Please be patient as the page loads
|
ANPRC_MOUSE
|
||||||
| SwissProt Accessions | P70180, P97804, Q9R025, Q9R027, Q9R028 | Gene names | Npr3 | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide clearance receptor precursor (ANP-C) (ANPRC) (NPR-C) (Atrial natriuretic peptide C-type receptor) (EF-2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ANPRC_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.996091 (rank : 2) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P17342 | Gene names | NPR3, ANPRC | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide clearance receptor precursor (ANP-C) (ANPRC) (NPR-C) (Atrial natriuretic peptide C-type receptor). | |||||
|
ANPRC_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P70180, P97804, Q9R025, Q9R027, Q9R028 | Gene names | Npr3 | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide clearance receptor precursor (ANP-C) (ANPRC) (NPR-C) (Atrial natriuretic peptide C-type receptor) (EF-2). | |||||
|
ANPRA_MOUSE
|
||||||
| θ value | 7.49467e-55 (rank : 3) | NC score | 0.379782 (rank : 5) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 600 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P18293 | Gene names | Npr1, Npra | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide receptor A precursor (ANP-A) (ANPRA) (GC-A) (Guanylate cyclase) (EC 4.6.1.2) (NPR-A) (Atrial natriuretic peptide A-type receptor). | |||||
|
ANPRA_HUMAN
|
||||||
| θ value | 4.85792e-54 (rank : 4) | NC score | 0.380231 (rank : 4) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 612 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P16066, Q6P4Q3 | Gene names | NPR1, ANPRA | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide receptor A precursor (ANP-A) (ANPRA) (GC-A) (Guanylate cyclase) (EC 4.6.1.2) (NPR-A) (Atrial natriuretic peptide A-type receptor). | |||||
|
ANPRB_MOUSE
|
||||||
| θ value | 2.3352e-48 (rank : 5) | NC score | 0.386577 (rank : 3) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6VVW5, Q6VVW3, Q6VVW4, Q8CGA9 | Gene names | Npr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Atrial natriuretic peptide receptor B precursor (ANP-B) (ANPRB) (GC-B) (Guanylate cyclase B) (EC 4.6.1.2) (NPR-B) (Atrial natriuretic peptide B-type receptor). | |||||
|
ANPRB_HUMAN
|
||||||
| θ value | 6.7944e-48 (rank : 6) | NC score | 0.372398 (rank : 6) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P20594, O60871, Q9UQ50 | Gene names | NPR2, ANPRB | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide receptor B precursor (ANP-B) (ANPRB) (GC-B) (Guanylate cyclase B) (EC 4.6.1.2) (NPR-B) (Atrial natriuretic peptide B-type receptor). | |||||
|
GUC2G_MOUSE
|
||||||
| θ value | 1.99529e-15 (rank : 7) | NC score | 0.309747 (rank : 9) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6TL19, Q8BWU7 | Gene names | Gucy2g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanylate cyclase 2G precursor (EC 4.6.1.2) (Guanylyl cyclase receptor G) (mGC-G). | |||||
|
GUC2F_HUMAN
|
||||||
| θ value | 7.59969e-07 (rank : 8) | NC score | 0.244596 (rank : 18) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 740 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P51841, Q9UJF1 | Gene names | GUCY2F, GUC2F, RETGC2 | |||
|
Domain Architecture |

|
|||||
| Description | Retinal guanylyl cyclase 2 precursor (EC 4.6.1.2) (Guanylate cyclase 2F, retinal) (RETGC-2) (Rod outer segment membrane guanylate cyclase 2) (ROS-GC2) (Guanylate cyclase F) (GC-F). | |||||
|
MGR1_MOUSE
|
||||||
| θ value | 2.21117e-06 (rank : 9) | NC score | 0.157327 (rank : 25) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P97772, Q6AXG4, Q9EPV6 | Gene names | Grm1, Gprc1a, Mglur1 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 1 precursor (mGluR1). | |||||
|
MGR1_HUMAN
|
||||||
| θ value | 3.77169e-06 (rank : 10) | NC score | 0.155895 (rank : 26) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13255, Q13256, Q14757, Q14758, Q5VTF7, Q5VTF8, Q9NU10, Q9UGS9, Q9UGT0 | Gene names | GRM1, GPRC1A, MGLUR1 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 1 precursor (mGluR1). | |||||
|
GABR1_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 11) | NC score | 0.349740 (rank : 7) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9WV18, Q9WU48, Q9WV15, Q9WV16, Q9WV17 | Gene names | Gabbr1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
GABR1_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 12) | NC score | 0.343154 (rank : 8) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UBS5, O95375, O95468, O95975, O96022, Q86W60, Q9UQQ0 | Gene names | GABBR1, GPRC3A | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
GABR2_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 13) | NC score | 0.247306 (rank : 17) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O75899, O75974, O75975, Q5VXZ2, Q8WX04, Q9P1R2, Q9UNR1, Q9UNS9 | Gene names | GABBR2, GPR51, GPRC3B | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 2 precursor (GABA-B receptor 2) (GABA-B-R2) (Gb2) (GABABR2) (G-protein coupled receptor 51) (HG20). | |||||
|
MGR5_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 14) | NC score | 0.146310 (rank : 37) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P41594 | Gene names | GRM5, GPRC1E, MGLUR5 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 5 precursor (mGluR5). | |||||
|
NMDZ1_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 15) | NC score | 0.086693 (rank : 59) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q05586, P35437, Q12867, Q12868 | Gene names | GRIN1, NMDAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit zeta 1 precursor (N-methyl-D- aspartate receptor subunit NR1). | |||||
|
NMDZ1_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 16) | NC score | 0.087017 (rank : 57) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P35438, Q8CFS4 | Gene names | Grin1, Glurz1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit zeta 1 precursor (N-methyl-D- aspartate receptor subunit NR1). | |||||
|
MGR4_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 17) | NC score | 0.123566 (rank : 40) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14833, Q5SZ84 | Gene names | GRM4, GPRC1D, MGLUR4 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 4 precursor (mGluR4). | |||||
|
GUC2D_HUMAN
|
||||||
| θ value | 0.125558 (rank : 18) | NC score | 0.250806 (rank : 16) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q02846, Q6LEA7 | Gene names | GUCY2D, CORD6, GUC1A4, GUC2D, RETGC, RETGC1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinal guanylyl cyclase 1 precursor (EC 4.6.1.2) (Guanylate cyclase 2D, retinal) (RETGC-1) (Rod outer segment membrane guanylate cyclase) (ROS-GC). | |||||
|
GRIK1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 19) | NC score | 0.040299 (rank : 94) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P39086, Q13001, Q86SU9 | Gene names | GRIK1, GLUR5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 1 precursor (Glutamate receptor 5) (GluR-5) (GluR5) (Excitatory amino acid receptor 3) (EAA3). | |||||
|
TS1R2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 20) | NC score | 0.112788 (rank : 42) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q925I4, Q923J8 | Gene names | Tas1r2, Gpr71, T1r2, Tr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 2 precursor (G-protein coupled receptor 71) (Sweet taste receptor T1R2). | |||||
|
MGR6_MOUSE
|
||||||
| θ value | 2.36792 (rank : 21) | NC score | 0.107534 (rank : 45) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5NCH9 | Gene names | Grm6, Gprc1f, Mglur6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metabotropic glutamate receptor 6 precursor (mGluR6). | |||||
|
GRIK1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 22) | NC score | 0.037689 (rank : 95) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q60934 | Gene names | Grik1, Glur5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 1 precursor (Glutamate receptor 5) (GluR-5) (GluR5). | |||||
|
MGR6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 23) | NC score | 0.107027 (rank : 46) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O15303 | Gene names | GRM6, GPRC1F, MGLUR6 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 6 precursor (mGluR6). | |||||
|
GUC2E_MOUSE
|
||||||
| θ value | 4.03905 (rank : 24) | NC score | 0.241782 (rank : 19) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P52785 | Gene names | Gucy2e, Guc2e | |||
|
Domain Architecture |

|
|||||
| Description | Guanylyl cyclase GC-E precursor (EC 4.6.1.2) (Guanylate cyclase 2E). | |||||
|
MGR8_HUMAN
|
||||||
| θ value | 4.03905 (rank : 25) | NC score | 0.108421 (rank : 43) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O00222, O15493, O95945, O95946, Q3MIV9, Q52M02, Q6J165 | Gene names | GRM8, GPRC1H, MGLUR8 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 8 precursor (mGluR8). | |||||
|
MGR8_MOUSE
|
||||||
| θ value | 4.03905 (rank : 26) | NC score | 0.108274 (rank : 44) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P47743, Q6B964 | Gene names | Grm8, Gprc1h, Mglur8 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 8 precursor (mGluR8). | |||||
|
TS1R1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 27) | NC score | 0.115279 (rank : 41) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q7RTX1, Q8NGZ7, Q8TDJ7, Q8TDJ8, Q8TDJ9, Q8TDK0 | Gene names | TAS1R1, GPR70, T1R1, TR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 1 precursor (G-protein coupled receptor 70) (Gm148). | |||||
|
ADCY1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 28) | NC score | 0.154128 (rank : 29) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q08828, Q75MI1 | Gene names | ADCY1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenylate cyclase type 1 (EC 4.6.1.1) (Adenylate cyclase type I) (ATP pyrophosphate-lyase 1) (Ca(2+)/calmodulin-activated adenylyl cyclase). | |||||
|
ADCY1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 29) | NC score | 0.152746 (rank : 32) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O88444, Q5SS89 | Gene names | Adcy1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenylate cyclase type 1 (EC 4.6.1.1) (Adenylate cyclase type I) (ATP pyrophosphate-lyase 1) (Ca(2+)/calmodulin-activated adenylyl cyclase). | |||||
|
ADCY2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 30) | NC score | 0.145947 (rank : 38) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q08462, Q9UPU2 | Gene names | ADCY2, KIAA1060 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 2 (EC 4.6.1.1) (Adenylate cyclase type II) (ATP pyrophosphate-lyase 2) (Adenylyl cyclase 2). | |||||
|
ADCY2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 31) | NC score | 0.143895 (rank : 39) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80TL1, Q8CFU6 | Gene names | Adcy2, Kiaa1060 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenylate cyclase type 2 (EC 4.6.1.1) (Adenylate cyclase type II) (ATP pyrophosphate-lyase 2) (Adenylyl cyclase 2). | |||||
|
ADCY3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 32) | NC score | 0.160455 (rank : 24) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60266, Q9UDB1 | Gene names | ADCY3, KIAA0511 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 3 (EC 4.6.1.1) (Adenylate cyclase type III) (Adenylate cyclase, olfactive type) (ATP pyrophosphate-lyase 3) (Adenylyl cyclase 3) (AC-III) (AC3). | |||||
|
ADCY3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 33) | NC score | 0.162850 (rank : 23) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VHH7 | Gene names | Adcy3 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 3 (EC 4.6.1.1) (Adenylate cyclase type III) (Adenylate cyclase, olfactive type) (ATP pyrophosphate-lyase 3) (Adenylyl cyclase 3) (AC-III) (AC3). | |||||
|
ADCY4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 34) | NC score | 0.149652 (rank : 36) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NFM4, Q96ML7 | Gene names | ADCY4 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 4 (EC 4.6.1.1) (Adenylate cyclase type IV) (ATP pyrophosphate-lyase 4) (Adenylyl cyclase 4). | |||||
|
ADCY4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 35) | NC score | 0.151798 (rank : 33) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q91WF3 | Gene names | Adcy4 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 4 (EC 4.6.1.1) (Adenylate cyclase type IV) (ATP pyrophosphate-lyase 4) (Adenylyl cyclase 4). | |||||
|
ADCY6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 36) | NC score | 0.153913 (rank : 31) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43306, Q9NR75 | Gene names | ADCY6, KIAA0422 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 6 (EC 4.6.1.1) (Adenylate cyclase type VI) (ATP pyrophosphate-lyase 6) (Ca(2+)-inhibitable adenylyl cyclase). | |||||
|
ADCY6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 37) | NC score | 0.154065 (rank : 30) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q01341 | Gene names | Adcy6 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 6 (EC 4.6.1.1) (Adenylate cyclase type VI) (ATP pyrophosphate-lyase 6) (Ca(2+)-inhibitable adenylyl cyclase). | |||||
|
ADCY7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 38) | NC score | 0.150192 (rank : 35) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P51828 | Gene names | ADCY7, KIAA0037 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenylate cyclase type 7 (EC 4.6.1.1) (Adenylate cyclase type VII) (ATP pyrophosphate-lyase 7) (Adenylyl cyclase 7). | |||||
|
ADCY7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 39) | NC score | 0.150685 (rank : 34) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P51829 | Gene names | Adcy7 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 7 (EC 4.6.1.1) (Adenylate cyclase type VII) (ATP pyrophosphate-lyase 7) (Adenylyl cyclase 7). | |||||
|
ADCY8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 40) | NC score | 0.154504 (rank : 27) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P40145 | Gene names | ADCY8 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 8 (EC 4.6.1.1) (Adenylate cyclase type VIII) (ATP pyrophosphate-lyase 8) (Ca(2+)/calmodulin-activated adenylyl cyclase). | |||||
|
ADCY8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 41) | NC score | 0.154500 (rank : 28) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P97490 | Gene names | Adcy8 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 8 (EC 4.6.1.1) (Adenylate cyclase type VIII) (ATP pyrophosphate-lyase 8) (Ca(2+)/calmodulin-activated adenylyl cyclase). | |||||
|
ADCY9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 42) | NC score | 0.179721 (rank : 21) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60503, O60273, Q4ZHT9, Q4ZIR5, Q9BWT4, Q9UGP2 | Gene names | ADCY9, KIAA0520 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 9 (EC 4.6.1.1) (Adenylate cyclase type IX) (ATP pyrophosphate-lyase 9) (Adenylyl cyclase 9). | |||||
|
ADCY9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 43) | NC score | 0.179237 (rank : 22) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P51830, Q61279 | Gene names | Adcy9 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 9 (EC 4.6.1.1) (Adenylate cyclase type IX) (ATP pyrophosphate-lyase 9) (Adenylyl cyclase 9) (Adenylyl cyclase type 10) (ACTP10). | |||||
|
ANKK1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 44) | NC score | 0.054697 (rank : 83) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 1075 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NFD2 | Gene names | ANKK1, PKK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and protein kinase domain-containing protein 1 (EC 2.7.11.1) (Protein kinase PKK2) (X-kinase) (SgK288). | |||||
|
ANKK1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 45) | NC score | 0.050193 (rank : 93) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BZ25 | Gene names | Ankk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and protein kinase domain-containing protein 1 (EC 2.7.11.1). | |||||
|
CASR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 46) | NC score | 0.091967 (rank : 54) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P41180, Q13912, Q16108, Q16109, Q16110, Q16379, Q4PJ19 | Gene names | CASR, GPRC2A, PCAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular calcium-sensing receptor precursor (CaSR) (Parathyroid Cell calcium-sensing receptor). | |||||
|
CASR_MOUSE
|
||||||
| θ value | θ > 10 (rank : 47) | NC score | 0.091685 (rank : 55) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9QY96, O08968, O88519, Q9QY95, Q9QZU8, Q9R1D6, Q9R1Y2 | Gene names | Casr, Gprc2a | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular calcium-sensing receptor precursor (CaSR) (Parathyroid Cell calcium-sensing receptor). | |||||
|
GCYA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 48) | NC score | 0.260992 (rank : 11) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P33402 | Gene names | GUCY1A2, GUC1A2, GUCSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate cyclase soluble subunit alpha-2 (EC 4.6.1.2) (GCS-alpha-2). | |||||
|
GCYA3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.254069 (rank : 15) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q02108, O43843, Q8TAH3 | Gene names | GUCY1A3, GUC1A3, GUCSA3, GUCY1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate cyclase soluble subunit alpha-3 (EC 4.6.1.2) (GCS-alpha-3) (Soluble guanylate cyclase large subunit) (GCS-alpha-1). | |||||
|
GCYA3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.254607 (rank : 14) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9ERL9, Q9DBQ3 | Gene names | Gucy1a3, Gucy1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate cyclase soluble subunit alpha-3 (EC 4.6.1.2) (GCS-alpha-3) (Soluble guanylate cyclase large subunit) (GCS-alpha-1). | |||||
|
GCYB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.258534 (rank : 13) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q02153, Q86WY5 | Gene names | GUCY1B3, GUC1B3, GUCSB3, GUCY1B1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate cyclase soluble subunit beta-1 (EC 4.6.1.2) (GCS-beta-1) (Soluble guanylate cyclase small subunit) (GCS-beta-3). | |||||
|
GCYB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.258829 (rank : 12) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O54865 | Gene names | Gucy1b3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanylate cyclase soluble subunit beta-1 (EC 4.6.1.2) (GCS-beta-1) (Soluble guanylate cyclase small subunit). | |||||
|
GCYB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.284352 (rank : 10) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O75343, Q9NZ64 | Gene names | GUCY1B2 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate cyclase soluble subunit beta-2 (EC 4.6.1.2) (GCS-beta-2). | |||||
|
GP156_HUMAN
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.057407 (rank : 79) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NFN8, Q86SN6 | Gene names | GPR156, GABABL, PGR28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 156 (GABAB-related G-protein coupled receptor) (G-protein coupled receptor PGR28). | |||||
|
GPC6A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.082124 (rank : 61) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5T6X5, Q6JK43, Q6JK44, Q8NGU8, Q8NHZ9, Q8TDT6 | Gene names | GPRC6A, GPCR33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor family C group 6 member A precursor (hGPRC6A) (G-protein coupled receptor 33) (hGPCR33). | |||||
|
GPC6A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.081381 (rank : 62) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8K4Z6, Q5DK51, Q5DK52 | Gene names | Gprc6a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor family C group 6 member A precursor. | |||||
|
GUC2C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.217898 (rank : 20) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P25092 | Gene names | GUCY2C, GUC2C, STAR | |||
|
Domain Architecture |

|
|||||
| Description | Heat-stable enterotoxin receptor precursor (GC-C) (Intestinal guanylate cyclase) (EC 4.6.1.2) (STA receptor) (hSTAR). | |||||
|
ILK1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.066408 (rank : 73) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 857 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13418 | Gene names | ILK, ILK1 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin-linked protein kinase 1 (EC 2.7.11.1) (ILK-1) (59 kDa serine/threonine-protein kinase) (p59ILK). | |||||
|
ILK_MOUSE
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.066050 (rank : 74) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 858 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O55222 | Gene names | Ilk | |||
|
Domain Architecture |

|
|||||
| Description | Integrin-linked protein kinase (EC 2.7.11.1). | |||||
|
IRAK1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.053302 (rank : 89) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P51617, Q7Z5V4, Q96RL2 | Gene names | IRAK1, IRAK | |||
|
Domain Architecture |
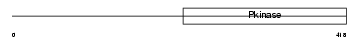
|
|||||
| Description | Interleukin-1 receptor-associated kinase 1 (EC 2.7.11.1) (IRAK-1). | |||||
|
IRAK1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.053370 (rank : 88) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62406, Q6Y3Z5, Q6Y3Z6 | Gene names | Irak1, Il1rak | |||
|
Domain Architecture |
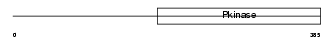
|
|||||
| Description | Interleukin-1 receptor-associated kinase 1 (EC 2.7.11.1) (IRAK-1) (IRAK) (Pelle-like protein kinase) (mPLK). | |||||
|
IRAK3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.075976 (rank : 65) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y616 | Gene names | IRAK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-1 receptor-associated kinase 3 (EC 2.7.11.1) (IRAK-3) (IL- 1 receptor-associated kinase M) (IRAK-M). | |||||
|
IRAK3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.073688 (rank : 69) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 628 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K4B2, Q8C7U8, Q8CE40, Q8K1S8 | Gene names | Irak3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-1 receptor-associated kinase 3 (EC 2.7.11.1) (IRAK-3) (IL- 1 receptor-associated kinase M) (IRAK-M). | |||||
|
IRAK4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.053473 (rank : 87) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 816 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NWZ3, Q8TDF7, Q9Y589 | Gene names | IRAK4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-1 receptor-associated kinase 4 (EC 2.7.11.1) (IRAK-4) (NY- REN-64 antigen). | |||||
|
IRAK4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.053489 (rank : 86) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R4K2, Q80WW1 | Gene names | Irak4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-1 receptor-associated kinase 4 (EC 2.7.11.1) (IRAK-4). | |||||
|
KSR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.074918 (rank : 67) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IVT5, Q13476 | Gene names | KSR1, KSR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinase suppressor of ras-1 (Kinase suppressor of ras). | |||||
|
KSR1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.074854 (rank : 68) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61097, Q61648, Q78DX8 | Gene names | Ksr1, Ksr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinase suppressor of ras-1 (Kinase suppressor of ras) (mKSR1) (Hb protein). | |||||
|
KSR2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.067970 (rank : 72) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6VAB6, Q8N775 | Gene names | KSR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinase suppressor of ras-2 (hKSR2). | |||||
|
MGR2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.098873 (rank : 52) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14416, Q52MC6, Q9H3N6 | Gene names | GRM2, GPRC1B, MGLUR2 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 2 precursor (mGluR2). | |||||
|
MGR3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.102685 (rank : 48) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q14832, Q75MV4, Q75N17, Q86YG6, Q8TBH9 | Gene names | GRM3, GPRC1C, MGLUR3 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 3 precursor (mGluR3). | |||||
|
MGR3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.105970 (rank : 47) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9QYS2 | Gene names | Grm3, Gprc1c, Mglur3 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 3 precursor (mGluR3). | |||||
|
MGR7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.099699 (rank : 51) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q14831, Q8NFS2, Q8NFS3, Q8NFS4 | Gene names | GRM7, GPRC1G, MGLUR7 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 7 precursor (mGluR7). | |||||
|
MGR7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.099753 (rank : 50) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q68ED2 | Gene names | Grm7, Gprc1g, Mglur7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metabotropic glutamate receptor 7 precursor (mGluR7). | |||||
|
MLKL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.077311 (rank : 63) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 600 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NB16, Q8N6V0 | Gene names | MLKL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mixed lineage kinase domain-like protein. | |||||
|
MLKL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.076139 (rank : 64) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 707 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D2Y4, Q8CIJ5 | Gene names | Mlkl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mixed lineage kinase domain-like protein. | |||||
|
MLTK_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.054848 (rank : 82) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 911 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NYL2, Q53SX1, Q580W8, Q59GY5, Q86YW8, Q9HCC4, Q9HCC5, Q9HDD2, Q9NYE9 | Gene names | MLTK, ZAK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase MLT (EC 2.7.11.25) (MLK-like mitogen-activated protein triple kinase) (Leucine zipper- and sterile alpha motif-containing kinase) (Sterile alpha motif- and leucine zipper-containing kinase AZK) (Mixed lineage kinase-related kinase) (MLK-related kinase) (MRK) (Cervical cancer suppressor gene 4 protein). | |||||
|
MLTK_MOUSE
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.054980 (rank : 81) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9ESL4, Q3V1X8, Q8BR73, Q9ESL3 | Gene names | Mltk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase MLT (EC 2.7.11.25) (MLK-like mitogen-activated protein triple kinase) (Leucine zipper- and sterile alpha motif kinase ZAK) (Sterile alpha motif- and leucine zipper-containing kinase AZK) (Mixed lineage kinase-related kinase) (MLK-related kinase) (MRK). | |||||
|
MOS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.050589 (rank : 92) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P00540 | Gene names | MOS | |||
|
Domain Architecture |
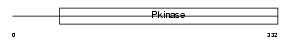
|
|||||
| Description | Proto-oncogene serine/threonine-protein kinase mos (EC 2.7.11.1) (c- mos) (Oocyte maturation factor mos). | |||||
|
RIPK1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.058528 (rank : 77) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 891 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13546, Q13180 | Gene names | RIPK1, RIP | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 1 (EC 2.7.11.1) (Serine/threonine-protein kinase RIP) (Cell death protein RIP) (Receptor-interacting protein). | |||||
|
RIPK1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.061657 (rank : 75) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q60855, Q3U0J3, Q8CD90 | Gene names | Ripk1, Rinp, Rip | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 1 (EC 2.7.11.1) (Serine/threonine-protein kinase RIP) (Cell death protein RIP) (Receptor-interacting protein). | |||||
|
RIPK2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.075690 (rank : 66) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43353, Q6UWF0 | Gene names | RIPK2, CARDIAK, RICK, RIP2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 2 (EC 2.7.11.1) (RIP-like-interacting CLARP kinase) (Receptor-interacting protein 2) (RIP-2) (CARD-containing interleukin-1 beta-converting enzyme- associated kinase) (CARD-containing IL-1 beta ICE-kinase). | |||||
|
RIPK2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.073112 (rank : 70) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 776 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P58801 | Gene names | Ripk2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
RIPK3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.058403 (rank : 78) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y572 | Gene names | RIPK3, RIP3 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 3 (EC 2.7.11.1) (RIP-like protein kinase 3) (Receptor-interacting protein 3) (RIP-3). | |||||
|
RIPK3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.059300 (rank : 76) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9QZL0 | Gene names | Ripk3, Rip3 | |||
|
Domain Architecture |
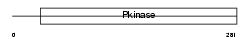
|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 3 (EC 2.7.11.1) (RIP-like protein kinase 3) (Receptor-interacting protein 3) (RIP-3) (mRIP3). | |||||
|
RIPK4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.054323 (rank : 85) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 1094 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P57078, Q96KH0 | Gene names | RIPK4, ANKRD3, DIK | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 4 (EC 2.7.11.1) (Ankyrin repeat domain protein 3) (PKC-delta-interacting protein kinase). | |||||
|
TESK2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.051443 (rank : 90) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96S53, Q8N520, Q9Y3Q6 | Gene names | TESK2 | |||
|
Domain Architecture |
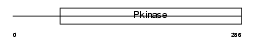
|
|||||
| Description | Dual specificity testis-specific protein kinase 2 (EC 2.7.12.1) (Testicular protein kinase 2). | |||||
|
TESK2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.051024 (rank : 91) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 833 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VCT9 | Gene names | Tesk2 | |||
|
Domain Architecture |
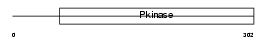
|
|||||
| Description | Dual specificity testis-specific protein kinase 2 (EC 2.7.12.1) (Testicular protein kinase 2). | |||||
|
TOPK_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.055335 (rank : 80) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 605 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96KB5, Q9NPD9, Q9NYL7, Q9NZK6 | Gene names | PBK, TOPK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-lymphokine-activated killer cell-originated protein kinase (EC 2.7.12.2) (T-LAK cell-originated protein kinase) (PDZ-binding kinase) (Spermatogenesis-related protein kinase) (SPK) (MAPKK-like protein kinase) (Nori-3). | |||||
|
TOPK_MOUSE
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.054414 (rank : 84) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JJ78, Q922V2, Q9D184 | Gene names | Pbk, Topk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-lymphokine-activated killer cell-originated protein kinase (EC 2.7.12.2) (T-LAK cell-originated protein kinase) (PDZ-binding kinase). | |||||
|
TS1R1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.086971 (rank : 58) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99PG6, Q6NS58, Q923J9, Q925I5, Q99PG5 | Gene names | Tas1r1, Gpr70, T1r1, Tr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 1 precursor (G-protein coupled receptor 70). | |||||
|
TS1R2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.093291 (rank : 53) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8TE23, Q5TZ19 | Gene names | TAS1R2, GPR71, T1R2, TR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 2 precursor (G-protein coupled receptor 71) (Sweet taste receptor T1R2). | |||||
|
TS1R3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.085439 (rank : 60) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7RTX0, Q5TA49, Q8NGW9 | Gene names | TAS1R3, T1R3, TR3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 3 precursor (Sweet taste receptor T1R3). | |||||
|
TS1R3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.101571 (rank : 49) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q925D8, Q91VA4, Q923K0, Q925A4, Q925D9 | Gene names | Tas1r3, Sac, T1r3, Tr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 3 precursor (Sweet taste receptor T1R3) (Saccharin preference protein). | |||||
|
V2R1B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.071156 (rank : 71) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6TAC4, O70409 | Gene names | V2r1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vomeronasal type-2 receptor 1b precursor (Putative pheromone receptor V2R1b). | |||||
|
V2RX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.089506 (rank : 56) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O70410 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vomeronasal type-2 receptor X precursor (Putative pheromone receptor V2RX). | |||||
|
ANPRC_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P70180, P97804, Q9R025, Q9R027, Q9R028 | Gene names | Npr3 | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide clearance receptor precursor (ANP-C) (ANPRC) (NPR-C) (Atrial natriuretic peptide C-type receptor) (EF-2). | |||||
|
ANPRC_HUMAN
|
||||||
| NC score | 0.996091 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P17342 | Gene names | NPR3, ANPRC | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide clearance receptor precursor (ANP-C) (ANPRC) (NPR-C) (Atrial natriuretic peptide C-type receptor). | |||||
|
ANPRB_MOUSE
|
||||||
| NC score | 0.386577 (rank : 3) | θ value | 2.3352e-48 (rank : 5) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6VVW5, Q6VVW3, Q6VVW4, Q8CGA9 | Gene names | Npr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Atrial natriuretic peptide receptor B precursor (ANP-B) (ANPRB) (GC-B) (Guanylate cyclase B) (EC 4.6.1.2) (NPR-B) (Atrial natriuretic peptide B-type receptor). | |||||
|
ANPRA_HUMAN
|
||||||
| NC score | 0.380231 (rank : 4) | θ value | 4.85792e-54 (rank : 4) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 612 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P16066, Q6P4Q3 | Gene names | NPR1, ANPRA | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide receptor A precursor (ANP-A) (ANPRA) (GC-A) (Guanylate cyclase) (EC 4.6.1.2) (NPR-A) (Atrial natriuretic peptide A-type receptor). | |||||
|
ANPRA_MOUSE
|
||||||
| NC score | 0.379782 (rank : 5) | θ value | 7.49467e-55 (rank : 3) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 600 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P18293 | Gene names | Npr1, Npra | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide receptor A precursor (ANP-A) (ANPRA) (GC-A) (Guanylate cyclase) (EC 4.6.1.2) (NPR-A) (Atrial natriuretic peptide A-type receptor). | |||||
|
ANPRB_HUMAN
|
||||||
| NC score | 0.372398 (rank : 6) | θ value | 6.7944e-48 (rank : 6) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P20594, O60871, Q9UQ50 | Gene names | NPR2, ANPRB | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide receptor B precursor (ANP-B) (ANPRB) (GC-B) (Guanylate cyclase B) (EC 4.6.1.2) (NPR-B) (Atrial natriuretic peptide B-type receptor). | |||||
|
GABR1_MOUSE
|
||||||
| NC score | 0.349740 (rank : 7) | θ value | 6.43352e-06 (rank : 11) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9WV18, Q9WU48, Q9WV15, Q9WV16, Q9WV17 | Gene names | Gabbr1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
GABR1_HUMAN
|
||||||
| NC score | 0.343154 (rank : 8) | θ value | 1.09739e-05 (rank : 12) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UBS5, O95375, O95468, O95975, O96022, Q86W60, Q9UQQ0 | Gene names | GABBR1, GPRC3A | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
GUC2G_MOUSE
|
||||||
| NC score | 0.309747 (rank : 9) | θ value | 1.99529e-15 (rank : 7) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6TL19, Q8BWU7 | Gene names | Gucy2g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanylate cyclase 2G precursor (EC 4.6.1.2) (Guanylyl cyclase receptor G) (mGC-G). | |||||
|
GCYB2_HUMAN
|
||||||
| NC score | 0.284352 (rank : 10) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O75343, Q9NZ64 | Gene names | GUCY1B2 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate cyclase soluble subunit beta-2 (EC 4.6.1.2) (GCS-beta-2). | |||||
|
GCYA2_HUMAN
|
||||||
| NC score | 0.260992 (rank : 11) | θ value | θ > 10 (rank : 48) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P33402 | Gene names | GUCY1A2, GUC1A2, GUCSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate cyclase soluble subunit alpha-2 (EC 4.6.1.2) (GCS-alpha-2). | |||||
|
GCYB1_MOUSE
|
||||||
| NC score | 0.258829 (rank : 12) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O54865 | Gene names | Gucy1b3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanylate cyclase soluble subunit beta-1 (EC 4.6.1.2) (GCS-beta-1) (Soluble guanylate cyclase small subunit). | |||||
|
GCYB1_HUMAN
|
||||||
| NC score | 0.258534 (rank : 13) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q02153, Q86WY5 | Gene names | GUCY1B3, GUC1B3, GUCSB3, GUCY1B1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate cyclase soluble subunit beta-1 (EC 4.6.1.2) (GCS-beta-1) (Soluble guanylate cyclase small subunit) (GCS-beta-3). | |||||
|
GCYA3_MOUSE
|
||||||
| NC score | 0.254607 (rank : 14) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9ERL9, Q9DBQ3 | Gene names | Gucy1a3, Gucy1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate cyclase soluble subunit alpha-3 (EC 4.6.1.2) (GCS-alpha-3) (Soluble guanylate cyclase large subunit) (GCS-alpha-1). | |||||
|
GCYA3_HUMAN
|
||||||
| NC score | 0.254069 (rank : 15) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q02108, O43843, Q8TAH3 | Gene names | GUCY1A3, GUC1A3, GUCSA3, GUCY1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate cyclase soluble subunit alpha-3 (EC 4.6.1.2) (GCS-alpha-3) (Soluble guanylate cyclase large subunit) (GCS-alpha-1). | |||||
|
GUC2D_HUMAN
|
||||||
| NC score | 0.250806 (rank : 16) | θ value | 0.125558 (rank : 18) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q02846, Q6LEA7 | Gene names | GUCY2D, CORD6, GUC1A4, GUC2D, RETGC, RETGC1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinal guanylyl cyclase 1 precursor (EC 4.6.1.2) (Guanylate cyclase 2D, retinal) (RETGC-1) (Rod outer segment membrane guanylate cyclase) (ROS-GC). | |||||
|
GABR2_HUMAN
|
||||||
| NC score | 0.247306 (rank : 17) | θ value | 3.19293e-05 (rank : 13) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O75899, O75974, O75975, Q5VXZ2, Q8WX04, Q9P1R2, Q9UNR1, Q9UNS9 | Gene names | GABBR2, GPR51, GPRC3B | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 2 precursor (GABA-B receptor 2) (GABA-B-R2) (Gb2) (GABABR2) (G-protein coupled receptor 51) (HG20). | |||||
|
GUC2F_HUMAN
|
||||||
| NC score | 0.244596 (rank : 18) | θ value | 7.59969e-07 (rank : 8) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 740 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P51841, Q9UJF1 | Gene names | GUCY2F, GUC2F, RETGC2 | |||
|
Domain Architecture |

|
|||||
| Description | Retinal guanylyl cyclase 2 precursor (EC 4.6.1.2) (Guanylate cyclase 2F, retinal) (RETGC-2) (Rod outer segment membrane guanylate cyclase 2) (ROS-GC2) (Guanylate cyclase F) (GC-F). | |||||
|
GUC2E_MOUSE
|
||||||
| NC score | 0.241782 (rank : 19) | θ value | 4.03905 (rank : 24) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P52785 | Gene names | Gucy2e, Guc2e | |||
|
Domain Architecture |

|
|||||
| Description | Guanylyl cyclase GC-E precursor (EC 4.6.1.2) (Guanylate cyclase 2E). | |||||
|
GUC2C_HUMAN
|
||||||
| NC score | 0.217898 (rank : 20) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P25092 | Gene names | GUCY2C, GUC2C, STAR | |||
|
Domain Architecture |

|
|||||
| Description | Heat-stable enterotoxin receptor precursor (GC-C) (Intestinal guanylate cyclase) (EC 4.6.1.2) (STA receptor) (hSTAR). | |||||
|
ADCY9_HUMAN
|
||||||
| NC score | 0.179721 (rank : 21) | θ value | θ > 10 (rank : 42) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60503, O60273, Q4ZHT9, Q4ZIR5, Q9BWT4, Q9UGP2 | Gene names | ADCY9, KIAA0520 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 9 (EC 4.6.1.1) (Adenylate cyclase type IX) (ATP pyrophosphate-lyase 9) (Adenylyl cyclase 9). | |||||
|
ADCY9_MOUSE
|
||||||
| NC score | 0.179237 (rank : 22) | θ value | θ > 10 (rank : 43) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P51830, Q61279 | Gene names | Adcy9 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 9 (EC 4.6.1.1) (Adenylate cyclase type IX) (ATP pyrophosphate-lyase 9) (Adenylyl cyclase 9) (Adenylyl cyclase type 10) (ACTP10). | |||||
|
ADCY3_MOUSE
|
||||||
| NC score | 0.162850 (rank : 23) | θ value | θ > 10 (rank : 33) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VHH7 | Gene names | Adcy3 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 3 (EC 4.6.1.1) (Adenylate cyclase type III) (Adenylate cyclase, olfactive type) (ATP pyrophosphate-lyase 3) (Adenylyl cyclase 3) (AC-III) (AC3). | |||||
|
ADCY3_HUMAN
|
||||||
| NC score | 0.160455 (rank : 24) | θ value | θ > 10 (rank : 32) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60266, Q9UDB1 | Gene names | ADCY3, KIAA0511 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 3 (EC 4.6.1.1) (Adenylate cyclase type III) (Adenylate cyclase, olfactive type) (ATP pyrophosphate-lyase 3) (Adenylyl cyclase 3) (AC-III) (AC3). | |||||
|
MGR1_MOUSE
|
||||||
| NC score | 0.157327 (rank : 25) | θ value | 2.21117e-06 (rank : 9) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P97772, Q6AXG4, Q9EPV6 | Gene names | Grm1, Gprc1a, Mglur1 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 1 precursor (mGluR1). | |||||
|
MGR1_HUMAN
|
||||||
| NC score | 0.155895 (rank : 26) | θ value | 3.77169e-06 (rank : 10) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13255, Q13256, Q14757, Q14758, Q5VTF7, Q5VTF8, Q9NU10, Q9UGS9, Q9UGT0 | Gene names | GRM1, GPRC1A, MGLUR1 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 1 precursor (mGluR1). | |||||
|
ADCY8_HUMAN
|
||||||
| NC score | 0.154504 (rank : 27) | θ value | θ > 10 (rank : 40) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P40145 | Gene names | ADCY8 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 8 (EC 4.6.1.1) (Adenylate cyclase type VIII) (ATP pyrophosphate-lyase 8) (Ca(2+)/calmodulin-activated adenylyl cyclase). | |||||
|
ADCY8_MOUSE
|
||||||
| NC score | 0.154500 (rank : 28) | θ value | θ > 10 (rank : 41) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P97490 | Gene names | Adcy8 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 8 (EC 4.6.1.1) (Adenylate cyclase type VIII) (ATP pyrophosphate-lyase 8) (Ca(2+)/calmodulin-activated adenylyl cyclase). | |||||
|
ADCY1_HUMAN
|
||||||
| NC score | 0.154128 (rank : 29) | θ value | θ > 10 (rank : 28) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q08828, Q75MI1 | Gene names | ADCY1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenylate cyclase type 1 (EC 4.6.1.1) (Adenylate cyclase type I) (ATP pyrophosphate-lyase 1) (Ca(2+)/calmodulin-activated adenylyl cyclase). | |||||
|
ADCY6_MOUSE
|
||||||
| NC score | 0.154065 (rank : 30) | θ value | θ > 10 (rank : 37) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q01341 | Gene names | Adcy6 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 6 (EC 4.6.1.1) (Adenylate cyclase type VI) (ATP pyrophosphate-lyase 6) (Ca(2+)-inhibitable adenylyl cyclase). | |||||
|
ADCY6_HUMAN
|
||||||
| NC score | 0.153913 (rank : 31) | θ value | θ > 10 (rank : 36) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43306, Q9NR75 | Gene names | ADCY6, KIAA0422 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 6 (EC 4.6.1.1) (Adenylate cyclase type VI) (ATP pyrophosphate-lyase 6) (Ca(2+)-inhibitable adenylyl cyclase). | |||||
|
ADCY1_MOUSE
|
||||||
| NC score | 0.152746 (rank : 32) | θ value | θ > 10 (rank : 29) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O88444, Q5SS89 | Gene names | Adcy1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenylate cyclase type 1 (EC 4.6.1.1) (Adenylate cyclase type I) (ATP pyrophosphate-lyase 1) (Ca(2+)/calmodulin-activated adenylyl cyclase). | |||||
|
ADCY4_MOUSE
|
||||||
| NC score | 0.151798 (rank : 33) | θ value | θ > 10 (rank : 35) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q91WF3 | Gene names | Adcy4 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 4 (EC 4.6.1.1) (Adenylate cyclase type IV) (ATP pyrophosphate-lyase 4) (Adenylyl cyclase 4). | |||||
|
ADCY7_MOUSE
|
||||||
| NC score | 0.150685 (rank : 34) | θ value | θ > 10 (rank : 39) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P51829 | Gene names | Adcy7 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 7 (EC 4.6.1.1) (Adenylate cyclase type VII) (ATP pyrophosphate-lyase 7) (Adenylyl cyclase 7). | |||||
|
ADCY7_HUMAN
|
||||||
| NC score | 0.150192 (rank : 35) | θ value | θ > 10 (rank : 38) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P51828 | Gene names | ADCY7, KIAA0037 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenylate cyclase type 7 (EC 4.6.1.1) (Adenylate cyclase type VII) (ATP pyrophosphate-lyase 7) (Adenylyl cyclase 7). | |||||
|
ADCY4_HUMAN
|
||||||
| NC score | 0.149652 (rank : 36) | θ value | θ > 10 (rank : 34) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NFM4, Q96ML7 | Gene names | ADCY4 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 4 (EC 4.6.1.1) (Adenylate cyclase type IV) (ATP pyrophosphate-lyase 4) (Adenylyl cyclase 4). | |||||
|
MGR5_HUMAN
|
||||||
| NC score | 0.146310 (rank : 37) | θ value | 0.000158464 (rank : 14) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P41594 | Gene names | GRM5, GPRC1E, MGLUR5 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 5 precursor (mGluR5). | |||||
|
ADCY2_HUMAN
|
||||||
| NC score | 0.145947 (rank : 38) | θ value | θ > 10 (rank : 30) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q08462, Q9UPU2 | Gene names | ADCY2, KIAA1060 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 2 (EC 4.6.1.1) (Adenylate cyclase type II) (ATP pyrophosphate-lyase 2) (Adenylyl cyclase 2). | |||||
|
ADCY2_MOUSE
|
||||||
| NC score | 0.143895 (rank : 39) | θ value | θ > 10 (rank : 31) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80TL1, Q8CFU6 | Gene names | Adcy2, Kiaa1060 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenylate cyclase type 2 (EC 4.6.1.1) (Adenylate cyclase type II) (ATP pyrophosphate-lyase 2) (Adenylyl cyclase 2). | |||||
|
MGR4_HUMAN
|
||||||
| NC score | 0.123566 (rank : 40) | θ value | 0.0736092 (rank : 17) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14833, Q5SZ84 | Gene names | GRM4, GPRC1D, MGLUR4 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 4 precursor (mGluR4). | |||||
|
TS1R1_HUMAN
|
||||||
| NC score | 0.115279 (rank : 41) | θ value | 4.03905 (rank : 27) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q7RTX1, Q8NGZ7, Q8TDJ7, Q8TDJ8, Q8TDJ9, Q8TDK0 | Gene names | TAS1R1, GPR70, T1R1, TR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 1 precursor (G-protein coupled receptor 70) (Gm148). | |||||
|
TS1R2_MOUSE
|
||||||
| NC score | 0.112788 (rank : 42) | θ value | 1.38821 (rank : 20) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q925I4, Q923J8 | Gene names | Tas1r2, Gpr71, T1r2, Tr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 2 precursor (G-protein coupled receptor 71) (Sweet taste receptor T1R2). | |||||
|
MGR8_HUMAN
|
||||||
| NC score | 0.108421 (rank : 43) | θ value | 4.03905 (rank : 25) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O00222, O15493, O95945, O95946, Q3MIV9, Q52M02, Q6J165 | Gene names | GRM8, GPRC1H, MGLUR8 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 8 precursor (mGluR8). | |||||
|
MGR8_MOUSE
|
||||||
| NC score | 0.108274 (rank : 44) | θ value | 4.03905 (rank : 26) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P47743, Q6B964 | Gene names | Grm8, Gprc1h, Mglur8 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 8 precursor (mGluR8). | |||||
|
MGR6_MOUSE
|
||||||
| NC score | 0.107534 (rank : 45) | θ value | 2.36792 (rank : 21) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5NCH9 | Gene names | Grm6, Gprc1f, Mglur6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metabotropic glutamate receptor 6 precursor (mGluR6). | |||||
|
MGR6_HUMAN
|
||||||
| NC score | 0.107027 (rank : 46) | θ value | 3.0926 (rank : 23) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O15303 | Gene names | GRM6, GPRC1F, MGLUR6 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 6 precursor (mGluR6). | |||||
|
MGR3_MOUSE
|
||||||
| NC score | 0.105970 (rank : 47) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9QYS2 | Gene names | Grm3, Gprc1c, Mglur3 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 3 precursor (mGluR3). | |||||
|
MGR3_HUMAN
|
||||||
| NC score | 0.102685 (rank : 48) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q14832, Q75MV4, Q75N17, Q86YG6, Q8TBH9 | Gene names | GRM3, GPRC1C, MGLUR3 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 3 precursor (mGluR3). | |||||
|
TS1R3_MOUSE
|
||||||
| NC score | 0.101571 (rank : 49) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q925D8, Q91VA4, Q923K0, Q925A4, Q925D9 | Gene names | Tas1r3, Sac, T1r3, Tr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 3 precursor (Sweet taste receptor T1R3) (Saccharin preference protein). | |||||
|
MGR7_MOUSE
|
||||||
| NC score | 0.099753 (rank : 50) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q68ED2 | Gene names | Grm7, Gprc1g, Mglur7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metabotropic glutamate receptor 7 precursor (mGluR7). | |||||
|
MGR7_HUMAN
|
||||||
| NC score | 0.099699 (rank : 51) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q14831, Q8NFS2, Q8NFS3, Q8NFS4 | Gene names | GRM7, GPRC1G, MGLUR7 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 7 precursor (mGluR7). | |||||
|
MGR2_HUMAN
|
||||||
| NC score | 0.098873 (rank : 52) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14416, Q52MC6, Q9H3N6 | Gene names | GRM2, GPRC1B, MGLUR2 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 2 precursor (mGluR2). | |||||
|
TS1R2_HUMAN
|
||||||
| NC score | 0.093291 (rank : 53) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8TE23, Q5TZ19 | Gene names | TAS1R2, GPR71, T1R2, TR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 2 precursor (G-protein coupled receptor 71) (Sweet taste receptor T1R2). | |||||
|
CASR_HUMAN
|
||||||
| NC score | 0.091967 (rank : 54) | θ value | θ > 10 (rank : 46) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P41180, Q13912, Q16108, Q16109, Q16110, Q16379, Q4PJ19 | Gene names | CASR, GPRC2A, PCAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular calcium-sensing receptor precursor (CaSR) (Parathyroid Cell calcium-sensing receptor). | |||||
|
CASR_MOUSE
|
||||||
| NC score | 0.091685 (rank : 55) | θ value | θ > 10 (rank : 47) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9QY96, O08968, O88519, Q9QY95, Q9QZU8, Q9R1D6, Q9R1Y2 | Gene names | Casr, Gprc2a | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular calcium-sensing receptor precursor (CaSR) (Parathyroid Cell calcium-sensing receptor). | |||||
|
V2RX_MOUSE
|
||||||
| NC score | 0.089506 (rank : 56) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O70410 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vomeronasal type-2 receptor X precursor (Putative pheromone receptor V2RX). | |||||
|
NMDZ1_MOUSE
|
||||||
| NC score | 0.087017 (rank : 57) | θ value | 0.00869519 (rank : 16) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P35438, Q8CFS4 | Gene names | Grin1, Glurz1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit zeta 1 precursor (N-methyl-D- aspartate receptor subunit NR1). | |||||
|
TS1R1_MOUSE
|
||||||
| NC score | 0.086971 (rank : 58) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99PG6, Q6NS58, Q923J9, Q925I5, Q99PG5 | Gene names | Tas1r1, Gpr70, T1r1, Tr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 1 precursor (G-protein coupled receptor 70). | |||||
|
NMDZ1_HUMAN
|
||||||
| NC score | 0.086693 (rank : 59) | θ value | 0.00869519 (rank : 15) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q05586, P35437, Q12867, Q12868 | Gene names | GRIN1, NMDAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit zeta 1 precursor (N-methyl-D- aspartate receptor subunit NR1). | |||||
|
TS1R3_HUMAN
|
||||||
| NC score | 0.085439 (rank : 60) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7RTX0, Q5TA49, Q8NGW9 | Gene names | TAS1R3, T1R3, TR3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 3 precursor (Sweet taste receptor T1R3). | |||||
|
GPC6A_HUMAN
|
||||||
| NC score | 0.082124 (rank : 61) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5T6X5, Q6JK43, Q6JK44, Q8NGU8, Q8NHZ9, Q8TDT6 | Gene names | GPRC6A, GPCR33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor family C group 6 member A precursor (hGPRC6A) (G-protein coupled receptor 33) (hGPCR33). | |||||
|
GPC6A_MOUSE
|
||||||
| NC score | 0.081381 (rank : 62) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8K4Z6, Q5DK51, Q5DK52 | Gene names | Gprc6a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor family C group 6 member A precursor. | |||||
|
MLKL_HUMAN
|
||||||
| NC score | 0.077311 (rank : 63) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 600 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NB16, Q8N6V0 | Gene names | MLKL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mixed lineage kinase domain-like protein. | |||||
|
MLKL_MOUSE
|
||||||
| NC score | 0.076139 (rank : 64) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 707 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D2Y4, Q8CIJ5 | Gene names | Mlkl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mixed lineage kinase domain-like protein. | |||||
|
IRAK3_HUMAN
|
||||||
| NC score | 0.075976 (rank : 65) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y616 | Gene names | IRAK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-1 receptor-associated kinase 3 (EC 2.7.11.1) (IRAK-3) (IL- 1 receptor-associated kinase M) (IRAK-M). | |||||
|
RIPK2_HUMAN
|
||||||
| NC score | 0.075690 (rank : 66) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43353, Q6UWF0 | Gene names | RIPK2, CARDIAK, RICK, RIP2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 2 (EC 2.7.11.1) (RIP-like-interacting CLARP kinase) (Receptor-interacting protein 2) (RIP-2) (CARD-containing interleukin-1 beta-converting enzyme- associated kinase) (CARD-containing IL-1 beta ICE-kinase). | |||||
|
KSR1_HUMAN
|
||||||
| NC score | 0.074918 (rank : 67) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IVT5, Q13476 | Gene names | KSR1, KSR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinase suppressor of ras-1 (Kinase suppressor of ras). | |||||
|
KSR1_MOUSE
|
||||||
| NC score | 0.074854 (rank : 68) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61097, Q61648, Q78DX8 | Gene names | Ksr1, Ksr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinase suppressor of ras-1 (Kinase suppressor of ras) (mKSR1) (Hb protein). | |||||
|
IRAK3_MOUSE
|
||||||
| NC score | 0.073688 (rank : 69) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 628 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K4B2, Q8C7U8, Q8CE40, Q8K1S8 | Gene names | Irak3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-1 receptor-associated kinase 3 (EC 2.7.11.1) (IRAK-3) (IL- 1 receptor-associated kinase M) (IRAK-M). | |||||
|
RIPK2_MOUSE
|
||||||
| NC score | 0.073112 (rank : 70) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 776 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P58801 | Gene names | Ripk2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
V2R1B_MOUSE
|
||||||
| NC score | 0.071156 (rank : 71) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6TAC4, O70409 | Gene names | V2r1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vomeronasal type-2 receptor 1b precursor (Putative pheromone receptor V2R1b). | |||||
|
KSR2_HUMAN
|
||||||
| NC score | 0.067970 (rank : 72) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6VAB6, Q8N775 | Gene names | KSR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinase suppressor of ras-2 (hKSR2). | |||||
|
ILK1_HUMAN
|
||||||
| NC score | 0.066408 (rank : 73) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 857 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13418 | Gene names | ILK, ILK1 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin-linked protein kinase 1 (EC 2.7.11.1) (ILK-1) (59 kDa serine/threonine-protein kinase) (p59ILK). | |||||
|
ILK_MOUSE
|
||||||
| NC score | 0.066050 (rank : 74) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 858 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O55222 | Gene names | Ilk | |||
|
Domain Architecture |

|
|||||
| Description | Integrin-linked protein kinase (EC 2.7.11.1). | |||||
|
RIPK1_MOUSE
|
||||||
| NC score | 0.061657 (rank : 75) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q60855, Q3U0J3, Q8CD90 | Gene names | Ripk1, Rinp, Rip | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 1 (EC 2.7.11.1) (Serine/threonine-protein kinase RIP) (Cell death protein RIP) (Receptor-interacting protein). | |||||
|
RIPK3_MOUSE
|
||||||
| NC score | 0.059300 (rank : 76) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9QZL0 | Gene names | Ripk3, Rip3 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 3 (EC 2.7.11.1) (RIP-like protein kinase 3) (Receptor-interacting protein 3) (RIP-3) (mRIP3). | |||||
|
RIPK1_HUMAN
|
||||||
| NC score | 0.058528 (rank : 77) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 891 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13546, Q13180 | Gene names | RIPK1, RIP | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 1 (EC 2.7.11.1) (Serine/threonine-protein kinase RIP) (Cell death protein RIP) (Receptor-interacting protein). | |||||
|
RIPK3_HUMAN
|
||||||
| NC score | 0.058403 (rank : 78) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y572 | Gene names | RIPK3, RIP3 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 3 (EC 2.7.11.1) (RIP-like protein kinase 3) (Receptor-interacting protein 3) (RIP-3). | |||||
|
GP156_HUMAN
|
||||||
| NC score | 0.057407 (rank : 79) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NFN8, Q86SN6 | Gene names | GPR156, GABABL, PGR28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 156 (GABAB-related G-protein coupled receptor) (G-protein coupled receptor PGR28). | |||||
|
TOPK_HUMAN
|
||||||
| NC score | 0.055335 (rank : 80) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 605 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96KB5, Q9NPD9, Q9NYL7, Q9NZK6 | Gene names | PBK, TOPK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-lymphokine-activated killer cell-originated protein kinase (EC 2.7.12.2) (T-LAK cell-originated protein kinase) (PDZ-binding kinase) (Spermatogenesis-related protein kinase) (SPK) (MAPKK-like protein kinase) (Nori-3). | |||||
|
MLTK_MOUSE
|
||||||
| NC score | 0.054980 (rank : 81) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9ESL4, Q3V1X8, Q8BR73, Q9ESL3 | Gene names | Mltk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase MLT (EC 2.7.11.25) (MLK-like mitogen-activated protein triple kinase) (Leucine zipper- and sterile alpha motif kinase ZAK) (Sterile alpha motif- and leucine zipper-containing kinase AZK) (Mixed lineage kinase-related kinase) (MLK-related kinase) (MRK). | |||||
|
MLTK_HUMAN
|
||||||
| NC score | 0.054848 (rank : 82) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 911 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NYL2, Q53SX1, Q580W8, Q59GY5, Q86YW8, Q9HCC4, Q9HCC5, Q9HDD2, Q9NYE9 | Gene names | MLTK, ZAK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase MLT (EC 2.7.11.25) (MLK-like mitogen-activated protein triple kinase) (Leucine zipper- and sterile alpha motif-containing kinase) (Sterile alpha motif- and leucine zipper-containing kinase AZK) (Mixed lineage kinase-related kinase) (MLK-related kinase) (MRK) (Cervical cancer suppressor gene 4 protein). | |||||
|
ANKK1_HUMAN
|
||||||
| NC score | 0.054697 (rank : 83) | θ value | θ > 10 (rank : 44) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 1075 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NFD2 | Gene names | ANKK1, PKK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and protein kinase domain-containing protein 1 (EC 2.7.11.1) (Protein kinase PKK2) (X-kinase) (SgK288). | |||||
|
TOPK_MOUSE
|
||||||
| NC score | 0.054414 (rank : 84) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JJ78, Q922V2, Q9D184 | Gene names | Pbk, Topk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-lymphokine-activated killer cell-originated protein kinase (EC 2.7.12.2) (T-LAK cell-originated protein kinase) (PDZ-binding kinase). | |||||
|
RIPK4_HUMAN
|
||||||
| NC score | 0.054323 (rank : 85) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 1094 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P57078, Q96KH0 | Gene names | RIPK4, ANKRD3, DIK | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 4 (EC 2.7.11.1) (Ankyrin repeat domain protein 3) (PKC-delta-interacting protein kinase). | |||||
|
IRAK4_MOUSE
|
||||||
| NC score | 0.053489 (rank : 86) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R4K2, Q80WW1 | Gene names | Irak4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-1 receptor-associated kinase 4 (EC 2.7.11.1) (IRAK-4). | |||||
|
IRAK4_HUMAN
|
||||||
| NC score | 0.053473 (rank : 87) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 816 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NWZ3, Q8TDF7, Q9Y589 | Gene names | IRAK4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-1 receptor-associated kinase 4 (EC 2.7.11.1) (IRAK-4) (NY- REN-64 antigen). | |||||
|
IRAK1_MOUSE
|
||||||
| NC score | 0.053370 (rank : 88) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62406, Q6Y3Z5, Q6Y3Z6 | Gene names | Irak1, Il1rak | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-1 receptor-associated kinase 1 (EC 2.7.11.1) (IRAK-1) (IRAK) (Pelle-like protein kinase) (mPLK). | |||||
|
IRAK1_HUMAN
|
||||||
| NC score | 0.053302 (rank : 89) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P51617, Q7Z5V4, Q96RL2 | Gene names | IRAK1, IRAK | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-1 receptor-associated kinase 1 (EC 2.7.11.1) (IRAK-1). | |||||
|
TESK2_HUMAN
|
||||||
| NC score | 0.051443 (rank : 90) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96S53, Q8N520, Q9Y3Q6 | Gene names | TESK2 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity testis-specific protein kinase 2 (EC 2.7.12.1) (Testicular protein kinase 2). | |||||
|
TESK2_MOUSE
|
||||||
| NC score | 0.051024 (rank : 91) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 833 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VCT9 | Gene names | Tesk2 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity testis-specific protein kinase 2 (EC 2.7.12.1) (Testicular protein kinase 2). | |||||
|
MOS_HUMAN
|
||||||
| NC score | 0.050589 (rank : 92) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P00540 | Gene names | MOS | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene serine/threonine-protein kinase mos (EC 2.7.11.1) (c- mos) (Oocyte maturation factor mos). | |||||
|
ANKK1_MOUSE
|
||||||
| NC score | 0.050193 (rank : 93) | θ value | θ > 10 (rank : 45) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BZ25 | Gene names | Ankk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and protein kinase domain-containing protein 1 (EC 2.7.11.1). | |||||
|
GRIK1_HUMAN
|
||||||
| NC score | 0.040299 (rank : 94) | θ value | 1.06291 (rank : 19) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P39086, Q13001, Q86SU9 | Gene names | GRIK1, GLUR5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 1 precursor (Glutamate receptor 5) (GluR-5) (GluR5) (Excitatory amino acid receptor 3) (EAA3). | |||||
|
GRIK1_MOUSE
|
||||||
| NC score | 0.037689 (rank : 95) | θ value | 3.0926 (rank : 22) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q60934 | Gene names | Grik1, Glur5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 1 precursor (Glutamate receptor 5) (GluR-5) (GluR5). | |||||