Please be patient as the page loads
|
GR65_MOUSE
|
||||||
| SwissProt Accessions | Q91X51, Q9D3L9 | Gene names | Gorasp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 1 (Golgi reassembly-stacking protein of 65 kDa) (GRASP65) (Golgi peripheral membrane protein p65). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
GR65_MOUSE
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q91X51, Q9D3L9 | Gene names | Gorasp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 1 (Golgi reassembly-stacking protein of 65 kDa) (GRASP65) (Golgi peripheral membrane protein p65). | |||||
|
GR65_HUMAN
|
||||||
| θ value | 1.77876e-181 (rank : 2) | NC score | 0.957825 (rank : 2) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9BQQ3, Q8N272, Q96H42 | Gene names | GORASP1, GRASP65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 1 (Golgi reassembly-stacking protein of 65 kDa) (GRASP65) (Golgi peripheral membrane protein p65) (Golgi phosphoprotein 5) (GOLPH5). | |||||
|
GORS2_HUMAN
|
||||||
| θ value | 1.71185e-83 (rank : 3) | NC score | 0.920037 (rank : 3) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9H8Y8, Q96I74, Q96K84, Q9H946, Q9UFW4 | Gene names | GORASP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 2 (GRS2) (Golgi reassembly-stacking protein of 55 kDa) (GRASP55) (p59) (Golgi phosphoprotein 6) (GOLPH6). | |||||
|
GORS2_MOUSE
|
||||||
| θ value | 1.71185e-83 (rank : 4) | NC score | 0.913370 (rank : 4) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99JX3, Q3U9D2, Q8CCD0, Q91W68 | Gene names | Gorasp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 2 (GRS2) (Golgi reassembly-stacking protein of 55 kDa) (GRASP55). | |||||
|
PSMD9_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 5) | NC score | 0.223808 (rank : 5) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CR00, Q3TG66 | Gene names | Psmd9 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 9 (26S proteasome regulatory subunit p27). | |||||
|
PSMD9_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 6) | NC score | 0.209755 (rank : 6) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O00233, Q9BQ42 | Gene names | PSMD9 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 9 (26S proteasome regulatory subunit p27). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 7) | NC score | 0.061260 (rank : 10) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MGR1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 8) | NC score | 0.021052 (rank : 34) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97772, Q6AXG4, Q9EPV6 | Gene names | Grm1, Gprc1a, Mglur1 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 1 precursor (mGluR1). | |||||
|
PDCD7_MOUSE
|
||||||
| θ value | 0.163984 (rank : 9) | NC score | 0.075325 (rank : 7) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9WTY1, Q8R5D9 | Gene names | Pdcd7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Programmed cell death protein 7 (ES18). | |||||
|
WASIP_HUMAN
|
||||||
| θ value | 0.163984 (rank : 10) | NC score | 0.071230 (rank : 8) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
LONM_HUMAN
|
||||||
| θ value | 0.21417 (rank : 11) | NC score | 0.057639 (rank : 11) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P36776, P36777, Q9UQ95 | Gene names | PRSS15 | |||
|
Domain Architecture |

|
|||||
| Description | Lon protease homolog, mitochondrial precursor (EC 3.4.21.-) (Lon protease-like protein) (LONP) (LONHs). | |||||
|
OBSCN_HUMAN
|
||||||
| θ value | 0.21417 (rank : 12) | NC score | 0.008933 (rank : 49) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
IL3RB_MOUSE
|
||||||
| θ value | 0.47712 (rank : 13) | NC score | 0.035892 (rank : 19) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P26955 | Gene names | Csf2rb, Aic2b, Csf2rb1, Il3rb1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytokine receptor common beta chain precursor (GM-CSF/IL-3/IL-5 receptor common beta-chain) (CD131 antigen). | |||||
|
WWP1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 14) | NC score | 0.015833 (rank : 40) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H0M0, O00307, Q96BP4 | Gene names | WWP1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1) (Atropin-1-interacting protein 5) (AIP5). | |||||
|
SNPC4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 15) | NC score | 0.040295 (rank : 16) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5SXM2, Q9Y6P7 | Gene names | SNAPC4, SNAP190 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor subunit alpha) (PSE-binding factor subunit alpha) (PTF subunit alpha). | |||||
|
BSN_HUMAN
|
||||||
| θ value | 0.813845 (rank : 16) | NC score | 0.032792 (rank : 25) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
PDCD7_HUMAN
|
||||||
| θ value | 0.813845 (rank : 17) | NC score | 0.062994 (rank : 9) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N8D1, Q96AK8, Q9Y6D7 | Gene names | PDCD7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Programmed cell death protein 7 (ES18) (HES18). | |||||
|
SYN1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 18) | NC score | 0.031572 (rank : 26) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P17600, O75825, Q5H9A9 | Gene names | SYN1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I) (Brain protein 4.1). | |||||
|
ENAM_HUMAN
|
||||||
| θ value | 1.06291 (rank : 19) | NC score | 0.052294 (rank : 13) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NRM1, Q9H3D1 | Gene names | ENAM | |||
|
Domain Architecture |

|
|||||
| Description | Enamelin precursor. | |||||
|
PDLI7_HUMAN
|
||||||
| θ value | 1.38821 (rank : 20) | NC score | 0.019337 (rank : 35) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 547 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NR12, Q14250, Q5XG82, Q6NVZ5, Q96C91, Q9BXB8, Q9BXB9 | Gene names | PDLIM7, ENIGMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
SMRC2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 21) | NC score | 0.039884 (rank : 17) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8TAQ2, Q92923, Q96E12, Q96GY4 | Gene names | SMARCC2, BAF170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
CBX2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 22) | NC score | 0.034541 (rank : 22) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14781, Q9BTB1 | Gene names | CBX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromobox protein homolog 2. | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 23) | NC score | 0.034476 (rank : 23) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
TRI66_HUMAN
|
||||||
| θ value | 1.81305 (rank : 24) | NC score | 0.037340 (rank : 18) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O15016, Q9BQQ4 | Gene names | TRIM66, KIAA0298 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
AB1IP_HUMAN
|
||||||
| θ value | 2.36792 (rank : 25) | NC score | 0.021988 (rank : 33) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7Z5R6, Q8IWS8, Q8IZZ7 | Gene names | APBB1IP, PREL1, RARP1, RIAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 1- interacting protein (APBB1-interacting protein 1) (Rap1-GTP- interacting adapter molecule) (RIAM) (Proline-rich EVH1 ligand 1) (PREL-1) (Proline-rich protein 73) (Retinoic acid-responsive proline- rich protein 1) (RARP-1). | |||||
|
REL_MOUSE
|
||||||
| θ value | 2.36792 (rank : 26) | NC score | 0.017026 (rank : 38) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P15307 | Gene names | Rel | |||
|
Domain Architecture |

|
|||||
| Description | C-Rel proto-oncogene protein (C-Rel protein). | |||||
|
SPAG8_HUMAN
|
||||||
| θ value | 2.36792 (rank : 27) | NC score | 0.049291 (rank : 14) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99932, Q12937, Q5TCV8, Q8WWB4 | Gene names | SPAG8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 8 (Sperm membrane protein 1) (SMP-1) (HSD-1) (Sperm membrane protein BS-84). | |||||
|
CPSF6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 28) | NC score | 0.034908 (rank : 20) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q16630, Q53ES1, Q9BSJ7, Q9BW18 | Gene names | CPSF6, CFIM68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6 (Cleavage and polyadenylation specificity factor 68 kDa subunit) (CPSF 68 kDa subunit) (Pre-mRNA cleavage factor Im 68 kDa subunit) (Protein HPBRII- 4/7). | |||||
|
CPSF6_MOUSE
|
||||||
| θ value | 3.0926 (rank : 29) | NC score | 0.034905 (rank : 21) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6NVF9, Q8BX86, Q8BXI8 | Gene names | Cpsf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6. | |||||
|
DNJC1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 30) | NC score | 0.016384 (rank : 39) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96KC8 | Gene names | DNAJC1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 1. | |||||
|
HXA10_HUMAN
|
||||||
| θ value | 3.0926 (rank : 31) | NC score | 0.006387 (rank : 51) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P31260, O43370, O43605, Q15949 | Gene names | HOXA10, HOX1H | |||
|
Domain Architecture |
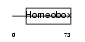
|
|||||
| Description | Homeobox protein Hox-A10 (Hox-1H) (Hox-1.8) (PL). | |||||
|
SF01_HUMAN
|
||||||
| θ value | 3.0926 (rank : 32) | NC score | 0.043187 (rank : 15) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15637, Q14818, Q14819, Q15913, Q8IY00, Q92744, Q92745, Q969H7, Q9BW01, Q9UEI0 | Gene names | SF1, ZFM1, ZNF162 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 1 (Zinc finger protein 162) (Transcription factor ZFM1) (Zinc finger gene in MEN1 locus) (Mammalian branch point-binding protein mBBP) (BBP). | |||||
|
3BP2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 33) | NC score | 0.028600 (rank : 27) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q06649 | Gene names | Sh3bp2, 3bp2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
ATF5_MOUSE
|
||||||
| θ value | 4.03905 (rank : 34) | NC score | 0.025271 (rank : 30) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O70191, Q58E51, Q99NH6 | Gene names | Atf5, Atfx, Nap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-5 (Activating transcription factor 5-alpha/beta) (Transcription factor ATFx) (Transcription factor-like protein ODA-10) (BZIP protein ATF7) (NRIF3- associated protein) (NAP1). | |||||
|
HCFC1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 35) | NC score | 0.026155 (rank : 28) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P51610, Q6P4G5 | Gene names | HCFC1, HCF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) (VP16 accessory protein) (VCAF) (CFF) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N-terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C- terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
MICA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 36) | NC score | 0.013467 (rank : 43) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8TDZ2, Q7Z633, Q8IVS9, Q96G47, Q9H6X6, Q9H7I0, Q9HAA1, Q9UFF7 | Gene names | MICAL1, MICAL, NICAL | |||
|
Domain Architecture |

|
|||||
| Description | NEDD9-interacting protein with calponin homology and LIM domains (Molecule interacting with CasL protein 1). | |||||
|
TIE1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 37) | NC score | 0.006189 (rank : 52) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35590 | Gene names | TIE1, TIE | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase receptor Tie-1 precursor (EC 2.7.10.1). | |||||
|
UBXD6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 38) | NC score | 0.025057 (rank : 31) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00124, Q7Z6F2 | Gene names | UBXD6, D8S2298E, REP8 | |||
|
Domain Architecture |

|
|||||
| Description | UBX domain-containing protein 6 (Reproduction 8 protein) (Protein Rep- 8). | |||||
|
DUS4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 39) | NC score | 0.008033 (rank : 50) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BFV3 | Gene names | Dusp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 4 (EC 3.1.3.48) (EC 3.1.3.16). | |||||
|
HCFC1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 40) | NC score | 0.025338 (rank : 29) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61191, Q684R1, Q7TSB0, Q8C2D0, Q9QWH2 | Gene names | Hcfc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N- terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C-terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
PDLI7_MOUSE
|
||||||
| θ value | 5.27518 (rank : 41) | NC score | 0.013380 (rank : 44) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3TJD7, Q80ZY6, Q810S3, Q8C1S4, Q9CRA1 | Gene names | Pdlim7, Enigma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
TIE1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 42) | NC score | 0.004969 (rank : 53) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1266 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q06806, Q811F4 | Gene names | Tie1, Tie, Tie-1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase receptor Tie-1 precursor (EC 2.7.10.1). | |||||
|
WASIP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 43) | NC score | 0.056583 (rank : 12) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 44) | NC score | 0.032844 (rank : 24) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
GLIS1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 45) | NC score | 0.000247 (rank : 54) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K1M4, Q6NZF6, Q8R4Z3 | Gene names | Glis1, Gli5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLIS1 (GLI-similar 1) (Gli homologous protein 1) (GliH1). | |||||
|
GRIP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.014459 (rank : 41) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9C0E4, Q8TEH9, Q9H7H3 | Gene names | GRIP2, KIAA1719 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor-interacting protein 2 (GRIP2 protein). | |||||
|
LR37B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.010299 (rank : 46) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96QE4, Q5YKG6 | Gene names | LRRC37B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37B precursor (C66 SLIT-like testicular protein). | |||||
|
RUSC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | 0.011385 (rank : 45) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BVN2, Q9UPY4, Q9Y4T5 | Gene names | RUSC1, NESCA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 1 (New molecule containing SH3 at the carboxy-terminus) (Nesca). | |||||
|
GLGB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | 0.018118 (rank : 37) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q04446, Q96EN0 | Gene names | GBE1 | |||
|
Domain Architecture |
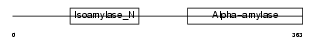
|
|||||
| Description | 1,4-alpha-glucan branching enzyme (EC 2.4.1.18) (Glycogen branching enzyme) (Brancher enzyme). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.009400 (rank : 47) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
S23IP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 51) | NC score | 0.013930 (rank : 42) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6NZC7, Q8BXY6 | Gene names | Sec23ip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SEC23-interacting protein. | |||||
|
SOX13_MOUSE
|
||||||
| θ value | 8.99809 (rank : 52) | NC score | 0.009199 (rank : 48) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q04891, Q922L3 | Gene names | Sox13, Sox-13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein. | |||||
|
SSBP4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | 0.024952 (rank : 32) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
ZN385_HUMAN
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.018241 (rank : 36) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96PM9, Q5VH53, Q9H7R6, Q9UFU3 | Gene names | ZNF385, HZF, RZF | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 385 (Hematopoietic zinc finger protein) (Retinal zinc finger protein). | |||||
|
GR65_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q91X51, Q9D3L9 | Gene names | Gorasp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 1 (Golgi reassembly-stacking protein of 65 kDa) (GRASP65) (Golgi peripheral membrane protein p65). | |||||
|
GR65_HUMAN
|
||||||
| NC score | 0.957825 (rank : 2) | θ value | 1.77876e-181 (rank : 2) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9BQQ3, Q8N272, Q96H42 | Gene names | GORASP1, GRASP65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 1 (Golgi reassembly-stacking protein of 65 kDa) (GRASP65) (Golgi peripheral membrane protein p65) (Golgi phosphoprotein 5) (GOLPH5). | |||||
|
GORS2_HUMAN
|
||||||
| NC score | 0.920037 (rank : 3) | θ value | 1.71185e-83 (rank : 3) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9H8Y8, Q96I74, Q96K84, Q9H946, Q9UFW4 | Gene names | GORASP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 2 (GRS2) (Golgi reassembly-stacking protein of 55 kDa) (GRASP55) (p59) (Golgi phosphoprotein 6) (GOLPH6). | |||||
|
GORS2_MOUSE
|
||||||
| NC score | 0.913370 (rank : 4) | θ value | 1.71185e-83 (rank : 4) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99JX3, Q3U9D2, Q8CCD0, Q91W68 | Gene names | Gorasp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 2 (GRS2) (Golgi reassembly-stacking protein of 55 kDa) (GRASP55). | |||||
|
PSMD9_MOUSE
|
||||||
| NC score | 0.223808 (rank : 5) | θ value | 0.0113563 (rank : 5) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CR00, Q3TG66 | Gene names | Psmd9 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 9 (26S proteasome regulatory subunit p27). | |||||
|
PSMD9_HUMAN
|
||||||
| NC score | 0.209755 (rank : 6) | θ value | 0.0330416 (rank : 6) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O00233, Q9BQ42 | Gene names | PSMD9 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 9 (26S proteasome regulatory subunit p27). | |||||
|
PDCD7_MOUSE
|
||||||
| NC score | 0.075325 (rank : 7) | θ value | 0.163984 (rank : 9) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9WTY1, Q8R5D9 | Gene names | Pdcd7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Programmed cell death protein 7 (ES18). | |||||
|
WASIP_HUMAN
|
||||||
| NC score | 0.071230 (rank : 8) | θ value | 0.163984 (rank : 10) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
PDCD7_HUMAN
|
||||||
| NC score | 0.062994 (rank : 9) | θ value | 0.813845 (rank : 17) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N8D1, Q96AK8, Q9Y6D7 | Gene names | PDCD7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Programmed cell death protein 7 (ES18) (HES18). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.061260 (rank : 10) | θ value | 0.125558 (rank : 7) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
LONM_HUMAN
|
||||||
| NC score | 0.057639 (rank : 11) | θ value | 0.21417 (rank : 11) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P36776, P36777, Q9UQ95 | Gene names | PRSS15 | |||
|
Domain Architecture |

|
|||||
| Description | Lon protease homolog, mitochondrial precursor (EC 3.4.21.-) (Lon protease-like protein) (LONP) (LONHs). | |||||
|
WASIP_MOUSE
|
||||||
| NC score | 0.056583 (rank : 12) | θ value | 5.27518 (rank : 43) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
ENAM_HUMAN
|
||||||
| NC score | 0.052294 (rank : 13) | θ value | 1.06291 (rank : 19) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NRM1, Q9H3D1 | Gene names | ENAM | |||
|
Domain Architecture |

|
|||||
| Description | Enamelin precursor. | |||||
|
SPAG8_HUMAN
|
||||||
| NC score | 0.049291 (rank : 14) | θ value | 2.36792 (rank : 27) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99932, Q12937, Q5TCV8, Q8WWB4 | Gene names | SPAG8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 8 (Sperm membrane protein 1) (SMP-1) (HSD-1) (Sperm membrane protein BS-84). | |||||
|
SF01_HUMAN
|
||||||
| NC score | 0.043187 (rank : 15) | θ value | 3.0926 (rank : 32) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15637, Q14818, Q14819, Q15913, Q8IY00, Q92744, Q92745, Q969H7, Q9BW01, Q9UEI0 | Gene names | SF1, ZFM1, ZNF162 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 1 (Zinc finger protein 162) (Transcription factor ZFM1) (Zinc finger gene in MEN1 locus) (Mammalian branch point-binding protein mBBP) (BBP). | |||||
|
SNPC4_HUMAN
|
||||||
| NC score | 0.040295 (rank : 16) | θ value | 0.62314 (rank : 15) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5SXM2, Q9Y6P7 | Gene names | SNAPC4, SNAP190 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor subunit alpha) (PSE-binding factor subunit alpha) (PTF subunit alpha). | |||||
|
SMRC2_HUMAN
|
||||||
| NC score | 0.039884 (rank : 17) | θ value | 1.38821 (rank : 21) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8TAQ2, Q92923, Q96E12, Q96GY4 | Gene names | SMARCC2, BAF170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
TRI66_HUMAN
|
||||||
| NC score | 0.037340 (rank : 18) | θ value | 1.81305 (rank : 24) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O15016, Q9BQQ4 | Gene names | TRIM66, KIAA0298 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
IL3RB_MOUSE
|
||||||
| NC score | 0.035892 (rank : 19) | θ value | 0.47712 (rank : 13) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P26955 | Gene names | Csf2rb, Aic2b, Csf2rb1, Il3rb1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytokine receptor common beta chain precursor (GM-CSF/IL-3/IL-5 receptor common beta-chain) (CD131 antigen). | |||||
|
CPSF6_HUMAN
|
||||||
| NC score | 0.034908 (rank : 20) | θ value | 3.0926 (rank : 28) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q16630, Q53ES1, Q9BSJ7, Q9BW18 | Gene names | CPSF6, CFIM68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6 (Cleavage and polyadenylation specificity factor 68 kDa subunit) (CPSF 68 kDa subunit) (Pre-mRNA cleavage factor Im 68 kDa subunit) (Protein HPBRII- 4/7). | |||||
|
CPSF6_MOUSE
|
||||||
| NC score | 0.034905 (rank : 21) | θ value | 3.0926 (rank : 29) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6NVF9, Q8BX86, Q8BXI8 | Gene names | Cpsf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6. | |||||
|
CBX2_HUMAN
|
||||||
| NC score | 0.034541 (rank : 22) | θ value | 1.81305 (rank : 22) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14781, Q9BTB1 | Gene names | CBX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromobox protein homolog 2. | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.034476 (rank : 23) | θ value | 1.81305 (rank : 23) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.032844 (rank : 24) | θ value | 6.88961 (rank : 44) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.032792 (rank : 25) | θ value | 0.813845 (rank : 16) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
SYN1_HUMAN
|
||||||
| NC score | 0.031572 (rank : 26) | θ value | 0.813845 (rank : 18) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P17600, O75825, Q5H9A9 | Gene names | SYN1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I) (Brain protein 4.1). | |||||
|
3BP2_MOUSE
|
||||||
| NC score | 0.028600 (rank : 27) | θ value | 4.03905 (rank : 33) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q06649 | Gene names | Sh3bp2, 3bp2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
HCFC1_HUMAN
|
||||||
| NC score | 0.026155 (rank : 28) | θ value | 4.03905 (rank : 35) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P51610, Q6P4G5 | Gene names | HCFC1, HCF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) (VP16 accessory protein) (VCAF) (CFF) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N-terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C- terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
HCFC1_MOUSE
|
||||||
| NC score | 0.025338 (rank : 29) | θ value | 5.27518 (rank : 40) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61191, Q684R1, Q7TSB0, Q8C2D0, Q9QWH2 | Gene names | Hcfc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N- terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C-terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
ATF5_MOUSE
|
||||||
| NC score | 0.025271 (rank : 30) | θ value | 4.03905 (rank : 34) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O70191, Q58E51, Q99NH6 | Gene names | Atf5, Atfx, Nap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-5 (Activating transcription factor 5-alpha/beta) (Transcription factor ATFx) (Transcription factor-like protein ODA-10) (BZIP protein ATF7) (NRIF3- associated protein) (NAP1). | |||||
|
UBXD6_HUMAN
|
||||||
| NC score | 0.025057 (rank : 31) | θ value | 4.03905 (rank : 38) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00124, Q7Z6F2 | Gene names | UBXD6, D8S2298E, REP8 | |||
|
Domain Architecture |

|
|||||
| Description | UBX domain-containing protein 6 (Reproduction 8 protein) (Protein Rep- 8). | |||||
|
SSBP4_HUMAN
|
||||||
| NC score | 0.024952 (rank : 32) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
AB1IP_HUMAN
|
||||||
| NC score | 0.021988 (rank : 33) | θ value | 2.36792 (rank : 25) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7Z5R6, Q8IWS8, Q8IZZ7 | Gene names | APBB1IP, PREL1, RARP1, RIAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 1- interacting protein (APBB1-interacting protein 1) (Rap1-GTP- interacting adapter molecule) (RIAM) (Proline-rich EVH1 ligand 1) (PREL-1) (Proline-rich protein 73) (Retinoic acid-responsive proline- rich protein 1) (RARP-1). | |||||
|
MGR1_MOUSE
|
||||||
| NC score | 0.021052 (rank : 34) | θ value | 0.163984 (rank : 8) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97772, Q6AXG4, Q9EPV6 | Gene names | Grm1, Gprc1a, Mglur1 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 1 precursor (mGluR1). | |||||
|
PDLI7_HUMAN
|
||||||
| NC score | 0.019337 (rank : 35) | θ value | 1.38821 (rank : 20) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 547 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NR12, Q14250, Q5XG82, Q6NVZ5, Q96C91, Q9BXB8, Q9BXB9 | Gene names | PDLIM7, ENIGMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
ZN385_HUMAN
|
||||||
| NC score | 0.018241 (rank : 36) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96PM9, Q5VH53, Q9H7R6, Q9UFU3 | Gene names | ZNF385, HZF, RZF | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 385 (Hematopoietic zinc finger protein) (Retinal zinc finger protein). | |||||
|
GLGB_HUMAN
|
||||||
| NC score | 0.018118 (rank : 37) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q04446, Q96EN0 | Gene names | GBE1 | |||
|
Domain Architecture |

|
|||||
| Description | 1,4-alpha-glucan branching enzyme (EC 2.4.1.18) (Glycogen branching enzyme) (Brancher enzyme). | |||||
|
REL_MOUSE
|
||||||
| NC score | 0.017026 (rank : 38) | θ value | 2.36792 (rank : 26) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P15307 | Gene names | Rel | |||
|
Domain Architecture |

|
|||||
| Description | C-Rel proto-oncogene protein (C-Rel protein). | |||||
|
DNJC1_HUMAN
|
||||||
| NC score | 0.016384 (rank : 39) | θ value | 3.0926 (rank : 30) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96KC8 | Gene names | DNAJC1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 1. | |||||
|
WWP1_HUMAN
|
||||||
| NC score | 0.015833 (rank : 40) | θ value | 0.47712 (rank : 14) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H0M0, O00307, Q96BP4 | Gene names | WWP1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1) (Atropin-1-interacting protein 5) (AIP5). | |||||
|
GRIP2_HUMAN
|
||||||
| NC score | 0.014459 (rank : 41) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9C0E4, Q8TEH9, Q9H7H3 | Gene names | GRIP2, KIAA1719 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor-interacting protein 2 (GRIP2 protein). | |||||
|
S23IP_MOUSE
|
||||||
| NC score | 0.013930 (rank : 42) | θ value | 8.99809 (rank : 51) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6NZC7, Q8BXY6 | Gene names | Sec23ip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SEC23-interacting protein. | |||||
|
MICA1_HUMAN
|
||||||
| NC score | 0.013467 (rank : 43) | θ value | 4.03905 (rank : 36) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8TDZ2, Q7Z633, Q8IVS9, Q96G47, Q9H6X6, Q9H7I0, Q9HAA1, Q9UFF7 | Gene names | MICAL1, MICAL, NICAL | |||
|
Domain Architecture |

|
|||||
| Description | NEDD9-interacting protein with calponin homology and LIM domains (Molecule interacting with CasL protein 1). | |||||
|
PDLI7_MOUSE
|
||||||
| NC score | 0.013380 (rank : 44) | θ value | 5.27518 (rank : 41) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3TJD7, Q80ZY6, Q810S3, Q8C1S4, Q9CRA1 | Gene names | Pdlim7, Enigma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
RUSC1_HUMAN
|
||||||
| NC score | 0.011385 (rank : 45) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BVN2, Q9UPY4, Q9Y4T5 | Gene names | RUSC1, NESCA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 1 (New molecule containing SH3 at the carboxy-terminus) (Nesca). | |||||
|
LR37B_HUMAN
|
||||||
| NC score | 0.010299 (rank : 46) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96QE4, Q5YKG6 | Gene names | LRRC37B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37B precursor (C66 SLIT-like testicular protein). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.009400 (rank : 47) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
SOX13_MOUSE
|
||||||
| NC score | 0.009199 (rank : 48) | θ value | 8.99809 (rank : 52) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q04891, Q922L3 | Gene names | Sox13, Sox-13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein. | |||||
|
OBSCN_HUMAN
|
||||||
| NC score | 0.008933 (rank : 49) | θ value | 0.21417 (rank : 12) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
DUS4_MOUSE
|
||||||
| NC score | 0.008033 (rank : 50) | θ value | 5.27518 (rank : 39) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BFV3 | Gene names | Dusp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 4 (EC 3.1.3.48) (EC 3.1.3.16). | |||||
|
HXA10_HUMAN
|
||||||
| NC score | 0.006387 (rank : 51) | θ value | 3.0926 (rank : 31) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P31260, O43370, O43605, Q15949 | Gene names | HOXA10, HOX1H | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A10 (Hox-1H) (Hox-1.8) (PL). | |||||
|
TIE1_HUMAN
|
||||||
| NC score | 0.006189 (rank : 52) | θ value | 4.03905 (rank : 37) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35590 | Gene names | TIE1, TIE | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase receptor Tie-1 precursor (EC 2.7.10.1). | |||||
|
TIE1_MOUSE
|
||||||
| NC score | 0.004969 (rank : 53) | θ value | 5.27518 (rank : 42) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1266 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q06806, Q811F4 | Gene names | Tie1, Tie, Tie-1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase receptor Tie-1 precursor (EC 2.7.10.1). | |||||
|
GLIS1_MOUSE
|
||||||
| NC score | 0.000247 (rank : 54) | θ value | 6.88961 (rank : 45) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K1M4, Q6NZF6, Q8R4Z3 | Gene names | Glis1, Gli5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLIS1 (GLI-similar 1) (Gli homologous protein 1) (GliH1). | |||||