Please be patient as the page loads
|
NMDE4_MOUSE
|
||||||
| SwissProt Accessions | Q03391 | Gene names | Grin2d | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 4 precursor (N-methyl D- aspartate receptor subtype 2D) (NR2D) (NMDAR2D). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NMDE1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.981840 (rank : 8) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q12879, O00669 | Gene names | GRIN2A | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 1 precursor (N-methyl D- aspartate receptor subtype 2A) (NR2A) (NMDAR2A) (hNR2A). | |||||
|
NMDE1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.982499 (rank : 5) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P35436 | Gene names | Grin2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 1 precursor (N-methyl D- aspartate receptor subtype 2A) (NR2A) (NMDAR2A). | |||||
|
NMDE2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.981957 (rank : 6) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q13224, Q12919, Q13220, Q13225, Q9UM56 | Gene names | GRIN2B | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 2 precursor (N-methyl D- aspartate receptor subtype 2B) (NR2B) (NMDAR2B) (N-methyl-D-aspartate receptor subunit 3) (NR3) (hNR3). | |||||
|
NMDE2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.981888 (rank : 7) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q01097, Q9DCB2 | Gene names | Grin2b | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 2 precursor (N-methyl D- aspartate receptor subtype 2B) (NR2B) (NMDAR2B). | |||||
|
NMDE3_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.986900 (rank : 3) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q14957 | Gene names | GRIN2C | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 3 precursor (N-methyl D- aspartate receptor subtype 2C) (NR2C) (NMDAR2C). | |||||
|
NMDE3_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.986577 (rank : 4) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q01098 | Gene names | Grin2c | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 3 precursor (N-methyl D- aspartate receptor subtype 2C) (NR2C) (NMDAR2C). | |||||
|
NMDE4_HUMAN
|
||||||
| θ value | 0 (rank : 7) | NC score | 0.995669 (rank : 2) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | O15399 | Gene names | GRIN2D | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 4 precursor (N-methyl D- aspartate receptor subtype 2D) (NR2D) (NMDAR2D) (EB11). | |||||
|
NMDE4_MOUSE
|
||||||
| θ value | 0 (rank : 8) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 158 | |
| SwissProt Accessions | Q03391 | Gene names | Grin2d | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 4 precursor (N-methyl D- aspartate receptor subtype 2D) (NR2D) (NMDAR2D). | |||||
|
NMD3A_HUMAN
|
||||||
| θ value | 1.11217e-74 (rank : 9) | NC score | 0.932969 (rank : 9) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8TCU5, Q8TF29, Q8WXI6 | Gene names | GRIN3A, KIAA1973 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit 3A precursor (N-methyl-D-aspartate receptor subtype NR3A) (NMDAR-L). | |||||
|
NMDZ1_MOUSE
|
||||||
| θ value | 2.3998e-69 (rank : 10) | NC score | 0.907391 (rank : 13) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P35438, Q8CFS4 | Gene names | Grin1, Glurz1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit zeta 1 precursor (N-methyl-D- aspartate receptor subunit NR1). | |||||
|
NMDZ1_HUMAN
|
||||||
| θ value | 6.98233e-69 (rank : 11) | NC score | 0.907764 (rank : 12) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q05586, P35437, Q12867, Q12868 | Gene names | GRIN1, NMDAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit zeta 1 precursor (N-methyl-D- aspartate receptor subunit NR1). | |||||
|
NMD3B_MOUSE
|
||||||
| θ value | 1.5555e-68 (rank : 12) | NC score | 0.925554 (rank : 10) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q91ZU9, Q8VHV0 | Gene names | Grin3b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit 3B precursor (N-methyl-D-aspartate receptor subunit NR3B) (NR3B) (NMDAR3B) (NMDA receptor NR4) (Nr4). | |||||
|
NMD3B_HUMAN
|
||||||
| θ value | 1.11475e-66 (rank : 13) | NC score | 0.924590 (rank : 11) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O60391, Q5EAK7, Q7RTW9 | Gene names | GRIN3B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit 3B precursor (N-methyl-D-aspartate receptor subtype NR3B) (NR3B) (NMDAR3B). | |||||
|
GRIK3_HUMAN
|
||||||
| θ value | 2.58187e-47 (rank : 14) | NC score | 0.844964 (rank : 17) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q13003, Q13004, Q16136 | Gene names | GRIK3, GLUR7 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 3 precursor (Glutamate receptor 7) (GluR-7) (GluR7) (Excitatory amino acid receptor 5) (EAA5). | |||||
|
GRIK5_MOUSE
|
||||||
| θ value | 6.35935e-46 (rank : 15) | NC score | 0.842927 (rank : 23) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q61626 | Gene names | Grik5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 5 precursor (Glutamate receptor KA-2) (KA2) (Glutamate receptor gamma-2) (GluR gamma-2). | |||||
|
GRIK5_HUMAN
|
||||||
| θ value | 2.41655e-45 (rank : 16) | NC score | 0.843202 (rank : 22) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q16478 | Gene names | GRIK5, GRIK2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 5 precursor (Glutamate receptor KA-2) (KA2) (Excitatory amino acid receptor 2) (EAA2). | |||||
|
GRIK1_HUMAN
|
||||||
| θ value | 9.18288e-45 (rank : 17) | NC score | 0.843848 (rank : 20) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P39086, Q13001, Q86SU9 | Gene names | GRIK1, GLUR5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 1 precursor (Glutamate receptor 5) (GluR-5) (GluR5) (Excitatory amino acid receptor 3) (EAA3). | |||||
|
GRIK4_HUMAN
|
||||||
| θ value | 1.01529e-43 (rank : 18) | NC score | 0.844153 (rank : 18) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q16099 | Gene names | GRIK4, GRIK | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 4 precursor (Glutamate receptor KA-1) (KA1) (Excitatory amino acid receptor 1) (EAA1). | |||||
|
GRIK4_MOUSE
|
||||||
| θ value | 1.01529e-43 (rank : 19) | NC score | 0.843853 (rank : 19) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BMF5 | Gene names | Grik4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate receptor, ionotropic kainate 4 precursor. | |||||
|
GRIA1_HUMAN
|
||||||
| θ value | 1.73182e-43 (rank : 20) | NC score | 0.836427 (rank : 26) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P42261 | Gene names | GRIA1, GLUH1, GLUR1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 1 precursor (GluR-1) (GluR-A) (GluR-K1) (Glutamate receptor ionotropic, AMPA 1) (AMPA-selective glutamate receptor 1). | |||||
|
GRIA1_MOUSE
|
||||||
| θ value | 1.73182e-43 (rank : 21) | NC score | 0.837693 (rank : 25) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P23818 | Gene names | Gria1, Glur1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 1 precursor (GluR-1) (GluR-A) (GluR-K1) (Glutamate receptor ionotropic, AMPA 1) (AMPA-selective glutamate receptor 1). | |||||
|
GRIK2_HUMAN
|
||||||
| θ value | 1.73182e-43 (rank : 22) | NC score | 0.842119 (rank : 24) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q13002 | Gene names | GRIK2, GLUR6 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 2 precursor (Glutamate receptor 6) (GluR-6) (GluR6) (Excitatory amino acid receptor 4) (EAA4). | |||||
|
GRIA4_HUMAN
|
||||||
| θ value | 3.85808e-43 (rank : 23) | NC score | 0.833793 (rank : 27) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P48058 | Gene names | GRIA4, GLUR4 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 4 precursor (GluR-4) (GluR4) (GluR-D) (Glutamate receptor ionotropic, AMPA 4) (AMPA-selective glutamate receptor 4). | |||||
|
GRIA2_HUMAN
|
||||||
| θ value | 1.46607e-42 (rank : 24) | NC score | 0.832645 (rank : 28) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P42262, Q96FP6 | Gene names | GRIA2, GLUR2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 2 precursor (GluR-2) (GluR-B) (GluR-K2) (Glutamate receptor ionotropic, AMPA 2) (AMPA-selective glutamate receptor 2). | |||||
|
GRIA2_MOUSE
|
||||||
| θ value | 4.2656e-42 (rank : 25) | NC score | 0.831969 (rank : 29) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P23819, Q61604, Q61605, Q8BG69, Q8BXU3, Q9D6D3 | Gene names | Gria2, Glur2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 2 precursor (GluR-2) (GluR-B) (GluR-K2) (Glutamate receptor ionotropic, AMPA 2) (AMPA-selective glutamate receptor 2). | |||||
|
GRID2_MOUSE
|
||||||
| θ value | 7.27602e-42 (rank : 26) | NC score | 0.848802 (rank : 14) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61625 | Gene names | Grid2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-2 subunit precursor (GluR delta-2). | |||||
|
GRIA3_MOUSE
|
||||||
| θ value | 1.62093e-41 (rank : 27) | NC score | 0.831671 (rank : 30) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Z2W9, Q5DTJ0 | Gene names | Gria3, Glur3, Kiaa4184 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate receptor 3 precursor (GluR-3) (GluR-C) (GluR-K3) (Glutamate receptor ionotropic, AMPA 3) (AMPA-selective glutamate receptor 3). | |||||
|
GRIA4_MOUSE
|
||||||
| θ value | 2.11701e-41 (rank : 28) | NC score | 0.831612 (rank : 31) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Z2W8 | Gene names | Gria4, Glur4 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 4 precursor (GluR-4) (GluR4) (GluR-D) (Glutamate receptor ionotropic, AMPA 4) (AMPA-selective glutamate receptor 4). | |||||
|
GRID1_MOUSE
|
||||||
| θ value | 2.11701e-41 (rank : 29) | NC score | 0.845243 (rank : 16) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61627 | Gene names | Grid1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-1 subunit precursor (GluR delta-1). | |||||
|
GRID2_HUMAN
|
||||||
| θ value | 2.11701e-41 (rank : 30) | NC score | 0.847698 (rank : 15) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O43424 | Gene names | GRID2, GLURD2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-2 subunit precursor (GluR delta-2). | |||||
|
GRIA3_HUMAN
|
||||||
| θ value | 8.04462e-41 (rank : 31) | NC score | 0.831478 (rank : 32) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P42263, Q4VXD5, Q4VXD6, Q9HDA0, Q9HDA1, Q9HDA2 | Gene names | GRIA3, GLUR3, GLURC | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 3 precursor (GluR-3) (GluR-C) (GluR-K3) (Glutamate receptor ionotropic, AMPA 3) (AMPA-selective glutamate receptor 3). | |||||
|
GRID1_HUMAN
|
||||||
| θ value | 2.34062e-40 (rank : 32) | NC score | 0.843568 (rank : 21) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9ULK0, Q8IXT3 | Gene names | GRID1, KIAA1220 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-1 subunit precursor (GluR delta-1). | |||||
|
GRIK1_MOUSE
|
||||||
| θ value | 1.12514e-34 (rank : 33) | NC score | 0.825882 (rank : 33) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q60934 | Gene names | Grik1, Glur5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 1 precursor (Glutamate receptor 5) (GluR-5) (GluR5). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 5.81887e-07 (rank : 34) | NC score | 0.043752 (rank : 42) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 35) | NC score | 0.044746 (rank : 41) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 36) | NC score | 0.043321 (rank : 43) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
GNAS1_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 37) | NC score | 0.021312 (rank : 73) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6R0H7, Q6R0H4, Q6R0H5, Q6R2J5, Q9JJX0, Q9Z1N8 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 38) | NC score | 0.032484 (rank : 47) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 39) | NC score | 0.022198 (rank : 67) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
K1802_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 40) | NC score | 0.018342 (rank : 86) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 41) | NC score | 0.047407 (rank : 39) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 42) | NC score | 0.040018 (rank : 45) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
MEGF9_MOUSE
|
||||||
| θ value | 0.125558 (rank : 43) | NC score | 0.016812 (rank : 88) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.125558 (rank : 44) | NC score | 0.031816 (rank : 48) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
TS1R3_HUMAN
|
||||||
| θ value | 0.125558 (rank : 45) | NC score | 0.030011 (rank : 49) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7RTX0, Q5TA49, Q8NGW9 | Gene names | TAS1R3, T1R3, TR3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 3 precursor (Sweet taste receptor T1R3). | |||||
|
ERB1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 46) | NC score | 0.132871 (rank : 34) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P60509 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-R(b)_3p24.3 provirus ancestral Env polyprotein precursor (Envelope polyprotein) (HERV-R(b) Env protein) [Includes: Surface protein domain (SU); Transmembrane protein domain (TM)]. | |||||
|
SEPT9_HUMAN
|
||||||
| θ value | 0.21417 (rank : 47) | NC score | 0.014587 (rank : 96) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
GABR1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 48) | NC score | 0.070125 (rank : 35) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UBS5, O95375, O95468, O95975, O96022, Q86W60, Q9UQQ0 | Gene names | GABBR1, GPRC3A | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
GABR1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 49) | NC score | 0.070049 (rank : 36) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9WV18, Q9WU48, Q9WV15, Q9WV16, Q9WV17 | Gene names | Gabbr1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
IASPP_MOUSE
|
||||||
| θ value | 0.279714 (rank : 50) | NC score | 0.021948 (rank : 68) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
FOXH1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 51) | NC score | 0.012150 (rank : 104) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O75593 | Gene names | FOXH1, FAST1, FAST2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein H1 (Forkhead activin signal transducer 1) (Fast- 1) (hFAST-1) (Forkhead activin signal transducer 2) (Fast-2). | |||||
|
PER3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 52) | NC score | 0.019337 (rank : 79) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
CO039_MOUSE
|
||||||
| θ value | 0.47712 (rank : 53) | NC score | 0.029494 (rank : 50) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q3TEI4, Q3TMF2, Q3U253, Q3UM75, Q9DA93 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39 homolog. | |||||
|
ELL_HUMAN
|
||||||
| θ value | 0.47712 (rank : 54) | NC score | 0.023569 (rank : 60) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P55199 | Gene names | ELL, C19orf17 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II elongation factor ELL (Eleven-nineteen lysine-rich leukemia protein). | |||||
|
GAK5_HUMAN
|
||||||
| θ value | 0.47712 (rank : 55) | NC score | 0.022591 (rank : 63) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P62684 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19p13.11 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K113 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
ZN516_MOUSE
|
||||||
| θ value | 0.47712 (rank : 56) | NC score | 0.001056 (rank : 147) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q7TSH3 | Gene names | Znf516, Kiaa0222, Zfp516 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 516. | |||||
|
BAZ2A_HUMAN
|
||||||
| θ value | 0.62314 (rank : 57) | NC score | 0.013359 (rank : 100) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
CEL_HUMAN
|
||||||
| θ value | 0.62314 (rank : 58) | NC score | 0.018031 (rank : 87) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
EDNRB_HUMAN
|
||||||
| θ value | 0.62314 (rank : 59) | NC score | 0.000007 (rank : 149) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 598 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P24530, O15343, Q9UQK3 | Gene names | EDNRB, ETRB | |||
|
Domain Architecture |

|
|||||
| Description | Endothelin B receptor precursor (ET-B) (Endothelin receptor Non- selective type). | |||||
|
SON_HUMAN
|
||||||
| θ value | 0.62314 (rank : 60) | NC score | 0.023587 (rank : 59) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
HXD3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 61) | NC score | 0.004383 (rank : 130) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P31249, Q99955, Q9BSC5 | Gene names | HOXD3, HOX4A | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D3 (Hox-4A). | |||||
|
JPH2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 62) | NC score | 0.015497 (rank : 92) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
MKL1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 63) | NC score | 0.015375 (rank : 94) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 64) | NC score | 0.015420 (rank : 93) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.813845 (rank : 65) | NC score | 0.015637 (rank : 90) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
APC_HUMAN
|
||||||
| θ value | 1.06291 (rank : 66) | NC score | 0.018881 (rank : 82) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P25054, Q15162, Q15163, Q93042 | Gene names | APC, DP2.5 | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC). | |||||
|
GAK11_HUMAN
|
||||||
| θ value | 1.06291 (rank : 67) | NC score | 0.021414 (rank : 72) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P63145, Q9UKI1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.21 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K101 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK12_HUMAN
|
||||||
| θ value | 1.06291 (rank : 68) | NC score | 0.021612 (rank : 70) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P63130, Q9UKI0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K102 Gag protein) (HERV-K(III) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 69) | NC score | 0.021142 (rank : 74) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q7LDI9, Q9UKH5, Q9Y6I1, Q9YNA6, Q9YNB0 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(HML-2.HOM) Gag protein) (HERV-K108 Gag protein) (HERV-K(C7) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 70) | NC score | 0.021469 (rank : 71) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P63126, Q9UKH4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_6q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K109 Gag protein) (HERV-K(C6) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
MEF2B_MOUSE
|
||||||
| θ value | 1.06291 (rank : 71) | NC score | 0.015511 (rank : 91) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O55087, O55088, O55089, O55090, O55231, Q61843 | Gene names | Mef2b | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2B. | |||||
|
PCDA9_HUMAN
|
||||||
| θ value | 1.06291 (rank : 72) | NC score | 0.001767 (rank : 145) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y5H5, O15053 | Gene names | PCDHA9, KIAA0345 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 9 precursor (PCDH-alpha9). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 73) | NC score | 0.022369 (rank : 66) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
ATS13_HUMAN
|
||||||
| θ value | 1.38821 (rank : 74) | NC score | 0.007392 (rank : 122) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q76LX8, Q6UY16, Q710F6, Q711T8, Q96L37, Q9H0G3 | Gene names | ADAMTS13, C9orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAMTS-13 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 13) (ADAM-TS 13) (ADAM-TS13) (von Willebrand factor-cleaving protease) (vWF-cleaving protease) (vWF-CP). | |||||
|
FX4L1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 75) | NC score | 0.009817 (rank : 110) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NU39 | Gene names | FOXD4L1 | |||
|
Domain Architecture |
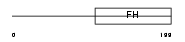
|
|||||
| Description | Forkhead box protein D4-like 1. | |||||
|
IL3RB_HUMAN
|
||||||
| θ value | 1.38821 (rank : 76) | NC score | 0.021758 (rank : 69) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P32927 | Gene names | CSF2RB, IL3RB, IL5RB | |||
|
Domain Architecture |

|
|||||
| Description | Cytokine receptor common beta chain precursor (GM-CSF/IL-3/IL-5 receptor common beta-chain) (CD131 antigen) (CDw131). | |||||
|
PDLI7_HUMAN
|
||||||
| θ value | 1.38821 (rank : 77) | NC score | 0.009671 (rank : 112) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 547 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9NR12, Q14250, Q5XG82, Q6NVZ5, Q96C91, Q9BXB8, Q9BXB9 | Gene names | PDLIM7, ENIGMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
UBP6_HUMAN
|
||||||
| θ value | 1.38821 (rank : 78) | NC score | 0.005878 (rank : 126) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35125, Q15634, Q86WP6, Q8IWT4 | Gene names | USP6, TRE2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 6 (EC 3.1.2.15) (Ubiquitin thioesterase 6) (Ubiquitin-specific-processing protease 6) (Deubiquitinating enzyme 6) (Proto-oncogene TRE-2). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 79) | NC score | 0.004274 (rank : 131) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
ANPRB_HUMAN
|
||||||
| θ value | 1.81305 (rank : 80) | NC score | 0.003721 (rank : 134) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P20594, O60871, Q9UQ50 | Gene names | NPR2, ANPRB | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide receptor B precursor (ANP-B) (ANPRB) (GC-B) (Guanylate cyclase B) (EC 4.6.1.2) (NPR-B) (Atrial natriuretic peptide B-type receptor). | |||||
|
CABIN_HUMAN
|
||||||
| θ value | 1.81305 (rank : 81) | NC score | 0.027717 (rank : 52) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Y6J0, Q9Y460 | Gene names | CABIN1, KIAA0330 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin-binding protein Cabin 1 (Calcineurin inhibitor) (CAIN). | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 82) | NC score | 0.020503 (rank : 76) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
RGS12_HUMAN
|
||||||
| θ value | 1.81305 (rank : 83) | NC score | 0.012618 (rank : 103) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O14924, O14922, O14923, O43510, O75338 | Gene names | RGS12 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 12 (RGS12). | |||||
|
TS13_MOUSE
|
||||||
| θ value | 1.81305 (rank : 84) | NC score | 0.026463 (rank : 54) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q01755 | Gene names | Tcp11 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific protein PBS13. | |||||
|
ZO3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 85) | NC score | 0.008760 (rank : 117) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95049 | Gene names | TJP3, ZO3 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-3 (Zonula occludens 3 protein) (Zona occludens 3 protein) (Tight junction protein 3). | |||||
|
CBP_HUMAN
|
||||||
| θ value | 2.36792 (rank : 86) | NC score | 0.028460 (rank : 51) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CK024_HUMAN
|
||||||
| θ value | 2.36792 (rank : 87) | NC score | 0.042952 (rank : 44) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
F120C_MOUSE
|
||||||
| θ value | 2.36792 (rank : 88) | NC score | 0.009644 (rank : 113) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C3F2 | Gene names | Fam120c, ORF34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UPF0318 protein FAM120C. | |||||
|
FUT4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 89) | NC score | 0.007707 (rank : 120) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P22083 | Gene names | FUT4, ELFT | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-(1,3)-fucosyltransferase (EC 2.4.1.-) (Galactoside 3-L- fucosyltransferase) (Fucosyltransferase 4) (FUCT-IV) (Fuc-TIV) (ELAM-1 ligand fucosyltransferase). | |||||
|
NRX2A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 90) | NC score | 0.012789 (rank : 102) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
NRX2B_HUMAN
|
||||||
| θ value | 2.36792 (rank : 91) | NC score | 0.018555 (rank : 84) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 2.36792 (rank : 92) | NC score | 0.019787 (rank : 78) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
FOXD4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 93) | NC score | 0.009847 (rank : 109) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q12950, Q5VVK1, Q8WXT6 | Gene names | FOXD4, FKHL9, FOXD4A, FREAC5 | |||
|
Domain Architecture |
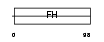
|
|||||
| Description | Forkhead box protein D4 (Forkhead-related protein FKHL9) (Forkhead- related transcription factor 5) (FREAC-5) (Myeloid factor-alpha). | |||||
|
HSF1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 94) | NC score | 0.010101 (rank : 108) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P38532, O70462 | Gene names | Hsf1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
HXD3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 95) | NC score | 0.002959 (rank : 140) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P09027, Q3UUD3 | Gene names | Hoxd3, Hox-4.1, Hoxd-3 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D3 (Hox-4.1) (Homeobox protein MH-19). | |||||
|
MEGF8_HUMAN
|
||||||
| θ value | 3.0926 (rank : 96) | NC score | 0.009012 (rank : 115) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7Z7M0, O75097 | Gene names | MEGF8, EGFL4, KIAA0817 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 8 (EGF-like domain- containing protein 4) (Multiple EGF-like domain protein 4). | |||||
|
MEGF8_MOUSE
|
||||||
| θ value | 3.0926 (rank : 97) | NC score | 0.010240 (rank : 107) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 520 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P60882, Q80TR3, Q80V41, Q8BMN9, Q8JZW7, Q8K0J3 | Gene names | Megf8, Egfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 8 (EGF-like domain- containing protein 4) (Multiple EGF-like domain protein 4). | |||||
|
POLS2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 98) | NC score | -0.000251 (rank : 153) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5K4E3, Q8NBY4 | Gene names | PRSS36 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyserase-2 precursor (EC 3.4.21.-) (Polyserine protease 2) (Protease serine 36). | |||||
|
S23IP_MOUSE
|
||||||
| θ value | 3.0926 (rank : 99) | NC score | 0.018945 (rank : 81) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6NZC7, Q8BXY6 | Gene names | Sec23ip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SEC23-interacting protein. | |||||
|
SSBP4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 100) | NC score | 0.018462 (rank : 85) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
ANPRB_MOUSE
|
||||||
| θ value | 4.03905 (rank : 101) | NC score | 0.003275 (rank : 139) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6VVW5, Q6VVW3, Q6VVW4, Q8CGA9 | Gene names | Npr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Atrial natriuretic peptide receptor B precursor (ANP-B) (ANPRB) (GC-B) (Guanylate cyclase B) (EC 4.6.1.2) (NPR-B) (Atrial natriuretic peptide B-type receptor). | |||||
|
BIM_MOUSE
|
||||||
| θ value | 4.03905 (rank : 102) | NC score | 0.012962 (rank : 101) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O54918, O54919, O54920 | Gene names | Bcl2l11, Bim | |||
|
Domain Architecture |

|
|||||
| Description | Bcl-2-like protein 11 (Bcl2-interacting mediator of cell death). | |||||
|
CNTRB_MOUSE
|
||||||
| θ value | 4.03905 (rank : 103) | NC score | 0.000305 (rank : 148) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8CB62, Q5NCF3 | Gene names | Cntrob, Lip8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
ECM1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 104) | NC score | 0.014176 (rank : 99) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q16610 | Gene names | ECM1 | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular matrix protein 1 precursor (Secretory component p85). | |||||
|
FOXD3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 105) | NC score | 0.007524 (rank : 121) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61060 | Gene names | Foxd3, Hfh2 | |||
|
Domain Architecture |
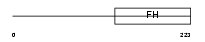
|
|||||
| Description | Forkhead box protein D3 (HNF3/FH transcription factor genesis) (Hepatocyte nuclear factor 3 forkhead homolog 2) (HFH-2). | |||||
|
GAK_MOUSE
|
||||||
| θ value | 4.03905 (rank : 106) | NC score | -0.002512 (rank : 155) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99KY4, Q6P1I8, Q6P9S5, Q8BM74, Q8K0Q4 | Gene names | Gak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||
|
GTSE1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 107) | NC score | 0.020429 (rank : 77) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NYZ3, Q53GX5, Q5R3I6, Q6DHX4, Q9BRE0, Q9UGZ9, Q9Y557 | Gene names | GTSE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G2 and S phase expressed protein 1 (B99 homolog). | |||||
|
SC24C_HUMAN
|
||||||
| θ value | 4.03905 (rank : 108) | NC score | 0.022432 (rank : 64) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
SOX13_MOUSE
|
||||||
| θ value | 4.03905 (rank : 109) | NC score | 0.005325 (rank : 129) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q04891, Q922L3 | Gene names | Sox13, Sox-13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein. | |||||
|
SPRE3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 110) | NC score | 0.011245 (rank : 106) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6P6N5, Q7TNJ8 | Gene names | Spred3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sprouty-related, EVH1 domain-containing protein 3 (Spred-3). | |||||
|
SRGEF_MOUSE
|
||||||
| θ value | 4.03905 (rank : 111) | NC score | 0.009704 (rank : 111) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80YD6, Q9QXB7 | Gene names | Sergef, Delgef, Gnefr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secretion-regulating guanine nucleotide exchange factor (Guanine nucleotide exchange factor-related protein) (Deafness locus-associated putative guanine nucleotide exchange factor) (DelGEF). | |||||
|
TS1R1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 112) | NC score | 0.049180 (rank : 38) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99PG6, Q6NS58, Q923J9, Q925I5, Q99PG5 | Gene names | Tas1r1, Gpr70, T1r1, Tr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 1 precursor (G-protein coupled receptor 70). | |||||
|
VEGFB_HUMAN
|
||||||
| θ value | 4.03905 (rank : 113) | NC score | 0.024317 (rank : 57) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P49765, Q16528 | Gene names | VEGFB, VRF | |||
|
Domain Architecture |
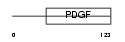
|
|||||
| Description | Vascular endothelial growth factor B precursor (VEGF-B) (VEGF-related factor) (VRF). | |||||
|
ZEP2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 114) | NC score | -0.000195 (rank : 152) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q3UHF7, O55140, Q3UVD4, Q3UVH5 | Gene names | Hivep2, Mibp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Human immunodeficiency virus type I enhancer-binding protein 2 homolog (Myc intron-binding protein 1) (MIBP-1). | |||||
|
CIC_HUMAN
|
||||||
| θ value | 5.27518 (rank : 115) | NC score | 0.026191 (rank : 55) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
EGR4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 116) | NC score | -0.004048 (rank : 158) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 710 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9WUF2 | Gene names | Egr4 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 4 (EGR-4). | |||||
|
FA47A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 117) | NC score | 0.014655 (rank : 95) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5JRC9, Q8TAA0 | Gene names | FAM47A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47A. | |||||
|
FOXN1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 118) | NC score | 0.016169 (rank : 89) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q61575 | Gene names | Foxn1, Fkh19, Hfh11, Whn | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein N1 (Transcription factor winged-helix nude) (Hepatocyte nuclear factor 3 forkhead homolog 11) (HNF-3/forkhead homolog 11) (HFH-11). | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 119) | NC score | 0.047121 (rank : 40) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
LHX2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 120) | NC score | 0.003328 (rank : 138) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P50458, O95860, Q8N1Z3 | Gene names | LHX2, LH2 | |||
|
Domain Architecture |
|
|||||
| Description | LIM/homeobox protein Lhx2 (Homeobox protein LH-2). | |||||
|
LHX2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 121) | NC score | 0.003343 (rank : 137) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z0S2 | Gene names | Lhx2 | |||
|
Domain Architecture |
|
|||||
| Description | LIM/homeobox protein Lhx2. | |||||
|
LRC56_HUMAN
|
||||||
| θ value | 5.27518 (rank : 122) | NC score | 0.008528 (rank : 118) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IYG6, Q8N3Q4 | Gene names | LRRC56 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 56. | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 123) | NC score | 0.025014 (rank : 56) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
NOTC3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 124) | NC score | -0.000096 (rank : 151) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1062 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UM47, Q9UEB3, Q9UPL3, Q9Y6L8 | Gene names | NOTCH3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
PDXL2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 125) | NC score | 0.011359 (rank : 105) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8CAE9, Q8CFW3 | Gene names | Podxl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Podocalyxin-like protein 2 precursor (Endoglycan). | |||||
|
PGBM_MOUSE
|
||||||
| θ value | 5.27518 (rank : 126) | NC score | -0.000071 (rank : 150) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q05793 | Gene names | Hspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
PSL1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 127) | NC score | 0.026915 (rank : 53) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
RBPSL_HUMAN
|
||||||
| θ value | 5.27518 (rank : 128) | NC score | 0.006492 (rank : 124) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UBG7, O95723, Q5QPU9, Q9ULV9 | Gene names | RBPSUHL, RBPL | |||
|
Domain Architecture |

|
|||||
| Description | Recombining binding protein suppressor of hairless-like protein (Transcription factor RBP-L). | |||||
|
TAU_MOUSE
|
||||||
| θ value | 5.27518 (rank : 129) | NC score | 0.020735 (rank : 75) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
ARSJ_HUMAN
|
||||||
| θ value | 6.88961 (rank : 130) | NC score | 0.005604 (rank : 128) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5FYB0, Q5FWE4, Q6UWT9 | Gene names | ARSJ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arylsulfatase J precursor (EC 3.1.6.-) (ASJ). | |||||
|
ARSJ_MOUSE
|
||||||
| θ value | 6.88961 (rank : 131) | NC score | 0.005612 (rank : 127) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BM89, Q8BKG5, Q8BL46 | Gene names | Arsj | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arylsulfatase J precursor (EC 3.1.6.-) (ASJ). | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 132) | NC score | 0.037963 (rank : 46) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
CELR2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 133) | NC score | -0.000672 (rank : 154) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9HCU4, Q92566 | Gene names | CELSR2, CDHF10, EGFL2, KIAA0279, MEGF3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 2 precursor (Epidermal growth factor-like 2) (Multiple epidermal growth factor-like domains 3) (Flamingo 1). | |||||
|
GAK10_HUMAN
|
||||||
| θ value | 6.88961 (rank : 134) | NC score | 0.018869 (rank : 83) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P87889, P10263, P10264, Q69385, Q9UKH6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q33.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K10 Gag protein) (HERV-K107 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GRIN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 135) | NC score | 0.019203 (rank : 80) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
M3K14_HUMAN
|
||||||
| θ value | 6.88961 (rank : 136) | NC score | -0.003352 (rank : 157) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 887 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99558, Q8IYN1 | Gene names | MAP3K14, NIK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 14 (EC 2.7.11.25) (NF- kappa beta-inducing kinase) (Serine/threonine-protein kinase NIK) (HsNIK). | |||||
|
MILK1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 137) | NC score | 0.022880 (rank : 61) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
MUC5B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 138) | NC score | 0.014259 (rank : 98) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
NDF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 139) | NC score | 0.002798 (rank : 141) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q60867, Q545N9, Q60897 | Gene names | Neurod1, Neurod | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 1 (NeuroD1). | |||||
|
RCOR2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 140) | NC score | 0.008472 (rank : 119) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8IZ40, Q96FP3 | Gene names | RCOR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 2. | |||||
|
SEM6B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 141) | NC score | 0.002327 (rank : 143) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H3T3, Q9NRK9 | Gene names | SEMA6B, SEMAZ | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6B precursor (Semaphorin Z) (Sema Z). | |||||
|
SYNJ1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 142) | NC score | 0.023858 (rank : 58) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O43426, O43425, O94984 | Gene names | SYNJ1, KIAA0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
TREF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 143) | NC score | 0.009467 (rank : 114) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BXJ2, Q6PCQ4, Q80X25, Q810H8, Q8BY31 | Gene names | Trerf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132). | |||||
|
BCOR_HUMAN
|
||||||
| θ value | 8.99809 (rank : 144) | NC score | 0.001798 (rank : 144) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6W2J9, Q6P4B6, Q7Z2K7, Q8TEB4, Q96DB3, Q9H232, Q9H233, Q9HCJ7, Q9NXF2 | Gene names | BCOR, KIAA1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
BLNK_MOUSE
|
||||||
| θ value | 8.99809 (rank : 145) | NC score | 0.008988 (rank : 116) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9QUN3, O88504 | Gene names | Blnk, Bash, Ly57, Slp65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell linker protein (Cytoplasmic adapter protein) (B-cell adapter containing SH2 domain protein) (B-cell adapter containing Src homology 2 domain protein) (Src homology 2 domain-containing leukocyte protein of 65 kDa) (Slp-65) (Lymphocyte antigen 57). | |||||
|
CBP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 146) | NC score | 0.022417 (rank : 65) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CIAS1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 147) | NC score | 0.002516 (rank : 142) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96P20, O75434, Q59H68, Q5JQS8, Q5JQS9, Q6TG35, Q8TCW0, Q8TEU9, Q8WXH9 | Gene names | CIAS1, NALP3, PYPAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Cold autoinflammatory syndrome 1 protein (Cryopyrin) (NACHT-, LRR- and PYD-containing protein 3) (PYRIN-containing APAF1-like protein 1) (Angiotensin/vasopressin receptor AII/AVP-like). | |||||
|
COX11_MOUSE
|
||||||
| θ value | 8.99809 (rank : 148) | NC score | 0.006531 (rank : 123) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6P8I6 | Gene names | Cox11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytochrome c oxidase assembly protein COX11, mitochondrial precursor. | |||||
|
DRD4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 149) | NC score | -0.002925 (rank : 156) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 852 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P51436, O35838 | Gene names | Drd4 | |||
|
Domain Architecture |

|
|||||
| Description | D(4) dopamine receptor (Dopamine D4 receptor) (D(2C) dopamine receptor). | |||||
|
EYA1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 150) | NC score | 0.003483 (rank : 135) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P97767, O08818 | Gene names | Eya1 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 1 (EC 3.1.3.48). | |||||
|
IF4G1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 151) | NC score | 0.014528 (rank : 97) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 8.99809 (rank : 152) | NC score | 0.022662 (rank : 62) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PCOC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 153) | NC score | 0.001589 (rank : 146) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15113, O14550 | Gene names | PCOLCE, PCPE1 | |||
|
Domain Architecture |

|
|||||
| Description | Procollagen C-endopeptidase enhancer 1 precursor (Procollagen COOH- terminal proteinase enhancer 1) (Procollagen C-proteinase enhancer 1) (PCPE-1) (Type I procollagen COOH-terminal proteinase enhancer) (Type 1 procollagen C-proteinase enhancer protein). | |||||
|
S19A1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 154) | NC score | 0.004154 (rank : 132) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P41440, O00553, O60227, Q13026 | Gene names | SLC19A1, FLOT1, RFC1 | |||
|
Domain Architecture |

|
|||||
| Description | Folate transporter 1 (Solute carrier family 19 member 1) (Placental folate transporter) (FOLT) (Reduced folate carrier protein) (RFC) (Intestinal folate carrier) (IFC-1). | |||||
|
SH22A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 155) | NC score | 0.003991 (rank : 133) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QXK9, Q9JHP6 | Gene names | Sh2d2a, Lad, Ribp | |||
|
Domain Architecture |

|
|||||
| Description | SH2 domain protein 2A (Lck-associated adapter protein) (Lad) (Rlk/Itk- binding protein) (Ribp). | |||||
|
SRBS1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 156) | NC score | 0.003419 (rank : 136) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q62417, Q80TF8, Q8BZI3, Q8K3Y2, Q921F8, Q9Z0Z8, Q9Z0Z9 | Gene names | Sorbs1, Kiaa1296, Sh3d5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorbin and SH3 domain-containing protein 1 (Ponsin) (c-Cbl-associated protein) (CAP) (SH3 domain protein 5) (SH3P12). | |||||
|
THAP7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 157) | NC score | 0.006085 (rank : 125) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BT49 | Gene names | THAP7 | |||
|
Domain Architecture |
|
|||||
| Description | THAP domain-containing protein 7. | |||||
|
TYK2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 158) | NC score | -0.004941 (rank : 159) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 850 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9R117, O88431, Q8VE41 | Gene names | Tyk2 | |||
|
Domain Architecture |

|
|||||
| Description | Non-receptor tyrosine-protein kinase TYK2 (EC 2.7.10.2). | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.054236 (rank : 37) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
NMDE4_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 8) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 158 | |
| SwissProt Accessions | Q03391 | Gene names | Grin2d | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 4 precursor (N-methyl D- aspartate receptor subtype 2D) (NR2D) (NMDAR2D). | |||||
|
NMDE4_HUMAN
|
||||||
| NC score | 0.995669 (rank : 2) | θ value | 0 (rank : 7) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | O15399 | Gene names | GRIN2D | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 4 precursor (N-methyl D- aspartate receptor subtype 2D) (NR2D) (NMDAR2D) (EB11). | |||||
|
NMDE3_HUMAN
|
||||||
| NC score | 0.986900 (rank : 3) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q14957 | Gene names | GRIN2C | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 3 precursor (N-methyl D- aspartate receptor subtype 2C) (NR2C) (NMDAR2C). | |||||
|
NMDE3_MOUSE
|
||||||
| NC score | 0.986577 (rank : 4) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q01098 | Gene names | Grin2c | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 3 precursor (N-methyl D- aspartate receptor subtype 2C) (NR2C) (NMDAR2C). | |||||
|
NMDE1_MOUSE
|
||||||
| NC score | 0.982499 (rank : 5) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P35436 | Gene names | Grin2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 1 precursor (N-methyl D- aspartate receptor subtype 2A) (NR2A) (NMDAR2A). | |||||
|
NMDE2_HUMAN
|
||||||
| NC score | 0.981957 (rank : 6) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q13224, Q12919, Q13220, Q13225, Q9UM56 | Gene names | GRIN2B | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 2 precursor (N-methyl D- aspartate receptor subtype 2B) (NR2B) (NMDAR2B) (N-methyl-D-aspartate receptor subunit 3) (NR3) (hNR3). | |||||
|
NMDE2_MOUSE
|
||||||
| NC score | 0.981888 (rank : 7) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q01097, Q9DCB2 | Gene names | Grin2b | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 2 precursor (N-methyl D- aspartate receptor subtype 2B) (NR2B) (NMDAR2B). | |||||
|
NMDE1_HUMAN
|
||||||
| NC score | 0.981840 (rank : 8) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q12879, O00669 | Gene names | GRIN2A | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 1 precursor (N-methyl D- aspartate receptor subtype 2A) (NR2A) (NMDAR2A) (hNR2A). | |||||
|
NMD3A_HUMAN
|
||||||
| NC score | 0.932969 (rank : 9) | θ value | 1.11217e-74 (rank : 9) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8TCU5, Q8TF29, Q8WXI6 | Gene names | GRIN3A, KIAA1973 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit 3A precursor (N-methyl-D-aspartate receptor subtype NR3A) (NMDAR-L). | |||||
|
NMD3B_MOUSE
|
||||||
| NC score | 0.925554 (rank : 10) | θ value | 1.5555e-68 (rank : 12) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q91ZU9, Q8VHV0 | Gene names | Grin3b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit 3B precursor (N-methyl-D-aspartate receptor subunit NR3B) (NR3B) (NMDAR3B) (NMDA receptor NR4) (Nr4). | |||||
|
NMD3B_HUMAN
|
||||||
| NC score | 0.924590 (rank : 11) | θ value | 1.11475e-66 (rank : 13) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O60391, Q5EAK7, Q7RTW9 | Gene names | GRIN3B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit 3B precursor (N-methyl-D-aspartate receptor subtype NR3B) (NR3B) (NMDAR3B). | |||||
|
NMDZ1_HUMAN
|
||||||
| NC score | 0.907764 (rank : 12) | θ value | 6.98233e-69 (rank : 11) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q05586, P35437, Q12867, Q12868 | Gene names | GRIN1, NMDAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit zeta 1 precursor (N-methyl-D- aspartate receptor subunit NR1). | |||||
|
NMDZ1_MOUSE
|
||||||
| NC score | 0.907391 (rank : 13) | θ value | 2.3998e-69 (rank : 10) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P35438, Q8CFS4 | Gene names | Grin1, Glurz1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit zeta 1 precursor (N-methyl-D- aspartate receptor subunit NR1). | |||||
|
GRID2_MOUSE
|
||||||
| NC score | 0.848802 (rank : 14) | θ value | 7.27602e-42 (rank : 26) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61625 | Gene names | Grid2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-2 subunit precursor (GluR delta-2). | |||||
|
GRID2_HUMAN
|
||||||
| NC score | 0.847698 (rank : 15) | θ value | 2.11701e-41 (rank : 30) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O43424 | Gene names | GRID2, GLURD2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-2 subunit precursor (GluR delta-2). | |||||
|
GRID1_MOUSE
|
||||||
| NC score | 0.845243 (rank : 16) | θ value | 2.11701e-41 (rank : 29) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61627 | Gene names | Grid1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-1 subunit precursor (GluR delta-1). | |||||
|
GRIK3_HUMAN
|
||||||
| NC score | 0.844964 (rank : 17) | θ value | 2.58187e-47 (rank : 14) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q13003, Q13004, Q16136 | Gene names | GRIK3, GLUR7 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 3 precursor (Glutamate receptor 7) (GluR-7) (GluR7) (Excitatory amino acid receptor 5) (EAA5). | |||||
|
GRIK4_HUMAN
|
||||||
| NC score | 0.844153 (rank : 18) | θ value | 1.01529e-43 (rank : 18) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q16099 | Gene names | GRIK4, GRIK | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 4 precursor (Glutamate receptor KA-1) (KA1) (Excitatory amino acid receptor 1) (EAA1). | |||||
|
GRIK4_MOUSE
|
||||||
| NC score | 0.843853 (rank : 19) | θ value | 1.01529e-43 (rank : 19) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BMF5 | Gene names | Grik4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate receptor, ionotropic kainate 4 precursor. | |||||
|
GRIK1_HUMAN
|
||||||
| NC score | 0.843848 (rank : 20) | θ value | 9.18288e-45 (rank : 17) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P39086, Q13001, Q86SU9 | Gene names | GRIK1, GLUR5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 1 precursor (Glutamate receptor 5) (GluR-5) (GluR5) (Excitatory amino acid receptor 3) (EAA3). | |||||
|
GRID1_HUMAN
|
||||||
| NC score | 0.843568 (rank : 21) | θ value | 2.34062e-40 (rank : 32) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9ULK0, Q8IXT3 | Gene names | GRID1, KIAA1220 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-1 subunit precursor (GluR delta-1). | |||||
|
GRIK5_HUMAN
|
||||||
| NC score | 0.843202 (rank : 22) | θ value | 2.41655e-45 (rank : 16) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q16478 | Gene names | GRIK5, GRIK2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 5 precursor (Glutamate receptor KA-2) (KA2) (Excitatory amino acid receptor 2) (EAA2). | |||||
|
GRIK5_MOUSE
|
||||||
| NC score | 0.842927 (rank : 23) | θ value | 6.35935e-46 (rank : 15) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q61626 | Gene names | Grik5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 5 precursor (Glutamate receptor KA-2) (KA2) (Glutamate receptor gamma-2) (GluR gamma-2). | |||||
|
GRIK2_HUMAN
|
||||||
| NC score | 0.842119 (rank : 24) | θ value | 1.73182e-43 (rank : 22) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q13002 | Gene names | GRIK2, GLUR6 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 2 precursor (Glutamate receptor 6) (GluR-6) (GluR6) (Excitatory amino acid receptor 4) (EAA4). | |||||
|
GRIA1_MOUSE
|
||||||
| NC score | 0.837693 (rank : 25) | θ value | 1.73182e-43 (rank : 21) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P23818 | Gene names | Gria1, Glur1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 1 precursor (GluR-1) (GluR-A) (GluR-K1) (Glutamate receptor ionotropic, AMPA 1) (AMPA-selective glutamate receptor 1). | |||||
|
GRIA1_HUMAN
|
||||||
| NC score | 0.836427 (rank : 26) | θ value | 1.73182e-43 (rank : 20) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P42261 | Gene names | GRIA1, GLUH1, GLUR1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 1 precursor (GluR-1) (GluR-A) (GluR-K1) (Glutamate receptor ionotropic, AMPA 1) (AMPA-selective glutamate receptor 1). | |||||
|
GRIA4_HUMAN
|
||||||
| NC score | 0.833793 (rank : 27) | θ value | 3.85808e-43 (rank : 23) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P48058 | Gene names | GRIA4, GLUR4 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 4 precursor (GluR-4) (GluR4) (GluR-D) (Glutamate receptor ionotropic, AMPA 4) (AMPA-selective glutamate receptor 4). | |||||
|
GRIA2_HUMAN
|
||||||
| NC score | 0.832645 (rank : 28) | θ value | 1.46607e-42 (rank : 24) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P42262, Q96FP6 | Gene names | GRIA2, GLUR2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 2 precursor (GluR-2) (GluR-B) (GluR-K2) (Glutamate receptor ionotropic, AMPA 2) (AMPA-selective glutamate receptor 2). | |||||
|
GRIA2_MOUSE
|
||||||
| NC score | 0.831969 (rank : 29) | θ value | 4.2656e-42 (rank : 25) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P23819, Q61604, Q61605, Q8BG69, Q8BXU3, Q9D6D3 | Gene names | Gria2, Glur2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 2 precursor (GluR-2) (GluR-B) (GluR-K2) (Glutamate receptor ionotropic, AMPA 2) (AMPA-selective glutamate receptor 2). | |||||
|
GRIA3_MOUSE
|
||||||
| NC score | 0.831671 (rank : 30) | θ value | 1.62093e-41 (rank : 27) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Z2W9, Q5DTJ0 | Gene names | Gria3, Glur3, Kiaa4184 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate receptor 3 precursor (GluR-3) (GluR-C) (GluR-K3) (Glutamate receptor ionotropic, AMPA 3) (AMPA-selective glutamate receptor 3). | |||||
|
GRIA4_MOUSE
|
||||||
| NC score | 0.831612 (rank : 31) | θ value | 2.11701e-41 (rank : 28) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Z2W8 | Gene names | Gria4, Glur4 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 4 precursor (GluR-4) (GluR4) (GluR-D) (Glutamate receptor ionotropic, AMPA 4) (AMPA-selective glutamate receptor 4). | |||||
|
GRIA3_HUMAN
|
||||||
| NC score | 0.831478 (rank : 32) | θ value | 8.04462e-41 (rank : 31) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P42263, Q4VXD5, Q4VXD6, Q9HDA0, Q9HDA1, Q9HDA2 | Gene names | GRIA3, GLUR3, GLURC | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 3 precursor (GluR-3) (GluR-C) (GluR-K3) (Glutamate receptor ionotropic, AMPA 3) (AMPA-selective glutamate receptor 3). | |||||
|
GRIK1_MOUSE
|
||||||
| NC score | 0.825882 (rank : 33) | θ value | 1.12514e-34 (rank : 33) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q60934 | Gene names | Grik1, Glur5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 1 precursor (Glutamate receptor 5) (GluR-5) (GluR5). | |||||
|
ERB1_HUMAN
|
||||||
| NC score | 0.132871 (rank : 34) | θ value | 0.21417 (rank : 46) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P60509 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-R(b)_3p24.3 provirus ancestral Env polyprotein precursor (Envelope polyprotein) (HERV-R(b) Env protein) [Includes: Surface protein domain (SU); Transmembrane protein domain (TM)]. | |||||
|
GABR1_HUMAN
|
||||||
| NC score | 0.070125 (rank : 35) | θ value | 0.279714 (rank : 48) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UBS5, O95375, O95468, O95975, O96022, Q86W60, Q9UQQ0 | Gene names | GABBR1, GPRC3A | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
GABR1_MOUSE
|
||||||
| NC score | 0.070049 (rank : 36) | θ value | 0.279714 (rank : 49) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9WV18, Q9WU48, Q9WV15, Q9WV16, Q9WV17 | Gene names | Gabbr1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.054236 (rank : 37) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
TS1R1_MOUSE
|
||||||
| NC score | 0.049180 (rank : 38) | θ value | 4.03905 (rank : 112) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99PG6, Q6NS58, Q923J9, Q925I5, Q99PG5 | Gene names | Tas1r1, Gpr70, T1r1, Tr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 1 precursor (G-protein coupled receptor 70). | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.047407 (rank : 39) | θ value | 0.0563607 (rank : 41) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.047121 (rank : 40) | θ value | 5.27518 (rank : 119) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.044746 (rank : 41) | θ value | 1.09739e-05 (rank : 35) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.043752 (rank : 42) | θ value | 5.81887e-07 (rank : 34) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.043321 (rank : 43) | θ value | 0.00509761 (rank : 36) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
CK024_HUMAN
|
||||||
| NC score | 0.042952 (rank : 44) | θ value | 2.36792 (rank : 87) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.040018 (rank : 45) | θ value | 0.0961366 (rank : 42) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.037963 (rank : 46) | θ value | 6.88961 (rank : 132) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.032484 (rank : 47) | θ value | 0.0330416 (rank : 38) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.031816 (rank : 48) | θ value | 0.125558 (rank : 44) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
TS1R3_HUMAN
|
||||||
| NC score | 0.030011 (rank : 49) | θ value | 0.125558 (rank : 45) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7RTX0, Q5TA49, Q8NGW9 | Gene names | TAS1R3, T1R3, TR3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 3 precursor (Sweet taste receptor T1R3). | |||||
|
CO039_MOUSE
|
||||||
| NC score | 0.029494 (rank : 50) | θ value | 0.47712 (rank : 53) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q3TEI4, Q3TMF2, Q3U253, Q3UM75, Q9DA93 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39 homolog. | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.028460 (rank : 51) | θ value | 2.36792 (rank : 86) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CABIN_HUMAN
|
||||||
| NC score | 0.027717 (rank : 52) | θ value | 1.81305 (rank : 81) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Y6J0, Q9Y460 | Gene names | CABIN1, KIAA0330 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin-binding protein Cabin 1 (Calcineurin inhibitor) (CAIN). | |||||
|
PSL1_HUMAN
|
||||||
| NC score | 0.026915 (rank : 53) | θ value | 5.27518 (rank : 127) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
TS13_MOUSE
|
||||||
| NC score | 0.026463 (rank : 54) | θ value | 1.81305 (rank : 84) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q01755 | Gene names | Tcp11 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific protein PBS13. | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.026191 (rank : 55) | θ value | 5.27518 (rank : 115) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.025014 (rank : 56) | θ value | 5.27518 (rank : 123) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
VEGFB_HUMAN
|
||||||
| NC score | 0.024317 (rank : 57) | θ value | 4.03905 (rank : 113) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P49765, Q16528 | Gene names | VEGFB, VRF | |||
|
Domain Architecture |

|
|||||
| Description | Vascular endothelial growth factor B precursor (VEGF-B) (VEGF-related factor) (VRF). | |||||
|
SYNJ1_HUMAN
|
||||||
| NC score | 0.023858 (rank : 58) | θ value | 6.88961 (rank : 142) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O43426, O43425, O94984 | Gene names | SYNJ1, KIAA0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.023587 (rank : 59) | θ value | 0.62314 (rank : 60) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
ELL_HUMAN
|
||||||
| NC score | 0.023569 (rank : 60) | θ value | 0.47712 (rank : 54) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P55199 | Gene names | ELL, C19orf17 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II elongation factor ELL (Eleven-nineteen lysine-rich leukemia protein). | |||||
|
MILK1_MOUSE
|
||||||
| NC score | 0.022880 (rank : 61) | θ value | 6.88961 (rank : 137) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.022662 (rank : 62) | θ value | 8.99809 (rank : 152) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
GAK5_HUMAN
|
||||||
| NC score | 0.022591 (rank : 63) | θ value | 0.47712 (rank : 55) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P62684 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19p13.11 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K113 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
SC24C_HUMAN
|
||||||
| NC score | 0.022432 (rank : 64) | θ value | 4.03905 (rank : 108) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.022417 (rank : 65) | θ value | 8.99809 (rank : 146) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.022369 (rank : 66) | θ value | 1.06291 (rank : 73) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.022198 (rank : 67) | θ value | 0.0330416 (rank : 39) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
IASPP_MOUSE
|
||||||
| NC score | 0.021948 (rank : 68) | θ value | 0.279714 (rank : 50) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
IL3RB_HUMAN
|
||||||
| NC score | 0.021758 (rank : 69) | θ value | 1.38821 (rank : 76) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P32927 | Gene names | CSF2RB, IL3RB, IL5RB | |||
|
Domain Architecture |

|
|||||
| Description | Cytokine receptor common beta chain precursor (GM-CSF/IL-3/IL-5 receptor common beta-chain) (CD131 antigen) (CDw131). | |||||
|
GAK12_HUMAN
|
||||||
| NC score | 0.021612 (rank : 70) | θ value | 1.06291 (rank : 68) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P63130, Q9UKI0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K102 Gag protein) (HERV-K(III) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK4_HUMAN
|
||||||
| NC score | 0.021469 (rank : 71) | θ value | 1.06291 (rank : 70) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P63126, Q9UKH4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_6q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K109 Gag protein) (HERV-K(C6) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK11_HUMAN
|
||||||
| NC score | 0.021414 (rank : 72) | θ value | 1.06291 (rank : 67) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P63145, Q9UKI1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.21 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K101 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GNAS1_MOUSE
|
||||||
| NC score | 0.021312 (rank : 73) | θ value | 0.0330416 (rank : 37) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6R0H7, Q6R0H4, Q6R0H5, Q6R2J5, Q9JJX0, Q9Z1N8 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
GAK2_HUMAN
|
||||||
| NC score | 0.021142 (rank : 74) | θ value | 1.06291 (rank : 69) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q7LDI9, Q9UKH5, Q9Y6I1, Q9YNA6, Q9YNB0 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(HML-2.HOM) Gag protein) (HERV-K108 Gag protein) (HERV-K(C7) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
TAU_MOUSE
|
||||||
| NC score | 0.020735 (rank : 75) | θ value | 5.27518 (rank : 129) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.020503 (rank : 76) | θ value | 1.81305 (rank : 82) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
GTSE1_HUMAN
|
||||||
| NC score | 0.020429 (rank : 77) | θ value | 4.03905 (rank : 107) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NYZ3, Q53GX5, Q5R3I6, Q6DHX4, Q9BRE0, Q9UGZ9, Q9Y557 | Gene names | GTSE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G2 and S phase expressed protein 1 (B99 homolog). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.019787 (rank : 78) | θ value | 2.36792 (rank : 92) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
PER3_HUMAN
|
||||||
| NC score | 0.019337 (rank : 79) | θ value | 0.365318 (rank : 52) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
GRIN1_HUMAN
|
||||||
| NC score | 0.019203 (rank : 80) | θ value | 6.88961 (rank : 135) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
S23IP_MOUSE
|
||||||
| NC score | 0.018945 (rank : 81) | θ value | 3.0926 (rank : 99) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6NZC7, Q8BXY6 | Gene names | Sec23ip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SEC23-interacting protein. | |||||
|
APC_HUMAN
|
||||||
| NC score | 0.018881 (rank : 82) | θ value | 1.06291 (rank : 66) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P25054, Q15162, Q15163, Q93042 | Gene names | APC, DP2.5 | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC). | |||||
|
GAK10_HUMAN
|
||||||
| NC score | 0.018869 (rank : 83) | θ value | 6.88961 (rank : 134) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P87889, P10263, P10264, Q69385, Q9UKH6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q33.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K10 Gag protein) (HERV-K107 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
NRX2B_HUMAN
|
||||||
| NC score | 0.018555 (rank : 84) | θ value | 2.36792 (rank : 91) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
SSBP4_HUMAN
|
||||||
| NC score | 0.018462 (rank : 85) | θ value | 3.0926 (rank : 100) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.018342 (rank : 86) | θ value | 0.0563607 (rank : 40) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.018031 (rank : 87) | θ value | 0.62314 (rank : 58) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
MEGF9_MOUSE
|
||||||
| NC score | 0.016812 (rank : 88) | θ value | 0.125558 (rank : 43) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
FOXN1_MOUSE
|
||||||
| NC score | 0.016169 (rank : 89) | θ value | 5.27518 (rank : 118) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q61575 | Gene names | Foxn1, Fkh19, Hfh11, Whn | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein N1 (Transcription factor winged-helix nude) (Hepatocyte nuclear factor 3 forkhead homolog 11) (HNF-3/forkhead homolog 11) (HFH-11). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.015637 (rank : 90) | θ value | 0.813845 (rank : 65) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
MEF2B_MOUSE
|
||||||
| NC score | 0.015511 (rank : 91) | θ value | 1.06291 (rank : 71) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O55087, O55088, O55089, O55090, O55231, Q61843 | Gene names | Mef2b | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2B. | |||||
|
JPH2_HUMAN
|
||||||
| NC score | 0.015497 (rank : 92) | θ value | 0.813845 (rank : 62) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.015420 (rank : 93) | θ value | 0.813845 (rank : 64) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
MKL1_MOUSE
|
||||||
| NC score | 0.015375 (rank : 94) | θ value | 0.813845 (rank : 63) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
FA47A_HUMAN
|
||||||
| NC score | 0.014655 (rank : 95) | θ value | 5.27518 (rank : 117) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5JRC9, Q8TAA0 | Gene names | FAM47A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47A. | |||||
|
SEPT9_HUMAN
|
||||||
| NC score | 0.014587 (rank : 96) | θ value | 0.21417 (rank : 47) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
IF4G1_MOUSE
|
||||||
| NC score | 0.014528 (rank : 97) | θ value | 8.99809 (rank : 151) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
MUC5B_HUMAN
|
||||||
| NC score | 0.014259 (rank : 98) | θ value | 6.88961 (rank : 138) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
ECM1_HUMAN
|
||||||
| NC score | 0.014176 (rank : 99) | θ value | 4.03905 (rank : 104) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q16610 | Gene names | ECM1 | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular matrix protein 1 precursor (Secretory component p85). | |||||
|
BAZ2A_HUMAN
|
||||||
| NC score | 0.013359 (rank : 100) | θ value | 0.62314 (rank : 57) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
BIM_MOUSE
|
||||||
| NC score | 0.012962 (rank : 101) | θ value | 4.03905 (rank : 102) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O54918, O54919, O54920 | Gene names | Bcl2l11, Bim | |||
|
Domain Architecture |

|
|||||
| Description | Bcl-2-like protein 11 (Bcl2-interacting mediator of cell death). | |||||
|
NRX2A_HUMAN
|
||||||
| NC score | 0.012789 (rank : 102) | θ value | 2.36792 (rank : 90) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
RGS12_HUMAN
|
||||||
| NC score | 0.012618 (rank : 103) | θ value | 1.81305 (rank : 83) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O14924, O14922, O14923, O43510, O75338 | Gene names | RGS12 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 12 (RGS12). | |||||
|
FOXH1_HUMAN
|
||||||
| NC score | 0.012150 (rank : 104) | θ value | 0.365318 (rank : 51) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O75593 | Gene names | FOXH1, FAST1, FAST2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein H1 (Forkhead activin signal transducer 1) (Fast- 1) (hFAST-1) (Forkhead activin signal transducer 2) (Fast-2). | |||||
|
PDXL2_MOUSE
|
||||||
| NC score | 0.011359 (rank : 105) | θ value | 5.27518 (rank : 125) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8CAE9, Q8CFW3 | Gene names | Podxl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Podocalyxin-like protein 2 precursor (Endoglycan). | |||||
|
SPRE3_MOUSE
|
||||||
| NC score | 0.011245 (rank : 106) | θ value | 4.03905 (rank : 110) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6P6N5, Q7TNJ8 | Gene names | Spred3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sprouty-related, EVH1 domain-containing protein 3 (Spred-3). | |||||
|
MEGF8_MOUSE
|
||||||
| NC score | 0.010240 (rank : 107) | θ value | 3.0926 (rank : 97) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 520 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P60882, Q80TR3, Q80V41, Q8BMN9, Q8JZW7, Q8K0J3 | Gene names | Megf8, Egfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 8 (EGF-like domain- containing protein 4) (Multiple EGF-like domain protein 4). | |||||
|
HSF1_MOUSE
|
||||||
| NC score | 0.010101 (rank : 108) | θ value | 3.0926 (rank : 94) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P38532, O70462 | Gene names | Hsf1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
FOXD4_HUMAN
|
||||||
| NC score | 0.009847 (rank : 109) | θ value | 3.0926 (rank : 93) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q12950, Q5VVK1, Q8WXT6 | Gene names | FOXD4, FKHL9, FOXD4A, FREAC5 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein D4 (Forkhead-related protein FKHL9) (Forkhead- related transcription factor 5) (FREAC-5) (Myeloid factor-alpha). | |||||
|
FX4L1_HUMAN
|
||||||
| NC score | 0.009817 (rank : 110) | θ value | 1.38821 (rank : 75) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NU39 | Gene names | FOXD4L1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein D4-like 1. | |||||
|
SRGEF_MOUSE
|
||||||
| NC score | 0.009704 (rank : 111) | θ value | 4.03905 (rank : 111) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80YD6, Q9QXB7 | Gene names | Sergef, Delgef, Gnefr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secretion-regulating guanine nucleotide exchange factor (Guanine nucleotide exchange factor-related protein) (Deafness locus-associated putative guanine nucleotide exchange factor) (DelGEF). | |||||
|
PDLI7_HUMAN
|
||||||
| NC score | 0.009671 (rank : 112) | θ value | 1.38821 (rank : 77) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 547 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9NR12, Q14250, Q5XG82, Q6NVZ5, Q96C91, Q9BXB8, Q9BXB9 | Gene names | PDLIM7, ENIGMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
F120C_MOUSE
|
||||||
| NC score | 0.009644 (rank : 113) | θ value | 2.36792 (rank : 88) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C3F2 | Gene names | Fam120c, ORF34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UPF0318 protein FAM120C. | |||||
|
TREF1_MOUSE
|
||||||
| NC score | 0.009467 (rank : 114) | θ value | 6.88961 (rank : 143) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BXJ2, Q6PCQ4, Q80X25, Q810H8, Q8BY31 | Gene names | Trerf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132). | |||||
|
MEGF8_HUMAN
|
||||||
| NC score | 0.009012 (rank : 115) | θ value | 3.0926 (rank : 96) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7Z7M0, O75097 | Gene names | MEGF8, EGFL4, KIAA0817 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 8 (EGF-like domain- containing protein 4) (Multiple EGF-like domain protein 4). | |||||
|
BLNK_MOUSE
|
||||||
| NC score | 0.008988 (rank : 116) | θ value | 8.99809 (rank : 145) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9QUN3, O88504 | Gene names | Blnk, Bash, Ly57, Slp65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell linker protein (Cytoplasmic adapter protein) (B-cell adapter containing SH2 domain protein) (B-cell adapter containing Src homology 2 domain protein) (Src homology 2 domain-containing leukocyte protein of 65 kDa) (Slp-65) (Lymphocyte antigen 57). | |||||
|
ZO3_HUMAN
|
||||||
| NC score | 0.008760 (rank : 117) | θ value | 1.81305 (rank : 85) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95049 | Gene names | TJP3, ZO3 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-3 (Zonula occludens 3 protein) (Zona occludens 3 protein) (Tight junction protein 3). | |||||
|
LRC56_HUMAN
|
||||||
| NC score | 0.008528 (rank : 118) | θ value | 5.27518 (rank : 122) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IYG6, Q8N3Q4 | Gene names | LRRC56 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 56. | |||||
|
RCOR2_HUMAN
|
||||||
| NC score | 0.008472 (rank : 119) | θ value | 6.88961 (rank : 140) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8IZ40, Q96FP3 | Gene names | RCOR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 2. | |||||
|
FUT4_HUMAN
|
||||||
| NC score | 0.007707 (rank : 120) | θ value | 2.36792 (rank : 89) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P22083 | Gene names | FUT4, ELFT | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-(1,3)-fucosyltransferase (EC 2.4.1.-) (Galactoside 3-L- fucosyltransferase) (Fucosyltransferase 4) (FUCT-IV) (Fuc-TIV) (ELAM-1 ligand fucosyltransferase). | |||||
|
FOXD3_MOUSE
|
||||||
| NC score | 0.007524 (rank : 121) | θ value | 4.03905 (rank : 105) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61060 | Gene names | Foxd3, Hfh2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein D3 (HNF3/FH transcription factor genesis) (Hepatocyte nuclear factor 3 forkhead homolog 2) (HFH-2). | |||||
|
ATS13_HUMAN
|
||||||
| NC score | 0.007392 (rank : 122) | θ value | 1.38821 (rank : 74) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q76LX8, Q6UY16, Q710F6, Q711T8, Q96L37, Q9H0G3 | Gene names | ADAMTS13, C9orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAMTS-13 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 13) (ADAM-TS 13) (ADAM-TS13) (von Willebrand factor-cleaving protease) (vWF-cleaving protease) (vWF-CP). | |||||
|
COX11_MOUSE
|
||||||
| NC score | 0.006531 (rank : 123) | θ value | 8.99809 (rank : 148) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6P8I6 | Gene names | Cox11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytochrome c oxidase assembly protein COX11, mitochondrial precursor. | |||||
|
RBPSL_HUMAN
|
||||||
| NC score | 0.006492 (rank : 124) | θ value | 5.27518 (rank : 128) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UBG7, O95723, Q5QPU9, Q9ULV9 | Gene names | RBPSUHL, RBPL | |||
|
Domain Architecture |

|
|||||
| Description | Recombining binding protein suppressor of hairless-like protein (Transcription factor RBP-L). | |||||
|
THAP7_HUMAN
|
||||||
| NC score | 0.006085 (rank : 125) | θ value | 8.99809 (rank : 157) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BT49 | Gene names | THAP7 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 7. | |||||
|
UBP6_HUMAN
|
||||||
| NC score | 0.005878 (rank : 126) | θ value | 1.38821 (rank : 78) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35125, Q15634, Q86WP6, Q8IWT4 | Gene names | USP6, TRE2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 6 (EC 3.1.2.15) (Ubiquitin thioesterase 6) (Ubiquitin-specific-processing protease 6) (Deubiquitinating enzyme 6) (Proto-oncogene TRE-2). | |||||
|
ARSJ_MOUSE
|
||||||
| NC score | 0.005612 (rank : 127) | θ value | 6.88961 (rank : 131) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BM89, Q8BKG5, Q8BL46 | Gene names | Arsj | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arylsulfatase J precursor (EC 3.1.6.-) (ASJ). | |||||
|
ARSJ_HUMAN
|
||||||
| NC score | 0.005604 (rank : 128) | θ value | 6.88961 (rank : 130) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5FYB0, Q5FWE4, Q6UWT9 | Gene names | ARSJ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arylsulfatase J precursor (EC 3.1.6.-) (ASJ). | |||||
|
SOX13_MOUSE
|
||||||
| NC score | 0.005325 (rank : 129) | θ value | 4.03905 (rank : 109) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q04891, Q922L3 | Gene names | Sox13, Sox-13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein. | |||||
|
HXD3_HUMAN
|
||||||
| NC score | 0.004383 (rank : 130) | θ value | 0.813845 (rank : 61) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P31249, Q99955, Q9BSC5 | Gene names | HOXD3, HOX4A | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D3 (Hox-4A). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.004274 (rank : 131) | θ value | 1.81305 (rank : 79) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
S19A1_HUMAN
|
||||||
| NC score | 0.004154 (rank : 132) | θ value | 8.99809 (rank : 154) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P41440, O00553, O60227, Q13026 | Gene names | SLC19A1, FLOT1, RFC1 | |||
|
Domain Architecture |

|
|||||
| Description | Folate transporter 1 (Solute carrier family 19 member 1) (Placental folate transporter) (FOLT) (Reduced folate carrier protein) (RFC) (Intestinal folate carrier) (IFC-1). | |||||
|
SH22A_MOUSE
|
||||||
| NC score | 0.003991 (rank : 133) | θ value | 8.99809 (rank : 155) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QXK9, Q9JHP6 | Gene names | Sh2d2a, Lad, Ribp | |||
|
Domain Architecture |

|
|||||
| Description | SH2 domain protein 2A (Lck-associated adapter protein) (Lad) (Rlk/Itk- binding protein) (Ribp). | |||||
|
ANPRB_HUMAN
|
||||||
| NC score | 0.003721 (rank : 134) | θ value | 1.81305 (rank : 80) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P20594, O60871, Q9UQ50 | Gene names | NPR2, ANPRB | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide receptor B precursor (ANP-B) (ANPRB) (GC-B) (Guanylate cyclase B) (EC 4.6.1.2) (NPR-B) (Atrial natriuretic peptide B-type receptor). | |||||
|
EYA1_MOUSE
|
||||||
| NC score | 0.003483 (rank : 135) | θ value | 8.99809 (rank : 150) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P97767, O08818 | Gene names | Eya1 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 1 (EC 3.1.3.48). | |||||
|
SRBS1_MOUSE
|
||||||
| NC score | 0.003419 (rank : 136) | θ value | 8.99809 (rank : 156) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q62417, Q80TF8, Q8BZI3, Q8K3Y2, Q921F8, Q9Z0Z8, Q9Z0Z9 | Gene names | Sorbs1, Kiaa1296, Sh3d5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorbin and SH3 domain-containing protein 1 (Ponsin) (c-Cbl-associated protein) (CAP) (SH3 domain protein 5) (SH3P12). | |||||
|
LHX2_MOUSE
|
||||||
| NC score | 0.003343 (rank : 137) | θ value | 5.27518 (rank : 121) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z0S2 | Gene names | Lhx2 | |||
|
Domain Architecture |

|
|||||
| Description | LIM/homeobox protein Lhx2. | |||||
|
LHX2_HUMAN
|
||||||
| NC score | 0.003328 (rank : 138) | θ value | 5.27518 (rank : 120) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P50458, O95860, Q8N1Z3 | Gene names | LHX2, LH2 | |||
|
Domain Architecture |

|
|||||
| Description | LIM/homeobox protein Lhx2 (Homeobox protein LH-2). | |||||
|
ANPRB_MOUSE
|
||||||
| NC score | 0.003275 (rank : 139) | θ value | 4.03905 (rank : 101) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6VVW5, Q6VVW3, Q6VVW4, Q8CGA9 | Gene names | Npr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Atrial natriuretic peptide receptor B precursor (ANP-B) (ANPRB) (GC-B) (Guanylate cyclase B) (EC 4.6.1.2) (NPR-B) (Atrial natriuretic peptide B-type receptor). | |||||
|
HXD3_MOUSE
|
||||||
| NC score | 0.002959 (rank : 140) | θ value | 3.0926 (rank : 95) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P09027, Q3UUD3 | Gene names | Hoxd3, Hox-4.1, Hoxd-3 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D3 (Hox-4.1) (Homeobox protein MH-19). | |||||
|
NDF1_MOUSE
|
||||||
| NC score | 0.002798 (rank : 141) | θ value | 6.88961 (rank : 139) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q60867, Q545N9, Q60897 | Gene names | Neurod1, Neurod | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 1 (NeuroD1). | |||||
|
CIAS1_HUMAN
|
||||||
| NC score | 0.002516 (rank : 142) | θ value | 8.99809 (rank : 147) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96P20, O75434, Q59H68, Q5JQS8, Q5JQS9, Q6TG35, Q8TCW0, Q8TEU9, Q8WXH9 | Gene names | CIAS1, NALP3, PYPAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Cold autoinflammatory syndrome 1 protein (Cryopyrin) (NACHT-, LRR- and PYD-containing protein 3) (PYRIN-containing APAF1-like protein 1) (Angiotensin/vasopressin receptor AII/AVP-like). | |||||
|
SEM6B_HUMAN
|
||||||
| NC score | 0.002327 (rank : 143) | θ value | 6.88961 (rank : 141) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H3T3, Q9NRK9 | Gene names | SEMA6B, SEMAZ | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6B precursor (Semaphorin Z) (Sema Z). | |||||
|
BCOR_HUMAN
|
||||||
| NC score | 0.001798 (rank : 144) | θ value | 8.99809 (rank : 144) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6W2J9, Q6P4B6, Q7Z2K7, Q8TEB4, Q96DB3, Q9H232, Q9H233, Q9HCJ7, Q9NXF2 | Gene names | BCOR, KIAA1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
PCDA9_HUMAN
|
||||||
| NC score | 0.001767 (rank : 145) | θ value | 1.06291 (rank : 72) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y5H5, O15053 | Gene names | PCDHA9, KIAA0345 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 9 precursor (PCDH-alpha9). | |||||
|
PCOC1_HUMAN
|
||||||
| NC score | 0.001589 (rank : 146) | θ value | 8.99809 (rank : 153) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15113, O14550 | Gene names | PCOLCE, PCPE1 | |||
|
Domain Architecture |

|
|||||
| Description | Procollagen C-endopeptidase enhancer 1 precursor (Procollagen COOH- terminal proteinase enhancer 1) (Procollagen C-proteinase enhancer 1) (PCPE-1) (Type I procollagen COOH-terminal proteinase enhancer) (Type 1 procollagen C-proteinase enhancer protein). | |||||
|
ZN516_MOUSE
|
||||||
| NC score | 0.001056 (rank : 147) | θ value | 0.47712 (rank : 56) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q7TSH3 | Gene names | Znf516, Kiaa0222, Zfp516 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 516. | |||||
|
CNTRB_MOUSE
|
||||||
| NC score | 0.000305 (rank : 148) | θ value | 4.03905 (rank : 103) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8CB62, Q5NCF3 | Gene names | Cntrob, Lip8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
EDNRB_HUMAN
|
||||||
| NC score | 0.000007 (rank : 149) | θ value | 0.62314 (rank : 59) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 598 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P24530, O15343, Q9UQK3 | Gene names | EDNRB, ETRB | |||
|
Domain Architecture |

|
|||||
| Description | Endothelin B receptor precursor (ET-B) (Endothelin receptor Non- selective type). | |||||
|
PGBM_MOUSE
|
||||||
| NC score | -0.000071 (rank : 150) | θ value | 5.27518 (rank : 126) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q05793 | Gene names | Hspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
NOTC3_HUMAN
|
||||||
| NC score | -0.000096 (rank : 151) | θ value | 5.27518 (rank : 124) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1062 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UM47, Q9UEB3, Q9UPL3, Q9Y6L8 | Gene names | NOTCH3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
ZEP2_MOUSE
|
||||||
| NC score | -0.000195 (rank : 152) | θ value | 4.03905 (rank : 114) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q3UHF7, O55140, Q3UVD4, Q3UVH5 | Gene names | Hivep2, Mibp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Human immunodeficiency virus type I enhancer-binding protein 2 homolog (Myc intron-binding protein 1) (MIBP-1). | |||||
|
POLS2_HUMAN
|
||||||
| NC score | -0.000251 (rank : 153) | θ value | 3.0926 (rank : 98) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5K4E3, Q8NBY4 | Gene names | PRSS36 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyserase-2 precursor (EC 3.4.21.-) (Polyserine protease 2) (Protease serine 36). | |||||
|
CELR2_HUMAN
|
||||||
| NC score | -0.000672 (rank : 154) | θ value | 6.88961 (rank : 133) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9HCU4, Q92566 | Gene names | CELSR2, CDHF10, EGFL2, KIAA0279, MEGF3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 2 precursor (Epidermal growth factor-like 2) (Multiple epidermal growth factor-like domains 3) (Flamingo 1). | |||||
|
GAK_MOUSE
|
||||||
| NC score | -0.002512 (rank : 155) | θ value | 4.03905 (rank : 106) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99KY4, Q6P1I8, Q6P9S5, Q8BM74, Q8K0Q4 | Gene names | Gak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||
|
DRD4_MOUSE
|
||||||
| NC score | -0.002925 (rank : 156) | θ value | 8.99809 (rank : 149) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 852 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P51436, O35838 | Gene names | Drd4 | |||
|
Domain Architecture |

|
|||||
| Description | D(4) dopamine receptor (Dopamine D4 receptor) (D(2C) dopamine receptor). | |||||
|
M3K14_HUMAN
|
||||||
| NC score | -0.003352 (rank : 157) | θ value | 6.88961 (rank : 136) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 887 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99558, Q8IYN1 | Gene names | MAP3K14, NIK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 14 (EC 2.7.11.25) (NF- kappa beta-inducing kinase) (Serine/threonine-protein kinase NIK) (HsNIK). | |||||
|
EGR4_MOUSE
|
||||||
| NC score | -0.004048 (rank : 158) | θ value | 5.27518 (rank : 116) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 710 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9WUF2 | Gene names | Egr4 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 4 (EGR-4). | |||||
|
TYK2_MOUSE
|
||||||
| NC score | -0.004941 (rank : 159) | θ value | 8.99809 (rank : 158) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 850 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9R117, O88431, Q8VE41 | Gene names | Tyk2 | |||
|
Domain Architecture |

|
|||||
| Description | Non-receptor tyrosine-protein kinase TYK2 (EC 2.7.10.2). | |||||