Please be patient as the page loads
|
NMDE2_HUMAN
|
||||||
| SwissProt Accessions | Q13224, Q12919, Q13220, Q13225, Q9UM56 | Gene names | GRIN2B | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 2 precursor (N-methyl D- aspartate receptor subtype 2B) (NR2B) (NMDAR2B) (N-methyl-D-aspartate receptor subunit 3) (NR3) (hNR3). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NMDE1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.992042 (rank : 4) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q12879, O00669 | Gene names | GRIN2A | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 1 precursor (N-methyl D- aspartate receptor subtype 2A) (NR2A) (NMDAR2A) (hNR2A). | |||||
|
NMDE1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.992656 (rank : 3) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P35436 | Gene names | Grin2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 1 precursor (N-methyl D- aspartate receptor subtype 2A) (NR2A) (NMDAR2A). | |||||
|
NMDE2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q13224, Q12919, Q13220, Q13225, Q9UM56 | Gene names | GRIN2B | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 2 precursor (N-methyl D- aspartate receptor subtype 2B) (NR2B) (NMDAR2B) (N-methyl-D-aspartate receptor subunit 3) (NR3) (hNR3). | |||||
|
NMDE2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.999778 (rank : 2) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q01097, Q9DCB2 | Gene names | Grin2b | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 2 precursor (N-methyl D- aspartate receptor subtype 2B) (NR2B) (NMDAR2B). | |||||
|
NMDE3_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.987058 (rank : 6) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q14957 | Gene names | GRIN2C | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 3 precursor (N-methyl D- aspartate receptor subtype 2C) (NR2C) (NMDAR2C). | |||||
|
NMDE3_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.987393 (rank : 5) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q01098 | Gene names | Grin2c | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 3 precursor (N-methyl D- aspartate receptor subtype 2C) (NR2C) (NMDAR2C). | |||||
|
NMDE4_HUMAN
|
||||||
| θ value | 0 (rank : 7) | NC score | 0.984420 (rank : 7) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O15399 | Gene names | GRIN2D | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 4 precursor (N-methyl D- aspartate receptor subtype 2D) (NR2D) (NMDAR2D) (EB11). | |||||
|
NMDE4_MOUSE
|
||||||
| θ value | 0 (rank : 8) | NC score | 0.981957 (rank : 8) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q03391 | Gene names | Grin2d | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 4 precursor (N-methyl D- aspartate receptor subtype 2D) (NR2D) (NMDAR2D). | |||||
|
NMDZ1_HUMAN
|
||||||
| θ value | 6.95003e-85 (rank : 9) | NC score | 0.911492 (rank : 12) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q05586, P35437, Q12867, Q12868 | Gene names | GRIN1, NMDAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit zeta 1 precursor (N-methyl-D- aspartate receptor subunit NR1). | |||||
|
NMDZ1_MOUSE
|
||||||
| θ value | 2.02215e-84 (rank : 10) | NC score | 0.911108 (rank : 13) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P35438, Q8CFS4 | Gene names | Grin1, Glurz1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit zeta 1 precursor (N-methyl-D- aspartate receptor subunit NR1). | |||||
|
NMD3A_HUMAN
|
||||||
| θ value | 9.39325e-82 (rank : 11) | NC score | 0.934870 (rank : 9) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8TCU5, Q8TF29, Q8WXI6 | Gene names | GRIN3A, KIAA1973 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit 3A precursor (N-methyl-D-aspartate receptor subtype NR3A) (NMDAR-L). | |||||
|
NMD3B_MOUSE
|
||||||
| θ value | 9.11921e-69 (rank : 12) | NC score | 0.925481 (rank : 10) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q91ZU9, Q8VHV0 | Gene names | Grin3b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit 3B precursor (N-methyl-D-aspartate receptor subunit NR3B) (NR3B) (NMDAR3B) (NMDA receptor NR4) (Nr4). | |||||
|
NMD3B_HUMAN
|
||||||
| θ value | 2.4834e-66 (rank : 13) | NC score | 0.924399 (rank : 11) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O60391, Q5EAK7, Q7RTW9 | Gene names | GRIN3B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit 3B precursor (N-methyl-D-aspartate receptor subtype NR3B) (NR3B) (NMDAR3B). | |||||
|
GRIK2_HUMAN
|
||||||
| θ value | 8.02596e-49 (rank : 14) | NC score | 0.839485 (rank : 19) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q13002 | Gene names | GRIK2, GLUR6 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 2 precursor (Glutamate receptor 6) (GluR-6) (GluR6) (Excitatory amino acid receptor 4) (EAA4). | |||||
|
GRIK1_HUMAN
|
||||||
| θ value | 3.98324e-48 (rank : 15) | NC score | 0.841230 (rank : 16) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P39086, Q13001, Q86SU9 | Gene names | GRIK1, GLUR5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 1 precursor (Glutamate receptor 5) (GluR-5) (GluR5) (Excitatory amino acid receptor 3) (EAA3). | |||||
|
GRIA4_HUMAN
|
||||||
| θ value | 1.15895e-47 (rank : 16) | NC score | 0.831684 (rank : 27) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P48058 | Gene names | GRIA4, GLUR4 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 4 precursor (GluR-4) (GluR4) (GluR-D) (Glutamate receptor ionotropic, AMPA 4) (AMPA-selective glutamate receptor 4). | |||||
|
GRIA4_MOUSE
|
||||||
| θ value | 9.81109e-47 (rank : 17) | NC score | 0.829582 (rank : 28) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Z2W8 | Gene names | Gria4, Glur4 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 4 precursor (GluR-4) (GluR4) (GluR-D) (Glutamate receptor ionotropic, AMPA 4) (AMPA-selective glutamate receptor 4). | |||||
|
GRIA1_MOUSE
|
||||||
| θ value | 1.28137e-46 (rank : 18) | NC score | 0.834654 (rank : 25) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P23818 | Gene names | Gria1, Glur1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 1 precursor (GluR-1) (GluR-A) (GluR-K1) (Glutamate receptor ionotropic, AMPA 1) (AMPA-selective glutamate receptor 1). | |||||
|
GRIA1_HUMAN
|
||||||
| θ value | 1.08474e-45 (rank : 19) | NC score | 0.833234 (rank : 26) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P42261 | Gene names | GRIA1, GLUH1, GLUR1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 1 precursor (GluR-1) (GluR-A) (GluR-K1) (Glutamate receptor ionotropic, AMPA 1) (AMPA-selective glutamate receptor 1). | |||||
|
GRIK3_HUMAN
|
||||||
| θ value | 1.08474e-45 (rank : 20) | NC score | 0.841197 (rank : 17) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q13003, Q13004, Q16136 | Gene names | GRIK3, GLUR7 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 3 precursor (Glutamate receptor 7) (GluR-7) (GluR7) (Excitatory amino acid receptor 5) (EAA5). | |||||
|
GRIA2_HUMAN
|
||||||
| θ value | 3.15612e-45 (rank : 21) | NC score | 0.829526 (rank : 29) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P42262, Q96FP6 | Gene names | GRIA2, GLUR2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 2 precursor (GluR-2) (GluR-B) (GluR-K2) (Glutamate receptor ionotropic, AMPA 2) (AMPA-selective glutamate receptor 2). | |||||
|
GRIA3_HUMAN
|
||||||
| θ value | 5.38352e-45 (rank : 22) | NC score | 0.829179 (rank : 30) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P42263, Q4VXD5, Q4VXD6, Q9HDA0, Q9HDA1, Q9HDA2 | Gene names | GRIA3, GLUR3, GLURC | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 3 precursor (GluR-3) (GluR-C) (GluR-K3) (Glutamate receptor ionotropic, AMPA 3) (AMPA-selective glutamate receptor 3). | |||||
|
GRIA2_MOUSE
|
||||||
| θ value | 9.18288e-45 (rank : 23) | NC score | 0.828856 (rank : 32) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P23819, Q61604, Q61605, Q8BG69, Q8BXU3, Q9D6D3 | Gene names | Gria2, Glur2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 2 precursor (GluR-2) (GluR-B) (GluR-K2) (Glutamate receptor ionotropic, AMPA 2) (AMPA-selective glutamate receptor 2). | |||||
|
GRIA3_MOUSE
|
||||||
| θ value | 9.18288e-45 (rank : 24) | NC score | 0.829169 (rank : 31) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Z2W9, Q5DTJ0 | Gene names | Gria3, Glur3, Kiaa4184 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate receptor 3 precursor (GluR-3) (GluR-C) (GluR-K3) (Glutamate receptor ionotropic, AMPA 3) (AMPA-selective glutamate receptor 3). | |||||
|
GRIK4_HUMAN
|
||||||
| θ value | 2.26182e-43 (rank : 25) | NC score | 0.838709 (rank : 21) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q16099 | Gene names | GRIK4, GRIK | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 4 precursor (Glutamate receptor KA-1) (KA1) (Excitatory amino acid receptor 1) (EAA1). | |||||
|
GRIK4_MOUSE
|
||||||
| θ value | 2.26182e-43 (rank : 26) | NC score | 0.838451 (rank : 22) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BMF5 | Gene names | Grik4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate receptor, ionotropic kainate 4 precursor. | |||||
|
GRID2_MOUSE
|
||||||
| θ value | 2.11701e-41 (rank : 27) | NC score | 0.844270 (rank : 14) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q61625 | Gene names | Grid2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-2 subunit precursor (GluR delta-2). | |||||
|
GRIK5_MOUSE
|
||||||
| θ value | 6.15952e-41 (rank : 28) | NC score | 0.836829 (rank : 24) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61626 | Gene names | Grik5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 5 precursor (Glutamate receptor KA-2) (KA2) (Glutamate receptor gamma-2) (GluR gamma-2). | |||||
|
GRIK5_HUMAN
|
||||||
| θ value | 1.05066e-40 (rank : 29) | NC score | 0.837246 (rank : 23) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q16478 | Gene names | GRIK5, GRIK2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 5 precursor (Glutamate receptor KA-2) (KA2) (Excitatory amino acid receptor 2) (EAA2). | |||||
|
GRID2_HUMAN
|
||||||
| θ value | 1.3722e-40 (rank : 30) | NC score | 0.842922 (rank : 15) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O43424 | Gene names | GRID2, GLURD2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-2 subunit precursor (GluR delta-2). | |||||
|
GRID1_MOUSE
|
||||||
| θ value | 6.81017e-40 (rank : 31) | NC score | 0.840349 (rank : 18) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61627 | Gene names | Grid1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-1 subunit precursor (GluR delta-1). | |||||
|
GRID1_HUMAN
|
||||||
| θ value | 8.89434e-40 (rank : 32) | NC score | 0.838904 (rank : 20) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9ULK0, Q8IXT3 | Gene names | GRID1, KIAA1220 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-1 subunit precursor (GluR delta-1). | |||||
|
GRIK1_MOUSE
|
||||||
| θ value | 1.12514e-34 (rank : 33) | NC score | 0.822514 (rank : 33) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q60934 | Gene names | Grik1, Glur5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 1 precursor (Glutamate receptor 5) (GluR-5) (GluR5). | |||||
|
FBLN1_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 34) | NC score | 0.016463 (rank : 42) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P23142, P23143, P23144, P37888, Q5TIC4, Q8TBH8, Q9HBQ5, Q9UC21, Q9UGR4, Q9UH41 | Gene names | FBLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-1 precursor. | |||||
|
ERB1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 35) | NC score | 0.139651 (rank : 34) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P60509 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-R(b)_3p24.3 provirus ancestral Env polyprotein precursor (Envelope polyprotein) (HERV-R(b) Env protein) [Includes: Surface protein domain (SU); Transmembrane protein domain (TM)]. | |||||
|
FBLN1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 36) | NC score | 0.013106 (rank : 46) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q08879, Q08878, Q8C3B1, Q91ZC9, Q922K8 | Gene names | Fbln1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-1 precursor (Basement-membrane protein 90) (BM-90). | |||||
|
MGR5_HUMAN
|
||||||
| θ value | 0.21417 (rank : 37) | NC score | 0.036051 (rank : 38) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P41594 | Gene names | GRM5, GPRC1E, MGLUR5 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 5 precursor (mGluR5). | |||||
|
ZN473_MOUSE
|
||||||
| θ value | 0.365318 (rank : 38) | NC score | -0.001340 (rank : 61) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 824 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BI67, Q8BI98, Q8BIB7 | Gene names | Znf473, Zfp100, Zfp473 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 473 homolog (Zinc finger protein 100) (Zfp-100). | |||||
|
GABR2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 39) | NC score | 0.045638 (rank : 37) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75899, O75974, O75975, Q5VXZ2, Q8WX04, Q9P1R2, Q9UNR1, Q9UNS9 | Gene names | GABBR2, GPR51, GPRC3B | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 2 precursor (GABA-B receptor 2) (GABA-B-R2) (Gb2) (GABABR2) (G-protein coupled receptor 51) (HG20). | |||||
|
BLK_HUMAN
|
||||||
| θ value | 1.06291 (rank : 40) | NC score | -0.000814 (rank : 60) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 994 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P51451, Q16291 | Gene names | BLK | |||
|
Domain Architecture |
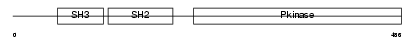
|
|||||
| Description | Tyrosine-protein kinase BLK (EC 2.7.10.2) (B lymphocyte kinase) (p55- BLK). | |||||
|
RBM25_HUMAN
|
||||||
| θ value | 1.38821 (rank : 41) | NC score | 0.009192 (rank : 51) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P49756, Q9UEQ5, Q9UIE9 | Gene names | RBM25, RNPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable RNA-binding protein 25 (RNA-binding motif protein 25) (RNA- binding region-containing protein 7) (Protein S164). | |||||
|
U383_MOUSE
|
||||||
| θ value | 1.38821 (rank : 42) | NC score | 0.023509 (rank : 41) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D2Q2, Q3V1K3 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative methyltransferase UPF0383 (EC 2.1.1.-). | |||||
|
MGR1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.034109 (rank : 40) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13255, Q13256, Q14757, Q14758, Q5VTF7, Q5VTF8, Q9NU10, Q9UGS9, Q9UGT0 | Gene names | GRM1, GPRC1A, MGLUR1 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 1 precursor (mGluR1). | |||||
|
MGR1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.034134 (rank : 39) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P97772, Q6AXG4, Q9EPV6 | Gene names | Grm1, Gprc1a, Mglur1 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 1 precursor (mGluR1). | |||||
|
RAD50_MOUSE
|
||||||
| θ value | 2.36792 (rank : 45) | NC score | 0.000210 (rank : 58) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
YLPM1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.010998 (rank : 48) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
RNF12_HUMAN
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.013106 (rank : 45) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NVW2, Q9Y598 | Gene names | RNF12, RLIM | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 12 (LIM domain-interacting RING finger protein) (RING finger LIM domain-binding protein) (R-LIM) (NY-REN-43 antigen). | |||||
|
RNF12_MOUSE
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.012424 (rank : 47) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WTV7 | Gene names | Rnf12, Rlim | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 12 (LIM domain-interacting RING finger protein) (RING finger LIM domain-binding protein) (R-LIM). | |||||
|
CD2L2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 49) | NC score | -0.004465 (rank : 67) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
CUTL2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 50) | NC score | 0.004850 (rank : 53) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14529 | Gene names | CUTL2, KIAA0293 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
PIR_MOUSE
|
||||||
| θ value | 6.88961 (rank : 51) | NC score | 0.010772 (rank : 49) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D711, Q3UAN0 | Gene names | Pir | |||
|
Domain Architecture |
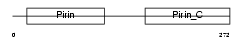
|
|||||
| Description | Pirin. | |||||
|
SFR16_HUMAN
|
||||||
| θ value | 6.88961 (rank : 52) | NC score | 0.014482 (rank : 44) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N2M8, O96026, Q6UW71, Q96DX2 | Gene names | SFRS16, SWAP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 16 (Suppressor of white-apricot homolog 2). | |||||
|
SFR16_MOUSE
|
||||||
| θ value | 6.88961 (rank : 53) | NC score | 0.014650 (rank : 43) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CFC7, Q8CFC8, Q9Z2N3 | Gene names | Sfrs16, Clasp, Swap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 16 (Suppressor of white-apricot homolog 2) (Clk4-associating SR-related protein). | |||||
|
UBP50_MOUSE
|
||||||
| θ value | 6.88961 (rank : 54) | NC score | 0.001569 (rank : 57) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6P8X6, Q9D558, Q9D9E7 | Gene names | Usp50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ubiquitin carboxyl-terminal hydrolase 50 (EC 3.1.2.15) (Ubiquitin thioesterase 50) (Ubiquitin-specific-processing protease 50) (Deubiquitinating enzyme 50). | |||||
|
ZNF23_HUMAN
|
||||||
| θ value | 6.88961 (rank : 55) | NC score | -0.003260 (rank : 65) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P17027, Q96IT3, Q9UG42 | Gene names | ZNF23, KOX16, ZNF359 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 23 (Zinc finger protein 359) (Zinc finger protein KOX16). | |||||
|
ADA11_HUMAN
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.003357 (rank : 54) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75078, Q14808, Q14809, Q14810 | Gene names | ADAM11, MDC | |||
|
Domain Architecture |
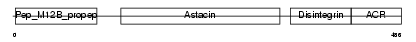
|
|||||
| Description | ADAM 11 precursor (A disintegrin and metalloproteinase domain 11) (Metalloproteinase-like, disintegrin-like, and cysteine-rich protein) (MDC). | |||||
|
ADA11_MOUSE
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.003252 (rank : 55) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9R1V4 | Gene names | Adam11, Mdc | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 11 precursor (A disintegrin and metalloproteinase domain 11) (Metalloproteinase-like, disintegrin-like, and cysteine-rich protein) (MDC). | |||||
|
FYN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | -0.002960 (rank : 64) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1078 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P06241 | Gene names | FYN | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase Fyn (EC 2.7.10.2) (p59-Fyn) (Protooncogene Syn) (SLK). | |||||
|
FYN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | -0.002889 (rank : 63) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P39688 | Gene names | Fyn | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase Fyn (EC 2.7.10.2) (p59-Fyn). | |||||
|
K0195_HUMAN
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.007716 (rank : 52) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12767, O75536, Q86XF1 | Gene names | KIAA0195, TMEM94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0195 (Transmembrane protein 94). | |||||
|
MYPT1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 61) | NC score | -0.001954 (rank : 62) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14974, Q86WU3, Q8NFR6, Q9BYH0 | Gene names | PPP1R12A, MBS, MYPT1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1) (Protein phosphatase myosin-binding subunit). | |||||
|
PIR_HUMAN
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | 0.009809 (rank : 50) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00625, Q5U0G0 | Gene names | PIR | |||
|
Domain Architecture |
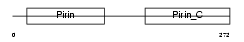
|
|||||
| Description | Pirin. | |||||
|
PTPRA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.000157 (rank : 59) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P18433, Q14513, Q7Z2I2, Q96TD9 | Gene names | PTPRA, PTPA | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase alpha precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase alpha) (R-PTP-alpha). | |||||
|
ZBT38_HUMAN
|
||||||
| θ value | 8.99809 (rank : 64) | NC score | -0.003356 (rank : 66) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 885 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NAP3 | Gene names | ZBTB38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and BTB domain-containing protein 38. | |||||
|
ZMYM3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 65) | NC score | 0.003036 (rank : 56) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14202, O15089 | Gene names | ZMYM3, DXS6673E, KIAA0385, ZNF261 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 3 (Zinc finger protein 261). | |||||
|
GABR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.054574 (rank : 35) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UBS5, O95375, O95468, O95975, O96022, Q86W60, Q9UQQ0 | Gene names | GABBR1, GPRC3A | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
GABR1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.054460 (rank : 36) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9WV18, Q9WU48, Q9WV15, Q9WV16, Q9WV17 | Gene names | Gabbr1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
NMDE2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q13224, Q12919, Q13220, Q13225, Q9UM56 | Gene names | GRIN2B | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 2 precursor (N-methyl D- aspartate receptor subtype 2B) (NR2B) (NMDAR2B) (N-methyl-D-aspartate receptor subunit 3) (NR3) (hNR3). | |||||
|
NMDE2_MOUSE
|
||||||
| NC score | 0.999778 (rank : 2) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q01097, Q9DCB2 | Gene names | Grin2b | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 2 precursor (N-methyl D- aspartate receptor subtype 2B) (NR2B) (NMDAR2B). | |||||
|
NMDE1_MOUSE
|
||||||
| NC score | 0.992656 (rank : 3) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P35436 | Gene names | Grin2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 1 precursor (N-methyl D- aspartate receptor subtype 2A) (NR2A) (NMDAR2A). | |||||
|
NMDE1_HUMAN
|
||||||
| NC score | 0.992042 (rank : 4) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q12879, O00669 | Gene names | GRIN2A | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 1 precursor (N-methyl D- aspartate receptor subtype 2A) (NR2A) (NMDAR2A) (hNR2A). | |||||
|
NMDE3_MOUSE
|
||||||
| NC score | 0.987393 (rank : 5) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q01098 | Gene names | Grin2c | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 3 precursor (N-methyl D- aspartate receptor subtype 2C) (NR2C) (NMDAR2C). | |||||
|
NMDE3_HUMAN
|
||||||
| NC score | 0.987058 (rank : 6) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q14957 | Gene names | GRIN2C | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 3 precursor (N-methyl D- aspartate receptor subtype 2C) (NR2C) (NMDAR2C). | |||||
|
NMDE4_HUMAN
|
||||||
| NC score | 0.984420 (rank : 7) | θ value | 0 (rank : 7) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O15399 | Gene names | GRIN2D | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 4 precursor (N-methyl D- aspartate receptor subtype 2D) (NR2D) (NMDAR2D) (EB11). | |||||
|
NMDE4_MOUSE
|
||||||
| NC score | 0.981957 (rank : 8) | θ value | 0 (rank : 8) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q03391 | Gene names | Grin2d | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 4 precursor (N-methyl D- aspartate receptor subtype 2D) (NR2D) (NMDAR2D). | |||||
|
NMD3A_HUMAN
|
||||||
| NC score | 0.934870 (rank : 9) | θ value | 9.39325e-82 (rank : 11) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8TCU5, Q8TF29, Q8WXI6 | Gene names | GRIN3A, KIAA1973 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit 3A precursor (N-methyl-D-aspartate receptor subtype NR3A) (NMDAR-L). | |||||
|
NMD3B_MOUSE
|
||||||
| NC score | 0.925481 (rank : 10) | θ value | 9.11921e-69 (rank : 12) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q91ZU9, Q8VHV0 | Gene names | Grin3b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit 3B precursor (N-methyl-D-aspartate receptor subunit NR3B) (NR3B) (NMDAR3B) (NMDA receptor NR4) (Nr4). | |||||
|
NMD3B_HUMAN
|
||||||
| NC score | 0.924399 (rank : 11) | θ value | 2.4834e-66 (rank : 13) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O60391, Q5EAK7, Q7RTW9 | Gene names | GRIN3B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit 3B precursor (N-methyl-D-aspartate receptor subtype NR3B) (NR3B) (NMDAR3B). | |||||
|
NMDZ1_HUMAN
|
||||||
| NC score | 0.911492 (rank : 12) | θ value | 6.95003e-85 (rank : 9) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q05586, P35437, Q12867, Q12868 | Gene names | GRIN1, NMDAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit zeta 1 precursor (N-methyl-D- aspartate receptor subunit NR1). | |||||
|
NMDZ1_MOUSE
|
||||||
| NC score | 0.911108 (rank : 13) | θ value | 2.02215e-84 (rank : 10) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P35438, Q8CFS4 | Gene names | Grin1, Glurz1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit zeta 1 precursor (N-methyl-D- aspartate receptor subunit NR1). | |||||
|
GRID2_MOUSE
|
||||||
| NC score | 0.844270 (rank : 14) | θ value | 2.11701e-41 (rank : 27) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q61625 | Gene names | Grid2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-2 subunit precursor (GluR delta-2). | |||||
|
GRID2_HUMAN
|
||||||
| NC score | 0.842922 (rank : 15) | θ value | 1.3722e-40 (rank : 30) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O43424 | Gene names | GRID2, GLURD2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-2 subunit precursor (GluR delta-2). | |||||
|
GRIK1_HUMAN
|
||||||
| NC score | 0.841230 (rank : 16) | θ value | 3.98324e-48 (rank : 15) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P39086, Q13001, Q86SU9 | Gene names | GRIK1, GLUR5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 1 precursor (Glutamate receptor 5) (GluR-5) (GluR5) (Excitatory amino acid receptor 3) (EAA3). | |||||
|
GRIK3_HUMAN
|
||||||
| NC score | 0.841197 (rank : 17) | θ value | 1.08474e-45 (rank : 20) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q13003, Q13004, Q16136 | Gene names | GRIK3, GLUR7 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 3 precursor (Glutamate receptor 7) (GluR-7) (GluR7) (Excitatory amino acid receptor 5) (EAA5). | |||||
|
GRID1_MOUSE
|
||||||
| NC score | 0.840349 (rank : 18) | θ value | 6.81017e-40 (rank : 31) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61627 | Gene names | Grid1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-1 subunit precursor (GluR delta-1). | |||||
|
GRIK2_HUMAN
|
||||||
| NC score | 0.839485 (rank : 19) | θ value | 8.02596e-49 (rank : 14) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q13002 | Gene names | GRIK2, GLUR6 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 2 precursor (Glutamate receptor 6) (GluR-6) (GluR6) (Excitatory amino acid receptor 4) (EAA4). | |||||
|
GRID1_HUMAN
|
||||||
| NC score | 0.838904 (rank : 20) | θ value | 8.89434e-40 (rank : 32) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9ULK0, Q8IXT3 | Gene names | GRID1, KIAA1220 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-1 subunit precursor (GluR delta-1). | |||||
|
GRIK4_HUMAN
|
||||||
| NC score | 0.838709 (rank : 21) | θ value | 2.26182e-43 (rank : 25) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q16099 | Gene names | GRIK4, GRIK | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 4 precursor (Glutamate receptor KA-1) (KA1) (Excitatory amino acid receptor 1) (EAA1). | |||||
|
GRIK4_MOUSE
|
||||||
| NC score | 0.838451 (rank : 22) | θ value | 2.26182e-43 (rank : 26) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BMF5 | Gene names | Grik4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate receptor, ionotropic kainate 4 precursor. | |||||
|
GRIK5_HUMAN
|
||||||
| NC score | 0.837246 (rank : 23) | θ value | 1.05066e-40 (rank : 29) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q16478 | Gene names | GRIK5, GRIK2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 5 precursor (Glutamate receptor KA-2) (KA2) (Excitatory amino acid receptor 2) (EAA2). | |||||
|
GRIK5_MOUSE
|
||||||
| NC score | 0.836829 (rank : 24) | θ value | 6.15952e-41 (rank : 28) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61626 | Gene names | Grik5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 5 precursor (Glutamate receptor KA-2) (KA2) (Glutamate receptor gamma-2) (GluR gamma-2). | |||||
|
GRIA1_MOUSE
|
||||||
| NC score | 0.834654 (rank : 25) | θ value | 1.28137e-46 (rank : 18) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P23818 | Gene names | Gria1, Glur1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 1 precursor (GluR-1) (GluR-A) (GluR-K1) (Glutamate receptor ionotropic, AMPA 1) (AMPA-selective glutamate receptor 1). | |||||
|
GRIA1_HUMAN
|
||||||
| NC score | 0.833234 (rank : 26) | θ value | 1.08474e-45 (rank : 19) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P42261 | Gene names | GRIA1, GLUH1, GLUR1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 1 precursor (GluR-1) (GluR-A) (GluR-K1) (Glutamate receptor ionotropic, AMPA 1) (AMPA-selective glutamate receptor 1). | |||||
|
GRIA4_HUMAN
|
||||||
| NC score | 0.831684 (rank : 27) | θ value | 1.15895e-47 (rank : 16) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P48058 | Gene names | GRIA4, GLUR4 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 4 precursor (GluR-4) (GluR4) (GluR-D) (Glutamate receptor ionotropic, AMPA 4) (AMPA-selective glutamate receptor 4). | |||||
|
GRIA4_MOUSE
|
||||||
| NC score | 0.829582 (rank : 28) | θ value | 9.81109e-47 (rank : 17) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Z2W8 | Gene names | Gria4, Glur4 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 4 precursor (GluR-4) (GluR4) (GluR-D) (Glutamate receptor ionotropic, AMPA 4) (AMPA-selective glutamate receptor 4). | |||||
|
GRIA2_HUMAN
|
||||||
| NC score | 0.829526 (rank : 29) | θ value | 3.15612e-45 (rank : 21) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P42262, Q96FP6 | Gene names | GRIA2, GLUR2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 2 precursor (GluR-2) (GluR-B) (GluR-K2) (Glutamate receptor ionotropic, AMPA 2) (AMPA-selective glutamate receptor 2). | |||||
|
GRIA3_HUMAN
|
||||||
| NC score | 0.829179 (rank : 30) | θ value | 5.38352e-45 (rank : 22) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P42263, Q4VXD5, Q4VXD6, Q9HDA0, Q9HDA1, Q9HDA2 | Gene names | GRIA3, GLUR3, GLURC | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 3 precursor (GluR-3) (GluR-C) (GluR-K3) (Glutamate receptor ionotropic, AMPA 3) (AMPA-selective glutamate receptor 3). | |||||
|
GRIA3_MOUSE
|
||||||
| NC score | 0.829169 (rank : 31) | θ value | 9.18288e-45 (rank : 24) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Z2W9, Q5DTJ0 | Gene names | Gria3, Glur3, Kiaa4184 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate receptor 3 precursor (GluR-3) (GluR-C) (GluR-K3) (Glutamate receptor ionotropic, AMPA 3) (AMPA-selective glutamate receptor 3). | |||||
|
GRIA2_MOUSE
|
||||||
| NC score | 0.828856 (rank : 32) | θ value | 9.18288e-45 (rank : 23) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P23819, Q61604, Q61605, Q8BG69, Q8BXU3, Q9D6D3 | Gene names | Gria2, Glur2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 2 precursor (GluR-2) (GluR-B) (GluR-K2) (Glutamate receptor ionotropic, AMPA 2) (AMPA-selective glutamate receptor 2). | |||||
|
GRIK1_MOUSE
|
||||||
| NC score | 0.822514 (rank : 33) | θ value | 1.12514e-34 (rank : 33) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q60934 | Gene names | Grik1, Glur5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 1 precursor (Glutamate receptor 5) (GluR-5) (GluR5). | |||||
|
ERB1_HUMAN
|
||||||
| NC score | 0.139651 (rank : 34) | θ value | 0.0563607 (rank : 35) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P60509 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-R(b)_3p24.3 provirus ancestral Env polyprotein precursor (Envelope polyprotein) (HERV-R(b) Env protein) [Includes: Surface protein domain (SU); Transmembrane protein domain (TM)]. | |||||
|
GABR1_HUMAN
|
||||||
| NC score | 0.054574 (rank : 35) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UBS5, O95375, O95468, O95975, O96022, Q86W60, Q9UQQ0 | Gene names | GABBR1, GPRC3A | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
GABR1_MOUSE
|
||||||
| NC score | 0.054460 (rank : 36) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9WV18, Q9WU48, Q9WV15, Q9WV16, Q9WV17 | Gene names | Gabbr1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
GABR2_HUMAN
|
||||||
| NC score | 0.045638 (rank : 37) | θ value | 0.47712 (rank : 39) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75899, O75974, O75975, Q5VXZ2, Q8WX04, Q9P1R2, Q9UNR1, Q9UNS9 | Gene names | GABBR2, GPR51, GPRC3B | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 2 precursor (GABA-B receptor 2) (GABA-B-R2) (Gb2) (GABABR2) (G-protein coupled receptor 51) (HG20). | |||||
|
MGR5_HUMAN
|
||||||
| NC score | 0.036051 (rank : 38) | θ value | 0.21417 (rank : 37) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P41594 | Gene names | GRM5, GPRC1E, MGLUR5 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 5 precursor (mGluR5). | |||||
|
MGR1_MOUSE
|
||||||
| NC score | 0.034134 (rank : 39) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P97772, Q6AXG4, Q9EPV6 | Gene names | Grm1, Gprc1a, Mglur1 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 1 precursor (mGluR1). | |||||
|
MGR1_HUMAN
|
||||||
| NC score | 0.034109 (rank : 40) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13255, Q13256, Q14757, Q14758, Q5VTF7, Q5VTF8, Q9NU10, Q9UGS9, Q9UGT0 | Gene names | GRM1, GPRC1A, MGLUR1 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 1 precursor (mGluR1). | |||||
|
U383_MOUSE
|
||||||
| NC score | 0.023509 (rank : 41) | θ value | 1.38821 (rank : 42) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D2Q2, Q3V1K3 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative methyltransferase UPF0383 (EC 2.1.1.-). | |||||
|
FBLN1_HUMAN
|
||||||
| NC score | 0.016463 (rank : 42) | θ value | 0.0148317 (rank : 34) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P23142, P23143, P23144, P37888, Q5TIC4, Q8TBH8, Q9HBQ5, Q9UC21, Q9UGR4, Q9UH41 | Gene names | FBLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-1 precursor. | |||||
|
SFR16_MOUSE
|
||||||
| NC score | 0.014650 (rank : 43) | θ value | 6.88961 (rank : 53) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CFC7, Q8CFC8, Q9Z2N3 | Gene names | Sfrs16, Clasp, Swap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 16 (Suppressor of white-apricot homolog 2) (Clk4-associating SR-related protein). | |||||
|
SFR16_HUMAN
|
||||||
| NC score | 0.014482 (rank : 44) | θ value | 6.88961 (rank : 52) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N2M8, O96026, Q6UW71, Q96DX2 | Gene names | SFRS16, SWAP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 16 (Suppressor of white-apricot homolog 2). | |||||
|
RNF12_HUMAN
|
||||||
| NC score | 0.013106 (rank : 45) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NVW2, Q9Y598 | Gene names | RNF12, RLIM | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 12 (LIM domain-interacting RING finger protein) (RING finger LIM domain-binding protein) (R-LIM) (NY-REN-43 antigen). | |||||
|
FBLN1_MOUSE
|
||||||
| NC score | 0.013106 (rank : 46) | θ value | 0.163984 (rank : 36) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q08879, Q08878, Q8C3B1, Q91ZC9, Q922K8 | Gene names | Fbln1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-1 precursor (Basement-membrane protein 90) (BM-90). | |||||
|
RNF12_MOUSE
|
||||||
| NC score | 0.012424 (rank : 47) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WTV7 | Gene names | Rnf12, Rlim | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 12 (LIM domain-interacting RING finger protein) (RING finger LIM domain-binding protein) (R-LIM). | |||||
|
YLPM1_MOUSE
|
||||||
| NC score | 0.010998 (rank : 48) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
PIR_MOUSE
|
||||||
| NC score | 0.010772 (rank : 49) | θ value | 6.88961 (rank : 51) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D711, Q3UAN0 | Gene names | Pir | |||
|
Domain Architecture |

|
|||||
| Description | Pirin. | |||||
|
PIR_HUMAN
|
||||||
| NC score | 0.009809 (rank : 50) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00625, Q5U0G0 | Gene names | PIR | |||
|
Domain Architecture |

|
|||||
| Description | Pirin. | |||||
|
RBM25_HUMAN
|
||||||
| NC score | 0.009192 (rank : 51) | θ value | 1.38821 (rank : 41) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P49756, Q9UEQ5, Q9UIE9 | Gene names | RBM25, RNPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable RNA-binding protein 25 (RNA-binding motif protein 25) (RNA- binding region-containing protein 7) (Protein S164). | |||||
|
K0195_HUMAN
|
||||||
| NC score | 0.007716 (rank : 52) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12767, O75536, Q86XF1 | Gene names | KIAA0195, TMEM94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0195 (Transmembrane protein 94). | |||||
|
CUTL2_HUMAN
|
||||||
| NC score | 0.004850 (rank : 53) | θ value | 6.88961 (rank : 50) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14529 | Gene names | CUTL2, KIAA0293 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
ADA11_HUMAN
|
||||||
| NC score | 0.003357 (rank : 54) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75078, Q14808, Q14809, Q14810 | Gene names | ADAM11, MDC | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 11 precursor (A disintegrin and metalloproteinase domain 11) (Metalloproteinase-like, disintegrin-like, and cysteine-rich protein) (MDC). | |||||
|
ADA11_MOUSE
|
||||||
| NC score | 0.003252 (rank : 55) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9R1V4 | Gene names | Adam11, Mdc | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 11 precursor (A disintegrin and metalloproteinase domain 11) (Metalloproteinase-like, disintegrin-like, and cysteine-rich protein) (MDC). | |||||
|
ZMYM3_HUMAN
|
||||||
| NC score | 0.003036 (rank : 56) | θ value | 8.99809 (rank : 65) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14202, O15089 | Gene names | ZMYM3, DXS6673E, KIAA0385, ZNF261 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 3 (Zinc finger protein 261). | |||||
|
UBP50_MOUSE
|
||||||
| NC score | 0.001569 (rank : 57) | θ value | 6.88961 (rank : 54) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6P8X6, Q9D558, Q9D9E7 | Gene names | Usp50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ubiquitin carboxyl-terminal hydrolase 50 (EC 3.1.2.15) (Ubiquitin thioesterase 50) (Ubiquitin-specific-processing protease 50) (Deubiquitinating enzyme 50). | |||||
|
RAD50_MOUSE
|
||||||
| NC score | 0.000210 (rank : 58) | θ value | 2.36792 (rank : 45) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
PTPRA_HUMAN
|
||||||
| NC score | 0.000157 (rank : 59) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P18433, Q14513, Q7Z2I2, Q96TD9 | Gene names | PTPRA, PTPA | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase alpha precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase alpha) (R-PTP-alpha). | |||||
|
BLK_HUMAN
|
||||||
| NC score | -0.000814 (rank : 60) | θ value | 1.06291 (rank : 40) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 994 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P51451, Q16291 | Gene names | BLK | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase BLK (EC 2.7.10.2) (B lymphocyte kinase) (p55- BLK). | |||||
|
ZN473_MOUSE
|
||||||
| NC score | -0.001340 (rank : 61) | θ value | 0.365318 (rank : 38) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 824 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BI67, Q8BI98, Q8BIB7 | Gene names | Znf473, Zfp100, Zfp473 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 473 homolog (Zinc finger protein 100) (Zfp-100). | |||||
|
MYPT1_HUMAN
|
||||||
| NC score | -0.001954 (rank : 62) | θ value | 8.99809 (rank : 61) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14974, Q86WU3, Q8NFR6, Q9BYH0 | Gene names | PPP1R12A, MBS, MYPT1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1) (Protein phosphatase myosin-binding subunit). | |||||
|
FYN_MOUSE
|
||||||
| NC score | -0.002889 (rank : 63) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P39688 | Gene names | Fyn | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase Fyn (EC 2.7.10.2) (p59-Fyn). | |||||
|
FYN_HUMAN
|
||||||
| NC score | -0.002960 (rank : 64) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1078 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P06241 | Gene names | FYN | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase Fyn (EC 2.7.10.2) (p59-Fyn) (Protooncogene Syn) (SLK). | |||||
|
ZNF23_HUMAN
|
||||||
| NC score | -0.003260 (rank : 65) | θ value | 6.88961 (rank : 55) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P17027, Q96IT3, Q9UG42 | Gene names | ZNF23, KOX16, ZNF359 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 23 (Zinc finger protein 359) (Zinc finger protein KOX16). | |||||
|
ZBT38_HUMAN
|
||||||
| NC score | -0.003356 (rank : 66) | θ value | 8.99809 (rank : 64) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 885 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NAP3 | Gene names | ZBTB38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and BTB domain-containing protein 38. | |||||
|
CD2L2_HUMAN
|
||||||
| NC score | -0.004465 (rank : 67) | θ value | 5.27518 (rank : 49) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||