Please be patient as the page loads
|
ELF1_HUMAN
|
||||||
| SwissProt Accessions | P32519, Q9UDE1 | Gene names | ELF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ELF1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | P32519, Q9UDE1 | Gene names | ELF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
ELF1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.983833 (rank : 2) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q60775 | Gene names | Elf1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
ELF4_HUMAN
|
||||||
| θ value | 2.73302e-81 (rank : 3) | NC score | 0.960186 (rank : 3) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q99607, O60435 | Gene names | ELF4, ELFR, MEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-4 (E74-like factor 4) (Myeloid Elf-1-like factor). | |||||
|
ELF4_MOUSE
|
||||||
| θ value | 3.69378e-78 (rank : 4) | NC score | 0.934190 (rank : 6) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9Z2U4 | Gene names | Elf4, Mef | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-4 (E74-like factor 4) (Myeloid Elf-1-like factor). | |||||
|
ELF2_HUMAN
|
||||||
| θ value | 3.70236e-70 (rank : 5) | NC score | 0.952596 (rank : 4) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q15723, Q15724, Q15725, Q6P1K5 | Gene names | ELF2, NERF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-2 (E74-like factor 2) (New ETS- related factor). | |||||
|
ELF2_MOUSE
|
||||||
| θ value | 1.6097e-65 (rank : 6) | NC score | 0.950645 (rank : 5) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9JHC9, Q6NST2, Q8BTX8, Q9JHC7, Q9JHC8, Q9JHD0 | Gene names | Elf2, Nerf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-2 (E74-like factor 2) (New ETS- related factor). | |||||
|
ELK1_HUMAN
|
||||||
| θ value | 4.43474e-23 (rank : 7) | NC score | 0.861675 (rank : 7) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P19419, O75606, O95058, Q969X8, Q9UJM4 | Gene names | ELK1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-1. | |||||
|
ELK1_MOUSE
|
||||||
| θ value | 4.15078e-21 (rank : 8) | NC score | 0.856675 (rank : 10) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P41969 | Gene names | Elk1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-1. | |||||
|
SPDEF_MOUSE
|
||||||
| θ value | 2.97466e-19 (rank : 9) | NC score | 0.857733 (rank : 9) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9WTP3 | Gene names | Spdef, Pdef, Pse | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAM pointed domain-containing Ets transcription factor (Prostate- derived Ets factor) (Prostate epithelium-specific Ets transcription factor) (Prostate-specific Ets). | |||||
|
SPDEF_HUMAN
|
||||||
| θ value | 3.88503e-19 (rank : 10) | NC score | 0.859314 (rank : 8) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O95238 | Gene names | SPDEF, PDEF, PSE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAM pointed domain-containing Ets transcription factor (Prostate- derived Ets factor) (Prostate epithelium-specific Ets transcription factor) (Prostate-specific Ets). | |||||
|
ETV4_HUMAN
|
||||||
| θ value | 2.5182e-18 (rank : 11) | NC score | 0.787509 (rank : 33) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P43268, Q96AW9 | Gene names | ETV4, E1AF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 4 (Adenovirus E1A enhancer-binding protein) (E1A-F). | |||||
|
ELK4_MOUSE
|
||||||
| θ value | 3.28887e-18 (rank : 12) | NC score | 0.836177 (rank : 15) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P41158 | Gene names | Elk4, Sap1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
|
ETS2_HUMAN
|
||||||
| θ value | 5.60996e-18 (rank : 13) | NC score | 0.820367 (rank : 22) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P15036 | Gene names | ETS2 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-2 protein. | |||||
|
ETS2_MOUSE
|
||||||
| θ value | 5.60996e-18 (rank : 14) | NC score | 0.818695 (rank : 25) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P15037 | Gene names | Ets2 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-2 protein. | |||||
|
ETV4_MOUSE
|
||||||
| θ value | 5.60996e-18 (rank : 15) | NC score | 0.787370 (rank : 34) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P28322 | Gene names | Etv4, Pea-3, Pea3 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 4 (Polyomavirus enhancer activator 3) (Protein PEA3). | |||||
|
ELK4_HUMAN
|
||||||
| θ value | 7.32683e-18 (rank : 16) | NC score | 0.825566 (rank : 19) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P28324, P28323 | Gene names | ELK4, SAP1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
|
ETS1_HUMAN
|
||||||
| θ value | 1.63225e-17 (rank : 17) | NC score | 0.817683 (rank : 26) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P14921, Q14278, Q16080 | Gene names | ETS1 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-1 protein (p54). | |||||
|
ETS1_MOUSE
|
||||||
| θ value | 1.63225e-17 (rank : 18) | NC score | 0.817534 (rank : 27) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P27577, Q61403 | Gene names | Ets1, Ets-1 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-1 protein (p54). | |||||
|
ETV6_HUMAN
|
||||||
| θ value | 2.7842e-17 (rank : 19) | NC score | 0.818915 (rank : 24) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P41212, Q9UMF6 | Gene names | ETV6, TEL, TEL1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV6 (ETS-related protein Tel1) (Tel) (ETS translocation variant 6). | |||||
|
ETV6_MOUSE
|
||||||
| θ value | 2.7842e-17 (rank : 20) | NC score | 0.819931 (rank : 23) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P97360 | Gene names | Etv6, Tel, Tel1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV6 (ETS-related protein Tel1) (Tel) (ETS translocation variant 6). | |||||
|
ETV2_MOUSE
|
||||||
| θ value | 4.74913e-17 (rank : 21) | NC score | 0.844315 (rank : 12) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P41163 | Gene names | Etv2, Er71, Etsrp71 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 2 (Ets-related protein 71). | |||||
|
ELK3_HUMAN
|
||||||
| θ value | 6.20254e-17 (rank : 22) | NC score | 0.837569 (rank : 14) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P41970, Q6FG57, Q6GU29, Q9UD17 | Gene names | ELK3, NET, SAP2 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-3 (ETS-related protein NET) (ETS- related protein ERP) (SRF accessory protein 2) (SAP-2). | |||||
|
ETV1_HUMAN
|
||||||
| θ value | 6.20254e-17 (rank : 23) | NC score | 0.785513 (rank : 36) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P50549, O75849, Q9UQ71, Q9Y636 | Gene names | ETV1, ER81 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 1 (ER81 protein). | |||||
|
ETV1_MOUSE
|
||||||
| θ value | 6.20254e-17 (rank : 24) | NC score | 0.786362 (rank : 35) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P41164 | Gene names | Etv1, Er81, Etsrp81 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 1 (ER81 protein). | |||||
|
ETV5_MOUSE
|
||||||
| θ value | 6.20254e-17 (rank : 25) | NC score | 0.762776 (rank : 41) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9CXC9, Q3TG49, Q8C0F3, Q9JHB1 | Gene names | Etv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 5. | |||||
|
ETV7_HUMAN
|
||||||
| θ value | 8.10077e-17 (rank : 26) | NC score | 0.855163 (rank : 11) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9Y603, Q9NZ65, Q9NZ66, Q9NZ68, Q9NZR8, Q9UNJ7, Q9Y5K4, Q9Y604 | Gene names | ETV7, TEL2, TELB, TREF | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV7 (Transcription factor Tel-2) (ETS-related protein Tel2) (Tel-related Ets factor). | |||||
|
ETV5_HUMAN
|
||||||
| θ value | 1.058e-16 (rank : 27) | NC score | 0.779159 (rank : 39) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P41161 | Gene names | ETV5, ERM | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 5 (Ets-related protein ERM). | |||||
|
GABPA_HUMAN
|
||||||
| θ value | 1.058e-16 (rank : 28) | NC score | 0.823769 (rank : 21) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q06546, Q12939 | Gene names | GABPA, E4TF1A | |||
|
Domain Architecture |

|
|||||
| Description | GA-binding protein alpha chain (GABP-subunit alpha) (Transcription factor E4TF1-60) (Nuclear respiratory factor 2 subunit alpha). | |||||
|
GABPA_MOUSE
|
||||||
| θ value | 1.058e-16 (rank : 29) | NC score | 0.825141 (rank : 20) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q00422 | Gene names | Gabpa, E4tf1a | |||
|
Domain Architecture |

|
|||||
| Description | GA-binding protein alpha chain (GABP-subunit alpha). | |||||
|
FLI1_MOUSE
|
||||||
| θ value | 2.35696e-16 (rank : 30) | NC score | 0.801260 (rank : 28) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P26323 | Gene names | Fli1, Fli-1 | |||
|
Domain Architecture |

|
|||||
| Description | Friend leukemia integration 1 transcription factor (Retroviral integration site protein Fli-1). | |||||
|
FLI1_HUMAN
|
||||||
| θ value | 3.07829e-16 (rank : 31) | NC score | 0.799817 (rank : 29) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q01543, Q14319, Q92480, Q9UE07 | Gene names | FLI1 | |||
|
Domain Architecture |

|
|||||
| Description | Friend leukemia integration 1 transcription factor (Fli-1 proto- oncogene) (ERGB transcription factor). | |||||
|
ELK3_MOUSE
|
||||||
| θ value | 4.02038e-16 (rank : 32) | NC score | 0.833822 (rank : 16) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P41971, P97747, Q62346 | Gene names | Elk3, Erp, Net | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-3 (ETS-related protein NET) (ETS- related protein ERP). | |||||
|
ETV3_HUMAN
|
||||||
| θ value | 4.02038e-16 (rank : 33) | NC score | 0.774297 (rank : 40) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P41162, Q8TAC8, Q9BX30 | Gene names | ETV3, METS, PE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
ETV3_MOUSE
|
||||||
| θ value | 6.85773e-16 (rank : 34) | NC score | 0.796039 (rank : 30) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8R4Z4, Q9QZW1 | Gene names | Etv3, Mets, Pe1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
ETV2_HUMAN
|
||||||
| θ value | 1.16975e-15 (rank : 35) | NC score | 0.839779 (rank : 13) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O00321, Q9UEA0 | Gene names | ETV2, ER71, ETSRP71 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 2 (Ets-related protein 71). | |||||
|
ERG_HUMAN
|
||||||
| θ value | 1.99529e-15 (rank : 36) | NC score | 0.792988 (rank : 31) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P11308, Q16113 | Gene names | ERG | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ERG (Transforming protein ERG). | |||||
|
ERG_MOUSE
|
||||||
| θ value | 1.99529e-15 (rank : 37) | NC score | 0.791372 (rank : 32) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P81270, Q8C5L4, Q920K7, Q920K8, Q920K9 | Gene names | Erg, Erg-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ERG. | |||||
|
ELF5_HUMAN
|
||||||
| θ value | 9.90251e-15 (rank : 38) | NC score | 0.829007 (rank : 18) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UKW6, O95175, Q8N2K9, Q96QY3, Q9UKW5 | Gene names | ELF5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-5 (E74-like factor 5) (Epithelium-specific Ets transcription factor 2) (ESE-2) (Epithelium- restricted ESE-1-related Ets factor). | |||||
|
ELF5_MOUSE
|
||||||
| θ value | 9.90251e-15 (rank : 39) | NC score | 0.830105 (rank : 17) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8VDK3, Q921H5, Q9Z2K6 | Gene names | Elf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-5 (E74-like factor 5). | |||||
|
ERF_HUMAN
|
||||||
| θ value | 1.86753e-13 (rank : 40) | NC score | 0.784232 (rank : 37) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P50548, Q9UPI7 | Gene names | ERF | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing transcription factor ERF (Ets2 repressor factor). | |||||
|
ERF_MOUSE
|
||||||
| θ value | 1.86753e-13 (rank : 41) | NC score | 0.783837 (rank : 38) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P70459 | Gene names | Erf | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing transcription factor ERF. | |||||
|
SPIC_HUMAN
|
||||||
| θ value | 3.08544e-08 (rank : 42) | NC score | 0.649312 (rank : 42) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8N5J4 | Gene names | SPIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-C. | |||||
|
SPIC_MOUSE
|
||||||
| θ value | 4.45536e-07 (rank : 43) | NC score | 0.617846 (rank : 46) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6P3D7, Q9Z0Y0 | Gene names | Spic | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-C (Pu.1-related factor) (Prf). | |||||
|
SPI1_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 44) | NC score | 0.637590 (rank : 44) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P17947 | Gene names | SPI1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor PU.1 (31 kDa transforming protein). | |||||
|
SPI1_MOUSE
|
||||||
| θ value | 1.09739e-05 (rank : 45) | NC score | 0.639745 (rank : 43) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P17433, Q99L57 | Gene names | Spi1, Sfpi-1, Sfpi1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor PU.1 (31 kDa transforming protein) (SFFV proviral integration 1 protein). | |||||
|
SPIB_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 46) | NC score | 0.627058 (rank : 45) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q01892, Q15359 | Gene names | SPIB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Spi-B. | |||||
|
SPIB_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 47) | NC score | 0.614669 (rank : 47) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O35906, O35907, O35909, O55199 | Gene names | Spib | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-B. | |||||
|
ZN750_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 48) | NC score | 0.050040 (rank : 52) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BH05, Q66JP3, Q8C0L1 | Gene names | Znf750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 49) | NC score | 0.056441 (rank : 48) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
CSPG2_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 50) | NC score | 0.015510 (rank : 73) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P13611, P20754, Q13010, Q13189, Q15123, Q9UCL9, Q9UNW5 | Gene names | CSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M) (Glial hyaluronate-binding protein) (GHAP). | |||||
|
SRBP1_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 51) | NC score | 0.046190 (rank : 53) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
TAF4_HUMAN
|
||||||
| θ value | 0.163984 (rank : 52) | NC score | 0.035491 (rank : 58) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
APC_HUMAN
|
||||||
| θ value | 0.21417 (rank : 53) | NC score | 0.043082 (rank : 55) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P25054, Q15162, Q15163, Q93042 | Gene names | APC, DP2.5 | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC). | |||||
|
APC_MOUSE
|
||||||
| θ value | 0.365318 (rank : 54) | NC score | 0.044667 (rank : 54) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q61315, Q62044 | Gene names | Apc | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC) (mAPC). | |||||
|
ARID2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 55) | NC score | 0.052564 (rank : 50) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q68CP9, Q5EB51, Q645I3, Q6ZRY5, Q7Z3I5, Q86T28, Q96SJ6, Q9HCL5 | Gene names | ARID2, KIAA1557 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 2 (ARID domain- containing protein 2) (BRG1-associated factor 200) (BAF200). | |||||
|
EMSY_MOUSE
|
||||||
| θ value | 0.62314 (rank : 56) | NC score | 0.051286 (rank : 51) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BMB0, Q5FWK5, Q80XU1, Q8VDW9 | Gene names | Emsy | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
EMSY_HUMAN
|
||||||
| θ value | 0.813845 (rank : 57) | NC score | 0.056229 (rank : 49) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q7Z589, Q4G109, Q8NBU6, Q8TE50, Q9H8I9, Q9NRH0 | Gene names | EMSY, C11orf30 | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
MCA1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 58) | NC score | 0.030894 (rank : 60) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P31230, Q60659 | Gene names | Scye1, Emap2 | |||
|
Domain Architecture |

|
|||||
| Description | Multisynthetase complex auxiliary component p43 [Contains: Endothelial monocyte-activating polypeptide 2 (EMAP-II) (Small inducible cytokine subfamily E member 1)]. | |||||
|
PHC3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 59) | NC score | 0.027712 (rank : 65) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8NDX5, Q5HYF0, Q6NSG2, Q8NFT7, Q8NFZ1, Q8TBM2, Q9H971, Q9H9I4 | Gene names | PHC3, EDR3, PH3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 3 (hPH3) (Homolog of polyhomeotic 3) (Early development regulatory protein 3). | |||||
|
SP4_MOUSE
|
||||||
| θ value | 1.06291 (rank : 60) | NC score | 0.000928 (rank : 99) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q62445 | Gene names | Sp4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp4. | |||||
|
STMN3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 61) | NC score | 0.017233 (rank : 69) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NZ72, O75527, Q969Y4 | Gene names | STMN3, SCLIP | |||
|
Domain Architecture |
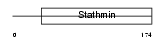
|
|||||
| Description | Stathmin-3 (SCG10-like protein). | |||||
|
K1210_HUMAN
|
||||||
| θ value | 1.38821 (rank : 62) | NC score | 0.029001 (rank : 63) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9ULL0, Q5JPN4 | Gene names | KIAA1210 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1210. | |||||
|
MYLK_MOUSE
|
||||||
| θ value | 1.38821 (rank : 63) | NC score | -0.002282 (rank : 102) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1489 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6PDN3, Q3TSJ7, Q80UX0, Q80YN7, Q80YN8, Q8K026, Q924D2, Q9ERD3 | Gene names | Mylk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin light chain kinase, smooth muscle (EC 2.7.11.18) (MLCK) (Telokin) (Kinase-related protein) (KRP). | |||||
|
PRG4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 64) | NC score | 0.041566 (rank : 56) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
LRC50_HUMAN
|
||||||
| θ value | 1.81305 (rank : 65) | NC score | 0.013801 (rank : 77) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NEP3, Q69YI8, Q69YJ0, Q96LP3 | Gene names | LRRC50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 50. | |||||
|
PODXL_HUMAN
|
||||||
| θ value | 1.81305 (rank : 66) | NC score | 0.035072 (rank : 59) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O00592 | Gene names | PODXL, PCLP, PCLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Podocalyxin-like protein 1 precursor. | |||||
|
SOX30_HUMAN
|
||||||
| θ value | 1.81305 (rank : 67) | NC score | 0.016197 (rank : 72) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O94993, O94995, Q8IYX6 | Gene names | SOX30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
HCN1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 68) | NC score | 0.006437 (rank : 87) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O88704, O54899, Q9D613 | Gene names | Hcn1, Bcng1, Hac2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 (Brain cyclic nucleotide-gated channel 1) (BCNG-1) (Hyperpolarization-activated cation channel 2) (HAC-2). | |||||
|
NOL8_HUMAN
|
||||||
| θ value | 2.36792 (rank : 69) | NC score | 0.014179 (rank : 76) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q76FK4, Q5TCC7, Q5TCC8, Q5TCD3, Q5TCD5, Q5TCD6, Q5TCD7, Q76D35, Q7L3E2, Q9H586, Q9H795, Q9H7W7, Q9H9J6, Q9NWA4, Q9NWM4 | Gene names | NOL8, NOP132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 8 (Nucleolar protein Nop132). | |||||
|
NU214_HUMAN
|
||||||
| θ value | 2.36792 (rank : 70) | NC score | 0.035847 (rank : 57) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
STMN1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 71) | NC score | 0.013481 (rank : 78) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P16949 | Gene names | STMN1, LAP18, OP18 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Protein Pr22). | |||||
|
TCRG1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 72) | NC score | 0.016995 (rank : 70) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 73) | NC score | 0.027919 (rank : 64) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
STMN1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 74) | NC score | 0.013078 (rank : 80) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P54227 | Gene names | Stmn1, Lag, Lap18, Pr22 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Pr22 protein) (Leukemia-associated gene protein). | |||||
|
WAPL_HUMAN
|
||||||
| θ value | 3.0926 (rank : 75) | NC score | 0.014891 (rank : 75) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7Z5K2, Q5VSK5, Q8IX10, Q92549 | Gene names | WAPAL, FOE, KIAA0261, WAPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wings apart-like protein homolog (Friend of EBNA2 protein). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | 0.013161 (rank : 79) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
NLGN3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.006664 (rank : 86) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BYM5, Q8BXR4 | Gene names | Nlgn3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-3 precursor (Gliotactin homolog). | |||||
|
ANK3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.004691 (rank : 93) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
BOC_HUMAN
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.006038 (rank : 88) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BWV1, Q6UXJ5, Q8N2P7, Q8NF26 | Gene names | BOC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brother of CDO precursor (Protein BOC). | |||||
|
CADH3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | -0.000055 (rank : 100) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P22223 | Gene names | CDH3, CDHP | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-3 precursor (Placental-cadherin) (P-cadherin). | |||||
|
CBP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.016198 (rank : 71) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CHK2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | -0.003928 (rank : 104) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O96017, Q6QA03, Q6QA04, Q6QA05, Q6QA06, Q6QA07, Q6QA08, Q6QA10, Q6QA11, Q6QA12, Q6QA13, Q9HCQ8, Q9UGF0, Q9UGF1 | Gene names | CHEK2, CHK2 | |||
|
Domain Architecture |
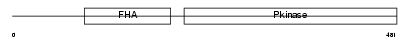
|
|||||
| Description | Serine/threonine-protein kinase Chk2 (EC 2.7.11.1) (Cds1). | |||||
|
CK024_MOUSE
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.023692 (rank : 68) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9D8N1, Q8VCP2 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 homolog precursor. | |||||
|
PADI6_MOUSE
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | 0.005698 (rank : 89) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K3V4, Q75WC6 | Gene names | Padi6, Padi5 | |||
|
Domain Architecture |

|
|||||
| Description | Protein-arginine deiminase type-6 (EC 3.5.3.15) (Protein-arginine deiminase type VI) (Peptidylarginine deiminase VI) (Arginine deiminase-like protein) (Egg and embryo abundant PAD) (ePAD). | |||||
|
PCOC1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.003965 (rank : 95) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15113, O14550 | Gene names | PCOLCE, PCPE1 | |||
|
Domain Architecture |

|
|||||
| Description | Procollagen C-endopeptidase enhancer 1 precursor (Procollagen COOH- terminal proteinase enhancer 1) (Procollagen C-proteinase enhancer 1) (PCPE-1) (Type I procollagen COOH-terminal proteinase enhancer) (Type 1 procollagen C-proteinase enhancer protein). | |||||
|
PO121_HUMAN
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.025425 (rank : 66) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
PO2F1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.003977 (rank : 94) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P14859, Q5TBT7, Q6PK46, Q8NEU9, Q9BPV1 | Gene names | POU2F1, OCT1, OTF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 2, transcription factor 1 (Octamer-binding transcription factor 1) (Oct-1) (OTF-1) (NF-A1). | |||||
|
KLF4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | -0.003689 (rank : 103) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43474, P78338, Q9UNP3 | Gene names | KLF4, EZF, GKLF | |||
|
Domain Architecture |

|
|||||
| Description | Krueppel-like factor 4 (Epithelial zinc-finger protein EZF) (Gut- enriched krueppel-like factor). | |||||
|
MAST1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | -0.002208 (rank : 101) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1146 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y2H9, O00114, Q8N6X0 | Gene names | MAST1, KIAA0973 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
NFYC_MOUSE
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.010585 (rank : 83) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P70353, O35088, Q6GU21 | Gene names | Nfyc | |||
|
Domain Architecture |
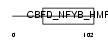
|
|||||
| Description | Nuclear transcription factor Y subunit gamma (Nuclear transcription factor Y subunit C) (NF-YC) (CAAT-box DNA-binding protein subunit C). | |||||
|
PHF20_MOUSE
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.005217 (rank : 91) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
RFX1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.024042 (rank : 67) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P22670 | Gene names | RFX1 | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II regulatory factor RFX1 (RFX) (Enhancer factor C) (EF-C). | |||||
|
SON_HUMAN
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.029514 (rank : 62) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
AAK1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.005046 (rank : 92) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q3UHJ0, Q3TY53, Q6ZPZ6, Q80XP6 | Gene names | Aak1, Kiaa1048 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP2-associated protein kinase 1 (EC 2.7.11.1) (Adaptor-associated kinase 1). | |||||
|
AT10B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.002902 (rank : 96) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O94823, Q9H725 | Gene names | ATP10B, ATPVB, KIAA0715 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase VB (EC 3.6.3.1). | |||||
|
AT8B3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.008102 (rank : 85) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60423, Q8IVB8, Q8N4Y8, Q96M22 | Gene names | ATP8B3, ATP1K, FOS37502_2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IK (EC 3.6.3.1) (ATPase class I type 8B member 3). | |||||
|
CAC1E_MOUSE
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.001506 (rank : 98) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61290 | Gene names | Cacna1e, Cach6, Cacnl1a6, Cchra1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
MUC5B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.030657 (rank : 61) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.015473 (rank : 74) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
RAI2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.010561 (rank : 84) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QVY8 | Gene names | Rai2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 2 (3f8). | |||||
|
RIOK1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.013059 (rank : 81) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BRS2, Q8NDC8, Q96NV9 | Gene names | RIOK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase RIO1 (EC 2.7.11.1) (RIO kinase 1). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.011407 (rank : 82) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
WNK1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.001989 (rank : 97) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1179 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P83741 | Gene names | Wnk1, Prkwnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1). | |||||
|
ZMYM3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.005685 (rank : 90) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14202, O15089 | Gene names | ZMYM3, DXS6673E, KIAA0385, ZNF261 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 3 (Zinc finger protein 261). | |||||
|
ELF1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | P32519, Q9UDE1 | Gene names | ELF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
ELF1_MOUSE
|
||||||
| NC score | 0.983833 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q60775 | Gene names | Elf1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
ELF4_HUMAN
|
||||||
| NC score | 0.960186 (rank : 3) | θ value | 2.73302e-81 (rank : 3) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q99607, O60435 | Gene names | ELF4, ELFR, MEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-4 (E74-like factor 4) (Myeloid Elf-1-like factor). | |||||
|
ELF2_HUMAN
|
||||||
| NC score | 0.952596 (rank : 4) | θ value | 3.70236e-70 (rank : 5) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q15723, Q15724, Q15725, Q6P1K5 | Gene names | ELF2, NERF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-2 (E74-like factor 2) (New ETS- related factor). | |||||
|
ELF2_MOUSE
|
||||||
| NC score | 0.950645 (rank : 5) | θ value | 1.6097e-65 (rank : 6) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9JHC9, Q6NST2, Q8BTX8, Q9JHC7, Q9JHC8, Q9JHD0 | Gene names | Elf2, Nerf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-2 (E74-like factor 2) (New ETS- related factor). | |||||
|
ELF4_MOUSE
|
||||||
| NC score | 0.934190 (rank : 6) | θ value | 3.69378e-78 (rank : 4) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9Z2U4 | Gene names | Elf4, Mef | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-4 (E74-like factor 4) (Myeloid Elf-1-like factor). | |||||
|
ELK1_HUMAN
|
||||||
| NC score | 0.861675 (rank : 7) | θ value | 4.43474e-23 (rank : 7) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P19419, O75606, O95058, Q969X8, Q9UJM4 | Gene names | ELK1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-1. | |||||
|
SPDEF_HUMAN
|
||||||
| NC score | 0.859314 (rank : 8) | θ value | 3.88503e-19 (rank : 10) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O95238 | Gene names | SPDEF, PDEF, PSE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAM pointed domain-containing Ets transcription factor (Prostate- derived Ets factor) (Prostate epithelium-specific Ets transcription factor) (Prostate-specific Ets). | |||||
|
SPDEF_MOUSE
|
||||||
| NC score | 0.857733 (rank : 9) | θ value | 2.97466e-19 (rank : 9) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9WTP3 | Gene names | Spdef, Pdef, Pse | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAM pointed domain-containing Ets transcription factor (Prostate- derived Ets factor) (Prostate epithelium-specific Ets transcription factor) (Prostate-specific Ets). | |||||
|
ELK1_MOUSE
|
||||||
| NC score | 0.856675 (rank : 10) | θ value | 4.15078e-21 (rank : 8) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P41969 | Gene names | Elk1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-1. | |||||
|
ETV7_HUMAN
|
||||||
| NC score | 0.855163 (rank : 11) | θ value | 8.10077e-17 (rank : 26) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9Y603, Q9NZ65, Q9NZ66, Q9NZ68, Q9NZR8, Q9UNJ7, Q9Y5K4, Q9Y604 | Gene names | ETV7, TEL2, TELB, TREF | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV7 (Transcription factor Tel-2) (ETS-related protein Tel2) (Tel-related Ets factor). | |||||
|
ETV2_MOUSE
|
||||||
| NC score | 0.844315 (rank : 12) | θ value | 4.74913e-17 (rank : 21) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P41163 | Gene names | Etv2, Er71, Etsrp71 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 2 (Ets-related protein 71). | |||||
|
ETV2_HUMAN
|
||||||
| NC score | 0.839779 (rank : 13) | θ value | 1.16975e-15 (rank : 35) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O00321, Q9UEA0 | Gene names | ETV2, ER71, ETSRP71 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 2 (Ets-related protein 71). | |||||
|
ELK3_HUMAN
|
||||||
| NC score | 0.837569 (rank : 14) | θ value | 6.20254e-17 (rank : 22) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P41970, Q6FG57, Q6GU29, Q9UD17 | Gene names | ELK3, NET, SAP2 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-3 (ETS-related protein NET) (ETS- related protein ERP) (SRF accessory protein 2) (SAP-2). | |||||
|
ELK4_MOUSE
|
||||||
| NC score | 0.836177 (rank : 15) | θ value | 3.28887e-18 (rank : 12) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P41158 | Gene names | Elk4, Sap1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
|
ELK3_MOUSE
|
||||||
| NC score | 0.833822 (rank : 16) | θ value | 4.02038e-16 (rank : 32) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P41971, P97747, Q62346 | Gene names | Elk3, Erp, Net | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-3 (ETS-related protein NET) (ETS- related protein ERP). | |||||
|
ELF5_MOUSE
|
||||||
| NC score | 0.830105 (rank : 17) | θ value | 9.90251e-15 (rank : 39) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8VDK3, Q921H5, Q9Z2K6 | Gene names | Elf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-5 (E74-like factor 5). | |||||
|
ELF5_HUMAN
|
||||||
| NC score | 0.829007 (rank : 18) | θ value | 9.90251e-15 (rank : 38) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UKW6, O95175, Q8N2K9, Q96QY3, Q9UKW5 | Gene names | ELF5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-5 (E74-like factor 5) (Epithelium-specific Ets transcription factor 2) (ESE-2) (Epithelium- restricted ESE-1-related Ets factor). | |||||
|
ELK4_HUMAN
|
||||||
| NC score | 0.825566 (rank : 19) | θ value | 7.32683e-18 (rank : 16) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P28324, P28323 | Gene names | ELK4, SAP1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
|
GABPA_MOUSE
|
||||||
| NC score | 0.825141 (rank : 20) | θ value | 1.058e-16 (rank : 29) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q00422 | Gene names | Gabpa, E4tf1a | |||
|
Domain Architecture |

|
|||||
| Description | GA-binding protein alpha chain (GABP-subunit alpha). | |||||
|
GABPA_HUMAN
|
||||||
| NC score | 0.823769 (rank : 21) | θ value | 1.058e-16 (rank : 28) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q06546, Q12939 | Gene names | GABPA, E4TF1A | |||
|
Domain Architecture |

|
|||||
| Description | GA-binding protein alpha chain (GABP-subunit alpha) (Transcription factor E4TF1-60) (Nuclear respiratory factor 2 subunit alpha). | |||||
|
ETS2_HUMAN
|
||||||
| NC score | 0.820367 (rank : 22) | θ value | 5.60996e-18 (rank : 13) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P15036 | Gene names | ETS2 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-2 protein. | |||||
|
ETV6_MOUSE
|
||||||
| NC score | 0.819931 (rank : 23) | θ value | 2.7842e-17 (rank : 20) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P97360 | Gene names | Etv6, Tel, Tel1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV6 (ETS-related protein Tel1) (Tel) (ETS translocation variant 6). | |||||
|
ETV6_HUMAN
|
||||||
| NC score | 0.818915 (rank : 24) | θ value | 2.7842e-17 (rank : 19) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P41212, Q9UMF6 | Gene names | ETV6, TEL, TEL1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV6 (ETS-related protein Tel1) (Tel) (ETS translocation variant 6). | |||||
|
ETS2_MOUSE
|
||||||
| NC score | 0.818695 (rank : 25) | θ value | 5.60996e-18 (rank : 14) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P15037 | Gene names | Ets2 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-2 protein. | |||||
|
ETS1_HUMAN
|
||||||
| NC score | 0.817683 (rank : 26) | θ value | 1.63225e-17 (rank : 17) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P14921, Q14278, Q16080 | Gene names | ETS1 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-1 protein (p54). | |||||
|
ETS1_MOUSE
|
||||||
| NC score | 0.817534 (rank : 27) | θ value | 1.63225e-17 (rank : 18) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P27577, Q61403 | Gene names | Ets1, Ets-1 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-1 protein (p54). | |||||
|
FLI1_MOUSE
|
||||||
| NC score | 0.801260 (rank : 28) | θ value | 2.35696e-16 (rank : 30) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P26323 | Gene names | Fli1, Fli-1 | |||
|
Domain Architecture |

|
|||||
| Description | Friend leukemia integration 1 transcription factor (Retroviral integration site protein Fli-1). | |||||
|
FLI1_HUMAN
|
||||||
| NC score | 0.799817 (rank : 29) | θ value | 3.07829e-16 (rank : 31) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q01543, Q14319, Q92480, Q9UE07 | Gene names | FLI1 | |||
|
Domain Architecture |

|
|||||
| Description | Friend leukemia integration 1 transcription factor (Fli-1 proto- oncogene) (ERGB transcription factor). | |||||
|
ETV3_MOUSE
|
||||||
| NC score | 0.796039 (rank : 30) | θ value | 6.85773e-16 (rank : 34) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8R4Z4, Q9QZW1 | Gene names | Etv3, Mets, Pe1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
ERG_HUMAN
|
||||||
| NC score | 0.792988 (rank : 31) | θ value | 1.99529e-15 (rank : 36) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P11308, Q16113 | Gene names | ERG | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ERG (Transforming protein ERG). | |||||
|
ERG_MOUSE
|
||||||
| NC score | 0.791372 (rank : 32) | θ value | 1.99529e-15 (rank : 37) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P81270, Q8C5L4, Q920K7, Q920K8, Q920K9 | Gene names | Erg, Erg-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ERG. | |||||
|
ETV4_HUMAN
|
||||||
| NC score | 0.787509 (rank : 33) | θ value | 2.5182e-18 (rank : 11) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P43268, Q96AW9 | Gene names | ETV4, E1AF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 4 (Adenovirus E1A enhancer-binding protein) (E1A-F). | |||||
|
ETV4_MOUSE
|
||||||
| NC score | 0.787370 (rank : 34) | θ value | 5.60996e-18 (rank : 15) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P28322 | Gene names | Etv4, Pea-3, Pea3 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 4 (Polyomavirus enhancer activator 3) (Protein PEA3). | |||||
|
ETV1_MOUSE
|
||||||
| NC score | 0.786362 (rank : 35) | θ value | 6.20254e-17 (rank : 24) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P41164 | Gene names | Etv1, Er81, Etsrp81 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 1 (ER81 protein). | |||||
|
ETV1_HUMAN
|
||||||
| NC score | 0.785513 (rank : 36) | θ value | 6.20254e-17 (rank : 23) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P50549, O75849, Q9UQ71, Q9Y636 | Gene names | ETV1, ER81 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 1 (ER81 protein). | |||||
|
ERF_HUMAN
|
||||||
| NC score | 0.784232 (rank : 37) | θ value | 1.86753e-13 (rank : 40) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P50548, Q9UPI7 | Gene names | ERF | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing transcription factor ERF (Ets2 repressor factor). | |||||
|
ERF_MOUSE
|
||||||
| NC score | 0.783837 (rank : 38) | θ value | 1.86753e-13 (rank : 41) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P70459 | Gene names | Erf | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing transcription factor ERF. | |||||
|
ETV5_HUMAN
|
||||||
| NC score | 0.779159 (rank : 39) | θ value | 1.058e-16 (rank : 27) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P41161 | Gene names | ETV5, ERM | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 5 (Ets-related protein ERM). | |||||
|
ETV3_HUMAN
|
||||||
| NC score | 0.774297 (rank : 40) | θ value | 4.02038e-16 (rank : 33) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P41162, Q8TAC8, Q9BX30 | Gene names | ETV3, METS, PE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
ETV5_MOUSE
|
||||||
| NC score | 0.762776 (rank : 41) | θ value | 6.20254e-17 (rank : 25) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9CXC9, Q3TG49, Q8C0F3, Q9JHB1 | Gene names | Etv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 5. | |||||
|
SPIC_HUMAN
|
||||||
| NC score | 0.649312 (rank : 42) | θ value | 3.08544e-08 (rank : 42) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8N5J4 | Gene names | SPIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-C. | |||||
|
SPI1_MOUSE
|
||||||
| NC score | 0.639745 (rank : 43) | θ value | 1.09739e-05 (rank : 45) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P17433, Q99L57 | Gene names | Spi1, Sfpi-1, Sfpi1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor PU.1 (31 kDa transforming protein) (SFFV proviral integration 1 protein). | |||||
|
SPI1_HUMAN
|
||||||
| NC score | 0.637590 (rank : 44) | θ value | 1.09739e-05 (rank : 44) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P17947 | Gene names | SPI1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor PU.1 (31 kDa transforming protein). | |||||
|
SPIB_HUMAN
|
||||||
| NC score | 0.627058 (rank : 45) | θ value | 1.87187e-05 (rank : 46) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q01892, Q15359 | Gene names | SPIB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Spi-B. | |||||
|
SPIC_MOUSE
|
||||||
| NC score | 0.617846 (rank : 46) | θ value | 4.45536e-07 (rank : 43) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6P3D7, Q9Z0Y0 | Gene names | Spic | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-C (Pu.1-related factor) (Prf). | |||||
|
SPIB_MOUSE
|
||||||
| NC score | 0.614669 (rank : 47) | θ value | 9.29e-05 (rank : 47) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O35906, O35907, O35909, O55199 | Gene names | Spib | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-B. | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.056441 (rank : 48) | θ value | 0.00665767 (rank : 49) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
EMSY_HUMAN
|
||||||
| NC score | 0.056229 (rank : 49) | θ value | 0.813845 (rank : 57) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q7Z589, Q4G109, Q8NBU6, Q8TE50, Q9H8I9, Q9NRH0 | Gene names | EMSY, C11orf30 | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
ARID2_HUMAN
|
||||||
| NC score | 0.052564 (rank : 50) | θ value | 0.62314 (rank : 55) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q68CP9, Q5EB51, Q645I3, Q6ZRY5, Q7Z3I5, Q86T28, Q96SJ6, Q9HCL5 | Gene names | ARID2, KIAA1557 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 2 (ARID domain- containing protein 2) (BRG1-associated factor 200) (BAF200). | |||||
|
EMSY_MOUSE
|
||||||
| NC score | 0.051286 (rank : 51) | θ value | 0.62314 (rank : 56) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BMB0, Q5FWK5, Q80XU1, Q8VDW9 | Gene names | Emsy | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
ZN750_MOUSE
|
||||||
| NC score | 0.050040 (rank : 52) | θ value | 0.00509761 (rank : 48) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BH05, Q66JP3, Q8C0L1 | Gene names | Znf750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
SRBP1_MOUSE
|
||||||
| NC score | 0.046190 (rank : 53) | θ value | 0.0252991 (rank : 51) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
APC_MOUSE
|
||||||
| NC score | 0.044667 (rank : 54) | θ value | 0.365318 (rank : 54) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q61315, Q62044 | Gene names | Apc | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC) (mAPC). | |||||
|
APC_HUMAN
|
||||||
| NC score | 0.043082 (rank : 55) | θ value | 0.21417 (rank : 53) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P25054, Q15162, Q15163, Q93042 | Gene names | APC, DP2.5 | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC). | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.041566 (rank : 56) | θ value | 1.38821 (rank : 64) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
NU214_HUMAN
|
||||||
| NC score | 0.035847 (rank : 57) | θ value | 2.36792 (rank : 70) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.035491 (rank : 58) | θ value | 0.163984 (rank : 52) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
PODXL_HUMAN
|
||||||
| NC score | 0.035072 (rank : 59) | θ value | 1.81305 (rank : 66) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O00592 | Gene names | PODXL, PCLP, PCLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Podocalyxin-like protein 1 precursor. | |||||
|
MCA1_MOUSE
|
||||||
| NC score | 0.030894 (rank : 60) | θ value | 0.813845 (rank : 58) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P31230, Q60659 | Gene names | Scye1, Emap2 | |||
|
Domain Architecture |

|
|||||
| Description | Multisynthetase complex auxiliary component p43 [Contains: Endothelial monocyte-activating polypeptide 2 (EMAP-II) (Small inducible cytokine subfamily E member 1)]. | |||||
|
MUC5B_HUMAN
|
||||||
| NC score | 0.030657 (rank : 61) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.029514 (rank : 62) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
K1210_HUMAN
|
||||||
| NC score | 0.029001 (rank : 63) | θ value | 1.38821 (rank : 62) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9ULL0, Q5JPN4 | Gene names | KIAA1210 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1210. | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.027919 (rank : 64) | θ value | 3.0926 (rank : 73) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
PHC3_HUMAN
|
||||||
| NC score | 0.027712 (rank : 65) | θ value | 1.06291 (rank : 59) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8NDX5, Q5HYF0, Q6NSG2, Q8NFT7, Q8NFZ1, Q8TBM2, Q9H971, Q9H9I4 | Gene names | PHC3, EDR3, PH3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 3 (hPH3) (Homolog of polyhomeotic 3) (Early development regulatory protein 3). | |||||
|
PO121_HUMAN
|
||||||
| NC score | 0.025425 (rank : 66) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
RFX1_HUMAN
|
||||||
| NC score | 0.024042 (rank : 67) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P22670 | Gene names | RFX1 | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II regulatory factor RFX1 (RFX) (Enhancer factor C) (EF-C). | |||||
|
CK024_MOUSE
|
||||||
| NC score | 0.023692 (rank : 68) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9D8N1, Q8VCP2 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 homolog precursor. | |||||
|
STMN3_HUMAN
|
||||||
| NC score | 0.017233 (rank : 69) | θ value | 1.06291 (rank : 61) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NZ72, O75527, Q969Y4 | Gene names | STMN3, SCLIP | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin-3 (SCG10-like protein). | |||||
|
TCRG1_HUMAN
|
||||||
| NC score | 0.016995 (rank : 70) | θ value | 2.36792 (rank : 72) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.016198 (rank : 71) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
SOX30_HUMAN
|
||||||
| NC score | 0.016197 (rank : 72) | θ value | 1.81305 (rank : 67) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O94993, O94995, Q8IYX6 | Gene names | SOX30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
CSPG2_HUMAN
|
||||||
| NC score | 0.015510 (rank : 73) | θ value | 0.0148317 (rank : 50) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P13611, P20754, Q13010, Q13189, Q15123, Q9UCL9, Q9UNW5 | Gene names | CSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M) (Glial hyaluronate-binding protein) (GHAP). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.015473 (rank : 74) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
WAPL_HUMAN
|
||||||
| NC score | 0.014891 (rank : 75) | θ value | 3.0926 (rank : 75) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7Z5K2, Q5VSK5, Q8IX10, Q92549 | Gene names | WAPAL, FOE, KIAA0261, WAPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wings apart-like protein homolog (Friend of EBNA2 protein). | |||||
|
NOL8_HUMAN
|
||||||
| NC score | 0.014179 (rank : 76) | θ value | 2.36792 (rank : 69) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q76FK4, Q5TCC7, Q5TCC8, Q5TCD3, Q5TCD5, Q5TCD6, Q5TCD7, Q76D35, Q7L3E2, Q9H586, Q9H795, Q9H7W7, Q9H9J6, Q9NWA4, Q9NWM4 | Gene names | NOL8, NOP132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 8 (Nucleolar protein Nop132). | |||||
|
LRC50_HUMAN
|
||||||
| NC score | 0.013801 (rank : 77) | θ value | 1.81305 (rank : 65) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NEP3, Q69YI8, Q69YJ0, Q96LP3 | Gene names | LRRC50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 50. | |||||
|
STMN1_HUMAN
|
||||||
| NC score | 0.013481 (rank : 78) | θ value | 2.36792 (rank : 71) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P16949 | Gene names | STMN1, LAP18, OP18 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Protein Pr22). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.013161 (rank : 79) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
STMN1_MOUSE
|
||||||
| NC score | 0.013078 (rank : 80) | θ value | 3.0926 (rank : 74) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P54227 | Gene names | Stmn1, Lag, Lap18, Pr22 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Pr22 protein) (Leukemia-associated gene protein). | |||||
|
RIOK1_HUMAN
|
||||||
| NC score | 0.013059 (rank : 81) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BRS2, Q8NDC8, Q96NV9 | Gene names | RIOK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase RIO1 (EC 2.7.11.1) (RIO kinase 1). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.011407 (rank : 82) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
NFYC_MOUSE
|
||||||
| NC score | 0.010585 (rank : 83) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P70353, O35088, Q6GU21 | Gene names | Nfyc | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear transcription factor Y subunit gamma (Nuclear transcription factor Y subunit C) (NF-YC) (CAAT-box DNA-binding protein subunit C). | |||||
|
RAI2_MOUSE
|
||||||
| NC score | 0.010561 (rank : 84) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QVY8 | Gene names | Rai2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 2 (3f8). | |||||
|
AT8B3_HUMAN
|
||||||
| NC score | 0.008102 (rank : 85) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60423, Q8IVB8, Q8N4Y8, Q96M22 | Gene names | ATP8B3, ATP1K, FOS37502_2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IK (EC 3.6.3.1) (ATPase class I type 8B member 3). | |||||
|
NLGN3_MOUSE
|
||||||
| NC score | 0.006664 (rank : 86) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BYM5, Q8BXR4 | Gene names | Nlgn3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-3 precursor (Gliotactin homolog). | |||||
|
HCN1_MOUSE
|
||||||
| NC score | 0.006437 (rank : 87) | θ value | 2.36792 (rank : 68) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O88704, O54899, Q9D613 | Gene names | Hcn1, Bcng1, Hac2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 (Brain cyclic nucleotide-gated channel 1) (BCNG-1) (Hyperpolarization-activated cation channel 2) (HAC-2). | |||||
|
BOC_HUMAN
|
||||||
| NC score | 0.006038 (rank : 88) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BWV1, Q6UXJ5, Q8N2P7, Q8NF26 | Gene names | BOC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brother of CDO precursor (Protein BOC). | |||||
|
PADI6_MOUSE
|
||||||
| NC score | 0.005698 (rank : 89) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K3V4, Q75WC6 | Gene names | Padi6, Padi5 | |||
|
Domain Architecture |

|
|||||
| Description | Protein-arginine deiminase type-6 (EC 3.5.3.15) (Protein-arginine deiminase type VI) (Peptidylarginine deiminase VI) (Arginine deiminase-like protein) (Egg and embryo abundant PAD) (ePAD). | |||||
|
ZMYM3_HUMAN
|
||||||
| NC score | 0.005685 (rank : 90) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14202, O15089 | Gene names | ZMYM3, DXS6673E, KIAA0385, ZNF261 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 3 (Zinc finger protein 261). | |||||
|
PHF20_MOUSE
|
||||||
| NC score | 0.005217 (rank : 91) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
AAK1_MOUSE
|
||||||
| NC score | 0.005046 (rank : 92) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q3UHJ0, Q3TY53, Q6ZPZ6, Q80XP6 | Gene names | Aak1, Kiaa1048 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP2-associated protein kinase 1 (EC 2.7.11.1) (Adaptor-associated kinase 1). | |||||
|
ANK3_HUMAN
|
||||||
| NC score | 0.004691 (rank : 93) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
PO2F1_HUMAN
|
||||||
| NC score | 0.003977 (rank : 94) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P14859, Q5TBT7, Q6PK46, Q8NEU9, Q9BPV1 | Gene names | POU2F1, OCT1, OTF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 2, transcription factor 1 (Octamer-binding transcription factor 1) (Oct-1) (OTF-1) (NF-A1). | |||||
|
PCOC1_HUMAN
|
||||||
| NC score | 0.003965 (rank : 95) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15113, O14550 | Gene names | PCOLCE, PCPE1 | |||
|
Domain Architecture |

|
|||||
| Description | Procollagen C-endopeptidase enhancer 1 precursor (Procollagen COOH- terminal proteinase enhancer 1) (Procollagen C-proteinase enhancer 1) (PCPE-1) (Type I procollagen COOH-terminal proteinase enhancer) (Type 1 procollagen C-proteinase enhancer protein). | |||||
|
AT10B_HUMAN
|
||||||
| NC score | 0.002902 (rank : 96) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O94823, Q9H725 | Gene names | ATP10B, ATPVB, KIAA0715 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase VB (EC 3.6.3.1). | |||||
|
WNK1_MOUSE
|
||||||
| NC score | 0.001989 (rank : 97) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1179 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P83741 | Gene names | Wnk1, Prkwnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1). | |||||
|
CAC1E_MOUSE
|
||||||
| NC score | 0.001506 (rank : 98) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61290 | Gene names | Cacna1e, Cach6, Cacnl1a6, Cchra1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
SP4_MOUSE
|
||||||
| NC score | 0.000928 (rank : 99) | θ value | 1.06291 (rank : 60) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q62445 | Gene names | Sp4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp4. | |||||
|
CADH3_HUMAN
|
||||||
| NC score | -0.000055 (rank : 100) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P22223 | Gene names | CDH3, CDHP | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-3 precursor (Placental-cadherin) (P-cadherin). | |||||
|
MAST1_HUMAN
|
||||||
| NC score | -0.002208 (rank : 101) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1146 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y2H9, O00114, Q8N6X0 | Gene names | MAST1, KIAA0973 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
MYLK_MOUSE
|
||||||
| NC score | -0.002282 (rank : 102) | θ value | 1.38821 (rank : 63) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1489 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6PDN3, Q3TSJ7, Q80UX0, Q80YN7, Q80YN8, Q8K026, Q924D2, Q9ERD3 | Gene names | Mylk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin light chain kinase, smooth muscle (EC 2.7.11.18) (MLCK) (Telokin) (Kinase-related protein) (KRP). | |||||
|
KLF4_HUMAN
|
||||||
| NC score | -0.003689 (rank : 103) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43474, P78338, Q9UNP3 | Gene names | KLF4, EZF, GKLF | |||
|
Domain Architecture |

|
|||||
| Description | Krueppel-like factor 4 (Epithelial zinc-finger protein EZF) (Gut- enriched krueppel-like factor). | |||||
|
CHK2_HUMAN
|
||||||
| NC score | -0.003928 (rank : 104) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O96017, Q6QA03, Q6QA04, Q6QA05, Q6QA06, Q6QA07, Q6QA08, Q6QA10, Q6QA11, Q6QA12, Q6QA13, Q9HCQ8, Q9UGF0, Q9UGF1 | Gene names | CHEK2, CHK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Chk2 (EC 2.7.11.1) (Cds1). | |||||