Please be patient as the page loads
|
GCYB2_HUMAN
|
||||||
| SwissProt Accessions | O75343, Q9NZ64 | Gene names | GUCY1B2 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate cyclase soluble subunit beta-2 (EC 4.6.1.2) (GCS-beta-2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
GCYB2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O75343, Q9NZ64 | Gene names | GUCY1B2 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate cyclase soluble subunit beta-2 (EC 4.6.1.2) (GCS-beta-2). | |||||
|
GCYB1_HUMAN
|
||||||
| θ value | 1.71583e-75 (rank : 2) | NC score | 0.964164 (rank : 4) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q02153, Q86WY5 | Gene names | GUCY1B3, GUC1B3, GUCSB3, GUCY1B1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate cyclase soluble subunit beta-1 (EC 4.6.1.2) (GCS-beta-1) (Soluble guanylate cyclase small subunit) (GCS-beta-3). | |||||
|
GCYB1_MOUSE
|
||||||
| θ value | 1.71583e-75 (rank : 3) | NC score | 0.964452 (rank : 3) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O54865 | Gene names | Gucy1b3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanylate cyclase soluble subunit beta-1 (EC 4.6.1.2) (GCS-beta-1) (Soluble guanylate cyclase small subunit). | |||||
|
GCYA2_HUMAN
|
||||||
| θ value | 3.23591e-74 (rank : 4) | NC score | 0.967677 (rank : 2) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P33402 | Gene names | GUCY1A2, GUC1A2, GUCSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate cyclase soluble subunit alpha-2 (EC 4.6.1.2) (GCS-alpha-2). | |||||
|
GCYA3_HUMAN
|
||||||
| θ value | 2.3998e-69 (rank : 5) | NC score | 0.961599 (rank : 5) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q02108, O43843, Q8TAH3 | Gene names | GUCY1A3, GUC1A3, GUCSA3, GUCY1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate cyclase soluble subunit alpha-3 (EC 4.6.1.2) (GCS-alpha-3) (Soluble guanylate cyclase large subunit) (GCS-alpha-1). | |||||
|
GCYA3_MOUSE
|
||||||
| θ value | 2.3998e-69 (rank : 6) | NC score | 0.960985 (rank : 6) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9ERL9, Q9DBQ3 | Gene names | Gucy1a3, Gucy1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate cyclase soluble subunit alpha-3 (EC 4.6.1.2) (GCS-alpha-3) (Soluble guanylate cyclase large subunit) (GCS-alpha-1). | |||||
|
GUC2G_MOUSE
|
||||||
| θ value | 2.041e-52 (rank : 7) | NC score | 0.569097 (rank : 25) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6TL19, Q8BWU7 | Gene names | Gucy2g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanylate cyclase 2G precursor (EC 4.6.1.2) (Guanylyl cyclase receptor G) (mGC-G). | |||||
|
ANPRB_HUMAN
|
||||||
| θ value | 2.25659e-51 (rank : 8) | NC score | 0.549701 (rank : 29) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P20594, O60871, Q9UQ50 | Gene names | NPR2, ANPRB | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide receptor B precursor (ANP-B) (ANPRB) (GC-B) (Guanylate cyclase B) (EC 4.6.1.2) (NPR-B) (Atrial natriuretic peptide B-type receptor). | |||||
|
ANPRA_MOUSE
|
||||||
| θ value | 2.94719e-51 (rank : 9) | NC score | 0.556064 (rank : 27) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 600 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P18293 | Gene names | Npr1, Npra | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide receptor A precursor (ANP-A) (ANPRA) (GC-A) (Guanylate cyclase) (EC 4.6.1.2) (NPR-A) (Atrial natriuretic peptide A-type receptor). | |||||
|
ANPRA_HUMAN
|
||||||
| θ value | 6.56568e-51 (rank : 10) | NC score | 0.555240 (rank : 28) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 612 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P16066, Q6P4Q3 | Gene names | NPR1, ANPRA | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide receptor A precursor (ANP-A) (ANPRA) (GC-A) (Guanylate cyclase) (EC 4.6.1.2) (NPR-A) (Atrial natriuretic peptide A-type receptor). | |||||
|
ANPRB_MOUSE
|
||||||
| θ value | 1.11993e-50 (rank : 11) | NC score | 0.566875 (rank : 26) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6VVW5, Q6VVW3, Q6VVW4, Q8CGA9 | Gene names | Npr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Atrial natriuretic peptide receptor B precursor (ANP-B) (ANPRB) (GC-B) (Guanylate cyclase B) (EC 4.6.1.2) (NPR-B) (Atrial natriuretic peptide B-type receptor). | |||||
|
GUC2F_HUMAN
|
||||||
| θ value | 4.25575e-50 (rank : 12) | NC score | 0.518341 (rank : 31) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 740 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P51841, Q9UJF1 | Gene names | GUCY2F, GUC2F, RETGC2 | |||
|
Domain Architecture |

|
|||||
| Description | Retinal guanylyl cyclase 2 precursor (EC 4.6.1.2) (Guanylate cyclase 2F, retinal) (RETGC-2) (Rod outer segment membrane guanylate cyclase 2) (ROS-GC2) (Guanylate cyclase F) (GC-F). | |||||
|
GUC2E_MOUSE
|
||||||
| θ value | 9.48078e-50 (rank : 13) | NC score | 0.613792 (rank : 23) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P52785 | Gene names | Gucy2e, Guc2e | |||
|
Domain Architecture |

|
|||||
| Description | Guanylyl cyclase GC-E precursor (EC 4.6.1.2) (Guanylate cyclase 2E). | |||||
|
GUC2D_HUMAN
|
||||||
| θ value | 6.7944e-48 (rank : 14) | NC score | 0.611661 (rank : 24) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q02846, Q6LEA7 | Gene names | GUCY2D, CORD6, GUC1A4, GUC2D, RETGC, RETGC1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinal guanylyl cyclase 1 precursor (EC 4.6.1.2) (Guanylate cyclase 2D, retinal) (RETGC-1) (Rod outer segment membrane guanylate cyclase) (ROS-GC). | |||||
|
GUC2C_HUMAN
|
||||||
| θ value | 5.38352e-45 (rank : 15) | NC score | 0.535120 (rank : 30) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P25092 | Gene names | GUCY2C, GUC2C, STAR | |||
|
Domain Architecture |

|
|||||
| Description | Heat-stable enterotoxin receptor precursor (GC-C) (Intestinal guanylate cyclase) (EC 4.6.1.2) (STA receptor) (hSTAR). | |||||
|
ADCY2_MOUSE
|
||||||
| θ value | 1.02001e-27 (rank : 16) | NC score | 0.803935 (rank : 22) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q80TL1, Q8CFU6 | Gene names | Adcy2, Kiaa1060 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenylate cyclase type 2 (EC 4.6.1.1) (Adenylate cyclase type II) (ATP pyrophosphate-lyase 2) (Adenylyl cyclase 2). | |||||
|
ADCY2_HUMAN
|
||||||
| θ value | 2.27234e-27 (rank : 17) | NC score | 0.806484 (rank : 21) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q08462, Q9UPU2 | Gene names | ADCY2, KIAA1060 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 2 (EC 4.6.1.1) (Adenylate cyclase type II) (ATP pyrophosphate-lyase 2) (Adenylyl cyclase 2). | |||||
|
ADCY4_MOUSE
|
||||||
| θ value | 2.51237e-26 (rank : 18) | NC score | 0.811655 (rank : 14) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q91WF3 | Gene names | Adcy4 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 4 (EC 4.6.1.1) (Adenylate cyclase type IV) (ATP pyrophosphate-lyase 4) (Adenylyl cyclase 4). | |||||
|
ADCY4_HUMAN
|
||||||
| θ value | 3.28125e-26 (rank : 19) | NC score | 0.809726 (rank : 18) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8NFM4, Q96ML7 | Gene names | ADCY4 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 4 (EC 4.6.1.1) (Adenylate cyclase type IV) (ATP pyrophosphate-lyase 4) (Adenylyl cyclase 4). | |||||
|
ADCY7_MOUSE
|
||||||
| θ value | 5.59698e-26 (rank : 20) | NC score | 0.811164 (rank : 15) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P51829 | Gene names | Adcy7 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 7 (EC 4.6.1.1) (Adenylate cyclase type VII) (ATP pyrophosphate-lyase 7) (Adenylyl cyclase 7). | |||||
|
ADCY7_HUMAN
|
||||||
| θ value | 7.30988e-26 (rank : 21) | NC score | 0.810188 (rank : 16) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P51828 | Gene names | ADCY7, KIAA0037 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenylate cyclase type 7 (EC 4.6.1.1) (Adenylate cyclase type VII) (ATP pyrophosphate-lyase 7) (Adenylyl cyclase 7). | |||||
|
ADCY6_HUMAN
|
||||||
| θ value | 1.24688e-25 (rank : 22) | NC score | 0.811877 (rank : 13) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O43306, Q9NR75 | Gene names | ADCY6, KIAA0422 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 6 (EC 4.6.1.1) (Adenylate cyclase type VI) (ATP pyrophosphate-lyase 6) (Ca(2+)-inhibitable adenylyl cyclase). | |||||
|
ADCY6_MOUSE
|
||||||
| θ value | 1.24688e-25 (rank : 23) | NC score | 0.809979 (rank : 17) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q01341 | Gene names | Adcy6 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 6 (EC 4.6.1.1) (Adenylate cyclase type VI) (ATP pyrophosphate-lyase 6) (Ca(2+)-inhibitable adenylyl cyclase). | |||||
|
ADCY3_HUMAN
|
||||||
| θ value | 4.01107e-24 (rank : 24) | NC score | 0.814987 (rank : 12) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O60266, Q9UDB1 | Gene names | ADCY3, KIAA0511 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 3 (EC 4.6.1.1) (Adenylate cyclase type III) (Adenylate cyclase, olfactive type) (ATP pyrophosphate-lyase 3) (Adenylyl cyclase 3) (AC-III) (AC3). | |||||
|
ADCY8_HUMAN
|
||||||
| θ value | 4.01107e-24 (rank : 25) | NC score | 0.806976 (rank : 19) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P40145 | Gene names | ADCY8 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 8 (EC 4.6.1.1) (Adenylate cyclase type VIII) (ATP pyrophosphate-lyase 8) (Ca(2+)/calmodulin-activated adenylyl cyclase). | |||||
|
ADCY8_MOUSE
|
||||||
| θ value | 4.01107e-24 (rank : 26) | NC score | 0.806589 (rank : 20) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P97490 | Gene names | Adcy8 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 8 (EC 4.6.1.1) (Adenylate cyclase type VIII) (ATP pyrophosphate-lyase 8) (Ca(2+)/calmodulin-activated adenylyl cyclase). | |||||
|
ADCY3_MOUSE
|
||||||
| θ value | 5.23862e-24 (rank : 27) | NC score | 0.815798 (rank : 11) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8VHH7 | Gene names | Adcy3 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 3 (EC 4.6.1.1) (Adenylate cyclase type III) (Adenylate cyclase, olfactive type) (ATP pyrophosphate-lyase 3) (Adenylyl cyclase 3) (AC-III) (AC3). | |||||
|
ADCY1_HUMAN
|
||||||
| θ value | 8.93572e-24 (rank : 28) | NC score | 0.818614 (rank : 9) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q08828, Q75MI1 | Gene names | ADCY1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenylate cyclase type 1 (EC 4.6.1.1) (Adenylate cyclase type I) (ATP pyrophosphate-lyase 1) (Ca(2+)/calmodulin-activated adenylyl cyclase). | |||||
|
ADCY1_MOUSE
|
||||||
| θ value | 2.59989e-23 (rank : 29) | NC score | 0.816085 (rank : 10) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O88444, Q5SS89 | Gene names | Adcy1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenylate cyclase type 1 (EC 4.6.1.1) (Adenylate cyclase type I) (ATP pyrophosphate-lyase 1) (Ca(2+)/calmodulin-activated adenylyl cyclase). | |||||
|
ADCY9_HUMAN
|
||||||
| θ value | 4.15078e-21 (rank : 30) | NC score | 0.822817 (rank : 7) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O60503, O60273, Q4ZHT9, Q4ZIR5, Q9BWT4, Q9UGP2 | Gene names | ADCY9, KIAA0520 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 9 (EC 4.6.1.1) (Adenylate cyclase type IX) (ATP pyrophosphate-lyase 9) (Adenylyl cyclase 9). | |||||
|
ADCY9_MOUSE
|
||||||
| θ value | 4.15078e-21 (rank : 31) | NC score | 0.822409 (rank : 8) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P51830, Q61279 | Gene names | Adcy9 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 9 (EC 4.6.1.1) (Adenylate cyclase type IX) (ATP pyrophosphate-lyase 9) (Adenylyl cyclase 9) (Adenylyl cyclase type 10) (ACTP10). | |||||
|
FGD3_HUMAN
|
||||||
| θ value | 0.163984 (rank : 32) | NC score | 0.016409 (rank : 71) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5JSP0, Q7Z7D9, Q8N5G1 | Gene names | FGD3, ZFYVE5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 3 (Zinc finger FYVE domain-containing protein 5). | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 0.21417 (rank : 33) | NC score | 0.007193 (rank : 89) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
MD1L1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 34) | NC score | 0.013161 (rank : 78) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y6D9, Q13312, Q75MI0, Q86UM4, Q9UNH0 | Gene names | MAD1L1, MAD1, TXBP181 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1) (Mitotic checkpoint MAD1 protein-homolog) (HsMAD1) (hMAD1) (Tax-binding protein 181). | |||||
|
FGD3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 35) | NC score | 0.013799 (rank : 77) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88842, Q8BQ72 | Gene names | Fgd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 3. | |||||
|
TLK2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 36) | NC score | 0.012848 (rank : 80) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1044 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q86UE8, Q9UKI7, Q9Y4F7 | Gene names | TLK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase tousled-like 2 (EC 2.7.11.1) (Tousled- like kinase 2) (PKU-alpha). | |||||
|
TLK2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 37) | NC score | 0.013048 (rank : 79) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O55047, Q9D5Y5 | Gene names | Tlk2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase tousled-like 2 (EC 2.7.11.1) (Tousled- like kinase 2). | |||||
|
ERC2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 38) | NC score | 0.010371 (rank : 83) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O15083, Q86TK4 | Gene names | ERC2, KIAA0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2. | |||||
|
ERC2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.010458 (rank : 82) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1166 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6PH08, Q80U20, Q8CCP1 | Gene names | Erc2, Cast1, D14Ertd171e, Kiaa0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2 (CAZ-associated structural protein 1) (CAST1). | |||||
|
CR034_HUMAN
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.015770 (rank : 72) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5BJE1, Q6ZP67, Q6ZU20 | Gene names | C18orf34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C18orf34. | |||||
|
STAG1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.014157 (rank : 76) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WVM7, O00539 | Gene names | STAG1, SA1 | |||
|
Domain Architecture |

|
|||||
| Description | Cohesin subunit SA-1 (Stromal antigen 1) (SCC3 homolog 1). | |||||
|
STAG1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.014267 (rank : 75) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D3E6, O08982 | Gene names | Stag1, Sa1 | |||
|
Domain Architecture |

|
|||||
| Description | Cohesin subunit SA-1 (Stromal antigen 1) (SCC3 homolog 1). | |||||
|
EST1A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.017470 (rank : 69) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86US8, O94837, Q86VH6, Q9UF60 | Gene names | EST1A, C17orf31, KIAA0732 | |||
|
Domain Architecture |

|
|||||
| Description | Telomerase-binding protein EST1A (Ever shorter telomeres 1A) (Telomerase subunit EST1A) (EST1-like protein A) (hSmg5/7a). | |||||
|
GCC2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.008603 (rank : 87) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
NIN_MOUSE
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.009852 (rank : 84) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
RBP24_HUMAN
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.008638 (rank : 86) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
EST1A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.016472 (rank : 70) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P61406, Q5NC64 | Gene names | Est1a, Kiaa0732 | |||
|
Domain Architecture |

|
|||||
| Description | Telomerase-binding protein EST1A (Ever shorter telomeres 1A) (Telomerase subunit EST1A) (EST1-like protein A). | |||||
|
GOGA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.006610 (rank : 92) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1222 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q92805, Q8IYZ9 | Gene names | GOLGA1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
KCMA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 49) | NC score | 0.014335 (rank : 74) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12791, Q12886, Q12917, Q12921, Q12960, Q13150, Q96LG8, Q9UBB0, Q9UQK6 | Gene names | KCNMA1, KCNMA | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit alpha 1 (Calcium-activated potassium channel, subfamily M subunit alpha 1) (Maxi K channel) (MaxiK) (BK channel) (K(VCA)alpha) (BKCA alpha) (KCa1.1) (Slowpoke homolog) (Slo homolog) (Slo-alpha) (Slo1) (hSlo). | |||||
|
KCMA1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.014339 (rank : 73) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q08460, Q64703, Q8VHF1, Q9R196 | Gene names | Kcnma1, Kcnma | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit alpha 1 (Calcium-activated potassium channel, subfamily M subunit alpha 1) (Maxi K channel) (MaxiK) (BK channel) (K(VCA)alpha) (BKCA alpha) (KCa1.1) (Slowpoke homolog) (Slo homolog) (Slo-alpha) (Slo1) (mSlo) (mSlo1). | |||||
|
CENPF_HUMAN
|
||||||
| θ value | 5.27518 (rank : 51) | NC score | 0.005677 (rank : 94) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
MY18A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.004041 (rank : 97) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
MY18A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.004988 (rank : 95) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
CEP55_MOUSE
|
||||||
| θ value | 6.88961 (rank : 54) | NC score | 0.009125 (rank : 85) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BT07, Q8C2J0, Q8K2I8, Q8R2Y4, Q9DBZ8 | Gene names | Cep55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome protein 55. | |||||
|
INVO_MOUSE
|
||||||
| θ value | 6.88961 (rank : 55) | NC score | 0.012257 (rank : 81) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P48997 | Gene names | Ivl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Involucrin. | |||||
|
KI21B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 56) | NC score | 0.003614 (rank : 98) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1017 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O75037, Q5T4J3 | Gene names | KIF21B, KIAA0449 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21B. | |||||
|
GCC2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.007640 (rank : 88) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.004955 (rank : 96) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
MAFK_HUMAN
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.007041 (rank : 91) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60675 | Gene names | MAFK | |||
|
Domain Architecture |
|
|||||
| Description | Transcription factor MafK (Erythroid transcription factor NF-E2 p18 subunit). | |||||
|
MAFK_MOUSE
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.007060 (rank : 90) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61827, Q60600 | Gene names | Mafk, Nfe2u | |||
|
Domain Architecture |
|
|||||
| Description | Transcription factor MafK (Erythroid transcription factor NF-E2 p18 subunit). | |||||
|
OPTN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 61) | NC score | 0.005992 (rank : 93) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1120 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8K3K8 | Gene names | Optn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin. | |||||
|
ANKK1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.054055 (rank : 58) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1075 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8NFD2 | Gene names | ANKK1, PKK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and protein kinase domain-containing protein 1 (EC 2.7.11.1) (Protein kinase PKK2) (X-kinase) (SgK288). | |||||
|
ANKK1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.051575 (rank : 63) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BZ25 | Gene names | Ankk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and protein kinase domain-containing protein 1 (EC 2.7.11.1). | |||||
|
ANPRC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.290611 (rank : 32) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P17342 | Gene names | NPR3, ANPRC | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide clearance receptor precursor (ANP-C) (ANPRC) (NPR-C) (Atrial natriuretic peptide C-type receptor). | |||||
|
ANPRC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.284352 (rank : 33) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70180, P97804, Q9R025, Q9R027, Q9R028 | Gene names | Npr3 | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide clearance receptor precursor (ANP-C) (ANPRC) (NPR-C) (Atrial natriuretic peptide C-type receptor) (EF-2). | |||||
|
GABR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.081117 (rank : 35) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UBS5, O95375, O95468, O95975, O96022, Q86W60, Q9UQQ0 | Gene names | GABBR1, GPRC3A | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
GABR1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.083557 (rank : 34) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9WV18, Q9WU48, Q9WV15, Q9WV16, Q9WV17 | Gene names | Gabbr1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
ILK1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.064934 (rank : 48) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 857 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13418 | Gene names | ILK, ILK1 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin-linked protein kinase 1 (EC 2.7.11.1) (ILK-1) (59 kDa serine/threonine-protein kinase) (p59ILK). | |||||
|
ILK_MOUSE
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.065258 (rank : 47) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 858 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O55222 | Gene names | Ilk | |||
|
Domain Architecture |

|
|||||
| Description | Integrin-linked protein kinase (EC 2.7.11.1). | |||||
|
IRAK1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.052427 (rank : 59) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P51617, Q7Z5V4, Q96RL2 | Gene names | IRAK1, IRAK | |||
|
Domain Architecture |
|
|||||
| Description | Interleukin-1 receptor-associated kinase 1 (EC 2.7.11.1) (IRAK-1). | |||||
|
IRAK1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.051815 (rank : 61) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q62406, Q6Y3Z5, Q6Y3Z6 | Gene names | Irak1, Il1rak | |||
|
Domain Architecture |
|
|||||
| Description | Interleukin-1 receptor-associated kinase 1 (EC 2.7.11.1) (IRAK-1) (IRAK) (Pelle-like protein kinase) (mPLK). | |||||
|
IRAK3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.080464 (rank : 36) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y616 | Gene names | IRAK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-1 receptor-associated kinase 3 (EC 2.7.11.1) (IRAK-3) (IL- 1 receptor-associated kinase M) (IRAK-M). | |||||
|
IRAK3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.078040 (rank : 39) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 628 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K4B2, Q8C7U8, Q8CE40, Q8K1S8 | Gene names | Irak3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-1 receptor-associated kinase 3 (EC 2.7.11.1) (IRAK-3) (IL- 1 receptor-associated kinase M) (IRAK-M). | |||||
|
IRAK4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.051434 (rank : 64) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 816 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NWZ3, Q8TDF7, Q9Y589 | Gene names | IRAK4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-1 receptor-associated kinase 4 (EC 2.7.11.1) (IRAK-4) (NY- REN-64 antigen). | |||||
|
IRAK4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.050314 (rank : 68) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8R4K2, Q80WW1 | Gene names | Irak4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-1 receptor-associated kinase 4 (EC 2.7.11.1) (IRAK-4). | |||||
|
KSR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.077573 (rank : 40) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IVT5, Q13476 | Gene names | KSR1, KSR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinase suppressor of ras-1 (Kinase suppressor of ras). | |||||
|
KSR1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.076591 (rank : 42) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61097, Q61648, Q78DX8 | Gene names | Ksr1, Ksr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinase suppressor of ras-1 (Kinase suppressor of ras) (mKSR1) (Hb protein). | |||||
|
KSR2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.069803 (rank : 44) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6VAB6, Q8N775 | Gene names | KSR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinase suppressor of ras-2 (hKSR2). | |||||
|
M3K12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.050374 (rank : 66) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q12852, Q86VQ5, Q8WY25 | Gene names | MAP3K12, ZPK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 12 (EC 2.7.11.25) (Mixed lineage kinase) (Leucine-zipper protein kinase) (ZPK) (Dual leucine zipper bearing kinase) (DLK) (MAPK-upstream kinase) (MUK). | |||||
|
M3K12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.050365 (rank : 67) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q60700, P70286, Q3TLL7, Q8C4N7, Q8CBX3, Q8CDL6 | Gene names | Map3k12, Zpk | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 12 (EC 2.7.11.25) (Mixed lineage kinase) (Leucine-zipper protein kinase) (ZPK) (Dual leucine zipper bearing kinase) (DLK) (MAPK-upstream kinase) (MUK). | |||||
|
MLKL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.077263 (rank : 41) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 600 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8NB16, Q8N6V0 | Gene names | MLKL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mixed lineage kinase domain-like protein. | |||||
|
MLKL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.075939 (rank : 43) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 707 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9D2Y4, Q8CIJ5 | Gene names | Mlkl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mixed lineage kinase domain-like protein. | |||||
|
MLTK_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.057222 (rank : 54) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 911 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NYL2, Q53SX1, Q580W8, Q59GY5, Q86YW8, Q9HCC4, Q9HCC5, Q9HDD2, Q9NYE9 | Gene names | MLTK, ZAK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase MLT (EC 2.7.11.25) (MLK-like mitogen-activated protein triple kinase) (Leucine zipper- and sterile alpha motif-containing kinase) (Sterile alpha motif- and leucine zipper-containing kinase AZK) (Mixed lineage kinase-related kinase) (MLK-related kinase) (MRK) (Cervical cancer suppressor gene 4 protein). | |||||
|
MLTK_MOUSE
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.057627 (rank : 53) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9ESL4, Q3V1X8, Q8BR73, Q9ESL3 | Gene names | Mltk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase MLT (EC 2.7.11.25) (MLK-like mitogen-activated protein triple kinase) (Leucine zipper- and sterile alpha motif kinase ZAK) (Sterile alpha motif- and leucine zipper-containing kinase AZK) (Mixed lineage kinase-related kinase) (MLK-related kinase) (MRK). | |||||
|
MOS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.050522 (rank : 65) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P00540 | Gene names | MOS | |||
|
Domain Architecture |
|
|||||
| Description | Proto-oncogene serine/threonine-protein kinase mos (EC 2.7.11.1) (c- mos) (Oocyte maturation factor mos). | |||||
|
RIPK1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.066946 (rank : 46) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 891 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13546, Q13180 | Gene names | RIPK1, RIP | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 1 (EC 2.7.11.1) (Serine/threonine-protein kinase RIP) (Cell death protein RIP) (Receptor-interacting protein). | |||||
|
RIPK1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.069105 (rank : 45) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q60855, Q3U0J3, Q8CD90 | Gene names | Ripk1, Rinp, Rip | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 1 (EC 2.7.11.1) (Serine/threonine-protein kinase RIP) (Cell death protein RIP) (Receptor-interacting protein). | |||||
|
RIPK2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.079751 (rank : 37) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O43353, Q6UWF0 | Gene names | RIPK2, CARDIAK, RICK, RIP2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 2 (EC 2.7.11.1) (RIP-like-interacting CLARP kinase) (Receptor-interacting protein 2) (RIP-2) (CARD-containing interleukin-1 beta-converting enzyme- associated kinase) (CARD-containing IL-1 beta ICE-kinase). | |||||
|
RIPK2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.078082 (rank : 38) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 776 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P58801 | Gene names | Ripk2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
RIPK3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.057851 (rank : 52) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y572 | Gene names | RIPK3, RIP3 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 3 (EC 2.7.11.1) (RIP-like protein kinase 3) (Receptor-interacting protein 3) (RIP-3). | |||||
|
RIPK3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.059548 (rank : 51) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9QZL0 | Gene names | Ripk3, Rip3 | |||
|
Domain Architecture |
|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 3 (EC 2.7.11.1) (RIP-like protein kinase 3) (Receptor-interacting protein 3) (RIP-3) (mRIP3). | |||||
|
RIPK4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.055120 (rank : 57) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1094 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P57078, Q96KH0 | Gene names | RIPK4, ANKRD3, DIK | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 4 (EC 2.7.11.1) (Ankyrin repeat domain protein 3) (PKC-delta-interacting protein kinase). | |||||
|
TESK1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.052404 (rank : 60) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15569 | Gene names | TESK1 | |||
|
Domain Architecture |
|
|||||
| Description | Dual specificity testis-specific protein kinase 1 (EC 2.7.12.1) (Testicular protein kinase 1). | |||||
|
TESK1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.051709 (rank : 62) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 862 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O70146, O70147 | Gene names | Tesk1 | |||
|
Domain Architecture |
|
|||||
| Description | Dual specificity testis-specific protein kinase 1 (EC 2.7.12.1) (Testicular protein kinase 1). | |||||
|
TESK2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.055849 (rank : 56) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96S53, Q8N520, Q9Y3Q6 | Gene names | TESK2 | |||
|
Domain Architecture |
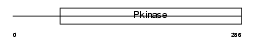
|
|||||
| Description | Dual specificity testis-specific protein kinase 2 (EC 2.7.12.1) (Testicular protein kinase 2). | |||||
|
TESK2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.055963 (rank : 55) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 833 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VCT9 | Gene names | Tesk2 | |||
|
Domain Architecture |
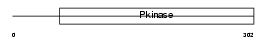
|
|||||
| Description | Dual specificity testis-specific protein kinase 2 (EC 2.7.12.1) (Testicular protein kinase 2). | |||||
|
TOPK_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.062649 (rank : 49) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 605 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96KB5, Q9NPD9, Q9NYL7, Q9NZK6 | Gene names | PBK, TOPK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-lymphokine-activated killer cell-originated protein kinase (EC 2.7.12.2) (T-LAK cell-originated protein kinase) (PDZ-binding kinase) (Spermatogenesis-related protein kinase) (SPK) (MAPKK-like protein kinase) (Nori-3). | |||||
|
TOPK_MOUSE
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.059729 (rank : 50) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JJ78, Q922V2, Q9D184 | Gene names | Pbk, Topk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-lymphokine-activated killer cell-originated protein kinase (EC 2.7.12.2) (T-LAK cell-originated protein kinase) (PDZ-binding kinase). | |||||
|
GCYB2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O75343, Q9NZ64 | Gene names | GUCY1B2 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate cyclase soluble subunit beta-2 (EC 4.6.1.2) (GCS-beta-2). | |||||
|
GCYA2_HUMAN
|
||||||
| NC score | 0.967677 (rank : 2) | θ value | 3.23591e-74 (rank : 4) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P33402 | Gene names | GUCY1A2, GUC1A2, GUCSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate cyclase soluble subunit alpha-2 (EC 4.6.1.2) (GCS-alpha-2). | |||||
|
GCYB1_MOUSE
|
||||||
| NC score | 0.964452 (rank : 3) | θ value | 1.71583e-75 (rank : 3) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O54865 | Gene names | Gucy1b3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanylate cyclase soluble subunit beta-1 (EC 4.6.1.2) (GCS-beta-1) (Soluble guanylate cyclase small subunit). | |||||
|
GCYB1_HUMAN
|
||||||
| NC score | 0.964164 (rank : 4) | θ value | 1.71583e-75 (rank : 2) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q02153, Q86WY5 | Gene names | GUCY1B3, GUC1B3, GUCSB3, GUCY1B1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate cyclase soluble subunit beta-1 (EC 4.6.1.2) (GCS-beta-1) (Soluble guanylate cyclase small subunit) (GCS-beta-3). | |||||
|
GCYA3_HUMAN
|
||||||
| NC score | 0.961599 (rank : 5) | θ value | 2.3998e-69 (rank : 5) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q02108, O43843, Q8TAH3 | Gene names | GUCY1A3, GUC1A3, GUCSA3, GUCY1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate cyclase soluble subunit alpha-3 (EC 4.6.1.2) (GCS-alpha-3) (Soluble guanylate cyclase large subunit) (GCS-alpha-1). | |||||
|
GCYA3_MOUSE
|
||||||
| NC score | 0.960985 (rank : 6) | θ value | 2.3998e-69 (rank : 6) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9ERL9, Q9DBQ3 | Gene names | Gucy1a3, Gucy1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate cyclase soluble subunit alpha-3 (EC 4.6.1.2) (GCS-alpha-3) (Soluble guanylate cyclase large subunit) (GCS-alpha-1). | |||||
|
ADCY9_HUMAN
|
||||||
| NC score | 0.822817 (rank : 7) | θ value | 4.15078e-21 (rank : 30) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O60503, O60273, Q4ZHT9, Q4ZIR5, Q9BWT4, Q9UGP2 | Gene names | ADCY9, KIAA0520 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 9 (EC 4.6.1.1) (Adenylate cyclase type IX) (ATP pyrophosphate-lyase 9) (Adenylyl cyclase 9). | |||||
|
ADCY9_MOUSE
|
||||||
| NC score | 0.822409 (rank : 8) | θ value | 4.15078e-21 (rank : 31) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P51830, Q61279 | Gene names | Adcy9 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 9 (EC 4.6.1.1) (Adenylate cyclase type IX) (ATP pyrophosphate-lyase 9) (Adenylyl cyclase 9) (Adenylyl cyclase type 10) (ACTP10). | |||||
|
ADCY1_HUMAN
|
||||||
| NC score | 0.818614 (rank : 9) | θ value | 8.93572e-24 (rank : 28) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q08828, Q75MI1 | Gene names | ADCY1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenylate cyclase type 1 (EC 4.6.1.1) (Adenylate cyclase type I) (ATP pyrophosphate-lyase 1) (Ca(2+)/calmodulin-activated adenylyl cyclase). | |||||
|
ADCY1_MOUSE
|
||||||
| NC score | 0.816085 (rank : 10) | θ value | 2.59989e-23 (rank : 29) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O88444, Q5SS89 | Gene names | Adcy1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenylate cyclase type 1 (EC 4.6.1.1) (Adenylate cyclase type I) (ATP pyrophosphate-lyase 1) (Ca(2+)/calmodulin-activated adenylyl cyclase). | |||||
|
ADCY3_MOUSE
|
||||||
| NC score | 0.815798 (rank : 11) | θ value | 5.23862e-24 (rank : 27) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8VHH7 | Gene names | Adcy3 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 3 (EC 4.6.1.1) (Adenylate cyclase type III) (Adenylate cyclase, olfactive type) (ATP pyrophosphate-lyase 3) (Adenylyl cyclase 3) (AC-III) (AC3). | |||||
|
ADCY3_HUMAN
|
||||||
| NC score | 0.814987 (rank : 12) | θ value | 4.01107e-24 (rank : 24) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O60266, Q9UDB1 | Gene names | ADCY3, KIAA0511 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 3 (EC 4.6.1.1) (Adenylate cyclase type III) (Adenylate cyclase, olfactive type) (ATP pyrophosphate-lyase 3) (Adenylyl cyclase 3) (AC-III) (AC3). | |||||
|
ADCY6_HUMAN
|
||||||
| NC score | 0.811877 (rank : 13) | θ value | 1.24688e-25 (rank : 22) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O43306, Q9NR75 | Gene names | ADCY6, KIAA0422 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 6 (EC 4.6.1.1) (Adenylate cyclase type VI) (ATP pyrophosphate-lyase 6) (Ca(2+)-inhibitable adenylyl cyclase). | |||||
|
ADCY4_MOUSE
|
||||||
| NC score | 0.811655 (rank : 14) | θ value | 2.51237e-26 (rank : 18) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q91WF3 | Gene names | Adcy4 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 4 (EC 4.6.1.1) (Adenylate cyclase type IV) (ATP pyrophosphate-lyase 4) (Adenylyl cyclase 4). | |||||
|
ADCY7_MOUSE
|
||||||
| NC score | 0.811164 (rank : 15) | θ value | 5.59698e-26 (rank : 20) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P51829 | Gene names | Adcy7 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 7 (EC 4.6.1.1) (Adenylate cyclase type VII) (ATP pyrophosphate-lyase 7) (Adenylyl cyclase 7). | |||||
|
ADCY7_HUMAN
|
||||||
| NC score | 0.810188 (rank : 16) | θ value | 7.30988e-26 (rank : 21) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P51828 | Gene names | ADCY7, KIAA0037 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenylate cyclase type 7 (EC 4.6.1.1) (Adenylate cyclase type VII) (ATP pyrophosphate-lyase 7) (Adenylyl cyclase 7). | |||||
|
ADCY6_MOUSE
|
||||||
| NC score | 0.809979 (rank : 17) | θ value | 1.24688e-25 (rank : 23) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q01341 | Gene names | Adcy6 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 6 (EC 4.6.1.1) (Adenylate cyclase type VI) (ATP pyrophosphate-lyase 6) (Ca(2+)-inhibitable adenylyl cyclase). | |||||
|
ADCY4_HUMAN
|
||||||
| NC score | 0.809726 (rank : 18) | θ value | 3.28125e-26 (rank : 19) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8NFM4, Q96ML7 | Gene names | ADCY4 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 4 (EC 4.6.1.1) (Adenylate cyclase type IV) (ATP pyrophosphate-lyase 4) (Adenylyl cyclase 4). | |||||
|
ADCY8_HUMAN
|
||||||
| NC score | 0.806976 (rank : 19) | θ value | 4.01107e-24 (rank : 25) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P40145 | Gene names | ADCY8 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 8 (EC 4.6.1.1) (Adenylate cyclase type VIII) (ATP pyrophosphate-lyase 8) (Ca(2+)/calmodulin-activated adenylyl cyclase). | |||||
|
ADCY8_MOUSE
|
||||||
| NC score | 0.806589 (rank : 20) | θ value | 4.01107e-24 (rank : 26) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P97490 | Gene names | Adcy8 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 8 (EC 4.6.1.1) (Adenylate cyclase type VIII) (ATP pyrophosphate-lyase 8) (Ca(2+)/calmodulin-activated adenylyl cyclase). | |||||
|
ADCY2_HUMAN
|
||||||
| NC score | 0.806484 (rank : 21) | θ value | 2.27234e-27 (rank : 17) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q08462, Q9UPU2 | Gene names | ADCY2, KIAA1060 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 2 (EC 4.6.1.1) (Adenylate cyclase type II) (ATP pyrophosphate-lyase 2) (Adenylyl cyclase 2). | |||||
|
ADCY2_MOUSE
|
||||||
| NC score | 0.803935 (rank : 22) | θ value | 1.02001e-27 (rank : 16) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q80TL1, Q8CFU6 | Gene names | Adcy2, Kiaa1060 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenylate cyclase type 2 (EC 4.6.1.1) (Adenylate cyclase type II) (ATP pyrophosphate-lyase 2) (Adenylyl cyclase 2). | |||||
|
GUC2E_MOUSE
|
||||||
| NC score | 0.613792 (rank : 23) | θ value | 9.48078e-50 (rank : 13) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P52785 | Gene names | Gucy2e, Guc2e | |||
|
Domain Architecture |

|
|||||
| Description | Guanylyl cyclase GC-E precursor (EC 4.6.1.2) (Guanylate cyclase 2E). | |||||
|
GUC2D_HUMAN
|
||||||
| NC score | 0.611661 (rank : 24) | θ value | 6.7944e-48 (rank : 14) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q02846, Q6LEA7 | Gene names | GUCY2D, CORD6, GUC1A4, GUC2D, RETGC, RETGC1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinal guanylyl cyclase 1 precursor (EC 4.6.1.2) (Guanylate cyclase 2D, retinal) (RETGC-1) (Rod outer segment membrane guanylate cyclase) (ROS-GC). | |||||
|
GUC2G_MOUSE
|
||||||
| NC score | 0.569097 (rank : 25) | θ value | 2.041e-52 (rank : 7) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6TL19, Q8BWU7 | Gene names | Gucy2g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanylate cyclase 2G precursor (EC 4.6.1.2) (Guanylyl cyclase receptor G) (mGC-G). | |||||
|
ANPRB_MOUSE
|
||||||
| NC score | 0.566875 (rank : 26) | θ value | 1.11993e-50 (rank : 11) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6VVW5, Q6VVW3, Q6VVW4, Q8CGA9 | Gene names | Npr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Atrial natriuretic peptide receptor B precursor (ANP-B) (ANPRB) (GC-B) (Guanylate cyclase B) (EC 4.6.1.2) (NPR-B) (Atrial natriuretic peptide B-type receptor). | |||||
|
ANPRA_MOUSE
|
||||||
| NC score | 0.556064 (rank : 27) | θ value | 2.94719e-51 (rank : 9) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 600 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P18293 | Gene names | Npr1, Npra | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide receptor A precursor (ANP-A) (ANPRA) (GC-A) (Guanylate cyclase) (EC 4.6.1.2) (NPR-A) (Atrial natriuretic peptide A-type receptor). | |||||
|
ANPRA_HUMAN
|
||||||
| NC score | 0.555240 (rank : 28) | θ value | 6.56568e-51 (rank : 10) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 612 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P16066, Q6P4Q3 | Gene names | NPR1, ANPRA | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide receptor A precursor (ANP-A) (ANPRA) (GC-A) (Guanylate cyclase) (EC 4.6.1.2) (NPR-A) (Atrial natriuretic peptide A-type receptor). | |||||
|
ANPRB_HUMAN
|
||||||
| NC score | 0.549701 (rank : 29) | θ value | 2.25659e-51 (rank : 8) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P20594, O60871, Q9UQ50 | Gene names | NPR2, ANPRB | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide receptor B precursor (ANP-B) (ANPRB) (GC-B) (Guanylate cyclase B) (EC 4.6.1.2) (NPR-B) (Atrial natriuretic peptide B-type receptor). | |||||
|
GUC2C_HUMAN
|
||||||
| NC score | 0.535120 (rank : 30) | θ value | 5.38352e-45 (rank : 15) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P25092 | Gene names | GUCY2C, GUC2C, STAR | |||
|
Domain Architecture |

|
|||||
| Description | Heat-stable enterotoxin receptor precursor (GC-C) (Intestinal guanylate cyclase) (EC 4.6.1.2) (STA receptor) (hSTAR). | |||||
|
GUC2F_HUMAN
|
||||||
| NC score | 0.518341 (rank : 31) | θ value | 4.25575e-50 (rank : 12) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 740 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P51841, Q9UJF1 | Gene names | GUCY2F, GUC2F, RETGC2 | |||
|
Domain Architecture |

|
|||||
| Description | Retinal guanylyl cyclase 2 precursor (EC 4.6.1.2) (Guanylate cyclase 2F, retinal) (RETGC-2) (Rod outer segment membrane guanylate cyclase 2) (ROS-GC2) (Guanylate cyclase F) (GC-F). | |||||
|
ANPRC_HUMAN
|
||||||
| NC score | 0.290611 (rank : 32) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P17342 | Gene names | NPR3, ANPRC | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide clearance receptor precursor (ANP-C) (ANPRC) (NPR-C) (Atrial natriuretic peptide C-type receptor). | |||||
|
ANPRC_MOUSE
|
||||||
| NC score | 0.284352 (rank : 33) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70180, P97804, Q9R025, Q9R027, Q9R028 | Gene names | Npr3 | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide clearance receptor precursor (ANP-C) (ANPRC) (NPR-C) (Atrial natriuretic peptide C-type receptor) (EF-2). | |||||
|
GABR1_MOUSE
|
||||||
| NC score | 0.083557 (rank : 34) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9WV18, Q9WU48, Q9WV15, Q9WV16, Q9WV17 | Gene names | Gabbr1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
GABR1_HUMAN
|
||||||
| NC score | 0.081117 (rank : 35) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UBS5, O95375, O95468, O95975, O96022, Q86W60, Q9UQQ0 | Gene names | GABBR1, GPRC3A | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
IRAK3_HUMAN
|
||||||
| NC score | 0.080464 (rank : 36) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y616 | Gene names | IRAK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-1 receptor-associated kinase 3 (EC 2.7.11.1) (IRAK-3) (IL- 1 receptor-associated kinase M) (IRAK-M). | |||||
|
RIPK2_HUMAN
|
||||||
| NC score | 0.079751 (rank : 37) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O43353, Q6UWF0 | Gene names | RIPK2, CARDIAK, RICK, RIP2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 2 (EC 2.7.11.1) (RIP-like-interacting CLARP kinase) (Receptor-interacting protein 2) (RIP-2) (CARD-containing interleukin-1 beta-converting enzyme- associated kinase) (CARD-containing IL-1 beta ICE-kinase). | |||||
|
RIPK2_MOUSE
|
||||||
| NC score | 0.078082 (rank : 38) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 776 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P58801 | Gene names | Ripk2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
IRAK3_MOUSE
|
||||||
| NC score | 0.078040 (rank : 39) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 628 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K4B2, Q8C7U8, Q8CE40, Q8K1S8 | Gene names | Irak3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-1 receptor-associated kinase 3 (EC 2.7.11.1) (IRAK-3) (IL- 1 receptor-associated kinase M) (IRAK-M). | |||||
|
KSR1_HUMAN
|
||||||
| NC score | 0.077573 (rank : 40) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IVT5, Q13476 | Gene names | KSR1, KSR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinase suppressor of ras-1 (Kinase suppressor of ras). | |||||
|
MLKL_HUMAN
|
||||||
| NC score | 0.077263 (rank : 41) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 600 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8NB16, Q8N6V0 | Gene names | MLKL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mixed lineage kinase domain-like protein. | |||||
|
KSR1_MOUSE
|
||||||
| NC score | 0.076591 (rank : 42) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61097, Q61648, Q78DX8 | Gene names | Ksr1, Ksr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinase suppressor of ras-1 (Kinase suppressor of ras) (mKSR1) (Hb protein). | |||||
|
MLKL_MOUSE
|
||||||
| NC score | 0.075939 (rank : 43) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 707 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9D2Y4, Q8CIJ5 | Gene names | Mlkl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mixed lineage kinase domain-like protein. | |||||
|
KSR2_HUMAN
|
||||||
| NC score | 0.069803 (rank : 44) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6VAB6, Q8N775 | Gene names | KSR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinase suppressor of ras-2 (hKSR2). | |||||
|
RIPK1_MOUSE
|
||||||
| NC score | 0.069105 (rank : 45) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q60855, Q3U0J3, Q8CD90 | Gene names | Ripk1, Rinp, Rip | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 1 (EC 2.7.11.1) (Serine/threonine-protein kinase RIP) (Cell death protein RIP) (Receptor-interacting protein). | |||||
|
RIPK1_HUMAN
|
||||||
| NC score | 0.066946 (rank : 46) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 891 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13546, Q13180 | Gene names | RIPK1, RIP | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 1 (EC 2.7.11.1) (Serine/threonine-protein kinase RIP) (Cell death protein RIP) (Receptor-interacting protein). | |||||
|
ILK_MOUSE
|
||||||
| NC score | 0.065258 (rank : 47) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 858 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O55222 | Gene names | Ilk | |||
|
Domain Architecture |

|
|||||
| Description | Integrin-linked protein kinase (EC 2.7.11.1). | |||||
|
ILK1_HUMAN
|
||||||
| NC score | 0.064934 (rank : 48) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 857 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13418 | Gene names | ILK, ILK1 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin-linked protein kinase 1 (EC 2.7.11.1) (ILK-1) (59 kDa serine/threonine-protein kinase) (p59ILK). | |||||
|
TOPK_HUMAN
|
||||||
| NC score | 0.062649 (rank : 49) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 605 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96KB5, Q9NPD9, Q9NYL7, Q9NZK6 | Gene names | PBK, TOPK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-lymphokine-activated killer cell-originated protein kinase (EC 2.7.12.2) (T-LAK cell-originated protein kinase) (PDZ-binding kinase) (Spermatogenesis-related protein kinase) (SPK) (MAPKK-like protein kinase) (Nori-3). | |||||
|
TOPK_MOUSE
|
||||||
| NC score | 0.059729 (rank : 50) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JJ78, Q922V2, Q9D184 | Gene names | Pbk, Topk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-lymphokine-activated killer cell-originated protein kinase (EC 2.7.12.2) (T-LAK cell-originated protein kinase) (PDZ-binding kinase). | |||||
|
RIPK3_MOUSE
|
||||||
| NC score | 0.059548 (rank : 51) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9QZL0 | Gene names | Ripk3, Rip3 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 3 (EC 2.7.11.1) (RIP-like protein kinase 3) (Receptor-interacting protein 3) (RIP-3) (mRIP3). | |||||
|
RIPK3_HUMAN
|
||||||
| NC score | 0.057851 (rank : 52) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y572 | Gene names | RIPK3, RIP3 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 3 (EC 2.7.11.1) (RIP-like protein kinase 3) (Receptor-interacting protein 3) (RIP-3). | |||||
|
MLTK_MOUSE
|
||||||
| NC score | 0.057627 (rank : 53) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9ESL4, Q3V1X8, Q8BR73, Q9ESL3 | Gene names | Mltk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase MLT (EC 2.7.11.25) (MLK-like mitogen-activated protein triple kinase) (Leucine zipper- and sterile alpha motif kinase ZAK) (Sterile alpha motif- and leucine zipper-containing kinase AZK) (Mixed lineage kinase-related kinase) (MLK-related kinase) (MRK). | |||||
|
MLTK_HUMAN
|
||||||
| NC score | 0.057222 (rank : 54) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 911 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NYL2, Q53SX1, Q580W8, Q59GY5, Q86YW8, Q9HCC4, Q9HCC5, Q9HDD2, Q9NYE9 | Gene names | MLTK, ZAK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase MLT (EC 2.7.11.25) (MLK-like mitogen-activated protein triple kinase) (Leucine zipper- and sterile alpha motif-containing kinase) (Sterile alpha motif- and leucine zipper-containing kinase AZK) (Mixed lineage kinase-related kinase) (MLK-related kinase) (MRK) (Cervical cancer suppressor gene 4 protein). | |||||
|
TESK2_MOUSE
|
||||||
| NC score | 0.055963 (rank : 55) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 833 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VCT9 | Gene names | Tesk2 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity testis-specific protein kinase 2 (EC 2.7.12.1) (Testicular protein kinase 2). | |||||
|
TESK2_HUMAN
|
||||||
| NC score | 0.055849 (rank : 56) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96S53, Q8N520, Q9Y3Q6 | Gene names | TESK2 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity testis-specific protein kinase 2 (EC 2.7.12.1) (Testicular protein kinase 2). | |||||
|
RIPK4_HUMAN
|
||||||
| NC score | 0.055120 (rank : 57) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1094 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P57078, Q96KH0 | Gene names | RIPK4, ANKRD3, DIK | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 4 (EC 2.7.11.1) (Ankyrin repeat domain protein 3) (PKC-delta-interacting protein kinase). | |||||
|
ANKK1_HUMAN
|
||||||
| NC score | 0.054055 (rank : 58) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1075 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8NFD2 | Gene names | ANKK1, PKK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and protein kinase domain-containing protein 1 (EC 2.7.11.1) (Protein kinase PKK2) (X-kinase) (SgK288). | |||||
|
IRAK1_HUMAN
|
||||||
| NC score | 0.052427 (rank : 59) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P51617, Q7Z5V4, Q96RL2 | Gene names | IRAK1, IRAK | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-1 receptor-associated kinase 1 (EC 2.7.11.1) (IRAK-1). | |||||
|
TESK1_HUMAN
|
||||||
| NC score | 0.052404 (rank : 60) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15569 | Gene names | TESK1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity testis-specific protein kinase 1 (EC 2.7.12.1) (Testicular protein kinase 1). | |||||
|
IRAK1_MOUSE
|
||||||
| NC score | 0.051815 (rank : 61) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q62406, Q6Y3Z5, Q6Y3Z6 | Gene names | Irak1, Il1rak | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-1 receptor-associated kinase 1 (EC 2.7.11.1) (IRAK-1) (IRAK) (Pelle-like protein kinase) (mPLK). | |||||
|
TESK1_MOUSE
|
||||||
| NC score | 0.051709 (rank : 62) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 862 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O70146, O70147 | Gene names | Tesk1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity testis-specific protein kinase 1 (EC 2.7.12.1) (Testicular protein kinase 1). | |||||
|
ANKK1_MOUSE
|
||||||
| NC score | 0.051575 (rank : 63) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BZ25 | Gene names | Ankk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and protein kinase domain-containing protein 1 (EC 2.7.11.1). | |||||
|
IRAK4_HUMAN
|
||||||
| NC score | 0.051434 (rank : 64) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 816 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NWZ3, Q8TDF7, Q9Y589 | Gene names | IRAK4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-1 receptor-associated kinase 4 (EC 2.7.11.1) (IRAK-4) (NY- REN-64 antigen). | |||||
|
MOS_HUMAN
|
||||||
| NC score | 0.050522 (rank : 65) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P00540 | Gene names | MOS | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene serine/threonine-protein kinase mos (EC 2.7.11.1) (c- mos) (Oocyte maturation factor mos). | |||||
|
M3K12_HUMAN
|
||||||
| NC score | 0.050374 (rank : 66) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q12852, Q86VQ5, Q8WY25 | Gene names | MAP3K12, ZPK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 12 (EC 2.7.11.25) (Mixed lineage kinase) (Leucine-zipper protein kinase) (ZPK) (Dual leucine zipper bearing kinase) (DLK) (MAPK-upstream kinase) (MUK). | |||||
|
M3K12_MOUSE
|
||||||
| NC score | 0.050365 (rank : 67) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q60700, P70286, Q3TLL7, Q8C4N7, Q8CBX3, Q8CDL6 | Gene names | Map3k12, Zpk | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 12 (EC 2.7.11.25) (Mixed lineage kinase) (Leucine-zipper protein kinase) (ZPK) (Dual leucine zipper bearing kinase) (DLK) (MAPK-upstream kinase) (MUK). | |||||
|
IRAK4_MOUSE
|
||||||
| NC score | 0.050314 (rank : 68) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8R4K2, Q80WW1 | Gene names | Irak4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-1 receptor-associated kinase 4 (EC 2.7.11.1) (IRAK-4). | |||||
|
EST1A_HUMAN
|
||||||
| NC score | 0.017470 (rank : 69) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86US8, O94837, Q86VH6, Q9UF60 | Gene names | EST1A, C17orf31, KIAA0732 | |||
|
Domain Architecture |

|
|||||
| Description | Telomerase-binding protein EST1A (Ever shorter telomeres 1A) (Telomerase subunit EST1A) (EST1-like protein A) (hSmg5/7a). | |||||
|
EST1A_MOUSE
|
||||||
| NC score | 0.016472 (rank : 70) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P61406, Q5NC64 | Gene names | Est1a, Kiaa0732 | |||
|
Domain Architecture |

|
|||||
| Description | Telomerase-binding protein EST1A (Ever shorter telomeres 1A) (Telomerase subunit EST1A) (EST1-like protein A). | |||||
|
FGD3_HUMAN
|
||||||
| NC score | 0.016409 (rank : 71) | θ value | 0.163984 (rank : 32) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5JSP0, Q7Z7D9, Q8N5G1 | Gene names | FGD3, ZFYVE5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 3 (Zinc finger FYVE domain-containing protein 5). | |||||
|
CR034_HUMAN
|
||||||
| NC score | 0.015770 (rank : 72) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5BJE1, Q6ZP67, Q6ZU20 | Gene names | C18orf34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C18orf34. | |||||
|
KCMA1_MOUSE
|
||||||
| NC score | 0.014339 (rank : 73) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q08460, Q64703, Q8VHF1, Q9R196 | Gene names | Kcnma1, Kcnma | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit alpha 1 (Calcium-activated potassium channel, subfamily M subunit alpha 1) (Maxi K channel) (MaxiK) (BK channel) (K(VCA)alpha) (BKCA alpha) (KCa1.1) (Slowpoke homolog) (Slo homolog) (Slo-alpha) (Slo1) (mSlo) (mSlo1). | |||||
|
KCMA1_HUMAN
|
||||||
| NC score | 0.014335 (rank : 74) | θ value | 4.03905 (rank : 49) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12791, Q12886, Q12917, Q12921, Q12960, Q13150, Q96LG8, Q9UBB0, Q9UQK6 | Gene names | KCNMA1, KCNMA | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit alpha 1 (Calcium-activated potassium channel, subfamily M subunit alpha 1) (Maxi K channel) (MaxiK) (BK channel) (K(VCA)alpha) (BKCA alpha) (KCa1.1) (Slowpoke homolog) (Slo homolog) (Slo-alpha) (Slo1) (hSlo). | |||||
|
STAG1_MOUSE
|
||||||
| NC score | 0.014267 (rank : 75) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D3E6, O08982 | Gene names | Stag1, Sa1 | |||
|
Domain Architecture |

|
|||||
| Description | Cohesin subunit SA-1 (Stromal antigen 1) (SCC3 homolog 1). | |||||
|
STAG1_HUMAN
|
||||||
| NC score | 0.014157 (rank : 76) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WVM7, O00539 | Gene names | STAG1, SA1 | |||
|
Domain Architecture |

|
|||||
| Description | Cohesin subunit SA-1 (Stromal antigen 1) (SCC3 homolog 1). | |||||
|
FGD3_MOUSE
|
||||||
| NC score | 0.013799 (rank : 77) | θ value | 1.06291 (rank : 35) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88842, Q8BQ72 | Gene names | Fgd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 3. | |||||
|
MD1L1_HUMAN
|
||||||
| NC score | 0.013161 (rank : 78) | θ value | 0.813845 (rank : 34) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y6D9, Q13312, Q75MI0, Q86UM4, Q9UNH0 | Gene names | MAD1L1, MAD1, TXBP181 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1) (Mitotic checkpoint MAD1 protein-homolog) (HsMAD1) (hMAD1) (Tax-binding protein 181). | |||||
|
TLK2_MOUSE
|
||||||
| NC score | 0.013048 (rank : 79) | θ value | 1.38821 (rank : 37) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O55047, Q9D5Y5 | Gene names | Tlk2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase tousled-like 2 (EC 2.7.11.1) (Tousled- like kinase 2). | |||||
|
TLK2_HUMAN
|
||||||
| NC score | 0.012848 (rank : 80) | θ value | 1.38821 (rank : 36) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1044 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q86UE8, Q9UKI7, Q9Y4F7 | Gene names | TLK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase tousled-like 2 (EC 2.7.11.1) (Tousled- like kinase 2) (PKU-alpha). | |||||
|
INVO_MOUSE
|
||||||
| NC score | 0.012257 (rank : 81) | θ value | 6.88961 (rank : 55) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P48997 | Gene names | Ivl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Involucrin. | |||||
|
ERC2_MOUSE
|
||||||
| NC score | 0.010458 (rank : 82) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1166 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6PH08, Q80U20, Q8CCP1 | Gene names | Erc2, Cast1, D14Ertd171e, Kiaa0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2 (CAZ-associated structural protein 1) (CAST1). | |||||
|
ERC2_HUMAN
|
||||||
| NC score | 0.010371 (rank : 83) | θ value | 1.81305 (rank : 38) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O15083, Q86TK4 | Gene names | ERC2, KIAA0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2. | |||||
|
NIN_MOUSE
|
||||||
| NC score | 0.009852 (rank : 84) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
CEP55_MOUSE
|
||||||
| NC score | 0.009125 (rank : 85) | θ value | 6.88961 (rank : 54) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BT07, Q8C2J0, Q8K2I8, Q8R2Y4, Q9DBZ8 | Gene names | Cep55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome protein 55. | |||||
|
RBP24_HUMAN
|
||||||
| NC score | 0.008638 (rank : 86) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
GCC2_HUMAN
|
||||||
| NC score | 0.008603 (rank : 87) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
GCC2_MOUSE
|
||||||
| NC score | 0.007640 (rank : 88) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.007193 (rank : 89) | θ value | 0.21417 (rank : 33) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
MAFK_MOUSE
|
||||||
| NC score | 0.007060 (rank : 90) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61827, Q60600 | Gene names | Mafk, Nfe2u | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafK (Erythroid transcription factor NF-E2 p18 subunit). | |||||
|
MAFK_HUMAN
|
||||||
| NC score | 0.007041 (rank : 91) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60675 | Gene names | MAFK | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafK (Erythroid transcription factor NF-E2 p18 subunit). | |||||
|
GOGA1_HUMAN
|
||||||
| NC score | 0.006610 (rank : 92) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1222 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q92805, Q8IYZ9 | Gene names | GOLGA1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
OPTN_MOUSE
|
||||||
| NC score | 0.005992 (rank : 93) | θ value | 8.99809 (rank : 61) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1120 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8K3K8 | Gene names | Optn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin. | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.005677 (rank : 94) | θ value | 5.27518 (rank : 51) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
MY18A_MOUSE
|
||||||
| NC score | 0.004988 (rank : 95) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.004955 (rank : 96) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
MY18A_HUMAN
|
||||||
| NC score | 0.004041 (rank : 97) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
KI21B_HUMAN
|
||||||
| NC score | 0.003614 (rank : 98) | θ value | 6.88961 (rank : 56) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 1017 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O75037, Q5T4J3 | Gene names | KIF21B, KIAA0449 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21B. | |||||