Please be patient as the page loads
|
K0195_HUMAN
|
||||||
| SwissProt Accessions | Q12767, O75536, Q86XF1 | Gene names | KIAA0195, TMEM94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0195 (Transmembrane protein 94). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
K0195_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q12767, O75536, Q86XF1 | Gene names | KIAA0195, TMEM94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0195 (Transmembrane protein 94). | |||||
|
K0195_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.994255 (rank : 2) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q7TSH8, Q80U66, Q8BL05, Q8K0Q0, Q91VY1 | Gene names | Kiaa0195 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0195. | |||||
|
AT2A3_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 3) | NC score | 0.085823 (rank : 4) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q93084, O60900, O60901, O75501, O75502, Q16115, Q8TEX5, Q8TEX6 | Gene names | ATP2A3 | |||
|
Domain Architecture |

|
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 3 (EC 3.6.3.8) (Calcium pump 3) (SERCA3) (SR Ca(2+)-ATPase 3). | |||||
|
AT2A3_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 4) | NC score | 0.084807 (rank : 6) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q64518, O70625, Q64517 | Gene names | Atp2a3 | |||
|
Domain Architecture |

|
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 3 (EC 3.6.3.8) (Calcium pump 3) (SERCA3) (SR Ca(2+)-ATPase 3). | |||||
|
AT2B1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 5) | NC score | 0.078390 (rank : 9) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P20020, Q12992, Q12993, Q13819, Q13820, Q13821, Q16504, Q93082 | Gene names | ATP2B1, PMCA1 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 1 (EC 3.6.3.8) (PMCA1) (Plasma membrane calcium pump isoform 1) (Plasma membrane calcium ATPase isoform 1). | |||||
|
AT2A1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 6) | NC score | 0.085011 (rank : 5) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O14983, O14984 | Gene names | ATP2A1 | |||
|
Domain Architecture |

|
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 (EC 3.6.3.8) (Calcium pump 1) (SERCA1) (SR Ca(2+)-ATPase 1) (Calcium-transporting ATPase sarcoplasmic reticulum type, fast twitch skeletal muscle isoform) (Endoplasmic reticulum class 1/2 Ca(2+) ATPase). | |||||
|
AT2A1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 7) | NC score | 0.083255 (rank : 7) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8R429 | Gene names | Atp2a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 (EC 3.6.3.8) (Calcium pump 1) (SERCA1) (SR Ca(2+)-ATPase 1) (Calcium-transporting ATPase sarcoplasmic reticulum type, fast twitch skeletal muscle isoform) (Endoplasmic reticulum class 1/2 Ca(2+) ATPase). | |||||
|
SPRL1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 8) | NC score | 0.048675 (rank : 22) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14515, Q14800 | Gene names | SPARCL1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (High endothelial venule protein) (Hevin) (MAST 9). | |||||
|
AP3B2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 9) | NC score | 0.037112 (rank : 30) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
AT2A2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 10) | NC score | 0.081242 (rank : 8) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O55143, Q9R2A9, Q9WUT5 | Gene names | Atp2a2 | |||
|
Domain Architecture |

|
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 (EC 3.6.3.8) (Calcium pump 2) (SERCA2) (SR Ca(2+)-ATPase 2) (Calcium-transporting ATPase sarcoplasmic reticulum type, slow twitch skeletal muscle isoform) (Endoplasmic reticulum class 1/2 Ca(2+) ATPase). | |||||
|
BNC2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 11) | NC score | 0.059140 (rank : 14) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6ZN30, Q6T3A3, Q8NAR2, Q9H6J0, Q9NXV0 | Gene names | BNC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein basonuclin-2. | |||||
|
BNC2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 12) | NC score | 0.058739 (rank : 15) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BMQ3, Q6T3A4 | Gene names | Bnc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein basonuclin-2. | |||||
|
DOCK7_HUMAN
|
||||||
| θ value | 0.47712 (rank : 13) | NC score | 0.030682 (rank : 31) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96N67, Q6ZV32, Q8TB82, Q96NG6, Q9C092 | Gene names | DOCK7, KIAA1771 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dedicator of cytokinesis protein 7. | |||||
|
AT2B3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 14) | NC score | 0.072713 (rank : 12) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q16720, Q12995, Q16858 | Gene names | ATP2B3 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 3 (EC 3.6.3.8) (PMCA3) (Plasma membrane calcium pump isoform 3) (Plasma membrane calcium ATPase isoform 3). | |||||
|
AT2B4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 15) | NC score | 0.072746 (rank : 11) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P23634, Q13450, Q13452, Q13455, Q16817 | Gene names | ATP2B4 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 4 (EC 3.6.3.8) (PMCA4) (Plasma membrane calcium pump isoform 4) (Plasma membrane calcium ATPase isoform 4). | |||||
|
FNDC3_MOUSE
|
||||||
| θ value | 0.62314 (rank : 16) | NC score | 0.043783 (rank : 24) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BX90, Q2VEY7, Q6ZQ15, Q811D3, Q8BTM3 | Gene names | Fndc3a, D14Ertd453e, Fndc3, Kiaa0970 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibronectin type-III domain-containing protein 3a. | |||||
|
K1802_HUMAN
|
||||||
| θ value | 0.62314 (rank : 17) | NC score | 0.017475 (rank : 40) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
AP3B2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 18) | NC score | 0.037204 (rank : 29) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13367, O14808 | Gene names | AP3B2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (AP-3 complex beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain) (Neuron-specific vesicle coat protein beta-NAP). | |||||
|
AT2A2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 19) | NC score | 0.078258 (rank : 10) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P16615, P16614 | Gene names | ATP2A2, ATP2B | |||
|
Domain Architecture |

|
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 (EC 3.6.3.8) (Calcium pump 2) (SERCA2) (SR Ca(2+)-ATPase 2) (Calcium-transporting ATPase sarcoplasmic reticulum type, slow twitch skeletal muscle isoform) (Endoplasmic reticulum class 1/2 Ca(2+) ATPase). | |||||
|
CR2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 20) | NC score | 0.018299 (rank : 39) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P20023, Q13866, Q14212 | Gene names | CR2, C3DR | |||
|
Domain Architecture |

|
|||||
| Description | Complement receptor type 2 precursor (Cr2) (Complement C3d receptor) (Epstein-Barr virus receptor) (EBV receptor) (CD21 antigen). | |||||
|
FNDC3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 21) | NC score | 0.038924 (rank : 28) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y2H6, Q5JVF9, Q6EVH3, Q6N020, Q6P9D5, Q9H1W1 | Gene names | FNDC3A, FNDC3, KIAA0970 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibronectin type-III domain-containing protein 3a. | |||||
|
CIC_MOUSE
|
||||||
| θ value | 1.81305 (rank : 22) | NC score | 0.016647 (rank : 41) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CBX4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 23) | NC score | 0.026858 (rank : 34) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O55187 | Gene names | Cbx4, Pc2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 4 (Polycomb 2 homolog) (Pc2) (mPc2). | |||||
|
CN037_MOUSE
|
||||||
| θ value | 2.36792 (rank : 24) | NC score | 0.029225 (rank : 33) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BT18, Q8BFQ7, Q8R1Q5, Q9D6C5 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf37 homolog precursor. | |||||
|
FOXJ3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 25) | NC score | 0.010798 (rank : 51) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UPW0, Q9NSS7 | Gene names | FOXJ3, KIAA1041 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein J3. | |||||
|
ZN281_HUMAN
|
||||||
| θ value | 2.36792 (rank : 26) | NC score | 0.003352 (rank : 64) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2X9, Q9NY92 | Gene names | ZNF281, GZP1, ZBP99 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 281 (Zinc finger DNA-binding protein 99) (Transcription factor ZBP-99) (GC-box-binding zinc finger protein 1). | |||||
|
ELK4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 27) | NC score | 0.012546 (rank : 46) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P28324, P28323 | Gene names | ELK4, SAP1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
|
ROBO1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 28) | NC score | 0.009033 (rank : 56) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y6N7, Q7Z300, Q9BUS7 | Gene names | ROBO1, DUTT1 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 1 precursor (H-Robo-1) (Deleted in U twenty twenty). | |||||
|
SEM6A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 29) | NC score | 0.011063 (rank : 50) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35464, Q6P5A8, Q6PCN9, Q9EQ71 | Gene names | Sema6a, Semaq | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6A precursor (Semaphorin VIA) (Sema VIA) (Semaphorin 6A-1) (SEMA6A-1) (Semaphorin Q) (Sema Q). | |||||
|
SFXN4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 30) | NC score | 0.042129 (rank : 25) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q925N1, Q5FW99 | Gene names | Sfxn4 | |||
|
Domain Architecture |
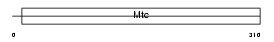
|
|||||
| Description | Sideroflexin-4. | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 31) | NC score | 0.030338 (rank : 32) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
FAF1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 32) | NC score | 0.039649 (rank : 26) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UNN5, Q9UF34, Q9UNT3, Q9Y2Z3 | Gene names | FAF1 | |||
|
Domain Architecture |

|
|||||
| Description | FAS-associated factor 1 (Protein FAF1) (hFAF1). | |||||
|
FAF1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 33) | NC score | 0.039431 (rank : 27) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P54731 | Gene names | Faf1 | |||
|
Domain Architecture |

|
|||||
| Description | FAS-associated factor 1 (Protein FAF1). | |||||
|
IQEC1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 34) | NC score | 0.011522 (rank : 48) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6DN90, O94863, Q96D85 | Gene names | IQSEC1, ARFGEP100, BRAG2, KIAA0763 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 1 (ADP-ribosylation factors guanine nucleotide-exchange protein 100) (ADP-ribosylation factors guanine nucleotide-exchange protein 2) (Brefeldin-resistant Arf-GEF 2 protein). | |||||
|
MTR1L_HUMAN
|
||||||
| θ value | 4.03905 (rank : 35) | NC score | 0.002838 (rank : 65) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1180 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13585 | Gene names | GPR50 | |||
|
Domain Architecture |

|
|||||
| Description | Melatonin-related receptor (G protein-coupled receptor 50) (H9). | |||||
|
MYST3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 36) | NC score | 0.011609 (rank : 47) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
PCD12_MOUSE
|
||||||
| θ value | 4.03905 (rank : 37) | NC score | 0.006786 (rank : 62) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O55134 | Gene names | Pcdh12 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-12 precursor (Vascular cadherin-2) (Vascular endothelial cadherin-2) (VE-cadherin-2) (VE-cad-2). | |||||
|
PTPRS_HUMAN
|
||||||
| θ value | 4.03905 (rank : 38) | NC score | 0.009537 (rank : 54) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13332, O75255, O75870, Q15718, Q16341 | Gene names | PTPRS | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase S precursor (EC 3.1.3.48) (R-PTP-S) (Protein-tyrosine phosphatase sigma) (R-PTP-sigma). | |||||
|
RFTN1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 39) | NC score | 0.020489 (rank : 37) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14699, Q496Y2, Q4QQI7, Q5JB48, Q7Z7P2 | Gene names | RFTN1, KIAA0084 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Raftlin (Raft-linking protein) (Cell migration-inducing gene 2 protein). | |||||
|
VCX3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 40) | NC score | 0.064513 (rank : 13) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NNX9, Q9P0H4 | Gene names | VCX3A, VCX3, VCX8R, VCXA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 3 (VCX-A protein) (Variably charged protein X-A) (Variable charge protein on X with eight repeats) (VCX- 8r). | |||||
|
VCXC_HUMAN
|
||||||
| θ value | 4.03905 (rank : 41) | NC score | 0.055995 (rank : 16) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H321 | Gene names | VCXC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | VCX-C protein (Variably charged protein X-C). | |||||
|
VCY1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 42) | NC score | 0.114851 (rank : 3) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O14598 | Gene names | VCY, BPY1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-specific basic protein Y 1 (Variably charged protein Y). | |||||
|
ZN337_HUMAN
|
||||||
| θ value | 4.03905 (rank : 43) | NC score | 0.000751 (rank : 66) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 781 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y3M9, Q9Y3Y5 | Gene names | ZNF337 | |||
|
Domain Architecture |
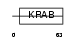
|
|||||
| Description | Zinc finger protein 337. | |||||
|
AFF1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 44) | NC score | 0.014513 (rank : 45) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
PTPRZ_HUMAN
|
||||||
| θ value | 5.27518 (rank : 45) | NC score | 0.010715 (rank : 52) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P23471 | Gene names | PTPRZ1, PTPRZ, PTPZ | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase zeta precursor (EC 3.1.3.48) (R-PTP-zeta). | |||||
|
BRD4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.008816 (rank : 57) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
CBX4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.021656 (rank : 36) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O00257, Q96C04 | Gene names | CBX4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 4 (Polycomb 2 homolog) (Pc2) (hPc2). | |||||
|
CN102_HUMAN
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | 0.026485 (rank : 35) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H7Z3, Q9NWH6 | Gene names | C14orf102 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf102. | |||||
|
FACD2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | 0.015466 (rank : 44) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80V62, Q9CWC7 | Gene names | Fancd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group D2 protein homolog (Protein FACD2). | |||||
|
RND1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 50) | NC score | 0.011481 (rank : 49) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92730 | Gene names | RND1, RHO6 | |||
|
Domain Architecture |
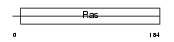
|
|||||
| Description | Rho-related GTP-binding protein Rho6 (Rho family GTPase 1) (Rnd1). | |||||
|
SYNPO_MOUSE
|
||||||
| θ value | 6.88961 (rank : 51) | NC score | 0.015762 (rank : 42) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CC35, Q99JI0 | Gene names | Synpo, Kiaa1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
VCX1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 52) | NC score | 0.044954 (rank : 23) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H320, Q9P0H3 | Gene names | VCX, VCX1, VCX10R, VCXB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 1 (VCX-B1 protein) (Variably charged protein X-B1) (Variable charge protein on X with ten repeats) (VCX- 10r). | |||||
|
C4BP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | 0.015614 (rank : 43) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P08607 | Gene names | C4bpa, C4bp | |||
|
Domain Architecture |
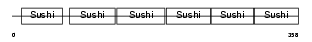
|
|||||
| Description | C4b-binding protein precursor (C4bp). | |||||
|
CP2DA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.006923 (rank : 61) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P24456, Q64490 | Gene names | Cyp2d10, Cyp2d-10 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 2D10 (EC 1.14.14.1) (CYPIID10) (P450-16-alpha) (P450CB) (Testosterone 16-alpha hydroxylase). | |||||
|
HUWE1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.005943 (rank : 63) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
MYPC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.009048 (rank : 55) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q00872, Q15497 | Gene names | MYBPC1, MYBPCS | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-binding protein C, slow-type (Slow MyBP-C) (C-protein, skeletal muscle slow isoform). | |||||
|
NMDE2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.007716 (rank : 59) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13224, Q12919, Q13220, Q13225, Q9UM56 | Gene names | GRIN2B | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 2 precursor (N-methyl D- aspartate receptor subtype 2B) (NR2B) (NMDAR2B) (N-methyl-D-aspartate receptor subunit 3) (NR3) (hNR3). | |||||
|
NMDE2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.007715 (rank : 60) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q01097, Q9DCB2 | Gene names | Grin2b | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 2 precursor (N-methyl D- aspartate receptor subtype 2B) (NR2B) (NMDAR2B). | |||||
|
PPP6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.009726 (rank : 53) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00743 | Gene names | PPP6C, PPP6 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 6 (EC 3.1.3.16) (PP6). | |||||
|
SEM6A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.008780 (rank : 58) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H2E6, Q9P2H9 | Gene names | SEMA6A, KIAA1368 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6A precursor (Semaphorin VIA) (Sema VIA) (Semaphorin 6A-1) (SEMA6A-1). | |||||
|
TXND2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 61) | NC score | 0.020338 (rank : 38) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
AT2B2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.054789 (rank : 18) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q01814, O00766, Q12994, Q16818 | Gene names | ATP2B2, PMCA2 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 2 (EC 3.6.3.8) (PMCA2) (Plasma membrane calcium pump isoform 2) (Plasma membrane calcium ATPase isoform 2). | |||||
|
AT2B2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.055348 (rank : 17) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9R0K7, O88863 | Gene names | Atp2b2, Pmca2 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 2 (EC 3.6.3.8) (PMCA2) (Plasma membrane calcium pump isoform 2) (Plasma membrane calcium ATPase isoform 2). | |||||
|
AT2C1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.051219 (rank : 21) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P98194, O76005, Q86V72, Q86V73, Q8N6V1, Q8NCJ7 | Gene names | ATP2C1, KIAA1347, PMR1L | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-transporting ATPase type 2C member 1 (EC 3.6.3.8) (ATPase 2C1) (ATP-dependent Ca(2+) pump PMR1). | |||||
|
AT2C1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.051383 (rank : 20) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80XR2, Q80YZ2 | Gene names | Atp2c1, Pmr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-transporting ATPase type 2C member 1 (EC 3.6.3.8) (ATPase 2C1) (ATP-dependent Ca(2+) pump PMR1). | |||||
|
AT2C2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.051430 (rank : 19) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75185 | Gene names | KIAA0703 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable calcium-transporting ATPase KIAA0703 (EC 3.6.3.8). | |||||
|
K0195_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q12767, O75536, Q86XF1 | Gene names | KIAA0195, TMEM94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0195 (Transmembrane protein 94). | |||||
|
K0195_MOUSE
|
||||||
| NC score | 0.994255 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q7TSH8, Q80U66, Q8BL05, Q8K0Q0, Q91VY1 | Gene names | Kiaa0195 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0195. | |||||
|
VCY1_HUMAN
|
||||||
| NC score | 0.114851 (rank : 3) | θ value | 4.03905 (rank : 42) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O14598 | Gene names | VCY, BPY1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-specific basic protein Y 1 (Variably charged protein Y). | |||||
|
AT2A3_HUMAN
|
||||||
| NC score | 0.085823 (rank : 4) | θ value | 0.0961366 (rank : 3) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q93084, O60900, O60901, O75501, O75502, Q16115, Q8TEX5, Q8TEX6 | Gene names | ATP2A3 | |||
|
Domain Architecture |

|
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 3 (EC 3.6.3.8) (Calcium pump 3) (SERCA3) (SR Ca(2+)-ATPase 3). | |||||
|
AT2A1_HUMAN
|
||||||
| NC score | 0.085011 (rank : 5) | θ value | 0.365318 (rank : 6) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O14983, O14984 | Gene names | ATP2A1 | |||
|
Domain Architecture |

|
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 (EC 3.6.3.8) (Calcium pump 1) (SERCA1) (SR Ca(2+)-ATPase 1) (Calcium-transporting ATPase sarcoplasmic reticulum type, fast twitch skeletal muscle isoform) (Endoplasmic reticulum class 1/2 Ca(2+) ATPase). | |||||
|
AT2A3_MOUSE
|
||||||
| NC score | 0.084807 (rank : 6) | θ value | 0.0961366 (rank : 4) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q64518, O70625, Q64517 | Gene names | Atp2a3 | |||
|
Domain Architecture |

|
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 3 (EC 3.6.3.8) (Calcium pump 3) (SERCA3) (SR Ca(2+)-ATPase 3). | |||||
|
AT2A1_MOUSE
|
||||||
| NC score | 0.083255 (rank : 7) | θ value | 0.365318 (rank : 7) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8R429 | Gene names | Atp2a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 (EC 3.6.3.8) (Calcium pump 1) (SERCA1) (SR Ca(2+)-ATPase 1) (Calcium-transporting ATPase sarcoplasmic reticulum type, fast twitch skeletal muscle isoform) (Endoplasmic reticulum class 1/2 Ca(2+) ATPase). | |||||
|
AT2A2_MOUSE
|
||||||
| NC score | 0.081242 (rank : 8) | θ value | 0.47712 (rank : 10) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O55143, Q9R2A9, Q9WUT5 | Gene names | Atp2a2 | |||
|
Domain Architecture |

|
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 (EC 3.6.3.8) (Calcium pump 2) (SERCA2) (SR Ca(2+)-ATPase 2) (Calcium-transporting ATPase sarcoplasmic reticulum type, slow twitch skeletal muscle isoform) (Endoplasmic reticulum class 1/2 Ca(2+) ATPase). | |||||
|
AT2B1_HUMAN
|
||||||
| NC score | 0.078390 (rank : 9) | θ value | 0.21417 (rank : 5) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P20020, Q12992, Q12993, Q13819, Q13820, Q13821, Q16504, Q93082 | Gene names | ATP2B1, PMCA1 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 1 (EC 3.6.3.8) (PMCA1) (Plasma membrane calcium pump isoform 1) (Plasma membrane calcium ATPase isoform 1). | |||||
|
AT2A2_HUMAN
|
||||||
| NC score | 0.078258 (rank : 10) | θ value | 1.06291 (rank : 19) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P16615, P16614 | Gene names | ATP2A2, ATP2B | |||
|
Domain Architecture |

|
|||||
| Description | Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 (EC 3.6.3.8) (Calcium pump 2) (SERCA2) (SR Ca(2+)-ATPase 2) (Calcium-transporting ATPase sarcoplasmic reticulum type, slow twitch skeletal muscle isoform) (Endoplasmic reticulum class 1/2 Ca(2+) ATPase). | |||||
|
AT2B4_HUMAN
|
||||||
| NC score | 0.072746 (rank : 11) | θ value | 0.62314 (rank : 15) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P23634, Q13450, Q13452, Q13455, Q16817 | Gene names | ATP2B4 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 4 (EC 3.6.3.8) (PMCA4) (Plasma membrane calcium pump isoform 4) (Plasma membrane calcium ATPase isoform 4). | |||||
|
AT2B3_HUMAN
|
||||||
| NC score | 0.072713 (rank : 12) | θ value | 0.62314 (rank : 14) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q16720, Q12995, Q16858 | Gene names | ATP2B3 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 3 (EC 3.6.3.8) (PMCA3) (Plasma membrane calcium pump isoform 3) (Plasma membrane calcium ATPase isoform 3). | |||||
|
VCX3_HUMAN
|
||||||
| NC score | 0.064513 (rank : 13) | θ value | 4.03905 (rank : 40) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NNX9, Q9P0H4 | Gene names | VCX3A, VCX3, VCX8R, VCXA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 3 (VCX-A protein) (Variably charged protein X-A) (Variable charge protein on X with eight repeats) (VCX- 8r). | |||||
|
BNC2_HUMAN
|
||||||
| NC score | 0.059140 (rank : 14) | θ value | 0.47712 (rank : 11) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6ZN30, Q6T3A3, Q8NAR2, Q9H6J0, Q9NXV0 | Gene names | BNC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein basonuclin-2. | |||||
|
BNC2_MOUSE
|
||||||
| NC score | 0.058739 (rank : 15) | θ value | 0.47712 (rank : 12) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BMQ3, Q6T3A4 | Gene names | Bnc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein basonuclin-2. | |||||
|
VCXC_HUMAN
|
||||||
| NC score | 0.055995 (rank : 16) | θ value | 4.03905 (rank : 41) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H321 | Gene names | VCXC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | VCX-C protein (Variably charged protein X-C). | |||||
|
AT2B2_MOUSE
|
||||||
| NC score | 0.055348 (rank : 17) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9R0K7, O88863 | Gene names | Atp2b2, Pmca2 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 2 (EC 3.6.3.8) (PMCA2) (Plasma membrane calcium pump isoform 2) (Plasma membrane calcium ATPase isoform 2). | |||||
|
AT2B2_HUMAN
|
||||||
| NC score | 0.054789 (rank : 18) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q01814, O00766, Q12994, Q16818 | Gene names | ATP2B2, PMCA2 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 2 (EC 3.6.3.8) (PMCA2) (Plasma membrane calcium pump isoform 2) (Plasma membrane calcium ATPase isoform 2). | |||||
|
AT2C2_HUMAN
|
||||||
| NC score | 0.051430 (rank : 19) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75185 | Gene names | KIAA0703 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable calcium-transporting ATPase KIAA0703 (EC 3.6.3.8). | |||||
|
AT2C1_MOUSE
|
||||||
| NC score | 0.051383 (rank : 20) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80XR2, Q80YZ2 | Gene names | Atp2c1, Pmr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-transporting ATPase type 2C member 1 (EC 3.6.3.8) (ATPase 2C1) (ATP-dependent Ca(2+) pump PMR1). | |||||
|
AT2C1_HUMAN
|
||||||
| NC score | 0.051219 (rank : 21) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P98194, O76005, Q86V72, Q86V73, Q8N6V1, Q8NCJ7 | Gene names | ATP2C1, KIAA1347, PMR1L | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-transporting ATPase type 2C member 1 (EC 3.6.3.8) (ATPase 2C1) (ATP-dependent Ca(2+) pump PMR1). | |||||
|
SPRL1_HUMAN
|
||||||
| NC score | 0.048675 (rank : 22) | θ value | 0.365318 (rank : 8) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14515, Q14800 | Gene names | SPARCL1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (High endothelial venule protein) (Hevin) (MAST 9). | |||||
|
VCX1_HUMAN
|
||||||
| NC score | 0.044954 (rank : 23) | θ value | 6.88961 (rank : 52) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H320, Q9P0H3 | Gene names | VCX, VCX1, VCX10R, VCXB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 1 (VCX-B1 protein) (Variably charged protein X-B1) (Variable charge protein on X with ten repeats) (VCX- 10r). | |||||
|
FNDC3_MOUSE
|
||||||
| NC score | 0.043783 (rank : 24) | θ value | 0.62314 (rank : 16) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BX90, Q2VEY7, Q6ZQ15, Q811D3, Q8BTM3 | Gene names | Fndc3a, D14Ertd453e, Fndc3, Kiaa0970 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibronectin type-III domain-containing protein 3a. | |||||
|
SFXN4_MOUSE
|
||||||
| NC score | 0.042129 (rank : 25) | θ value | 3.0926 (rank : 30) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q925N1, Q5FW99 | Gene names | Sfxn4 | |||
|
Domain Architecture |

|
|||||
| Description | Sideroflexin-4. | |||||
|
FAF1_HUMAN
|
||||||
| NC score | 0.039649 (rank : 26) | θ value | 4.03905 (rank : 32) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UNN5, Q9UF34, Q9UNT3, Q9Y2Z3 | Gene names | FAF1 | |||
|
Domain Architecture |

|
|||||
| Description | FAS-associated factor 1 (Protein FAF1) (hFAF1). | |||||
|
FAF1_MOUSE
|
||||||
| NC score | 0.039431 (rank : 27) | θ value | 4.03905 (rank : 33) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P54731 | Gene names | Faf1 | |||
|
Domain Architecture |

|
|||||
| Description | FAS-associated factor 1 (Protein FAF1). | |||||
|
FNDC3_HUMAN
|
||||||
| NC score | 0.038924 (rank : 28) | θ value | 1.38821 (rank : 21) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y2H6, Q5JVF9, Q6EVH3, Q6N020, Q6P9D5, Q9H1W1 | Gene names | FNDC3A, FNDC3, KIAA0970 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibronectin type-III domain-containing protein 3a. | |||||
|
AP3B2_HUMAN
|
||||||
| NC score | 0.037204 (rank : 29) | θ value | 0.813845 (rank : 18) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13367, O14808 | Gene names | AP3B2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (AP-3 complex beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain) (Neuron-specific vesicle coat protein beta-NAP). | |||||
|
AP3B2_MOUSE
|
||||||
| NC score | 0.037112 (rank : 30) | θ value | 0.47712 (rank : 9) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
DOCK7_HUMAN
|
||||||
| NC score | 0.030682 (rank : 31) | θ value | 0.47712 (rank : 13) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96N67, Q6ZV32, Q8TB82, Q96NG6, Q9C092 | Gene names | DOCK7, KIAA1771 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dedicator of cytokinesis protein 7. | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.030338 (rank : 32) | θ value | 3.0926 (rank : 31) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
CN037_MOUSE
|
||||||
| NC score | 0.029225 (rank : 33) | θ value | 2.36792 (rank : 24) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BT18, Q8BFQ7, Q8R1Q5, Q9D6C5 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf37 homolog precursor. | |||||
|
CBX4_MOUSE
|
||||||
| NC score | 0.026858 (rank : 34) | θ value | 2.36792 (rank : 23) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O55187 | Gene names | Cbx4, Pc2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 4 (Polycomb 2 homolog) (Pc2) (mPc2). | |||||
|
CN102_HUMAN
|
||||||
| NC score | 0.026485 (rank : 35) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H7Z3, Q9NWH6 | Gene names | C14orf102 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf102. | |||||
|
CBX4_HUMAN
|
||||||
| NC score | 0.021656 (rank : 36) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O00257, Q96C04 | Gene names | CBX4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 4 (Polycomb 2 homolog) (Pc2) (hPc2). | |||||
|
RFTN1_HUMAN
|
||||||
| NC score | 0.020489 (rank : 37) | θ value | 4.03905 (rank : 39) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14699, Q496Y2, Q4QQI7, Q5JB48, Q7Z7P2 | Gene names | RFTN1, KIAA0084 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Raftlin (Raft-linking protein) (Cell migration-inducing gene 2 protein). | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.020338 (rank : 38) | θ value | 8.99809 (rank : 61) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
CR2_HUMAN
|
||||||
| NC score | 0.018299 (rank : 39) | θ value | 1.06291 (rank : 20) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P20023, Q13866, Q14212 | Gene names | CR2, C3DR | |||
|
Domain Architecture |

|
|||||
| Description | Complement receptor type 2 precursor (Cr2) (Complement C3d receptor) (Epstein-Barr virus receptor) (EBV receptor) (CD21 antigen). | |||||
|
K1802_HUMAN
|
||||||
| NC score | 0.017475 (rank : 40) | θ value | 0.62314 (rank : 17) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
CIC_MOUSE
|
||||||
| NC score | 0.016647 (rank : 41) | θ value | 1.81305 (rank : 22) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
SYNPO_MOUSE
|
||||||
| NC score | 0.015762 (rank : 42) | θ value | 6.88961 (rank : 51) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CC35, Q99JI0 | Gene names | Synpo, Kiaa1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
C4BP_MOUSE
|
||||||
| NC score | 0.015614 (rank : 43) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P08607 | Gene names | C4bpa, C4bp | |||
|
Domain Architecture |

|
|||||
| Description | C4b-binding protein precursor (C4bp). | |||||
|
FACD2_MOUSE
|
||||||
| NC score | 0.015466 (rank : 44) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80V62, Q9CWC7 | Gene names | Fancd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group D2 protein homolog (Protein FACD2). | |||||
|
AFF1_MOUSE
|
||||||
| NC score | 0.014513 (rank : 45) | θ value | 5.27518 (rank : 44) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
ELK4_HUMAN
|
||||||
| NC score | 0.012546 (rank : 46) | θ value | 3.0926 (rank : 27) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P28324, P28323 | Gene names | ELK4, SAP1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
|
MYST3_MOUSE
|
||||||
| NC score | 0.011609 (rank : 47) | θ value | 4.03905 (rank : 36) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
IQEC1_HUMAN
|
||||||
| NC score | 0.011522 (rank : 48) | θ value | 4.03905 (rank : 34) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6DN90, O94863, Q96D85 | Gene names | IQSEC1, ARFGEP100, BRAG2, KIAA0763 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 1 (ADP-ribosylation factors guanine nucleotide-exchange protein 100) (ADP-ribosylation factors guanine nucleotide-exchange protein 2) (Brefeldin-resistant Arf-GEF 2 protein). | |||||
|
RND1_HUMAN
|
||||||
| NC score | 0.011481 (rank : 49) | θ value | 6.88961 (rank : 50) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92730 | Gene names | RND1, RHO6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-related GTP-binding protein Rho6 (Rho family GTPase 1) (Rnd1). | |||||
|
SEM6A_MOUSE
|
||||||
| NC score | 0.011063 (rank : 50) | θ value | 3.0926 (rank : 29) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35464, Q6P5A8, Q6PCN9, Q9EQ71 | Gene names | Sema6a, Semaq | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6A precursor (Semaphorin VIA) (Sema VIA) (Semaphorin 6A-1) (SEMA6A-1) (Semaphorin Q) (Sema Q). | |||||
|
FOXJ3_HUMAN
|
||||||
| NC score | 0.010798 (rank : 51) | θ value | 2.36792 (rank : 25) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UPW0, Q9NSS7 | Gene names | FOXJ3, KIAA1041 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein J3. | |||||
|
PTPRZ_HUMAN
|
||||||
| NC score | 0.010715 (rank : 52) | θ value | 5.27518 (rank : 45) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P23471 | Gene names | PTPRZ1, PTPRZ, PTPZ | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase zeta precursor (EC 3.1.3.48) (R-PTP-zeta). | |||||
|
PPP6_HUMAN
|
||||||
| NC score | 0.009726 (rank : 53) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00743 | Gene names | PPP6C, PPP6 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 6 (EC 3.1.3.16) (PP6). | |||||
|
PTPRS_HUMAN
|
||||||
| NC score | 0.009537 (rank : 54) | θ value | 4.03905 (rank : 38) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13332, O75255, O75870, Q15718, Q16341 | Gene names | PTPRS | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase S precursor (EC 3.1.3.48) (R-PTP-S) (Protein-tyrosine phosphatase sigma) (R-PTP-sigma). | |||||
|
MYPC1_HUMAN
|
||||||
| NC score | 0.009048 (rank : 55) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q00872, Q15497 | Gene names | MYBPC1, MYBPCS | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-binding protein C, slow-type (Slow MyBP-C) (C-protein, skeletal muscle slow isoform). | |||||
|
ROBO1_HUMAN
|
||||||
| NC score | 0.009033 (rank : 56) | θ value | 3.0926 (rank : 28) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y6N7, Q7Z300, Q9BUS7 | Gene names | ROBO1, DUTT1 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 1 precursor (H-Robo-1) (Deleted in U twenty twenty). | |||||
|
BRD4_HUMAN
|
||||||
| NC score | 0.008816 (rank : 57) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
SEM6A_HUMAN
|
||||||
| NC score | 0.008780 (rank : 58) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H2E6, Q9P2H9 | Gene names | SEMA6A, KIAA1368 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6A precursor (Semaphorin VIA) (Sema VIA) (Semaphorin 6A-1) (SEMA6A-1). | |||||
|
NMDE2_HUMAN
|
||||||
| NC score | 0.007716 (rank : 59) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13224, Q12919, Q13220, Q13225, Q9UM56 | Gene names | GRIN2B | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 2 precursor (N-methyl D- aspartate receptor subtype 2B) (NR2B) (NMDAR2B) (N-methyl-D-aspartate receptor subunit 3) (NR3) (hNR3). | |||||
|
NMDE2_MOUSE
|
||||||
| NC score | 0.007715 (rank : 60) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q01097, Q9DCB2 | Gene names | Grin2b | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 2 precursor (N-methyl D- aspartate receptor subtype 2B) (NR2B) (NMDAR2B). | |||||
|
CP2DA_MOUSE
|
||||||
| NC score | 0.006923 (rank : 61) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P24456, Q64490 | Gene names | Cyp2d10, Cyp2d-10 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 2D10 (EC 1.14.14.1) (CYPIID10) (P450-16-alpha) (P450CB) (Testosterone 16-alpha hydroxylase). | |||||
|
PCD12_MOUSE
|
||||||
| NC score | 0.006786 (rank : 62) | θ value | 4.03905 (rank : 37) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O55134 | Gene names | Pcdh12 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-12 precursor (Vascular cadherin-2) (Vascular endothelial cadherin-2) (VE-cadherin-2) (VE-cad-2). | |||||
|
HUWE1_HUMAN
|
||||||
| NC score | 0.005943 (rank : 63) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
ZN281_HUMAN
|
||||||
| NC score | 0.003352 (rank : 64) | θ value | 2.36792 (rank : 26) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2X9, Q9NY92 | Gene names | ZNF281, GZP1, ZBP99 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 281 (Zinc finger DNA-binding protein 99) (Transcription factor ZBP-99) (GC-box-binding zinc finger protein 1). | |||||
|
MTR1L_HUMAN
|
||||||
| NC score | 0.002838 (rank : 65) | θ value | 4.03905 (rank : 35) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1180 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13585 | Gene names | GPR50 | |||
|
Domain Architecture |

|
|||||
| Description | Melatonin-related receptor (G protein-coupled receptor 50) (H9). | |||||
|
ZN337_HUMAN
|
||||||
| NC score | 0.000751 (rank : 66) | θ value | 4.03905 (rank : 43) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 781 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y3M9, Q9Y3Y5 | Gene names | ZNF337 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 337. | |||||