Please be patient as the page loads
|
ELK4_HUMAN
|
||||||
| SwissProt Accessions | P28324, P28323 | Gene names | ELK4, SAP1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ELK4_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 184 | |
| SwissProt Accessions | P28324, P28323 | Gene names | ELK4, SAP1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
|
ELK4_MOUSE
|
||||||
| θ value | 1.50581e-180 (rank : 2) | NC score | 0.983081 (rank : 2) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P41158 | Gene names | Elk4, Sap1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
|
ELK3_HUMAN
|
||||||
| θ value | 7.19216e-82 (rank : 3) | NC score | 0.971380 (rank : 3) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P41970, Q6FG57, Q6GU29, Q9UD17 | Gene names | ELK3, NET, SAP2 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-3 (ETS-related protein NET) (ETS- related protein ERP) (SRF accessory protein 2) (SAP-2). | |||||
|
ELK3_MOUSE
|
||||||
| θ value | 8.79181e-80 (rank : 4) | NC score | 0.969171 (rank : 4) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P41971, P97747, Q62346 | Gene names | Elk3, Erp, Net | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-3 (ETS-related protein NET) (ETS- related protein ERP). | |||||
|
ELK1_MOUSE
|
||||||
| θ value | 3.48141e-52 (rank : 5) | NC score | 0.948526 (rank : 5) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P41969 | Gene names | Elk1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-1. | |||||
|
ELK1_HUMAN
|
||||||
| θ value | 7.25919e-50 (rank : 6) | NC score | 0.944914 (rank : 6) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P19419, O75606, O95058, Q969X8, Q9UJM4 | Gene names | ELK1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-1. | |||||
|
ETV4_HUMAN
|
||||||
| θ value | 1.73987e-27 (rank : 7) | NC score | 0.860241 (rank : 22) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P43268, Q96AW9 | Gene names | ETV4, E1AF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 4 (Adenovirus E1A enhancer-binding protein) (E1A-F). | |||||
|
ETV4_MOUSE
|
||||||
| θ value | 1.73987e-27 (rank : 8) | NC score | 0.860734 (rank : 20) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P28322 | Gene names | Etv4, Pea-3, Pea3 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 4 (Polyomavirus enhancer activator 3) (Protein PEA3). | |||||
|
ETV1_HUMAN
|
||||||
| θ value | 1.47289e-26 (rank : 9) | NC score | 0.862539 (rank : 18) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P50549, O75849, Q9UQ71, Q9Y636 | Gene names | ETV1, ER81 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 1 (ER81 protein). | |||||
|
ETV1_MOUSE
|
||||||
| θ value | 1.47289e-26 (rank : 10) | NC score | 0.863745 (rank : 17) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P41164 | Gene names | Etv1, Er81, Etsrp81 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 1 (ER81 protein). | |||||
|
ETV5_HUMAN
|
||||||
| θ value | 5.59698e-26 (rank : 11) | NC score | 0.858141 (rank : 23) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P41161 | Gene names | ETV5, ERM | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 5 (Ets-related protein ERM). | |||||
|
ETV5_MOUSE
|
||||||
| θ value | 5.59698e-26 (rank : 12) | NC score | 0.839667 (rank : 30) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9CXC9, Q3TG49, Q8C0F3, Q9JHB1 | Gene names | Etv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 5. | |||||
|
ERG_MOUSE
|
||||||
| θ value | 6.84181e-24 (rank : 13) | NC score | 0.860650 (rank : 21) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P81270, Q8C5L4, Q920K7, Q920K8, Q920K9 | Gene names | Erg, Erg-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ERG. | |||||
|
ERG_HUMAN
|
||||||
| θ value | 1.16704e-23 (rank : 14) | NC score | 0.860759 (rank : 19) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P11308, Q16113 | Gene names | ERG | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ERG (Transforming protein ERG). | |||||
|
FLI1_HUMAN
|
||||||
| θ value | 1.99067e-23 (rank : 15) | NC score | 0.866528 (rank : 16) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q01543, Q14319, Q92480, Q9UE07 | Gene names | FLI1 | |||
|
Domain Architecture |

|
|||||
| Description | Friend leukemia integration 1 transcription factor (Fli-1 proto- oncogene) (ERGB transcription factor). | |||||
|
FLI1_MOUSE
|
||||||
| θ value | 1.99067e-23 (rank : 16) | NC score | 0.867836 (rank : 15) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P26323 | Gene names | Fli1, Fli-1 | |||
|
Domain Architecture |

|
|||||
| Description | Friend leukemia integration 1 transcription factor (Retroviral integration site protein Fli-1). | |||||
|
GABPA_HUMAN
|
||||||
| θ value | 2.20094e-22 (rank : 17) | NC score | 0.877550 (rank : 10) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q06546, Q12939 | Gene names | GABPA, E4TF1A | |||
|
Domain Architecture |

|
|||||
| Description | GA-binding protein alpha chain (GABP-subunit alpha) (Transcription factor E4TF1-60) (Nuclear respiratory factor 2 subunit alpha). | |||||
|
ETS2_HUMAN
|
||||||
| θ value | 3.75424e-22 (rank : 18) | NC score | 0.871973 (rank : 11) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P15036 | Gene names | ETS2 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-2 protein. | |||||
|
ETS2_MOUSE
|
||||||
| θ value | 3.75424e-22 (rank : 19) | NC score | 0.870902 (rank : 14) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P15037 | Gene names | Ets2 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-2 protein. | |||||
|
GABPA_MOUSE
|
||||||
| θ value | 4.9032e-22 (rank : 20) | NC score | 0.878589 (rank : 9) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q00422 | Gene names | Gabpa, E4tf1a | |||
|
Domain Architecture |

|
|||||
| Description | GA-binding protein alpha chain (GABP-subunit alpha). | |||||
|
ERF_HUMAN
|
||||||
| θ value | 1.09232e-21 (rank : 21) | NC score | 0.855993 (rank : 26) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P50548, Q9UPI7 | Gene names | ERF | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing transcription factor ERF (Ets2 repressor factor). | |||||
|
ERF_MOUSE
|
||||||
| θ value | 1.09232e-21 (rank : 22) | NC score | 0.856355 (rank : 25) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P70459 | Gene names | Erf | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing transcription factor ERF. | |||||
|
ETS1_HUMAN
|
||||||
| θ value | 1.42661e-21 (rank : 23) | NC score | 0.871028 (rank : 13) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P14921, Q14278, Q16080 | Gene names | ETS1 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-1 protein (p54). | |||||
|
ETS1_MOUSE
|
||||||
| θ value | 1.42661e-21 (rank : 24) | NC score | 0.871069 (rank : 12) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P27577, Q61403 | Gene names | Ets1, Ets-1 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-1 protein (p54). | |||||
|
ETV3_HUMAN
|
||||||
| θ value | 1.86321e-21 (rank : 25) | NC score | 0.833303 (rank : 31) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P41162, Q8TAC8, Q9BX30 | Gene names | ETV3, METS, PE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
ETV3_MOUSE
|
||||||
| θ value | 1.86321e-21 (rank : 26) | NC score | 0.852552 (rank : 29) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8R4Z4, Q9QZW1 | Gene names | Etv3, Mets, Pe1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
ETV2_HUMAN
|
||||||
| θ value | 2.06002e-20 (rank : 27) | NC score | 0.890583 (rank : 7) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O00321, Q9UEA0 | Gene names | ETV2, ER71, ETSRP71 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 2 (Ets-related protein 71). | |||||
|
ETV2_MOUSE
|
||||||
| θ value | 2.06002e-20 (rank : 28) | NC score | 0.888254 (rank : 8) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P41163 | Gene names | Etv2, Er71, Etsrp71 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 2 (Ets-related protein 71). | |||||
|
ELF1_MOUSE
|
||||||
| θ value | 2.27762e-19 (rank : 29) | NC score | 0.821536 (rank : 36) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q60775 | Gene names | Elf1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
ELF1_HUMAN
|
||||||
| θ value | 7.32683e-18 (rank : 30) | NC score | 0.825566 (rank : 35) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P32519, Q9UDE1 | Gene names | ELF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
ELF4_HUMAN
|
||||||
| θ value | 1.24977e-17 (rank : 31) | NC score | 0.825927 (rank : 34) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q99607, O60435 | Gene names | ELF4, ELFR, MEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-4 (E74-like factor 4) (Myeloid Elf-1-like factor). | |||||
|
ELF4_MOUSE
|
||||||
| θ value | 1.24977e-17 (rank : 32) | NC score | 0.807368 (rank : 41) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9Z2U4 | Gene names | Elf4, Mef | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-4 (E74-like factor 4) (Myeloid Elf-1-like factor). | |||||
|
ELF2_HUMAN
|
||||||
| θ value | 3.63628e-17 (rank : 33) | NC score | 0.826440 (rank : 33) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q15723, Q15724, Q15725, Q6P1K5 | Gene names | ELF2, NERF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-2 (E74-like factor 2) (New ETS- related factor). | |||||
|
ELF2_MOUSE
|
||||||
| θ value | 3.63628e-17 (rank : 34) | NC score | 0.828239 (rank : 32) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9JHC9, Q6NST2, Q8BTX8, Q9JHC7, Q9JHC8, Q9JHD0 | Gene names | Elf2, Nerf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-2 (E74-like factor 2) (New ETS- related factor). | |||||
|
SPDEF_HUMAN
|
||||||
| θ value | 8.10077e-17 (rank : 35) | NC score | 0.855434 (rank : 27) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O95238 | Gene names | SPDEF, PDEF, PSE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAM pointed domain-containing Ets transcription factor (Prostate- derived Ets factor) (Prostate epithelium-specific Ets transcription factor) (Prostate-specific Ets). | |||||
|
SPDEF_MOUSE
|
||||||
| θ value | 1.058e-16 (rank : 36) | NC score | 0.853688 (rank : 28) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9WTP3 | Gene names | Spdef, Pdef, Pse | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAM pointed domain-containing Ets transcription factor (Prostate- derived Ets factor) (Prostate epithelium-specific Ets transcription factor) (Prostate-specific Ets). | |||||
|
ETV6_HUMAN
|
||||||
| θ value | 2.20605e-14 (rank : 37) | NC score | 0.816083 (rank : 38) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P41212, Q9UMF6 | Gene names | ETV6, TEL, TEL1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV6 (ETS-related protein Tel1) (Tel) (ETS translocation variant 6). | |||||
|
ETV6_MOUSE
|
||||||
| θ value | 2.20605e-14 (rank : 38) | NC score | 0.818083 (rank : 37) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P97360 | Gene names | Etv6, Tel, Tel1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV6 (ETS-related protein Tel1) (Tel) (ETS translocation variant 6). | |||||
|
ETV7_HUMAN
|
||||||
| θ value | 2.20605e-14 (rank : 39) | NC score | 0.856920 (rank : 24) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9Y603, Q9NZ65, Q9NZ66, Q9NZ68, Q9NZR8, Q9UNJ7, Q9Y5K4, Q9Y604 | Gene names | ETV7, TEL2, TELB, TREF | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV7 (Transcription factor Tel-2) (ETS-related protein Tel2) (Tel-related Ets factor). | |||||
|
ELF5_HUMAN
|
||||||
| θ value | 1.74796e-11 (rank : 40) | NC score | 0.809226 (rank : 40) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UKW6, O95175, Q8N2K9, Q96QY3, Q9UKW5 | Gene names | ELF5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-5 (E74-like factor 5) (Epithelium-specific Ets transcription factor 2) (ESE-2) (Epithelium- restricted ESE-1-related Ets factor). | |||||
|
ELF5_MOUSE
|
||||||
| θ value | 1.74796e-11 (rank : 41) | NC score | 0.810465 (rank : 39) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8VDK3, Q921H5, Q9Z2K6 | Gene names | Elf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-5 (E74-like factor 5). | |||||
|
SPI1_HUMAN
|
||||||
| θ value | 1.17247e-07 (rank : 42) | NC score | 0.642632 (rank : 43) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P17947 | Gene names | SPI1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor PU.1 (31 kDa transforming protein). | |||||
|
SPI1_MOUSE
|
||||||
| θ value | 1.17247e-07 (rank : 43) | NC score | 0.643854 (rank : 42) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P17433, Q99L57 | Gene names | Spi1, Sfpi-1, Sfpi1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor PU.1 (31 kDa transforming protein) (SFFV proviral integration 1 protein). | |||||
|
SPIB_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 44) | NC score | 0.632105 (rank : 44) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q01892, Q15359 | Gene names | SPIB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Spi-B. | |||||
|
SPIB_MOUSE
|
||||||
| θ value | 1.09739e-05 (rank : 45) | NC score | 0.613341 (rank : 45) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O35906, O35907, O35909, O55199 | Gene names | Spib | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-B. | |||||
|
SPIC_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 46) | NC score | 0.584763 (rank : 46) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8N5J4 | Gene names | SPIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-C. | |||||
|
MAP4_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 47) | NC score | 0.066929 (rank : 51) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
PRDM2_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 48) | NC score | 0.018010 (rank : 128) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
ZAN_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 49) | NC score | 0.039397 (rank : 74) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
SPIC_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 50) | NC score | 0.548290 (rank : 47) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6P3D7, Q9Z0Y0 | Gene names | Spic | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-C (Pu.1-related factor) (Prf). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 51) | NC score | 0.044562 (rank : 67) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
K1802_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 52) | NC score | 0.046436 (rank : 63) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
FBLN2_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 53) | NC score | 0.010155 (rank : 149) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P98095 | Gene names | FBLN2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-2 precursor. | |||||
|
SON_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 54) | NC score | 0.061915 (rank : 55) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 55) | NC score | 0.077040 (rank : 49) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 112 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
IF4G2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 56) | NC score | 0.039802 (rank : 72) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P78344, O60877, P78404, Q53EU1, Q58EZ2, Q8NI71, Q96C16 | Gene names | EIF4G2, DAP5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 2 (eIF-4-gamma 2) (eIF-4G 2) (eIF4G 2) (p97) (Death-associated protein 5) (DAP-5). | |||||
|
IF4G2_MOUSE
|
||||||
| θ value | 0.125558 (rank : 57) | NC score | 0.039805 (rank : 71) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62448, P97867, Q921U3 | Gene names | Eif4g2, Nat1 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 2 (eIF-4-gamma 2) (eIF-4G 2) (eIF4G 2) (p97) (Novel APOBEC-1 target 1) (Translation repressor NAT1). | |||||
|
MLL4_HUMAN
|
||||||
| θ value | 0.125558 (rank : 58) | NC score | 0.039966 (rank : 70) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
SF3A1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 59) | NC score | 0.048084 (rank : 62) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8K4Z5, Q8C0M7, Q8C128, Q8C175, Q921T3 | Gene names | Sf3a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 3 subunit 1 (SF3a120). | |||||
|
GGA3_MOUSE
|
||||||
| θ value | 0.21417 (rank : 60) | NC score | 0.029029 (rank : 100) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BMI3, Q80U71 | Gene names | Gga3, Kiaa0154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
SF3A1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 61) | NC score | 0.048396 (rank : 61) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15459 | Gene names | SF3A1, SAP114 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3 subunit 1 (Spliceosome-associated protein 114) (SAP 114) (SF3a120). | |||||
|
EXO1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 62) | NC score | 0.024173 (rank : 115) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QZ11, Q3TLM4, Q923A5 | Gene names | Exo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exonuclease 1 (EC 3.1.-.-) (mExo1) (Exonuclease I). | |||||
|
GP112_HUMAN
|
||||||
| θ value | 0.365318 (rank : 63) | NC score | 0.029439 (rank : 99) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8IZF6, Q86SM6 | Gene names | GPR112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 112. | |||||
|
MNT_HUMAN
|
||||||
| θ value | 0.365318 (rank : 64) | NC score | 0.036489 (rank : 77) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | 0.365318 (rank : 65) | NC score | 0.058507 (rank : 56) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
RBM12_HUMAN
|
||||||
| θ value | 0.365318 (rank : 66) | NC score | 0.033202 (rank : 83) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NTZ6, O94865, Q8N3B1, Q9H196 | Gene names | RBM12, KIAA0765 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN). | |||||
|
SF3B2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 67) | NC score | 0.037357 (rank : 76) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13435 | Gene names | SF3B2, SAP145 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 2 (Spliceosome-associated protein 145) (SAP 145) (SF3b150) (Pre-mRNA-splicing factor SF3b 145 kDa subunit). | |||||
|
SON_HUMAN
|
||||||
| θ value | 0.365318 (rank : 68) | NC score | 0.078027 (rank : 48) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
CD2L7_HUMAN
|
||||||
| θ value | 0.47712 (rank : 69) | NC score | 0.001294 (rank : 174) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9NYV4, O94978 | Gene names | CRKRS, CRK7, KIAA0904 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle 2-related protein kinase 7 (EC 2.7.11.22) (CDC2- related protein kinase 7) (Cdc2-related kinase, arginine/serine-rich) (CrkRS). | |||||
|
MILK1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 70) | NC score | 0.028931 (rank : 101) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
PRG4_HUMAN
|
||||||
| θ value | 0.47712 (rank : 71) | NC score | 0.055917 (rank : 57) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
TACC3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 72) | NC score | 0.012595 (rank : 140) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JJ11, Q9WVK9 | Gene names | Tacc3, Aint | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ARNT-interacting protein). | |||||
|
UBP24_HUMAN
|
||||||
| θ value | 0.47712 (rank : 73) | NC score | 0.012012 (rank : 142) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UPU5, Q6ZSY2, Q8N2Y4, Q9NXD1 | Gene names | USP24, KIAA1057 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 24 (EC 3.1.2.15) (Ubiquitin thioesterase 24) (Ubiquitin-specific-processing protease 24) (Deubiquitinating enzyme 24). | |||||
|
CK024_HUMAN
|
||||||
| θ value | 0.62314 (rank : 74) | NC score | 0.072229 (rank : 50) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
LDB3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 75) | NC score | 0.016273 (rank : 133) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O75112, Q5K6N9, Q5K6P0, Q5K6P1, Q96FH2, Q9Y4Z3, Q9Y4Z4, Q9Y4Z5 | Gene names | LDB3, KIAA0613, ZASP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher). | |||||
|
MAML2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 76) | NC score | 0.034593 (rank : 81) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8IZL2, Q6AI23, Q6Y3A3, Q8IUL3, Q96JK6 | Gene names | MAML2, KIAA1819 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 2 (Mam-2). | |||||
|
MEGF9_HUMAN
|
||||||
| θ value | 0.62314 (rank : 77) | NC score | 0.010875 (rank : 147) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9H1U4, O75098 | Gene names | MEGF9, EGFL5, KIAA0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
MINT_HUMAN
|
||||||
| θ value | 0.62314 (rank : 78) | NC score | 0.028826 (rank : 102) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MK07_MOUSE
|
||||||
| θ value | 0.62314 (rank : 79) | NC score | 0.000629 (rank : 175) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1054 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9WVS8 | Gene names | Mapk7, Erk5 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase 7 (EC 2.7.11.24) (Extracellular signal-regulated kinase 5) (ERK-5) (BMK1 kinase). | |||||
|
SNPC4_MOUSE
|
||||||
| θ value | 0.62314 (rank : 80) | NC score | 0.025950 (rank : 110) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BP86, Q6PGG7, Q80UG9, Q810L1 | Gene names | Snapc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit). | |||||
|
GGA3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 81) | NC score | 0.027263 (rank : 107) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NZ52, Q15017, Q6IS16, Q9UJY3 | Gene names | GGA3, KIAA0154 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
NCOR2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 82) | NC score | 0.024185 (rank : 114) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
PRR12_HUMAN
|
||||||
| θ value | 0.813845 (rank : 83) | NC score | 0.049206 (rank : 60) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
RAPH1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 84) | NC score | 0.045467 (rank : 65) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 85) | NC score | 0.032100 (rank : 89) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
SYNJ1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 86) | NC score | 0.038852 (rank : 75) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O43426, O43425, O94984 | Gene names | SYNJ1, KIAA0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 87) | NC score | 0.028041 (rank : 103) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
ZBT12_HUMAN
|
||||||
| θ value | 0.813845 (rank : 88) | NC score | -0.001758 (rank : 178) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y330, Q5JQ98 | Gene names | ZBTB12, C6orf46, G10, NG35 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 12 (Protein G10). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 1.06291 (rank : 89) | NC score | 0.027824 (rank : 105) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
PDXL2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 90) | NC score | 0.023020 (rank : 118) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NZ53, Q6UVY4, Q8WUV6 | Gene names | PODXL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Podocalyxin-like protein 2 precursor (Endoglycan). | |||||
|
SRBS1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 91) | NC score | 0.017351 (rank : 129) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9BX66, Q7LBE5, Q8IVK0, Q8IVQ4, Q96KF3, Q96KF4, Q9BX64, Q9BX65, Q9P2Q0, Q9UFT2, Q9UHN7, Q9Y338 | Gene names | SORBS1, KIAA1296, SH3D5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorbin and SH3 domain-containing protein 1 (Ponsin) (c-Cbl-associated protein) (CAP) (SH3 domain protein 5) (SH3P12). | |||||
|
CD2AP_MOUSE
|
||||||
| θ value | 1.38821 (rank : 92) | NC score | 0.014197 (rank : 135) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9JLQ0, O88903, Q8K4Z1, Q8VCI9 | Gene names | Cd2ap, Mets1 | |||
|
Domain Architecture |
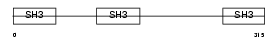
|
|||||
| Description | CD2-associated protein (Mesenchyme-to-epithelium transition protein with SH3 domains 1) (METS-1). | |||||
|
LAP2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 93) | NC score | 0.002080 (rank : 171) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80TH2, Q8BQ14, Q8CE41, Q8K171, Q99JU3, Q9JI47 | Gene names | Erbb2ip, Erbin, Kiaa1225, Lap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP2 (Erbb2-interacting protein) (Erbin) (Densin-180-like protein). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 94) | NC score | 0.064464 (rank : 52) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
RBM12_MOUSE
|
||||||
| θ value | 1.38821 (rank : 95) | NC score | 0.035100 (rank : 80) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8R4X3, Q8K373, Q8R302, Q9CS80 | Gene names | Rbm12 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN). | |||||
|
ZIM10_HUMAN
|
||||||
| θ value | 1.38821 (rank : 96) | NC score | 0.030268 (rank : 97) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
ZIM10_MOUSE
|
||||||
| θ value | 1.38821 (rank : 97) | NC score | 0.031992 (rank : 90) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6P1E1, Q6PDK9, Q6PF85, Q8BW47 | Gene names | Rai17, Zimp10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
BPTF_HUMAN
|
||||||
| θ value | 1.81305 (rank : 98) | NC score | 0.023678 (rank : 116) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
CS007_HUMAN
|
||||||
| θ value | 1.81305 (rank : 99) | NC score | 0.027313 (rank : 106) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
CS007_MOUSE
|
||||||
| θ value | 1.81305 (rank : 100) | NC score | 0.030997 (rank : 92) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
GNDS_MOUSE
|
||||||
| θ value | 1.81305 (rank : 101) | NC score | 0.016636 (rank : 132) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q03385 | Gene names | Ralgds, Rgds | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator (RalGEF) (RalGDS). | |||||
|
MBD5_HUMAN
|
||||||
| θ value | 1.81305 (rank : 102) | NC score | 0.025886 (rank : 111) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9P267, Q9NUV6 | Gene names | MBD5, KIAA1461 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 5 (Methyl-CpG-binding protein MBD5). | |||||
|
REPS1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 103) | NC score | 0.019239 (rank : 126) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96D71, Q8NDR7, Q8WU62, Q9BXY9 | Gene names | REPS1 | |||
|
Domain Architecture |

|
|||||
| Description | RalBP1-associated Eps domain-containing protein 1 (RalBP1-interacting protein 1). | |||||
|
REPS1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 104) | NC score | 0.019452 (rank : 125) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O54916, Q8C9J9, Q99LR8 | Gene names | Reps1 | |||
|
Domain Architecture |

|
|||||
| Description | RalBP1-associated Eps domain-containing protein 1 (RalBP1-interacting protein 1). | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 105) | NC score | 0.064076 (rank : 53) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
MAP4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 106) | NC score | 0.054552 (rank : 58) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
PHC3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 107) | NC score | 0.029681 (rank : 98) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8NDX5, Q5HYF0, Q6NSG2, Q8NFT7, Q8NFZ1, Q8TBM2, Q9H971, Q9H9I4 | Gene names | PHC3, EDR3, PH3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 3 (hPH3) (Homolog of polyhomeotic 3) (Early development regulatory protein 3). | |||||
|
PHLB2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 108) | NC score | 0.004130 (rank : 166) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q86SQ0, Q59EA8, Q68CY3, Q6NT98, Q8N8U8, Q8NAB1, Q8NCU5 | Gene names | PHLDB2, LL5B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
ANR11_HUMAN
|
||||||
| θ value | 3.0926 (rank : 109) | NC score | 0.008814 (rank : 153) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
ANTR1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 110) | NC score | 0.016984 (rank : 130) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9CZ52 | Gene names | Antxr1, Atr, Tem8 | |||
|
Domain Architecture |
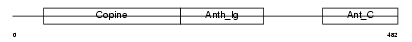
|
|||||
| Description | Anthrax toxin receptor 1 precursor (Tumor endothelial marker 8). | |||||
|
ATS12_HUMAN
|
||||||
| θ value | 3.0926 (rank : 111) | NC score | 0.003071 (rank : 168) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P58397, Q6UWL3 | Gene names | ADAMTS12 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 12) (ADAM-TS 12) (ADAM-TS12). | |||||
|
CD2AP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 112) | NC score | 0.016217 (rank : 134) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y5K6, Q9UG97 | Gene names | CD2AP | |||
|
Domain Architecture |
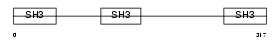
|
|||||
| Description | CD2-associated protein (Cas ligand with multiple SH3 domains) (Adapter protein CMS). | |||||
|
DYSF_HUMAN
|
||||||
| θ value | 3.0926 (rank : 113) | NC score | 0.011354 (rank : 145) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O75923, O75696, Q9UEN7 | Gene names | DYSF, FER1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Dysferlin (Dystrophy-associated fer-1-like protein) (Fer-1-like protein 1). | |||||
|
K0195_HUMAN
|
||||||
| θ value | 3.0926 (rank : 114) | NC score | 0.012546 (rank : 141) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12767, O75536, Q86XF1 | Gene names | KIAA0195, TMEM94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0195 (Transmembrane protein 94). | |||||
|
LAT_MOUSE
|
||||||
| θ value | 3.0926 (rank : 115) | NC score | 0.035784 (rank : 79) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O54957 | Gene names | Lat | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Linker for activation of T-cells family member 1 (36 kDa phospho- tyrosine adapter protein) (pp36) (p36-38). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 116) | NC score | 0.062659 (rank : 54) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MUC2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 117) | NC score | 0.040472 (rank : 69) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
NFAC1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 118) | NC score | 0.013189 (rank : 138) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O88942, O70345 | Gene names | Nfatc1, Nfat2, Nfatc | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 1 (NFAT transcription complex cytosolic component) (NF-ATc1) (NF-ATc). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 3.0926 (rank : 119) | NC score | 0.041962 (rank : 68) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
SRBP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 120) | NC score | 0.032255 (rank : 87) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
ACRO_HUMAN
|
||||||
| θ value | 4.03905 (rank : 121) | NC score | 0.001298 (rank : 173) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P10323, Q6ICK2 | Gene names | ACR, ACRS | |||
|
Domain Architecture |
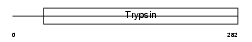
|
|||||
| Description | Acrosin precursor (EC 3.4.21.10) [Contains: Acrosin light chain; Acrosin heavy chain]. | |||||
|
ATBF1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 122) | NC score | 0.005884 (rank : 162) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
ATF6A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 123) | NC score | 0.018135 (rank : 127) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P18850, O15139, Q5VW62, Q6IPB5, Q9UEC9 | Gene names | ATF6 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-6 alpha (Activating transcription factor 6 alpha) (ATF6-alpha). | |||||
|
CSPG2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 124) | NC score | 0.008885 (rank : 152) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q62059, Q62058, Q9CUU0 | Gene names | Cspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M). | |||||
|
EFS_HUMAN
|
||||||
| θ value | 4.03905 (rank : 125) | NC score | 0.020124 (rank : 124) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O43281, O43282 | Gene names | EFS | |||
|
Domain Architecture |

|
|||||
| Description | Embryonal Fyn-associated substrate (HEFS). | |||||
|
ELAV2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 126) | NC score | 0.005268 (rank : 165) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12926, Q13235, Q59G15, Q8NEM4, Q9H1Q8 | Gene names | ELAVL2, HUB | |||
|
Domain Architecture |
|
|||||
| Description | ELAV-like protein 2 (Hu-antigen B) (HuB) (ELAV-like neuronal protein 1) (Nervous system-specific RNA-binding protein Hel-N1). | |||||
|
ELAV2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 127) | NC score | 0.005274 (rank : 164) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60899 | Gene names | Elavl2, Hub | |||
|
Domain Architecture |
|
|||||
| Description | ELAV-like protein 2 (Hu-antigen B) (HuB) (ELAV-like neuronal protein 1) (Nervous system-specific RNA-binding protein Mel-N1). | |||||
|
GGN_MOUSE
|
||||||
| θ value | 4.03905 (rank : 128) | NC score | 0.030580 (rank : 93) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q80WJ1, Q5EBP4, Q80WI9, Q80WJ0 | Gene names | Ggn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
GRB10_HUMAN
|
||||||
| θ value | 4.03905 (rank : 129) | NC score | 0.008097 (rank : 157) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13322, O00427, O00701, O75222, Q92606, Q92907, Q92948 | Gene names | GRB10, GRBIR, KIAA0207 | |||
|
Domain Architecture |

|
|||||
| Description | Growth factor receptor-bound protein 10 (GRB10 adaptor protein) (Insulin receptor-binding protein GRB-IR). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 4.03905 (rank : 130) | NC score | 0.039603 (rank : 73) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
PDC6I_HUMAN
|
||||||
| θ value | 4.03905 (rank : 131) | NC score | 0.026241 (rank : 109) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8WUM4, Q9BX86, Q9NUN0, Q9P2H2, Q9UKL5 | Gene names | PDCD6IP, AIP1, ALIX, KIAA1375 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death 6-interacting protein (PDCD6-interacting protein) (ALG-2-interacting protein 1) (Hp95). | |||||
|
PER3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 132) | NC score | 0.022391 (rank : 120) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
SFPQ_HUMAN
|
||||||
| θ value | 4.03905 (rank : 133) | NC score | 0.021604 (rank : 122) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
GTSE1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 134) | NC score | 0.032576 (rank : 85) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NYZ3, Q53GX5, Q5R3I6, Q6DHX4, Q9BRE0, Q9UGZ9, Q9Y557 | Gene names | GTSE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G2 and S phase expressed protein 1 (B99 homolog). | |||||
|
HELI_HUMAN
|
||||||
| θ value | 5.27518 (rank : 135) | NC score | -0.002890 (rank : 182) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UKS7, Q8N6S1 | Gene names | IKZF2, HELIOS, ZNFN1A2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein Helios (IKAROS family zinc finger protein 2). | |||||
|
HELI_MOUSE
|
||||||
| θ value | 5.27518 (rank : 136) | NC score | -0.002873 (rank : 181) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 746 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P81183 | Gene names | Ikzf2, Helios, Zfpn1a2, Znfn1a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein Helios (IKAROS family zinc finger protein 2). | |||||
|
K1210_HUMAN
|
||||||
| θ value | 5.27518 (rank : 137) | NC score | 0.030394 (rank : 95) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9ULL0, Q5JPN4 | Gene names | KIAA1210 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1210. | |||||
|
KIF2C_HUMAN
|
||||||
| θ value | 5.27518 (rank : 138) | NC score | 0.002385 (rank : 169) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99661, Q96C18, Q96HB8, Q9BWV8 | Gene names | KIF2C, KNSL6 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF2C (Mitotic centromere-associated kinesin) (MCAK) (Kinesin-like protein 6). | |||||
|
LATS1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 139) | NC score | -0.002236 (rank : 180) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1027 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BYR2, Q9Z0W4 | Gene names | Lats1, Warts | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS1 (EC 2.7.11.1) (Large tumor suppressor homolog 1) (WARTS protein kinase). | |||||
|
LAT_HUMAN
|
||||||
| θ value | 5.27518 (rank : 140) | NC score | 0.020989 (rank : 123) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43561, O43919 | Gene names | LAT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Linker for activation of T-cells family member 1 (36 kDa phospho- tyrosine adapter protein) (pp36) (p36-38). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 141) | NC score | 0.050557 (rank : 59) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
SCYL2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 142) | NC score | 0.001388 (rank : 172) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CFE4, Q3UT57, Q3UWU9, Q8K0M4 | Gene names | Scyl2, Cvak104, D10Ertd802e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCY1-like protein 2 (Coated vesicle-associated kinase of 104 kDa). | |||||
|
WNK2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 143) | NC score | 0.007596 (rank : 159) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
ATBF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 144) | NC score | 0.009249 (rank : 151) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
BRD4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 145) | NC score | 0.031356 (rank : 91) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
BRD4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 146) | NC score | 0.032142 (rank : 88) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
CASB_MOUSE
|
||||||
| θ value | 6.88961 (rank : 147) | NC score | 0.013631 (rank : 137) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P10598, Q543D9, Q8VCT6, Q8VCU8, Q91VI5, Q922Y5, Q9D1U6, Q9D1U7 | Gene names | Csn2, Csnb | |||
|
Domain Architecture |

|
|||||
| Description | Beta-casein precursor. | |||||
|
CEND3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 148) | NC score | 0.003424 (rank : 167) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8WWN8 | Gene names | CENTD3, ARAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3). | |||||
|
CORIN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 149) | NC score | -0.002060 (rank : 179) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z319 | Gene names | Corin, Crn, Lrp4 | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuteric peptide-converting enzyme (EC 3.4.21.-) (pro-ANP- converting enzyme) (Corin) (Low density lipoprotein receptor-related protein 4). | |||||
|
DGKK_HUMAN
|
||||||
| θ value | 6.88961 (rank : 150) | NC score | 0.023658 (rank : 117) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5KSL6 | Gene names | DGKK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase kappa (EC 2.7.1.107) (Diglyceride kinase kappa) (DGK-kappa) (DAG kinase kappa) (142 kDa diacylglycerol kinase). | |||||
|
DYSF_MOUSE
|
||||||
| θ value | 6.88961 (rank : 151) | NC score | 0.010474 (rank : 148) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ESD7, Q9QXC0 | Gene names | Dysf, Fer1l1 | |||
|
Domain Architecture |

|
|||||
| Description | Dysferlin (Dystrophy-associated fer-1-like protein) (Fer-1-like protein 1). | |||||
|
ELL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 152) | NC score | 0.009689 (rank : 150) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O08856 | Gene names | Ell | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II elongation factor ELL (Eleven-nineteen lysine-rich leukemia protein). | |||||
|
IL3RB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 153) | NC score | 0.011851 (rank : 143) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P32927 | Gene names | CSF2RB, IL3RB, IL5RB | |||
|
Domain Architecture |

|
|||||
| Description | Cytokine receptor common beta chain precursor (GM-CSF/IL-3/IL-5 receptor common beta-chain) (CD131 antigen) (CDw131). | |||||
|
LR37A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 154) | NC score | 0.032340 (rank : 86) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O60309, Q49A01, Q49A80, Q8NB33 | Gene names | LRRC37A, KIAA0563 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37A. | |||||
|
M3K10_HUMAN
|
||||||
| θ value | 6.88961 (rank : 155) | NC score | -0.004232 (rank : 184) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1071 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q02779, Q12761, Q14871 | Gene names | MAP3K10, MLK2, MST | |||
|
Domain Architecture |
|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 10 (EC 2.7.11.25) (Mixed lineage kinase 2) (Protein kinase MST). | |||||
|
MAST3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 156) | NC score | -0.003449 (rank : 183) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1128 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O60307, Q7LDZ8, Q9UPI0 | Gene names | MAST3, KIAA0561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 3 (EC 2.7.11.1). | |||||
|
NFAC4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 157) | NC score | 0.011465 (rank : 144) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14934 | Gene names | NFATC4, NFAT3 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 4 (NF-ATc4) (NFATc4) (T cell transcription factor NFAT3) (NF-AT3). | |||||
|
PCQAP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 158) | NC score | 0.022606 (rank : 119) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96RN5, O15413, Q6IC31, Q8NF16, Q96CT0, Q96IH7, Q9P1T3 | Gene names | PCQAP, ARC105, CTG7A, TIG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (TPA-inducible gene 1 protein) (TIG-1) (Activator-recruited cofactor 105 kDa component) (ARC105) (CTG repeat protein 7a). | |||||
|
SETBP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 159) | NC score | 0.022090 (rank : 121) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Z180, Q66JL8 | Gene names | Setbp1, Kiaa0437 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET-binding protein (SEB). | |||||
|
SMDF_HUMAN
|
||||||
| θ value | 6.88961 (rank : 160) | NC score | 0.012732 (rank : 139) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15491 | Gene names | NRG1, GGF, HGL, HRGA, NDF, SMDF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuregulin-1, sensory and motor neuron-derived factor isoform. | |||||
|
SYNJ1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 161) | NC score | 0.044870 (rank : 66) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
ZN638_HUMAN
|
||||||
| θ value | 6.88961 (rank : 162) | NC score | 0.008032 (rank : 158) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q14966, Q53R34, Q5XJ05, Q68DP3, Q6P2H2, Q7Z3T7, Q8NF92, Q8TCA1, Q9H2G1, Q9NP37 | Gene names | ZNF638, NP220, ZFML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein) (Cutaneous T-cell lymphoma-associated antigen se33-1) (CTCL tumor antigen se33-1). | |||||
|
ANKY1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 163) | NC score | 0.006249 (rank : 161) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9P2S6, Q8IYX5, Q8NDK5, Q9NX10 | Gene names | ANKMY1, TSAL1, ZMYND13 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and MYND domain-containing protein 1 (Testis-specific ankyrin-like protein 1) (Zinc-finger MYND domain-containing protein 13). | |||||
|
CELR3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 164) | NC score | -0.000945 (rank : 177) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 680 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NYQ7, O75092 | Gene names | CELSR3, CDHF11, EGFL1, FMI1, KIAA0812, MEGF2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 3 precursor (Flamingo homolog 1) (hFmi1) (Multiple epidermal growth factor-like domains 2) (Epidermal growth factor-like 1). | |||||
|
CN037_HUMAN
|
||||||
| θ value | 8.99809 (rank : 165) | NC score | 0.016680 (rank : 131) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q86TY3, Q6P5Q1, Q86TY1 | Gene names | C14orf37 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf37 precursor. | |||||
|
CS016_HUMAN
|
||||||
| θ value | 8.99809 (rank : 166) | NC score | 0.034457 (rank : 82) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8ND99 | Gene names | C19orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf16. | |||||
|
FGF5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 167) | NC score | 0.002121 (rank : 170) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P15656, O88825 | Gene names | Fgf5, Fgf-5 | |||
|
Domain Architecture |

|
|||||
| Description | Fibroblast growth factor 5 precursor (FGF-5) (HBGF-5). | |||||
|
FIP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 168) | NC score | 0.008097 (rank : 156) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6UN15, Q499Y4, Q49AU3, Q7Z608, Q8WVN3, Q96F80, Q9H077 | Gene names | FIP1L1, FIP1, RHE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA 3'-end-processing factor FIP1 (FIP1-like 1) (Factor interacting with PAP) (hFip1) (Rearranged in hypereosinophilia). | |||||
|
HNF3G_HUMAN
|
||||||
| θ value | 8.99809 (rank : 169) | NC score | -0.000029 (rank : 176) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P55318, Q9UMW9 | Gene names | FOXA3, HNF3G, TCF3G | |||
|
Domain Architecture |
|
|||||
| Description | Hepatocyte nuclear factor 3-gamma (HNF-3G) (Forkhead box protein A3) (Fork head-related protein FKH H3). | |||||
|
HRX_HUMAN
|
||||||
| θ value | 8.99809 (rank : 170) | NC score | 0.024759 (rank : 112) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
JPH1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 171) | NC score | 0.005403 (rank : 163) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9HDC5 | Gene names | JPH1, JP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-1 (Junctophilin type 1) (JP-1). | |||||
|
K1802_HUMAN
|
||||||
| θ value | 8.99809 (rank : 172) | NC score | 0.027934 (rank : 104) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
MKL2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 173) | NC score | 0.013980 (rank : 136) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P59759 | Gene names | Mkl2, Mrtfb | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 2 (Myocardin-related transcription factor B) (MRTF-B). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 174) | NC score | 0.036111 (rank : 78) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
NELFA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 175) | NC score | 0.026696 (rank : 108) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BG30, Q8BVE4, Q8VEI7, Q9CSJ9, Q9Z1V9 | Gene names | Whsc2, Nelfa, Whsc2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Negative elongation factor A (NELF-A) (Wolf-Hirschhorn syndrome candidate 2 homolog) (mWHSC2). | |||||
|
NUD19_MOUSE
|
||||||
| θ value | 8.99809 (rank : 176) | NC score | 0.011167 (rank : 146) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P11930 | Gene names | Nudt19, D7Rp2e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoside diphosphate-linked moiety X motif 19 (EC 3.-.-.-) (Nudix motif 19) (Testosterone-regulated RP2 protein) (Androgen-regulated protein RP2). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 8.99809 (rank : 177) | NC score | 0.033098 (rank : 84) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
PO121_HUMAN
|
||||||
| θ value | 8.99809 (rank : 178) | NC score | 0.030355 (rank : 96) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
PRG4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 179) | NC score | 0.046113 (rank : 64) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
RERE_HUMAN
|
||||||
| θ value | 8.99809 (rank : 180) | NC score | 0.024665 (rank : 113) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
TREF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 181) | NC score | 0.008105 (rank : 155) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96PN7, Q7Z6T2, Q7Z6T3, Q9NQ72, Q9NQ73, Q9NUN9 | Gene names | TRERF1, RAPA, TREP132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132) (Zinc finger protein rapa). | |||||
|
UBP10_MOUSE
|
||||||
| θ value | 8.99809 (rank : 182) | NC score | 0.006305 (rank : 160) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P52479, Q91VY7 | Gene names | Usp10, Ode-1, Uchrp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
UBQL3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 183) | NC score | 0.008231 (rank : 154) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9H347, Q9NRE0 | Gene names | UBQLN3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-3. | |||||
|
YLPM1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 184) | NC score | 0.030485 (rank : 94) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
ELK4_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 184 | |
| SwissProt Accessions | P28324, P28323 | Gene names | ELK4, SAP1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
|
ELK4_MOUSE
|
||||||
| NC score | 0.983081 (rank : 2) | θ value | 1.50581e-180 (rank : 2) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P41158 | Gene names | Elk4, Sap1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
|
ELK3_HUMAN
|
||||||
| NC score | 0.971380 (rank : 3) | θ value | 7.19216e-82 (rank : 3) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P41970, Q6FG57, Q6GU29, Q9UD17 | Gene names | ELK3, NET, SAP2 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-3 (ETS-related protein NET) (ETS- related protein ERP) (SRF accessory protein 2) (SAP-2). | |||||
|
ELK3_MOUSE
|
||||||
| NC score | 0.969171 (rank : 4) | θ value | 8.79181e-80 (rank : 4) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P41971, P97747, Q62346 | Gene names | Elk3, Erp, Net | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-3 (ETS-related protein NET) (ETS- related protein ERP). | |||||
|
ELK1_MOUSE
|
||||||
| NC score | 0.948526 (rank : 5) | θ value | 3.48141e-52 (rank : 5) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P41969 | Gene names | Elk1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-1. | |||||
|
ELK1_HUMAN
|
||||||
| NC score | 0.944914 (rank : 6) | θ value | 7.25919e-50 (rank : 6) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P19419, O75606, O95058, Q969X8, Q9UJM4 | Gene names | ELK1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-1. | |||||
|
ETV2_HUMAN
|
||||||
| NC score | 0.890583 (rank : 7) | θ value | 2.06002e-20 (rank : 27) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O00321, Q9UEA0 | Gene names | ETV2, ER71, ETSRP71 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 2 (Ets-related protein 71). | |||||
|
ETV2_MOUSE
|
||||||
| NC score | 0.888254 (rank : 8) | θ value | 2.06002e-20 (rank : 28) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P41163 | Gene names | Etv2, Er71, Etsrp71 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 2 (Ets-related protein 71). | |||||
|
GABPA_MOUSE
|
||||||
| NC score | 0.878589 (rank : 9) | θ value | 4.9032e-22 (rank : 20) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q00422 | Gene names | Gabpa, E4tf1a | |||
|
Domain Architecture |

|
|||||
| Description | GA-binding protein alpha chain (GABP-subunit alpha). | |||||
|
GABPA_HUMAN
|
||||||
| NC score | 0.877550 (rank : 10) | θ value | 2.20094e-22 (rank : 17) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q06546, Q12939 | Gene names | GABPA, E4TF1A | |||
|
Domain Architecture |

|
|||||
| Description | GA-binding protein alpha chain (GABP-subunit alpha) (Transcription factor E4TF1-60) (Nuclear respiratory factor 2 subunit alpha). | |||||
|
ETS2_HUMAN
|
||||||
| NC score | 0.871973 (rank : 11) | θ value | 3.75424e-22 (rank : 18) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P15036 | Gene names | ETS2 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-2 protein. | |||||
|
ETS1_MOUSE
|
||||||
| NC score | 0.871069 (rank : 12) | θ value | 1.42661e-21 (rank : 24) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P27577, Q61403 | Gene names | Ets1, Ets-1 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-1 protein (p54). | |||||
|
ETS1_HUMAN
|
||||||
| NC score | 0.871028 (rank : 13) | θ value | 1.42661e-21 (rank : 23) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P14921, Q14278, Q16080 | Gene names | ETS1 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-1 protein (p54). | |||||
|
ETS2_MOUSE
|
||||||
| NC score | 0.870902 (rank : 14) | θ value | 3.75424e-22 (rank : 19) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P15037 | Gene names | Ets2 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-2 protein. | |||||
|
FLI1_MOUSE
|
||||||
| NC score | 0.867836 (rank : 15) | θ value | 1.99067e-23 (rank : 16) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P26323 | Gene names | Fli1, Fli-1 | |||
|
Domain Architecture |

|
|||||
| Description | Friend leukemia integration 1 transcription factor (Retroviral integration site protein Fli-1). | |||||
|
FLI1_HUMAN
|
||||||
| NC score | 0.866528 (rank : 16) | θ value | 1.99067e-23 (rank : 15) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q01543, Q14319, Q92480, Q9UE07 | Gene names | FLI1 | |||
|
Domain Architecture |

|
|||||
| Description | Friend leukemia integration 1 transcription factor (Fli-1 proto- oncogene) (ERGB transcription factor). | |||||
|
ETV1_MOUSE
|
||||||
| NC score | 0.863745 (rank : 17) | θ value | 1.47289e-26 (rank : 10) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P41164 | Gene names | Etv1, Er81, Etsrp81 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 1 (ER81 protein). | |||||
|
ETV1_HUMAN
|
||||||
| NC score | 0.862539 (rank : 18) | θ value | 1.47289e-26 (rank : 9) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P50549, O75849, Q9UQ71, Q9Y636 | Gene names | ETV1, ER81 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 1 (ER81 protein). | |||||
|
ERG_HUMAN
|
||||||
| NC score | 0.860759 (rank : 19) | θ value | 1.16704e-23 (rank : 14) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P11308, Q16113 | Gene names | ERG | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ERG (Transforming protein ERG). | |||||
|
ETV4_MOUSE
|
||||||
| NC score | 0.860734 (rank : 20) | θ value | 1.73987e-27 (rank : 8) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P28322 | Gene names | Etv4, Pea-3, Pea3 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 4 (Polyomavirus enhancer activator 3) (Protein PEA3). | |||||
|
ERG_MOUSE
|
||||||
| NC score | 0.860650 (rank : 21) | θ value | 6.84181e-24 (rank : 13) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P81270, Q8C5L4, Q920K7, Q920K8, Q920K9 | Gene names | Erg, Erg-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ERG. | |||||
|
ETV4_HUMAN
|
||||||
| NC score | 0.860241 (rank : 22) | θ value | 1.73987e-27 (rank : 7) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P43268, Q96AW9 | Gene names | ETV4, E1AF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 4 (Adenovirus E1A enhancer-binding protein) (E1A-F). | |||||
|
ETV5_HUMAN
|
||||||
| NC score | 0.858141 (rank : 23) | θ value | 5.59698e-26 (rank : 11) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P41161 | Gene names | ETV5, ERM | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 5 (Ets-related protein ERM). | |||||
|
ETV7_HUMAN
|
||||||
| NC score | 0.856920 (rank : 24) | θ value | 2.20605e-14 (rank : 39) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9Y603, Q9NZ65, Q9NZ66, Q9NZ68, Q9NZR8, Q9UNJ7, Q9Y5K4, Q9Y604 | Gene names | ETV7, TEL2, TELB, TREF | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV7 (Transcription factor Tel-2) (ETS-related protein Tel2) (Tel-related Ets factor). | |||||
|
ERF_MOUSE
|
||||||
| NC score | 0.856355 (rank : 25) | θ value | 1.09232e-21 (rank : 22) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P70459 | Gene names | Erf | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing transcription factor ERF. | |||||
|
ERF_HUMAN
|
||||||
| NC score | 0.855993 (rank : 26) | θ value | 1.09232e-21 (rank : 21) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P50548, Q9UPI7 | Gene names | ERF | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing transcription factor ERF (Ets2 repressor factor). | |||||
|
SPDEF_HUMAN
|
||||||
| NC score | 0.855434 (rank : 27) | θ value | 8.10077e-17 (rank : 35) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O95238 | Gene names | SPDEF, PDEF, PSE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAM pointed domain-containing Ets transcription factor (Prostate- derived Ets factor) (Prostate epithelium-specific Ets transcription factor) (Prostate-specific Ets). | |||||
|
SPDEF_MOUSE
|
||||||
| NC score | 0.853688 (rank : 28) | θ value | 1.058e-16 (rank : 36) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9WTP3 | Gene names | Spdef, Pdef, Pse | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAM pointed domain-containing Ets transcription factor (Prostate- derived Ets factor) (Prostate epithelium-specific Ets transcription factor) (Prostate-specific Ets). | |||||
|
ETV3_MOUSE
|
||||||
| NC score | 0.852552 (rank : 29) | θ value | 1.86321e-21 (rank : 26) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8R4Z4, Q9QZW1 | Gene names | Etv3, Mets, Pe1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
ETV5_MOUSE
|
||||||
| NC score | 0.839667 (rank : 30) | θ value | 5.59698e-26 (rank : 12) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9CXC9, Q3TG49, Q8C0F3, Q9JHB1 | Gene names | Etv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 5. | |||||
|
ETV3_HUMAN
|
||||||
| NC score | 0.833303 (rank : 31) | θ value | 1.86321e-21 (rank : 25) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P41162, Q8TAC8, Q9BX30 | Gene names | ETV3, METS, PE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
ELF2_MOUSE
|
||||||
| NC score | 0.828239 (rank : 32) | θ value | 3.63628e-17 (rank : 34) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9JHC9, Q6NST2, Q8BTX8, Q9JHC7, Q9JHC8, Q9JHD0 | Gene names | Elf2, Nerf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-2 (E74-like factor 2) (New ETS- related factor). | |||||
|
ELF2_HUMAN
|
||||||
| NC score | 0.826440 (rank : 33) | θ value | 3.63628e-17 (rank : 33) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q15723, Q15724, Q15725, Q6P1K5 | Gene names | ELF2, NERF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-2 (E74-like factor 2) (New ETS- related factor). | |||||
|
ELF4_HUMAN
|
||||||
| NC score | 0.825927 (rank : 34) | θ value | 1.24977e-17 (rank : 31) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q99607, O60435 | Gene names | ELF4, ELFR, MEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-4 (E74-like factor 4) (Myeloid Elf-1-like factor). | |||||
|
ELF1_HUMAN
|
||||||
| NC score | 0.825566 (rank : 35) | θ value | 7.32683e-18 (rank : 30) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P32519, Q9UDE1 | Gene names | ELF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
ELF1_MOUSE
|
||||||
| NC score | 0.821536 (rank : 36) | θ value | 2.27762e-19 (rank : 29) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q60775 | Gene names | Elf1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
ETV6_MOUSE
|
||||||
| NC score | 0.818083 (rank : 37) | θ value | 2.20605e-14 (rank : 38) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P97360 | Gene names | Etv6, Tel, Tel1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV6 (ETS-related protein Tel1) (Tel) (ETS translocation variant 6). | |||||
|
ETV6_HUMAN
|
||||||
| NC score | 0.816083 (rank : 38) | θ value | 2.20605e-14 (rank : 37) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P41212, Q9UMF6 | Gene names | ETV6, TEL, TEL1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV6 (ETS-related protein Tel1) (Tel) (ETS translocation variant 6). | |||||
|
ELF5_MOUSE
|
||||||
| NC score | 0.810465 (rank : 39) | θ value | 1.74796e-11 (rank : 41) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8VDK3, Q921H5, Q9Z2K6 | Gene names | Elf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-5 (E74-like factor 5). | |||||
|
ELF5_HUMAN
|
||||||
| NC score | 0.809226 (rank : 40) | θ value | 1.74796e-11 (rank : 40) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UKW6, O95175, Q8N2K9, Q96QY3, Q9UKW5 | Gene names | ELF5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-5 (E74-like factor 5) (Epithelium-specific Ets transcription factor 2) (ESE-2) (Epithelium- restricted ESE-1-related Ets factor). | |||||
|
ELF4_MOUSE
|
||||||
| NC score | 0.807368 (rank : 41) | θ value | 1.24977e-17 (rank : 32) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9Z2U4 | Gene names | Elf4, Mef | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-4 (E74-like factor 4) (Myeloid Elf-1-like factor). | |||||
|
SPI1_MOUSE
|
||||||
| NC score | 0.643854 (rank : 42) | θ value | 1.17247e-07 (rank : 43) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P17433, Q99L57 | Gene names | Spi1, Sfpi-1, Sfpi1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor PU.1 (31 kDa transforming protein) (SFFV proviral integration 1 protein). | |||||
|
SPI1_HUMAN
|
||||||
| NC score | 0.642632 (rank : 43) | θ value | 1.17247e-07 (rank : 42) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P17947 | Gene names | SPI1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor PU.1 (31 kDa transforming protein). | |||||
|
SPIB_HUMAN
|
||||||
| NC score | 0.632105 (rank : 44) | θ value | 8.40245e-06 (rank : 44) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q01892, Q15359 | Gene names | SPIB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Spi-B. | |||||
|
SPIB_MOUSE
|
||||||
| NC score | 0.613341 (rank : 45) | θ value | 1.09739e-05 (rank : 45) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O35906, O35907, O35909, O55199 | Gene names | Spib | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-B. | |||||
|
SPIC_HUMAN
|
||||||
| NC score | 0.584763 (rank : 46) | θ value | 0.000121331 (rank : 46) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8N5J4 | Gene names | SPIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-C. | |||||
|
SPIC_MOUSE
|
||||||
| NC score | 0.548290 (rank : 47) | θ value | 0.0148317 (rank : 50) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6P3D7, Q9Z0Y0 | Gene names | Spic | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-C (Pu.1-related factor) (Prf). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.078027 (rank : 48) | θ value | 0.365318 (rank : 68) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.077040 (rank : 49) | θ value | 0.0736092 (rank : 55) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 112 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
CK024_HUMAN
|
||||||
| NC score | 0.072229 (rank : 50) | θ value | 0.62314 (rank : 74) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
MAP4_HUMAN
|
||||||
| NC score | 0.066929 (rank : 51) | θ value | 0.00665767 (rank : 47) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.064464 (rank : 52) | θ value | 1.38821 (rank : 94) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.064076 (rank : 53) | θ value | 2.36792 (rank : 105) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.062659 (rank : 54) | θ value | 3.0926 (rank : 116) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.061915 (rank : 55) | θ value | 0.0563607 (rank : 54) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.058507 (rank : 56) | θ value | 0.365318 (rank : 65) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.055917 (rank : 57) | θ value | 0.47712 (rank : 71) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
MAP4_MOUSE
|
||||||
| NC score | 0.054552 (rank : 58) | θ value | 2.36792 (rank : 106) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.050557 (rank : 59) | θ value | 5.27518 (rank : 141) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.049206 (rank : 60) | θ value | 0.813845 (rank : 83) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
SF3A1_HUMAN
|
||||||
| NC score | 0.048396 (rank : 61) | θ value | 0.21417 (rank : 61) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15459 | Gene names | SF3A1, SAP114 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3 subunit 1 (Spliceosome-associated protein 114) (SAP 114) (SF3a120). | |||||
|
SF3A1_MOUSE
|
||||||
| NC score | 0.048084 (rank : 62) | θ value | 0.163984 (rank : 59) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8K4Z5, Q8C0M7, Q8C128, Q8C175, Q921T3 | Gene names | Sf3a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 3 subunit 1 (SF3a120). | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.046436 (rank : 63) | θ value | 0.0330416 (rank : 52) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.046113 (rank : 64) | θ value | 8.99809 (rank : 179) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
RAPH1_HUMAN
|
||||||
| NC score | 0.045467 (rank : 65) | θ value | 0.813845 (rank : 84) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
SYNJ1_MOUSE
|
||||||
| NC score | 0.044870 (rank : 66) | θ value | 6.88961 (rank : 161) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.044562 (rank : 67) | θ value | 0.0193708 (rank : 51) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.041962 (rank : 68) | θ value | 3.0926 (rank : 119) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.040472 (rank : 69) | θ value | 3.0926 (rank : 117) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
MLL4_HUMAN
|
||||||
| NC score | 0.039966 (rank : 70) | θ value | 0.125558 (rank : 58) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
IF4G2_MOUSE
|
||||||
| NC score | 0.039805 (rank : 71) | θ value | 0.125558 (rank : 57) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62448, P97867, Q921U3 | Gene names | Eif4g2, Nat1 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 2 (eIF-4-gamma 2) (eIF-4G 2) (eIF4G 2) (p97) (Novel APOBEC-1 target 1) (Translation repressor NAT1). | |||||
|
IF4G2_HUMAN
|
||||||
| NC score | 0.039802 (rank : 72) | θ value | 0.125558 (rank : 56) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P78344, O60877, P78404, Q53EU1, Q58EZ2, Q8NI71, Q96C16 | Gene names | EIF4G2, DAP5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 2 (eIF-4-gamma 2) (eIF-4G 2) (eIF4G 2) (p97) (Death-associated protein 5) (DAP-5). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.039603 (rank : 73) | θ value | 4.03905 (rank : 130) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
ZAN_HUMAN
|
||||||
| NC score | 0.039397 (rank : 74) | θ value | 0.00869519 (rank : 49) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
SYNJ1_HUMAN
|
||||||
| NC score | 0.038852 (rank : 75) | θ value | 0.813845 (rank : 86) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O43426, O43425, O94984 | Gene names | SYNJ1, KIAA0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
SF3B2_HUMAN
|
||||||
| NC score | 0.037357 (rank : 76) | θ value | 0.365318 (rank : 67) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13435 | Gene names | SF3B2, SAP145 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 2 (Spliceosome-associated protein 145) (SAP 145) (SF3b150) (Pre-mRNA-splicing factor SF3b 145 kDa subunit). | |||||
|
MNT_HUMAN
|
||||||
| NC score | 0.036489 (rank : 77) | θ value | 0.365318 (rank : 64) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.036111 (rank : 78) | θ value | 8.99809 (rank : 174) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
LAT_MOUSE
|
||||||
| NC score | 0.035784 (rank : 79) | θ value | 3.0926 (rank : 115) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O54957 | Gene names | Lat | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Linker for activation of T-cells family member 1 (36 kDa phospho- tyrosine adapter protein) (pp36) (p36-38). | |||||
|
RBM12_MOUSE
|
||||||
| NC score | 0.035100 (rank : 80) | θ value | 1.38821 (rank : 95) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8R4X3, Q8K373, Q8R302, Q9CS80 | Gene names | Rbm12 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN). | |||||
|
MAML2_HUMAN
|
||||||
| NC score | 0.034593 (rank : 81) | θ value | 0.62314 (rank : 76) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8IZL2, Q6AI23, Q6Y3A3, Q8IUL3, Q96JK6 | Gene names | MAML2, KIAA1819 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 2 (Mam-2). | |||||
|
CS016_HUMAN
|
||||||
| NC score | 0.034457 (rank : 82) | θ value | 8.99809 (rank : 166) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8ND99 | Gene names | C19orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf16. | |||||
|
RBM12_HUMAN
|
||||||
| NC score | 0.033202 (rank : 83) | θ value | 0.365318 (rank : 66) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NTZ6, O94865, Q8N3B1, Q9H196 | Gene names | RBM12, KIAA0765 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.033098 (rank : 84) | θ value | 8.99809 (rank : 177) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
GTSE1_HUMAN
|
||||||
| NC score | 0.032576 (rank : 85) | θ value | 5.27518 (rank : 134) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NYZ3, Q53GX5, Q5R3I6, Q6DHX4, Q9BRE0, Q9UGZ9, Q9Y557 | Gene names | GTSE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G2 and S phase expressed protein 1 (B99 homolog). | |||||
|
LR37A_HUMAN
|
||||||
| NC score | 0.032340 (rank : 86) | θ value | 6.88961 (rank : 154) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O60309, Q49A01, Q49A80, Q8NB33 | Gene names | LRRC37A, KIAA0563 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37A. | |||||
|
SRBP1_MOUSE
|
||||||
| NC score | 0.032255 (rank : 87) | θ value | 3.0926 (rank : 120) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
BRD4_MOUSE
|
||||||
| NC score | 0.032142 (rank : 88) | θ value | 6.88961 (rank : 146) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.032100 (rank : 89) | θ value | 0.813845 (rank : 85) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
ZIM10_MOUSE
|
||||||
| NC score | 0.031992 (rank : 90) | θ value | 1.38821 (rank : 97) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6P1E1, Q6PDK9, Q6PF85, Q8BW47 | Gene names | Rai17, Zimp10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
BRD4_HUMAN
|
||||||
| NC score | 0.031356 (rank : 91) | θ value | 6.88961 (rank : 145) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
CS007_MOUSE
|
||||||
| NC score | 0.030997 (rank : 92) | θ value | 1.81305 (rank : 100) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
GGN_MOUSE
|
||||||
| NC score | 0.030580 (rank : 93) | θ value | 4.03905 (rank : 128) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q80WJ1, Q5EBP4, Q80WI9, Q80WJ0 | Gene names | Ggn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
YLPM1_MOUSE
|
||||||
| NC score | 0.030485 (rank : 94) | θ value | 8.99809 (rank : 184) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
K1210_HUMAN
|
||||||
| NC score | 0.030394 (rank : 95) | θ value | 5.27518 (rank : 137) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9ULL0, Q5JPN4 | Gene names | KIAA1210 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1210. | |||||
|
PO121_HUMAN
|
||||||
| NC score | 0.030355 (rank : 96) | θ value | 8.99809 (rank : 178) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
ZIM10_HUMAN
|
||||||
| NC score | 0.030268 (rank : 97) | θ value | 1.38821 (rank : 96) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
PHC3_HUMAN
|
||||||
| NC score | 0.029681 (rank : 98) | θ value | 2.36792 (rank : 107) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8NDX5, Q5HYF0, Q6NSG2, Q8NFT7, Q8NFZ1, Q8TBM2, Q9H971, Q9H9I4 | Gene names | PHC3, EDR3, PH3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 3 (hPH3) (Homolog of polyhomeotic 3) (Early development regulatory protein 3). | |||||
|
GP112_HUMAN
|
||||||
| NC score | 0.029439 (rank : 99) | θ value | 0.365318 (rank : 63) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8IZF6, Q86SM6 | Gene names | GPR112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 112. | |||||
|
GGA3_MOUSE
|
||||||
| NC score | 0.029029 (rank : 100) | θ value | 0.21417 (rank : 60) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BMI3, Q80U71 | Gene names | Gga3, Kiaa0154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
MILK1_MOUSE
|
||||||
| NC score | 0.028931 (rank : 101) | θ value | 0.47712 (rank : 70) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.028826 (rank : 102) | θ value | 0.62314 (rank : 78) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.028041 (rank : 103) | θ value | 0.813845 (rank : 87) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
K1802_HUMAN
|
||||||
| NC score | 0.027934 (rank : 104) | θ value | 8.99809 (rank : 172) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.027824 (rank : 105) | θ value | 1.06291 (rank : 89) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
CS007_HUMAN
|
||||||
| NC score | 0.027313 (rank : 106) | θ value | 1.81305 (rank : 99) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
GGA3_HUMAN
|
||||||
| NC score | 0.027263 (rank : 107) | θ value | 0.813845 (rank : 81) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NZ52, Q15017, Q6IS16, Q9UJY3 | Gene names | GGA3, KIAA0154 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
NELFA_MOUSE
|
||||||
| NC score | 0.026696 (rank : 108) | θ value | 8.99809 (rank : 175) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BG30, Q8BVE4, Q8VEI7, Q9CSJ9, Q9Z1V9 | Gene names | Whsc2, Nelfa, Whsc2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Negative elongation factor A (NELF-A) (Wolf-Hirschhorn syndrome candidate 2 homolog) (mWHSC2). | |||||
|
PDC6I_HUMAN
|
||||||
| NC score | 0.026241 (rank : 109) | θ value | 4.03905 (rank : 131) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8WUM4, Q9BX86, Q9NUN0, Q9P2H2, Q9UKL5 | Gene names | PDCD6IP, AIP1, ALIX, KIAA1375 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death 6-interacting protein (PDCD6-interacting protein) (ALG-2-interacting protein 1) (Hp95). | |||||
|
SNPC4_MOUSE
|
||||||
| NC score | 0.025950 (rank : 110) | θ value | 0.62314 (rank : 80) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BP86, Q6PGG7, Q80UG9, Q810L1 | Gene names | Snapc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit). | |||||
|
MBD5_HUMAN
|
||||||
| NC score | 0.025886 (rank : 111) | θ value | 1.81305 (rank : 102) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9P267, Q9NUV6 | Gene names | MBD5, KIAA1461 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 5 (Methyl-CpG-binding protein MBD5). | |||||
|
HRX_HUMAN
|
||||||
| NC score | 0.024759 (rank : 112) | θ value | 8.99809 (rank : 170) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
RERE_HUMAN
|
||||||
| NC score | 0.024665 (rank : 113) | θ value | 8.99809 (rank : 180) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
NCOR2_MOUSE
|
||||||
| NC score | 0.024185 (rank : 114) | θ value | 0.813845 (rank : 82) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
EXO1_MOUSE
|
||||||
| NC score | 0.024173 (rank : 115) | θ value | 0.365318 (rank : 62) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QZ11, Q3TLM4, Q923A5 | Gene names | Exo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exonuclease 1 (EC 3.1.-.-) (mExo1) (Exonuclease I). | |||||
|
BPTF_HUMAN
|
||||||
| NC score | 0.023678 (rank : 116) | θ value | 1.81305 (rank : 98) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
DGKK_HUMAN
|
||||||
| NC score | 0.023658 (rank : 117) | θ value | 6.88961 (rank : 150) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5KSL6 | Gene names | DGKK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase kappa (EC 2.7.1.107) (Diglyceride kinase kappa) (DGK-kappa) (DAG kinase kappa) (142 kDa diacylglycerol kinase). | |||||
|
PDXL2_HUMAN
|
||||||
| NC score | 0.023020 (rank : 118) | θ value | 1.06291 (rank : 90) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NZ53, Q6UVY4, Q8WUV6 | Gene names | PODXL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Podocalyxin-like protein 2 precursor (Endoglycan). | |||||
|
PCQAP_HUMAN
|
||||||
| NC score | 0.022606 (rank : 119) | θ value | 6.88961 (rank : 158) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96RN5, O15413, Q6IC31, Q8NF16, Q96CT0, Q96IH7, Q9P1T3 | Gene names | PCQAP, ARC105, CTG7A, TIG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (TPA-inducible gene 1 protein) (TIG-1) (Activator-recruited cofactor 105 kDa component) (ARC105) (CTG repeat protein 7a). | |||||
|
PER3_HUMAN
|
||||||
| NC score | 0.022391 (rank : 120) | θ value | 4.03905 (rank : 132) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
SETBP_MOUSE
|
||||||
| NC score | 0.022090 (rank : 121) | θ value | 6.88961 (rank : 159) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Z180, Q66JL8 | Gene names | Setbp1, Kiaa0437 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET-binding protein (SEB). | |||||
|
SFPQ_HUMAN
|
||||||
| NC score | 0.021604 (rank : 122) | θ value | 4.03905 (rank : 133) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
LAT_HUMAN
|
||||||
| NC score | 0.020989 (rank : 123) | θ value | 5.27518 (rank : 140) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43561, O43919 | Gene names | LAT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Linker for activation of T-cells family member 1 (36 kDa phospho- tyrosine adapter protein) (pp36) (p36-38). | |||||
|
EFS_HUMAN
|
||||||
| NC score | 0.020124 (rank : 124) | θ value | 4.03905 (rank : 125) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O43281, O43282 | Gene names | EFS | |||
|
Domain Architecture |

|
|||||
| Description | Embryonal Fyn-associated substrate (HEFS). | |||||
|
REPS1_MOUSE
|
||||||
| NC score | 0.019452 (rank : 125) | θ value | 1.81305 (rank : 104) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O54916, Q8C9J9, Q99LR8 | Gene names | Reps1 | |||
|
Domain Architecture |

|
|||||
| Description | RalBP1-associated Eps domain-containing protein 1 (RalBP1-interacting protein 1). | |||||
|
REPS1_HUMAN
|
||||||
| NC score | 0.019239 (rank : 126) | θ value | 1.81305 (rank : 103) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96D71, Q8NDR7, Q8WU62, Q9BXY9 | Gene names | REPS1 | |||
|
Domain Architecture |

|
|||||
| Description | RalBP1-associated Eps domain-containing protein 1 (RalBP1-interacting protein 1). | |||||
|
ATF6A_HUMAN
|
||||||
| NC score | 0.018135 (rank : 127) | θ value | 4.03905 (rank : 123) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P18850, O15139, Q5VW62, Q6IPB5, Q9UEC9 | Gene names | ATF6 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-6 alpha (Activating transcription factor 6 alpha) (ATF6-alpha). | |||||
|
PRDM2_HUMAN
|
||||||
| NC score | 0.018010 (rank : 128) | θ value | 0.00869519 (rank : 48) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
SRBS1_HUMAN
|
||||||
| NC score | 0.017351 (rank : 129) | θ value | 1.06291 (rank : 91) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9BX66, Q7LBE5, Q8IVK0, Q8IVQ4, Q96KF3, Q96KF4, Q9BX64, Q9BX65, Q9P2Q0, Q9UFT2, Q9UHN7, Q9Y338 | Gene names | SORBS1, KIAA1296, SH3D5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorbin and SH3 domain-containing protein 1 (Ponsin) (c-Cbl-associated protein) (CAP) (SH3 domain protein 5) (SH3P12). | |||||
|
ANTR1_MOUSE
|
||||||
| NC score | 0.016984 (rank : 130) | θ value | 3.0926 (rank : 110) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9CZ52 | Gene names | Antxr1, Atr, Tem8 | |||
|
Domain Architecture |

|
|||||
| Description | Anthrax toxin receptor 1 precursor (Tumor endothelial marker 8). | |||||
|
CN037_HUMAN
|
||||||
| NC score | 0.016680 (rank : 131) | θ value | 8.99809 (rank : 165) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q86TY3, Q6P5Q1, Q86TY1 | Gene names | C14orf37 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf37 precursor. | |||||
|
GNDS_MOUSE
|
||||||
| NC score | 0.016636 (rank : 132) | θ value | 1.81305 (rank : 101) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q03385 | Gene names | Ralgds, Rgds | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator (RalGEF) (RalGDS). | |||||
|
LDB3_HUMAN
|
||||||
| NC score | 0.016273 (rank : 133) | θ value | 0.62314 (rank : 75) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O75112, Q5K6N9, Q5K6P0, Q5K6P1, Q96FH2, Q9Y4Z3, Q9Y4Z4, Q9Y4Z5 | Gene names | LDB3, KIAA0613, ZASP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher). | |||||
|
CD2AP_HUMAN
|
||||||
| NC score | 0.016217 (rank : 134) | θ value | 3.0926 (rank : 112) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y5K6, Q9UG97 | Gene names | CD2AP | |||
|
Domain Architecture |

|
|||||
| Description | CD2-associated protein (Cas ligand with multiple SH3 domains) (Adapter protein CMS). | |||||
|
CD2AP_MOUSE
|
||||||
| NC score | 0.014197 (rank : 135) | θ value | 1.38821 (rank : 92) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9JLQ0, O88903, Q8K4Z1, Q8VCI9 | Gene names | Cd2ap, Mets1 | |||
|
Domain Architecture |

|
|||||
| Description | CD2-associated protein (Mesenchyme-to-epithelium transition protein with SH3 domains 1) (METS-1). | |||||
|
MKL2_MOUSE
|
||||||
| NC score | 0.013980 (rank : 136) | θ value | 8.99809 (rank : 173) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P59759 | Gene names | Mkl2, Mrtfb | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 2 (Myocardin-related transcription factor B) (MRTF-B). | |||||
|
CASB_MOUSE
|
||||||
| NC score | 0.013631 (rank : 137) | θ value | 6.88961 (rank : 147) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P10598, Q543D9, Q8VCT6, Q8VCU8, Q91VI5, Q922Y5, Q9D1U6, Q9D1U7 | Gene names | Csn2, Csnb | |||
|
Domain Architecture |

|
|||||
| Description | Beta-casein precursor. | |||||
|
NFAC1_MOUSE
|
||||||
| NC score | 0.013189 (rank : 138) | θ value | 3.0926 (rank : 118) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O88942, O70345 | Gene names | Nfatc1, Nfat2, Nfatc | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 1 (NFAT transcription complex cytosolic component) (NF-ATc1) (NF-ATc). | |||||
|
SMDF_HUMAN
|
||||||
| NC score | 0.012732 (rank : 139) | θ value | 6.88961 (rank : 160) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15491 | Gene names | NRG1, GGF, HGL, HRGA, NDF, SMDF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuregulin-1, sensory and motor neuron-derived factor isoform. | |||||
|
TACC3_MOUSE
|
||||||
| NC score | 0.012595 (rank : 140) | θ value | 0.47712 (rank : 72) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JJ11, Q9WVK9 | Gene names | Tacc3, Aint | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ARNT-interacting protein). | |||||
|
K0195_HUMAN
|
||||||
| NC score | 0.012546 (rank : 141) | θ value | 3.0926 (rank : 114) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12767, O75536, Q86XF1 | Gene names | KIAA0195, TMEM94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0195 (Transmembrane protein 94). | |||||
|
UBP24_HUMAN
|
||||||
| NC score | 0.012012 (rank : 142) | θ value | 0.47712 (rank : 73) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UPU5, Q6ZSY2, Q8N2Y4, Q9NXD1 | Gene names | USP24, KIAA1057 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 24 (EC 3.1.2.15) (Ubiquitin thioesterase 24) (Ubiquitin-specific-processing protease 24) (Deubiquitinating enzyme 24). | |||||
|
IL3RB_HUMAN
|
||||||
| NC score | 0.011851 (rank : 143) | θ value | 6.88961 (rank : 153) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P32927 | Gene names | CSF2RB, IL3RB, IL5RB | |||
|
Domain Architecture |

|
|||||
| Description | Cytokine receptor common beta chain precursor (GM-CSF/IL-3/IL-5 receptor common beta-chain) (CD131 antigen) (CDw131). | |||||
|
NFAC4_HUMAN
|
||||||
| NC score | 0.011465 (rank : 144) | θ value | 6.88961 (rank : 157) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14934 | Gene names | NFATC4, NFAT3 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 4 (NF-ATc4) (NFATc4) (T cell transcription factor NFAT3) (NF-AT3). | |||||
|
DYSF_HUMAN
|
||||||
| NC score | 0.011354 (rank : 145) | θ value | 3.0926 (rank : 113) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O75923, O75696, Q9UEN7 | Gene names | DYSF, FER1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Dysferlin (Dystrophy-associated fer-1-like protein) (Fer-1-like protein 1). | |||||
|
NUD19_MOUSE
|
||||||
| NC score | 0.011167 (rank : 146) | θ value | 8.99809 (rank : 176) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P11930 | Gene names | Nudt19, D7Rp2e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoside diphosphate-linked moiety X motif 19 (EC 3.-.-.-) (Nudix motif 19) (Testosterone-regulated RP2 protein) (Androgen-regulated protein RP2). | |||||
|
MEGF9_HUMAN
|
||||||
| NC score | 0.010875 (rank : 147) | θ value | 0.62314 (rank : 77) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9H1U4, O75098 | Gene names | MEGF9, EGFL5, KIAA0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
DYSF_MOUSE
|
||||||
| NC score | 0.010474 (rank : 148) | θ value | 6.88961 (rank : 151) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ESD7, Q9QXC0 | Gene names | Dysf, Fer1l1 | |||
|
Domain Architecture |

|
|||||
| Description | Dysferlin (Dystrophy-associated fer-1-like protein) (Fer-1-like protein 1). | |||||
|
FBLN2_HUMAN
|
||||||
| NC score | 0.010155 (rank : 149) | θ value | 0.0431538 (rank : 53) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P98095 | Gene names | FBLN2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-2 precursor. | |||||
|
ELL_MOUSE
|
||||||
| NC score | 0.009689 (rank : 150) | θ value | 6.88961 (rank : 152) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O08856 | Gene names | Ell | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II elongation factor ELL (Eleven-nineteen lysine-rich leukemia protein). | |||||
|
ATBF1_MOUSE
|
||||||
| NC score | 0.009249 (rank : 151) | θ value | 6.88961 (rank : 144) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
CSPG2_MOUSE
|
||||||
| NC score | 0.008885 (rank : 152) | θ value | 4.03905 (rank : 124) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q62059, Q62058, Q9CUU0 | Gene names | Cspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M). | |||||
|
ANR11_HUMAN
|
||||||
| NC score | 0.008814 (rank : 153) | θ value | 3.0926 (rank : 109) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
UBQL3_HUMAN
|
||||||
| NC score | 0.008231 (rank : 154) | θ value | 8.99809 (rank : 183) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9H347, Q9NRE0 | Gene names | UBQLN3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-3. | |||||
|
TREF1_HUMAN
|
||||||
| NC score | 0.008105 (rank : 155) | θ value | 8.99809 (rank : 181) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96PN7, Q7Z6T2, Q7Z6T3, Q9NQ72, Q9NQ73, Q9NUN9 | Gene names | TRERF1, RAPA, TREP132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132) (Zinc finger protein rapa). | |||||
|
FIP1_HUMAN
|
||||||
| NC score | 0.008097 (rank : 156) | θ value | 8.99809 (rank : 168) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6UN15, Q499Y4, Q49AU3, Q7Z608, Q8WVN3, Q96F80, Q9H077 | Gene names | FIP1L1, FIP1, RHE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA 3'-end-processing factor FIP1 (FIP1-like 1) (Factor interacting with PAP) (hFip1) (Rearranged in hypereosinophilia). | |||||
|
GRB10_HUMAN
|
||||||
| NC score | 0.008097 (rank : 157) | θ value | 4.03905 (rank : 129) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13322, O00427, O00701, O75222, Q92606, Q92907, Q92948 | Gene names | GRB10, GRBIR, KIAA0207 | |||
|
Domain Architecture |

|
|||||
| Description | Growth factor receptor-bound protein 10 (GRB10 adaptor protein) (Insulin receptor-binding protein GRB-IR). | |||||
|
ZN638_HUMAN
|
||||||
| NC score | 0.008032 (rank : 158) | θ value | 6.88961 (rank : 162) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q14966, Q53R34, Q5XJ05, Q68DP3, Q6P2H2, Q7Z3T7, Q8NF92, Q8TCA1, Q9H2G1, Q9NP37 | Gene names | ZNF638, NP220, ZFML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein) (Cutaneous T-cell lymphoma-associated antigen se33-1) (CTCL tumor antigen se33-1). | |||||
|
WNK2_HUMAN
|
||||||
| NC score | 0.007596 (rank : 159) | θ value | 5.27518 (rank : 143) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
UBP10_MOUSE
|
||||||
| NC score | 0.006305 (rank : 160) | θ value | 8.99809 (rank : 182) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P52479, Q91VY7 | Gene names | Usp10, Ode-1, Uchrp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
ANKY1_HUMAN
|
||||||
| NC score | 0.006249 (rank : 161) | θ value | 8.99809 (rank : 163) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9P2S6, Q8IYX5, Q8NDK5, Q9NX10 | Gene names | ANKMY1, TSAL1, ZMYND13 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and MYND domain-containing protein 1 (Testis-specific ankyrin-like protein 1) (Zinc-finger MYND domain-containing protein 13). | |||||
|
ATBF1_HUMAN
|
||||||
| NC score | 0.005884 (rank : 162) | θ value | 4.03905 (rank : 122) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
JPH1_HUMAN
|
||||||
| NC score | 0.005403 (rank : 163) | θ value | 8.99809 (rank : 171) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9HDC5 | Gene names | JPH1, JP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-1 (Junctophilin type 1) (JP-1). | |||||
|
ELAV2_MOUSE
|
||||||
| NC score | 0.005274 (rank : 164) | θ value | 4.03905 (rank : 127) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60899 | Gene names | Elavl2, Hub | |||
|
Domain Architecture |

|
|||||
| Description | ELAV-like protein 2 (Hu-antigen B) (HuB) (ELAV-like neuronal protein 1) (Nervous system-specific RNA-binding protein Mel-N1). | |||||
|
ELAV2_HUMAN
|
||||||
| NC score | 0.005268 (rank : 165) | θ value | 4.03905 (rank : 126) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12926, Q13235, Q59G15, Q8NEM4, Q9H1Q8 | Gene names | ELAVL2, HUB | |||
|
Domain Architecture |

|
|||||
| Description | ELAV-like protein 2 (Hu-antigen B) (HuB) (ELAV-like neuronal protein 1) (Nervous system-specific RNA-binding protein Hel-N1). | |||||
|
PHLB2_HUMAN
|
||||||
| NC score | 0.004130 (rank : 166) | θ value | 2.36792 (rank : 108) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q86SQ0, Q59EA8, Q68CY3, Q6NT98, Q8N8U8, Q8NAB1, Q8NCU5 | Gene names | PHLDB2, LL5B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
CEND3_HUMAN
|
||||||
| NC score | 0.003424 (rank : 167) | θ value | 6.88961 (rank : 148) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8WWN8 | Gene names | CENTD3, ARAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3). | |||||
|
ATS12_HUMAN
|
||||||
| NC score | 0.003071 (rank : 168) | θ value | 3.0926 (rank : 111) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P58397, Q6UWL3 | Gene names | ADAMTS12 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 12) (ADAM-TS 12) (ADAM-TS12). | |||||
|
KIF2C_HUMAN
|
||||||
| NC score | 0.002385 (rank : 169) | θ value | 5.27518 (rank : 138) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99661, Q96C18, Q96HB8, Q9BWV8 | Gene names | KIF2C, KNSL6 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF2C (Mitotic centromere-associated kinesin) (MCAK) (Kinesin-like protein 6). | |||||
|
FGF5_MOUSE
|
||||||
| NC score | 0.002121 (rank : 170) | θ value | 8.99809 (rank : 167) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P15656, O88825 | Gene names | Fgf5, Fgf-5 | |||
|
Domain Architecture |

|
|||||
| Description | Fibroblast growth factor 5 precursor (FGF-5) (HBGF-5). | |||||
|
LAP2_MOUSE
|
||||||
| NC score | 0.002080 (rank : 171) | θ value | 1.38821 (rank : 93) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80TH2, Q8BQ14, Q8CE41, Q8K171, Q99JU3, Q9JI47 | Gene names | Erbb2ip, Erbin, Kiaa1225, Lap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP2 (Erbb2-interacting protein) (Erbin) (Densin-180-like protein). | |||||
|
SCYL2_MOUSE
|
||||||
| NC score | 0.001388 (rank : 172) | θ value | 5.27518 (rank : 142) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CFE4, Q3UT57, Q3UWU9, Q8K0M4 | Gene names | Scyl2, Cvak104, D10Ertd802e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCY1-like protein 2 (Coated vesicle-associated kinase of 104 kDa). | |||||
|
ACRO_HUMAN
|
||||||
| NC score | 0.001298 (rank : 173) | θ value | 4.03905 (rank : 121) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P10323, Q6ICK2 | Gene names | ACR, ACRS | |||
|
Domain Architecture |

|
|||||
| Description | Acrosin precursor (EC 3.4.21.10) [Contains: Acrosin light chain; Acrosin heavy chain]. | |||||
|
CD2L7_HUMAN
|
||||||
| NC score | 0.001294 (rank : 174) | θ value | 0.47712 (rank : 69) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9NYV4, O94978 | Gene names | CRKRS, CRK7, KIAA0904 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle 2-related protein kinase 7 (EC 2.7.11.22) (CDC2- related protein kinase 7) (Cdc2-related kinase, arginine/serine-rich) (CrkRS). | |||||
|
MK07_MOUSE
|
||||||
| NC score | 0.000629 (rank : 175) | θ value | 0.62314 (rank : 79) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1054 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9WVS8 | Gene names | Mapk7, Erk5 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase 7 (EC 2.7.11.24) (Extracellular signal-regulated kinase 5) (ERK-5) (BMK1 kinase). | |||||
|
HNF3G_HUMAN
|
||||||
| NC score | -0.000029 (rank : 176) | θ value | 8.99809 (rank : 169) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P55318, Q9UMW9 | Gene names | FOXA3, HNF3G, TCF3G | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 3-gamma (HNF-3G) (Forkhead box protein A3) (Fork head-related protein FKH H3). | |||||
|
CELR3_HUMAN
|
||||||
| NC score | -0.000945 (rank : 177) | θ value | 8.99809 (rank : 164) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 680 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NYQ7, O75092 | Gene names | CELSR3, CDHF11, EGFL1, FMI1, KIAA0812, MEGF2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 3 precursor (Flamingo homolog 1) (hFmi1) (Multiple epidermal growth factor-like domains 2) (Epidermal growth factor-like 1). | |||||
|
ZBT12_HUMAN
|
||||||
| NC score | -0.001758 (rank : 178) | θ value | 0.813845 (rank : 88) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y330, Q5JQ98 | Gene names | ZBTB12, C6orf46, G10, NG35 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 12 (Protein G10). | |||||
|
CORIN_MOUSE
|
||||||
| NC score | -0.002060 (rank : 179) | θ value | 6.88961 (rank : 149) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z319 | Gene names | Corin, Crn, Lrp4 | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuteric peptide-converting enzyme (EC 3.4.21.-) (pro-ANP- converting enzyme) (Corin) (Low density lipoprotein receptor-related protein 4). | |||||
|
LATS1_MOUSE
|
||||||
| NC score | -0.002236 (rank : 180) | θ value | 5.27518 (rank : 139) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1027 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BYR2, Q9Z0W4 | Gene names | Lats1, Warts | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS1 (EC 2.7.11.1) (Large tumor suppressor homolog 1) (WARTS protein kinase). | |||||
|
HELI_MOUSE
|
||||||
| NC score | -0.002873 (rank : 181) | θ value | 5.27518 (rank : 136) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 746 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P81183 | Gene names | Ikzf2, Helios, Zfpn1a2, Znfn1a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein Helios (IKAROS family zinc finger protein 2). | |||||
|
HELI_HUMAN
|
||||||
| NC score | -0.002890 (rank : 182) | θ value | 5.27518 (rank : 135) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UKS7, Q8N6S1 | Gene names | IKZF2, HELIOS, ZNFN1A2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein Helios (IKAROS family zinc finger protein 2). | |||||
|
MAST3_HUMAN
|
||||||
| NC score | -0.003449 (rank : 183) | θ value | 6.88961 (rank : 156) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1128 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O60307, Q7LDZ8, Q9UPI0 | Gene names | MAST3, KIAA0561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 3 (EC 2.7.11.1). | |||||
|
M3K10_HUMAN
|
||||||
| NC score | -0.004232 (rank : 184) | θ value | 6.88961 (rank : 155) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1071 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q02779, Q12761, Q14871 | Gene names | MAP3K10, MLK2, MST | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 10 (EC 2.7.11.25) (Mixed lineage kinase 2) (Protein kinase MST). | |||||