Please be patient as the page loads
|
MGR1_MOUSE
|
||||||
| SwissProt Accessions | P97772, Q6AXG4, Q9EPV6 | Gene names | Grm1, Gprc1a, Mglur1 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 1 precursor (mGluR1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MGR1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.997411 (rank : 2) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q13255, Q13256, Q14757, Q14758, Q5VTF7, Q5VTF8, Q9NU10, Q9UGS9, Q9UGT0 | Gene names | GRM1, GPRC1A, MGLUR1 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 1 precursor (mGluR1). | |||||
|
MGR1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | P97772, Q6AXG4, Q9EPV6 | Gene names | Grm1, Gprc1a, Mglur1 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 1 precursor (mGluR1). | |||||
|
MGR2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.979508 (rank : 4) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q14416, Q52MC6, Q9H3N6 | Gene names | GRM2, GPRC1B, MGLUR2 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 2 precursor (mGluR2). | |||||
|
MGR5_HUMAN
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.992949 (rank : 3) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P41594 | Gene names | GRM5, GPRC1E, MGLUR5 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 5 precursor (mGluR5). | |||||
|
MGR3_HUMAN
|
||||||
| θ value | 8.53067e-184 (rank : 5) | NC score | 0.976398 (rank : 5) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q14832, Q75MV4, Q75N17, Q86YG6, Q8TBH9 | Gene names | GRM3, GPRC1C, MGLUR3 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 3 precursor (mGluR3). | |||||
|
MGR3_MOUSE
|
||||||
| θ value | 1.11414e-183 (rank : 6) | NC score | 0.976015 (rank : 6) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9QYS2 | Gene names | Grm3, Gprc1c, Mglur3 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 3 precursor (mGluR3). | |||||
|
MGR4_HUMAN
|
||||||
| θ value | 4.1007e-178 (rank : 7) | NC score | 0.975513 (rank : 7) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q14833, Q5SZ84 | Gene names | GRM4, GPRC1D, MGLUR4 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 4 precursor (mGluR4). | |||||
|
MGR8_HUMAN
|
||||||
| θ value | 5.01279e-176 (rank : 8) | NC score | 0.973701 (rank : 10) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O00222, O15493, O95945, O95946, Q3MIV9, Q52M02, Q6J165 | Gene names | GRM8, GPRC1H, MGLUR8 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 8 precursor (mGluR8). | |||||
|
MGR7_MOUSE
|
||||||
| θ value | 1.11673e-175 (rank : 9) | NC score | 0.972045 (rank : 13) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q68ED2 | Gene names | Grm7, Gprc1g, Mglur7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metabotropic glutamate receptor 7 precursor (mGluR7). | |||||
|
MGR8_MOUSE
|
||||||
| θ value | 1.11673e-175 (rank : 10) | NC score | 0.973737 (rank : 9) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P47743, Q6B964 | Gene names | Grm8, Gprc1h, Mglur8 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 8 precursor (mGluR8). | |||||
|
MGR7_HUMAN
|
||||||
| θ value | 1.4585e-175 (rank : 11) | NC score | 0.972153 (rank : 12) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q14831, Q8NFS2, Q8NFS3, Q8NFS4 | Gene names | GRM7, GPRC1G, MGLUR7 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 7 precursor (mGluR7). | |||||
|
MGR6_MOUSE
|
||||||
| θ value | 6.77497e-173 (rank : 12) | NC score | 0.972546 (rank : 11) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q5NCH9 | Gene names | Grm6, Gprc1f, Mglur6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metabotropic glutamate receptor 6 precursor (mGluR6). | |||||
|
MGR6_HUMAN
|
||||||
| θ value | 5.73535e-172 (rank : 13) | NC score | 0.973800 (rank : 8) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O15303 | Gene names | GRM6, GPRC1F, MGLUR6 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 6 precursor (mGluR6). | |||||
|
CASR_HUMAN
|
||||||
| θ value | 9.34977e-98 (rank : 14) | NC score | 0.943547 (rank : 14) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P41180, Q13912, Q16108, Q16109, Q16110, Q16379, Q4PJ19 | Gene names | CASR, GPRC2A, PCAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular calcium-sensing receptor precursor (CaSR) (Parathyroid Cell calcium-sensing receptor). | |||||
|
CASR_MOUSE
|
||||||
| θ value | 3.55291e-97 (rank : 15) | NC score | 0.943510 (rank : 15) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9QY96, O08968, O88519, Q9QY95, Q9QZU8, Q9R1D6, Q9R1Y2 | Gene names | Casr, Gprc2a | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular calcium-sensing receptor precursor (CaSR) (Parathyroid Cell calcium-sensing receptor). | |||||
|
V2RX_MOUSE
|
||||||
| θ value | 2.47765e-74 (rank : 16) | NC score | 0.933672 (rank : 16) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O70410 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vomeronasal type-2 receptor X precursor (Putative pheromone receptor V2RX). | |||||
|
TS1R1_MOUSE
|
||||||
| θ value | 1.2325e-65 (rank : 17) | NC score | 0.906602 (rank : 21) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q99PG6, Q6NS58, Q923J9, Q925I5, Q99PG5 | Gene names | Tas1r1, Gpr70, T1r1, Tr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 1 precursor (G-protein coupled receptor 70). | |||||
|
TS1R1_HUMAN
|
||||||
| θ value | 2.32439e-64 (rank : 18) | NC score | 0.906061 (rank : 22) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q7RTX1, Q8NGZ7, Q8TDJ7, Q8TDJ8, Q8TDJ9, Q8TDK0 | Gene names | TAS1R1, GPR70, T1R1, TR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 1 precursor (G-protein coupled receptor 70) (Gm148). | |||||
|
GPC6A_HUMAN
|
||||||
| θ value | 4.38363e-63 (rank : 19) | NC score | 0.915073 (rank : 18) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q5T6X5, Q6JK43, Q6JK44, Q8NGU8, Q8NHZ9, Q8TDT6 | Gene names | GPRC6A, GPCR33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor family C group 6 member A precursor (hGPRC6A) (G-protein coupled receptor 33) (hGPCR33). | |||||
|
GPC6A_MOUSE
|
||||||
| θ value | 2.84138e-62 (rank : 20) | NC score | 0.913057 (rank : 19) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8K4Z6, Q5DK51, Q5DK52 | Gene names | Gprc6a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor family C group 6 member A precursor. | |||||
|
TS1R2_HUMAN
|
||||||
| θ value | 7.01481e-53 (rank : 21) | NC score | 0.904562 (rank : 23) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8TE23, Q5TZ19 | Gene names | TAS1R2, GPR71, T1R2, TR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 2 precursor (G-protein coupled receptor 71) (Sweet taste receptor T1R2). | |||||
|
TS1R2_MOUSE
|
||||||
| θ value | 5.02714e-51 (rank : 22) | NC score | 0.908324 (rank : 20) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q925I4, Q923J8 | Gene names | Tas1r2, Gpr71, T1r2, Tr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 2 precursor (G-protein coupled receptor 71) (Sweet taste receptor T1R2). | |||||
|
TS1R3_MOUSE
|
||||||
| θ value | 8.57503e-51 (rank : 23) | NC score | 0.893618 (rank : 24) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q925D8, Q91VA4, Q923K0, Q925A4, Q925D9 | Gene names | Tas1r3, Sac, T1r3, Tr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 3 precursor (Sweet taste receptor T1R3) (Saccharin preference protein). | |||||
|
V2R1B_MOUSE
|
||||||
| θ value | 1.61718e-49 (rank : 24) | NC score | 0.920796 (rank : 17) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6TAC4, O70409 | Gene names | V2r1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vomeronasal type-2 receptor 1b precursor (Putative pheromone receptor V2R1b). | |||||
|
TS1R3_HUMAN
|
||||||
| θ value | 4.86918e-46 (rank : 25) | NC score | 0.888386 (rank : 25) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q7RTX0, Q5TA49, Q8NGW9 | Gene names | TAS1R3, T1R3, TR3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 3 precursor (Sweet taste receptor T1R3). | |||||
|
GABR2_HUMAN
|
||||||
| θ value | 1.29631e-06 (rank : 26) | NC score | 0.531294 (rank : 26) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O75899, O75974, O75975, Q5VXZ2, Q8WX04, Q9P1R2, Q9UNR1, Q9UNS9 | Gene names | GABBR2, GPR51, GPRC3B | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 2 precursor (GABA-B receptor 2) (GABA-B-R2) (Gb2) (GABABR2) (G-protein coupled receptor 51) (HG20). | |||||
|
ANPRC_MOUSE
|
||||||
| θ value | 2.21117e-06 (rank : 27) | NC score | 0.157327 (rank : 38) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P70180, P97804, Q9R025, Q9R027, Q9R028 | Gene names | Npr3 | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide clearance receptor precursor (ANP-C) (ANPRC) (NPR-C) (Atrial natriuretic peptide C-type receptor) (EF-2). | |||||
|
GP158_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 28) | NC score | 0.246178 (rank : 31) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5T848, Q6QR81, Q9ULT3 | Gene names | GPR158, KIAA1136 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 158 precursor. | |||||
|
GP179_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 29) | NC score | 0.198527 (rank : 35) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6PRD1 | Gene names | GPR179, GPR158L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 179 precursor (Probable G-protein coupled receptor 158-like 1). | |||||
|
GABR1_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 30) | NC score | 0.408627 (rank : 28) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UBS5, O95375, O95468, O95975, O96022, Q86W60, Q9UQQ0 | Gene names | GABBR1, GPRC3A | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
GABR1_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 31) | NC score | 0.408653 (rank : 27) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9WV18, Q9WU48, Q9WV15, Q9WV16, Q9WV17 | Gene names | Gabbr1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
ANPRC_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 32) | NC score | 0.136380 (rank : 39) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P17342 | Gene names | NPR3, ANPRC | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide clearance receptor precursor (ANP-C) (ANPRC) (NPR-C) (Atrial natriuretic peptide C-type receptor). | |||||
|
GPC5B_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 33) | NC score | 0.294890 (rank : 30) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NZH0, O75205, Q8NBZ8 | Gene names | GPRC5B, RAIG2 | |||
|
Domain Architecture |

|
|||||
| Description | G-protein coupled receptor family C group 5 member B precursor (Retinoic acid-induced gene 2 protein) (RAIG-2) (A-69G12.1). | |||||
|
GP158_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 34) | NC score | 0.225011 (rank : 32) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8C419, Q8CHB0 | Gene names | Gpr158, Kiaa1136 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 158 precursor. | |||||
|
GPC5B_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 35) | NC score | 0.320249 (rank : 29) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q923Z0, Q8CCV3 | Gene names | Gprc5b, Raig2 | |||
|
Domain Architecture |

|
|||||
| Description | G-protein coupled receptor family C group 5 member B precursor (Retinoic acid-induced gene 2 protein) (RAIG-2). | |||||
|
GPC5C_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 36) | NC score | 0.181898 (rank : 36) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8K3J9, Q8K0H0 | Gene names | Gprc5c, Raig3 | |||
|
Domain Architecture |

|
|||||
| Description | G-protein coupled receptor family C group 5 member C precursor (Retinoic acid-induced gene 3 protein) (RAIG-3). | |||||
|
GRIK1_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 37) | NC score | 0.068709 (rank : 48) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P39086, Q13001, Q86SU9 | Gene names | GRIK1, GLUR5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 1 precursor (Glutamate receptor 5) (GluR-5) (GluR5) (Excitatory amino acid receptor 3) (EAA3). | |||||
|
GUC2G_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 38) | NC score | 0.057305 (rank : 49) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6TL19, Q8BWU7 | Gene names | Gucy2g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanylate cyclase 2G precursor (EC 4.6.1.2) (Guanylyl cyclase receptor G) (mGC-G). | |||||
|
GRIK1_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 39) | NC score | 0.068897 (rank : 47) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q60934 | Gene names | Grik1, Glur5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 1 precursor (Glutamate receptor 5) (GluR-5) (GluR5). | |||||
|
NMDZ1_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 40) | NC score | 0.201881 (rank : 33) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P35438, Q8CFS4 | Gene names | Grin1, Glurz1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit zeta 1 precursor (N-methyl-D- aspartate receptor subunit NR1). | |||||
|
BCAR1_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 41) | NC score | 0.041177 (rank : 51) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61140, Q60869 | Gene names | Bcar1, Cas, Crkas | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
GRIK2_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 42) | NC score | 0.069019 (rank : 46) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q13002 | Gene names | GRIK2, GLUR6 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 2 precursor (Glutamate receptor 6) (GluR-6) (GluR6) (Excitatory amino acid receptor 4) (EAA4). | |||||
|
NMDZ1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 43) | NC score | 0.198650 (rank : 34) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q05586, P35437, Q12867, Q12868 | Gene names | GRIN1, NMDAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit zeta 1 precursor (N-methyl-D- aspartate receptor subunit NR1). | |||||
|
NOTC2_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 44) | NC score | 0.005021 (rank : 79) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1069 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O35516, Q06008, Q60941 | Gene names | Notch2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (Motch B) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
CSKI2_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 45) | NC score | 0.007813 (rank : 75) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 812 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8VHK1, Q6ZPX1, Q7TS77 | Gene names | Caskin2, Kiaa1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
PRD14_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 46) | NC score | -0.000869 (rank : 92) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 757 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9GZV8, Q86UX9 | Gene names | PRDM14 | |||
|
Domain Architecture |
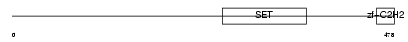
|
|||||
| Description | PR domain zinc finger protein 14 (PR domain-containing protein 14). | |||||
|
GR65_MOUSE
|
||||||
| θ value | 0.163984 (rank : 47) | NC score | 0.021052 (rank : 58) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91X51, Q9D3L9 | Gene names | Gorasp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 1 (Golgi reassembly-stacking protein of 65 kDa) (GRASP65) (Golgi peripheral membrane protein p65). | |||||
|
GPC5C_HUMAN
|
||||||
| θ value | 0.21417 (rank : 48) | NC score | 0.157541 (rank : 37) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NQ84, Q9NZG5 | Gene names | GPRC5C, RAIG3 | |||
|
Domain Architecture |
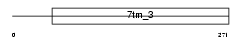
|
|||||
| Description | G-protein coupled receptor family C group 5 member C precursor (Retinoic acid-induced gene 3 protein) (RAIG-3). | |||||
|
LZTR1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 49) | NC score | 0.016567 (rank : 59) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CQ33 | Gene names | Lztr1, Tcfl2 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-zipper-like transcriptional regulator 1 (LZTR-1). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 0.279714 (rank : 50) | NC score | 0.010183 (rank : 72) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
BCAR1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 51) | NC score | 0.032286 (rank : 56) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P56945 | Gene names | BCAR1, CAS, CRKAS | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 0.365318 (rank : 52) | NC score | 0.010565 (rank : 71) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
NMD3A_HUMAN
|
||||||
| θ value | 0.365318 (rank : 53) | NC score | 0.039432 (rank : 52) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8TCU5, Q8TF29, Q8WXI6 | Gene names | GRIN3A, KIAA1973 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit 3A precursor (N-methyl-D-aspartate receptor subtype NR3A) (NMDAR-L). | |||||
|
HSF1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 54) | NC score | 0.013453 (rank : 66) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P38532, O70462 | Gene names | Hsf1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 0.47712 (rank : 55) | NC score | 0.016466 (rank : 60) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
LZTR1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 56) | NC score | 0.015106 (rank : 62) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N653, Q14776 | Gene names | LZTR1, TCFL2 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-zipper-like transcriptional regulator 1 (LZTR-1). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 0.62314 (rank : 57) | NC score | 0.014276 (rank : 64) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
ZN516_MOUSE
|
||||||
| θ value | 1.06291 (rank : 58) | NC score | -0.001269 (rank : 93) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7TSH3 | Gene names | Znf516, Kiaa0222, Zfp516 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 516. | |||||
|
ECM1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 59) | NC score | 0.014934 (rank : 63) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61508 | Gene names | Ecm1 | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular matrix protein 1 precursor (Secretory component p85). | |||||
|
GLIS3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 60) | NC score | -0.001723 (rank : 94) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 861 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6XP49, Q8BI60 | Gene names | Glis3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLIS3 (GLI-similar 3). | |||||
|
NFAC2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 61) | NC score | 0.013645 (rank : 65) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13469, Q13468, Q5TFW7, Q5TFW8, Q9NPX6, Q9NQH3, Q9UJR2 | Gene names | NFATC2, NFAT1, NFATP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 2 (T cell transcription factor NFAT1) (NFAT pre-existing subunit) (NF-ATp). | |||||
|
NFAC2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 62) | NC score | 0.013178 (rank : 67) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60591, Q60984, Q60985 | Gene names | Nfatc2, Nfat1, Nfatp | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 2 (T cell transcription factor NFAT1) (NFAT pre-existing subunit) (NF-ATp). | |||||
|
LTBP2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 63) | NC score | 0.011422 (rank : 70) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O08999, Q8C6W9 | Gene names | Ltbp2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
NPAS4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 64) | NC score | 0.012736 (rank : 68) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BGD7, Q3V3U3 | Gene names | Npas4, Nxf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF) (Limbic-enhanced PAS protein) (LE-PAS). | |||||
|
GRIK3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 65) | NC score | 0.043468 (rank : 50) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13003, Q13004, Q16136 | Gene names | GRIK3, GLUR7 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 3 precursor (Glutamate receptor 7) (GluR-7) (GluR7) (Excitatory amino acid receptor 5) (EAA5). | |||||
|
MKL1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 66) | NC score | 0.007885 (rank : 74) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
NMDE2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 67) | NC score | 0.034134 (rank : 54) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13224, Q12919, Q13220, Q13225, Q9UM56 | Gene names | GRIN2B | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 2 precursor (N-methyl D- aspartate receptor subtype 2B) (NR2B) (NMDAR2B) (N-methyl-D-aspartate receptor subunit 3) (NR3) (hNR3). | |||||
|
NMDE2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 68) | NC score | 0.034412 (rank : 53) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q01097, Q9DCB2 | Gene names | Grin2b | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 2 precursor (N-methyl D- aspartate receptor subtype 2B) (NR2B) (NMDAR2B). | |||||
|
NPAS4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 69) | NC score | 0.011815 (rank : 69) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IUM7, Q8N8S5, Q8N9Q9 | Gene names | NPAS4, NXF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF). | |||||
|
AFAD_HUMAN
|
||||||
| θ value | 3.0926 (rank : 70) | NC score | 0.003739 (rank : 82) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P55196, O75087, O75088, O75089, Q5TIG6, Q9NSN7, Q9NU92 | Gene names | MLLT4, AF6 | |||
|
Domain Architecture |

|
|||||
| Description | Afadin (Protein AF-6). | |||||
|
NMD3B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 71) | NC score | 0.032649 (rank : 55) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O60391, Q5EAK7, Q7RTW9 | Gene names | GRIN3B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit 3B precursor (N-methyl-D-aspartate receptor subtype NR3B) (NR3B) (NMDAR3B). | |||||
|
AKAP3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.007698 (rank : 76) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O88987 | Gene names | Akap3, Akap110 | |||
|
Domain Architecture |
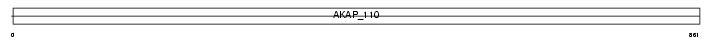
|
|||||
| Description | A-kinase anchor protein 3 (Protein kinase A-anchoring protein 3) (PRKA3) (A-kinase anchor protein 110 kDa) (AKAP 110). | |||||
|
MAST2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 73) | NC score | -0.003887 (rank : 96) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1189 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6P0Q8, O94899, Q5VT07, Q5VT08, Q7LGC4, Q8NDG1, Q96B94, Q9BYE8 | Gene names | MAST2, KIAA0807, MAST205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 74) | NC score | 0.003801 (rank : 81) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
CRSP3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.015915 (rank : 61) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ULK4, O95403, Q9H0J2, Q9NTT9, Q9NTU0, Q9Y5P7, Q9Y667 | Gene names | CRSP3, ARC130, DRIP130, KIAA1216, SUR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 3 (Cofactor required for Sp1 transcriptional activation subunit 3) (Transcriptional coactivator CRSP130) (Vitamin D3 receptor-interacting protein complex 130 kDa component) (DRIP130) (Activator-recruited cofactor 130 kDa component) (ARC130). | |||||
|
LTBP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.008258 (rank : 73) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 552 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14767, Q99907, Q9NS51 | Gene names | LTBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
NOTC2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.002902 (rank : 85) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q04721, Q99734, Q9H240 | Gene names | NOTCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (hN2) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
PAX3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | -0.000563 (rank : 90) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P24610, Q3UFQ9, Q8BRE7, Q9CXI6 | Gene names | Pax3, Pax-3 | |||
|
Domain Architecture |
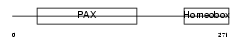
|
|||||
| Description | Paired box protein Pax-3. | |||||
|
ZIM10_HUMAN
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.005102 (rank : 78) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
CCD21_HUMAN
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | -0.002535 (rank : 95) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
CE152_HUMAN
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | -0.000650 (rank : 91) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94986, Q6NTA0 | Gene names | CEP152, KIAA0912 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 152 kDa (Cep152 protein). | |||||
|
CO5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.006952 (rank : 77) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P01031 | Gene names | C5 | |||
|
Domain Architecture |

|
|||||
| Description | Complement C5 precursor [Contains: Complement C5 beta chain; Complement C5 alpha chain; C5a anaphylatoxin; Complement C5 alpha' chain]. | |||||
|
COFA1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.003482 (rank : 83) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P39059, Q5T6J4, Q9Y4W4 | Gene names | COL15A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XV) chain precursor [Contains: Endostatin (Endostatin-XV) (Restin) (Related to endostatin)]. | |||||
|
LR37A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.002820 (rank : 86) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O60309, Q49A01, Q49A80, Q8NB33 | Gene names | LRRC37A, KIAA0563 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37A. | |||||
|
U258_HUMAN
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.021118 (rank : 57) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P59773 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UPF0258 protein. | |||||
|
CD248_MOUSE
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.002550 (rank : 87) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q91V98, Q3UMV6, Q91ZV1 | Gene names | Cd248, Tem1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
GAK19_HUMAN
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.003078 (rank : 84) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P62691 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_4p16.1 provirus ancestral Gag polyprotein (Gag polyprotein). | |||||
|
SSBP4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.001194 (rank : 89) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
UB7I1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.004145 (rank : 80) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P58283, Q3U493, Q3UE56, Q3UGM3, Q68FN0, Q6P1H8, Q6PWY5, Q8BN27, Q8C1U3 | Gene names | Ubce7ip1, Triad3, Uip83, Zin | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin ligase TRIAD3 (EC 6.3.2.-) (Ubiquitin-conjugating enzyme 7-interacting protein 1) (UbcM4-interacting protein 83) (Triad domain- containing protein 3). | |||||
|
ZF106_MOUSE
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.002025 (rank : 88) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88466, O55185, O88465, O88467, Q792M1, Q792P4, Q8CDZ8, Q8R3I4, Q9ESU3 | Gene names | Zfp106, H3a, Sh3bp3, Sirm, Znf474 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 106 (Zfp-106) (Zinc finger protein 474) (H3a minor histocompatibility antigen) (Son of insulin receptor mutant). | |||||
|
GP156_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.098249 (rank : 43) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8NFN8, Q86SN6 | Gene names | GPR156, GABABL, PGR28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 156 (GABAB-related G-protein coupled receptor) (G-protein coupled receptor PGR28). | |||||
|
GP156_MOUSE
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.118974 (rank : 40) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6PCP7, Q8K452 | Gene names | Gpr156, Gababl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 156 (GABAB-related G-protein coupled receptor). | |||||
|
GPC5D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.092498 (rank : 44) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NZD1, Q3KNV3, Q7Z5J9, Q8TDS6 | Gene names | GPRC5D | |||
|
Domain Architecture |
|
|||||
| Description | G-protein coupled receptor family C group 5 member D. | |||||
|
GPC5D_MOUSE
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.117791 (rank : 41) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9JIL6, Q8CJI0, Q8CJI1 | Gene names | Gprc5d | |||
|
Domain Architecture |
|
|||||
| Description | G-protein coupled receptor family C group 5 member D. | |||||
|
RAI3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.082972 (rank : 45) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NFJ5, O95357 | Gene names | GPRC5A, RAI3, RAIG1 | |||
|
Domain Architecture |
|
|||||
| Description | Retinoic acid-induced protein 3 (G-protein coupled receptor family C group 5 member A) (Retinoic acid-induced gene 1 protein) (RAIG-1) (Orphan G-protein coupling receptor PEIG-1). | |||||
|
RAI3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.110733 (rank : 42) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BHL4, Q8K067, Q8K1G5 | Gene names | Gprc5a, Rai3, Raig1 | |||
|
Domain Architecture |
|
|||||
| Description | Retinoic acid-induced protein 3 (G-protein coupled receptor family C group 5 member A) (Retinoic acid-induced gene 1 protein) (RAIG-1). | |||||
|
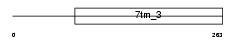
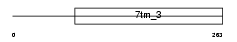
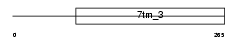
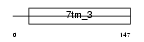
MGR1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | P97772, Q6AXG4, Q9EPV6 | Gene names | Grm1, Gprc1a, Mglur1 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 1 precursor (mGluR1). | |||||
|
MGR1_HUMAN
|
||||||
| NC score | 0.997411 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q13255, Q13256, Q14757, Q14758, Q5VTF7, Q5VTF8, Q9NU10, Q9UGS9, Q9UGT0 | Gene names | GRM1, GPRC1A, MGLUR1 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 1 precursor (mGluR1). | |||||
|
MGR5_HUMAN
|
||||||
| NC score | 0.992949 (rank : 3) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P41594 | Gene names | GRM5, GPRC1E, MGLUR5 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 5 precursor (mGluR5). | |||||
|
MGR2_HUMAN
|
||||||
| NC score | 0.979508 (rank : 4) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q14416, Q52MC6, Q9H3N6 | Gene names | GRM2, GPRC1B, MGLUR2 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 2 precursor (mGluR2). | |||||
|
MGR3_HUMAN
|
||||||
| NC score | 0.976398 (rank : 5) | θ value | 8.53067e-184 (rank : 5) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q14832, Q75MV4, Q75N17, Q86YG6, Q8TBH9 | Gene names | GRM3, GPRC1C, MGLUR3 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 3 precursor (mGluR3). | |||||
|
MGR3_MOUSE
|
||||||
| NC score | 0.976015 (rank : 6) | θ value | 1.11414e-183 (rank : 6) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9QYS2 | Gene names | Grm3, Gprc1c, Mglur3 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 3 precursor (mGluR3). | |||||
|
MGR4_HUMAN
|
||||||
| NC score | 0.975513 (rank : 7) | θ value | 4.1007e-178 (rank : 7) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q14833, Q5SZ84 | Gene names | GRM4, GPRC1D, MGLUR4 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 4 precursor (mGluR4). | |||||
|
MGR6_HUMAN
|
||||||
| NC score | 0.973800 (rank : 8) | θ value | 5.73535e-172 (rank : 13) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O15303 | Gene names | GRM6, GPRC1F, MGLUR6 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 6 precursor (mGluR6). | |||||
|
MGR8_MOUSE
|
||||||
| NC score | 0.973737 (rank : 9) | θ value | 1.11673e-175 (rank : 10) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P47743, Q6B964 | Gene names | Grm8, Gprc1h, Mglur8 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 8 precursor (mGluR8). | |||||
|
MGR8_HUMAN
|
||||||
| NC score | 0.973701 (rank : 10) | θ value | 5.01279e-176 (rank : 8) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O00222, O15493, O95945, O95946, Q3MIV9, Q52M02, Q6J165 | Gene names | GRM8, GPRC1H, MGLUR8 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 8 precursor (mGluR8). | |||||
|
MGR6_MOUSE
|
||||||
| NC score | 0.972546 (rank : 11) | θ value | 6.77497e-173 (rank : 12) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q5NCH9 | Gene names | Grm6, Gprc1f, Mglur6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metabotropic glutamate receptor 6 precursor (mGluR6). | |||||
|
MGR7_HUMAN
|
||||||
| NC score | 0.972153 (rank : 12) | θ value | 1.4585e-175 (rank : 11) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q14831, Q8NFS2, Q8NFS3, Q8NFS4 | Gene names | GRM7, GPRC1G, MGLUR7 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 7 precursor (mGluR7). | |||||
|
MGR7_MOUSE
|
||||||
| NC score | 0.972045 (rank : 13) | θ value | 1.11673e-175 (rank : 9) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q68ED2 | Gene names | Grm7, Gprc1g, Mglur7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metabotropic glutamate receptor 7 precursor (mGluR7). | |||||
|
CASR_HUMAN
|
||||||
| NC score | 0.943547 (rank : 14) | θ value | 9.34977e-98 (rank : 14) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P41180, Q13912, Q16108, Q16109, Q16110, Q16379, Q4PJ19 | Gene names | CASR, GPRC2A, PCAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular calcium-sensing receptor precursor (CaSR) (Parathyroid Cell calcium-sensing receptor). | |||||
|
CASR_MOUSE
|
||||||
| NC score | 0.943510 (rank : 15) | θ value | 3.55291e-97 (rank : 15) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9QY96, O08968, O88519, Q9QY95, Q9QZU8, Q9R1D6, Q9R1Y2 | Gene names | Casr, Gprc2a | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular calcium-sensing receptor precursor (CaSR) (Parathyroid Cell calcium-sensing receptor). | |||||
|
V2RX_MOUSE
|
||||||
| NC score | 0.933672 (rank : 16) | θ value | 2.47765e-74 (rank : 16) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O70410 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vomeronasal type-2 receptor X precursor (Putative pheromone receptor V2RX). | |||||
|
V2R1B_MOUSE
|
||||||
| NC score | 0.920796 (rank : 17) | θ value | 1.61718e-49 (rank : 24) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6TAC4, O70409 | Gene names | V2r1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vomeronasal type-2 receptor 1b precursor (Putative pheromone receptor V2R1b). | |||||
|
GPC6A_HUMAN
|
||||||
| NC score | 0.915073 (rank : 18) | θ value | 4.38363e-63 (rank : 19) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q5T6X5, Q6JK43, Q6JK44, Q8NGU8, Q8NHZ9, Q8TDT6 | Gene names | GPRC6A, GPCR33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor family C group 6 member A precursor (hGPRC6A) (G-protein coupled receptor 33) (hGPCR33). | |||||
|
GPC6A_MOUSE
|
||||||
| NC score | 0.913057 (rank : 19) | θ value | 2.84138e-62 (rank : 20) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8K4Z6, Q5DK51, Q5DK52 | Gene names | Gprc6a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor family C group 6 member A precursor. | |||||
|
TS1R2_MOUSE
|
||||||
| NC score | 0.908324 (rank : 20) | θ value | 5.02714e-51 (rank : 22) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q925I4, Q923J8 | Gene names | Tas1r2, Gpr71, T1r2, Tr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 2 precursor (G-protein coupled receptor 71) (Sweet taste receptor T1R2). | |||||
|
TS1R1_MOUSE
|
||||||
| NC score | 0.906602 (rank : 21) | θ value | 1.2325e-65 (rank : 17) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q99PG6, Q6NS58, Q923J9, Q925I5, Q99PG5 | Gene names | Tas1r1, Gpr70, T1r1, Tr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 1 precursor (G-protein coupled receptor 70). | |||||
|
TS1R1_HUMAN
|
||||||
| NC score | 0.906061 (rank : 22) | θ value | 2.32439e-64 (rank : 18) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q7RTX1, Q8NGZ7, Q8TDJ7, Q8TDJ8, Q8TDJ9, Q8TDK0 | Gene names | TAS1R1, GPR70, T1R1, TR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 1 precursor (G-protein coupled receptor 70) (Gm148). | |||||
|
TS1R2_HUMAN
|
||||||
| NC score | 0.904562 (rank : 23) | θ value | 7.01481e-53 (rank : 21) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8TE23, Q5TZ19 | Gene names | TAS1R2, GPR71, T1R2, TR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 2 precursor (G-protein coupled receptor 71) (Sweet taste receptor T1R2). | |||||
|
TS1R3_MOUSE
|
||||||
| NC score | 0.893618 (rank : 24) | θ value | 8.57503e-51 (rank : 23) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q925D8, Q91VA4, Q923K0, Q925A4, Q925D9 | Gene names | Tas1r3, Sac, T1r3, Tr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 3 precursor (Sweet taste receptor T1R3) (Saccharin preference protein). | |||||
|
TS1R3_HUMAN
|
||||||
| NC score | 0.888386 (rank : 25) | θ value | 4.86918e-46 (rank : 25) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q7RTX0, Q5TA49, Q8NGW9 | Gene names | TAS1R3, T1R3, TR3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 3 precursor (Sweet taste receptor T1R3). | |||||
|
GABR2_HUMAN
|
||||||
| NC score | 0.531294 (rank : 26) | θ value | 1.29631e-06 (rank : 26) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O75899, O75974, O75975, Q5VXZ2, Q8WX04, Q9P1R2, Q9UNR1, Q9UNS9 | Gene names | GABBR2, GPR51, GPRC3B | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 2 precursor (GABA-B receptor 2) (GABA-B-R2) (Gb2) (GABABR2) (G-protein coupled receptor 51) (HG20). | |||||
|
GABR1_MOUSE
|
||||||
| NC score | 0.408653 (rank : 27) | θ value | 1.43324e-05 (rank : 31) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9WV18, Q9WU48, Q9WV15, Q9WV16, Q9WV17 | Gene names | Gabbr1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
GABR1_HUMAN
|
||||||
| NC score | 0.408627 (rank : 28) | θ value | 1.43324e-05 (rank : 30) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UBS5, O95375, O95468, O95975, O96022, Q86W60, Q9UQQ0 | Gene names | GABBR1, GPRC3A | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
GPC5B_MOUSE
|
||||||
| NC score | 0.320249 (rank : 29) | θ value | 0.000786445 (rank : 35) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q923Z0, Q8CCV3 | Gene names | Gprc5b, Raig2 | |||
|
Domain Architecture |

|
|||||
| Description | G-protein coupled receptor family C group 5 member B precursor (Retinoic acid-induced gene 2 protein) (RAIG-2). | |||||
|
GPC5B_HUMAN
|
||||||
| NC score | 0.294890 (rank : 30) | θ value | 0.00035302 (rank : 33) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NZH0, O75205, Q8NBZ8 | Gene names | GPRC5B, RAIG2 | |||
|
Domain Architecture |

|
|||||
| Description | G-protein coupled receptor family C group 5 member B precursor (Retinoic acid-induced gene 2 protein) (RAIG-2) (A-69G12.1). | |||||
|
GP158_HUMAN
|
||||||
| NC score | 0.246178 (rank : 31) | θ value | 1.09739e-05 (rank : 28) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5T848, Q6QR81, Q9ULT3 | Gene names | GPR158, KIAA1136 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 158 precursor. | |||||
|
GP158_MOUSE
|
||||||
| NC score | 0.225011 (rank : 32) | θ value | 0.000786445 (rank : 34) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8C419, Q8CHB0 | Gene names | Gpr158, Kiaa1136 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 158 precursor. | |||||
|
NMDZ1_MOUSE
|
||||||
| NC score | 0.201881 (rank : 33) | θ value | 0.0113563 (rank : 40) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P35438, Q8CFS4 | Gene names | Grin1, Glurz1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit zeta 1 precursor (N-methyl-D- aspartate receptor subunit NR1). | |||||
|
NMDZ1_HUMAN
|
||||||
| NC score | 0.198650 (rank : 34) | θ value | 0.0431538 (rank : 43) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q05586, P35437, Q12867, Q12868 | Gene names | GRIN1, NMDAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit zeta 1 precursor (N-methyl-D- aspartate receptor subunit NR1). | |||||
|
GP179_HUMAN
|
||||||
| NC score | 0.198527 (rank : 35) | θ value | 1.09739e-05 (rank : 29) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6PRD1 | Gene names | GPR179, GPR158L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 179 precursor (Probable G-protein coupled receptor 158-like 1). | |||||
|
GPC5C_MOUSE
|
||||||
| NC score | 0.181898 (rank : 36) | θ value | 0.00134147 (rank : 36) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8K3J9, Q8K0H0 | Gene names | Gprc5c, Raig3 | |||
|
Domain Architecture |

|
|||||
| Description | G-protein coupled receptor family C group 5 member C precursor (Retinoic acid-induced gene 3 protein) (RAIG-3). | |||||
|
GPC5C_HUMAN
|
||||||
| NC score | 0.157541 (rank : 37) | θ value | 0.21417 (rank : 48) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NQ84, Q9NZG5 | Gene names | GPRC5C, RAIG3 | |||
|
Domain Architecture |

|
|||||
| Description | G-protein coupled receptor family C group 5 member C precursor (Retinoic acid-induced gene 3 protein) (RAIG-3). | |||||
|
ANPRC_MOUSE
|
||||||
| NC score | 0.157327 (rank : 38) | θ value | 2.21117e-06 (rank : 27) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P70180, P97804, Q9R025, Q9R027, Q9R028 | Gene names | Npr3 | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide clearance receptor precursor (ANP-C) (ANPRC) (NPR-C) (Atrial natriuretic peptide C-type receptor) (EF-2). | |||||
|
ANPRC_HUMAN
|
||||||
| NC score | 0.136380 (rank : 39) | θ value | 0.000270298 (rank : 32) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P17342 | Gene names | NPR3, ANPRC | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide clearance receptor precursor (ANP-C) (ANPRC) (NPR-C) (Atrial natriuretic peptide C-type receptor). | |||||
|
GP156_MOUSE
|
||||||
| NC score | 0.118974 (rank : 40) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6PCP7, Q8K452 | Gene names | Gpr156, Gababl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 156 (GABAB-related G-protein coupled receptor). | |||||
|
GPC5D_MOUSE
|
||||||
| NC score | 0.117791 (rank : 41) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9JIL6, Q8CJI0, Q8CJI1 | Gene names | Gprc5d | |||
|
Domain Architecture |

|
|||||
| Description | G-protein coupled receptor family C group 5 member D. | |||||
|
RAI3_MOUSE
|
||||||
| NC score | 0.110733 (rank : 42) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BHL4, Q8K067, Q8K1G5 | Gene names | Gprc5a, Rai3, Raig1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid-induced protein 3 (G-protein coupled receptor family C group 5 member A) (Retinoic acid-induced gene 1 protein) (RAIG-1). | |||||
|
GP156_HUMAN
|
||||||
| NC score | 0.098249 (rank : 43) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8NFN8, Q86SN6 | Gene names | GPR156, GABABL, PGR28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 156 (GABAB-related G-protein coupled receptor) (G-protein coupled receptor PGR28). | |||||
|
GPC5D_HUMAN
|
||||||
| NC score | 0.092498 (rank : 44) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NZD1, Q3KNV3, Q7Z5J9, Q8TDS6 | Gene names | GPRC5D | |||
|
Domain Architecture |

|
|||||
| Description | G-protein coupled receptor family C group 5 member D. | |||||
|
RAI3_HUMAN
|
||||||
| NC score | 0.082972 (rank : 45) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NFJ5, O95357 | Gene names | GPRC5A, RAI3, RAIG1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid-induced protein 3 (G-protein coupled receptor family C group 5 member A) (Retinoic acid-induced gene 1 protein) (RAIG-1) (Orphan G-protein coupling receptor PEIG-1). | |||||
|
GRIK2_HUMAN
|
||||||
| NC score | 0.069019 (rank : 46) | θ value | 0.0431538 (rank : 42) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q13002 | Gene names | GRIK2, GLUR6 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 2 precursor (Glutamate receptor 6) (GluR-6) (GluR6) (Excitatory amino acid receptor 4) (EAA4). | |||||
|
GRIK1_MOUSE
|
||||||
| NC score | 0.068897 (rank : 47) | θ value | 0.00228821 (rank : 39) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q60934 | Gene names | Grik1, Glur5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 1 precursor (Glutamate receptor 5) (GluR-5) (GluR5). | |||||
|
GRIK1_HUMAN
|
||||||
| NC score | 0.068709 (rank : 48) | θ value | 0.00134147 (rank : 37) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P39086, Q13001, Q86SU9 | Gene names | GRIK1, GLUR5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 1 precursor (Glutamate receptor 5) (GluR-5) (GluR5) (Excitatory amino acid receptor 3) (EAA3). | |||||
|
GUC2G_MOUSE
|
||||||
| NC score | 0.057305 (rank : 49) | θ value | 0.00175202 (rank : 38) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6TL19, Q8BWU7 | Gene names | Gucy2g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanylate cyclase 2G precursor (EC 4.6.1.2) (Guanylyl cyclase receptor G) (mGC-G). | |||||
|
GRIK3_HUMAN
|
||||||
| NC score | 0.043468 (rank : 50) | θ value | 2.36792 (rank : 65) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13003, Q13004, Q16136 | Gene names | GRIK3, GLUR7 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 3 precursor (Glutamate receptor 7) (GluR-7) (GluR7) (Excitatory amino acid receptor 5) (EAA5). | |||||
|
BCAR1_MOUSE
|
||||||
| NC score | 0.041177 (rank : 51) | θ value | 0.0252991 (rank : 41) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61140, Q60869 | Gene names | Bcar1, Cas, Crkas | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
NMD3A_HUMAN
|
||||||
| NC score | 0.039432 (rank : 52) | θ value | 0.365318 (rank : 53) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8TCU5, Q8TF29, Q8WXI6 | Gene names | GRIN3A, KIAA1973 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit 3A precursor (N-methyl-D-aspartate receptor subtype NR3A) (NMDAR-L). | |||||
|
NMDE2_MOUSE
|
||||||
| NC score | 0.034412 (rank : 53) | θ value | 2.36792 (rank : 68) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q01097, Q9DCB2 | Gene names | Grin2b | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 2 precursor (N-methyl D- aspartate receptor subtype 2B) (NR2B) (NMDAR2B). | |||||
|
NMDE2_HUMAN
|
||||||
| NC score | 0.034134 (rank : 54) | θ value | 2.36792 (rank : 67) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13224, Q12919, Q13220, Q13225, Q9UM56 | Gene names | GRIN2B | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 2 precursor (N-methyl D- aspartate receptor subtype 2B) (NR2B) (NMDAR2B) (N-methyl-D-aspartate receptor subunit 3) (NR3) (hNR3). | |||||
|
NMD3B_HUMAN
|
||||||
| NC score | 0.032649 (rank : 55) | θ value | 3.0926 (rank : 71) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O60391, Q5EAK7, Q7RTW9 | Gene names | GRIN3B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit 3B precursor (N-methyl-D-aspartate receptor subtype NR3B) (NR3B) (NMDAR3B). | |||||
|
BCAR1_HUMAN
|
||||||
| NC score | 0.032286 (rank : 56) | θ value | 0.365318 (rank : 51) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P56945 | Gene names | BCAR1, CAS, CRKAS | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
U258_HUMAN
|
||||||
| NC score | 0.021118 (rank : 57) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P59773 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UPF0258 protein. | |||||
|
GR65_MOUSE
|
||||||
| NC score | 0.021052 (rank : 58) | θ value | 0.163984 (rank : 47) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91X51, Q9D3L9 | Gene names | Gorasp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 1 (Golgi reassembly-stacking protein of 65 kDa) (GRASP65) (Golgi peripheral membrane protein p65). | |||||
|
LZTR1_MOUSE
|
||||||
| NC score | 0.016567 (rank : 59) | θ value | 0.21417 (rank : 49) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CQ33 | Gene names | Lztr1, Tcfl2 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-zipper-like transcriptional regulator 1 (LZTR-1). | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.016466 (rank : 60) | θ value | 0.47712 (rank : 55) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
CRSP3_HUMAN
|
||||||
| NC score | 0.015915 (rank : 61) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ULK4, O95403, Q9H0J2, Q9NTT9, Q9NTU0, Q9Y5P7, Q9Y667 | Gene names | CRSP3, ARC130, DRIP130, KIAA1216, SUR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 3 (Cofactor required for Sp1 transcriptional activation subunit 3) (Transcriptional coactivator CRSP130) (Vitamin D3 receptor-interacting protein complex 130 kDa component) (DRIP130) (Activator-recruited cofactor 130 kDa component) (ARC130). | |||||
|
LZTR1_HUMAN
|
||||||
| NC score | 0.015106 (rank : 62) | θ value | 0.62314 (rank : 56) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N653, Q14776 | Gene names | LZTR1, TCFL2 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-zipper-like transcriptional regulator 1 (LZTR-1). | |||||
|
ECM1_MOUSE
|
||||||
| NC score | 0.014934 (rank : 63) | θ value | 1.38821 (rank : 59) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61508 | Gene names | Ecm1 | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular matrix protein 1 precursor (Secretory component p85). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.014276 (rank : 64) | θ value | 0.62314 (rank : 57) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
NFAC2_HUMAN
|
||||||
| NC score | 0.013645 (rank : 65) | θ value | 1.38821 (rank : 61) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13469, Q13468, Q5TFW7, Q5TFW8, Q9NPX6, Q9NQH3, Q9UJR2 | Gene names | NFATC2, NFAT1, NFATP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 2 (T cell transcription factor NFAT1) (NFAT pre-existing subunit) (NF-ATp). | |||||
|
HSF1_MOUSE
|
||||||
| NC score | 0.013453 (rank : 66) | θ value | 0.47712 (rank : 54) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P38532, O70462 | Gene names | Hsf1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
NFAC2_MOUSE
|
||||||
| NC score | 0.013178 (rank : 67) | θ value | 1.38821 (rank : 62) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60591, Q60984, Q60985 | Gene names | Nfatc2, Nfat1, Nfatp | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 2 (T cell transcription factor NFAT1) (NFAT pre-existing subunit) (NF-ATp). | |||||
|
NPAS4_MOUSE
|
||||||
| NC score | 0.012736 (rank : 68) | θ value | 1.81305 (rank : 64) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BGD7, Q3V3U3 | Gene names | Npas4, Nxf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF) (Limbic-enhanced PAS protein) (LE-PAS). | |||||
|
NPAS4_HUMAN
|
||||||
| NC score | 0.011815 (rank : 69) | θ value | 2.36792 (rank : 69) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IUM7, Q8N8S5, Q8N9Q9 | Gene names | NPAS4, NXF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF). | |||||
|
LTBP2_MOUSE
|
||||||
| NC score | 0.011422 (rank : 70) | θ value | 1.81305 (rank : 63) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O08999, Q8C6W9 | Gene names | Ltbp2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.010565 (rank : 71) | θ value | 0.365318 (rank : 52) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.010183 (rank : 72) | θ value | 0.279714 (rank : 50) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
LTBP2_HUMAN
|
||||||
| NC score | 0.008258 (rank : 73) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 552 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14767, Q99907, Q9NS51 | Gene names | LTBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
MKL1_MOUSE
|
||||||
| NC score | 0.007885 (rank : 74) | θ value | 2.36792 (rank : 66) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
CSKI2_MOUSE
|
||||||
| NC score | 0.007813 (rank : 75) | θ value | 0.0736092 (rank : 45) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 812 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8VHK1, Q6ZPX1, Q7TS77 | Gene names | Caskin2, Kiaa1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
AKAP3_MOUSE
|
||||||
| NC score | 0.007698 (rank : 76) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O88987 | Gene names | Akap3, Akap110 | |||
|
Domain Architecture |
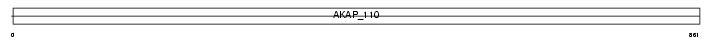
|
|||||
| Description | A-kinase anchor protein 3 (Protein kinase A-anchoring protein 3) (PRKA3) (A-kinase anchor protein 110 kDa) (AKAP 110). | |||||
|
CO5_HUMAN
|
||||||
| NC score | 0.006952 (rank : 77) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P01031 | Gene names | C5 | |||
|
Domain Architecture |

|
|||||
| Description | Complement C5 precursor [Contains: Complement C5 beta chain; Complement C5 alpha chain; C5a anaphylatoxin; Complement C5 alpha' chain]. | |||||
|
ZIM10_HUMAN
|
||||||
| NC score | 0.005102 (rank : 78) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
NOTC2_MOUSE
|
||||||
| NC score | 0.005021 (rank : 79) | θ value | 0.0563607 (rank : 44) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1069 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O35516, Q06008, Q60941 | Gene names | Notch2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (Motch B) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
UB7I1_MOUSE
|
||||||
| NC score | 0.004145 (rank : 80) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P58283, Q3U493, Q3UE56, Q3UGM3, Q68FN0, Q6P1H8, Q6PWY5, Q8BN27, Q8C1U3 | Gene names | Ubce7ip1, Triad3, Uip83, Zin | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin ligase TRIAD3 (EC 6.3.2.-) (Ubiquitin-conjugating enzyme 7-interacting protein 1) (UbcM4-interacting protein 83) (Triad domain- containing protein 3). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.003801 (rank : 81) | θ value | 4.03905 (rank : 74) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
AFAD_HUMAN
|
||||||
| NC score | 0.003739 (rank : 82) | θ value | 3.0926 (rank : 70) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P55196, O75087, O75088, O75089, Q5TIG6, Q9NSN7, Q9NU92 | Gene names | MLLT4, AF6 | |||
|
Domain Architecture |

|
|||||
| Description | Afadin (Protein AF-6). | |||||
|
COFA1_HUMAN
|
||||||
| NC score | 0.003482 (rank : 83) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P39059, Q5T6J4, Q9Y4W4 | Gene names | COL15A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XV) chain precursor [Contains: Endostatin (Endostatin-XV) (Restin) (Related to endostatin)]. | |||||
|
GAK19_HUMAN
|
||||||
| NC score | 0.003078 (rank : 84) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P62691 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_4p16.1 provirus ancestral Gag polyprotein (Gag polyprotein). | |||||
|
NOTC2_HUMAN
|
||||||
| NC score | 0.002902 (rank : 85) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q04721, Q99734, Q9H240 | Gene names | NOTCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (hN2) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
LR37A_HUMAN
|
||||||
| NC score | 0.002820 (rank : 86) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O60309, Q49A01, Q49A80, Q8NB33 | Gene names | LRRC37A, KIAA0563 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37A. | |||||
|
CD248_MOUSE
|
||||||
| NC score | 0.002550 (rank : 87) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q91V98, Q3UMV6, Q91ZV1 | Gene names | Cd248, Tem1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
ZF106_MOUSE
|
||||||
| NC score | 0.002025 (rank : 88) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88466, O55185, O88465, O88467, Q792M1, Q792P4, Q8CDZ8, Q8R3I4, Q9ESU3 | Gene names | Zfp106, H3a, Sh3bp3, Sirm, Znf474 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 106 (Zfp-106) (Zinc finger protein 474) (H3a minor histocompatibility antigen) (Son of insulin receptor mutant). | |||||
|
SSBP4_HUMAN
|
||||||
| NC score | 0.001194 (rank : 89) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
PAX3_MOUSE
|
||||||
| NC score | -0.000563 (rank : 90) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P24610, Q3UFQ9, Q8BRE7, Q9CXI6 | Gene names | Pax3, Pax-3 | |||
|
Domain Architecture |

|
|||||
| Description | Paired box protein Pax-3. | |||||
|
CE152_HUMAN
|
||||||
| NC score | -0.000650 (rank : 91) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94986, Q6NTA0 | Gene names | CEP152, KIAA0912 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 152 kDa (Cep152 protein). | |||||
|
PRD14_HUMAN
|
||||||
| NC score | -0.000869 (rank : 92) | θ value | 0.0961366 (rank : 46) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 757 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9GZV8, Q86UX9 | Gene names | PRDM14 | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 14 (PR domain-containing protein 14). | |||||
|
ZN516_MOUSE
|
||||||
| NC score | -0.001269 (rank : 93) | θ value | 1.06291 (rank : 58) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7TSH3 | Gene names | Znf516, Kiaa0222, Zfp516 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 516. | |||||
|
GLIS3_MOUSE
|
||||||
| NC score | -0.001723 (rank : 94) | θ value | 1.38821 (rank : 60) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 861 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6XP49, Q8BI60 | Gene names | Glis3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLIS3 (GLI-similar 3). | |||||
|
CCD21_HUMAN
|
||||||
| NC score | -0.002535 (rank : 95) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
MAST2_HUMAN
|
||||||
| NC score | -0.003887 (rank : 96) | θ value | 4.03905 (rank : 73) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1189 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6P0Q8, O94899, Q5VT07, Q5VT08, Q7LGC4, Q8NDG1, Q96B94, Q9BYE8 | Gene names | MAST2, KIAA0807, MAST205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||