Please be patient as the page loads
|
GABR1_HUMAN
|
||||||
| SwissProt Accessions | Q9UBS5, O95375, O95468, O95975, O96022, Q86W60, Q9UQQ0 | Gene names | GABBR1, GPRC3A | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
GABR1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9UBS5, O95375, O95468, O95975, O96022, Q86W60, Q9UQQ0 | Gene names | GABBR1, GPRC3A | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
GABR1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999599 (rank : 2) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9WV18, Q9WU48, Q9WV15, Q9WV16, Q9WV17 | Gene names | Gabbr1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
GABR2_HUMAN
|
||||||
| θ value | 1.02419e-128 (rank : 3) | NC score | 0.841848 (rank : 3) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O75899, O75974, O75975, Q5VXZ2, Q8WX04, Q9P1R2, Q9UNR1, Q9UNS9 | Gene names | GABBR2, GPR51, GPRC3B | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 2 precursor (GABA-B receptor 2) (GABA-B-R2) (Gb2) (GABABR2) (G-protein coupled receptor 51) (HG20). | |||||
|
GP156_HUMAN
|
||||||
| θ value | 2.20605e-14 (rank : 4) | NC score | 0.446619 (rank : 4) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8NFN8, Q86SN6 | Gene names | GPR156, GABABL, PGR28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 156 (GABAB-related G-protein coupled receptor) (G-protein coupled receptor PGR28). | |||||
|
ANPRB_MOUSE
|
||||||
| θ value | 2.28291e-11 (rank : 5) | NC score | 0.143530 (rank : 79) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6VVW5, Q6VVW3, Q6VVW4, Q8CGA9 | Gene names | Npr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Atrial natriuretic peptide receptor B precursor (ANP-B) (ANPRB) (GC-B) (Guanylate cyclase B) (EC 4.6.1.2) (NPR-B) (Atrial natriuretic peptide B-type receptor). | |||||
|
ANPRB_HUMAN
|
||||||
| θ value | 1.47974e-10 (rank : 6) | NC score | 0.136526 (rank : 81) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P20594, O60871, Q9UQ50 | Gene names | NPR2, ANPRB | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide receptor B precursor (ANP-B) (ANPRB) (GC-B) (Guanylate cyclase B) (EC 4.6.1.2) (NPR-B) (Atrial natriuretic peptide B-type receptor). | |||||
|
MGR8_MOUSE
|
||||||
| θ value | 6.21693e-09 (rank : 7) | NC score | 0.427172 (rank : 8) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P47743, Q6B964 | Gene names | Grm8, Gprc1h, Mglur8 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 8 precursor (mGluR8). | |||||
|
MGR8_HUMAN
|
||||||
| θ value | 8.11959e-09 (rank : 8) | NC score | 0.426484 (rank : 10) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O00222, O15493, O95945, O95946, Q3MIV9, Q52M02, Q6J165 | Gene names | GRM8, GPRC1H, MGLUR8 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 8 precursor (mGluR8). | |||||
|
TS1R3_HUMAN
|
||||||
| θ value | 1.38499e-08 (rank : 9) | NC score | 0.438617 (rank : 5) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q7RTX0, Q5TA49, Q8NGW9 | Gene names | TAS1R3, T1R3, TR3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 3 precursor (Sweet taste receptor T1R3). | |||||
|
MGR4_HUMAN
|
||||||
| θ value | 1.80886e-08 (rank : 10) | NC score | 0.430625 (rank : 7) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q14833, Q5SZ84 | Gene names | GRM4, GPRC1D, MGLUR4 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 4 precursor (mGluR4). | |||||
|
ANPRA_MOUSE
|
||||||
| θ value | 1.53129e-07 (rank : 11) | NC score | 0.127195 (rank : 86) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 600 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P18293 | Gene names | Npr1, Npra | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide receptor A precursor (ANP-A) (ANPRA) (GC-A) (Guanylate cyclase) (EC 4.6.1.2) (NPR-A) (Atrial natriuretic peptide A-type receptor). | |||||
|
MGR7_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 12) | NC score | 0.425525 (rank : 12) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q14831, Q8NFS2, Q8NFS3, Q8NFS4 | Gene names | GRM7, GPRC1G, MGLUR7 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 7 precursor (mGluR7). | |||||
|
MGR7_MOUSE
|
||||||
| θ value | 1.53129e-07 (rank : 13) | NC score | 0.425845 (rank : 11) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q68ED2 | Gene names | Grm7, Gprc1g, Mglur7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metabotropic glutamate receptor 7 precursor (mGluR7). | |||||
|
TS1R1_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 14) | NC score | 0.431342 (rank : 6) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q7RTX1, Q8NGZ7, Q8TDJ7, Q8TDJ8, Q8TDJ9, Q8TDK0 | Gene names | TAS1R1, GPR70, T1R1, TR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 1 precursor (G-protein coupled receptor 70) (Gm148). | |||||
|
GP156_MOUSE
|
||||||
| θ value | 1.99992e-07 (rank : 15) | NC score | 0.393056 (rank : 29) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6PCP7, Q8K452 | Gene names | Gpr156, Gababl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 156 (GABAB-related G-protein coupled receptor). | |||||
|
DAF_HUMAN
|
||||||
| θ value | 4.45536e-07 (rank : 16) | NC score | 0.354203 (rank : 30) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P08174, P09679, P78361 | Gene names | CD55, CR, DAF | |||
|
Domain Architecture |
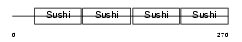
|
|||||
| Description | Complement decay-accelerating factor precursor (CD55 antigen). | |||||
|
ANPRA_HUMAN
|
||||||
| θ value | 5.81887e-07 (rank : 17) | NC score | 0.125362 (rank : 87) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 612 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P16066, Q6P4Q3 | Gene names | NPR1, ANPRA | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide receptor A precursor (ANP-A) (ANPRA) (GC-A) (Guanylate cyclase) (EC 4.6.1.2) (NPR-A) (Atrial natriuretic peptide A-type receptor). | |||||
|
CSMD3_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 18) | NC score | 0.266177 (rank : 59) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q7Z407, Q96PZ3 | Gene names | CSMD3, KIAA1894 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 3 precursor (CUB and sushi multiple domains protein 3). | |||||
|
CASR_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 19) | NC score | 0.412987 (rank : 16) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P41180, Q13912, Q16108, Q16109, Q16110, Q16379, Q4PJ19 | Gene names | CASR, GPRC2A, PCAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular calcium-sensing receptor precursor (CaSR) (Parathyroid Cell calcium-sensing receptor). | |||||
|
V2RX_MOUSE
|
||||||
| θ value | 4.92598e-06 (rank : 20) | NC score | 0.414804 (rank : 13) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O70410 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vomeronasal type-2 receptor X precursor (Putative pheromone receptor V2RX). | |||||
|
CSMD1_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 21) | NC score | 0.260964 (rank : 60) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q96PZ7, Q96QU9, Q96RM4 | Gene names | CSMD1, KIAA1890 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 1 precursor (CUB and sushi multiple domains protein 1). | |||||
|
MGR6_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 22) | NC score | 0.414452 (rank : 15) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O15303 | Gene names | GRM6, GPRC1F, MGLUR6 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 6 precursor (mGluR6). | |||||
|
TS1R1_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 23) | NC score | 0.411575 (rank : 18) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99PG6, Q6NS58, Q923J9, Q925I5, Q99PG5 | Gene names | Tas1r1, Gpr70, T1r1, Tr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 1 precursor (G-protein coupled receptor 70). | |||||
|
ANPRC_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 24) | NC score | 0.337098 (rank : 33) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P17342 | Gene names | NPR3, ANPRC | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide clearance receptor precursor (ANP-C) (ANPRC) (NPR-C) (Atrial natriuretic peptide C-type receptor). | |||||
|
ANPRC_MOUSE
|
||||||
| θ value | 1.09739e-05 (rank : 25) | NC score | 0.343154 (rank : 32) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P70180, P97804, Q9R025, Q9R027, Q9R028 | Gene names | Npr3 | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide clearance receptor precursor (ANP-C) (ANPRC) (NPR-C) (Atrial natriuretic peptide C-type receptor) (EF-2). | |||||
|
CASR_MOUSE
|
||||||
| θ value | 1.09739e-05 (rank : 26) | NC score | 0.411286 (rank : 19) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9QY96, O08968, O88519, Q9QY95, Q9QZU8, Q9R1D6, Q9R1Y2 | Gene names | Casr, Gprc2a | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular calcium-sensing receptor precursor (CaSR) (Parathyroid Cell calcium-sensing receptor). | |||||
|
LYAM2_MOUSE
|
||||||
| θ value | 1.09739e-05 (rank : 27) | NC score | 0.266742 (rank : 58) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q00690 | Gene names | Sele, Elam-1 | |||
|
Domain Architecture |

|
|||||
| Description | E-selectin precursor (Endothelial leukocyte adhesion molecule 1) (ELAM-1) (Leukocyte-endothelial cell adhesion molecule 2) (LECAM2) (CD62E antigen). | |||||
|
MGR1_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 28) | NC score | 0.409002 (rank : 21) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q13255, Q13256, Q14757, Q14758, Q5VTF7, Q5VTF8, Q9NU10, Q9UGS9, Q9UGT0 | Gene names | GRM1, GPRC1A, MGLUR1 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 1 precursor (mGluR1). | |||||
|
TS1R3_MOUSE
|
||||||
| θ value | 1.09739e-05 (rank : 29) | NC score | 0.426901 (rank : 9) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q925D8, Q91VA4, Q923K0, Q925A4, Q925D9 | Gene names | Tas1r3, Sac, T1r3, Tr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 3 precursor (Sweet taste receptor T1R3) (Saccharin preference protein). | |||||
|
GPC6A_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 30) | NC score | 0.408472 (rank : 23) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8K4Z6, Q5DK51, Q5DK52 | Gene names | Gprc6a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor family C group 6 member A precursor. | |||||
|
MGR1_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 31) | NC score | 0.408627 (rank : 22) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P97772, Q6AXG4, Q9EPV6 | Gene names | Grm1, Gprc1a, Mglur1 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 1 precursor (mGluR1). | |||||
|
CR1_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 32) | NC score | 0.328548 (rank : 34) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P17927, Q16745, Q9UQV2 | Gene names | CR1, C3BR | |||
|
Domain Architecture |

|
|||||
| Description | Complement receptor type 1 precursor (C3b/C4b receptor) (CD35 antigen). | |||||
|
MGR6_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 33) | NC score | 0.412963 (rank : 17) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q5NCH9 | Gene names | Grm6, Gprc1f, Mglur6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metabotropic glutamate receptor 6 precursor (mGluR6). | |||||
|
MGR3_MOUSE
|
||||||
| θ value | 7.1131e-05 (rank : 34) | NC score | 0.401067 (rank : 26) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9QYS2 | Gene names | Grm3, Gprc1c, Mglur3 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 3 precursor (mGluR3). | |||||
|
LYAM3_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 35) | NC score | 0.269442 (rank : 56) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q01102 | Gene names | Selp, Grmp | |||
|
Domain Architecture |

|
|||||
| Description | P-selectin precursor (Granule membrane protein 140) (GMP-140) (PADGEM) (Leukocyte-endothelial cell adhesion molecule 3) (LECAM3) (CD62P antigen). | |||||
|
TS1R2_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 36) | NC score | 0.414671 (rank : 14) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q925I4, Q923J8 | Gene names | Tas1r2, Gpr71, T1r2, Tr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 2 precursor (G-protein coupled receptor 71) (Sweet taste receptor T1R2). | |||||
|
CSMD2_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 37) | NC score | 0.252574 (rank : 63) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q7Z408, Q5VT59, Q8N963, Q96Q03, Q9H4V7, Q9H4V8, Q9H4V9, Q9H4W0, Q9H4W1, Q9H4W2, Q9H4W3, Q9H4W4, Q9HCY5, Q9HCY6, Q9HCY7 | Gene names | CSMD2, KIAA1884 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 2 (CUB and sushi multiple domains protein 2). | |||||
|
MGR5_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 38) | NC score | 0.410516 (rank : 20) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P41594 | Gene names | GRM5, GPRC1E, MGLUR5 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 5 precursor (mGluR5). | |||||
|
TS1R2_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 39) | NC score | 0.407079 (rank : 24) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8TE23, Q5TZ19 | Gene names | TAS1R2, GPR71, T1R2, TR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 2 precursor (G-protein coupled receptor 71) (Sweet taste receptor T1R2). | |||||
|
CFAH_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 40) | NC score | 0.304511 (rank : 43) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P08603, P78435, Q14570, Q38G77, Q5TFM3, Q8N708, Q9NU86 | Gene names | CFH, HF, HF1 | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor H precursor (H factor 1). | |||||
|
FHR1_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 41) | NC score | 0.292516 (rank : 49) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q03591, Q9UJ17 | Gene names | CFHL1, CFHL, FHR1, HFL1 | |||
|
Domain Architecture |
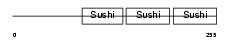
|
|||||
| Description | Complement factor H-related protein 1 precursor (FHR-1) (H factor-like protein 1) (H-factor-like 1) (H36). | |||||
|
GPC6A_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 42) | NC score | 0.402170 (rank : 25) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5T6X5, Q6JK43, Q6JK44, Q8NGU8, Q8NHZ9, Q8TDT6 | Gene names | GPRC6A, GPCR33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor family C group 6 member A precursor (hGPRC6A) (G-protein coupled receptor 33) (hGPCR33). | |||||
|
CSMD1_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 43) | NC score | 0.252453 (rank : 64) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q923L3, Q8BUV1, Q8BYQ3 | Gene names | Csmd1 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 1 precursor (CUB and sushi multiple domains protein 1). | |||||
|
LYAM3_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 44) | NC score | 0.269600 (rank : 55) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P16109, Q8IVD1 | Gene names | SELP, GMRP | |||
|
Domain Architecture |

|
|||||
| Description | P-selectin precursor (Granule membrane protein 140) (GMP-140) (PADGEM) (Leukocyte-endothelial cell adhesion molecule 3) (LECAM3) (CD62P antigen). | |||||
|
DAF1_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 45) | NC score | 0.310954 (rank : 40) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q61475, P97732, Q61397 | Gene names | Cd55a, Daf1 | |||
|
Domain Architecture |

|
|||||
| Description | Complement decay-accelerating factor, GPI-anchored precursor (DAF-GPI) (CD55 antigen). | |||||
|
MGR2_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 46) | NC score | 0.398100 (rank : 27) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q14416, Q52MC6, Q9H3N6 | Gene names | GRM2, GPRC1B, MGLUR2 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 2 precursor (mGluR2). | |||||
|
MGR3_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 47) | NC score | 0.395505 (rank : 28) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q14832, Q75MV4, Q75N17, Q86YG6, Q8TBH9 | Gene names | GRM3, GPRC1C, MGLUR3 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 3 precursor (mGluR3). | |||||
|
CR2_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 48) | NC score | 0.317294 (rank : 38) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P20023, Q13866, Q14212 | Gene names | CR2, C3DR | |||
|
Domain Architecture |

|
|||||
| Description | Complement receptor type 2 precursor (Cr2) (Complement C3d receptor) (Epstein-Barr virus receptor) (EBV receptor) (CD21 antigen). | |||||
|
F13B_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 49) | NC score | 0.306724 (rank : 41) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q07968 | Gene names | F13b, Cf13b | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor XIII B chain precursor (Protein-glutamine gamma- glutamyltransferase B chain) (Transglutaminase B chain). | |||||
|
CR2_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 50) | NC score | 0.317675 (rank : 37) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P19070 | Gene names | Cr2 | |||
|
Domain Architecture |

|
|||||
| Description | Complement receptor type 2 precursor (Cr2) (Complement C3d receptor) (CD21 antigen). | |||||
|
CRRY_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 51) | NC score | 0.323227 (rank : 36) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q64735, Q3TV30, Q58E68, Q61447, Q61449, Q8CF59, Q8K328 | Gene names | Crry, Cry | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement regulatory protein Crry precursor (Protein p65). | |||||
|
LYAM2_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 52) | NC score | 0.253976 (rank : 62) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P16581, P16111 | Gene names | SELE, ELAM1 | |||
|
Domain Architecture |

|
|||||
| Description | E-selectin precursor (Endothelial leukocyte adhesion molecule 1) (ELAM-1) (Leukocyte-endothelial cell adhesion molecule 2) (LECAM2) (CD62E antigen). | |||||
|
SUSD2_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 53) | NC score | 0.238124 (rank : 68) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9UGT4, Q9H5Y6 | Gene names | SUSD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 2 precursor. | |||||
|
ZP3R_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 54) | NC score | 0.312720 (rank : 39) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q60736 | Gene names | Zp3r, Sp56 | |||
|
Domain Architecture |

|
|||||
| Description | Zona pellucida sperm-binding protein 3 receptor precursor (Sperm fertilization protein 56) (sp56). | |||||
|
FHR5_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 55) | NC score | 0.293859 (rank : 48) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9BXR6 | Gene names | CFHL5, FHR5 | |||
|
Domain Architecture |
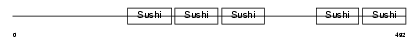
|
|||||
| Description | Complement factor H-related protein 5 precursor (FHR-5). | |||||
|
GRIA2_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 56) | NC score | 0.089487 (rank : 97) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P42262, Q96FP6 | Gene names | GRIA2, GLUR2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 2 precursor (GluR-2) (GluR-B) (GluR-K2) (Glutamate receptor ionotropic, AMPA 2) (AMPA-selective glutamate receptor 2). | |||||
|
GRIA2_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 57) | NC score | 0.089522 (rank : 96) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P23819, Q61604, Q61605, Q8BG69, Q8BXU3, Q9D6D3 | Gene names | Gria2, Glur2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 2 precursor (GluR-2) (GluR-B) (GluR-K2) (Glutamate receptor ionotropic, AMPA 2) (AMPA-selective glutamate receptor 2). | |||||
|
SUSD2_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 58) | NC score | 0.233876 (rank : 71) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9DBX3, Q8BY19, Q8BYN0, Q8BYZ3, Q8BYZ9, Q8BZQ4, Q8C179 | Gene names | Susd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 2 precursor. | |||||
|
C4BB_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 59) | NC score | 0.324147 (rank : 35) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P20851 | Gene names | C4BPB | |||
|
Domain Architecture |
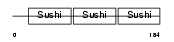
|
|||||
| Description | C4b-binding protein beta chain precursor. | |||||
|
MCP_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 60) | NC score | 0.306280 (rank : 42) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O88174, Q9R0R9 | Gene names | Cd46, Mcp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane cofactor protein precursor (CD46 antigen). | |||||
|
DAF2_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 61) | NC score | 0.299724 (rank : 46) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q61476 | Gene names | Cd55b, Daf2 | |||
|
Domain Architecture |
|
|||||
| Description | Complement decay-accelerating factor transmembrane isoform precursor (DAF-TM) (CD55 antigen). | |||||
|
GRIA1_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 62) | NC score | 0.086102 (rank : 99) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P42261 | Gene names | GRIA1, GLUH1, GLUR1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 1 precursor (GluR-1) (GluR-A) (GluR-K1) (Glutamate receptor ionotropic, AMPA 1) (AMPA-selective glutamate receptor 1). | |||||
|
GRIA1_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 63) | NC score | 0.086358 (rank : 98) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P23818 | Gene names | Gria1, Glur1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 1 precursor (GluR-1) (GluR-A) (GluR-K1) (Glutamate receptor ionotropic, AMPA 1) (AMPA-selective glutamate receptor 1). | |||||
|
FHR3_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 64) | NC score | 0.260557 (rank : 61) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q02985, Q9UJ16 | Gene names | CFHL3, FHR3 | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor H-related protein 3 precursor (FHR-3) (H factor-like protein 3) (DOWN16). | |||||
|
CO6_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 65) | NC score | 0.132321 (rank : 82) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P13671 | Gene names | C6 | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C6 precursor. | |||||
|
CFAH_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 66) | NC score | 0.287860 (rank : 52) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P06909 | Gene names | Cfh, Hf1 | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor H precursor (Protein beta-1-H). | |||||
|
CO2_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 67) | NC score | 0.165973 (rank : 74) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P06681, O19694, Q13904 | Gene names | C2 | |||
|
Domain Architecture |

|
|||||
| Description | Complement C2 precursor (EC 3.4.21.43) (C3/C5 convertase) [Contains: Complement C2b fragment; Complement C2a fragment]. | |||||
|
F13B_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 68) | NC score | 0.292252 (rank : 50) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P05160 | Gene names | F13B | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor XIII B chain precursor (Protein-glutamine gamma- glutamyltransferase B chain) (Transglutaminase B chain) (Fibrin- stabilizing factor B subunit). | |||||
|
CO2_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 69) | NC score | 0.143267 (rank : 80) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P21180 | Gene names | C2 | |||
|
Domain Architecture |

|
|||||
| Description | Complement C2 precursor (EC 3.4.21.43) (C3/C5 convertase) [Contains: Complement C2b fragment; Complement C2a fragment]. | |||||
|
SEZ6L_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 70) | NC score | 0.237703 (rank : 69) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9BYH1, O95917, Q5THY5, Q6IBZ4, Q6UXD4, Q9NUI3, Q9NUI4, Q9NUI5, Q9Y2E1, Q9Y3J6 | Gene names | SEZ6L, KIAA0927 | |||
|
Domain Architecture |

|
|||||
| Description | Seizure 6-like protein precursor. | |||||
|
SUSD4_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 71) | NC score | 0.272506 (rank : 54) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q5VX71, Q6UX62, Q9BSR0, Q9NWG0 | Gene names | SUSD4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 4 precursor. | |||||
|
C4BP_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 72) | NC score | 0.302951 (rank : 44) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P08607 | Gene names | C4bpa, C4bp | |||
|
Domain Architecture |
|
|||||
| Description | C4b-binding protein precursor (C4bp). | |||||
|
MCP_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 73) | NC score | 0.301708 (rank : 45) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P15529, Q15429, Q53GV9, Q5HY94, Q5VWS6, Q5VWS7, Q5VWS8, Q5VWS9, Q5VWT0, Q5VWT1, Q5VWT2, Q6N0A1, Q7Z3R5, Q9NNW2, Q9NNW3, Q9NNW4 | Gene names | CD46, MCP | |||
|
Domain Architecture |

|
|||||
| Description | Membrane cofactor protein precursor (Trophoblast leukocyte common antigen) (TLX) (CD46 antigen). | |||||
|
APOH_HUMAN
|
||||||
| θ value | 0.125558 (rank : 74) | NC score | 0.292136 (rank : 51) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P02749, Q9UCN7 | Gene names | APOH, B2G1 | |||
|
Domain Architecture |
|
|||||
| Description | Beta-2-glycoprotein 1 precursor (Beta-2-glycoprotein I) (Apolipoprotein H) (Apo-H) (B2GPI) (Beta(2)GPI) (Activated protein C- binding protein) (APC inhibitor) (Anticardiolipin cofactor). | |||||
|
LYAM1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 75) | NC score | 0.196092 (rank : 72) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P18337 | Gene names | Sell, Lnhr, Ly-22 | |||
|
Domain Architecture |
|
|||||
| Description | L-selectin precursor (Lymph node homing receptor) (Leukocyte adhesion molecule 1) (LAM-1) (Lymphocyte antigen 22) (Ly-22) (Lymphocyte surface MEL-14 antigen) (Leukocyte-endothelial cell adhesion molecule 1) (LECAM1) (CD62L antigen). | |||||
|
NMDE4_HUMAN
|
||||||
| θ value | 0.163984 (rank : 76) | NC score | 0.072748 (rank : 108) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O15399 | Gene names | GRIN2D | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 4 precursor (N-methyl D- aspartate receptor subtype 2D) (NR2D) (NMDAR2D) (EB11). | |||||
|
C4BP_HUMAN
|
||||||
| θ value | 0.21417 (rank : 77) | NC score | 0.295637 (rank : 47) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P04003 | Gene names | C4BPA, C4BP | |||
|
Domain Architecture |

|
|||||
| Description | C4b-binding protein alpha chain precursor (C4bp) (Proline-rich protein) (PRP). | |||||
|
CFAB_HUMAN
|
||||||
| θ value | 0.21417 (rank : 78) | NC score | 0.161694 (rank : 75) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P00751, O15006, Q29944, Q96HX6, Q9BTF5, Q9BX92 | Gene names | CFB, BF | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor B precursor (EC 3.4.21.47) (C3/C5 convertase) (Properdin factor B) (Glycine-rich beta glycoprotein) (GBG) (PBF2) [Contains: Complement factor B Ba fragment; Complement factor B Bb fragment]. | |||||
|
GUC2F_HUMAN
|
||||||
| θ value | 0.21417 (rank : 79) | NC score | 0.096785 (rank : 93) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 740 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P51841, Q9UJF1 | Gene names | GUCY2F, GUC2F, RETGC2 | |||
|
Domain Architecture |

|
|||||
| Description | Retinal guanylyl cyclase 2 precursor (EC 4.6.1.2) (Guanylate cyclase 2F, retinal) (RETGC-2) (Rod outer segment membrane guanylate cyclase 2) (ROS-GC2) (Guanylate cyclase F) (GC-F). | |||||
|
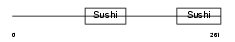
FHR4_HUMAN
|
||||||
| θ value | 0.279714 (rank : 80) | NC score | 0.246503 (rank : 65) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q92496, Q9UJY6 | Gene names | CFHL4, FHR4 | |||
|
Domain Architecture |
|
|||||
| Description | Complement factor H-related protein 4 precursor (FHR-4). | |||||
|
NMDE4_MOUSE
|
||||||
| θ value | 0.279714 (rank : 81) | NC score | 0.070125 (rank : 115) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q03391 | Gene names | Grin2d | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 4 precursor (N-methyl D- aspartate receptor subtype 2D) (NR2D) (NMDAR2D). | |||||
|
SUSD4_MOUSE
|
||||||
| θ value | 0.279714 (rank : 82) | NC score | 0.266962 (rank : 57) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8BH32, Q8VC43 | Gene names | Susd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 4 precursor. | |||||
|
CFAB_MOUSE
|
||||||
| θ value | 0.813845 (rank : 83) | NC score | 0.155331 (rank : 76) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P04186 | Gene names | Cfb, Bf, H2-Bf | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor B precursor (EC 3.4.21.47) (C3/C5 convertase) [Contains: Complement factor B Ba fragment; Complement factor B Bb fragment]. | |||||
|
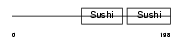
FHR2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 84) | NC score | 0.244172 (rank : 66) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P36980, Q14310 | Gene names | CFHL2, FHR2, HFL3 | |||
|
Domain Architecture |
|
|||||
| Description | Complement factor H-related protein 2 precursor (FHR-2) (H factor-like protein 2) (H factor-like 3) (DDESK59). | |||||
|
ATF3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 85) | NC score | 0.054351 (rank : 134) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P18847 | Gene names | ATF3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-3 (Activating transcription factor 3). | |||||
|
ATF3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 86) | NC score | 0.054136 (rank : 135) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q60765 | Gene names | Atf3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-3 (Activating transcription factor 3) (Transcription factor LRG-21). | |||||
|
CP250_HUMAN
|
||||||
| θ value | 1.06291 (rank : 87) | NC score | 0.013798 (rank : 159) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
GP158_HUMAN
|
||||||
| θ value | 1.06291 (rank : 88) | NC score | 0.116436 (rank : 90) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5T848, Q6QR81, Q9ULT3 | Gene names | GPR158, KIAA1136 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 158 precursor. | |||||
|
GRIK4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 89) | NC score | 0.071071 (rank : 112) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q16099 | Gene names | GRIK4, GRIK | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 4 precursor (Glutamate receptor KA-1) (KA1) (Excitatory amino acid receptor 1) (EAA1). | |||||
|
EVPL_HUMAN
|
||||||
| θ value | 1.38821 (rank : 90) | NC score | 0.019705 (rank : 149) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 895 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q92817 | Gene names | EVPL | |||
|
Domain Architecture |

|
|||||
| Description | Envoplakin (210 kDa paraneoplastic pemphigus antigen) (p210) (210 kDa cornified envelope precursor protein). | |||||
|
GRIA3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 91) | NC score | 0.074603 (rank : 105) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Z2W9, Q5DTJ0 | Gene names | Gria3, Glur3, Kiaa4184 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate receptor 3 precursor (GluR-3) (GluR-C) (GluR-K3) (Glutamate receptor ionotropic, AMPA 3) (AMPA-selective glutamate receptor 3). | |||||
|
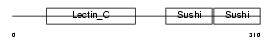
LYAM1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 92) | NC score | 0.189960 (rank : 73) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P14151, P15023 | Gene names | SELL, LNHR, LYAM1 | |||
|
Domain Architecture |
|
|||||
| Description | L-selectin precursor (Lymph node homing receptor) (Leukocyte adhesion molecule 1) (LAM-1) (Leukocyte surface antigen Leu-8) (TQ1) (gp90-MEL) (Leukocyte-endothelial cell adhesion molecule 1) (LECAM1) (CD62L antigen). | |||||
|
SRPX_MOUSE
|
||||||
| θ value | 1.38821 (rank : 93) | NC score | 0.243520 (rank : 67) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9R0M3, Q9R0M2 | Gene names | Srpx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi-repeat-containing protein SRPX precursor (DRS protein). | |||||
|
CCD46_HUMAN
|
||||||
| θ value | 1.81305 (rank : 94) | NC score | 0.015412 (rank : 156) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
GRID2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 95) | NC score | 0.067449 (rank : 119) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43424 | Gene names | GRID2, GLURD2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-2 subunit precursor (GluR delta-2). | |||||
|
PAPOB_MOUSE
|
||||||
| θ value | 1.81305 (rank : 96) | NC score | 0.035523 (rank : 143) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9WVP6, Q9R1R3 | Gene names | Papolb, Papt, Tpap | |||
|
Domain Architecture |

|
|||||
| Description | Poly(A) polymerase beta (EC 2.7.7.19) (PAP beta) (Polynucleotide adenylyltransferase beta) (Testis-specific poly(A) polymerase). | |||||
|
SRPX_HUMAN
|
||||||
| θ value | 1.81305 (rank : 97) | NC score | 0.237504 (rank : 70) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P78539, Q99652, Q99913 | Gene names | SRPX, ETX1 | |||
|
Domain Architecture |

|
|||||
| Description | Sushi repeat-containing protein SRPX precursor. | |||||
|
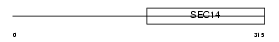
ATCAY_MOUSE
|
||||||
| θ value | 2.36792 (rank : 98) | NC score | 0.021450 (rank : 148) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BHE3, Q3TR94 | Gene names | Atcay | |||
|
Domain Architecture |
|
|||||
| Description | Caytaxin. | |||||
|
GRIK4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 99) | NC score | 0.068348 (rank : 118) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BMF5 | Gene names | Grik4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate receptor, ionotropic kainate 4 precursor. | |||||
|
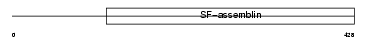
K1C14_MOUSE
|
||||||
| θ value | 2.36792 (rank : 100) | NC score | 0.013611 (rank : 160) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 501 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q61781, Q91VQ4 | Gene names | Krt14, Krt1-14 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type I cytoskeletal 14 (Cytokeratin-14) (CK-14) (Keratin-14) (K14). | |||||
|
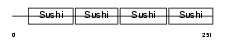
APOH_MOUSE
|
||||||
| θ value | 3.0926 (rank : 101) | NC score | 0.275144 (rank : 53) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q01339 | Gene names | Apoh, B2gp1 | |||
|
Domain Architecture |
|
|||||
| Description | Beta-2-glycoprotein 1 precursor (Beta-2-glycoprotein I) (Apolipoprotein H) (Apo-H) (B2GPI) (Beta(2)GPI) (Activated protein C- binding protein) (APC inhibitor). | |||||
|
GRIA3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 102) | NC score | 0.072167 (rank : 111) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P42263, Q4VXD5, Q4VXD6, Q9HDA0, Q9HDA1, Q9HDA2 | Gene names | GRIA3, GLUR3, GLURC | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 3 precursor (GluR-3) (GluR-C) (GluR-K3) (Glutamate receptor ionotropic, AMPA 3) (AMPA-selective glutamate receptor 3). | |||||
|
GRID2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 103) | NC score | 0.065810 (rank : 122) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61625 | Gene names | Grid2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-2 subunit precursor (GluR delta-2). | |||||
|
HAP1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 104) | NC score | 0.026558 (rank : 147) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 606 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P54257, O75358, Q9H4G3, Q9HA98, Q9NY90 | Gene names | HAP1, HLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-associated protein 1 (HAP-1) (Neuroan 1). | |||||
|
HEM0_HUMAN
|
||||||
| θ value | 3.0926 (rank : 105) | NC score | 0.019035 (rank : 151) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P22557, Q13735 | Gene names | ALAS2, ALASE, ASB | |||
|
Domain Architecture |

|
|||||
| Description | 5-aminolevulinate synthase, erythroid-specific, mitochondrial precursor (EC 2.3.1.37) (5-aminolevulinic acid synthase) (Delta- aminolevulinate synthase) (Delta-ALA synthetase) (ALAS-E). | |||||
|
HOME2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 106) | NC score | 0.016151 (rank : 154) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 882 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9QWW1, O89025, Q9Z0E4 | Gene names | Homer2, Vesl2 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 2 (Homer-2) (Cupidin) (VASP/Ena-related gene up- regulated during seizure and LTP 2) (Vesl-2). | |||||
|
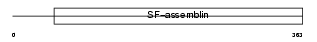
K1H4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 107) | NC score | 0.011581 (rank : 168) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 476 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O76011 | Gene names | KRTHA4, HHA4, HKA4 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type I cuticular Ha4 (Hair keratin, type I Ha4). | |||||
|
ROCK2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 108) | NC score | 0.007883 (rank : 179) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 2073 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O75116, Q9UQN5 | Gene names | ROCK2, KIAA0619 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2) (Rho kinase 2). | |||||
|
TPR_HUMAN
|
||||||
| θ value | 3.0926 (rank : 109) | NC score | 0.012475 (rank : 165) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
ANR26_MOUSE
|
||||||
| θ value | 4.03905 (rank : 110) | NC score | 0.010250 (rank : 174) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1554 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q811D2, Q69ZS2, Q9CS61 | Gene names | Ankrd26, Kiaa1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
CT2NL_HUMAN
|
||||||
| θ value | 4.03905 (rank : 111) | NC score | 0.017260 (rank : 153) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9P2B4, Q96B40 | Gene names | CTTNBP2NL, KIAA1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
ASPP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 112) | NC score | 0.005403 (rank : 183) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
CCD21_HUMAN
|
||||||
| θ value | 5.27518 (rank : 113) | NC score | 0.013400 (rank : 161) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
CT2NL_MOUSE
|
||||||
| θ value | 5.27518 (rank : 114) | NC score | 0.019628 (rank : 150) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 902 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99LJ0, Q6ZPR0, Q8BSV1 | Gene names | Cttnbp2nl, Kiaa1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
GUC2G_MOUSE
|
||||||
| θ value | 5.27518 (rank : 115) | NC score | 0.096647 (rank : 94) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6TL19, Q8BWU7 | Gene names | Gucy2g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanylate cyclase 2G precursor (EC 4.6.1.2) (Guanylyl cyclase receptor G) (mGC-G). | |||||
|
KI13A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 116) | NC score | 0.006518 (rank : 182) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9EQW7, O35062 | Gene names | Kif13a | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF13A. | |||||
|
MYH9_HUMAN
|
||||||
| θ value | 5.27518 (rank : 117) | NC score | 0.013147 (rank : 163) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MYH9_MOUSE
|
||||||
| θ value | 5.27518 (rank : 118) | NC score | 0.013158 (rank : 162) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
PAPOA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 119) | NC score | 0.028922 (rank : 146) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P51003, Q86SX4, Q86TV0, Q8IYF5, Q9BVU2 | Gene names | PAPOLA, PAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(A) polymerase alpha (EC 2.7.7.19) (PAP) (Polynucleotide adenylyltransferase alpha). | |||||
|
PAPOA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 120) | NC score | 0.030859 (rank : 144) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61183, Q61208, Q61209, Q8K4X2 | Gene names | Papola, Pap, Plap | |||
|
Domain Architecture |

|
|||||
| Description | Poly(A) polymerase alpha (EC 2.7.7.19) (PAP) (Polynucleotide adenylyltransferase). | |||||
|
PAPOB_HUMAN
|
||||||
| θ value | 5.27518 (rank : 121) | NC score | 0.029424 (rank : 145) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NRJ5, Q8NE14 | Gene names | PAPOLB, PAPT | |||
|
Domain Architecture |

|
|||||
| Description | Poly(A) polymerase beta (EC 2.7.7.19) (PAP beta) (Polynucleotide adenylyltransferase beta) (Testis-specific poly(A) polymerase). | |||||
|
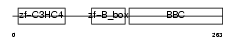
TRI63_HUMAN
|
||||||
| θ value | 5.27518 (rank : 122) | NC score | 0.008644 (rank : 178) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q969Q1, Q5T2I1, Q96BD3, Q96KD9, Q9BYV4 | Gene names | TRIM63, IRF, MURF1, RNF28, SMRZ | |||
|
Domain Architecture |
|
|||||
| Description | Ubiquitin ligase TRIM63 (EC 6.3.2.-) (Tripartite motif-containing protein 63) (Muscle-specific RING finger protein 1) (MuRF1) (MURF-1) (RING finger protein 28) (Striated muscle RING zinc finger protein) (Iris RING finger protein). | |||||
|
CALI_HUMAN
|
||||||
| θ value | 6.88961 (rank : 123) | NC score | 0.007627 (rank : 180) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13939, Q9BXG7 | Gene names | CCIN | |||
|
Domain Architecture |

|
|||||
| Description | Calicin. | |||||
|
GEMI5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 124) | NC score | 0.009118 (rank : 177) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 424 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BX17 | Gene names | Gemin5 | |||
|
Domain Architecture |

|
|||||
| Description | Gem-associated protein 5 (Gemin5). | |||||
|
HIP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 125) | NC score | 0.012426 (rank : 166) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O00291, O00328, Q8TDL4 | Gene names | HIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1 (HIP-I). | |||||
|
MYH2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 126) | NC score | 0.012618 (rank : 164) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
ROCK2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 127) | NC score | 0.006813 (rank : 181) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 2072 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P70336, Q8BR64, Q8CBR0 | Gene names | Rock2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2). | |||||
|
RTN3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 128) | NC score | 0.017671 (rank : 152) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95197, Q496K2, Q7RTM8 | Gene names | RTN3, NSPL2 | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-3 (Neuroendocrine-specific protein-like 2) (NSP-like protein II) (NSPLII). | |||||
|
SAS10_MOUSE
|
||||||
| θ value | 6.88961 (rank : 129) | NC score | 0.014455 (rank : 157) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JI13, Q3UL95, Q8R5C6, Q9JJ12 | Gene names | Crlz1, Crl1, Sas10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Something about silencing protein 10 (Disrupter of silencing SAS10) (Charged amino acid-rich leucine zipper 1) (Crl-1). | |||||
|
TRAK1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 130) | NC score | 0.014031 (rank : 158) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UPV9, Q96B69 | Gene names | TRAK1, KIAA1042, OIP106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trafficking kinesin-binding protein 1 (106 kDa O-GlcNAc transferase- interacting protein). | |||||
|
CP135_MOUSE
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.011431 (rank : 169) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
K1C14_HUMAN
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | 0.011376 (rank : 170) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P02533, Q14715, Q53XY3, Q9BUE3, Q9UBN2, Q9UBN3, Q9UCY4 | Gene names | KRT14 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type I cytoskeletal 14 (Cytokeratin-14) (CK-14) (Keratin-14) (K14). | |||||
|
K1C16_HUMAN
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | 0.009127 (rank : 176) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P08779, P30654, Q16402, Q9UBG8 | Gene names | KRT16, KRT16A | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 16 (Cytokeratin-16) (CK-16) (Keratin-16) (K16). | |||||
|
MASTL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 134) | NC score | 0.001241 (rank : 184) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 894 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96GX5, Q5T8D5, Q5T8D7, Q8NCD6, Q96SJ5 | Gene names | MASTL, THC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase-like (EC 2.7.11.1). | |||||
|
MYH10_HUMAN
|
||||||
| θ value | 8.99809 (rank : 135) | NC score | 0.011122 (rank : 171) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH10_MOUSE
|
||||||
| θ value | 8.99809 (rank : 136) | NC score | 0.010999 (rank : 172) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 137) | NC score | 0.010975 (rank : 173) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
MYH1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 138) | NC score | 0.012074 (rank : 167) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
NMDE2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 139) | NC score | 0.058557 (rank : 126) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q01097, Q9DCB2 | Gene names | Grin2b | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 2 precursor (N-methyl D- aspartate receptor subtype 2B) (NR2B) (NMDAR2B). | |||||
|
PCDBC_HUMAN
|
||||||
| θ value | 8.99809 (rank : 140) | NC score | 0.000156 (rank : 185) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y5F1 | Gene names | PCDHB12 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 12 precursor (PCDH-beta12). | |||||
|
RTN3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 141) | NC score | 0.015492 (rank : 155) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ES97 | Gene names | Rtn3 | |||
|
Domain Architecture |
|
|||||
| Description | Reticulon-3. | |||||
|
TROAP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 142) | NC score | 0.010210 (rank : 175) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12815 | Gene names | TROAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trophinin-associated protein (Tastin) (Trophinin-assisting protein). | |||||
|
ADCY9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.051089 (rank : 140) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60503, O60273, Q4ZHT9, Q4ZIR5, Q9BWT4, Q9UGP2 | Gene names | ADCY9, KIAA0520 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 9 (EC 4.6.1.1) (Adenylate cyclase type IX) (ATP pyrophosphate-lyase 9) (Adenylyl cyclase 9). | |||||
|
ADCY9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.050969 (rank : 141) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P51830, Q61279 | Gene names | Adcy9 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 9 (EC 4.6.1.1) (Adenylate cyclase type IX) (ATP pyrophosphate-lyase 9) (Adenylyl cyclase 9) (Adenylyl cyclase type 10) (ACTP10). | |||||
|
CO7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.119791 (rank : 88) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P10643, Q6P3T5, Q92489 | Gene names | C7 | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C7 precursor. | |||||
|
GCYA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.074837 (rank : 104) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P33402 | Gene names | GUCY1A2, GUC1A2, GUCSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate cyclase soluble subunit alpha-2 (EC 4.6.1.2) (GCS-alpha-2). | |||||
|
GCYA3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.072505 (rank : 110) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q02108, O43843, Q8TAH3 | Gene names | GUCY1A3, GUC1A3, GUCSA3, GUCY1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate cyclase soluble subunit alpha-3 (EC 4.6.1.2) (GCS-alpha-3) (Soluble guanylate cyclase large subunit) (GCS-alpha-1). | |||||
|
GCYA3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.072540 (rank : 109) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9ERL9, Q9DBQ3 | Gene names | Gucy1a3, Gucy1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate cyclase soluble subunit alpha-3 (EC 4.6.1.2) (GCS-alpha-3) (Soluble guanylate cyclase large subunit) (GCS-alpha-1). | |||||
|
GCYB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.074052 (rank : 107) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q02153, Q86WY5 | Gene names | GUCY1B3, GUC1B3, GUCSB3, GUCY1B1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate cyclase soluble subunit beta-1 (EC 4.6.1.2) (GCS-beta-1) (Soluble guanylate cyclase small subunit) (GCS-beta-3). | |||||
|
GCYB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.074179 (rank : 106) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O54865 | Gene names | Gucy1b3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanylate cyclase soluble subunit beta-1 (EC 4.6.1.2) (GCS-beta-1) (Soluble guanylate cyclase small subunit). | |||||
|
GCYB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.081117 (rank : 100) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75343, Q9NZ64 | Gene names | GUCY1B2 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate cyclase soluble subunit beta-2 (EC 4.6.1.2) (GCS-beta-2). | |||||
|
GP158_MOUSE
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.094516 (rank : 95) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8C419, Q8CHB0 | Gene names | Gpr158, Kiaa1136 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 158 precursor. | |||||
|
GP179_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.079502 (rank : 103) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6PRD1 | Gene names | GPR179, GPR158L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 179 precursor (Probable G-protein coupled receptor 158-like 1). | |||||
|
GPC5B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.103065 (rank : 92) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NZH0, O75205, Q8NBZ8 | Gene names | GPRC5B, RAIG2 | |||
|
Domain Architecture |

|
|||||
| Description | G-protein coupled receptor family C group 5 member B precursor (Retinoic acid-induced gene 2 protein) (RAIG-2) (A-69G12.1). | |||||
|
GPC5B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.106990 (rank : 91) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q923Z0, Q8CCV3 | Gene names | Gprc5b, Raig2 | |||
|
Domain Architecture |

|
|||||
| Description | G-protein coupled receptor family C group 5 member B precursor (Retinoic acid-induced gene 2 protein) (RAIG-2). | |||||
|
GRIA4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.065160 (rank : 124) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P48058 | Gene names | GRIA4, GLUR4 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 4 precursor (GluR-4) (GluR4) (GluR-D) (Glutamate receptor ionotropic, AMPA 4) (AMPA-selective glutamate receptor 4). | |||||
|
GRIA4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.065260 (rank : 123) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Z2W8 | Gene names | Gria4, Glur4 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 4 precursor (GluR-4) (GluR4) (GluR-D) (Glutamate receptor ionotropic, AMPA 4) (AMPA-selective glutamate receptor 4). | |||||
|
GRID1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.056664 (rank : 130) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9ULK0, Q8IXT3 | Gene names | GRID1, KIAA1220 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-1 subunit precursor (GluR delta-1). | |||||
|
GRID1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.056655 (rank : 131) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61627 | Gene names | Grid1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-1 subunit precursor (GluR delta-1). | |||||
|
GRIK1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.070159 (rank : 114) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P39086, Q13001, Q86SU9 | Gene names | GRIK1, GLUR5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 1 precursor (Glutamate receptor 5) (GluR-5) (GluR5) (Excitatory amino acid receptor 3) (EAA3). | |||||
|
GRIK1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.069648 (rank : 116) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q60934 | Gene names | Grik1, Glur5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 1 precursor (Glutamate receptor 5) (GluR-5) (GluR5). | |||||
|
GRIK2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.071028 (rank : 113) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13002 | Gene names | GRIK2, GLUR6 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 2 precursor (Glutamate receptor 6) (GluR-6) (GluR6) (Excitatory amino acid receptor 4) (EAA4). | |||||
|
GRIK3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.062153 (rank : 125) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q13003, Q13004, Q16136 | Gene names | GRIK3, GLUR7 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 3 precursor (Glutamate receptor 7) (GluR-7) (GluR7) (Excitatory amino acid receptor 5) (EAA5). | |||||
|
GRIK5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.057909 (rank : 128) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q16478 | Gene names | GRIK5, GRIK2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 5 precursor (Glutamate receptor KA-2) (KA2) (Excitatory amino acid receptor 2) (EAA2). | |||||
|
GRIK5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.057984 (rank : 127) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61626 | Gene names | Grik5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 5 precursor (Glutamate receptor KA-2) (KA2) (Glutamate receptor gamma-2) (GluR gamma-2). | |||||
|
GUC2C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.057227 (rank : 129) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P25092 | Gene names | GUCY2C, GUC2C, STAR | |||
|
Domain Architecture |

|
|||||
| Description | Heat-stable enterotoxin receptor precursor (GC-C) (Intestinal guanylate cyclase) (EC 4.6.1.2) (STA receptor) (hSTAR). | |||||
|
GUC2D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.065952 (rank : 121) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q02846, Q6LEA7 | Gene names | GUCY2D, CORD6, GUC1A4, GUC2D, RETGC, RETGC1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinal guanylyl cyclase 1 precursor (EC 4.6.1.2) (Guanylate cyclase 2D, retinal) (RETGC-1) (Rod outer segment membrane guanylate cyclase) (ROS-GC). | |||||
|
GUC2E_MOUSE
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.066121 (rank : 120) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P52785 | Gene names | Gucy2e, Guc2e | |||
|
Domain Architecture |

|
|||||
| Description | Guanylyl cyclase GC-E precursor (EC 4.6.1.2) (Guanylate cyclase 2E). | |||||
|
K0247_HUMAN
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.080914 (rank : 101) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q92537 | Gene names | KIAA0247 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0247 precursor. | |||||
|
MASP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.050486 (rank : 142) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O00187, O75754, Q5TEQ5, Q5TER0, Q96QG4, Q9BZH0, Q9H498, Q9H499, Q9UBP3, Q9ULC7, Q9UMV3, Q9Y270 | Gene names | MASP2 | |||
|
Domain Architecture |

|
|||||
| Description | Mannan-binding lectin serine protease 2 precursor (EC 3.4.21.104) (Mannose-binding protein-associated serine protease 2) (MASP-2) (MBL- associated serine protease 2) [Contains: Mannan-binding lectin serine protease 2 A chain; Mannan-binding lectin serine protease 2 B chain]. | |||||
|
MASP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.056380 (rank : 132) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q91WP0, Q9QXA4, Q9QXD2, Q9QXD5, Q9Z338 | Gene names | Masp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mannan-binding lectin serine protease 2 precursor (EC 3.4.21.104) (Mannose-binding protein-associated serine protease 2) (MASP-2) (MBL- associated serine protease 2) [Contains: Mannan-binding lectin serine protease 2 A chain; Mannan-binding lectin serine protease 2 B chain]. | |||||
|
NMD3A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.051155 (rank : 139) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8TCU5, Q8TF29, Q8WXI6 | Gene names | GRIN3A, KIAA1973 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit 3A precursor (N-methyl-D-aspartate receptor subtype NR3A) (NMDAR-L). | |||||
|
NMDE2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.054574 (rank : 133) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13224, Q12919, Q13220, Q13225, Q9UM56 | Gene names | GRIN2B | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 2 precursor (N-methyl D- aspartate receptor subtype 2B) (NR2B) (NMDAR2B) (N-methyl-D-aspartate receptor subunit 3) (NR3) (hNR3). | |||||
|
NMDE3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.052940 (rank : 137) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14957 | Gene names | GRIN2C | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 3 precursor (N-methyl D- aspartate receptor subtype 2C) (NR2C) (NMDAR2C). | |||||
|
NMDE3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.053306 (rank : 136) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q01098 | Gene names | Grin2c | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 3 precursor (N-methyl D- aspartate receptor subtype 2C) (NR2C) (NMDAR2C). | |||||
|
NMDZ1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.129999 (rank : 84) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q05586, P35437, Q12867, Q12868 | Gene names | GRIN1, NMDAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit zeta 1 precursor (N-methyl-D- aspartate receptor subunit NR1). | |||||
|
NMDZ1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.129583 (rank : 85) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P35438, Q8CFS4 | Gene names | Grin1, Glurz1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit zeta 1 precursor (N-methyl-D- aspartate receptor subunit NR1). | |||||
|
PAP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.130523 (rank : 83) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9BXP8, Q96PH7, Q96PH8, Q9H4C9 | Gene names | PAPPA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pappalysin-2 precursor (EC 3.4.24.-) (Pregnancy-associated plasma protein-A2) (PAPP-A2) (Pregnancy-associated plasma protein-E1) (PAPP- E). | |||||
|
PAPPA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.150800 (rank : 77) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q13219, Q08371, Q68G52, Q9UDK7 | Gene names | PAPPA | |||
|
Domain Architecture |

|
|||||
| Description | Pappalysin-1 precursor (EC 3.4.24.79) (Pregnancy-associated plasma protein-A) (PAPP-A) (Insulin-like growth factor-dependent IGF-binding protein 4 protease) (IGF-dependent IGFBP-4 protease) (IGFBP-4ase). | |||||
|
PAPPA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.149645 (rank : 78) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8R4K8, Q80VW3, Q80YY1, Q8K423, Q8R4K7, Q9ES06, Q9JK57 | Gene names | Pappa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pappalysin-1 precursor (EC 3.4.24.79) (Pregnancy-associated plasma protein-A) (PAPP-A) (Insulin-like growth factor-dependent IGF-binding protein 4 protease) (IGF-dependent IGFBP-4 protease) (IGFBP-4ase). | |||||
|
PERT_MOUSE
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.051287 (rank : 138) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P35419, Q8C8B1 | Gene names | Tpo | |||
|
Domain Architecture |
|
|||||
| Description | Thyroid peroxidase precursor (EC 1.11.1.8) (TPO). | |||||
|
SUSD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.080039 (rank : 102) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6UWL2, Q5T8V7, Q6P9G7, Q8WU83, Q9H6V2, Q9NTA7 | Gene names | SUSD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 1 precursor. | |||||
|
SUSD3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.068447 (rank : 117) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96L08, Q49AA6, Q6UXV7 | Gene names | SUSD3, UNQ9387/PRO34275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 3. | |||||
|
SUSD3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.116555 (rank : 89) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9D176, Q80XL5, Q9CYV1, Q9DA86 | Gene names | Susd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 3. | |||||
|
V2R1B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.343354 (rank : 31) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6TAC4, O70409 | Gene names | V2r1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vomeronasal type-2 receptor 1b precursor (Putative pheromone receptor V2R1b). | |||||
|
GABR1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9UBS5, O95375, O95468, O95975, O96022, Q86W60, Q9UQQ0 | Gene names | GABBR1, GPRC3A | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
GABR1_MOUSE
|
||||||
| NC score | 0.999599 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | Q9WV18, Q9WU48, Q9WV15, Q9WV16, Q9WV17 | Gene names | Gabbr1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
GABR2_HUMAN
|
||||||
| NC score | 0.841848 (rank : 3) | θ value | 1.02419e-128 (rank : 3) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O75899, O75974, O75975, Q5VXZ2, Q8WX04, Q9P1R2, Q9UNR1, Q9UNS9 | Gene names | GABBR2, GPR51, GPRC3B | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 2 precursor (GABA-B receptor 2) (GABA-B-R2) (Gb2) (GABABR2) (G-protein coupled receptor 51) (HG20). | |||||
|
GP156_HUMAN
|
||||||
| NC score | 0.446619 (rank : 4) | θ value | 2.20605e-14 (rank : 4) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8NFN8, Q86SN6 | Gene names | GPR156, GABABL, PGR28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 156 (GABAB-related G-protein coupled receptor) (G-protein coupled receptor PGR28). | |||||
|
TS1R3_HUMAN
|
||||||
| NC score | 0.438617 (rank : 5) | θ value | 1.38499e-08 (rank : 9) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q7RTX0, Q5TA49, Q8NGW9 | Gene names | TAS1R3, T1R3, TR3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 3 precursor (Sweet taste receptor T1R3). | |||||
|
TS1R1_HUMAN
|
||||||
| NC score | 0.431342 (rank : 6) | θ value | 1.53129e-07 (rank : 14) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q7RTX1, Q8NGZ7, Q8TDJ7, Q8TDJ8, Q8TDJ9, Q8TDK0 | Gene names | TAS1R1, GPR70, T1R1, TR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 1 precursor (G-protein coupled receptor 70) (Gm148). | |||||
|
MGR4_HUMAN
|
||||||
| NC score | 0.430625 (rank : 7) | θ value | 1.80886e-08 (rank : 10) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q14833, Q5SZ84 | Gene names | GRM4, GPRC1D, MGLUR4 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 4 precursor (mGluR4). | |||||
|
MGR8_MOUSE
|
||||||
| NC score | 0.427172 (rank : 8) | θ value | 6.21693e-09 (rank : 7) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P47743, Q6B964 | Gene names | Grm8, Gprc1h, Mglur8 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 8 precursor (mGluR8). | |||||
|
TS1R3_MOUSE
|
||||||
| NC score | 0.426901 (rank : 9) | θ value | 1.09739e-05 (rank : 29) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q925D8, Q91VA4, Q923K0, Q925A4, Q925D9 | Gene names | Tas1r3, Sac, T1r3, Tr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 3 precursor (Sweet taste receptor T1R3) (Saccharin preference protein). | |||||
|
MGR8_HUMAN
|
||||||
| NC score | 0.426484 (rank : 10) | θ value | 8.11959e-09 (rank : 8) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O00222, O15493, O95945, O95946, Q3MIV9, Q52M02, Q6J165 | Gene names | GRM8, GPRC1H, MGLUR8 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 8 precursor (mGluR8). | |||||
|
MGR7_MOUSE
|
||||||
| NC score | 0.425845 (rank : 11) | θ value | 1.53129e-07 (rank : 13) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q68ED2 | Gene names | Grm7, Gprc1g, Mglur7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metabotropic glutamate receptor 7 precursor (mGluR7). | |||||
|
MGR7_HUMAN
|
||||||
| NC score | 0.425525 (rank : 12) | θ value | 1.53129e-07 (rank : 12) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q14831, Q8NFS2, Q8NFS3, Q8NFS4 | Gene names | GRM7, GPRC1G, MGLUR7 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 7 precursor (mGluR7). | |||||
|
V2RX_MOUSE
|
||||||
| NC score | 0.414804 (rank : 13) | θ value | 4.92598e-06 (rank : 20) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O70410 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vomeronasal type-2 receptor X precursor (Putative pheromone receptor V2RX). | |||||
|
TS1R2_MOUSE
|
||||||
| NC score | 0.414671 (rank : 14) | θ value | 9.29e-05 (rank : 36) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q925I4, Q923J8 | Gene names | Tas1r2, Gpr71, T1r2, Tr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 2 precursor (G-protein coupled receptor 71) (Sweet taste receptor T1R2). | |||||
|
MGR6_HUMAN
|
||||||
| NC score | 0.414452 (rank : 15) | θ value | 6.43352e-06 (rank : 22) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O15303 | Gene names | GRM6, GPRC1F, MGLUR6 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 6 precursor (mGluR6). | |||||
|
CASR_HUMAN
|
||||||
| NC score | 0.412987 (rank : 16) | θ value | 4.92598e-06 (rank : 19) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P41180, Q13912, Q16108, Q16109, Q16110, Q16379, Q4PJ19 | Gene names | CASR, GPRC2A, PCAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular calcium-sensing receptor precursor (CaSR) (Parathyroid Cell calcium-sensing receptor). | |||||
|
MGR6_MOUSE
|
||||||
| NC score | 0.412963 (rank : 17) | θ value | 3.19293e-05 (rank : 33) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q5NCH9 | Gene names | Grm6, Gprc1f, Mglur6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metabotropic glutamate receptor 6 precursor (mGluR6). | |||||
|
TS1R1_MOUSE
|
||||||
| NC score | 0.411575 (rank : 18) | θ value | 6.43352e-06 (rank : 23) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99PG6, Q6NS58, Q923J9, Q925I5, Q99PG5 | Gene names | Tas1r1, Gpr70, T1r1, Tr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 1 precursor (G-protein coupled receptor 70). | |||||
|
CASR_MOUSE
|
||||||
| NC score | 0.411286 (rank : 19) | θ value | 1.09739e-05 (rank : 26) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9QY96, O08968, O88519, Q9QY95, Q9QZU8, Q9R1D6, Q9R1Y2 | Gene names | Casr, Gprc2a | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular calcium-sensing receptor precursor (CaSR) (Parathyroid Cell calcium-sensing receptor). | |||||
|
MGR5_HUMAN
|
||||||
| NC score | 0.410516 (rank : 20) | θ value | 0.000158464 (rank : 38) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P41594 | Gene names | GRM5, GPRC1E, MGLUR5 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 5 precursor (mGluR5). | |||||
|
MGR1_HUMAN
|
||||||
| NC score | 0.409002 (rank : 21) | θ value | 1.09739e-05 (rank : 28) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q13255, Q13256, Q14757, Q14758, Q5VTF7, Q5VTF8, Q9NU10, Q9UGS9, Q9UGT0 | Gene names | GRM1, GPRC1A, MGLUR1 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 1 precursor (mGluR1). | |||||
|
MGR1_MOUSE
|
||||||
| NC score | 0.408627 (rank : 22) | θ value | 1.43324e-05 (rank : 31) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P97772, Q6AXG4, Q9EPV6 | Gene names | Grm1, Gprc1a, Mglur1 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 1 precursor (mGluR1). | |||||
|
GPC6A_MOUSE
|
||||||
| NC score | 0.408472 (rank : 23) | θ value | 1.43324e-05 (rank : 30) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8K4Z6, Q5DK51, Q5DK52 | Gene names | Gprc6a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor family C group 6 member A precursor. | |||||
|
TS1R2_HUMAN
|
||||||
| NC score | 0.407079 (rank : 24) | θ value | 0.00020696 (rank : 39) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8TE23, Q5TZ19 | Gene names | TAS1R2, GPR71, T1R2, TR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 2 precursor (G-protein coupled receptor 71) (Sweet taste receptor T1R2). | |||||
|
GPC6A_HUMAN
|
||||||
| NC score | 0.402170 (rank : 25) | θ value | 0.000270298 (rank : 42) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5T6X5, Q6JK43, Q6JK44, Q8NGU8, Q8NHZ9, Q8TDT6 | Gene names | GPRC6A, GPCR33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor family C group 6 member A precursor (hGPRC6A) (G-protein coupled receptor 33) (hGPCR33). | |||||
|
MGR3_MOUSE
|
||||||
| NC score | 0.401067 (rank : 26) | θ value | 7.1131e-05 (rank : 34) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9QYS2 | Gene names | Grm3, Gprc1c, Mglur3 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 3 precursor (mGluR3). | |||||
|
MGR2_HUMAN
|
||||||
| NC score | 0.398100 (rank : 27) | θ value | 0.000786445 (rank : 46) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q14416, Q52MC6, Q9H3N6 | Gene names | GRM2, GPRC1B, MGLUR2 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 2 precursor (mGluR2). | |||||
|
MGR3_HUMAN
|
||||||
| NC score | 0.395505 (rank : 28) | θ value | 0.000786445 (rank : 47) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q14832, Q75MV4, Q75N17, Q86YG6, Q8TBH9 | Gene names | GRM3, GPRC1C, MGLUR3 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 3 precursor (mGluR3). | |||||
|
GP156_MOUSE
|
||||||
| NC score | 0.393056 (rank : 29) | θ value | 1.99992e-07 (rank : 15) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6PCP7, Q8K452 | Gene names | Gpr156, Gababl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 156 (GABAB-related G-protein coupled receptor). | |||||
|
DAF_HUMAN
|
||||||
| NC score | 0.354203 (rank : 30) | θ value | 4.45536e-07 (rank : 16) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P08174, P09679, P78361 | Gene names | CD55, CR, DAF | |||
|
Domain Architecture |

|
|||||
| Description | Complement decay-accelerating factor precursor (CD55 antigen). | |||||
|
V2R1B_MOUSE
|
||||||
| NC score | 0.343354 (rank : 31) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6TAC4, O70409 | Gene names | V2r1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vomeronasal type-2 receptor 1b precursor (Putative pheromone receptor V2R1b). | |||||
|
ANPRC_MOUSE
|
||||||
| NC score | 0.343154 (rank : 32) | θ value | 1.09739e-05 (rank : 25) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P70180, P97804, Q9R025, Q9R027, Q9R028 | Gene names | Npr3 | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide clearance receptor precursor (ANP-C) (ANPRC) (NPR-C) (Atrial natriuretic peptide C-type receptor) (EF-2). | |||||
|
ANPRC_HUMAN
|
||||||
| NC score | 0.337098 (rank : 33) | θ value | 1.09739e-05 (rank : 24) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P17342 | Gene names | NPR3, ANPRC | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide clearance receptor precursor (ANP-C) (ANPRC) (NPR-C) (Atrial natriuretic peptide C-type receptor). | |||||
|
CR1_HUMAN
|
||||||
| NC score | 0.328548 (rank : 34) | θ value | 3.19293e-05 (rank : 32) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P17927, Q16745, Q9UQV2 | Gene names | CR1, C3BR | |||
|
Domain Architecture |

|
|||||
| Description | Complement receptor type 1 precursor (C3b/C4b receptor) (CD35 antigen). | |||||
|
C4BB_HUMAN
|
||||||
| NC score | 0.324147 (rank : 35) | θ value | 0.0148317 (rank : 59) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P20851 | Gene names | C4BPB | |||
|
Domain Architecture |

|
|||||
| Description | C4b-binding protein beta chain precursor. | |||||
|
CRRY_MOUSE
|
||||||
| NC score | 0.323227 (rank : 36) | θ value | 0.00298849 (rank : 51) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q64735, Q3TV30, Q58E68, Q61447, Q61449, Q8CF59, Q8K328 | Gene names | Crry, Cry | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement regulatory protein Crry precursor (Protein p65). | |||||
|
CR2_MOUSE
|
||||||
| NC score | 0.317675 (rank : 37) | θ value | 0.00134147 (rank : 50) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P19070 | Gene names | Cr2 | |||
|
Domain Architecture |

|
|||||
| Description | Complement receptor type 2 precursor (Cr2) (Complement C3d receptor) (CD21 antigen). | |||||
|
CR2_HUMAN
|
||||||
| NC score | 0.317294 (rank : 38) | θ value | 0.00102713 (rank : 48) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P20023, Q13866, Q14212 | Gene names | CR2, C3DR | |||
|
Domain Architecture |

|
|||||
| Description | Complement receptor type 2 precursor (Cr2) (Complement C3d receptor) (Epstein-Barr virus receptor) (EBV receptor) (CD21 antigen). | |||||
|
ZP3R_MOUSE
|
||||||
| NC score | 0.312720 (rank : 39) | θ value | 0.00298849 (rank : 54) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q60736 | Gene names | Zp3r, Sp56 | |||
|
Domain Architecture |

|
|||||
| Description | Zona pellucida sperm-binding protein 3 receptor precursor (Sperm fertilization protein 56) (sp56). | |||||
|
DAF1_MOUSE
|
||||||
| NC score | 0.310954 (rank : 40) | θ value | 0.000602161 (rank : 45) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q61475, P97732, Q61397 | Gene names | Cd55a, Daf1 | |||
|
Domain Architecture |

|
|||||
| Description | Complement decay-accelerating factor, GPI-anchored precursor (DAF-GPI) (CD55 antigen). | |||||
|
F13B_MOUSE
|
||||||
| NC score | 0.306724 (rank : 41) | θ value | 0.00102713 (rank : 49) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q07968 | Gene names | F13b, Cf13b | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor XIII B chain precursor (Protein-glutamine gamma- glutamyltransferase B chain) (Transglutaminase B chain). | |||||
|
MCP_MOUSE
|
||||||
| NC score | 0.306280 (rank : 42) | θ value | 0.0148317 (rank : 60) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O88174, Q9R0R9 | Gene names | Cd46, Mcp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane cofactor protein precursor (CD46 antigen). | |||||
|
CFAH_HUMAN
|
||||||
| NC score | 0.304511 (rank : 43) | θ value | 0.000270298 (rank : 40) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P08603, P78435, Q14570, Q38G77, Q5TFM3, Q8N708, Q9NU86 | Gene names | CFH, HF, HF1 | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor H precursor (H factor 1). | |||||
|
C4BP_MOUSE
|
||||||
| NC score | 0.302951 (rank : 44) | θ value | 0.0961366 (rank : 72) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P08607 | Gene names | C4bpa, C4bp | |||
|
Domain Architecture |

|
|||||
| Description | C4b-binding protein precursor (C4bp). | |||||
|
MCP_HUMAN
|
||||||
| NC score | 0.301708 (rank : 45) | θ value | 0.0961366 (rank : 73) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P15529, Q15429, Q53GV9, Q5HY94, Q5VWS6, Q5VWS7, Q5VWS8, Q5VWS9, Q5VWT0, Q5VWT1, Q5VWT2, Q6N0A1, Q7Z3R5, Q9NNW2, Q9NNW3, Q9NNW4 | Gene names | CD46, MCP | |||
|
Domain Architecture |

|
|||||
| Description | Membrane cofactor protein precursor (Trophoblast leukocyte common antigen) (TLX) (CD46 antigen). | |||||
|
DAF2_MOUSE
|
||||||
| NC score | 0.299724 (rank : 46) | θ value | 0.0252991 (rank : 61) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q61476 | Gene names | Cd55b, Daf2 | |||
|
Domain Architecture |

|
|||||
| Description | Complement decay-accelerating factor transmembrane isoform precursor (DAF-TM) (CD55 antigen). | |||||
|
C4BP_HUMAN
|
||||||
| NC score | 0.295637 (rank : 47) | θ value | 0.21417 (rank : 77) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P04003 | Gene names | C4BPA, C4BP | |||
|
Domain Architecture |

|
|||||
| Description | C4b-binding protein alpha chain precursor (C4bp) (Proline-rich protein) (PRP). | |||||
|
FHR5_HUMAN
|
||||||
| NC score | 0.293859 (rank : 48) | θ value | 0.00390308 (rank : 55) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9BXR6 | Gene names | CFHL5, FHR5 | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor H-related protein 5 precursor (FHR-5). | |||||
|
FHR1_HUMAN
|
||||||
| NC score | 0.292516 (rank : 49) | θ value | 0.000270298 (rank : 41) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q03591, Q9UJ17 | Gene names | CFHL1, CFHL, FHR1, HFL1 | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor H-related protein 1 precursor (FHR-1) (H factor-like protein 1) (H-factor-like 1) (H36). | |||||
|
F13B_HUMAN
|
||||||
| NC score | 0.292252 (rank : 50) | θ value | 0.0563607 (rank : 68) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P05160 | Gene names | F13B | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor XIII B chain precursor (Protein-glutamine gamma- glutamyltransferase B chain) (Transglutaminase B chain) (Fibrin- stabilizing factor B subunit). | |||||
|
APOH_HUMAN
|
||||||
| NC score | 0.292136 (rank : 51) | θ value | 0.125558 (rank : 74) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P02749, Q9UCN7 | Gene names | APOH, B2G1 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-2-glycoprotein 1 precursor (Beta-2-glycoprotein I) (Apolipoprotein H) (Apo-H) (B2GPI) (Beta(2)GPI) (Activated protein C- binding protein) (APC inhibitor) (Anticardiolipin cofactor). | |||||
|
CFAH_MOUSE
|
||||||
| NC score | 0.287860 (rank : 52) | θ value | 0.0563607 (rank : 66) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P06909 | Gene names | Cfh, Hf1 | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor H precursor (Protein beta-1-H). | |||||
|
APOH_MOUSE
|
||||||
| NC score | 0.275144 (rank : 53) | θ value | 3.0926 (rank : 101) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q01339 | Gene names | Apoh, B2gp1 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-2-glycoprotein 1 precursor (Beta-2-glycoprotein I) (Apolipoprotein H) (Apo-H) (B2GPI) (Beta(2)GPI) (Activated protein C- binding protein) (APC inhibitor). | |||||
|
SUSD4_HUMAN
|
||||||
| NC score | 0.272506 (rank : 54) | θ value | 0.0736092 (rank : 71) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q5VX71, Q6UX62, Q9BSR0, Q9NWG0 | Gene names | SUSD4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 4 precursor. | |||||
|
LYAM3_HUMAN
|
||||||
| NC score | 0.269600 (rank : 55) | θ value | 0.00035302 (rank : 44) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P16109, Q8IVD1 | Gene names | SELP, GMRP | |||
|
Domain Architecture |

|
|||||
| Description | P-selectin precursor (Granule membrane protein 140) (GMP-140) (PADGEM) (Leukocyte-endothelial cell adhesion molecule 3) (LECAM3) (CD62P antigen). | |||||
|
LYAM3_MOUSE
|
||||||
| NC score | 0.269442 (rank : 56) | θ value | 9.29e-05 (rank : 35) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q01102 | Gene names | Selp, Grmp | |||
|
Domain Architecture |

|
|||||
| Description | P-selectin precursor (Granule membrane protein 140) (GMP-140) (PADGEM) (Leukocyte-endothelial cell adhesion molecule 3) (LECAM3) (CD62P antigen). | |||||
|
SUSD4_MOUSE
|
||||||
| NC score | 0.266962 (rank : 57) | θ value | 0.279714 (rank : 82) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8BH32, Q8VC43 | Gene names | Susd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 4 precursor. | |||||
|
LYAM2_MOUSE
|
||||||
| NC score | 0.266742 (rank : 58) | θ value | 1.09739e-05 (rank : 27) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q00690 | Gene names | Sele, Elam-1 | |||
|
Domain Architecture |

|
|||||
| Description | E-selectin precursor (Endothelial leukocyte adhesion molecule 1) (ELAM-1) (Leukocyte-endothelial cell adhesion molecule 2) (LECAM2) (CD62E antigen). | |||||
|
CSMD3_HUMAN
|
||||||
| NC score | 0.266177 (rank : 59) | θ value | 2.21117e-06 (rank : 18) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q7Z407, Q96PZ3 | Gene names | CSMD3, KIAA1894 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 3 precursor (CUB and sushi multiple domains protein 3). | |||||
|
CSMD1_HUMAN
|
||||||
| NC score | 0.260964 (rank : 60) | θ value | 6.43352e-06 (rank : 21) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q96PZ7, Q96QU9, Q96RM4 | Gene names | CSMD1, KIAA1890 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 1 precursor (CUB and sushi multiple domains protein 1). | |||||
|
FHR3_HUMAN
|
||||||
| NC score | 0.260557 (rank : 61) | θ value | 0.0330416 (rank : 64) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q02985, Q9UJ16 | Gene names | CFHL3, FHR3 | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor H-related protein 3 precursor (FHR-3) (H factor-like protein 3) (DOWN16). | |||||
|
LYAM2_HUMAN
|
||||||
| NC score | 0.253976 (rank : 62) | θ value | 0.00298849 (rank : 52) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P16581, P16111 | Gene names | SELE, ELAM1 | |||
|
Domain Architecture |

|
|||||
| Description | E-selectin precursor (Endothelial leukocyte adhesion molecule 1) (ELAM-1) (Leukocyte-endothelial cell adhesion molecule 2) (LECAM2) (CD62E antigen). | |||||
|
CSMD2_HUMAN
|
||||||
| NC score | 0.252574 (rank : 63) | θ value | 0.000121331 (rank : 37) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q7Z408, Q5VT59, Q8N963, Q96Q03, Q9H4V7, Q9H4V8, Q9H4V9, Q9H4W0, Q9H4W1, Q9H4W2, Q9H4W3, Q9H4W4, Q9HCY5, Q9HCY6, Q9HCY7 | Gene names | CSMD2, KIAA1884 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 2 (CUB and sushi multiple domains protein 2). | |||||
|
CSMD1_MOUSE
|
||||||
| NC score | 0.252453 (rank : 64) | θ value | 0.00035302 (rank : 43) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q923L3, Q8BUV1, Q8BYQ3 | Gene names | Csmd1 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 1 precursor (CUB and sushi multiple domains protein 1). | |||||
|
FHR4_HUMAN
|
||||||
| NC score | 0.246503 (rank : 65) | θ value | 0.279714 (rank : 80) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q92496, Q9UJY6 | Gene names | CFHL4, FHR4 | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor H-related protein 4 precursor (FHR-4). | |||||
|
FHR2_HUMAN
|
||||||
| NC score | 0.244172 (rank : 66) | θ value | 0.813845 (rank : 84) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P36980, Q14310 | Gene names | CFHL2, FHR2, HFL3 | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor H-related protein 2 precursor (FHR-2) (H factor-like protein 2) (H factor-like 3) (DDESK59). | |||||
|
SRPX_MOUSE
|
||||||
| NC score | 0.243520 (rank : 67) | θ value | 1.38821 (rank : 93) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9R0M3, Q9R0M2 | Gene names | Srpx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi-repeat-containing protein SRPX precursor (DRS protein). | |||||
|
SUSD2_HUMAN
|
||||||
| NC score | 0.238124 (rank : 68) | θ value | 0.00298849 (rank : 53) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9UGT4, Q9H5Y6 | Gene names | SUSD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 2 precursor. | |||||
|
SEZ6L_HUMAN
|
||||||
| NC score | 0.237703 (rank : 69) | θ value | 0.0736092 (rank : 70) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9BYH1, O95917, Q5THY5, Q6IBZ4, Q6UXD4, Q9NUI3, Q9NUI4, Q9NUI5, Q9Y2E1, Q9Y3J6 | Gene names | SEZ6L, KIAA0927 | |||
|
Domain Architecture |

|
|||||
| Description | Seizure 6-like protein precursor. | |||||
|
SRPX_HUMAN
|
||||||
| NC score | 0.237504 (rank : 70) | θ value | 1.81305 (rank : 97) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P78539, Q99652, Q99913 | Gene names | SRPX, ETX1 | |||
|
Domain Architecture |

|
|||||
| Description | Sushi repeat-containing protein SRPX precursor. | |||||
|
SUSD2_MOUSE
|
||||||
| NC score | 0.233876 (rank : 71) | θ value | 0.00869519 (rank : 58) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9DBX3, Q8BY19, Q8BYN0, Q8BYZ3, Q8BYZ9, Q8BZQ4, Q8C179 | Gene names | Susd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 2 precursor. | |||||
|
LYAM1_MOUSE
|
||||||
| NC score | 0.196092 (rank : 72) | θ value | 0.125558 (rank : 75) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P18337 | Gene names | Sell, Lnhr, Ly-22 | |||
|
Domain Architecture |

|
|||||
| Description | L-selectin precursor (Lymph node homing receptor) (Leukocyte adhesion molecule 1) (LAM-1) (Lymphocyte antigen 22) (Ly-22) (Lymphocyte surface MEL-14 antigen) (Leukocyte-endothelial cell adhesion molecule 1) (LECAM1) (CD62L antigen). | |||||
|
LYAM1_HUMAN
|
||||||
| NC score | 0.189960 (rank : 73) | θ value | 1.38821 (rank : 92) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P14151, P15023 | Gene names | SELL, LNHR, LYAM1 | |||
|
Domain Architecture |

|
|||||
| Description | L-selectin precursor (Lymph node homing receptor) (Leukocyte adhesion molecule 1) (LAM-1) (Leukocyte surface antigen Leu-8) (TQ1) (gp90-MEL) (Leukocyte-endothelial cell adhesion molecule 1) (LECAM1) (CD62L antigen). | |||||
|
CO2_HUMAN
|
||||||
| NC score | 0.165973 (rank : 74) | θ value | 0.0563607 (rank : 67) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P06681, O19694, Q13904 | Gene names | C2 | |||
|
Domain Architecture |

|
|||||
| Description | Complement C2 precursor (EC 3.4.21.43) (C3/C5 convertase) [Contains: Complement C2b fragment; Complement C2a fragment]. | |||||
|
CFAB_HUMAN
|
||||||
| NC score | 0.161694 (rank : 75) | θ value | 0.21417 (rank : 78) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P00751, O15006, Q29944, Q96HX6, Q9BTF5, Q9BX92 | Gene names | CFB, BF | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor B precursor (EC 3.4.21.47) (C3/C5 convertase) (Properdin factor B) (Glycine-rich beta glycoprotein) (GBG) (PBF2) [Contains: Complement factor B Ba fragment; Complement factor B Bb fragment]. | |||||
|
CFAB_MOUSE
|
||||||
| NC score | 0.155331 (rank : 76) | θ value | 0.813845 (rank : 83) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P04186 | Gene names | Cfb, Bf, H2-Bf | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor B precursor (EC 3.4.21.47) (C3/C5 convertase) [Contains: Complement factor B Ba fragment; Complement factor B Bb fragment]. | |||||
|
PAPPA_HUMAN
|
||||||
| NC score | 0.150800 (rank : 77) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q13219, Q08371, Q68G52, Q9UDK7 | Gene names | PAPPA | |||
|
Domain Architecture |

|
|||||
| Description | Pappalysin-1 precursor (EC 3.4.24.79) (Pregnancy-associated plasma protein-A) (PAPP-A) (Insulin-like growth factor-dependent IGF-binding protein 4 protease) (IGF-dependent IGFBP-4 protease) (IGFBP-4ase). | |||||
|
PAPPA_MOUSE
|
||||||
| NC score | 0.149645 (rank : 78) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8R4K8, Q80VW3, Q80YY1, Q8K423, Q8R4K7, Q9ES06, Q9JK57 | Gene names | Pappa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pappalysin-1 precursor (EC 3.4.24.79) (Pregnancy-associated plasma protein-A) (PAPP-A) (Insulin-like growth factor-dependent IGF-binding protein 4 protease) (IGF-dependent IGFBP-4 protease) (IGFBP-4ase). | |||||
|
ANPRB_MOUSE
|
||||||
| NC score | 0.143530 (rank : 79) | θ value | 2.28291e-11 (rank : 5) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6VVW5, Q6VVW3, Q6VVW4, Q8CGA9 | Gene names | Npr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Atrial natriuretic peptide receptor B precursor (ANP-B) (ANPRB) (GC-B) (Guanylate cyclase B) (EC 4.6.1.2) (NPR-B) (Atrial natriuretic peptide B-type receptor). | |||||
|
CO2_MOUSE
|
||||||
| NC score | 0.143267 (rank : 80) | θ value | 0.0736092 (rank : 69) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P21180 | Gene names | C2 | |||
|
Domain Architecture |

|
|||||
| Description | Complement C2 precursor (EC 3.4.21.43) (C3/C5 convertase) [Contains: Complement C2b fragment; Complement C2a fragment]. | |||||
|
ANPRB_HUMAN
|
||||||
| NC score | 0.136526 (rank : 81) | θ value | 1.47974e-10 (rank : 6) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P20594, O60871, Q9UQ50 | Gene names | NPR2, ANPRB | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide receptor B precursor (ANP-B) (ANPRB) (GC-B) (Guanylate cyclase B) (EC 4.6.1.2) (NPR-B) (Atrial natriuretic peptide B-type receptor). | |||||
|
CO6_HUMAN
|
||||||
| NC score | 0.132321 (rank : 82) | θ value | 0.0431538 (rank : 65) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P13671 | Gene names | C6 | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C6 precursor. | |||||
|
PAP2_HUMAN
|
||||||
| NC score | 0.130523 (rank : 83) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9BXP8, Q96PH7, Q96PH8, Q9H4C9 | Gene names | PAPPA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pappalysin-2 precursor (EC 3.4.24.-) (Pregnancy-associated plasma protein-A2) (PAPP-A2) (Pregnancy-associated plasma protein-E1) (PAPP- E). | |||||
|
NMDZ1_HUMAN
|
||||||
| NC score | 0.129999 (rank : 84) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q05586, P35437, Q12867, Q12868 | Gene names | GRIN1, NMDAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit zeta 1 precursor (N-methyl-D- aspartate receptor subunit NR1). | |||||
|
NMDZ1_MOUSE
|
||||||
| NC score | 0.129583 (rank : 85) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P35438, Q8CFS4 | Gene names | Grin1, Glurz1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit zeta 1 precursor (N-methyl-D- aspartate receptor subunit NR1). | |||||
|
ANPRA_MOUSE
|
||||||
| NC score | 0.127195 (rank : 86) | θ value | 1.53129e-07 (rank : 11) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 600 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P18293 | Gene names | Npr1, Npra | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide receptor A precursor (ANP-A) (ANPRA) (GC-A) (Guanylate cyclase) (EC 4.6.1.2) (NPR-A) (Atrial natriuretic peptide A-type receptor). | |||||
|
ANPRA_HUMAN
|
||||||
| NC score | 0.125362 (rank : 87) | θ value | 5.81887e-07 (rank : 17) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 612 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P16066, Q6P4Q3 | Gene names | NPR1, ANPRA | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide receptor A precursor (ANP-A) (ANPRA) (GC-A) (Guanylate cyclase) (EC 4.6.1.2) (NPR-A) (Atrial natriuretic peptide A-type receptor). | |||||
|
CO7_HUMAN
|
||||||
| NC score | 0.119791 (rank : 88) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P10643, Q6P3T5, Q92489 | Gene names | C7 | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C7 precursor. | |||||
|
SUSD3_MOUSE
|
||||||
| NC score | 0.116555 (rank : 89) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9D176, Q80XL5, Q9CYV1, Q9DA86 | Gene names | Susd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 3. | |||||
|
GP158_HUMAN
|
||||||
| NC score | 0.116436 (rank : 90) | θ value | 1.06291 (rank : 88) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5T848, Q6QR81, Q9ULT3 | Gene names | GPR158, KIAA1136 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 158 precursor. | |||||
|
GPC5B_MOUSE
|
||||||
| NC score | 0.106990 (rank : 91) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q923Z0, Q8CCV3 | Gene names | Gprc5b, Raig2 | |||
|
Domain Architecture |

|
|||||
| Description | G-protein coupled receptor family C group 5 member B precursor (Retinoic acid-induced gene 2 protein) (RAIG-2). | |||||
|
GPC5B_HUMAN
|
||||||
| NC score | 0.103065 (rank : 92) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NZH0, O75205, Q8NBZ8 | Gene names | GPRC5B, RAIG2 | |||
|
Domain Architecture |

|
|||||
| Description | G-protein coupled receptor family C group 5 member B precursor (Retinoic acid-induced gene 2 protein) (RAIG-2) (A-69G12.1). | |||||
|
GUC2F_HUMAN
|
||||||
| NC score | 0.096785 (rank : 93) | θ value | 0.21417 (rank : 79) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 740 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P51841, Q9UJF1 | Gene names | GUCY2F, GUC2F, RETGC2 | |||
|
Domain Architecture |

|
|||||
| Description | Retinal guanylyl cyclase 2 precursor (EC 4.6.1.2) (Guanylate cyclase 2F, retinal) (RETGC-2) (Rod outer segment membrane guanylate cyclase 2) (ROS-GC2) (Guanylate cyclase F) (GC-F). | |||||
|
GUC2G_MOUSE
|
||||||
| NC score | 0.096647 (rank : 94) | θ value | 5.27518 (rank : 115) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6TL19, Q8BWU7 | Gene names | Gucy2g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanylate cyclase 2G precursor (EC 4.6.1.2) (Guanylyl cyclase receptor G) (mGC-G). | |||||
|
GP158_MOUSE
|
||||||
| NC score | 0.094516 (rank : 95) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8C419, Q8CHB0 | Gene names | Gpr158, Kiaa1136 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 158 precursor. | |||||
|
GRIA2_MOUSE
|
||||||
| NC score | 0.089522 (rank : 96) | θ value | 0.00665767 (rank : 57) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P23819, Q61604, Q61605, Q8BG69, Q8BXU3, Q9D6D3 | Gene names | Gria2, Glur2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 2 precursor (GluR-2) (GluR-B) (GluR-K2) (Glutamate receptor ionotropic, AMPA 2) (AMPA-selective glutamate receptor 2). | |||||
|
GRIA2_HUMAN
|
||||||
| NC score | 0.089487 (rank : 97) | θ value | 0.00665767 (rank : 56) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P42262, Q96FP6 | Gene names | GRIA2, GLUR2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 2 precursor (GluR-2) (GluR-B) (GluR-K2) (Glutamate receptor ionotropic, AMPA 2) (AMPA-selective glutamate receptor 2). | |||||
|
GRIA1_MOUSE
|
||||||
| NC score | 0.086358 (rank : 98) | θ value | 0.0252991 (rank : 63) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P23818 | Gene names | Gria1, Glur1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 1 precursor (GluR-1) (GluR-A) (GluR-K1) (Glutamate receptor ionotropic, AMPA 1) (AMPA-selective glutamate receptor 1). | |||||
|
GRIA1_HUMAN
|
||||||
| NC score | 0.086102 (rank : 99) | θ value | 0.0252991 (rank : 62) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P42261 | Gene names | GRIA1, GLUH1, GLUR1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 1 precursor (GluR-1) (GluR-A) (GluR-K1) (Glutamate receptor ionotropic, AMPA 1) (AMPA-selective glutamate receptor 1). | |||||
|
GCYB2_HUMAN
|
||||||
| NC score | 0.081117 (rank : 100) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75343, Q9NZ64 | Gene names | GUCY1B2 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate cyclase soluble subunit beta-2 (EC 4.6.1.2) (GCS-beta-2). | |||||
|
K0247_HUMAN
|
||||||
| NC score | 0.080914 (rank : 101) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q92537 | Gene names | KIAA0247 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0247 precursor. | |||||
|
SUSD1_HUMAN
|
||||||
| NC score | 0.080039 (rank : 102) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6UWL2, Q5T8V7, Q6P9G7, Q8WU83, Q9H6V2, Q9NTA7 | Gene names | SUSD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 1 precursor. | |||||
|
GP179_HUMAN
|
||||||
| NC score | 0.079502 (rank : 103) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6PRD1 | Gene names | GPR179, GPR158L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 179 precursor (Probable G-protein coupled receptor 158-like 1). | |||||
|
GCYA2_HUMAN
|
||||||
| NC score | 0.074837 (rank : 104) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P33402 | Gene names | GUCY1A2, GUC1A2, GUCSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate cyclase soluble subunit alpha-2 (EC 4.6.1.2) (GCS-alpha-2). | |||||
|
GRIA3_MOUSE
|
||||||
| NC score | 0.074603 (rank : 105) | θ value | 1.38821 (rank : 91) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Z2W9, Q5DTJ0 | Gene names | Gria3, Glur3, Kiaa4184 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate receptor 3 precursor (GluR-3) (GluR-C) (GluR-K3) (Glutamate receptor ionotropic, AMPA 3) (AMPA-selective glutamate receptor 3). | |||||
|
GCYB1_MOUSE
|
||||||
| NC score | 0.074179 (rank : 106) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O54865 | Gene names | Gucy1b3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanylate cyclase soluble subunit beta-1 (EC 4.6.1.2) (GCS-beta-1) (Soluble guanylate cyclase small subunit). | |||||
|
GCYB1_HUMAN
|
||||||
| NC score | 0.074052 (rank : 107) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q02153, Q86WY5 | Gene names | GUCY1B3, GUC1B3, GUCSB3, GUCY1B1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate cyclase soluble subunit beta-1 (EC 4.6.1.2) (GCS-beta-1) (Soluble guanylate cyclase small subunit) (GCS-beta-3). | |||||
|
NMDE4_HUMAN
|
||||||
| NC score | 0.072748 (rank : 108) | θ value | 0.163984 (rank : 76) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O15399 | Gene names | GRIN2D | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 4 precursor (N-methyl D- aspartate receptor subtype 2D) (NR2D) (NMDAR2D) (EB11). | |||||
|
GCYA3_MOUSE
|
||||||
| NC score | 0.072540 (rank : 109) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9ERL9, Q9DBQ3 | Gene names | Gucy1a3, Gucy1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate cyclase soluble subunit alpha-3 (EC 4.6.1.2) (GCS-alpha-3) (Soluble guanylate cyclase large subunit) (GCS-alpha-1). | |||||
|
GCYA3_HUMAN
|
||||||
| NC score | 0.072505 (rank : 110) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q02108, O43843, Q8TAH3 | Gene names | GUCY1A3, GUC1A3, GUCSA3, GUCY1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate cyclase soluble subunit alpha-3 (EC 4.6.1.2) (GCS-alpha-3) (Soluble guanylate cyclase large subunit) (GCS-alpha-1). | |||||
|
GRIA3_HUMAN
|
||||||
| NC score | 0.072167 (rank : 111) | θ value | 3.0926 (rank : 102) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P42263, Q4VXD5, Q4VXD6, Q9HDA0, Q9HDA1, Q9HDA2 | Gene names | GRIA3, GLUR3, GLURC | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 3 precursor (GluR-3) (GluR-C) (GluR-K3) (Glutamate receptor ionotropic, AMPA 3) (AMPA-selective glutamate receptor 3). | |||||
|
GRIK4_HUMAN
|
||||||
| NC score | 0.071071 (rank : 112) | θ value | 1.06291 (rank : 89) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q16099 | Gene names | GRIK4, GRIK | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 4 precursor (Glutamate receptor KA-1) (KA1) (Excitatory amino acid receptor 1) (EAA1). | |||||
|
GRIK2_HUMAN
|
||||||
| NC score | 0.071028 (rank : 113) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13002 | Gene names | GRIK2, GLUR6 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 2 precursor (Glutamate receptor 6) (GluR-6) (GluR6) (Excitatory amino acid receptor 4) (EAA4). | |||||
|
GRIK1_HUMAN
|
||||||
| NC score | 0.070159 (rank : 114) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P39086, Q13001, Q86SU9 | Gene names | GRIK1, GLUR5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 1 precursor (Glutamate receptor 5) (GluR-5) (GluR5) (Excitatory amino acid receptor 3) (EAA3). | |||||
|
NMDE4_MOUSE
|
||||||
| NC score | 0.070125 (rank : 115) | θ value | 0.279714 (rank : 81) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q03391 | Gene names | Grin2d | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 4 precursor (N-methyl D- aspartate receptor subtype 2D) (NR2D) (NMDAR2D). | |||||
|
GRIK1_MOUSE
|
||||||
| NC score | 0.069648 (rank : 116) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q60934 | Gene names | Grik1, Glur5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 1 precursor (Glutamate receptor 5) (GluR-5) (GluR5). | |||||
|
SUSD3_HUMAN
|
||||||
| NC score | 0.068447 (rank : 117) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96L08, Q49AA6, Q6UXV7 | Gene names | SUSD3, UNQ9387/PRO34275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 3. | |||||
|
GRIK4_MOUSE
|
||||||
| NC score | 0.068348 (rank : 118) | θ value | 2.36792 (rank : 99) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BMF5 | Gene names | Grik4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate receptor, ionotropic kainate 4 precursor. | |||||
|
GRID2_HUMAN
|
||||||
| NC score | 0.067449 (rank : 119) | θ value | 1.81305 (rank : 95) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43424 | Gene names | GRID2, GLURD2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-2 subunit precursor (GluR delta-2). | |||||
|
GUC2E_MOUSE
|
||||||
| NC score | 0.066121 (rank : 120) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P52785 | Gene names | Gucy2e, Guc2e | |||
|
Domain Architecture |

|
|||||
| Description | Guanylyl cyclase GC-E precursor (EC 4.6.1.2) (Guanylate cyclase 2E). | |||||
|
GUC2D_HUMAN
|
||||||
| NC score | 0.065952 (rank : 121) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q02846, Q6LEA7 | Gene names | GUCY2D, CORD6, GUC1A4, GUC2D, RETGC, RETGC1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinal guanylyl cyclase 1 precursor (EC 4.6.1.2) (Guanylate cyclase 2D, retinal) (RETGC-1) (Rod outer segment membrane guanylate cyclase) (ROS-GC). | |||||
|
GRID2_MOUSE
|
||||||
| NC score | 0.065810 (rank : 122) | θ value | 3.0926 (rank : 103) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61625 | Gene names | Grid2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-2 subunit precursor (GluR delta-2). | |||||
|
GRIA4_MOUSE
|
||||||
| NC score | 0.065260 (rank : 123) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Z2W8 | Gene names | Gria4, Glur4 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 4 precursor (GluR-4) (GluR4) (GluR-D) (Glutamate receptor ionotropic, AMPA 4) (AMPA-selective glutamate receptor 4). | |||||
|
GRIA4_HUMAN
|
||||||
| NC score | 0.065160 (rank : 124) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P48058 | Gene names | GRIA4, GLUR4 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor 4 precursor (GluR-4) (GluR4) (GluR-D) (Glutamate receptor ionotropic, AMPA 4) (AMPA-selective glutamate receptor 4). | |||||
|
GRIK3_HUMAN
|
||||||
| NC score | 0.062153 (rank : 125) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q13003, Q13004, Q16136 | Gene names | GRIK3, GLUR7 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 3 precursor (Glutamate receptor 7) (GluR-7) (GluR7) (Excitatory amino acid receptor 5) (EAA5). | |||||
|
NMDE2_MOUSE
|
||||||
| NC score | 0.058557 (rank : 126) | θ value | 8.99809 (rank : 139) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q01097, Q9DCB2 | Gene names | Grin2b | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 2 precursor (N-methyl D- aspartate receptor subtype 2B) (NR2B) (NMDAR2B). | |||||
|
GRIK5_MOUSE
|
||||||
| NC score | 0.057984 (rank : 127) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61626 | Gene names | Grik5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 5 precursor (Glutamate receptor KA-2) (KA2) (Glutamate receptor gamma-2) (GluR gamma-2). | |||||
|
GRIK5_HUMAN
|
||||||
| NC score | 0.057909 (rank : 128) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q16478 | Gene names | GRIK5, GRIK2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 5 precursor (Glutamate receptor KA-2) (KA2) (Excitatory amino acid receptor 2) (EAA2). | |||||
|
GUC2C_HUMAN
|
||||||
| NC score | 0.057227 (rank : 129) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P25092 | Gene names | GUCY2C, GUC2C, STAR | |||
|
Domain Architecture |

|
|||||
| Description | Heat-stable enterotoxin receptor precursor (GC-C) (Intestinal guanylate cyclase) (EC 4.6.1.2) (STA receptor) (hSTAR). | |||||
|
GRID1_HUMAN
|
||||||
| NC score | 0.056664 (rank : 130) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9ULK0, Q8IXT3 | Gene names | GRID1, KIAA1220 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-1 subunit precursor (GluR delta-1). | |||||
|
GRID1_MOUSE
|
||||||
| NC score | 0.056655 (rank : 131) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61627 | Gene names | Grid1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor delta-1 subunit precursor (GluR delta-1). | |||||
|
MASP2_MOUSE
|
||||||
| NC score | 0.056380 (rank : 132) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q91WP0, Q9QXA4, Q9QXD2, Q9QXD5, Q9Z338 | Gene names | Masp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mannan-binding lectin serine protease 2 precursor (EC 3.4.21.104) (Mannose-binding protein-associated serine protease 2) (MASP-2) (MBL- associated serine protease 2) [Contains: Mannan-binding lectin serine protease 2 A chain; Mannan-binding lectin serine protease 2 B chain]. | |||||
|
NMDE2_HUMAN
|
||||||
| NC score | 0.054574 (rank : 133) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13224, Q12919, Q13220, Q13225, Q9UM56 | Gene names | GRIN2B | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 2 precursor (N-methyl D- aspartate receptor subtype 2B) (NR2B) (NMDAR2B) (N-methyl-D-aspartate receptor subunit 3) (NR3) (hNR3). | |||||
|
ATF3_HUMAN
|
||||||
| NC score | 0.054351 (rank : 134) | θ value | 1.06291 (rank : 85) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P18847 | Gene names | ATF3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-3 (Activating transcription factor 3). | |||||
|
ATF3_MOUSE
|
||||||
| NC score | 0.054136 (rank : 135) | θ value | 1.06291 (rank : 86) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q60765 | Gene names | Atf3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-3 (Activating transcription factor 3) (Transcription factor LRG-21). | |||||
|
NMDE3_MOUSE
|
||||||
| NC score | 0.053306 (rank : 136) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q01098 | Gene names | Grin2c | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 3 precursor (N-methyl D- aspartate receptor subtype 2C) (NR2C) (NMDAR2C). | |||||
|
NMDE3_HUMAN
|
||||||
| NC score | 0.052940 (rank : 137) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14957 | Gene names | GRIN2C | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 3 precursor (N-methyl D- aspartate receptor subtype 2C) (NR2C) (NMDAR2C). | |||||
|
PERT_MOUSE
|
||||||
| NC score | 0.051287 (rank : 138) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P35419, Q8C8B1 | Gene names | Tpo | |||
|
Domain Architecture |
|
|||||
| Description | Thyroid peroxidase precursor (EC 1.11.1.8) (TPO). | |||||
|
NMD3A_HUMAN
|
||||||
| NC score | 0.051155 (rank : 139) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8TCU5, Q8TF29, Q8WXI6 | Gene names | GRIN3A, KIAA1973 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit 3A precursor (N-methyl-D-aspartate receptor subtype NR3A) (NMDAR-L). | |||||
|
ADCY9_HUMAN
|
||||||
| NC score | 0.051089 (rank : 140) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60503, O60273, Q4ZHT9, Q4ZIR5, Q9BWT4, Q9UGP2 | Gene names | ADCY9, KIAA0520 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 9 (EC 4.6.1.1) (Adenylate cyclase type IX) (ATP pyrophosphate-lyase 9) (Adenylyl cyclase 9). | |||||
|
ADCY9_MOUSE
|
||||||
| NC score | 0.050969 (rank : 141) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P51830, Q61279 | Gene names | Adcy9 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 9 (EC 4.6.1.1) (Adenylate cyclase type IX) (ATP pyrophosphate-lyase 9) (Adenylyl cyclase 9) (Adenylyl cyclase type 10) (ACTP10). | |||||
|
MASP2_HUMAN
|
||||||
| NC score | 0.050486 (rank : 142) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O00187, O75754, Q5TEQ5, Q5TER0, Q96QG4, Q9BZH0, Q9H498, Q9H499, Q9UBP3, Q9ULC7, Q9UMV3, Q9Y270 | Gene names | MASP2 | |||
|
Domain Architecture |

|
|||||
| Description | Mannan-binding lectin serine protease 2 precursor (EC 3.4.21.104) (Mannose-binding protein-associated serine protease 2) (MASP-2) (MBL- associated serine protease 2) [Contains: Mannan-binding lectin serine protease 2 A chain; Mannan-binding lectin serine protease 2 B chain]. | |||||
|
PAPOB_MOUSE
|
||||||
| NC score | 0.035523 (rank : 143) | θ value | 1.81305 (rank : 96) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9WVP6, Q9R1R3 | Gene names | Papolb, Papt, Tpap | |||
|
Domain Architecture |

|
|||||
| Description | Poly(A) polymerase beta (EC 2.7.7.19) (PAP beta) (Polynucleotide adenylyltransferase beta) (Testis-specific poly(A) polymerase). | |||||
|
PAPOA_MOUSE
|
||||||
| NC score | 0.030859 (rank : 144) | θ value | 5.27518 (rank : 120) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61183, Q61208, Q61209, Q8K4X2 | Gene names | Papola, Pap, Plap | |||
|
Domain Architecture |

|
|||||
| Description | Poly(A) polymerase alpha (EC 2.7.7.19) (PAP) (Polynucleotide adenylyltransferase). | |||||
|
PAPOB_HUMAN
|
||||||
| NC score | 0.029424 (rank : 145) | θ value | 5.27518 (rank : 121) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NRJ5, Q8NE14 | Gene names | PAPOLB, PAPT | |||
|
Domain Architecture |

|
|||||
| Description | Poly(A) polymerase beta (EC 2.7.7.19) (PAP beta) (Polynucleotide adenylyltransferase beta) (Testis-specific poly(A) polymerase). | |||||
|
PAPOA_HUMAN
|
||||||
| NC score | 0.028922 (rank : 146) | θ value | 5.27518 (rank : 119) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P51003, Q86SX4, Q86TV0, Q8IYF5, Q9BVU2 | Gene names | PAPOLA, PAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(A) polymerase alpha (EC 2.7.7.19) (PAP) (Polynucleotide adenylyltransferase alpha). | |||||
|
HAP1_HUMAN
|
||||||
| NC score | 0.026558 (rank : 147) | θ value | 3.0926 (rank : 104) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 606 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P54257, O75358, Q9H4G3, Q9HA98, Q9NY90 | Gene names | HAP1, HLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-associated protein 1 (HAP-1) (Neuroan 1). | |||||
|
ATCAY_MOUSE
|
||||||
| NC score | 0.021450 (rank : 148) | θ value | 2.36792 (rank : 98) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BHE3, Q3TR94 | Gene names | Atcay | |||
|
Domain Architecture |

|
|||||
| Description | Caytaxin. | |||||
|
EVPL_HUMAN
|
||||||
| NC score | 0.019705 (rank : 149) | θ value | 1.38821 (rank : 90) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 895 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q92817 | Gene names | EVPL | |||
|
Domain Architecture |

|
|||||
| Description | Envoplakin (210 kDa paraneoplastic pemphigus antigen) (p210) (210 kDa cornified envelope precursor protein). | |||||
|
CT2NL_MOUSE
|
||||||
| NC score | 0.019628 (rank : 150) | θ value | 5.27518 (rank : 114) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 902 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99LJ0, Q6ZPR0, Q8BSV1 | Gene names | Cttnbp2nl, Kiaa1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
HEM0_HUMAN
|
||||||
| NC score | 0.019035 (rank : 151) | θ value | 3.0926 (rank : 105) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P22557, Q13735 | Gene names | ALAS2, ALASE, ASB | |||
|
Domain Architecture |

|
|||||
| Description | 5-aminolevulinate synthase, erythroid-specific, mitochondrial precursor (EC 2.3.1.37) (5-aminolevulinic acid synthase) (Delta- aminolevulinate synthase) (Delta-ALA synthetase) (ALAS-E). | |||||
|
RTN3_HUMAN
|
||||||
| NC score | 0.017671 (rank : 152) | θ value | 6.88961 (rank : 128) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95197, Q496K2, Q7RTM8 | Gene names | RTN3, NSPL2 | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-3 (Neuroendocrine-specific protein-like 2) (NSP-like protein II) (NSPLII). | |||||
|
CT2NL_HUMAN
|
||||||
| NC score | 0.017260 (rank : 153) | θ value | 4.03905 (rank : 111) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9P2B4, Q96B40 | Gene names | CTTNBP2NL, KIAA1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
HOME2_MOUSE
|
||||||
| NC score | 0.016151 (rank : 154) | θ value | 3.0926 (rank : 106) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 882 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9QWW1, O89025, Q9Z0E4 | Gene names | Homer2, Vesl2 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 2 (Homer-2) (Cupidin) (VASP/Ena-related gene up- regulated during seizure and LTP 2) (Vesl-2). | |||||
|
RTN3_MOUSE
|
||||||
| NC score | 0.015492 (rank : 155) | θ value | 8.99809 (rank : 141) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ES97 | Gene names | Rtn3 | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-3. | |||||
|
CCD46_HUMAN
|
||||||
| NC score | 0.015412 (rank : 156) | θ value | 1.81305 (rank : 94) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
SAS10_MOUSE
|
||||||
| NC score | 0.014455 (rank : 157) | θ value | 6.88961 (rank : 129) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JI13, Q3UL95, Q8R5C6, Q9JJ12 | Gene names | Crlz1, Crl1, Sas10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Something about silencing protein 10 (Disrupter of silencing SAS10) (Charged amino acid-rich leucine zipper 1) (Crl-1). | |||||
|
TRAK1_HUMAN
|
||||||
| NC score | 0.014031 (rank : 158) | θ value | 6.88961 (rank : 130) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UPV9, Q96B69 | Gene names | TRAK1, KIAA1042, OIP106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trafficking kinesin-binding protein 1 (106 kDa O-GlcNAc transferase- interacting protein). | |||||
|
CP250_HUMAN
|
||||||
| NC score | 0.013798 (rank : 159) | θ value | 1.06291 (rank : 87) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
K1C14_MOUSE
|
||||||
| NC score | 0.013611 (rank : 160) | θ value | 2.36792 (rank : 100) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 501 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q61781, Q91VQ4 | Gene names | Krt14, Krt1-14 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 14 (Cytokeratin-14) (CK-14) (Keratin-14) (K14). | |||||
|
CCD21_HUMAN
|
||||||
| NC score | 0.013400 (rank : 161) | θ value | 5.27518 (rank : 113) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
MYH9_MOUSE
|
||||||
| NC score | 0.013158 (rank : 162) | θ value | 5.27518 (rank : 118) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MYH9_HUMAN
|
||||||
| NC score | 0.013147 (rank : 163) | θ value | 5.27518 (rank : 117) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MYH2_HUMAN
|
||||||
| NC score | 0.012618 (rank : 164) | θ value | 6.88961 (rank : 126) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.012475 (rank : 165) | θ value | 3.0926 (rank : 109) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
HIP1_HUMAN
|
||||||
| NC score | 0.012426 (rank : 166) | θ value | 6.88961 (rank : 125) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O00291, O00328, Q8TDL4 | Gene names | HIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1 (HIP-I). | |||||
|
MYH1_MOUSE
|
||||||
| NC score | 0.012074 (rank : 167) | θ value | 8.99809 (rank : 138) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
K1H4_HUMAN
|
||||||
| NC score | 0.011581 (rank : 168) | θ value | 3.0926 (rank : 107) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 476 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O76011 | Gene names | KRTHA4, HHA4, HKA4 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cuticular Ha4 (Hair keratin, type I Ha4). | |||||
|
CP135_MOUSE
|
||||||
| NC score | 0.011431 (rank : 169) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
K1C14_HUMAN
|
||||||
| NC score | 0.011376 (rank : 170) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P02533, Q14715, Q53XY3, Q9BUE3, Q9UBN2, Q9UBN3, Q9UCY4 | Gene names | KRT14 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 14 (Cytokeratin-14) (CK-14) (Keratin-14) (K14). | |||||
|
MYH10_HUMAN
|
||||||
| NC score | 0.011122 (rank : 171) | θ value | 8.99809 (rank : 135) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH10_MOUSE
|
||||||
| NC score | 0.010999 (rank : 172) | θ value | 8.99809 (rank : 136) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH1_HUMAN
|
||||||
| NC score | 0.010975 (rank : 173) | θ value | 8.99809 (rank : 137) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
ANR26_MOUSE
|
||||||
| NC score | 0.010250 (rank : 174) | θ value | 4.03905 (rank : 110) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1554 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q811D2, Q69ZS2, Q9CS61 | Gene names | Ankrd26, Kiaa1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
TROAP_HUMAN
|
||||||
| NC score | 0.010210 (rank : 175) | θ value | 8.99809 (rank : 142) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12815 | Gene names | TROAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trophinin-associated protein (Tastin) (Trophinin-assisting protein). | |||||
|
K1C16_HUMAN
|
||||||
| NC score | 0.009127 (rank : 176) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P08779, P30654, Q16402, Q9UBG8 | Gene names | KRT16, KRT16A | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 16 (Cytokeratin-16) (CK-16) (Keratin-16) (K16). | |||||
|
GEMI5_MOUSE
|
||||||
| NC score | 0.009118 (rank : 177) | θ value | 6.88961 (rank : 124) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 424 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BX17 | Gene names | Gemin5 | |||
|
Domain Architecture |

|
|||||
| Description | Gem-associated protein 5 (Gemin5). | |||||
|
TRI63_HUMAN
|
||||||
| NC score | 0.008644 (rank : 178) | θ value | 5.27518 (rank : 122) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q969Q1, Q5T2I1, Q96BD3, Q96KD9, Q9BYV4 | Gene names | TRIM63, IRF, MURF1, RNF28, SMRZ | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin ligase TRIM63 (EC 6.3.2.-) (Tripartite motif-containing protein 63) (Muscle-specific RING finger protein 1) (MuRF1) (MURF-1) (RING finger protein 28) (Striated muscle RING zinc finger protein) (Iris RING finger protein). | |||||
|
ROCK2_HUMAN
|
||||||
| NC score | 0.007883 (rank : 179) | θ value | 3.0926 (rank : 108) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 2073 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O75116, Q9UQN5 | Gene names | ROCK2, KIAA0619 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2) (Rho kinase 2). | |||||
|
CALI_HUMAN
|
||||||
| NC score | 0.007627 (rank : 180) | θ value | 6.88961 (rank : 123) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13939, Q9BXG7 | Gene names | CCIN | |||
|
Domain Architecture |

|
|||||
| Description | Calicin. | |||||
|
ROCK2_MOUSE
|
||||||
| NC score | 0.006813 (rank : 181) | θ value | 6.88961 (rank : 127) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 2072 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P70336, Q8BR64, Q8CBR0 | Gene names | Rock2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2). | |||||
|
KI13A_MOUSE
|
||||||
| NC score | 0.006518 (rank : 182) | θ value | 5.27518 (rank : 116) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9EQW7, O35062 | Gene names | Kif13a | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF13A. | |||||
|
ASPP1_MOUSE
|
||||||
| NC score | 0.005403 (rank : 183) | θ value | 5.27518 (rank : 112) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
MASTL_HUMAN
|
||||||
| NC score | 0.001241 (rank : 184) | θ value | 8.99809 (rank : 134) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 894 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96GX5, Q5T8D5, Q5T8D7, Q8NCD6, Q96SJ5 | Gene names | MASTL, THC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase-like (EC 2.7.11.1). | |||||
|
PCDBC_HUMAN
|
||||||
| NC score | 0.000156 (rank : 185) | θ value | 8.99809 (rank : 140) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y5F1 | Gene names | PCDHB12 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 12 precursor (PCDH-beta12). | |||||