) (SAP-2). | |||||
|
GABPA_MOUSE
|
||||||
| θ value | 4.01107e-24 (rank : 21) | NC score | 0.855809 (rank : 19) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q00422 | Gene names | Gabpa, E4tf1a | |||
|
Domain Architecture |

|
|||||
| Description | GA-binding protein alpha chain (GABP-subunit alpha). | |||||
|
GABPA_HUMAN
|
||||||
| θ value | 8.93572e-24 (rank : 22) | NC score | 0.854304 (rank : 20) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q06546, Q12939 | Gene names | GABPA, E4TF1A | |||
|
Domain Architecture |

|
|||||
| Description | GA-binding protein alpha chain (GABP-subunit alpha) (Transcription factor E4TF1-60) (Nuclear respiratory factor 2 subunit alpha). | |||||
|
ETV2_HUMAN
|
||||||
| θ value | 1.16704e-23 (rank : 23) | NC score | 0.874352 (rank : 7) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O00321, Q9UEA0 | Gene names | ETV2, ER71, ETSRP71 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 2 (Ets-related protein 71). | |||||
|
ETV2_MOUSE
|
||||||
| θ value | 2.20094e-22 (rank : 24) | NC score | 0.869288 (rank : 15) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P41163 | Gene names | Etv2, Er71, Etsrp71 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 2 (Ets-related protein 71). | |||||
|
ERF_HUMAN
|
||||||
| θ value | 5.07402e-19 (rank : 25) | NC score | 0.815421 (rank : 26) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P50548, Q9UPI7 | Gene names | ERF | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing transcription factor ERF (Ets2 repressor factor). | |||||
|
ERF_MOUSE
|
||||||
| θ value | 5.07402e-19 (rank : 26) | NC score | 0.815328 (rank : 27) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P70459 | Gene names | Erf | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing transcription factor ERF. | |||||
|
ETV3_HUMAN
|
||||||
| θ value | 1.92812e-18 (rank : 27) | NC score | 0.783542 (rank : 31) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P41162, Q8TAC8, Q9BX30 | Gene names | ETV3, METS, PE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
ETV3_MOUSE
|
||||||
| θ value | 3.28887e-18 (rank : 28) | NC score | 0.804401 (rank : 29) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8R4Z4, Q9QZW1 | Gene names | Etv3, Mets, Pe1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
ELF1_MOUSE
|
||||||
| θ value | 4.74913e-17 (rank : 29) | NC score | 0.772808 (rank : 38) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q60775 | Gene names | Elf1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
ELF1_HUMAN
|
||||||
| θ value | 1.058e-16 (rank : 30) | NC score | 0.779159 (rank : 35) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P32519, Q9UDE1 | Gene names | ELF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
ELF4_HUMAN
|
||||||
| θ value | 4.02038e-16 (rank : 31) | NC score | 0.779643 (rank : 34) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q99607, O60435 | Gene names | ELF4, ELFR, MEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-4 (E74-like factor 4) (Myeloid Elf-1-like factor). | |||||
|
ELF4_MOUSE
|
||||||
| θ value | 4.02038e-16 (rank : 32) | NC score | 0.760137 (rank : 41) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9Z2U4 | Gene names | Elf4, Mef | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-4 (E74-like factor 4) (Myeloid Elf-1-like factor). | |||||
|
SPDEF_HUMAN
|
||||||
| θ value | 6.85773e-16 (rank : 33) | NC score | 0.816911 (rank : 25) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O95238 | Gene names | SPDEF, PDEF, PSE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAM pointed domain-containing Ets transcription factor (Prostate- derived Ets factor) (Prostate epithelium-specific Ets transcription factor) (Prostate-specific Ets). | |||||
|
ELF2_HUMAN
|
||||||
| θ value | 8.95645e-16 (rank : 34) | NC score | 0.780779 (rank : 32) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q15723, Q15724, Q15725, Q6P1K5 | Gene names | ELF2, NERF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-2 (E74-like factor 2) (New ETS- related factor). | |||||
|
SPDEF_MOUSE
|
||||||
| θ value | 8.95645e-16 (rank : 35) | NC score | 0.815071 (rank : 28) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9WTP3 | Gene names | Spdef, Pdef, Pse | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAM pointed domain-containing Ets transcription factor (Prostate- derived Ets factor) (Prostate epithelium-specific Ets transcription factor) (Prostate-specific Ets). | |||||
|
ELF2_MOUSE
|
||||||
| θ value | 1.99529e-15 (rank : 36) | NC score | 0.780309 (rank : 33) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9JHC9, Q6NST2, Q8BTX8, Q9JHC7, Q9JHC8, Q9JHD0 | Gene names | Elf2, Nerf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-2 (E74-like factor 2) (New ETS- related factor). | |||||
|
ETV6_HUMAN
|
||||||
| θ value | 2.88119e-14 (rank : 37) | NC score | 0.777889 (rank : 37) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P41212, Q9UMF6 | Gene names | ETV6, TEL, TEL1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV6 (ETS-related protein Tel1) (Tel) (ETS translocation variant 6). | |||||
|
ETV6_MOUSE
|
||||||
| θ value | 2.88119e-14 (rank : 38) | NC score | 0.778423 (rank : 36) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P97360 | Gene names | Etv6, Tel, Tel1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV6 (ETS-related protein Tel1) (Tel) (ETS translocation variant 6). | |||||
|
ETV7_HUMAN
|
||||||
| θ value | 2.28291e-11 (rank : 39) | NC score | 0.799106 (rank : 30) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9Y603, Q9NZ65, Q9NZ66, Q9NZ68, Q9NZR8, Q9UNJ7, Q9Y5K4, Q9Y604 | Gene names | ETV7, TEL2, TELB, TREF | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV7 (Transcription factor Tel-2) (ETS-related protein Tel2) (Tel-related Ets factor). | |||||
|
ELF5_HUMAN
|
||||||
| θ value | 1.133e-10 (rank : 40) | NC score | 0.760577 (rank : 39) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9UKW6, O95175, Q8N2K9, Q96QY3, Q9UKW5 | Gene names | ELF5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-5 (E74-like factor 5) (Epithelium-specific Ets transcription factor 2) (ESE-2) (Epithelium- restricted ESE-1-related Ets factor). | |||||
|
ELF5_MOUSE
|
||||||
| θ value | 1.133e-10 (rank : 41) | NC score | 0.760432 (rank : 40) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8VDK3, Q921H5, Q9Z2K6 | Gene names | Elf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-5 (E74-like factor 5). | |||||
|
SPIB_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 42) | NC score | 0.578466 (rank : 42) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q01892, Q15359 | Gene names | SPIB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Spi-B. | |||||
|
SPI1_MOUSE
|
||||||
| θ value | 8.40245e-06 (rank : 43) | NC score | 0.573041 (rank : 43) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P17433, Q99L57 | Gene names | Spi1, Sfpi-1, Sfpi1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor PU.1 (31 kDa transforming protein) (SFFV proviral integration 1 protein). | |||||
|
SPI1_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 44) | NC score | 0.571829 (rank : 44) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P17947 | Gene names | SPI1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor PU.1 (31 kDa transforming protein). | |||||
|
SPIB_MOUSE
|
||||||
| θ value | 1.09739e-05 (rank : 45) | NC score | 0.562597 (rank : 45) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O35906, O35907, O35909, O55199 | Gene names | Spib | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-B. | |||||
|
SPR1B_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 46) | NC score | 0.070995 (rank : 49) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q62267 | Gene names | Sprr1b | |||
|
Domain Architecture |

|
|||||
| Description | Cornifin B (Small proline-rich protein 1B) (SPR1B) (SPR1 B). | |||||
|
PTN23_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 47) | NC score | 0.048516 (rank : 67) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9H3S7, Q7KZF8, Q8N6Z5, Q9BSR5, Q9P257, Q9UG03, Q9UMZ4 | Gene names | PTPN23, KIAA1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His- domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14). | |||||
|
ZIM10_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 48) | NC score | 0.066416 (rank : 53) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
ZIM10_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 49) | NC score | 0.067310 (rank : 52) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q6P1E1, Q6PDK9, Q6PF85, Q8BW47 | Gene names | Rai17, Zimp10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
SCAP_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 50) | NC score | 0.042710 (rank : 77) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q12770, Q8N2E0, Q8WUA1 | Gene names | SCAP, KIAA0199 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein cleavage-activating protein (SREBP cleavage-activating protein) (SCAP). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 51) | NC score | 0.033875 (rank : 94) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 52) | NC score | 0.044752 (rank : 72) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 108 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 53) | NC score | 0.038103 (rank : 88) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
SSBP4_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 54) | NC score | 0.045901 (rank : 70) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 55) | NC score | 0.039371 (rank : 86) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
COKA1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 56) | NC score | 0.023220 (rank : 122) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9P218, Q4VXQ4, Q8WUT2, Q96CY9, Q9BQU6, Q9BQU7 | Gene names | COL20A1, KIAA1510 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XX) chain precursor. | |||||
|
PO6F2_MOUSE
|
||||||
| θ value | 0.125558 (rank : 57) | NC score | 0.035546 (rank : 93) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BJI4 | Gene names | Pou6f2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | POU domain, class 6, transcription factor 2. | |||||
|
SOX13_MOUSE
|
||||||
| θ value | 0.21417 (rank : 58) | NC score | 0.027129 (rank : 114) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q04891, Q922L3 | Gene names | Sox13, Sox-13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein. | |||||
|
PTN23_MOUSE
|
||||||
| θ value | 0.279714 (rank : 59) | NC score | 0.044140 (rank : 73) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
SPR1B_HUMAN
|
||||||
| θ value | 0.279714 (rank : 60) | NC score | 0.065402 (rank : 54) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P22528, P22529, P22530, Q5T524 | Gene names | SPRR1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin B (Small proline-rich protein IB) (SPR-IB) (14.9 kDa pancornulin). | |||||
|
SPRR3_HUMAN
|
||||||
| θ value | 0.279714 (rank : 61) | NC score | 0.061856 (rank : 56) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UBC9, O75597, Q4ZGI7, Q5T525, Q8NET7, Q9UDG3 | Gene names | SPRR3, SPRC | |||
|
Domain Architecture |

|
|||||
| Description | Small proline-rich protein 3 (Cornifin beta) (Esophagin) (22 kDa pancornulin). | |||||
|
TPO_MOUSE
|
||||||
| θ value | 0.365318 (rank : 62) | NC score | 0.041343 (rank : 80) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P40226 | Gene names | Thpo | |||
|
Domain Architecture |

|
|||||
| Description | Thrombopoietin precursor (Megakaryocyte colony-stimulating factor) (Myeloproliferative leukemia virus oncogene ligand) (C-mpl ligand) (ML) (Megakaryocyte growth and development factor) (MGDF). | |||||
|
ZN438_HUMAN
|
||||||
| θ value | 0.365318 (rank : 63) | NC score | 0.007310 (rank : 177) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q7Z4V0, Q5T426, Q658Q4, Q6ZN65 | Gene names | ZNF438 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 438. | |||||
|
PHC2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 64) | NC score | 0.035773 (rank : 91) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8IXK0, Q5T0C1, Q6NUJ6, Q6ZQR1, Q8N306, Q8TAG8, Q96BL4, Q9Y4Y7 | Gene names | PHC2, EDR2, PH2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 2 (hPH2) (Early development regulatory protein 2). | |||||
|
PHLA1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 65) | NC score | 0.044896 (rank : 71) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8WV24, Q15184, Q2TAN2, Q9NZ17 | Gene names | PHLDA1, PHRIP, TDAG51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family A member 1 (T-cell death- associated gene 51 protein) (Apoptosis-associated nuclear protein) (Proline- and histidine-rich protein) (Proline- and glutamine-rich protein) (PQ-rich protein). | |||||
|
SOX13_HUMAN
|
||||||
| θ value | 0.47712 (rank : 66) | NC score | 0.030594 (rank : 106) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9UN79, O95275, O95826, Q9UHW7 | Gene names | SOX13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein (Type 1 diabetes autoantigen ICA12) (Islet cell antigen 12). | |||||
|
FOXJ3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 67) | NC score | 0.019154 (rank : 129) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UPW0, Q9NSS7 | Gene names | FOXJ3, KIAA1041 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein J3. | |||||
|
LTBP4_MOUSE
|
||||||
| θ value | 0.62314 (rank : 68) | NC score | 0.008012 (rank : 174) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8K4G1, Q8K4G0 | Gene names | Ltbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 4 precursor (LTBP-4). | |||||
|
PELP1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 69) | NC score | 0.041574 (rank : 79) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
SPR1A_HUMAN
|
||||||
| θ value | 0.62314 (rank : 70) | NC score | 0.059124 (rank : 58) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P35321, Q9UDG4 | Gene names | SPRR1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein IA) (SPR-IA) (SPRK) (19 kDa pancornulin). | |||||
|
BRD4_MOUSE
|
||||||
| θ value | 0.813845 (rank : 71) | NC score | 0.040424 (rank : 83) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
BSN_HUMAN
|
||||||
| θ value | 0.813845 (rank : 72) | NC score | 0.048644 (rank : 66) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
MYST3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 73) | NC score | 0.028688 (rank : 112) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
PININ_MOUSE
|
||||||
| θ value | 0.813845 (rank : 74) | NC score | 0.010300 (rank : 160) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
RNZ2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 75) | NC score | 0.031840 (rank : 98) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80Y81, Q99MF0, Q99MF1, Q9CTA2, Q9D1A8, Q9EPZ2 | Gene names | Elac2 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc phosphodiesterase ELAC protein 2 (EC 3.1.26.11) (Ribonuclease Z 2) (RNase Z 2) (tRNase Z 2) (tRNA 3 endonuclease 2) (ElaC homolog protein 2). | |||||
|
SFPQ_HUMAN
|
||||||
| θ value | 0.813845 (rank : 76) | NC score | 0.038768 (rank : 87) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
SYNJ1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 77) | NC score | 0.031274 (rank : 101) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O43426, O43425, O94984 | Gene names | SYNJ1, KIAA0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
WDR33_HUMAN
|
||||||
| θ value | 0.813845 (rank : 78) | NC score | 0.013538 (rank : 147) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 638 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9C0J8 | Gene names | WDR33, WDC146 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 33 (WD repeat protein WDC146). | |||||
|
ATBF1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 79) | NC score | 0.011480 (rank : 155) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
CG1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 80) | NC score | 0.031537 (rank : 100) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13495 | Gene names | CXorf6, CG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CG1 protein (F18). | |||||
|
K0355_HUMAN
|
||||||
| θ value | 1.06291 (rank : 81) | NC score | 0.048274 (rank : 68) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O15063 | Gene names | KIAA0355 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0355. | |||||
|
KCNN3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 82) | NC score | 0.016020 (rank : 135) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P58391 | Gene names | Kcnn3, Sk3 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 3 (SK3). | |||||
|
PRR8_HUMAN
|
||||||
| θ value | 1.06291 (rank : 83) | NC score | 0.056987 (rank : 59) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9NSV0 | Gene names | PRR8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 8. | |||||
|
RBP2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 84) | NC score | 0.015202 (rank : 138) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P49792, Q13074, Q15280, Q53TE2, Q59FH7 | Gene names | RANBP2, NUP358 | |||
|
Domain Architecture |

|
|||||
| Description | Ran-binding protein 2 (RanBP2) (Nuclear pore complex protein Nup358) (Nucleoporin Nup358) (358 kDa nucleoporin) (P270). | |||||
|
CUTL2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 85) | NC score | 0.010207 (rank : 161) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O14529 | Gene names | CUTL2, KIAA0293 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
HAP1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 86) | NC score | 0.009232 (rank : 163) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 606 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P54257, O75358, Q9H4G3, Q9HA98, Q9NY90 | Gene names | HAP1, HLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-associated protein 1 (HAP-1) (Neuroan 1). | |||||
|
ZN746_MOUSE
|
||||||
| θ value | 1.38821 (rank : 87) | NC score | -0.003243 (rank : 188) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q3U133 | Gene names | Znf746, Zfp746 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 746. | |||||
|
DCC_MOUSE
|
||||||
| θ value | 1.81305 (rank : 88) | NC score | 0.008941 (rank : 165) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P70211 | Gene names | Dcc | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC). | |||||
|
IL4RA_HUMAN
|
||||||
| θ value | 1.81305 (rank : 89) | NC score | 0.029394 (rank : 111) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P24394, Q96P01, Q9H181, Q9H182, Q9H183, Q9H184, Q9H185, Q9H186, Q9H187, Q9H188 | Gene names | IL4R, 582J2.1, IL4RA | |||
|
Domain Architecture |
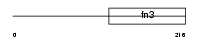
|
|||||
| Description | Interleukin-4 receptor alpha chain precursor (IL-4R-alpha) (CD124 antigen) [Contains: Soluble interleukin-4 receptor alpha chain (sIL4Ralpha/prot) (IL-4-binding protein) (IL4-BP)]. | |||||
|
PDC6I_HUMAN
|
||||||
| θ value | 1.81305 (rank : 90) | NC score | 0.056852 (rank : 60) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8WUM4, Q9BX86, Q9NUN0, Q9P2H2, Q9UKL5 | Gene names | PDCD6IP, AIP1, ALIX, KIAA1375 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death 6-interacting protein (PDCD6-interacting protein) (ALG-2-interacting protein 1) (Hp95). | |||||
|
PHLA1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 91) | NC score | 0.032900 (rank : 96) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q62392, Q3TY48, Q3UG87 | Gene names | Phlda1, Tdag51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family A member 1 (T-cell death- associated gene 51 protein) (Proline- and glutamine-rich protein) (PQR protein). | |||||
|
SSXT_MOUSE
|
||||||
| θ value | 1.81305 (rank : 92) | NC score | 0.045984 (rank : 69) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q62280 | Gene names | Ss18, Ssxt, Syt | |||
|
Domain Architecture |
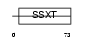
|
|||||
| Description | SSXT protein (SYT protein) (Synovial sarcoma-associated Ss18-alpha). | |||||
|
AP4M1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 93) | NC score | 0.019197 (rank : 128) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00189, Q9UHK9 | Gene names | AP4M1, MUARP2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-4 complex subunit mu-1 (Adapter-related protein complex 4 mu-1 subunit) (Mu subunit of AP-4) (AP-4 adapter complex mu subunit) (Mu- adaptin-related protein 2) (mu-ARP2) (mu4). | |||||
|
BRD4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 94) | NC score | 0.043699 (rank : 75) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
CD166_MOUSE
|
||||||
| θ value | 2.36792 (rank : 95) | NC score | 0.012170 (rank : 151) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61490, O70136, Q8CDA5, Q8R2T0 | Gene names | Alcam | |||
|
Domain Architecture |
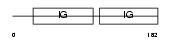
|
|||||
| Description | CD166 antigen precursor (Activated leukocyte-cell adhesion molecule) (ALCAM) (DM-GRASP protein). | |||||
|
CSP9_HUMAN
|
||||||
| θ value | 2.36792 (rank : 96) | NC score | 0.025203 (rank : 118) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43513 | Gene names | CRSP9, ARC34, MED7 | |||
|
Domain Architecture |
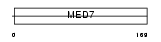
|
|||||
| Description | Cofactor required for Sp1 transcriptional activation subunit 9 (Transcriptional coactivator CRSP33) (RNA polymerase transcriptional regulation mediator subunit 7 homolog) (hMED7) (Activator-recruited cofactor 34 kDa component) (ARC34). | |||||
|
CSP9_MOUSE
|
||||||
| θ value | 2.36792 (rank : 97) | NC score | 0.025233 (rank : 117) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CZB6, Q3UGV1, Q91VN3, Q9CWV8, Q9D0V9 | Gene names | Crsp9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cofactor required for Sp1 transcriptional activation subunit 9. | |||||
|
ENAM_HUMAN
|
||||||
| θ value | 2.36792 (rank : 98) | NC score | 0.030445 (rank : 107) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9NRM1, Q9H3D1 | Gene names | ENAM | |||
|
Domain Architecture |

|
|||||
| Description | Enamelin precursor. | |||||
|
INADL_HUMAN
|
||||||
| θ value | 2.36792 (rank : 99) | NC score | 0.008290 (rank : 172) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NI35, O15249, O43742, O60833, Q5VUA5, Q5VUA6, Q5VUA7, Q5VUA8, Q5VUA9, Q5VUB0, Q8WU78, Q9H3N9 | Gene names | INADL, PATJ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | InaD-like protein (Inadl protein) (hINADL) (Pals1-associated tight junction protein) (Protein associated to tight junctions). | |||||
|
PCQAP_HUMAN
|
||||||
| θ value | 2.36792 (rank : 100) | NC score | 0.030808 (rank : 104) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96RN5, O15413, Q6IC31, Q8NF16, Q96CT0, Q96IH7, Q9P1T3 | Gene names | PCQAP, ARC105, CTG7A, TIG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (TPA-inducible gene 1 protein) (TIG-1) (Activator-recruited cofactor 105 kDa component) (ARC105) (CTG repeat protein 7a). | |||||
|
AMELY_HUMAN
|
||||||
| θ value | 3.0926 (rank : 101) | NC score | 0.030948 (rank : 103) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99218, Q6RWT1 | Gene names | AMELY, AMGL, AMGY | |||
|
Domain Architecture |

|
|||||
| Description | Amelogenin, Y isoform precursor. | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 3.0926 (rank : 102) | NC score | 0.040749 (rank : 82) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
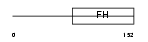
FOXJ3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 103) | NC score | 0.008690 (rank : 169) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BUR3, Q3UTD8 | Gene names | Foxj3, Kiaa1041 | |||
|
Domain Architecture |
|
|||||
| Description | Forkhead box protein J3. | |||||
|
GAK10_HUMAN
|
||||||
| θ value | 3.0926 (rank : 104) | NC score | 0.010499 (rank : 159) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P87889, P10263, P10264, Q69385, Q9UKH6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q33.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K10 Gag protein) (HERV-K107 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GNAS1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 105) | NC score | 0.014485 (rank : 144) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q6R0H7, Q6R0H4, Q6R0H5, Q6R2J5, Q9JJX0, Q9Z1N8 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
IASPP_MOUSE
|
||||||
| θ value | 3.0926 (rank : 106) | NC score | 0.020577 (rank : 125) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
MAML2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 107) | NC score | 0.025485 (rank : 115) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8IZL2, Q6AI23, Q6Y3A3, Q8IUL3, Q96JK6 | Gene names | MAML2, KIAA1819 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 2 (Mam-2). | |||||
|
MCR_MOUSE
|
||||||
| θ value | 3.0926 (rank : 108) | NC score | 0.008289 (rank : 173) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VII8, Q8VII9 | Gene names | Nr3c2, Mlr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mineralocorticoid receptor (MR). | |||||
|
NALDL_HUMAN
|
||||||
| θ value | 3.0926 (rank : 109) | NC score | 0.011421 (rank : 156) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UQQ1, O43176 | Gene names | NAALADL1, NAALADASEL, NAALADL | |||
|
Domain Architecture |

|
|||||
| Description | N-acetylated-alpha-linked acidic dipeptidase-like protein (EC 3.4.17.21) (NAALADase L) (Ileal dipeptidylpeptidase) (100 kDa ileum brush border membrane protein) (I100). | |||||
|
NALDL_MOUSE
|
||||||
| θ value | 3.0926 (rank : 110) | NC score | 0.011602 (rank : 154) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7M758 | Gene names | Naaladl1, Naaladasel | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylated-alpha-linked acidic dipeptidase-like protein (EC 3.4.17.21) (NAALADase L). | |||||
|
PDC6I_MOUSE
|
||||||
| θ value | 3.0926 (rank : 111) | NC score | 0.043754 (rank : 74) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9WU78, O88695, O89014, Q8BSL8, Q8R0H5, Q99LR3, Q9QZN8 | Gene names | Pdcd6ip, Aip1, Alix | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death 6-interacting protein (ALG-2-interacting protein X) (ALG-2-interacting protein 1) (E2F1-inducible protein) (Eig2). | |||||
|
SFPQ_MOUSE
|
||||||
| θ value | 3.0926 (rank : 112) | NC score | 0.035731 (rank : 92) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
SON_HUMAN
|
||||||
| θ value | 3.0926 (rank : 113) | NC score | 0.040239 (rank : 84) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
SPR1A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 114) | NC score | 0.063782 (rank : 55) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q62266 | Gene names | Sprr1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein 1A) (SPR1A) (SPR1 A). | |||||
|
SPRR3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 115) | NC score | 0.067716 (rank : 51) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O09116 | Gene names | Sprr3 | |||
|
Domain Architecture |

|
|||||
| Description | Small proline-rich protein 3 (Cornifin beta). | |||||
|
SSXT_HUMAN
|
||||||
| θ value | 3.0926 (rank : 116) | NC score | 0.049143 (rank : 64) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q15532, Q16404, Q9BXC6 | Gene names | SS18, SSXT, SYT | |||
|
Domain Architecture |
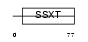
|
|||||
| Description | SSXT protein (Synovial sarcoma, translocated to X chromosome) (SYT protein). | |||||
|
TAXB1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 117) | NC score | -0.000257 (rank : 185) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86VP1, O60398, O95770, Q13311, Q9BQG5, Q9UI88 | Gene names | TAX1BP1, T6BP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 1 (TRAF6-binding protein). | |||||
|
TCF7_MOUSE
|
||||||
| θ value | 3.0926 (rank : 118) | NC score | 0.023584 (rank : 121) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q00417 | Gene names | Tcf7, Tcf-1, Tcf1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7 (T-cell-specific transcription factor 1) (TCF- 1) (T-cell factor 1). | |||||
|
ARI1A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 119) | NC score | 0.043678 (rank : 76) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
C1QC_HUMAN
|
||||||
| θ value | 4.03905 (rank : 120) | NC score | 0.008884 (rank : 167) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P02747, Q7Z502, Q96DL2, Q96H05 | Gene names | C1QC, C1QG | |||
|
Domain Architecture |
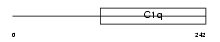
|
|||||
| Description | Complement C1q subcomponent subunit C precursor. | |||||
|
CCNK_HUMAN
|
||||||
| θ value | 4.03905 (rank : 121) | NC score | 0.025301 (rank : 116) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O75909, Q96B63, Q9NNY9 | Gene names | CCNK, CPR4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-K. | |||||
|
DAZP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 122) | NC score | 0.014273 (rank : 145) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96EP5, Q96MJ3, Q9NRR9 | Gene names | DAZAP1 | |||
|
Domain Architecture |
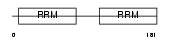
|
|||||
| Description | DAZ-associated protein 1 (Deleted in azoospermia-associated protein 1). | |||||
|
IASPP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 123) | NC score | 0.017459 (rank : 134) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8WUF5, Q2PNZ9, Q5DU71, Q5I1X4, Q6P1R7, Q6PKF8, Q9Y290 | Gene names | PPP1R13L, IASPP, NKIP1, PPP1R13BL, RAI | |||
|
Domain Architecture |

|
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
IQEC2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 124) | NC score | 0.029850 (rank : 109) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q5JU85, O60275 | Gene names | IQSEC2, KIAA0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
IQEC2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 125) | NC score | 0.029840 (rank : 110) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q5DU25 | Gene names | Iqsec2, Kiaa0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
PHLPP_MOUSE
|
||||||
| θ value | 4.03905 (rank : 126) | NC score | 0.002868 (rank : 182) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CHE4, Q6PCN0, Q8QZU8, Q8R5E5, Q99KL6 | Gene names | Phlpp, Kiaa0606, Plekhe1, Scop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein). | |||||
|
PO6F2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 127) | NC score | 0.024675 (rank : 120) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P78424, P78425, Q75ME8, Q86UM6, Q9UDS7 | Gene names | POU6F2, RPF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 2 (Retina-derived POU-domain factor 1) (RPF-1). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 128) | NC score | 0.038001 (rank : 89) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
PSL1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 129) | NC score | 0.020402 (rank : 126) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
STAT2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 130) | NC score | 0.011854 (rank : 153) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9WVL2, Q64188, Q64189, Q64250 | Gene names | Stat2 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 2. | |||||
|
TTC28_HUMAN
|
||||||
| θ value | 4.03905 (rank : 131) | NC score | 0.011407 (rank : 157) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96AY4, O95928, O95929, Q9NTE4, Q9UG31, Q9UGG5, Q9UPV8, Q9Y3S5 | Gene names | TTC28, KIAA1043 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
ZN385_HUMAN
|
||||||
| θ value | 4.03905 (rank : 132) | NC score | 0.031024 (rank : 102) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q96PM9, Q5VH53, Q9H7R6, Q9UFU3 | Gene names | ZNF385, HZF, RZF | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 385 (Hematopoietic zinc finger protein) (Retinal zinc finger protein). | |||||
|
ZO1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 133) | NC score | 0.013952 (rank : 146) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q07157, Q4ZGJ6 | Gene names | TJP1, ZO1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 134) | NC score | 0.007666 (rank : 175) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
CD166_HUMAN
|
||||||
| θ value | 5.27518 (rank : 135) | NC score | 0.008343 (rank : 171) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13740, O60892 | Gene names | ALCAM, MEMD | |||
|
Domain Architecture |

|
|||||
| Description | CD166 antigen precursor (Activated leukocyte-cell adhesion molecule) (ALCAM). | |||||
|
CO1A1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 136) | NC score | 0.015024 (rank : 139) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P11087, Q53WT0, Q60635, Q61367, Q61427, Q63919, Q6PCL3, Q810J9 | Gene names | Col1a1, Cola1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(I) chain precursor. | |||||
|
CO4A3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 137) | NC score | 0.018866 (rank : 130) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q01955, Q9BQT2 | Gene names | COL4A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(IV) chain precursor (Goodpasture antigen). | |||||
|
COLQ_HUMAN
|
||||||
| θ value | 5.27518 (rank : 138) | NC score | 0.017487 (rank : 133) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y215, Q6DK18, Q6YH18, Q6YH19, Q6YH20, Q6YH21, Q9NP18, Q9NP19, Q9NP20, Q9NP21, Q9NP22, Q9NP23, Q9NP24, Q9UP88 | Gene names | COLQ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acetylcholinesterase collagenic tail peptide precursor (AChE Q subunit) (Acetylcholinesterase-associated collagen). | |||||
|
ENAM_MOUSE
|
||||||
| θ value | 5.27518 (rank : 139) | NC score | 0.030752 (rank : 105) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O55196 | Gene names | Enam | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enamelin precursor. | |||||
|
GLIS1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 140) | NC score | -0.003184 (rank : 187) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8NBF1, Q8N9V9 | Gene names | GLIS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLIS1 (GLI-similar 1). | |||||
|
HELZ_HUMAN
|
||||||
| θ value | 5.27518 (rank : 141) | NC score | 0.016008 (rank : 136) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P42694 | Gene names | HELZ, KIAA0054 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable helicase with zinc-finger domain (EC 3.6.1.-). | |||||
|
MOT8_HUMAN
|
||||||
| θ value | 5.27518 (rank : 142) | NC score | 0.008749 (rank : 168) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P36021, Q7Z797 | Gene names | SLC16A2, MCT8, XPCT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Monocarboxylate transporter 8 (MCT 8) (MCT 7) (Solute carrier family 16 member 2) (X-linked PEST-containing transporter). | |||||
|
MYST3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 143) | NC score | 0.029888 (rank : 108) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
PYGO1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 144) | NC score | 0.015651 (rank : 137) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y3Y4 | Gene names | PYGO1 | |||
|
Domain Architecture |

|
|||||
| Description | Pygopus homolog 1. | |||||
|
RBP2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 145) | NC score | 0.009030 (rank : 164) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9ERU9, Q61992, Q8C9K9 | Gene names | Ranbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2 (RanBP2). | |||||
|
SHC1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 146) | NC score | 0.003896 (rank : 180) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P98083, Q3U2Q7, Q8BFY3, Q8K4C6, Q8K4C7 | Gene names | Shc1, Shc, ShcA | |||
|
Domain Architecture |

|
|||||
| Description | SHC-transforming protein 1 (SH2 domain protein C1) (Src homology 2 domain-containing-transforming protein C1). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 147) | NC score | 0.012166 (rank : 152) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TROAP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 148) | NC score | 0.048763 (rank : 65) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q12815 | Gene names | TROAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trophinin-associated protein (Tastin) (Trophinin-assisting protein). | |||||
|
ZCH11_HUMAN
|
||||||
| θ value | 5.27518 (rank : 149) | NC score | 0.020774 (rank : 124) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5TAX3, Q12764, Q5TAX4, Q86XZ3 | Gene names | ZCCHC11, KIAA0191 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 11. | |||||
|
AMELX_HUMAN
|
||||||
| θ value | 6.88961 (rank : 150) | NC score | 0.036079 (rank : 90) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q99217, Q96NW6, Q9UCA7 | Gene names | AMELX, AMG, AMGX | |||
|
Domain Architecture |

|
|||||
| Description | Amelogenin, X isoform precursor. | |||||
|
CC026_HUMAN
|
||||||
| θ value | 6.88961 (rank : 151) | NC score | 0.008927 (rank : 166) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BQ75 | Gene names | C3orf26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf26. | |||||
|
CD248_HUMAN
|
||||||
| θ value | 6.88961 (rank : 152) | NC score | 0.014912 (rank : 141) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9HCU0, Q3SX55, Q96KB6 | Gene names | CD248, CD164L1, TEM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
CO8A1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 153) | NC score | 0.018242 (rank : 131) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q00780, Q9D2V4 | Gene names | Col8a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VIII) chain precursor. | |||||
|
EP300_HUMAN
|
||||||
| θ value | 6.88961 (rank : 154) | NC score | 0.032011 (rank : 97) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
HD_HUMAN
|
||||||
| θ value | 6.88961 (rank : 155) | NC score | 0.020091 (rank : 127) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P42858, Q9UQB7 | Gene names | HD, IT15 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin (Huntington disease protein) (HD protein). | |||||
|
HD_MOUSE
|
||||||
| θ value | 6.88961 (rank : 156) | NC score | 0.012641 (rank : 150) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P42859 | Gene names | Hd, Hdh | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin (Huntington disease protein homolog) (HD protein). | |||||
|
HNRL1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 157) | NC score | 0.014698 (rank : 143) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9BUJ2, O76022, Q6ZSZ0, Q7L8P4, Q8N6Z4, Q96G37, Q9HAL3, Q9UG75 | Gene names | HNRPUL1, E1BAP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein U-like protein 1 (Adenovirus early region 1B-associated protein 5) (E1B-55 kDa-associated protein 5) (E1B-AP5). | |||||
|
K0141_HUMAN
|
||||||
| θ value | 6.88961 (rank : 158) | NC score | 0.008460 (rank : 170) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14154, Q969R4, Q96EU9 | Gene names | KIAA0141 | |||
|
Domain Architecture |

|
|||||
| Description | Protein KIAA0141. | |||||
|
MCR_HUMAN
|
||||||
| θ value | 6.88961 (rank : 159) | NC score | 0.006260 (rank : 179) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P08235, Q96KQ8, Q96KQ9 | Gene names | NR3C2, MCR, MLR | |||
|
Domain Architecture |

|
|||||
| Description | Mineralocorticoid receptor (MR). | |||||
|
MKL1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 160) | NC score | 0.011037 (rank : 158) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q969V6, Q8TCL1, Q96SC5, Q96SC6, Q9P2B0 | Gene names | MKL1, KIAA1438, MAL | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein) (Megakaryocytic acute leukemia protein). | |||||
|
PB1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 161) | NC score | 0.013118 (rank : 148) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q86U86, Q9H2T3, Q9H2T4, Q9H2T5, Q9H301, Q9H314 | Gene names | PB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein polybromo-1 (hPB1) (Polybromo-1D) (BRG1-associated factor 180) (BAF180). | |||||
|
R3HD2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 162) | NC score | 0.028347 (rank : 113) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q80TM6, Q80YB1, Q8BLS5, Q9CW50 | Gene names | R3hdm2, Kiaa1002 | |||
|
Domain Architecture |

|
|||||
| Description | R3H domain-containing protein 2. | |||||
|
RERE_HUMAN
|
||||||
| θ value | 6.88961 (rank : 163) | NC score | 0.032938 (rank : 95) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
RFX1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 164) | NC score | 0.014892 (rank : 142) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P48377 | Gene names | Rfx1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein RFX1. | |||||
|
SFR15_HUMAN
|
||||||
| θ value | 6.88961 (rank : 165) | NC score | 0.040883 (rank : 81) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O95104, Q6P1M5, Q8N3I8, Q9UFM1, Q9ULP8 | Gene names | SFRS15, KIAA1172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 15 (CTD-binding SR-like protein RA4). | |||||
|
TCF19_HUMAN
|
||||||
| θ value | 6.88961 (rank : 166) | NC score | 0.012790 (rank : 149) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y242, Q13176, Q15967, Q5SQ89, Q5STD6, Q5STF5, Q9BUM2, Q9UBH7 | Gene names | TCF19, SC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 19 (Transcription factor SC1). | |||||
|
ZN687_HUMAN
|
||||||
| θ value | 6.88961 (rank : 167) | NC score | 0.003428 (rank : 181) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8N1G0, Q68DQ8, Q9H937, Q9P2A7 | Gene names | ZNF687, KIAA1441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
BSN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 168) | NC score | 0.041721 (rank : 78) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
C1QC_MOUSE
|
||||||
| θ value | 8.99809 (rank : 169) | NC score | 0.007320 (rank : 176) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q02105 | Gene names | C1qc, C1qg | |||
|
Domain Architecture |
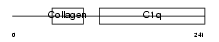
|
|||||
| Description | Complement C1q subcomponent subunit C precursor. | |||||
|
CAC1H_HUMAN
|
||||||
| θ value | 8.99809 (rank : 170) | NC score | 0.002322 (rank : 183) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
COGA1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 171) | NC score | 0.021193 (rank : 123) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q07092 | Gene names | COL16A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVI) chain precursor. | |||||
|
CSKI1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 172) | NC score | 0.009805 (rank : 162) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
FA61A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 173) | NC score | 0.006599 (rank : 178) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K2F8, Q9CTG8 | Gene names | Fam61a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM61A. | |||||
|
GLI1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 174) | NC score | -0.004100 (rank : 189) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P47806, Q9QYK1 | Gene names | Gli1, Gli | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI1 (Glioma-associated oncogene homolog). | |||||
|
NOTC2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 175) | NC score | -0.001029 (rank : 186) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1069 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O35516, Q06008, Q60941 | Gene names | Notch2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (Motch B) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
NUMBL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 176) | NC score | 0.014956 (rank : 140) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Y6R0, Q7Z4J9 | Gene names | NUMBL | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein (Numb-R). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 8.99809 (rank : 177) | NC score | 0.039413 (rank : 85) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PCQAP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 178) | NC score | 0.031761 (rank : 99) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q924H2 | Gene names | Pcqap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (mPcqap). | |||||
|
SET1A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 179) | NC score | 0.025144 (rank : 119) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
SPEG_HUMAN
|
||||||
| θ value | 8.99809 (rank : 180) | NC score | 0.001247 (rank : 184) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
TCF20_MOUSE
|
||||||
| θ value | 8.99809 (rank : 181) | NC score | 0.017899 (rank : 132) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.061735 (rank : 57) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.054155 (rank : 61) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PRP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.054023 (rank : 62) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.052433 (rank : 63) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRPC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.067910 (rank : 50) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P02810, Q4VBP2, Q53XA2, Q6P2F6 | Gene names | PRH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Salivary acidic proline-rich phosphoprotein 1/2 precursor (PRP-1/PRP- 2) (Parotid proline-rich protein 1/2) (Pr1/Pr2) (Protein C) (Parotid acidic protein) (Pa) (Parotid isoelectric focusing variant protein) (PIF-S) (Parotid double-band protein) (Db-s) [Contains: Salivary acidic proline-rich phosphoprotein 1/2; Salivary acidic proline-rich phosphoprotein 3/4 (PRP-3/PRP-4) (Protein A) (PIF-F) (Db-F); Peptide P-C]. | |||||
|
PRR13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.090966 (rank : 48) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NZ81, Q6FIG7, Q6MZP8, Q6NXQ6, Q6PKF9 | Gene names | PRR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
SPIC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.451316 (rank : 46) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8N5J4 | Gene names | SPIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-C. | |||||
|
SPIC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 189) | NC score | 0.427888 (rank : 47) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6P3D7, Q9Z0Y0 | Gene names | Spic | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-C (Pu.1-related factor) (Prf). | |||||
|
ETV5_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 181 | |
| SwissProt Accessions | P41161 | Gene names | ETV5, ERM | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 5 (Ets-related protein ERM). | |||||
|
ETV1_MOUSE
|
||||||
| NC score | 0.981219 (rank : 2) | θ value | 2.94559e-168 (rank : 4) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P41164 | Gene names | Etv1, Er81, Etsrp81 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 1 (ER81 protein). | |||||
|
ETV1_HUMAN
|
||||||
| NC score | 0.980633 (rank : 3) | θ value | 1.19589e-169 (rank : 3) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P50549, O75849, Q9UQ71, Q9Y636 | Gene names | ETV1, ER81 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 1 (ER81 protein). | |||||
|
ETV5_MOUSE
|
||||||
| NC score | 0.972469 (rank : 4) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 125 | |
| SwissProt Accessions | Q9CXC9, Q3TG49, Q8C0F3, Q9JHB1 | Gene names | Etv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 5. | |||||
|
ETV4_HUMAN
|
||||||
| NC score | 0.968104 (rank : 5) | θ value | 1.75107e-120 (rank : 5) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P43268, Q96AW9 | Gene names | ETV4, E1AF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 4 (Adenovirus E1A enhancer-binding protein) (E1A-F). | |||||
|
ETV4_MOUSE
|
||||||
| NC score | 0.966686 (rank : 6) | θ value | 4.03687e-117 (rank : 6) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P28322 | Gene names | Etv4, Pea-3, Pea3 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 4 (Polyomavirus enhancer activator 3) (Protein PEA3). | |||||
|
ETV2_HUMAN
|
||||||
| NC score | 0.874352 (rank : 7) | θ value | 1.16704e-23 (rank : 23) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O00321, Q9UEA0 | Gene names | ETV2, ER71, ETSRP71 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 2 (Ets-related protein 71). | |||||
|
ELK1_HUMAN
|
||||||
| NC score | 0.873017 (rank : 8) | θ value | 1.62847e-25 (rank : 17) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P19419, O75606, O95058, Q969X8, Q9UJM4 | Gene names | ELK1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-1. | |||||
|
ELK4_MOUSE
|
||||||
| NC score | 0.871634 (rank : 9) | θ value | 8.63488e-27 (rank : 11) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P41158 | Gene names | Elk4, Sap1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
|
ELK3_HUMAN
|
||||||
| NC score | 0.871632 (rank : 10) | θ value | 1.05554e-24 (rank : 20) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P41970, Q6FG57, Q6GU29, Q9UD17 | Gene names | ELK3, NET, SAP2 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-3 (ETS-related protein NET) (ETS- related protein ERP) (SRF accessory protein 2) (SAP-2). | |||||
|
ELK1_MOUSE
|
||||||
| NC score | 0.870566 (rank : 11) | θ value | 1.62847e-25 (rank : 18) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P41969 | Gene names | Elk1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-1. | |||||
|
ETS1_MOUSE
|
||||||
| NC score | 0.870085 (rank : 12) | θ value | 1.08979e-29 (rank : 8) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P27577, Q61403 | Gene names | Ets1, Ets-1 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-1 protein (p54). | |||||
|
ETS1_HUMAN
|
||||||
| NC score | 0.870027 (rank : 13) | θ value | 1.08979e-29 (rank : 7) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P14921, Q14278, Q16080 | Gene names | ETS1 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-1 protein (p54). | |||||
|
ELK3_MOUSE
|
||||||
| NC score | 0.869422 (rank : 14) | θ value | 8.08199e-25 (rank : 19) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P41971, P97747, Q62346 | Gene names | Elk3, Erp, Net | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-3 (ETS-related protein NET) (ETS- related protein ERP). | |||||
|
ETV2_MOUSE
|
||||||
| NC score | 0.869288 (rank : 15) | θ value | 2.20094e-22 (rank : 24) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P41163 | Gene names | Etv2, Er71, Etsrp71 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 2 (Ets-related protein 71). | |||||
|
ETS2_HUMAN
|
||||||
| NC score | 0.867608 (rank : 16) | θ value | 5.40856e-29 (rank : 9) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P15036 | Gene names | ETS2 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-2 protein. | |||||
|
ETS2_MOUSE
|
||||||
| NC score | 0.866953 (rank : 17) | θ value | 5.40856e-29 (rank : 10) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P15037 | Gene names | Ets2 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-2 protein. | |||||
|
ELK4_HUMAN
|
||||||
| NC score | 0.858141 (rank : 18) | θ value | 5.59698e-26 (rank : 14) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P28324, P28323 | Gene names | ELK4, SAP1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
|
GABPA_MOUSE
|
||||||
| NC score | 0.855809 (rank : 19) | θ value | 4.01107e-24 (rank : 21) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q00422 | Gene names | Gabpa, E4tf1a | |||
|
Domain Architecture |

|
|||||
| Description | GA-binding protein alpha chain (GABP-subunit alpha). | |||||
|
GABPA_HUMAN
|
||||||
| NC score | 0.854304 (rank : 20) | θ value | 8.93572e-24 (rank : 22) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q06546, Q12939 | Gene names | GABPA, E4TF1A | |||
|
Domain Architecture |

|
|||||
| Description | GA-binding protein alpha chain (GABP-subunit alpha) (Transcription factor E4TF1-60) (Nuclear respiratory factor 2 subunit alpha). | |||||
|
FLI1_MOUSE
|
||||||
| NC score | 0.845457 (rank : 21) | θ value | 2.51237e-26 (rank : 13) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P26323 | Gene names | Fli1, Fli-1 | |||
|
Domain Architecture |

|
|||||
| Description | Friend leukemia integration 1 transcription factor (Retroviral integration site protein Fli-1). | |||||
|
FLI1_HUMAN
|
||||||
| NC score | 0.844066 (rank : 22) | θ value | 1.92365e-26 (rank : 12) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q01543, Q14319, Q92480, Q9UE07 | Gene names | FLI1 | |||
|
Domain Architecture |

|
|||||
| Description | Friend leukemia integration 1 transcription factor (Fli-1 proto- oncogene) (ERGB transcription factor). | |||||
|
ERG_MOUSE
|
||||||
| NC score | 0.842520 (rank : 23) | θ value | 7.30988e-26 (rank : 15) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P81270, Q8C5L4, Q920K7, Q920K8, Q920K9 | Gene names | Erg, Erg-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ERG. | |||||
|
ERG_HUMAN
|
||||||
| NC score | 0.841862 (rank : 24) | θ value | 1.24688e-25 (rank : 16) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P11308, Q16113 | Gene names | ERG | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ERG (Transforming protein ERG). | |||||
|
SPDEF_HUMAN
|
||||||
| NC score | 0.816911 (rank : 25) | θ value | 6.85773e-16 (rank : 33) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O95238 | Gene names | SPDEF, PDEF, PSE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAM pointed domain-containing Ets transcription factor (Prostate- derived Ets factor) (Prostate epithelium-specific Ets transcription factor) (Prostate-specific Ets). | |||||
|
ERF_HUMAN
|
||||||
| NC score | 0.815421 (rank : 26) | θ value | 5.07402e-19 (rank : 25) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P50548, Q9UPI7 | Gene names | ERF | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing transcription factor ERF (Ets2 repressor factor). | |||||
|
ERF_MOUSE
|
||||||
| NC score | 0.815328 (rank : 27) | θ value | 5.07402e-19 (rank : 26) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P70459 | Gene names | Erf | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing transcription factor ERF. | |||||
|
SPDEF_MOUSE
|
||||||
| NC score | 0.815071 (rank : 28) | θ value | 8.95645e-16 (rank : 35) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9WTP3 | Gene names | Spdef, Pdef, Pse | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAM pointed domain-containing Ets transcription factor (Prostate- derived Ets factor) (Prostate epithelium-specific Ets transcription factor) (Prostate-specific Ets). | |||||
|
ETV3_MOUSE
|
||||||
| NC score | 0.804401 (rank : 29) | θ value | 3.28887e-18 (rank : 28) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8R4Z4, Q9QZW1 | Gene names | Etv3, Mets, Pe1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
ETV7_HUMAN
|
||||||
| NC score | 0.799106 (rank : 30) | θ value | 2.28291e-11 (rank : 39) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9Y603, Q9NZ65, Q9NZ66, Q9NZ68, Q9NZR8, Q9UNJ7, Q9Y5K4, Q9Y604 | Gene names | ETV7, TEL2, TELB, TREF | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV7 (Transcription factor Tel-2) (ETS-related protein Tel2) (Tel-related Ets factor). | |||||
|
ETV3_HUMAN
|
||||||
| NC score | 0.783542 (rank : 31) | θ value | 1.92812e-18 (rank : 27) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P41162, Q8TAC8, Q9BX30 | Gene names | ETV3, METS, PE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
ELF2_HUMAN
|
||||||
| NC score | 0.780779 (rank : 32) | θ value | 8.95645e-16 (rank : 34) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q15723, Q15724, Q15725, Q6P1K5 | Gene names | ELF2, NERF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-2 (E74-like factor 2) (New ETS- related factor). | |||||
|
ELF2_MOUSE
|
||||||
| NC score | 0.780309 (rank : 33) | θ value | 1.99529e-15 (rank : 36) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9JHC9, Q6NST2, Q8BTX8, Q9JHC7, Q9JHC8, Q9JHD0 | Gene names | Elf2, Nerf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-2 (E74-like factor 2) (New ETS- related factor). | |||||
|
ELF4_HUMAN
|
||||||
| NC score | 0.779643 (rank : 34) | θ value | 4.02038e-16 (rank : 31) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q99607, O60435 | Gene names | ELF4, ELFR, MEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-4 (E74-like factor 4) (Myeloid Elf-1-like factor). | |||||
|
ELF1_HUMAN
|
||||||
| NC score | 0.779159 (rank : 35) | θ value | 1.058e-16 (rank : 30) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P32519, Q9UDE1 | Gene names | ELF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
ETV6_MOUSE
|
||||||
| NC score | 0.778423 (rank : 36) | θ value | 2.88119e-14 (rank : 38) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P97360 | Gene names | Etv6, Tel, Tel1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV6 (ETS-related protein Tel1) (Tel) (ETS translocation variant 6). | |||||
|
ETV6_HUMAN
|
||||||
| NC score | 0.777889 (rank : 37) | θ value | 2.88119e-14 (rank : 37) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P41212, Q9UMF6 | Gene names | ETV6, TEL, TEL1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV6 (ETS-related protein Tel1) (Tel) (ETS translocation variant 6). | |||||
|
ELF1_MOUSE
|
||||||
| NC score | 0.772808 (rank : 38) | θ value | 4.74913e-17 (rank : 29) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q60775 | Gene names | Elf1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
ELF5_HUMAN
|
||||||
| NC score | 0.760577 (rank : 39) | θ value | 1.133e-10 (rank : 40) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9UKW6, O95175, Q8N2K9, Q96QY3, Q9UKW5 | Gene names | ELF5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-5 (E74-like factor 5) (Epithelium-specific Ets transcription factor 2) (ESE-2) (Epithelium- restricted ESE-1-related Ets factor). | |||||
|
ELF5_MOUSE
|
||||||
| NC score | 0.760432 (rank : 40) | θ value | 1.133e-10 (rank : 41) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8VDK3, Q921H5, Q9Z2K6 | Gene names | Elf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-5 (E74-like factor 5). | |||||
|
ELF4_MOUSE
|
||||||
| NC score | 0.760137 (rank : 41) | θ value | 4.02038e-16 (rank : 32) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9Z2U4 | Gene names | Elf4, Mef | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-4 (E74-like factor 4) (Myeloid Elf-1-like factor). | |||||
|
SPIB_HUMAN
|
||||||
| NC score | 0.578466 (rank : 42) | θ value | 4.92598e-06 (rank : 42) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q01892, Q15359 | Gene names | SPIB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Spi-B. | |||||
|
SPI1_MOUSE
|
||||||
| NC score | 0.573041 (rank : 43) | θ value | 8.40245e-06 (rank : 43) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P17433, Q99L57 | Gene names | Spi1, Sfpi-1, Sfpi1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor PU.1 (31 kDa transforming protein) (SFFV proviral integration 1 protein). | |||||
|
SPI1_HUMAN
|
||||||
| NC score | 0.571829 (rank : 44) | θ value | 1.09739e-05 (rank : 44) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P17947 | Gene names | SPI1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor PU.1 (31 kDa transforming protein). | |||||
|
SPIB_MOUSE
|
||||||
| NC score | 0.562597 (rank : 45) | θ value | 1.09739e-05 (rank : 45) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O35906, O35907, O35909, O55199 | Gene names | Spib | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-B. | |||||
|
SPIC_HUMAN
|
||||||
| NC score | 0.451316 (rank : 46) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8N5J4 | Gene names | SPIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-C. | |||||
|
SPIC_MOUSE
|
||||||
| NC score | 0.427888 (rank : 47) | θ value | θ > 10 (rank : 189) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6P3D7, Q9Z0Y0 | Gene names | Spic | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-C (Pu.1-related factor) (Prf). | |||||
|
PRR13_HUMAN
|
||||||
| NC score | 0.090966 (rank : 48) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NZ81, Q6FIG7, Q6MZP8, Q6NXQ6, Q6PKF9 | Gene names | PRR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
SPR1B_MOUSE
|
||||||
| NC score | 0.070995 (rank : 49) | θ value | 0.000461057 (rank : 46) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q62267 | Gene names | Sprr1b | |||
|
Domain Architecture |

|
|||||
| Description | Cornifin B (Small proline-rich protein 1B) (SPR1B) (SPR1 B). | |||||
|
PRPC_HUMAN
|
||||||
| NC score | 0.067910 (rank : 50) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P02810, Q4VBP2, Q53XA2, Q6P2F6 | Gene names | PRH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Salivary acidic proline-rich phosphoprotein 1/2 precursor (PRP-1/PRP- 2) (Parotid proline-rich protein 1/2) (Pr1/Pr2) (Protein C) (Parotid acidic protein) (Pa) (Parotid isoelectric focusing variant protein) (PIF-S) (Parotid double-band protein) (Db-s) [Contains: Salivary acidic proline-rich phosphoprotein 1/2; Salivary acidic proline-rich phosphoprotein 3/4 (PRP-3/PRP-4) (Protein A) (PIF-F) (Db-F); Peptide P-C]. | |||||
|
SPRR3_MOUSE
|
||||||
| NC score | 0.067716 (rank : 51) | θ value | 3.0926 (rank : 115) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O09116 | Gene names | Sprr3 | |||
|
Domain Architecture |

|
|||||
| Description | Small proline-rich protein 3 (Cornifin beta). | |||||
|
ZIM10_MOUSE
|
||||||
| NC score | 0.067310 (rank : 52) | θ value | 0.00869519 (rank : 49) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q6P1E1, Q6PDK9, Q6PF85, Q8BW47 | Gene names | Rai17, Zimp10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
ZIM10_HUMAN
|
||||||
| NC score | 0.066416 (rank : 53) | θ value | 0.00665767 (rank : 48) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
SPR1B_HUMAN
|
||||||
| NC score | 0.065402 (rank : 54) | θ value | 0.279714 (rank : 60) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P22528, P22529, P22530, Q5T524 | Gene names | SPRR1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin B (Small proline-rich protein IB) (SPR-IB) (14.9 kDa pancornulin). | |||||
|
SPR1A_MOUSE
|
||||||
| NC score | 0.063782 (rank : 55) | θ value | 3.0926 (rank : 114) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q62266 | Gene names | Sprr1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein 1A) (SPR1A) (SPR1 A). | |||||
|
SPRR3_HUMAN
|
||||||
| NC score | 0.061856 (rank : 56) | θ value | 0.279714 (rank : 61) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UBC9, O75597, Q4ZGI7, Q5T525, Q8NET7, Q9UDG3 | Gene names | SPRR3, SPRC | |||
|
Domain Architecture |

|
|||||
| Description | Small proline-rich protein 3 (Cornifin beta) (Esophagin) (22 kDa pancornulin). | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.061735 (rank : 57) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
SPR1A_HUMAN
|
||||||
| NC score | 0.059124 (rank : 58) | θ value | 0.62314 (rank : 70) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P35321, Q9UDG4 | Gene names | SPRR1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein IA) (SPR-IA) (SPRK) (19 kDa pancornulin). | |||||
|
PRR8_HUMAN
|
||||||
| NC score | 0.056987 (rank : 59) | θ value | 1.06291 (rank : 83) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9NSV0 | Gene names | PRR8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 8. | |||||
|
PDC6I_HUMAN
|
||||||
| NC score | 0.056852 (rank : 60) | θ value | 1.81305 (rank : 90) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8WUM4, Q9BX86, Q9NUN0, Q9P2H2, Q9UKL5 | Gene names | PDCD6IP, AIP1, ALIX, KIAA1375 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death 6-interacting protein (PDCD6-interacting protein) (ALG-2-interacting protein 1) (Hp95). | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.054155 (rank : 61) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.054023 (rank : 62) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.052433 (rank : 63) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
SSXT_HUMAN
|
||||||
| NC score | 0.049143 (rank : 64) | θ value | 3.0926 (rank : 116) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q15532, Q16404, Q9BXC6 | Gene names | SS18, SSXT, SYT | |||
|
Domain Architecture |

|
|||||
| Description | SSXT protein (Synovial sarcoma, translocated to X chromosome) (SYT protein). | |||||
|
TROAP_HUMAN
|
||||||
| NC score | 0.048763 (rank : 65) | θ value | 5.27518 (rank : 148) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q12815 | Gene names | TROAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trophinin-associated protein (Tastin) (Trophinin-assisting protein). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.048644 (rank : 66) | θ value | 0.813845 (rank : 72) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
PTN23_HUMAN
|
||||||
| NC score | 0.048516 (rank : 67) | θ value | 0.00665767 (rank : 47) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9H3S7, Q7KZF8, Q8N6Z5, Q9BSR5, Q9P257, Q9UG03, Q9UMZ4 | Gene names | PTPN23, KIAA1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His- domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14). | |||||
|
K0355_HUMAN
|
||||||
| NC score | 0.048274 (rank : 68) | θ value | 1.06291 (rank : 81) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O15063 | Gene names | KIAA0355 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0355. | |||||
|
SSXT_MOUSE
|
||||||
| NC score | 0.045984 (rank : 69) | θ value | 1.81305 (rank : 92) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q62280 | Gene names | Ss18, Ssxt, Syt | |||
|
Domain Architecture |

|
|||||
| Description | SSXT protein (SYT protein) (Synovial sarcoma-associated Ss18-alpha). | |||||
|
SSBP4_HUMAN
|
||||||
| NC score | 0.045901 (rank : 70) | θ value | 0.0736092 (rank : 54) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
PHLA1_HUMAN
|
||||||
| NC score | 0.044896 (rank : 71) | θ value | 0.47712 (rank : 65) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8WV24, Q15184, Q2TAN2, Q9NZ17 | Gene names | PHLDA1, PHRIP, TDAG51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family A member 1 (T-cell death- associated gene 51 protein) (Apoptosis-associated nuclear protein) (Proline- and histidine-rich protein) (Proline- and glutamine-rich protein) (PQ-rich protein). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.044752 (rank : 72) | θ value | 0.0431538 (rank : 52) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 108 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
PTN23_MOUSE
|
||||||
| NC score | 0.044140 (rank : 73) | θ value | 0.279714 (rank : 59) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
PDC6I_MOUSE
|
||||||
| NC score | 0.043754 (rank : 74) | θ value | 3.0926 (rank : 111) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9WU78, O88695, O89014, Q8BSL8, Q8R0H5, Q99LR3, Q9QZN8 | Gene names | Pdcd6ip, Aip1, Alix | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death 6-interacting protein (ALG-2-interacting protein X) (ALG-2-interacting protein 1) (E2F1-inducible protein) (Eig2). | |||||
|
BRD4_HUMAN
|
||||||
| NC score | 0.043699 (rank : 75) | θ value | 2.36792 (rank : 94) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
ARI1A_HUMAN
|
||||||
| NC score | 0.043678 (rank : 76) | θ value | 4.03905 (rank : 119) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
SCAP_HUMAN
|
||||||
| NC score | 0.042710 (rank : 77) | θ value | 0.0193708 (rank : 50) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q12770, Q8N2E0, Q8WUA1 | Gene names | SCAP, KIAA0199 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein cleavage-activating protein (SREBP cleavage-activating protein) (SCAP). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.041721 (rank : 78) | θ value | 8.99809 (rank : 168) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
PELP1_MOUSE
|
||||||
| NC score | 0.041574 (rank : 79) | θ value | 0.62314 (rank : 69) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
TPO_MOUSE
|
||||||
| NC score | 0.041343 (rank : 80) | θ value | 0.365318 (rank : 62) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P40226 | Gene names | Thpo | |||
|
Domain Architecture |

|
|||||
| Description | Thrombopoietin precursor (Megakaryocyte colony-stimulating factor) (Myeloproliferative leukemia virus oncogene ligand) (C-mpl ligand) (ML) (Megakaryocyte growth and development factor) (MGDF). | |||||
|
SFR15_HUMAN
|
||||||
| NC score | 0.040883 (rank : 81) | θ value | 6.88961 (rank : 165) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O95104, Q6P1M5, Q8N3I8, Q9UFM1, Q9ULP8 | Gene names | SFRS15, KIAA1172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 15 (CTD-binding SR-like protein RA4). | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.040749 (rank : 82) | θ value | 3.0926 (rank : 102) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
BRD4_MOUSE
|
||||||
| NC score | 0.040424 (rank : 83) | θ value | 0.813845 (rank : 71) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.040239 (rank : 84) | θ value | 3.0926 (rank : 113) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.039413 (rank : 85) | θ value | 8.99809 (rank : 177) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.039371 (rank : 86) | θ value | 0.0961366 (rank : 55) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
SFPQ_HUMAN
|
||||||
| NC score | 0.038768 (rank : 87) | θ value | 0.813845 (rank : 76) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.038103 (rank : 88) | θ value | 0.0736092 (rank : 53) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.038001 (rank : 89) | θ value | 4.03905 (rank : 128) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
AMELX_HUMAN
|
||||||
| NC score | 0.036079 (rank : 90) | θ value | 6.88961 (rank : 150) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q99217, Q96NW6, Q9UCA7 | Gene names | AMELX, AMG, AMGX | |||
|
Domain Architecture |

|
|||||
| Description | Amelogenin, X isoform precursor. | |||||
|
PHC2_HUMAN
|
||||||
| NC score | 0.035773 (rank : 91) | θ value | 0.47712 (rank : 64) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8IXK0, Q5T0C1, Q6NUJ6, Q6ZQR1, Q8N306, Q8TAG8, Q96BL4, Q9Y4Y7 | Gene names | PHC2, EDR2, PH2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 2 (hPH2) (Early development regulatory protein 2). | |||||
|
SFPQ_MOUSE
|
||||||
| NC score | 0.035731 (rank : 92) | θ value | 3.0926 (rank : 112) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
PO6F2_MOUSE
|
||||||
| NC score | 0.035546 (rank : 93) | θ value | 0.125558 (rank : 57) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BJI4 | Gene names | Pou6f2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | POU domain, class 6, transcription factor 2. | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.033875 (rank : 94) | θ value | 0.0330416 (rank : 51) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
RERE_HUMAN
|
||||||
| NC score | 0.032938 (rank : 95) | θ value | 6.88961 (rank : 163) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
PHLA1_MOUSE
|
||||||
| NC score | 0.032900 (rank : 96) | θ value | 1.81305 (rank : 91) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q62392, Q3TY48, Q3UG87 | Gene names | Phlda1, Tdag51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family A member 1 (T-cell death- associated gene 51 protein) (Proline- and glutamine-rich protein) (PQR protein). | |||||
|
EP300_HUMAN
|
||||||
| NC score | 0.032011 (rank : 97) | θ value | 6.88961 (rank : 154) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
RNZ2_MOUSE
|
||||||
| NC score | 0.031840 (rank : 98) | θ value | 0.813845 (rank : 75) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80Y81, Q99MF0, Q99MF1, Q9CTA2, Q9D1A8, Q9EPZ2 | Gene names | Elac2 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc phosphodiesterase ELAC protein 2 (EC 3.1.26.11) (Ribonuclease Z 2) (RNase Z 2) (tRNase Z 2) (tRNA 3 endonuclease 2) (ElaC homolog protein 2). | |||||
|
PCQAP_MOUSE
|
||||||
| NC score | 0.031761 (rank : 99) | θ value | 8.99809 (rank : 178) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q924H2 | Gene names | Pcqap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (mPcqap). | |||||
|
CG1_HUMAN
|
||||||
| NC score | 0.031537 (rank : 100) | θ value | 1.06291 (rank : 80) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13495 | Gene names | CXorf6, CG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CG1 protein (F18). | |||||
|
SYNJ1_HUMAN
|
||||||
| NC score | 0.031274 (rank : 101) | θ value | 0.813845 (rank : 77) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O43426, O43425, O94984 | Gene names | SYNJ1, KIAA0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
ZN385_HUMAN
|
||||||
| NC score | 0.031024 (rank : 102) | θ value | 4.03905 (rank : 132) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q96PM9, Q5VH53, Q9H7R6, Q9UFU3 | Gene names | ZNF385, HZF, RZF | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 385 (Hematopoietic zinc finger protein) (Retinal zinc finger protein). | |||||
|
AMELY_HUMAN
|
||||||
| NC score | 0.030948 (rank : 103) | θ value | 3.0926 (rank : 101) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99218, Q6RWT1 | Gene names | AMELY, AMGL, AMGY | |||
|
Domain Architecture |

|
|||||
| Description | Amelogenin, Y isoform precursor. | |||||
|
PCQAP_HUMAN
|
||||||
| NC score | 0.030808 (rank : 104) | θ value | 2.36792 (rank : 100) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96RN5, O15413, Q6IC31, Q8NF16, Q96CT0, Q96IH7, Q9P1T3 | Gene names | PCQAP, ARC105, CTG7A, TIG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (TPA-inducible gene 1 protein) (TIG-1) (Activator-recruited cofactor 105 kDa component) (ARC105) (CTG repeat protein 7a). | |||||
|
ENAM_MOUSE
|
||||||
| NC score | 0.030752 (rank : 105) | θ value | 5.27518 (rank : 139) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O55196 | Gene names | Enam | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enamelin precursor. | |||||
|
SOX13_HUMAN
|
||||||
| NC score | 0.030594 (rank : 106) | θ value | 0.47712 (rank : 66) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9UN79, O95275, O95826, Q9UHW7 | Gene names | SOX13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein (Type 1 diabetes autoantigen ICA12) (Islet cell antigen 12). | |||||
|
ENAM_HUMAN
|
||||||
| NC score | 0.030445 (rank : 107) | θ value | 2.36792 (rank : 98) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9NRM1, Q9H3D1 | Gene names | ENAM | |||
|
Domain Architecture |

|
|||||
| Description | Enamelin precursor. | |||||
|
MYST3_MOUSE
|
||||||
| NC score | 0.029888 (rank : 108) | θ value | 5.27518 (rank : 143) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
IQEC2_HUMAN
|
||||||
| NC score | 0.029850 (rank : 109) | θ value | 4.03905 (rank : 124) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q5JU85, O60275 | Gene names | IQSEC2, KIAA0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
IQEC2_MOUSE
|
||||||
| NC score | 0.029840 (rank : 110) | θ value | 4.03905 (rank : 125) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q5DU25 | Gene names | Iqsec2, Kiaa0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
IL4RA_HUMAN
|
||||||
| NC score | 0.029394 (rank : 111) | θ value | 1.81305 (rank : 89) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P24394, Q96P01, Q9H181, Q9H182, Q9H183, Q9H184, Q9H185, Q9H186, Q9H187, Q9H188 | Gene names | IL4R, 582J2.1, IL4RA | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-4 receptor alpha chain precursor (IL-4R-alpha) (CD124 antigen) [Contains: Soluble interleukin-4 receptor alpha chain (sIL4Ralpha/prot) (IL-4-binding protein) (IL4-BP)]. | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.028688 (rank : 112) | θ value | 0.813845 (rank : 73) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
R3HD2_MOUSE
|
||||||
| NC score | 0.028347 (rank : 113) | θ value | 6.88961 (rank : 162) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q80TM6, Q80YB1, Q8BLS5, Q9CW50 | Gene names | R3hdm2, Kiaa1002 | |||
|
Domain Architecture |

|
|||||
| Description | R3H domain-containing protein 2. | |||||
|
SOX13_MOUSE
|
||||||
| NC score | 0.027129 (rank : 114) | θ value | 0.21417 (rank : 58) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q04891, Q922L3 | Gene names | Sox13, Sox-13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein. | |||||
|
MAML2_HUMAN
|
||||||
| NC score | 0.025485 (rank : 115) | θ value | 3.0926 (rank : 107) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8IZL2, Q6AI23, Q6Y3A3, Q8IUL3, Q96JK6 | Gene names | MAML2, KIAA1819 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 2 (Mam-2). | |||||
|
CCNK_HUMAN
|
||||||
| NC score | 0.025301 (rank : 116) | θ value | 4.03905 (rank : 121) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O75909, Q96B63, Q9NNY9 | Gene names | CCNK, CPR4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-K. | |||||
|
CSP9_MOUSE
|
||||||
| NC score | 0.025233 (rank : 117) | θ value | 2.36792 (rank : 97) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CZB6, Q3UGV1, Q91VN3, Q9CWV8, Q9D0V9 | Gene names | Crsp9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cofactor required for Sp1 transcriptional activation subunit 9. | |||||
|
CSP9_HUMAN
|
||||||
| NC score | 0.025203 (rank : 118) | θ value | 2.36792 (rank : 96) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43513 | Gene names | CRSP9, ARC34, MED7 | |||
|
Domain Architecture |

|
|||||
| Description | Cofactor required for Sp1 transcriptional activation subunit 9 (Transcriptional coactivator CRSP33) (RNA polymerase transcriptional regulation mediator subunit 7 homolog) (hMED7) (Activator-recruited cofactor 34 kDa component) (ARC34). | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.025144 (rank : 119) | θ value | 8.99809 (rank : 179) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
PO6F2_HUMAN
|
||||||
| NC score | 0.024675 (rank : 120) | θ value | 4.03905 (rank : 127) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P78424, P78425, Q75ME8, Q86UM6, Q9UDS7 | Gene names | POU6F2, RPF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 2 (Retina-derived POU-domain factor 1) (RPF-1). | |||||
|
TCF7_MOUSE
|
||||||
| NC score | 0.023584 (rank : 121) | θ value | 3.0926 (rank : 118) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q00417 | Gene names | Tcf7, Tcf-1, Tcf1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7 (T-cell-specific transcription factor 1) (TCF- 1) (T-cell factor 1). | |||||
|
COKA1_HUMAN
|
||||||
| NC score | 0.023220 (rank : 122) | θ value | 0.125558 (rank : 56) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9P218, Q4VXQ4, Q8WUT2, Q96CY9, Q9BQU6, Q9BQU7 | Gene names | COL20A1, KIAA1510 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XX) chain precursor. | |||||
|
COGA1_HUMAN
|
||||||
| NC score | 0.021193 (rank : 123) | θ value | 8.99809 (rank : 171) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q07092 | Gene names | COL16A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVI) chain precursor. | |||||
|
ZCH11_HUMAN
|
||||||
| NC score | 0.020774 (rank : 124) | θ value | 5.27518 (rank : 149) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5TAX3, Q12764, Q5TAX4, Q86XZ3 | Gene names | ZCCHC11, KIAA0191 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 11. | |||||
|
IASPP_MOUSE
|
||||||
| NC score | 0.020577 (rank : 125) | θ value | 3.0926 (rank : 106) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
PSL1_HUMAN
|
||||||
| NC score | 0.020402 (rank : 126) | θ value | 4.03905 (rank : 129) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
HD_HUMAN
|
||||||
| NC score | 0.020091 (rank : 127) | θ value | 6.88961 (rank : 155) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P42858, Q9UQB7 | Gene names | HD, IT15 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin (Huntington disease protein) (HD protein). | |||||
|
AP4M1_HUMAN
|
||||||
| NC score | 0.019197 (rank : 128) | θ value | 2.36792 (rank : 93) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00189, Q9UHK9 | Gene names | AP4M1, MUARP2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-4 complex subunit mu-1 (Adapter-related protein complex 4 mu-1 subunit) (Mu subunit of AP-4) (AP-4 adapter complex mu subunit) (Mu- adaptin-related protein 2) (mu-ARP2) (mu4). | |||||
|
FOXJ3_HUMAN
|
||||||
| NC score | 0.019154 (rank : 129) | θ value | 0.62314 (rank : 67) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UPW0, Q9NSS7 | Gene names | FOXJ3, KIAA1041 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein J3. | |||||
|
CO4A3_HUMAN
|
||||||
| NC score | 0.018866 (rank : 130) | θ value | 5.27518 (rank : 137) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q01955, Q9BQT2 | Gene names | COL4A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(IV) chain precursor (Goodpasture antigen). | |||||
|
CO8A1_MOUSE
|
||||||
| NC score | 0.018242 (rank : 131) | θ value | 6.88961 (rank : 153) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q00780, Q9D2V4 | Gene names | Col8a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VIII) chain precursor. | |||||
|
TCF20_MOUSE
|
||||||
| NC score | 0.017899 (rank : 132) | θ value | 8.99809 (rank : 181) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
COLQ_HUMAN
|
||||||
| NC score | 0.017487 (rank : 133) | θ value | 5.27518 (rank : 138) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y215, Q6DK18, Q6YH18, Q6YH19, Q6YH20, Q6YH21, Q9NP18, Q9NP19, Q9NP20, Q9NP21, Q9NP22, Q9NP23, Q9NP24, Q9UP88 | Gene names | COLQ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acetylcholinesterase collagenic tail peptide precursor (AChE Q subunit) (Acetylcholinesterase-associated collagen). | |||||
|
IASPP_HUMAN
|
||||||
| NC score | 0.017459 (rank : 134) | θ value | 4.03905 (rank : 123) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8WUF5, Q2PNZ9, Q5DU71, Q5I1X4, Q6P1R7, Q6PKF8, Q9Y290 | Gene names | PPP1R13L, IASPP, NKIP1, PPP1R13BL, RAI | |||
|
Domain Architecture |

|
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
KCNN3_MOUSE
|
||||||
| NC score | 0.016020 (rank : 135) | θ value | 1.06291 (rank : 82) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P58391 | Gene names | Kcnn3, Sk3 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 3 (SK3). | |||||
|
HELZ_HUMAN
|
||||||
| NC score | 0.016008 (rank : 136) | θ value | 5.27518 (rank : 141) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P42694 | Gene names | HELZ, KIAA0054 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable helicase with zinc-finger domain (EC 3.6.1.-). | |||||
|
PYGO1_HUMAN
|
||||||
| NC score | 0.015651 (rank : 137) | θ value | 5.27518 (rank : 144) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y3Y4 | Gene names | PYGO1 | |||
|
Domain Architecture |

|
|||||
| Description | Pygopus homolog 1. | |||||
|
RBP2_HUMAN
|
||||||
| NC score | 0.015202 (rank : 138) | θ value | 1.06291 (rank : 84) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P49792, Q13074, Q15280, Q53TE2, Q59FH7 | Gene names | RANBP2, NUP358 | |||
|
Domain Architecture |

|
|||||
| Description | Ran-binding protein 2 (RanBP2) (Nuclear pore complex protein Nup358) (Nucleoporin Nup358) (358 kDa nucleoporin) (P270). | |||||
|
CO1A1_MOUSE
|
||||||
| NC score | 0.015024 (rank : 139) | θ value | 5.27518 (rank : 136) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P11087, Q53WT0, Q60635, Q61367, Q61427, Q63919, Q6PCL3, Q810J9 | Gene names | Col1a1, Cola1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(I) chain precursor. | |||||
|
NUMBL_HUMAN
|
||||||
| NC score | 0.014956 (rank : 140) | θ value | 8.99809 (rank : 176) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Y6R0, Q7Z4J9 | Gene names | NUMBL | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein (Numb-R). | |||||
|
CD248_HUMAN
|
||||||
| NC score | 0.014912 (rank : 141) | θ value | 6.88961 (rank : 152) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9HCU0, Q3SX55, Q96KB6 | Gene names | CD248, CD164L1, TEM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
RFX1_MOUSE
|
||||||
| NC score | 0.014892 (rank : 142) | θ value | 6.88961 (rank : 164) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P48377 | Gene names | Rfx1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein RFX1. | |||||
|
HNRL1_HUMAN
|
||||||
| NC score | 0.014698 (rank : 143) | θ value | 6.88961 (rank : 157) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9BUJ2, O76022, Q6ZSZ0, Q7L8P4, Q8N6Z4, Q96G37, Q9HAL3, Q9UG75 | Gene names | HNRPUL1, E1BAP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein U-like protein 1 (Adenovirus early region 1B-associated protein 5) (E1B-55 kDa-associated protein 5) (E1B-AP5). | |||||
|
GNAS1_MOUSE
|
||||||
| NC score | 0.014485 (rank : 144) | θ value | 3.0926 (rank : 105) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q6R0H7, Q6R0H4, Q6R0H5, Q6R2J5, Q9JJX0, Q9Z1N8 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
DAZP1_HUMAN
|
||||||
| NC score | 0.014273 (rank : 145) | θ value | 4.03905 (rank : 122) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96EP5, Q96MJ3, Q9NRR9 | Gene names | DAZAP1 | |||
|
Domain Architecture |

|
|||||
| Description | DAZ-associated protein 1 (Deleted in azoospermia-associated protein 1). | |||||
|
ZO1_HUMAN
|
||||||
| NC score | 0.013952 (rank : 146) | θ value | 4.03905 (rank : 133) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q07157, Q4ZGJ6 | Gene names | TJP1, ZO1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
WDR33_HUMAN
|
||||||
| NC score | 0.013538 (rank : 147) | θ value | 0.813845 (rank : 78) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 638 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9C0J8 | Gene names | WDR33, WDC146 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 33 (WD repeat protein WDC146). | |||||
|
PB1_HUMAN
|
||||||
| NC score | 0.013118 (rank : 148) | θ value | 6.88961 (rank : 161) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q86U86, Q9H2T3, Q9H2T4, Q9H2T5, Q9H301, Q9H314 | Gene names | PB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein polybromo-1 (hPB1) (Polybromo-1D) (BRG1-associated factor 180) (BAF180). | |||||
|
TCF19_HUMAN
|
||||||
| NC score | 0.012790 (rank : 149) | θ value | 6.88961 (rank : 166) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y242, Q13176, Q15967, Q5SQ89, Q5STD6, Q5STF5, Q9BUM2, Q9UBH7 | Gene names | TCF19, SC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 19 (Transcription factor SC1). | |||||
|
HD_MOUSE
|
||||||
| NC score | 0.012641 (rank : 150) | θ value | 6.88961 (rank : 156) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P42859 | Gene names | Hd, Hdh | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin (Huntington disease protein homolog) (HD protein). | |||||
|
CD166_MOUSE
|
||||||
| NC score | 0.012170 (rank : 151) | θ value | 2.36792 (rank : 95) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61490, O70136, Q8CDA5, Q8R2T0 | Gene names | Alcam | |||
|
Domain Architecture |

|
|||||
| Description | CD166 antigen precursor (Activated leukocyte-cell adhesion molecule) (ALCAM) (DM-GRASP protein). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.012166 (rank : 152) | θ value | 5.27518 (rank : 147) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
STAT2_MOUSE
|
||||||
| NC score | 0.011854 (rank : 153) | θ value | 4.03905 (rank : 130) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9WVL2, Q64188, Q64189, Q64250 | Gene names | Stat2 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 2. | |||||
|
NALDL_MOUSE
|
||||||
| NC score | 0.011602 (rank : 154) | θ value | 3.0926 (rank : 110) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7M758 | Gene names | Naaladl1, Naaladasel | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylated-alpha-linked acidic dipeptidase-like protein (EC 3.4.17.21) (NAALADase L). | |||||
|
ATBF1_MOUSE
|
||||||
| NC score | 0.011480 (rank : 155) | θ value | 1.06291 (rank : 79) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
NALDL_HUMAN
|
||||||
| NC score | 0.011421 (rank : 156) | θ value | 3.0926 (rank : 109) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UQQ1, O43176 | Gene names | NAALADL1, NAALADASEL, NAALADL | |||
|
Domain Architecture |

|
|||||
| Description | N-acetylated-alpha-linked acidic dipeptidase-like protein (EC 3.4.17.21) (NAALADase L) (Ileal dipeptidylpeptidase) (100 kDa ileum brush border membrane protein) (I100). | |||||
|
TTC28_HUMAN
|
||||||
| NC score | 0.011407 (rank : 157) | θ value | 4.03905 (rank : 131) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96AY4, O95928, O95929, Q9NTE4, Q9UG31, Q9UGG5, Q9UPV8, Q9Y3S5 | Gene names | TTC28, KIAA1043 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
MKL1_HUMAN
|
||||||
| NC score | 0.011037 (rank : 158) | θ value | 6.88961 (rank : 160) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q969V6, Q8TCL1, Q96SC5, Q96SC6, Q9P2B0 | Gene names | MKL1, KIAA1438, MAL | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein) (Megakaryocytic acute leukemia protein). | |||||
|
GAK10_HUMAN
|
||||||
| NC score | 0.010499 (rank : 159) | θ value | 3.0926 (rank : 104) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P87889, P10263, P10264, Q69385, Q9UKH6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q33.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K10 Gag protein) (HERV-K107 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
PININ_MOUSE
|
||||||
| NC score | 0.010300 (rank : 160) | θ value | 0.813845 (rank : 74) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
CUTL2_HUMAN
|
||||||
| NC score | 0.010207 (rank : 161) | θ value | 1.38821 (rank : 85) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O14529 | Gene names | CUTL2, KIAA0293 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
CSKI1_MOUSE
|
||||||
| NC score | 0.009805 (rank : 162) | θ value | 8.99809 (rank : 172) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
HAP1_HUMAN
|
||||||
| NC score | 0.009232 (rank : 163) | θ value | 1.38821 (rank : 86) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 606 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P54257, O75358, Q9H4G3, Q9HA98, Q9NY90 | Gene names | HAP1, HLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-associated protein 1 (HAP-1) (Neuroan 1). | |||||
|
RBP2_MOUSE
|
||||||
| NC score | 0.009030 (rank : 164) | θ value | 5.27518 (rank : 145) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9ERU9, Q61992, Q8C9K9 | Gene names | Ranbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2 (RanBP2). | |||||
|
DCC_MOUSE
|
||||||
| NC score | 0.008941 (rank : 165) | θ value | 1.81305 (rank : 88) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P70211 | Gene names | Dcc | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC). | |||||
|
CC026_HUMAN
|
||||||
| NC score | 0.008927 (rank : 166) | θ value | 6.88961 (rank : 151) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BQ75 | Gene names | C3orf26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf26. | |||||
|
C1QC_HUMAN
|
||||||
| NC score | 0.008884 (rank : 167) | θ value | 4.03905 (rank : 120) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P02747, Q7Z502, Q96DL2, Q96H05 | Gene names | C1QC, C1QG | |||
|
Domain Architecture |

|
|||||
| Description | Complement C1q subcomponent subunit C precursor. | |||||
|
MOT8_HUMAN
|
||||||
| NC score | 0.008749 (rank : 168) | θ value | 5.27518 (rank : 142) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P36021, Q7Z797 | Gene names | SLC16A2, MCT8, XPCT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Monocarboxylate transporter 8 (MCT 8) (MCT 7) (Solute carrier family 16 member 2) (X-linked PEST-containing transporter). | |||||
|
FOXJ3_MOUSE
|
||||||
| NC score | 0.008690 (rank : 169) | θ value | 3.0926 (rank : 103) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BUR3, Q3UTD8 | Gene names | Foxj3, Kiaa1041 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein J3. | |||||
|
K0141_HUMAN
|
||||||
| NC score | 0.008460 (rank : 170) | θ value | 6.88961 (rank : 158) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14154, Q969R4, Q96EU9 | Gene names | KIAA0141 | |||
|
Domain Architecture |

|
|||||
| Description | Protein KIAA0141. | |||||
|
CD166_HUMAN
|
||||||
| NC score | 0.008343 (rank : 171) | θ value | 5.27518 (rank : 135) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13740, O60892 | Gene names | ALCAM, MEMD | |||
|
Domain Architecture |

|
|||||
| Description | CD166 antigen precursor (Activated leukocyte-cell adhesion molecule) (ALCAM). | |||||
|
INADL_HUMAN
|
||||||
| NC score | 0.008290 (rank : 172) | θ value | 2.36792 (rank : 99) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NI35, O15249, O43742, O60833, Q5VUA5, Q5VUA6, Q5VUA7, Q5VUA8, Q5VUA9, Q5VUB0, Q8WU78, Q9H3N9 | Gene names | INADL, PATJ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | InaD-like protein (Inadl protein) (hINADL) (Pals1-associated tight junction protein) (Protein associated to tight junctions). | |||||
|
MCR_MOUSE
|
||||||
| NC score | 0.008289 (rank : 173) | θ value | 3.0926 (rank : 108) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VII8, Q8VII9 | Gene names | Nr3c2, Mlr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mineralocorticoid receptor (MR). | |||||
|
LTBP4_MOUSE
|
||||||
| NC score | 0.008012 (rank : 174) | θ value | 0.62314 (rank : 68) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8K4G1, Q8K4G0 | Gene names | Ltbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 4 precursor (LTBP-4). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.007666 (rank : 175) | θ value | 5.27518 (rank : 134) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
C1QC_MOUSE
|
||||||
| NC score | 0.007320 (rank : 176) | θ value | 8.99809 (rank : 169) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q02105 | Gene names | C1qc, C1qg | |||
|
Domain Architecture |

|
|||||
| Description | Complement C1q subcomponent subunit C precursor. | |||||
|
ZN438_HUMAN
|
||||||
| NC score | 0.007310 (rank : 177) | θ value | 0.365318 (rank : 63) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q7Z4V0, Q5T426, Q658Q4, Q6ZN65 | Gene names | ZNF438 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 438. | |||||
|
FA61A_MOUSE
|
||||||
| NC score | 0.006599 (rank : 178) | θ value | 8.99809 (rank : 173) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K2F8, Q9CTG8 | Gene names | Fam61a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM61A. | |||||
|
MCR_HUMAN
|
||||||
| NC score | 0.006260 (rank : 179) | θ value | 6.88961 (rank : 159) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P08235, Q96KQ8, Q96KQ9 | Gene names | NR3C2, MCR, MLR | |||
|
Domain Architecture |

|
|||||
| Description | Mineralocorticoid receptor (MR). | |||||
|
SHC1_MOUSE
|
||||||
| NC score | 0.003896 (rank : 180) | θ value | 5.27518 (rank : 146) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P98083, Q3U2Q7, Q8BFY3, Q8K4C6, Q8K4C7 | Gene names | Shc1, Shc, ShcA | |||
|
Domain Architecture |

|
|||||
| Description | SHC-transforming protein 1 (SH2 domain protein C1) (Src homology 2 domain-containing-transforming protein C1). | |||||
|
ZN687_HUMAN
|
||||||
| NC score | 0.003428 (rank : 181) | θ value | 6.88961 (rank : 167) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8N1G0, Q68DQ8, Q9H937, Q9P2A7 | Gene names | ZNF687, KIAA1441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
PHLPP_MOUSE
|
||||||
| NC score | 0.002868 (rank : 182) | θ value | 4.03905 (rank : 126) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CHE4, Q6PCN0, Q8QZU8, Q8R5E5, Q99KL6 | Gene names | Phlpp, Kiaa0606, Plekhe1, Scop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein). | |||||
|
CAC1H_HUMAN
|
||||||
| NC score | 0.002322 (rank : 183) | θ value | 8.99809 (rank : 170) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
SPEG_HUMAN
|
||||||
| NC score | 0.001247 (rank : 184) | θ value | 8.99809 (rank : 180) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
TAXB1_HUMAN
|
||||||
| NC score | -0.000257 (rank : 185) | θ value | 3.0926 (rank : 117) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86VP1, O60398, O95770, Q13311, Q9BQG5, Q9UI88 | Gene names | TAX1BP1, T6BP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 1 (TRAF6-binding protein). | |||||
|
NOTC2_MOUSE
|
||||||
| NC score | -0.001029 (rank : 186) | θ value | 8.99809 (rank : 175) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 1069 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O35516, Q06008, Q60941 | Gene names | Notch2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (Motch B) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
GLIS1_HUMAN
|
||||||
| NC score | -0.003184 (rank : 187) | θ value | 5.27518 (rank : 140) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8NBF1, Q8N9V9 | Gene names | GLIS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLIS1 (GLI-similar 1). | |||||
|
ZN746_MOUSE
|
||||||
| NC score | -0.003243 (rank : 188) | θ value | 1.38821 (rank : 87) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q3U133 | Gene names | Znf746, Zfp746 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 746. | |||||
|
GLI1_MOUSE
|
||||||
| NC score | -0.004100 (rank : 189) | θ value | 8.99809 (rank : 174) | |||
| Query Neighborhood Hits | 181 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P47806, Q9QYK1 | Gene names | Gli1, Gli | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI1 (Glioma-associated oncogene homolog). | |||||