Please be patient as the page loads
|
ADCY3_HUMAN
|
||||||
| SwissProt Accessions | O60266, Q9UDB1 | Gene names | ADCY3, KIAA0511 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 3 (EC 4.6.1.1) (Adenylate cyclase type III) (Adenylate cyclase, olfactive type) (ATP pyrophosphate-lyase 3) (Adenylyl cyclase 3) (AC-III) (AC3). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ADCY3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O60266, Q9UDB1 | Gene names | ADCY3, KIAA0511 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 3 (EC 4.6.1.1) (Adenylate cyclase type III) (Adenylate cyclase, olfactive type) (ATP pyrophosphate-lyase 3) (Adenylyl cyclase 3) (AC-III) (AC3). | |||||
|
ADCY3_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.995171 (rank : 2) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8VHH7 | Gene names | Adcy3 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 3 (EC 4.6.1.1) (Adenylate cyclase type III) (Adenylate cyclase, olfactive type) (ATP pyrophosphate-lyase 3) (Adenylyl cyclase 3) (AC-III) (AC3). | |||||
|
ADCY8_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.979728 (rank : 3) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P40145 | Gene names | ADCY8 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 8 (EC 4.6.1.1) (Adenylate cyclase type VIII) (ATP pyrophosphate-lyase 8) (Ca(2+)/calmodulin-activated adenylyl cyclase). | |||||
|
ADCY8_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.979628 (rank : 4) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P97490 | Gene names | Adcy8 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 8 (EC 4.6.1.1) (Adenylate cyclase type VIII) (ATP pyrophosphate-lyase 8) (Ca(2+)/calmodulin-activated adenylyl cyclase). | |||||
|
ADCY6_HUMAN
|
||||||
| θ value | 2.25536e-168 (rank : 5) | NC score | 0.977983 (rank : 5) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O43306, Q9NR75 | Gene names | ADCY6, KIAA0422 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 6 (EC 4.6.1.1) (Adenylate cyclase type VI) (ATP pyrophosphate-lyase 6) (Ca(2+)-inhibitable adenylyl cyclase). | |||||
|
ADCY6_MOUSE
|
||||||
| θ value | 2.25536e-168 (rank : 6) | NC score | 0.977346 (rank : 6) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q01341 | Gene names | Adcy6 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 6 (EC 4.6.1.1) (Adenylate cyclase type VI) (ATP pyrophosphate-lyase 6) (Ca(2+)-inhibitable adenylyl cyclase). | |||||
|
ADCY2_MOUSE
|
||||||
| θ value | 4.55495e-161 (rank : 7) | NC score | 0.974533 (rank : 7) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q80TL1, Q8CFU6 | Gene names | Adcy2, Kiaa1060 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenylate cyclase type 2 (EC 4.6.1.1) (Adenylate cyclase type II) (ATP pyrophosphate-lyase 2) (Adenylyl cyclase 2). | |||||
|
ADCY2_HUMAN
|
||||||
| θ value | 1.68038e-147 (rank : 8) | NC score | 0.968018 (rank : 10) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q08462, Q9UPU2 | Gene names | ADCY2, KIAA1060 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 2 (EC 4.6.1.1) (Adenylate cyclase type II) (ATP pyrophosphate-lyase 2) (Adenylyl cyclase 2). | |||||
|
ADCY7_MOUSE
|
||||||
| θ value | 3.06949e-141 (rank : 9) | NC score | 0.973711 (rank : 9) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P51829 | Gene names | Adcy7 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 7 (EC 4.6.1.1) (Adenylate cyclase type VII) (ATP pyrophosphate-lyase 7) (Adenylyl cyclase 7). | |||||
|
ADCY7_HUMAN
|
||||||
| θ value | 1.52337e-140 (rank : 10) | NC score | 0.974282 (rank : 8) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P51828 | Gene names | ADCY7, KIAA0037 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenylate cyclase type 7 (EC 4.6.1.1) (Adenylate cyclase type VII) (ATP pyrophosphate-lyase 7) (Adenylyl cyclase 7). | |||||
|
ADCY4_MOUSE
|
||||||
| θ value | 1.02182e-136 (rank : 11) | NC score | 0.967085 (rank : 11) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q91WF3 | Gene names | Adcy4 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 4 (EC 4.6.1.1) (Adenylate cyclase type IV) (ATP pyrophosphate-lyase 4) (Adenylyl cyclase 4). | |||||
|
ADCY4_HUMAN
|
||||||
| θ value | 8.9516e-133 (rank : 12) | NC score | 0.966242 (rank : 12) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8NFM4, Q96ML7 | Gene names | ADCY4 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 4 (EC 4.6.1.1) (Adenylate cyclase type IV) (ATP pyrophosphate-lyase 4) (Adenylyl cyclase 4). | |||||
|
ADCY1_HUMAN
|
||||||
| θ value | 8.79181e-80 (rank : 13) | NC score | 0.956498 (rank : 14) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q08828, Q75MI1 | Gene names | ADCY1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenylate cyclase type 1 (EC 4.6.1.1) (Adenylate cyclase type I) (ATP pyrophosphate-lyase 1) (Ca(2+)/calmodulin-activated adenylyl cyclase). | |||||
|
ADCY1_MOUSE
|
||||||
| θ value | 1.14825e-79 (rank : 14) | NC score | 0.963602 (rank : 13) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O88444, Q5SS89 | Gene names | Adcy1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenylate cyclase type 1 (EC 4.6.1.1) (Adenylate cyclase type I) (ATP pyrophosphate-lyase 1) (Ca(2+)/calmodulin-activated adenylyl cyclase). | |||||
|
ADCY9_MOUSE
|
||||||
| θ value | 1.51013e-55 (rank : 15) | NC score | 0.907819 (rank : 16) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P51830, Q61279 | Gene names | Adcy9 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 9 (EC 4.6.1.1) (Adenylate cyclase type IX) (ATP pyrophosphate-lyase 9) (Adenylyl cyclase 9) (Adenylyl cyclase type 10) (ACTP10). | |||||
|
ADCY9_HUMAN
|
||||||
| θ value | 1.97229e-55 (rank : 16) | NC score | 0.907891 (rank : 15) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O60503, O60273, Q4ZHT9, Q4ZIR5, Q9BWT4, Q9UGP2 | Gene names | ADCY9, KIAA0520 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 9 (EC 4.6.1.1) (Adenylate cyclase type IX) (ATP pyrophosphate-lyase 9) (Adenylyl cyclase 9). | |||||
|
ANPRB_HUMAN
|
||||||
| θ value | 8.63488e-27 (rank : 17) | NC score | 0.447701 (rank : 29) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P20594, O60871, Q9UQ50 | Gene names | NPR2, ANPRB | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide receptor B precursor (ANP-B) (ANPRB) (GC-B) (Guanylate cyclase B) (EC 4.6.1.2) (NPR-B) (Atrial natriuretic peptide B-type receptor). | |||||
|
ANPRA_MOUSE
|
||||||
| θ value | 1.12775e-26 (rank : 18) | NC score | 0.452511 (rank : 27) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 600 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P18293 | Gene names | Npr1, Npra | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide receptor A precursor (ANP-A) (ANPRA) (GC-A) (Guanylate cyclase) (EC 4.6.1.2) (NPR-A) (Atrial natriuretic peptide A-type receptor). | |||||
|
ANPRA_HUMAN
|
||||||
| θ value | 1.47289e-26 (rank : 19) | NC score | 0.451334 (rank : 28) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 612 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P16066, Q6P4Q3 | Gene names | NPR1, ANPRA | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide receptor A precursor (ANP-A) (ANPRA) (GC-A) (Guanylate cyclase) (EC 4.6.1.2) (NPR-A) (Atrial natriuretic peptide A-type receptor). | |||||
|
ANPRB_MOUSE
|
||||||
| θ value | 2.51237e-26 (rank : 20) | NC score | 0.460433 (rank : 26) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6VVW5, Q6VVW3, Q6VVW4, Q8CGA9 | Gene names | Npr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Atrial natriuretic peptide receptor B precursor (ANP-B) (ANPRB) (GC-B) (Guanylate cyclase B) (EC 4.6.1.2) (NPR-B) (Atrial natriuretic peptide B-type receptor). | |||||
|
GCYA3_HUMAN
|
||||||
| θ value | 4.28545e-26 (rank : 21) | NC score | 0.795078 (rank : 18) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q02108, O43843, Q8TAH3 | Gene names | GUCY1A3, GUC1A3, GUCSA3, GUCY1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate cyclase soluble subunit alpha-3 (EC 4.6.1.2) (GCS-alpha-3) (Soluble guanylate cyclase large subunit) (GCS-alpha-1). | |||||
|
GCYA3_MOUSE
|
||||||
| θ value | 1.24688e-25 (rank : 22) | NC score | 0.793173 (rank : 20) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9ERL9, Q9DBQ3 | Gene names | Gucy1a3, Gucy1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate cyclase soluble subunit alpha-3 (EC 4.6.1.2) (GCS-alpha-3) (Soluble guanylate cyclase large subunit) (GCS-alpha-1). | |||||
|
GUC2G_MOUSE
|
||||||
| θ value | 3.07116e-24 (rank : 23) | NC score | 0.467258 (rank : 25) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6TL19, Q8BWU7 | Gene names | Gucy2g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanylate cyclase 2G precursor (EC 4.6.1.2) (Guanylyl cyclase receptor G) (mGC-G). | |||||
|
GCYB2_HUMAN
|
||||||
| θ value | 4.01107e-24 (rank : 24) | NC score | 0.814987 (rank : 17) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O75343, Q9NZ64 | Gene names | GUCY1B2 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate cyclase soluble subunit beta-2 (EC 4.6.1.2) (GCS-beta-2). | |||||
|
GCYA2_HUMAN
|
||||||
| θ value | 1.16704e-23 (rank : 25) | NC score | 0.794950 (rank : 19) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P33402 | Gene names | GUCY1A2, GUC1A2, GUCSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate cyclase soluble subunit alpha-2 (EC 4.6.1.2) (GCS-alpha-2). | |||||
|
GCYB1_HUMAN
|
||||||
| θ value | 6.40375e-22 (rank : 26) | NC score | 0.784211 (rank : 22) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q02153, Q86WY5 | Gene names | GUCY1B3, GUC1B3, GUCSB3, GUCY1B1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate cyclase soluble subunit beta-1 (EC 4.6.1.2) (GCS-beta-1) (Soluble guanylate cyclase small subunit) (GCS-beta-3). | |||||
|
GUC2E_MOUSE
|
||||||
| θ value | 8.36355e-22 (rank : 27) | NC score | 0.496878 (rank : 23) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P52785 | Gene names | Gucy2e, Guc2e | |||
|
Domain Architecture |

|
|||||
| Description | Guanylyl cyclase GC-E precursor (EC 4.6.1.2) (Guanylate cyclase 2E). | |||||
|
GCYB1_MOUSE
|
||||||
| θ value | 1.09232e-21 (rank : 28) | NC score | 0.784381 (rank : 21) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O54865 | Gene names | Gucy1b3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanylate cyclase soluble subunit beta-1 (EC 4.6.1.2) (GCS-beta-1) (Soluble guanylate cyclase small subunit). | |||||
|
GUC2F_HUMAN
|
||||||
| θ value | 1.2077e-20 (rank : 29) | NC score | 0.420535 (rank : 31) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 740 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P51841, Q9UJF1 | Gene names | GUCY2F, GUC2F, RETGC2 | |||
|
Domain Architecture |

|
|||||
| Description | Retinal guanylyl cyclase 2 precursor (EC 4.6.1.2) (Guanylate cyclase 2F, retinal) (RETGC-2) (Rod outer segment membrane guanylate cyclase 2) (ROS-GC2) (Guanylate cyclase F) (GC-F). | |||||
|
GUC2C_HUMAN
|
||||||
| θ value | 3.51386e-20 (rank : 30) | NC score | 0.435917 (rank : 30) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P25092 | Gene names | GUCY2C, GUC2C, STAR | |||
|
Domain Architecture |

|
|||||
| Description | Heat-stable enterotoxin receptor precursor (GC-C) (Intestinal guanylate cyclase) (EC 4.6.1.2) (STA receptor) (hSTAR). | |||||
|
GUC2D_HUMAN
|
||||||
| θ value | 7.82807e-20 (rank : 31) | NC score | 0.493432 (rank : 24) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q02846, Q6LEA7 | Gene names | GUCY2D, CORD6, GUC1A4, GUC2D, RETGC, RETGC1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinal guanylyl cyclase 1 precursor (EC 4.6.1.2) (Guanylate cyclase 2D, retinal) (RETGC-1) (Rod outer segment membrane guanylate cyclase) (ROS-GC). | |||||
|
NUAK2_MOUSE
|
||||||
| θ value | 0.163984 (rank : 32) | NC score | 0.012872 (rank : 39) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 878 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BZN4, Q80ZW3, Q8CIC0, Q9DBV0 | Gene names | Nuak2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NUAK family SNF1-like kinase 2 (EC 2.7.11.1). | |||||
|
JHD2B_HUMAN
|
||||||
| θ value | 0.62314 (rank : 33) | NC score | 0.016923 (rank : 36) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7LBC6, Q9BVH6, Q9BW93, Q9BZ52, Q9NYF4, Q9UPS0 | Gene names | JMJD1B, C5orf7, JHDM2B, KIAA1082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2B (EC 1.14.11.-) (Jumonji domain-containing protein 1B) (Nuclear protein 5qNCA). | |||||
|
JHD2B_MOUSE
|
||||||
| θ value | 0.813845 (rank : 34) | NC score | 0.016832 (rank : 37) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZPY7, Q2VPQ5, Q5U5V7, Q6P9K3, Q8CCE2, Q8K2A5, Q9CU57 | Gene names | Jmjd1b, Jhdm2b, Kiaa1082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2B (EC 1.14.11.-) (Jumonji domain-containing protein 1B). | |||||
|
SETD2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 35) | NC score | 0.015762 (rank : 38) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
UBE4B_MOUSE
|
||||||
| θ value | 0.813845 (rank : 36) | NC score | 0.034602 (rank : 34) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ES00, Q9EQE0 | Gene names | Ube4b, Ufd2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin conjugation factor E4 B (Ubiquitin fusion degradation protein 2) (Ufd2a). | |||||
|
ASPN_MOUSE
|
||||||
| θ value | 1.06291 (rank : 37) | NC score | 0.005960 (rank : 48) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99MQ4, Q9D6A2 | Gene names | Aspn | |||
|
Domain Architecture |

|
|||||
| Description | Asporin precursor. | |||||
|
GNAS1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 38) | NC score | 0.004041 (rank : 50) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6R0H7, Q6R0H4, Q6R0H5, Q6R2J5, Q9JJX0, Q9Z1N8 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
UBE4B_HUMAN
|
||||||
| θ value | 1.06291 (rank : 39) | NC score | 0.031968 (rank : 35) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95155, O75169, O95338, Q5SZ12, Q5SZ16, Q96QD4, Q9BYI7 | Gene names | UBE4B, HDNB1, KIAA0684, UFD2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin conjugation factor E4 B (Ubiquitin fusion degradation protein 2) (Homozygously deleted in neuroblastoma 1). | |||||
|
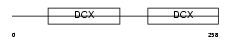
DCX_HUMAN
|
||||||
| θ value | 1.38821 (rank : 40) | NC score | 0.009400 (rank : 43) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43602, O43911 | Gene names | DCX, DBCN, LISX | |||
|
Domain Architecture |
|
|||||
| Description | Neuronal migration protein doublecortin (Lissencephalin-X) (Lis-X) (Doublin). | |||||
|
MBB1A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.007651 (rank : 45) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
MTFR1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 42) | NC score | 0.010947 (rank : 41) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99MB2, Q9CQZ4 | Gene names | Mtfr1, Kiaa0009 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial fission regulator 1. | |||||
|
PCNT_MOUSE
|
||||||
| θ value | 4.03905 (rank : 43) | NC score | 0.000111 (rank : 57) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1207 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P48725 | Gene names | Pcnt, Pcnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pericentrin. | |||||
|
PRLPM_MOUSE
|
||||||
| θ value | 4.03905 (rank : 44) | NC score | 0.010568 (rank : 42) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JHK0, Q9JI05 | Gene names | Prlpm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Placental prolactin-like protein M precursor (PRL-like protein M) (PLP-M). | |||||
|
SI1L3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 45) | NC score | 0.004765 (rank : 49) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60292, Q2TV87 | Gene names | SIPA1L3, KIAA0545, SPAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 3 (SPA-1-like protein 3). | |||||
|
ADA12_HUMAN
|
||||||
| θ value | 5.27518 (rank : 46) | NC score | 0.002391 (rank : 52) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43184, O60470 | Gene names | ADAM12, MLTN | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 12) (Meltrin alpha). | |||||
|
NAC1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 47) | NC score | 0.007387 (rank : 46) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70414 | Gene names | Slc8a1, Ncx | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/calcium exchanger 1 precursor (Na(+)/Ca(2+)-exchange protein 1). | |||||
|
NEK11_HUMAN
|
||||||
| θ value | 5.27518 (rank : 48) | NC score | 0.012837 (rank : 40) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1043 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8NG66, Q5JPC0, Q8NG65, Q8TBY1, Q9H5F4 | Gene names | NEK11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase Nek11 (EC 2.7.11.1) (NimA-related protein kinase 11) (Never in mitosis A-related kinase 11). | |||||
|
STEA1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 49) | NC score | 0.008172 (rank : 44) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CWR7, Q6P8X4 | Gene names | Steap1, Steap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Six transmembrane epithelial antigen of prostate 1. | |||||
|
NCOR2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 50) | NC score | 0.002084 (rank : 55) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
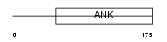
ANR10_HUMAN
|
||||||
| θ value | 8.99809 (rank : 51) | NC score | -0.000207 (rank : 58) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NXR5 | Gene names | ANKRD10 | |||
|
Domain Architecture |
|
|||||
| Description | Ankyrin repeat domain-containing protein 10. | |||||
|
BPTF_HUMAN
|
||||||
| θ value | 8.99809 (rank : 52) | NC score | 0.000170 (rank : 56) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
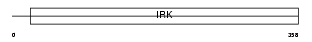
IRK4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | 0.002145 (rank : 54) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48050 | Gene names | KCNJ4 | |||
|
Domain Architecture |
|
|||||
| Description | Inward rectifier potassium channel 4 (Potassium channel, inwardly rectifying subfamily J member 4) (Inward rectifier K(+) channel Kir2.3) (Hippocampal inward rectifier) (HIR) (HRK1) (HIRK2). | |||||
|
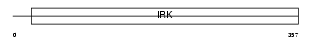
IRK4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.002147 (rank : 53) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P52189 | Gene names | Kcnj4, Irk3 | |||
|
Domain Architecture |
|
|||||
| Description | Inward rectifier potassium channel 4 (Potassium channel, inwardly rectifying subfamily J member 4) (Inward rectifier K(+) channel Kir2.3) (IRK3). | |||||
|
NAC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.006362 (rank : 47) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P32418, O95849, Q9UBL8, Q9UDN1, Q9UDN2, Q9UKX6 | Gene names | SLC8A1, CNC, NCX1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/calcium exchanger 1 precursor (Na(+)/Ca(2+)-exchange protein 1). | |||||
|
PCNT_HUMAN
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | -0.001769 (rank : 60) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | -0.001678 (rank : 59) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
ZF106_MOUSE
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.003155 (rank : 51) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O88466, O55185, O88465, O88467, Q792M1, Q792P4, Q8CDZ8, Q8R3I4, Q9ESU3 | Gene names | Zfp106, H3a, Sh3bp3, Sirm, Znf474 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 106 (Zfp-106) (Zinc finger protein 474) (H3a minor histocompatibility antigen) (Son of insulin receptor mutant). | |||||
|
ANPRC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.163886 (rank : 32) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P17342 | Gene names | NPR3, ANPRC | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide clearance receptor precursor (ANP-C) (ANPRC) (NPR-C) (Atrial natriuretic peptide C-type receptor). | |||||
|
ANPRC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.160455 (rank : 33) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70180, P97804, Q9R025, Q9R027, Q9R028 | Gene names | Npr3 | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide clearance receptor precursor (ANP-C) (ANPRC) (NPR-C) (Atrial natriuretic peptide C-type receptor) (EF-2). | |||||
|
ADCY3_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O60266, Q9UDB1 | Gene names | ADCY3, KIAA0511 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 3 (EC 4.6.1.1) (Adenylate cyclase type III) (Adenylate cyclase, olfactive type) (ATP pyrophosphate-lyase 3) (Adenylyl cyclase 3) (AC-III) (AC3). | |||||
|
ADCY3_MOUSE
|
||||||
| NC score | 0.995171 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8VHH7 | Gene names | Adcy3 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 3 (EC 4.6.1.1) (Adenylate cyclase type III) (Adenylate cyclase, olfactive type) (ATP pyrophosphate-lyase 3) (Adenylyl cyclase 3) (AC-III) (AC3). | |||||
|
ADCY8_HUMAN
|
||||||
| NC score | 0.979728 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P40145 | Gene names | ADCY8 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 8 (EC 4.6.1.1) (Adenylate cyclase type VIII) (ATP pyrophosphate-lyase 8) (Ca(2+)/calmodulin-activated adenylyl cyclase). | |||||
|
ADCY8_MOUSE
|
||||||
| NC score | 0.979628 (rank : 4) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P97490 | Gene names | Adcy8 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 8 (EC 4.6.1.1) (Adenylate cyclase type VIII) (ATP pyrophosphate-lyase 8) (Ca(2+)/calmodulin-activated adenylyl cyclase). | |||||
|
ADCY6_HUMAN
|
||||||
| NC score | 0.977983 (rank : 5) | θ value | 2.25536e-168 (rank : 5) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O43306, Q9NR75 | Gene names | ADCY6, KIAA0422 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 6 (EC 4.6.1.1) (Adenylate cyclase type VI) (ATP pyrophosphate-lyase 6) (Ca(2+)-inhibitable adenylyl cyclase). | |||||
|
ADCY6_MOUSE
|
||||||
| NC score | 0.977346 (rank : 6) | θ value | 2.25536e-168 (rank : 6) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q01341 | Gene names | Adcy6 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 6 (EC 4.6.1.1) (Adenylate cyclase type VI) (ATP pyrophosphate-lyase 6) (Ca(2+)-inhibitable adenylyl cyclase). | |||||
|
ADCY2_MOUSE
|
||||||
| NC score | 0.974533 (rank : 7) | θ value | 4.55495e-161 (rank : 7) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q80TL1, Q8CFU6 | Gene names | Adcy2, Kiaa1060 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenylate cyclase type 2 (EC 4.6.1.1) (Adenylate cyclase type II) (ATP pyrophosphate-lyase 2) (Adenylyl cyclase 2). | |||||
|
ADCY7_HUMAN
|
||||||
| NC score | 0.974282 (rank : 8) | θ value | 1.52337e-140 (rank : 10) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P51828 | Gene names | ADCY7, KIAA0037 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenylate cyclase type 7 (EC 4.6.1.1) (Adenylate cyclase type VII) (ATP pyrophosphate-lyase 7) (Adenylyl cyclase 7). | |||||
|
ADCY7_MOUSE
|
||||||
| NC score | 0.973711 (rank : 9) | θ value | 3.06949e-141 (rank : 9) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P51829 | Gene names | Adcy7 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 7 (EC 4.6.1.1) (Adenylate cyclase type VII) (ATP pyrophosphate-lyase 7) (Adenylyl cyclase 7). | |||||
|
ADCY2_HUMAN
|
||||||
| NC score | 0.968018 (rank : 10) | θ value | 1.68038e-147 (rank : 8) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q08462, Q9UPU2 | Gene names | ADCY2, KIAA1060 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 2 (EC 4.6.1.1) (Adenylate cyclase type II) (ATP pyrophosphate-lyase 2) (Adenylyl cyclase 2). | |||||
|
ADCY4_MOUSE
|
||||||
| NC score | 0.967085 (rank : 11) | θ value | 1.02182e-136 (rank : 11) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q91WF3 | Gene names | Adcy4 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 4 (EC 4.6.1.1) (Adenylate cyclase type IV) (ATP pyrophosphate-lyase 4) (Adenylyl cyclase 4). | |||||
|
ADCY4_HUMAN
|
||||||
| NC score | 0.966242 (rank : 12) | θ value | 8.9516e-133 (rank : 12) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8NFM4, Q96ML7 | Gene names | ADCY4 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 4 (EC 4.6.1.1) (Adenylate cyclase type IV) (ATP pyrophosphate-lyase 4) (Adenylyl cyclase 4). | |||||
|
ADCY1_MOUSE
|
||||||
| NC score | 0.963602 (rank : 13) | θ value | 1.14825e-79 (rank : 14) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O88444, Q5SS89 | Gene names | Adcy1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenylate cyclase type 1 (EC 4.6.1.1) (Adenylate cyclase type I) (ATP pyrophosphate-lyase 1) (Ca(2+)/calmodulin-activated adenylyl cyclase). | |||||
|
ADCY1_HUMAN
|
||||||
| NC score | 0.956498 (rank : 14) | θ value | 8.79181e-80 (rank : 13) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q08828, Q75MI1 | Gene names | ADCY1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenylate cyclase type 1 (EC 4.6.1.1) (Adenylate cyclase type I) (ATP pyrophosphate-lyase 1) (Ca(2+)/calmodulin-activated adenylyl cyclase). | |||||
|
ADCY9_HUMAN
|
||||||
| NC score | 0.907891 (rank : 15) | θ value | 1.97229e-55 (rank : 16) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O60503, O60273, Q4ZHT9, Q4ZIR5, Q9BWT4, Q9UGP2 | Gene names | ADCY9, KIAA0520 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 9 (EC 4.6.1.1) (Adenylate cyclase type IX) (ATP pyrophosphate-lyase 9) (Adenylyl cyclase 9). | |||||
|
ADCY9_MOUSE
|
||||||
| NC score | 0.907819 (rank : 16) | θ value | 1.51013e-55 (rank : 15) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P51830, Q61279 | Gene names | Adcy9 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 9 (EC 4.6.1.1) (Adenylate cyclase type IX) (ATP pyrophosphate-lyase 9) (Adenylyl cyclase 9) (Adenylyl cyclase type 10) (ACTP10). | |||||
|
GCYB2_HUMAN
|
||||||
| NC score | 0.814987 (rank : 17) | θ value | 4.01107e-24 (rank : 24) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O75343, Q9NZ64 | Gene names | GUCY1B2 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate cyclase soluble subunit beta-2 (EC 4.6.1.2) (GCS-beta-2). | |||||
|
GCYA3_HUMAN
|
||||||
| NC score | 0.795078 (rank : 18) | θ value | 4.28545e-26 (rank : 21) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q02108, O43843, Q8TAH3 | Gene names | GUCY1A3, GUC1A3, GUCSA3, GUCY1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate cyclase soluble subunit alpha-3 (EC 4.6.1.2) (GCS-alpha-3) (Soluble guanylate cyclase large subunit) (GCS-alpha-1). | |||||
|
GCYA2_HUMAN
|
||||||
| NC score | 0.794950 (rank : 19) | θ value | 1.16704e-23 (rank : 25) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P33402 | Gene names | GUCY1A2, GUC1A2, GUCSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate cyclase soluble subunit alpha-2 (EC 4.6.1.2) (GCS-alpha-2). | |||||
|
GCYA3_MOUSE
|
||||||
| NC score | 0.793173 (rank : 20) | θ value | 1.24688e-25 (rank : 22) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9ERL9, Q9DBQ3 | Gene names | Gucy1a3, Gucy1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate cyclase soluble subunit alpha-3 (EC 4.6.1.2) (GCS-alpha-3) (Soluble guanylate cyclase large subunit) (GCS-alpha-1). | |||||
|
GCYB1_MOUSE
|
||||||
| NC score | 0.784381 (rank : 21) | θ value | 1.09232e-21 (rank : 28) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O54865 | Gene names | Gucy1b3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanylate cyclase soluble subunit beta-1 (EC 4.6.1.2) (GCS-beta-1) (Soluble guanylate cyclase small subunit). | |||||
|
GCYB1_HUMAN
|
||||||
| NC score | 0.784211 (rank : 22) | θ value | 6.40375e-22 (rank : 26) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q02153, Q86WY5 | Gene names | GUCY1B3, GUC1B3, GUCSB3, GUCY1B1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate cyclase soluble subunit beta-1 (EC 4.6.1.2) (GCS-beta-1) (Soluble guanylate cyclase small subunit) (GCS-beta-3). | |||||
|
GUC2E_MOUSE
|
||||||
| NC score | 0.496878 (rank : 23) | θ value | 8.36355e-22 (rank : 27) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P52785 | Gene names | Gucy2e, Guc2e | |||
|
Domain Architecture |

|
|||||
| Description | Guanylyl cyclase GC-E precursor (EC 4.6.1.2) (Guanylate cyclase 2E). | |||||
|
GUC2D_HUMAN
|
||||||
| NC score | 0.493432 (rank : 24) | θ value | 7.82807e-20 (rank : 31) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q02846, Q6LEA7 | Gene names | GUCY2D, CORD6, GUC1A4, GUC2D, RETGC, RETGC1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinal guanylyl cyclase 1 precursor (EC 4.6.1.2) (Guanylate cyclase 2D, retinal) (RETGC-1) (Rod outer segment membrane guanylate cyclase) (ROS-GC). | |||||
|
GUC2G_MOUSE
|
||||||
| NC score | 0.467258 (rank : 25) | θ value | 3.07116e-24 (rank : 23) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6TL19, Q8BWU7 | Gene names | Gucy2g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanylate cyclase 2G precursor (EC 4.6.1.2) (Guanylyl cyclase receptor G) (mGC-G). | |||||
|
ANPRB_MOUSE
|
||||||
| NC score | 0.460433 (rank : 26) | θ value | 2.51237e-26 (rank : 20) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6VVW5, Q6VVW3, Q6VVW4, Q8CGA9 | Gene names | Npr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Atrial natriuretic peptide receptor B precursor (ANP-B) (ANPRB) (GC-B) (Guanylate cyclase B) (EC 4.6.1.2) (NPR-B) (Atrial natriuretic peptide B-type receptor). | |||||
|
ANPRA_MOUSE
|
||||||
| NC score | 0.452511 (rank : 27) | θ value | 1.12775e-26 (rank : 18) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 600 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P18293 | Gene names | Npr1, Npra | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide receptor A precursor (ANP-A) (ANPRA) (GC-A) (Guanylate cyclase) (EC 4.6.1.2) (NPR-A) (Atrial natriuretic peptide A-type receptor). | |||||
|
ANPRA_HUMAN
|
||||||
| NC score | 0.451334 (rank : 28) | θ value | 1.47289e-26 (rank : 19) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 612 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P16066, Q6P4Q3 | Gene names | NPR1, ANPRA | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide receptor A precursor (ANP-A) (ANPRA) (GC-A) (Guanylate cyclase) (EC 4.6.1.2) (NPR-A) (Atrial natriuretic peptide A-type receptor). | |||||
|
ANPRB_HUMAN
|
||||||
| NC score | 0.447701 (rank : 29) | θ value | 8.63488e-27 (rank : 17) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P20594, O60871, Q9UQ50 | Gene names | NPR2, ANPRB | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide receptor B precursor (ANP-B) (ANPRB) (GC-B) (Guanylate cyclase B) (EC 4.6.1.2) (NPR-B) (Atrial natriuretic peptide B-type receptor). | |||||
|
GUC2C_HUMAN
|
||||||
| NC score | 0.435917 (rank : 30) | θ value | 3.51386e-20 (rank : 30) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P25092 | Gene names | GUCY2C, GUC2C, STAR | |||
|
Domain Architecture |

|
|||||
| Description | Heat-stable enterotoxin receptor precursor (GC-C) (Intestinal guanylate cyclase) (EC 4.6.1.2) (STA receptor) (hSTAR). | |||||
|
GUC2F_HUMAN
|
||||||
| NC score | 0.420535 (rank : 31) | θ value | 1.2077e-20 (rank : 29) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 740 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P51841, Q9UJF1 | Gene names | GUCY2F, GUC2F, RETGC2 | |||
|
Domain Architecture |

|
|||||
| Description | Retinal guanylyl cyclase 2 precursor (EC 4.6.1.2) (Guanylate cyclase 2F, retinal) (RETGC-2) (Rod outer segment membrane guanylate cyclase 2) (ROS-GC2) (Guanylate cyclase F) (GC-F). | |||||
|
ANPRC_HUMAN
|
||||||
| NC score | 0.163886 (rank : 32) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P17342 | Gene names | NPR3, ANPRC | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide clearance receptor precursor (ANP-C) (ANPRC) (NPR-C) (Atrial natriuretic peptide C-type receptor). | |||||
|
ANPRC_MOUSE
|
||||||
| NC score | 0.160455 (rank : 33) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70180, P97804, Q9R025, Q9R027, Q9R028 | Gene names | Npr3 | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide clearance receptor precursor (ANP-C) (ANPRC) (NPR-C) (Atrial natriuretic peptide C-type receptor) (EF-2). | |||||
|
UBE4B_MOUSE
|
||||||
| NC score | 0.034602 (rank : 34) | θ value | 0.813845 (rank : 36) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ES00, Q9EQE0 | Gene names | Ube4b, Ufd2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin conjugation factor E4 B (Ubiquitin fusion degradation protein 2) (Ufd2a). | |||||
|
UBE4B_HUMAN
|
||||||
| NC score | 0.031968 (rank : 35) | θ value | 1.06291 (rank : 39) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95155, O75169, O95338, Q5SZ12, Q5SZ16, Q96QD4, Q9BYI7 | Gene names | UBE4B, HDNB1, KIAA0684, UFD2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin conjugation factor E4 B (Ubiquitin fusion degradation protein 2) (Homozygously deleted in neuroblastoma 1). | |||||
|
JHD2B_HUMAN
|
||||||
| NC score | 0.016923 (rank : 36) | θ value | 0.62314 (rank : 33) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7LBC6, Q9BVH6, Q9BW93, Q9BZ52, Q9NYF4, Q9UPS0 | Gene names | JMJD1B, C5orf7, JHDM2B, KIAA1082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2B (EC 1.14.11.-) (Jumonji domain-containing protein 1B) (Nuclear protein 5qNCA). | |||||
|
JHD2B_MOUSE
|
||||||
| NC score | 0.016832 (rank : 37) | θ value | 0.813845 (rank : 34) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZPY7, Q2VPQ5, Q5U5V7, Q6P9K3, Q8CCE2, Q8K2A5, Q9CU57 | Gene names | Jmjd1b, Jhdm2b, Kiaa1082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2B (EC 1.14.11.-) (Jumonji domain-containing protein 1B). | |||||
|
SETD2_HUMAN
|
||||||
| NC score | 0.015762 (rank : 38) | θ value | 0.813845 (rank : 35) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
NUAK2_MOUSE
|
||||||
| NC score | 0.012872 (rank : 39) | θ value | 0.163984 (rank : 32) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 878 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BZN4, Q80ZW3, Q8CIC0, Q9DBV0 | Gene names | Nuak2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NUAK family SNF1-like kinase 2 (EC 2.7.11.1). | |||||
|
NEK11_HUMAN
|
||||||
| NC score | 0.012837 (rank : 40) | θ value | 5.27518 (rank : 48) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1043 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8NG66, Q5JPC0, Q8NG65, Q8TBY1, Q9H5F4 | Gene names | NEK11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase Nek11 (EC 2.7.11.1) (NimA-related protein kinase 11) (Never in mitosis A-related kinase 11). | |||||
|
MTFR1_MOUSE
|
||||||
| NC score | 0.010947 (rank : 41) | θ value | 4.03905 (rank : 42) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99MB2, Q9CQZ4 | Gene names | Mtfr1, Kiaa0009 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial fission regulator 1. | |||||
|
PRLPM_MOUSE
|
||||||
| NC score | 0.010568 (rank : 42) | θ value | 4.03905 (rank : 44) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JHK0, Q9JI05 | Gene names | Prlpm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Placental prolactin-like protein M precursor (PRL-like protein M) (PLP-M). | |||||
|
DCX_HUMAN
|
||||||
| NC score | 0.009400 (rank : 43) | θ value | 1.38821 (rank : 40) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43602, O43911 | Gene names | DCX, DBCN, LISX | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal migration protein doublecortin (Lissencephalin-X) (Lis-X) (Doublin). | |||||
|
STEA1_MOUSE
|
||||||
| NC score | 0.008172 (rank : 44) | θ value | 5.27518 (rank : 49) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CWR7, Q6P8X4 | Gene names | Steap1, Steap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Six transmembrane epithelial antigen of prostate 1. | |||||
|
MBB1A_HUMAN
|
||||||
| NC score | 0.007651 (rank : 45) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
NAC1_MOUSE
|
||||||
| NC score | 0.007387 (rank : 46) | θ value | 5.27518 (rank : 47) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70414 | Gene names | Slc8a1, Ncx | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/calcium exchanger 1 precursor (Na(+)/Ca(2+)-exchange protein 1). | |||||
|
NAC1_HUMAN
|
||||||
| NC score | 0.006362 (rank : 47) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P32418, O95849, Q9UBL8, Q9UDN1, Q9UDN2, Q9UKX6 | Gene names | SLC8A1, CNC, NCX1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/calcium exchanger 1 precursor (Na(+)/Ca(2+)-exchange protein 1). | |||||
|
ASPN_MOUSE
|
||||||
| NC score | 0.005960 (rank : 48) | θ value | 1.06291 (rank : 37) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99MQ4, Q9D6A2 | Gene names | Aspn | |||
|
Domain Architecture |

|
|||||
| Description | Asporin precursor. | |||||
|
SI1L3_HUMAN
|
||||||
| NC score | 0.004765 (rank : 49) | θ value | 4.03905 (rank : 45) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60292, Q2TV87 | Gene names | SIPA1L3, KIAA0545, SPAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 3 (SPA-1-like protein 3). | |||||
|
GNAS1_MOUSE
|
||||||
| NC score | 0.004041 (rank : 50) | θ value | 1.06291 (rank : 38) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6R0H7, Q6R0H4, Q6R0H5, Q6R2J5, Q9JJX0, Q9Z1N8 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
ZF106_MOUSE
|
||||||
| NC score | 0.003155 (rank : 51) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O88466, O55185, O88465, O88467, Q792M1, Q792P4, Q8CDZ8, Q8R3I4, Q9ESU3 | Gene names | Zfp106, H3a, Sh3bp3, Sirm, Znf474 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 106 (Zfp-106) (Zinc finger protein 474) (H3a minor histocompatibility antigen) (Son of insulin receptor mutant). | |||||
|
ADA12_HUMAN
|
||||||
| NC score | 0.002391 (rank : 52) | θ value | 5.27518 (rank : 46) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43184, O60470 | Gene names | ADAM12, MLTN | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 12) (Meltrin alpha). | |||||
|
IRK4_MOUSE
|
||||||
| NC score | 0.002147 (rank : 53) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P52189 | Gene names | Kcnj4, Irk3 | |||
|
Domain Architecture |

|
|||||
| Description | Inward rectifier potassium channel 4 (Potassium channel, inwardly rectifying subfamily J member 4) (Inward rectifier K(+) channel Kir2.3) (IRK3). | |||||
|
IRK4_HUMAN
|
||||||
| NC score | 0.002145 (rank : 54) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48050 | Gene names | KCNJ4 | |||
|
Domain Architecture |

|
|||||
| Description | Inward rectifier potassium channel 4 (Potassium channel, inwardly rectifying subfamily J member 4) (Inward rectifier K(+) channel Kir2.3) (Hippocampal inward rectifier) (HIR) (HRK1) (HIRK2). | |||||
|
NCOR2_HUMAN
|
||||||
| NC score | 0.002084 (rank : 55) | θ value | 6.88961 (rank : 50) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
BPTF_HUMAN
|
||||||
| NC score | 0.000170 (rank : 56) | θ value | 8.99809 (rank : 52) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
PCNT_MOUSE
|
||||||
| NC score | 0.000111 (rank : 57) | θ value | 4.03905 (rank : 43) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1207 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P48725 | Gene names | Pcnt, Pcnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pericentrin. | |||||
|
ANR10_HUMAN
|
||||||
| NC score | -0.000207 (rank : 58) | θ value | 8.99809 (rank : 51) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NXR5 | Gene names | ANKRD10 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 10. | |||||
|
TARA_HUMAN
|
||||||
| NC score | -0.001678 (rank : 59) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
PCNT_HUMAN
|
||||||
| NC score | -0.001769 (rank : 60) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||