Please be patient as the page loads
|
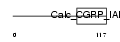
FAF1_HUMAN
|
||||||
| SwissProt Accessions | Q9UNN5, Q9UF34, Q9UNT3, Q9Y2Z3 | Gene names | FAF1 | |||
|
Domain Architecture |

|
|||||
| Description | FAS-associated factor 1 (Protein FAF1) (hFAF1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
FAF1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q9UNN5, Q9UF34, Q9UNT3, Q9Y2Z3 | Gene names | FAF1 | |||
|
Domain Architecture |

|
|||||
| Description | FAS-associated factor 1 (Protein FAF1) (hFAF1). | |||||
|
FAF1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.997369 (rank : 2) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | P54731 | Gene names | Faf1 | |||
|
Domain Architecture |

|
|||||
| Description | FAS-associated factor 1 (Protein FAF1). | |||||
|
UBXD8_HUMAN
|
||||||
| θ value | 2.12685e-25 (rank : 3) | NC score | 0.648660 (rank : 3) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96CS3, O94963, Q8IUF2, Q9BRP2, Q9BVM7 | Gene names | UBXD8, ETEA, KIAA0887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UBX domain-containing protein 8 (Protein ETEA). | |||||
|
UBXD8_MOUSE
|
||||||
| θ value | 2.77775e-25 (rank : 4) | NC score | 0.648591 (rank : 4) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q3TDN2, Q3U9W2, Q80TP7, Q80W42 | Gene names | Ubxd8, Kiaa0887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UBX domain-containing protein 8. | |||||
|
UBXD6_HUMAN
|
||||||
| θ value | 1.80886e-08 (rank : 5) | NC score | 0.422727 (rank : 6) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O00124, Q7Z6F2 | Gene names | UBXD6, D8S2298E, REP8 | |||
|
Domain Architecture |

|
|||||
| Description | UBX domain-containing protein 6 (Reproduction 8 protein) (Protein Rep- 8). | |||||
|
UBXD6_MOUSE
|
||||||
| θ value | 4.0297e-08 (rank : 6) | NC score | 0.437332 (rank : 5) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9QZ49 | Gene names | Ubxd6, D0H8S2298E, Rep8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UBX domain-containing protein 6 (Reproduction 8 protein) (Protein Rep- 8). | |||||
|
UBXD7_HUMAN
|
||||||
| θ value | 8.97725e-08 (rank : 7) | NC score | 0.383557 (rank : 7) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O94888, Q6ZP77, Q86X20, Q8N327 | Gene names | UBXD7, KIAA0794 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UBX domain-containing protein 7. | |||||
|
UBXD7_MOUSE
|
||||||
| θ value | 4.45536e-07 (rank : 8) | NC score | 0.372469 (rank : 8) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6P5G6, Q6ZQ44 | Gene names | Ubxd7, Kiaa0794 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UBX domain-containing protein 7. | |||||
|
SAKS1_MOUSE
|
||||||
| θ value | 1.09739e-05 (rank : 9) | NC score | 0.302098 (rank : 9) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q922Y1, Q3UCP8 | Gene names | Saks1, D19Ertd721e | |||
|
Domain Architecture |

|
|||||
| Description | SAPK substrate protein 1 (UBA/UBX 33.3 kDa protein) (mY33K) (Protein 2B28). | |||||
|
UBXD2_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 10) | NC score | 0.257323 (rank : 11) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q92575, Q8IYM5 | Gene names | UBXD2, KIAA0242, UBXDC1 | |||
|
Domain Architecture |

|
|||||
| Description | UBX domain-containing protein 2. | |||||
|
SAKS1_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 11) | NC score | 0.294295 (rank : 10) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q04323, Q9BV93, Q9BVV5 | Gene names | SAKS1 | |||
|
Domain Architecture |

|
|||||
| Description | SAPK substrate protein 1 (UBA/UBX 33.3 kDa protein). | |||||
|
UBXD1_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 12) | NC score | 0.230168 (rank : 13) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BZV1, Q96AH1, Q96IK9, Q9BZV0 | Gene names | UBXD1, UBXDC2 | |||
|
Domain Architecture |
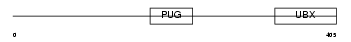
|
|||||
| Description | UBX domain-containing protein 1. | |||||
|
UBXD2_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 13) | NC score | 0.243579 (rank : 12) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8VCH8, Q8BW17 | Gene names | Ubxd2, Ubxdc1 | |||
|
Domain Architecture |

|
|||||
| Description | UBX domain-containing protein 2. | |||||
|
UBXD1_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 14) | NC score | 0.214298 (rank : 14) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99PL6, Q91W79 | Gene names | Ubxd1, Ubxdc2 | |||
|
Domain Architecture |
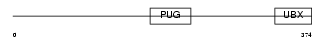
|
|||||
| Description | UBX domain-containing protein 1. | |||||
|
LIPA2_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 15) | NC score | 0.076930 (rank : 20) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1044 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O75334 | Gene names | PPFIA2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
CCD46_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 16) | NC score | 0.076796 (rank : 21) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q5PR68, Q80ZN6, Q99JS4, Q9D9T1 | Gene names | Ccdc46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled coil domain-containing protein 46. | |||||
|
CNTRB_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 17) | NC score | 0.094463 (rank : 16) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q8CB62, Q5NCF3 | Gene names | Cntrob, Lip8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
CAF1A_HUMAN
|
||||||
| θ value | 0.163984 (rank : 18) | NC score | 0.092316 (rank : 17) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q13111, Q7Z7K3, Q9UJY8 | Gene names | CHAF1A, CAF, CAF1P150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
OTU7B_HUMAN
|
||||||
| θ value | 0.163984 (rank : 19) | NC score | 0.073512 (rank : 23) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6GQQ9, Q8WWA7, Q9NQ53, Q9UFF4 | Gene names | OTUD7B, ZA20D1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | OTU domain-containing protein 7B (EC 3.-.-.-) (Zinc finger protein Cezanne) (Zinc finger A20 domain-containing protein 1) (Cellular zinc finger anti-NF-kappa B protein). | |||||
|
GCP60_HUMAN
|
||||||
| θ value | 0.21417 (rank : 20) | NC score | 0.101349 (rank : 15) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9H3P7, Q5VTJ0, Q6P9F1, Q8IZC5, Q8N4D6, Q9H6U3 | Gene names | ACBD3, GCP60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi resident protein GCP60 (Acyl-CoA-binding domain-containing protein 3) (Golgi phosphoprotein 1) (GOLPH1) (Golgi complex-associated protein 1) (GOCAP1) (PBR- and PKA-associated protein 7) (Peripheral benzodiazepine receptor-associated protein PAP7). | |||||
|
STK10_MOUSE
|
||||||
| θ value | 0.21417 (rank : 21) | NC score | 0.038553 (rank : 96) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
ITSN1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 22) | NC score | 0.061370 (rank : 34) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
CCD46_HUMAN
|
||||||
| θ value | 0.365318 (rank : 23) | NC score | 0.067656 (rank : 28) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
LIPA2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 24) | NC score | 0.073317 (rank : 24) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1282 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8BSS9, Q8BN73 | Gene names | Ppfia2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
MYH4_MOUSE
|
||||||
| θ value | 0.365318 (rank : 25) | NC score | 0.059908 (rank : 39) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
UACA_MOUSE
|
||||||
| θ value | 0.365318 (rank : 26) | NC score | 0.049862 (rank : 81) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8CGB3, Q69ZG3, Q7TN77, Q8BJC8 | Gene names | Uaca, Kiaa1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein (Nucling) (Nuclear membrane-binding protein). | |||||
|
AFAD_HUMAN
|
||||||
| θ value | 0.47712 (rank : 27) | NC score | 0.064035 (rank : 32) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P55196, O75087, O75088, O75089, Q5TIG6, Q9NSN7, Q9NU92 | Gene names | MLLT4, AF6 | |||
|
Domain Architecture |

|
|||||
| Description | Afadin (Protein AF-6). | |||||
|
ENAH_HUMAN
|
||||||
| θ value | 0.47712 (rank : 28) | NC score | 0.068544 (rank : 26) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8N8S7, Q502W5, Q5T5M7, Q5VTQ9, Q5VTR0, Q9NVF3, Q9UFB8 | Gene names | ENAH, MENA | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog. | |||||
|
MYH1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 29) | NC score | 0.060305 (rank : 37) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
MYH3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 30) | NC score | 0.058716 (rank : 44) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
MYH4_HUMAN
|
||||||
| θ value | 0.47712 (rank : 31) | NC score | 0.056312 (rank : 50) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
UBXD3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 32) | NC score | 0.081631 (rank : 19) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BG34, Q3TTR1, Q8BP58, Q8R0B6 | Gene names | Ubxd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UBX domain-containing protein 3. | |||||
|
MYH2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 33) | NC score | 0.060104 (rank : 38) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
PRDM1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 34) | NC score | 0.002057 (rank : 128) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 766 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q60636 | Gene names | Prdm1, Blimp1 | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 1 (PR domain-containing protein 1) (Beta-interferon gene positive-regulatory domain I-binding factor) (BLIMP-1). | |||||
|
ITSN1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 35) | NC score | 0.055198 (rank : 55) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
MYH1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 36) | NC score | 0.059623 (rank : 40) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
ECM2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 37) | NC score | 0.017952 (rank : 118) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94769 | Gene names | ECM2 | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular matrix protein 2 precursor (Matrix glycoprotein SC1/ECM2). | |||||
|
M4K4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 38) | NC score | 0.020582 (rank : 116) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1589 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O95819, O75172, Q9NST7 | Gene names | MAP4K4, HGK, KIAA0687, NIK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
M4K4_MOUSE
|
||||||
| θ value | 1.06291 (rank : 39) | NC score | 0.020242 (rank : 117) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1560 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P97820 | Gene names | Map4k4, Nik | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
RBP1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 40) | NC score | 0.047343 (rank : 88) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q62172, Q9CRE5 | Gene names | Ralbp1, Rip1 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
EEA1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 41) | NC score | 0.048941 (rank : 83) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
ENAH_MOUSE
|
||||||
| θ value | 1.38821 (rank : 42) | NC score | 0.052294 (rank : 61) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
GCP60_MOUSE
|
||||||
| θ value | 1.38821 (rank : 43) | NC score | 0.083057 (rank : 18) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BMP6, O35371 | Gene names | Acbd3, Gcp60, Pap7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi resident protein GCP60 (Acyl-CoA-binding domain-containing protein 3) (Golgi phosphoprotein 1) (GOLPH1) (Golgi complex-associated protein 1) (GOCAP1) (PBR- and PKA-associated protein 7) (Peripheral benzodiazepine receptor-associated protein PAP7). | |||||
|
CA082_MOUSE
|
||||||
| θ value | 1.81305 (rank : 44) | NC score | 0.032027 (rank : 102) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VC34, Q3TLC8, Q3TSP8, Q3UKX0, Q8C7M5, Q8CBW8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UPF0385 protein C1orf82 homolog. | |||||
|
CAF1A_MOUSE
|
||||||
| θ value | 1.81305 (rank : 45) | NC score | 0.074574 (rank : 22) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9QWF0 | Gene names | Chaf1a, Caip150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
EVPL_MOUSE
|
||||||
| θ value | 1.81305 (rank : 46) | NC score | 0.056101 (rank : 52) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 952 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9D952 | Gene names | Evpl | |||
|
Domain Architecture |

|
|||||
| Description | Envoplakin (p210) (210 kDa cornified envelope precursor protein). | |||||
|
STIM2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 47) | NC score | 0.069561 (rank : 25) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9P246, Q96BF1, Q9BQH2, Q9H8R1 | Gene names | STIM2, KIAA1482 | |||
|
Domain Architecture |
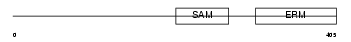
|
|||||
| Description | Stromal interaction molecule 2 precursor. | |||||
|
STIM2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.068479 (rank : 27) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P83093, Q69ZI3, Q80VF4 | Gene names | Stim2, Kiaa1482 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal interaction molecule 2 precursor. | |||||
|
CROCC_MOUSE
|
||||||
| θ value | 2.36792 (rank : 49) | NC score | 0.057821 (rank : 47) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
OTU7A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 50) | NC score | 0.059012 (rank : 43) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TE49, Q8IWK5 | Gene names | OTUD7A, C15orf16, OTUD7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | OTU domain-containing protein 7A (EC 3.-.-.-) (Zinc finger protein Cezanne 2). | |||||
|
OTU7A_MOUSE
|
||||||
| θ value | 2.36792 (rank : 51) | NC score | 0.058633 (rank : 45) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R554 | Gene names | Otud7a, Otud7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | OTU domain-containing protein 7A (EC 3.-.-.-) (Zinc finger protein Cezanne 2). | |||||
|
PLSB_MOUSE
|
||||||
| θ value | 2.36792 (rank : 52) | NC score | 0.036668 (rank : 97) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61586 | Gene names | Gpam | |||
|
Domain Architecture |

|
|||||
| Description | Glycerol-3-phosphate acyltransferase, mitochondrial precursor (EC 2.3.1.15) (GPAT) (P90). | |||||
|
RHG05_MOUSE
|
||||||
| θ value | 2.36792 (rank : 53) | NC score | 0.034705 (rank : 100) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P97393 | Gene names | Arhgap5, Rhogap5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
ZRF1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 54) | NC score | 0.055931 (rank : 53) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q99543 | Gene names | ZRF1, DNAJC2, MPHOSPH11, MPP11 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1 (M-phase phosphoprotein 11). | |||||
|
ANKZ1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.036490 (rank : 98) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q80UU1, Q9CZF5 | Gene names | Ankzf1, D1Ertd161e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and zinc finger domain-containing protein 1. | |||||
|
CALD1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | 0.057568 (rank : 48) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
GP108_MOUSE
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.029723 (rank : 105) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91WD0, Q8BXE9, Q925B1 | Gene names | Gpr108, Lustr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein GPR108 precursor (Lung seven transmembrane receptor 2). | |||||
|
MYO6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.048747 (rank : 84) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UM54, Q9BZZ7, Q9UEG2 | Gene names | MYO6, KIAA0389 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-6 (Myosin VI). | |||||
|
SMCA4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.021993 (rank : 114) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P51532, O95052, Q9HBD3 | Gene names | SMARCA4, BRG1, SNF2B, SNF2L4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L4 (EC 3.6.1.-) (ATP- dependent helicase SMARCA4) (SNF2-beta) (BRG-1 protein) (Mitotic growth and transcription activator) (Brahma protein homolog 1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4). | |||||
|
DREB_MOUSE
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.048552 (rank : 86) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9QXS6, Q922X1, Q9QXS5 | Gene names | Dbn1, Drba | |||
|
Domain Architecture |

|
|||||
| Description | Drebrin (Developmentally-regulated brain protein). | |||||
|
INCE_HUMAN
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.056564 (rank : 49) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
INCE_MOUSE
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.058518 (rank : 46) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9WU62, Q7TN28, Q8BGN4, Q8CGI4 | Gene names | Incenp | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
K0195_HUMAN
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.039649 (rank : 95) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12767, O75536, Q86XF1 | Gene names | KIAA0195, TMEM94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0195 (Transmembrane protein 94). | |||||
|
K0195_MOUSE
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.039869 (rank : 94) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TSH8, Q80U66, Q8BL05, Q8K0Q0, Q91VY1 | Gene names | Kiaa0195 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0195. | |||||
|
REST_HUMAN
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.052034 (rank : 64) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
RYR1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 66) | NC score | 0.015629 (rank : 119) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P21817, Q16314, Q16368, Q9NPK1, Q9P1U4 | Gene names | RYR1, RYDR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 1 (Skeletal muscle-type ryanodine receptor) (RyR1) (RYR-1) (Skeletal muscle calcium release channel). | |||||
|
SPTA2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.033382 (rank : 101) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 888 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q13813, Q13186, Q15324, Q16606, Q59EF1, Q5VXV5, Q5VXV6, Q7Z6M5, Q9P0V0 | Gene names | SPTAN1, SPTA2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, brain (Spectrin, non-erythroid alpha chain) (Alpha-II spectrin) (Fodrin alpha chain). | |||||
|
IF3A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.065435 (rank : 30) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q14152, O00653 | Gene names | EIF3S10, KIAA0139 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a). | |||||
|
IF3A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.065612 (rank : 29) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P23116, Q60697, Q62162 | Gene names | Eif3s10, Csma, Eif3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a) (p162 protein) (Centrosomin). | |||||
|
K1949_MOUSE
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.030984 (rank : 103) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BQ30, Q3UDS6, Q8CEY9, Q8R3D8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1949 homolog. | |||||
|
PELP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.036259 (rank : 99) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8IZL8, O15450, Q5EGN3, Q6NTE6, Q96FT1, Q9BU60 | Gene names | PELP1, HMX3, MNAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3). | |||||
|
RABE2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.048054 (rank : 87) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 545 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9H5N1 | Gene names | RABEP2, RABPT5B | |||
|
Domain Architecture |
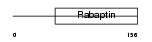
|
|||||
| Description | Rab GTPase-binding effector protein 2 (Rabaptin-5beta). | |||||
|
RBP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.039946 (rank : 93) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q15311 | Gene names | RALBP1, RLIP1, RLIP76 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (76 kDa Ral-interacting protein) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
RHG05_HUMAN
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.029416 (rank : 106) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13017 | Gene names | ARHGAP5, RHOGAP5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
SNX11_MOUSE
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.015348 (rank : 120) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91WL6, Q3V3A6 | Gene names | Snx11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorting nexin-11. | |||||
|
ARBK2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.002840 (rank : 127) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 849 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35626, Q9UGW9 | Gene names | ADRBK2, BARK2, GRK3 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adrenergic receptor kinase 2 (EC 2.7.11.15) (Beta-ARK-2) (G- protein-coupled receptor kinase 3). | |||||
|
CALC_MOUSE
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.027561 (rank : 107) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70160, Q8K1K5 | Gene names | Calca, Calc | |||
|
Domain Architecture |
|
|||||
| Description | Calcitonin precursor. | |||||
|
CCD11_HUMAN
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.059506 (rank : 41) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q96M91 | Gene names | CCDC11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 11. | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.048703 (rank : 85) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
GOGA5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.049009 (rank : 82) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1168 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8TBA6, O95287, Q03962, Q9UQQ7 | Gene names | GOLGA5, RETII, RFG5 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (RET-fused gene 5 protein) (Ret-II protein). | |||||
|
MY18A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.053273 (rank : 58) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
PELP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.029897 (rank : 104) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
REST_MOUSE
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.049944 (rank : 79) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
RGP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.023548 (rank : 110) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P46060 | Gene names | RANGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
SRC8_MOUSE
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.021546 (rank : 115) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q60598 | Gene names | Cttn, Ems1 | |||
|
Domain Architecture |
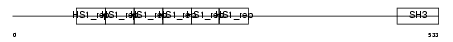
|
|||||
| Description | Src substrate cortactin. | |||||
|
UBP8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.022110 (rank : 113) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P40818, Q7Z3U2, Q86VA0, Q8IWI7 | Gene names | USP8, KIAA0055, UBPY | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (hUBPy). | |||||
|
ZRF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.049875 (rank : 80) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P54103 | Gene names | Zrf1, Dnajc2 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1. | |||||
|
ABTB1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.009614 (rank : 124) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99LJ2, Q8C2K7, Q91XU4 | Gene names | Abtb1, Bpoz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and BTB/POZ domain-containing protein 1. | |||||
|
CROCC_HUMAN
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.055179 (rank : 56) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
CROP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.043600 (rank : 90) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O95232, Q6PHR9, Q9NUY0, Q9P2S7 | Gene names | CROP, CREAP1, O48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cisplatin resistance-associated overexpressed protein (cAMP regulatory element-associated protein 1) (CRE-associated protein 1) (CREAP-1) (Luc7A) (Okadaic acid-inducible phosphoprotein OA48-18). | |||||
|
CROP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.040800 (rank : 91) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5SUF2, Q3U9D5, Q8BUJ5, Q921Z3, Q9CRS7, Q9CTY4 | Gene names | Crop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cisplatin resistance-associated overexpressed protein. | |||||
|
FYB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.023379 (rank : 111) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O15117, O00359 | Gene names | FYB, SLAP130 | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
FYB_MOUSE
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.025627 (rank : 108) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O35601, Q9Z2H3 | Gene names | Fyb | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
MYO6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.044582 (rank : 89) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q64331 | Gene names | Myo6, Sv | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-6 (Myosin VI). | |||||
|
MYT1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.013357 (rank : 121) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8CFC2, O08995, Q8CFH1 | Gene names | Myt1, Kiaa0835, Nzf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1 (MyT1) (Neural zinc finger factor 2) (NZF-2). | |||||
|
NIN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.051965 (rank : 66) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
PSA7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.012262 (rank : 122) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14818, Q9BR53, Q9H4K5, Q9UEU8 | Gene names | PSMA7 | |||
|
Domain Architecture |

|
|||||
| Description | Proteasome subunit alpha type 7 (EC 3.4.25.1) (Proteasome subunit RC6- 1) (Proteasome subunit XAPC7). | |||||
|
RENT2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.040439 (rank : 92) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
ROCK2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.024458 (rank : 109) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 2073 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O75116, Q9UQN5 | Gene names | ROCK2, KIAA0619 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2) (Rho kinase 2). | |||||
|
SRC8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.022533 (rank : 112) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q14247 | Gene names | CTTN, EMS1 | |||
|
Domain Architecture |
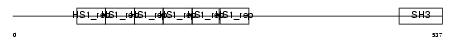
|
|||||
| Description | Src substrate cortactin (Amplaxin) (Oncogene EMS1). | |||||
|
STK38_HUMAN
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.005289 (rank : 126) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 840 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15208 | Gene names | STK38, NDR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase 38 (EC 2.7.11.1) (NDR1 protein kinase) (Nuclear Dbf2-related kinase 1). | |||||
|
STK38_MOUSE
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.005325 (rank : 125) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 838 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91VJ4 | Gene names | Stk38, Ndr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase 38 (EC 2.7.11.1) (NDR1 protein kinase) (Nuclear Dbf2-related kinase 1). | |||||
|
STMN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.060893 (rank : 35) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P16949 | Gene names | STMN1, LAP18, OP18 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Protein Pr22). | |||||
|
STMN1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.060795 (rank : 36) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P54227 | Gene names | Stmn1, Lag, Lap18, Pr22 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Pr22 protein) (Leukemia-associated gene protein). | |||||
|
WHRN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.010117 (rank : 123) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80VW5, Q80TC2, Q80VW4 | Gene names | Whrn, Kiaa1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin. | |||||
|
AZI1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.055803 (rank : 54) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1000 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9UPN4, Q96F50 | Gene names | AZI1, CEP131, KIAA1118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5-azacytidine-induced protein 1 (Pre-acrosome localization protein 1) (Centrosomal protein of 131 kDa) (Cep131 protein). | |||||
|
CCD91_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.052167 (rank : 62) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q7Z6B0, Q68D43, Q6IA78, Q8NEN7, Q9NUW9 | Gene names | CCDC91, GGABP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner) (p56 accessory protein). | |||||
|
CCHCR_MOUSE
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.050677 (rank : 73) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 932 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8K2I2 | Gene names | Cchcr1, Hcr | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein). | |||||
|
CI093_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.050308 (rank : 78) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q6TFL3, Q5SU58, Q6P1W1, Q6ZR13, Q8N1Z4, Q8N8L3, Q8TBR2 | Gene names | C9orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf93. | |||||
|
CING_MOUSE
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.050661 (rank : 74) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
CNTRB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.052802 (rank : 60) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8N137, Q331K3, Q69YV7, Q8NCB8, Q8WXV3, Q96CQ7, Q9C060 | Gene names | CNTROB, LIP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
CP250_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.050573 (rank : 76) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
CYLN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.051322 (rank : 70) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1004 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9UDT6, O14527, O43611 | Gene names | CYLN2, KIAA0291, WBSCR4, WSCR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115) (Williams-Beuren syndrome chromosome region 4 protein). | |||||
|
DNJC8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.051878 (rank : 67) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O75937, Q6IBA4, Q8N4Z5, Q9P051, Q9P067 | Gene names | DNAJC8, SPF31 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 8 (Splicing protein spf31). | |||||
|
DNJC8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.051259 (rank : 71) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6NZB0, Q6PG72, Q8C2M6 | Gene names | Dnajc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 8. | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.050588 (rank : 75) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
GRAP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.054378 (rank : 57) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1234 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q8VD04, O35693, Q3T9C3, Q69ZP9 | Gene names | Gripap1, DXImx47e, Kiaa1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1) (HCMV-interacting protein). | |||||
|
LIPA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.059385 (rank : 42) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1294 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q13136, Q13135, Q14567, Q8N4I2 | Gene names | PPFIA1, LIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-1) (PTPRF-interacting protein alpha-1) (LAR-interacting protein 1) (LIP.1). | |||||
|
MY18A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.052862 (rank : 59) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
MYH13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.052139 (rank : 63) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
MYH14_MOUSE
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.051254 (rank : 72) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
MYH7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.051352 (rank : 69) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
MYH8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.052008 (rank : 65) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
MYH8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.050568 (rank : 77) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
STIM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.063416 (rank : 33) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q13586 | Gene names | STIM1, GOK | |||
|
Domain Architecture |

|
|||||
| Description | Stromal interaction molecule 1 precursor. | |||||
|
STIM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.064864 (rank : 31) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P70302 | Gene names | Stim1, Sim | |||
|
Domain Architecture |

|
|||||
| Description | Stromal interaction molecule 1 precursor. | |||||
|
STMN3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.056110 (rank : 51) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9NZ72, O75527, Q969Y4 | Gene names | STMN3, SCLIP | |||
|
Domain Architecture |
|
|||||
| Description | Stathmin-3 (SCG10-like protein). | |||||
|
SWP70_MOUSE
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.051393 (rank : 68) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1097 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q6A028, O88443, Q3TQR6, Q3UCA3, Q6P1D0 | Gene names | Swap70, Kiaa0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
FAF1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q9UNN5, Q9UF34, Q9UNT3, Q9Y2Z3 | Gene names | FAF1 | |||
|
Domain Architecture |

|
|||||
| Description | FAS-associated factor 1 (Protein FAF1) (hFAF1). | |||||
|
FAF1_MOUSE
|
||||||
| NC score | 0.997369 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | P54731 | Gene names | Faf1 | |||
|
Domain Architecture |

|
|||||
| Description | FAS-associated factor 1 (Protein FAF1). | |||||
|
UBXD8_HUMAN
|
||||||
| NC score | 0.648660 (rank : 3) | θ value | 2.12685e-25 (rank : 3) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96CS3, O94963, Q8IUF2, Q9BRP2, Q9BVM7 | Gene names | UBXD8, ETEA, KIAA0887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UBX domain-containing protein 8 (Protein ETEA). | |||||
|
UBXD8_MOUSE
|
||||||
| NC score | 0.648591 (rank : 4) | θ value | 2.77775e-25 (rank : 4) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q3TDN2, Q3U9W2, Q80TP7, Q80W42 | Gene names | Ubxd8, Kiaa0887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UBX domain-containing protein 8. | |||||
|
UBXD6_MOUSE
|
||||||
| NC score | 0.437332 (rank : 5) | θ value | 4.0297e-08 (rank : 6) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9QZ49 | Gene names | Ubxd6, D0H8S2298E, Rep8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UBX domain-containing protein 6 (Reproduction 8 protein) (Protein Rep- 8). | |||||
|
UBXD6_HUMAN
|
||||||
| NC score | 0.422727 (rank : 6) | θ value | 1.80886e-08 (rank : 5) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O00124, Q7Z6F2 | Gene names | UBXD6, D8S2298E, REP8 | |||
|
Domain Architecture |

|
|||||
| Description | UBX domain-containing protein 6 (Reproduction 8 protein) (Protein Rep- 8). | |||||
|
UBXD7_HUMAN
|
||||||
| NC score | 0.383557 (rank : 7) | θ value | 8.97725e-08 (rank : 7) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O94888, Q6ZP77, Q86X20, Q8N327 | Gene names | UBXD7, KIAA0794 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UBX domain-containing protein 7. | |||||
|
UBXD7_MOUSE
|
||||||
| NC score | 0.372469 (rank : 8) | θ value | 4.45536e-07 (rank : 8) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6P5G6, Q6ZQ44 | Gene names | Ubxd7, Kiaa0794 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UBX domain-containing protein 7. | |||||
|
SAKS1_MOUSE
|
||||||
| NC score | 0.302098 (rank : 9) | θ value | 1.09739e-05 (rank : 9) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q922Y1, Q3UCP8 | Gene names | Saks1, D19Ertd721e | |||
|
Domain Architecture |

|
|||||
| Description | SAPK substrate protein 1 (UBA/UBX 33.3 kDa protein) (mY33K) (Protein 2B28). | |||||
|
SAKS1_HUMAN
|
||||||
| NC score | 0.294295 (rank : 10) | θ value | 2.44474e-05 (rank : 11) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q04323, Q9BV93, Q9BVV5 | Gene names | SAKS1 | |||
|
Domain Architecture |

|
|||||
| Description | SAPK substrate protein 1 (UBA/UBX 33.3 kDa protein). | |||||
|
UBXD2_HUMAN
|
||||||
| NC score | 0.257323 (rank : 11) | θ value | 1.87187e-05 (rank : 10) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q92575, Q8IYM5 | Gene names | UBXD2, KIAA0242, UBXDC1 | |||
|
Domain Architecture |

|
|||||
| Description | UBX domain-containing protein 2. | |||||
|
UBXD2_MOUSE
|
||||||
| NC score | 0.243579 (rank : 12) | θ value | 0.00102713 (rank : 13) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8VCH8, Q8BW17 | Gene names | Ubxd2, Ubxdc1 | |||
|
Domain Architecture |

|
|||||
| Description | UBX domain-containing protein 2. | |||||
|
UBXD1_HUMAN
|
||||||
| NC score | 0.230168 (rank : 13) | θ value | 0.000602161 (rank : 12) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BZV1, Q96AH1, Q96IK9, Q9BZV0 | Gene names | UBXD1, UBXDC2 | |||
|
Domain Architecture |

|
|||||
| Description | UBX domain-containing protein 1. | |||||
|
UBXD1_MOUSE
|
||||||
| NC score | 0.214298 (rank : 14) | θ value | 0.00134147 (rank : 14) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99PL6, Q91W79 | Gene names | Ubxd1, Ubxdc2 | |||
|
Domain Architecture |

|
|||||
| Description | UBX domain-containing protein 1. | |||||
|
GCP60_HUMAN
|
||||||
| NC score | 0.101349 (rank : 15) | θ value | 0.21417 (rank : 20) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9H3P7, Q5VTJ0, Q6P9F1, Q8IZC5, Q8N4D6, Q9H6U3 | Gene names | ACBD3, GCP60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi resident protein GCP60 (Acyl-CoA-binding domain-containing protein 3) (Golgi phosphoprotein 1) (GOLPH1) (Golgi complex-associated protein 1) (GOCAP1) (PBR- and PKA-associated protein 7) (Peripheral benzodiazepine receptor-associated protein PAP7). | |||||
|
CNTRB_MOUSE
|
||||||
| NC score | 0.094463 (rank : 16) | θ value | 0.0961366 (rank : 17) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q8CB62, Q5NCF3 | Gene names | Cntrob, Lip8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
CAF1A_HUMAN
|
||||||
| NC score | 0.092316 (rank : 17) | θ value | 0.163984 (rank : 18) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q13111, Q7Z7K3, Q9UJY8 | Gene names | CHAF1A, CAF, CAF1P150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
GCP60_MOUSE
|
||||||
| NC score | 0.083057 (rank : 18) | θ value | 1.38821 (rank : 43) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BMP6, O35371 | Gene names | Acbd3, Gcp60, Pap7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi resident protein GCP60 (Acyl-CoA-binding domain-containing protein 3) (Golgi phosphoprotein 1) (GOLPH1) (Golgi complex-associated protein 1) (GOCAP1) (PBR- and PKA-associated protein 7) (Peripheral benzodiazepine receptor-associated protein PAP7). | |||||
|
UBXD3_MOUSE
|
||||||
| NC score | 0.081631 (rank : 19) | θ value | 0.47712 (rank : 32) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BG34, Q3TTR1, Q8BP58, Q8R0B6 | Gene names | Ubxd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UBX domain-containing protein 3. | |||||
|
LIPA2_HUMAN
|
||||||
| NC score | 0.076930 (rank : 20) | θ value | 0.0252991 (rank : 15) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1044 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O75334 | Gene names | PPFIA2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
CCD46_MOUSE
|
||||||
| NC score | 0.076796 (rank : 21) | θ value | 0.0330416 (rank : 16) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q5PR68, Q80ZN6, Q99JS4, Q9D9T1 | Gene names | Ccdc46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled coil domain-containing protein 46. | |||||
|
CAF1A_MOUSE
|
||||||
| NC score | 0.074574 (rank : 22) | θ value | 1.81305 (rank : 45) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9QWF0 | Gene names | Chaf1a, Caip150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
OTU7B_HUMAN
|
||||||
| NC score | 0.073512 (rank : 23) | θ value | 0.163984 (rank : 19) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6GQQ9, Q8WWA7, Q9NQ53, Q9UFF4 | Gene names | OTUD7B, ZA20D1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | OTU domain-containing protein 7B (EC 3.-.-.-) (Zinc finger protein Cezanne) (Zinc finger A20 domain-containing protein 1) (Cellular zinc finger anti-NF-kappa B protein). | |||||
|
LIPA2_MOUSE
|
||||||
| NC score | 0.073317 (rank : 24) | θ value | 0.365318 (rank : 24) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1282 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8BSS9, Q8BN73 | Gene names | Ppfia2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
STIM2_HUMAN
|
||||||
| NC score | 0.069561 (rank : 25) | θ value | 1.81305 (rank : 47) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9P246, Q96BF1, Q9BQH2, Q9H8R1 | Gene names | STIM2, KIAA1482 | |||
|
Domain Architecture |

|
|||||
| Description | Stromal interaction molecule 2 precursor. | |||||
|
ENAH_HUMAN
|
||||||
| NC score | 0.068544 (rank : 26) | θ value | 0.47712 (rank : 28) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8N8S7, Q502W5, Q5T5M7, Q5VTQ9, Q5VTR0, Q9NVF3, Q9UFB8 | Gene names | ENAH, MENA | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog. | |||||
|
STIM2_MOUSE
|
||||||
| NC score | 0.068479 (rank : 27) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P83093, Q69ZI3, Q80VF4 | Gene names | Stim2, Kiaa1482 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal interaction molecule 2 precursor. | |||||
|
CCD46_HUMAN
|
||||||
| NC score | 0.067656 (rank : 28) | θ value | 0.365318 (rank : 23) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
IF3A_MOUSE
|
||||||
| NC score | 0.065612 (rank : 29) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P23116, Q60697, Q62162 | Gene names | Eif3s10, Csma, Eif3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a) (p162 protein) (Centrosomin). | |||||
|
IF3A_HUMAN
|
||||||
| NC score | 0.065435 (rank : 30) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q14152, O00653 | Gene names | EIF3S10, KIAA0139 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a). | |||||
|
STIM1_MOUSE
|
||||||
| NC score | 0.064864 (rank : 31) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P70302 | Gene names | Stim1, Sim | |||
|
Domain Architecture |

|
|||||
| Description | Stromal interaction molecule 1 precursor. | |||||
|
AFAD_HUMAN
|
||||||
| NC score | 0.064035 (rank : 32) | θ value | 0.47712 (rank : 27) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P55196, O75087, O75088, O75089, Q5TIG6, Q9NSN7, Q9NU92 | Gene names | MLLT4, AF6 | |||
|
Domain Architecture |

|
|||||
| Description | Afadin (Protein AF-6). | |||||
|
STIM1_HUMAN
|
||||||
| NC score | 0.063416 (rank : 33) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q13586 | Gene names | STIM1, GOK | |||
|
Domain Architecture |

|
|||||
| Description | Stromal interaction molecule 1 precursor. | |||||
|
ITSN1_MOUSE
|
||||||
| NC score | 0.061370 (rank : 34) | θ value | 0.279714 (rank : 22) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
STMN1_HUMAN
|
||||||
| NC score | 0.060893 (rank : 35) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P16949 | Gene names | STMN1, LAP18, OP18 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Protein Pr22). | |||||
|
STMN1_MOUSE
|
||||||
| NC score | 0.060795 (rank : 36) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P54227 | Gene names | Stmn1, Lag, Lap18, Pr22 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Pr22 protein) (Leukemia-associated gene protein). | |||||
|
MYH1_MOUSE
|
||||||
| NC score | 0.060305 (rank : 37) | θ value | 0.47712 (rank : 29) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
MYH2_HUMAN
|
||||||
| NC score | 0.060104 (rank : 38) | θ value | 0.62314 (rank : 33) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
MYH4_MOUSE
|
||||||
| NC score | 0.059908 (rank : 39) | θ value | 0.365318 (rank : 25) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
MYH1_HUMAN
|
||||||
| NC score | 0.059623 (rank : 40) | θ value | 0.813845 (rank : 36) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
CCD11_HUMAN
|
||||||
| NC score | 0.059506 (rank : 41) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q96M91 | Gene names | CCDC11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 11. | |||||
|
LIPA1_HUMAN
|
||||||
| NC score | 0.059385 (rank : 42) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1294 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q13136, Q13135, Q14567, Q8N4I2 | Gene names | PPFIA1, LIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-1) (PTPRF-interacting protein alpha-1) (LAR-interacting protein 1) (LIP.1). | |||||
|
OTU7A_HUMAN
|
||||||
| NC score | 0.059012 (rank : 43) | θ value | 2.36792 (rank : 50) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TE49, Q8IWK5 | Gene names | OTUD7A, C15orf16, OTUD7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | OTU domain-containing protein 7A (EC 3.-.-.-) (Zinc finger protein Cezanne 2). | |||||
|
MYH3_HUMAN
|
||||||
| NC score | 0.058716 (rank : 44) | θ value | 0.47712 (rank : 30) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
OTU7A_MOUSE
|
||||||
| NC score | 0.058633 (rank : 45) | θ value | 2.36792 (rank : 51) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R554 | Gene names | Otud7a, Otud7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | OTU domain-containing protein 7A (EC 3.-.-.-) (Zinc finger protein Cezanne 2). | |||||
|
INCE_MOUSE
|
||||||
| NC score | 0.058518 (rank : 46) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9WU62, Q7TN28, Q8BGN4, Q8CGI4 | Gene names | Incenp | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
CROCC_MOUSE
|
||||||
| NC score | 0.057821 (rank : 47) | θ value | 2.36792 (rank : 49) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
CALD1_HUMAN
|
||||||
| NC score | 0.057568 (rank : 48) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
INCE_HUMAN
|
||||||
| NC score | 0.056564 (rank : 49) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
MYH4_HUMAN
|
||||||
| NC score | 0.056312 (rank : 50) | θ value | 0.47712 (rank : 31) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
STMN3_HUMAN
|
||||||
| NC score | 0.056110 (rank : 51) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9NZ72, O75527, Q969Y4 | Gene names | STMN3, SCLIP | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin-3 (SCG10-like protein). | |||||
|
EVPL_MOUSE
|
||||||
| NC score | 0.056101 (rank : 52) | θ value | 1.81305 (rank : 46) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 952 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9D952 | Gene names | Evpl | |||
|
Domain Architecture |

|
|||||
| Description | Envoplakin (p210) (210 kDa cornified envelope precursor protein). | |||||
|
ZRF1_HUMAN
|
||||||
| NC score | 0.055931 (rank : 53) | θ value | 2.36792 (rank : 54) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q99543 | Gene names | ZRF1, DNAJC2, MPHOSPH11, MPP11 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1 (M-phase phosphoprotein 11). | |||||
|
AZI1_HUMAN
|
||||||
| NC score | 0.055803 (rank : 54) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1000 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9UPN4, Q96F50 | Gene names | AZI1, CEP131, KIAA1118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5-azacytidine-induced protein 1 (Pre-acrosome localization protein 1) (Centrosomal protein of 131 kDa) (Cep131 protein). | |||||
|
ITSN1_HUMAN
|
||||||
| NC score | 0.055198 (rank : 55) | θ value | 0.813845 (rank : 35) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
CROCC_HUMAN
|
||||||
| NC score | 0.055179 (rank : 56) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
GRAP1_MOUSE
|
||||||
| NC score | 0.054378 (rank : 57) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1234 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q8VD04, O35693, Q3T9C3, Q69ZP9 | Gene names | Gripap1, DXImx47e, Kiaa1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1) (HCMV-interacting protein). | |||||
|
MY18A_MOUSE
|
||||||
| NC score | 0.053273 (rank : 58) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
MY18A_HUMAN
|
||||||
| NC score | 0.052862 (rank : 59) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
CNTRB_HUMAN
|
||||||
| NC score | 0.052802 (rank : 60) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8N137, Q331K3, Q69YV7, Q8NCB8, Q8WXV3, Q96CQ7, Q9C060 | Gene names | CNTROB, LIP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
ENAH_MOUSE
|
||||||
| NC score | 0.052294 (rank : 61) | θ value | 1.38821 (rank : 42) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
CCD91_HUMAN
|
||||||
| NC score | 0.052167 (rank : 62) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q7Z6B0, Q68D43, Q6IA78, Q8NEN7, Q9NUW9 | Gene names | CCDC91, GGABP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner) (p56 accessory protein). | |||||
|
MYH13_HUMAN
|
||||||
| NC score | 0.052139 (rank : 63) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
REST_HUMAN
|
||||||
| NC score | 0.052034 (rank : 64) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
MYH8_HUMAN
|
||||||
| NC score | 0.052008 (rank : 65) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
NIN_HUMAN
|
||||||
| NC score | 0.051965 (rank : 66) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
DNJC8_HUMAN
|
||||||
| NC score | 0.051878 (rank : 67) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O75937, Q6IBA4, Q8N4Z5, Q9P051, Q9P067 | Gene names | DNAJC8, SPF31 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 8 (Splicing protein spf31). | |||||
|
SWP70_MOUSE
|
||||||
| NC score | 0.051393 (rank : 68) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1097 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q6A028, O88443, Q3TQR6, Q3UCA3, Q6P1D0 | Gene names | Swap70, Kiaa0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
MYH7_HUMAN
|
||||||
| NC score | 0.051352 (rank : 69) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
CYLN2_HUMAN
|
||||||
| NC score | 0.051322 (rank : 70) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1004 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9UDT6, O14527, O43611 | Gene names | CYLN2, KIAA0291, WBSCR4, WSCR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115) (Williams-Beuren syndrome chromosome region 4 protein). | |||||
|
DNJC8_MOUSE
|
||||||
| NC score | 0.051259 (rank : 71) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6NZB0, Q6PG72, Q8C2M6 | Gene names | Dnajc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 8. | |||||
|
MYH14_MOUSE
|
||||||
| NC score | 0.051254 (rank : 72) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
CCHCR_MOUSE
|
||||||
| NC score | 0.050677 (rank : 73) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 932 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8K2I2 | Gene names | Cchcr1, Hcr | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein). | |||||
|
CING_MOUSE
|
||||||
| NC score | 0.050661 (rank : 74) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.050588 (rank : 75) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
CP250_HUMAN
|
||||||
| NC score | 0.050573 (rank : 76) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
MYH8_MOUSE
|
||||||
| NC score | 0.050568 (rank : 77) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
CI093_HUMAN
|
||||||
| NC score | 0.050308 (rank : 78) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q6TFL3, Q5SU58, Q6P1W1, Q6ZR13, Q8N1Z4, Q8N8L3, Q8TBR2 | Gene names | C9orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf93. | |||||
|
REST_MOUSE
|
||||||
| NC score | 0.049944 (rank : 79) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
ZRF1_MOUSE
|
||||||
| NC score | 0.049875 (rank : 80) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P54103 | Gene names | Zrf1, Dnajc2 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1. | |||||
|
UACA_MOUSE
|
||||||
| NC score | 0.049862 (rank : 81) | θ value | 0.365318 (rank : 26) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8CGB3, Q69ZG3, Q7TN77, Q8BJC8 | Gene names | Uaca, Kiaa1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein (Nucling) (Nuclear membrane-binding protein). | |||||
|
GOGA5_HUMAN
|
||||||
| NC score | 0.049009 (rank : 82) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1168 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8TBA6, O95287, Q03962, Q9UQQ7 | Gene names | GOLGA5, RETII, RFG5 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (RET-fused gene 5 protein) (Ret-II protein). | |||||
|
EEA1_MOUSE
|
||||||
| NC score | 0.048941 (rank : 83) | θ value | 1.38821 (rank : 41) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
MYO6_HUMAN
|
||||||
| NC score | 0.048747 (rank : 84) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UM54, Q9BZZ7, Q9UEG2 | Gene names | MYO6, KIAA0389 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-6 (Myosin VI). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.048703 (rank : 85) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
DREB_MOUSE
|
||||||
| NC score | 0.048552 (rank : 86) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9QXS6, Q922X1, Q9QXS5 | Gene names | Dbn1, Drba | |||
|
Domain Architecture |

|
|||||
| Description | Drebrin (Developmentally-regulated brain protein). | |||||
|
RABE2_HUMAN
|
||||||
| NC score | 0.048054 (rank : 87) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 545 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9H5N1 | Gene names | RABEP2, RABPT5B | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 2 (Rabaptin-5beta). | |||||
|
RBP1_MOUSE
|
||||||
| NC score | 0.047343 (rank : 88) | θ value | 1.06291 (rank : 40) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q62172, Q9CRE5 | Gene names | Ralbp1, Rip1 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
MYO6_MOUSE
|
||||||
| NC score | 0.044582 (rank : 89) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q64331 | Gene names | Myo6, Sv | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-6 (Myosin VI). | |||||
|
CROP_HUMAN
|
||||||
| NC score | 0.043600 (rank : 90) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O95232, Q6PHR9, Q9NUY0, Q9P2S7 | Gene names | CROP, CREAP1, O48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cisplatin resistance-associated overexpressed protein (cAMP regulatory element-associated protein 1) (CRE-associated protein 1) (CREAP-1) (Luc7A) (Okadaic acid-inducible phosphoprotein OA48-18). | |||||
|
CROP_MOUSE
|
||||||
| NC score | 0.040800 (rank : 91) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5SUF2, Q3U9D5, Q8BUJ5, Q921Z3, Q9CRS7, Q9CTY4 | Gene names | Crop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cisplatin resistance-associated overexpressed protein. | |||||
|
RENT2_HUMAN
|
||||||
| NC score | 0.040439 (rank : 92) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
RBP1_HUMAN
|
||||||
| NC score | 0.039946 (rank : 93) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q15311 | Gene names | RALBP1, RLIP1, RLIP76 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (76 kDa Ral-interacting protein) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
K0195_MOUSE
|
||||||
| NC score | 0.039869 (rank : 94) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TSH8, Q80U66, Q8BL05, Q8K0Q0, Q91VY1 | Gene names | Kiaa0195 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0195. | |||||
|
K0195_HUMAN
|
||||||
| NC score | 0.039649 (rank : 95) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12767, O75536, Q86XF1 | Gene names | KIAA0195, TMEM94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0195 (Transmembrane protein 94). | |||||
|
STK10_MOUSE
|
||||||
| NC score | 0.038553 (rank : 96) | θ value | 0.21417 (rank : 21) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
PLSB_MOUSE
|
||||||
| NC score | 0.036668 (rank : 97) | θ value | 2.36792 (rank : 52) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61586 | Gene names | Gpam | |||
|
Domain Architecture |

|
|||||
| Description | Glycerol-3-phosphate acyltransferase, mitochondrial precursor (EC 2.3.1.15) (GPAT) (P90). | |||||
|
ANKZ1_MOUSE
|
||||||
| NC score | 0.036490 (rank : 98) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q80UU1, Q9CZF5 | Gene names | Ankzf1, D1Ertd161e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and zinc finger domain-containing protein 1. | |||||
|
PELP1_HUMAN
|
||||||
| NC score | 0.036259 (rank : 99) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8IZL8, O15450, Q5EGN3, Q6NTE6, Q96FT1, Q9BU60 | Gene names | PELP1, HMX3, MNAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3). | |||||
|
RHG05_MOUSE
|
||||||
| NC score | 0.034705 (rank : 100) | θ value | 2.36792 (rank : 53) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P97393 | Gene names | Arhgap5, Rhogap5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
SPTA2_HUMAN
|
||||||
| NC score | 0.033382 (rank : 101) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 888 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q13813, Q13186, Q15324, Q16606, Q59EF1, Q5VXV5, Q5VXV6, Q7Z6M5, Q9P0V0 | Gene names | SPTAN1, SPTA2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, brain (Spectrin, non-erythroid alpha chain) (Alpha-II spectrin) (Fodrin alpha chain). | |||||
|
CA082_MOUSE
|
||||||
| NC score | 0.032027 (rank : 102) | θ value | 1.81305 (rank : 44) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VC34, Q3TLC8, Q3TSP8, Q3UKX0, Q8C7M5, Q8CBW8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UPF0385 protein C1orf82 homolog. | |||||
|
K1949_MOUSE
|
||||||
| NC score | 0.030984 (rank : 103) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BQ30, Q3UDS6, Q8CEY9, Q8R3D8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1949 homolog. | |||||
|
PELP1_MOUSE
|
||||||
| NC score | 0.029897 (rank : 104) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
GP108_MOUSE
|
||||||
| NC score | 0.029723 (rank : 105) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91WD0, Q8BXE9, Q925B1 | Gene names | Gpr108, Lustr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein GPR108 precursor (Lung seven transmembrane receptor 2). | |||||
|
RHG05_HUMAN
|
||||||
| NC score | 0.029416 (rank : 106) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13017 | Gene names | ARHGAP5, RHOGAP5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
CALC_MOUSE
|
||||||
| NC score | 0.027561 (rank : 107) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70160, Q8K1K5 | Gene names | Calca, Calc | |||
|
Domain Architecture |

|
|||||
| Description | Calcitonin precursor. | |||||
|
FYB_MOUSE
|
||||||
| NC score | 0.025627 (rank : 108) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O35601, Q9Z2H3 | Gene names | Fyb | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
ROCK2_HUMAN
|
||||||
| NC score | 0.024458 (rank : 109) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 2073 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O75116, Q9UQN5 | Gene names | ROCK2, KIAA0619 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2) (Rho kinase 2). | |||||
|
RGP1_HUMAN
|
||||||
| NC score | 0.023548 (rank : 110) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P46060 | Gene names | RANGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
FYB_HUMAN
|
||||||
| NC score | 0.023379 (rank : 111) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O15117, O00359 | Gene names | FYB, SLAP130 | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
SRC8_HUMAN
|
||||||
| NC score | 0.022533 (rank : 112) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q14247 | Gene names | CTTN, EMS1 | |||
|
Domain Architecture |
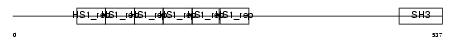
|
|||||
| Description | Src substrate cortactin (Amplaxin) (Oncogene EMS1). | |||||
|
UBP8_HUMAN
|
||||||
| NC score | 0.022110 (rank : 113) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P40818, Q7Z3U2, Q86VA0, Q8IWI7 | Gene names | USP8, KIAA0055, UBPY | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (hUBPy). | |||||
|
SMCA4_HUMAN
|
||||||
| NC score | 0.021993 (rank : 114) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P51532, O95052, Q9HBD3 | Gene names | SMARCA4, BRG1, SNF2B, SNF2L4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L4 (EC 3.6.1.-) (ATP- dependent helicase SMARCA4) (SNF2-beta) (BRG-1 protein) (Mitotic growth and transcription activator) (Brahma protein homolog 1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4). | |||||
|
SRC8_MOUSE
|
||||||
| NC score | 0.021546 (rank : 115) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q60598 | Gene names | Cttn, Ems1 | |||
|
Domain Architecture |
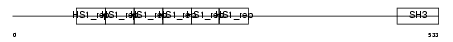
|
|||||
| Description | Src substrate cortactin. | |||||
|
M4K4_HUMAN
|
||||||
| NC score | 0.020582 (rank : 116) | θ value | 1.06291 (rank : 38) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1589 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O95819, O75172, Q9NST7 | Gene names | MAP4K4, HGK, KIAA0687, NIK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
M4K4_MOUSE
|
||||||
| NC score | 0.020242 (rank : 117) | θ value | 1.06291 (rank : 39) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1560 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P97820 | Gene names | Map4k4, Nik | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
ECM2_HUMAN
|
||||||
| NC score | 0.017952 (rank : 118) | θ value | 1.06291 (rank : 37) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94769 | Gene names | ECM2 | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular matrix protein 2 precursor (Matrix glycoprotein SC1/ECM2). | |||||
|
RYR1_HUMAN
|
||||||
| NC score | 0.015629 (rank : 119) | θ value | 4.03905 (rank : 66) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P21817, Q16314, Q16368, Q9NPK1, Q9P1U4 | Gene names | RYR1, RYDR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 1 (Skeletal muscle-type ryanodine receptor) (RyR1) (RYR-1) (Skeletal muscle calcium release channel). | |||||
|
SNX11_MOUSE
|
||||||
| NC score | 0.015348 (rank : 120) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91WL6, Q3V3A6 | Gene names | Snx11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorting nexin-11. | |||||
|
MYT1_MOUSE
|
||||||
| NC score | 0.013357 (rank : 121) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8CFC2, O08995, Q8CFH1 | Gene names | Myt1, Kiaa0835, Nzf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1 (MyT1) (Neural zinc finger factor 2) (NZF-2). | |||||
|
PSA7_HUMAN
|
||||||
| NC score | 0.012262 (rank : 122) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14818, Q9BR53, Q9H4K5, Q9UEU8 | Gene names | PSMA7 | |||
|
Domain Architecture |

|
|||||
| Description | Proteasome subunit alpha type 7 (EC 3.4.25.1) (Proteasome subunit RC6- 1) (Proteasome subunit XAPC7). | |||||
|
WHRN_MOUSE
|
||||||
| NC score | 0.010117 (rank : 123) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80VW5, Q80TC2, Q80VW4 | Gene names | Whrn, Kiaa1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin. | |||||
|
ABTB1_MOUSE
|
||||||
| NC score | 0.009614 (rank : 124) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99LJ2, Q8C2K7, Q91XU4 | Gene names | Abtb1, Bpoz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and BTB/POZ domain-containing protein 1. | |||||
|
STK38_MOUSE
|
||||||
| NC score | 0.005325 (rank : 125) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 838 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91VJ4 | Gene names | Stk38, Ndr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase 38 (EC 2.7.11.1) (NDR1 protein kinase) (Nuclear Dbf2-related kinase 1). | |||||
|
STK38_HUMAN
|
||||||
| NC score | 0.005289 (rank : 126) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 840 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15208 | Gene names | STK38, NDR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase 38 (EC 2.7.11.1) (NDR1 protein kinase) (Nuclear Dbf2-related kinase 1). | |||||
|
ARBK2_HUMAN
|
||||||
| NC score | 0.002840 (rank : 127) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 849 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35626, Q9UGW9 | Gene names | ADRBK2, BARK2, GRK3 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adrenergic receptor kinase 2 (EC 2.7.11.15) (Beta-ARK-2) (G- protein-coupled receptor kinase 3). | |||||
|
PRDM1_MOUSE
|
||||||
| NC score | 0.002057 (rank : 128) | θ value | 0.62314 (rank : 34) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 766 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q60636 | Gene names | Prdm1, Blimp1 | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 1 (PR domain-containing protein 1) (Beta-interferon gene positive-regulatory domain I-binding factor) (BLIMP-1). | |||||