Please be patient as the page loads
|
GR65_HUMAN
|
||||||
| SwissProt Accessions | Q9BQQ3, Q8N272, Q96H42 | Gene names | GORASP1, GRASP65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 1 (Golgi reassembly-stacking protein of 65 kDa) (GRASP65) (Golgi peripheral membrane protein p65) (Golgi phosphoprotein 5) (GOLPH5). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
GR65_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 148 | |
| SwissProt Accessions | Q9BQQ3, Q8N272, Q96H42 | Gene names | GORASP1, GRASP65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 1 (Golgi reassembly-stacking protein of 65 kDa) (GRASP65) (Golgi peripheral membrane protein p65) (Golgi phosphoprotein 5) (GOLPH5). | |||||
|
GR65_MOUSE
|
||||||
| θ value | 1.77876e-181 (rank : 2) | NC score | 0.957825 (rank : 2) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91X51, Q9D3L9 | Gene names | Gorasp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 1 (Golgi reassembly-stacking protein of 65 kDa) (GRASP65) (Golgi peripheral membrane protein p65). | |||||
|
GORS2_HUMAN
|
||||||
| θ value | 1.54831e-84 (rank : 3) | NC score | 0.904480 (rank : 3) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H8Y8, Q96I74, Q96K84, Q9H946, Q9UFW4 | Gene names | GORASP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 2 (GRS2) (Golgi reassembly-stacking protein of 55 kDa) (GRASP55) (p59) (Golgi phosphoprotein 6) (GOLPH6). | |||||
|
GORS2_MOUSE
|
||||||
| θ value | 1.54831e-84 (rank : 4) | NC score | 0.897445 (rank : 4) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99JX3, Q3U9D2, Q8CCD0, Q91W68 | Gene names | Gorasp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 2 (GRS2) (Golgi reassembly-stacking protein of 55 kDa) (GRASP55). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 5) | NC score | 0.097254 (rank : 8) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
PSMD9_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 6) | NC score | 0.227977 (rank : 5) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CR00, Q3TG66 | Gene names | Psmd9 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 9 (26S proteasome regulatory subunit p27). | |||||
|
PSMD9_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 7) | NC score | 0.214607 (rank : 6) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O00233, Q9BQ42 | Gene names | PSMD9 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 9 (26S proteasome regulatory subunit p27). | |||||
|
FMNL_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 8) | NC score | 0.080221 (rank : 11) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O95466, Q6DKG5, Q6IBP3, Q86UH1, Q8N671, Q8TDH1, Q96H10 | Gene names | FMNL1, C17orf1, C17orf1B, FMNL | |||
|
Domain Architecture |

|
|||||
| Description | Formin-like protein 1 (Leukocyte formin) (CLL-associated antigen KW- 13). | |||||
|
SN1L2_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 9) | NC score | 0.013918 (rank : 134) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8CFH6 | Gene names | Snf1lk2, Sik2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SNF1-like kinase 2 (EC 2.7.11.1) (Salt-inducible kinase 2). | |||||
|
ABLM1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 10) | NC score | 0.022340 (rank : 94) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O14639, Q15039, Q68CQ9, Q9BUP1 | Gene names | ABLIM1, ABLIM, KIAA0059 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding LIM protein 1 (Actin-binding LIM protein family member 1) (abLIM-1) (Actin-binding double-zinc-finger protein) (LIMAB1) (Limatin). | |||||
|
PDXL2_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 11) | NC score | 0.070774 (rank : 15) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8CAE9, Q8CFW3 | Gene names | Podxl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Podocalyxin-like protein 2 precursor (Endoglycan). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 12) | NC score | 0.082661 (rank : 10) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
TAF6L_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 13) | NC score | 0.055239 (rank : 26) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8R2K4, Q8C5T2 | Gene names | Taf6l, Paf65a | |||
|
Domain Architecture |
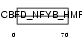
|
|||||
| Description | TAF6-like RNA polymerase II p300/CBP-associated factor-associated factor 65 kDa subunit 6L (PCAF-associated factor 65 alpha) (PAF65- alpha). | |||||
|
NF2L2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 14) | NC score | 0.038794 (rank : 48) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q16236, Q96F71 | Gene names | NFE2L2, NRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 2 (NF-E2-related factor 2) (NFE2-related factor 2) (Nuclear factor, erythroid derived 2, like 2) (HEBP1). | |||||
|
SMRC2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 15) | NC score | 0.065895 (rank : 19) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8TAQ2, Q92923, Q96E12, Q96GY4 | Gene names | SMARCC2, BAF170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
PGCA_MOUSE
|
||||||
| θ value | 0.279714 (rank : 16) | NC score | 0.024445 (rank : 88) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
SN1L2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 17) | NC score | 0.012823 (rank : 138) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H0K1, O94878, Q6AZE2, Q76N03, Q8NCV7, Q96CZ8 | Gene names | SNF1LK2, KIAA0781, QIK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SNF1-like kinase 2 (EC 2.7.11.1) (Qin- induced kinase). | |||||
|
TIE1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 18) | NC score | 0.010984 (rank : 141) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P35590 | Gene names | TIE1, TIE | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase receptor Tie-1 precursor (EC 2.7.10.1). | |||||
|
ALMS1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 19) | NC score | 0.043464 (rank : 43) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8TCU4, Q53S05, Q580Q8, Q86VP9, Q9Y4G4 | Gene names | ALMS1, KIAA0328 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alstrom syndrome protein 1. | |||||
|
IRF3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 20) | NC score | 0.035774 (rank : 60) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q14653 | Gene names | IRF3 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 3 (IRF-3). | |||||
|
KBTB9_MOUSE
|
||||||
| θ value | 0.365318 (rank : 21) | NC score | 0.015034 (rank : 125) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80T74 | Gene names | Kbtbd9, Kiaa1921 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch repeat and BTB domain-containing protein 9. | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 22) | NC score | 0.073164 (rank : 14) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
APXL_HUMAN
|
||||||
| θ value | 0.47712 (rank : 23) | NC score | 0.049729 (rank : 36) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13796 | Gene names | APXL | |||
|
Domain Architecture |

|
|||||
| Description | Apical-like protein (Protein APXL). | |||||
|
ENAM_HUMAN
|
||||||
| θ value | 0.47712 (rank : 24) | NC score | 0.065498 (rank : 20) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NRM1, Q9H3D1 | Gene names | ENAM | |||
|
Domain Architecture |

|
|||||
| Description | Enamelin precursor. | |||||
|
RX_HUMAN
|
||||||
| θ value | 0.47712 (rank : 25) | NC score | 0.016308 (rank : 120) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y2V3 | Gene names | RAX, RX | |||
|
Domain Architecture |

|
|||||
| Description | Retinal homeobox protein Rx (Retina and anterior neural fold homeobox protein). | |||||
|
WASIP_HUMAN
|
||||||
| θ value | 0.47712 (rank : 26) | NC score | 0.101180 (rank : 7) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
HXB3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 27) | NC score | 0.021676 (rank : 95) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P14651, O95615, P17484 | Gene names | HOXB3, HOX2G | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B3 (Hox-2G) (Hox-2.7). | |||||
|
SF01_HUMAN
|
||||||
| θ value | 0.62314 (rank : 28) | NC score | 0.070199 (rank : 17) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q15637, Q14818, Q14819, Q15913, Q8IY00, Q92744, Q92745, Q969H7, Q9BW01, Q9UEI0 | Gene names | SF1, ZFM1, ZNF162 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 1 (Zinc finger protein 162) (Transcription factor ZFM1) (Zinc finger gene in MEN1 locus) (Mammalian branch point-binding protein mBBP) (BBP). | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | 0.813845 (rank : 29) | NC score | 0.045913 (rank : 41) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
WASIP_MOUSE
|
||||||
| θ value | 0.813845 (rank : 30) | NC score | 0.087678 (rank : 9) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
CEP41_HUMAN
|
||||||
| θ value | 1.06291 (rank : 31) | NC score | 0.037653 (rank : 54) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BYV8, Q7Z496, Q86TM1, Q8NFU8, Q9H6A3, Q9NPV3 | Gene names | TSGA14, CEP41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 41 kDa (Cep41 protein) (Testis-specific gene A14 protein). | |||||
|
CU118_HUMAN
|
||||||
| θ value | 1.06291 (rank : 32) | NC score | 0.063891 (rank : 22) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 2 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96PM8 | Gene names | C21orf118, PRED85 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B lymphocyte activation-related protein BC-1514. | |||||
|
ENAH_HUMAN
|
||||||
| θ value | 1.06291 (rank : 33) | NC score | 0.038731 (rank : 49) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8N8S7, Q502W5, Q5T5M7, Q5VTQ9, Q5VTR0, Q9NVF3, Q9UFB8 | Gene names | ENAH, MENA | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog. | |||||
|
FCRLA_HUMAN
|
||||||
| θ value | 1.06291 (rank : 34) | NC score | 0.026650 (rank : 84) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7L513, Q5VXA1, Q5VXA2, Q5VXA3, Q5VXB0, Q8NEW4, Q8WXH3, Q96PC6, Q96PJ0, Q96PJ1, Q96PJ2, Q96PJ4, Q9BR57 | Gene names | FCRLA, FCRL, FCRL1, FCRLM1, FCRX, FREB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fc receptor-like and mucin-like 1 precursor (Fc receptor-like A) (Fc receptor homolog expressed in B-cells) (Fc receptor-related protein X) (FcRX) (Fc receptor-like protein). | |||||
|
FMN2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 35) | NC score | 0.056583 (rank : 25) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9JL04 | Gene names | Fmn2 | |||
|
Domain Architecture |

|
|||||
| Description | Formin-2. | |||||
|
FREM2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 36) | NC score | 0.017256 (rank : 116) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5SZK8, Q4QQG1, Q5H9N8, Q5T6Q1, Q6N057, Q6ZSB4, Q7Z305, Q7Z341 | Gene names | FREM2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FRAS1-related extracellular matrix protein 2 precursor (ECM3 homolog). | |||||
|
K1849_MOUSE
|
||||||
| θ value | 1.06291 (rank : 37) | NC score | 0.026078 (rank : 86) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q69Z89, Q8BNL0, Q8C549, Q8CAL1 | Gene names | Kiaa1849 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1849. | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 38) | NC score | 0.079546 (rank : 12) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
NHS_HUMAN
|
||||||
| θ value | 1.06291 (rank : 39) | NC score | 0.052547 (rank : 30) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6T4R5, Q5J7Q0, Q5J7Q1, Q68DR5 | Gene names | NHS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nance-Horan syndrome protein (Congenital cataracts and dental anomalies protein). | |||||
|
TCRG1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 40) | NC score | 0.034226 (rank : 67) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
ZDHC8_MOUSE
|
||||||
| θ value | 1.06291 (rank : 41) | NC score | 0.028901 (rank : 80) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5Y5T5, Q7TNF7 | Gene names | Zdhhc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable palmitoyltransferase ZDHHC8 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 8) (DHHC-8). | |||||
|
ENAH_MOUSE
|
||||||
| θ value | 1.38821 (rank : 42) | NC score | 0.052743 (rank : 29) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
MILK1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 43) | NC score | 0.039387 (rank : 47) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 1.38821 (rank : 44) | NC score | 0.056618 (rank : 24) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
TIE1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 45) | NC score | 0.009786 (rank : 144) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1266 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q06806, Q811F4 | Gene names | Tie1, Tie, Tie-1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase receptor Tie-1 precursor (EC 2.7.10.1). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 1.81305 (rank : 46) | NC score | 0.068793 (rank : 18) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
OTU7B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 47) | NC score | 0.028151 (rank : 82) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6GQQ9, Q8WWA7, Q9NQ53, Q9UFF4 | Gene names | OTUD7B, ZA20D1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | OTU domain-containing protein 7B (EC 3.-.-.-) (Zinc finger protein Cezanne) (Zinc finger A20 domain-containing protein 1) (Cellular zinc finger anti-NF-kappa B protein). | |||||
|
TCRG1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.033037 (rank : 72) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
ZDHC8_HUMAN
|
||||||
| θ value | 1.81305 (rank : 49) | NC score | 0.030043 (rank : 79) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9ULC8, Q6ICL1, Q6ZNF5, Q7Z6L9 | Gene names | ZDHHC8, KIAA1292, ZDHHCL1, ZNF378 | |||
|
Domain Architecture |
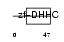
|
|||||
| Description | Probable palmitoyltransferase ZDHHC8 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 8) (DHHC-8) (Zinc finger protein 378). | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 2.36792 (rank : 50) | NC score | 0.061731 (rank : 23) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
CO8A2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 51) | NC score | 0.017636 (rank : 112) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P25318, Q68ED0, Q6KAQ4, Q6P1C4 | Gene names | Col8a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-2(VIII) chain precursor (Endothelial collagen). | |||||
|
CSKI1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 52) | NC score | 0.032802 (rank : 73) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
CSKI1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 53) | NC score | 0.035681 (rank : 61) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
CSKI2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 54) | NC score | 0.030502 (rank : 77) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8WXE0, Q7LG69, Q9ULT1 | Gene names | CASKIN2, KIAA1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
CSKI2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 55) | NC score | 0.031310 (rank : 76) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 812 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8VHK1, Q6ZPX1, Q7TS77 | Gene names | Caskin2, Kiaa1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
DIAP2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 56) | NC score | 0.036469 (rank : 57) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O60879, O60878, Q8WX06, Q8WX48, Q9UJL2 | Gene names | DIAPH2, DIA | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 2 (Diaphanous-related formin-2) (DRF2). | |||||
|
MTSS1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 57) | NC score | 0.038127 (rank : 52) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O43312, Q8TCA2, Q96RX2 | Gene names | MTSS1, KIAA0429, MIM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein) (Metastasis suppressor YGL-1). | |||||
|
NUFP1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 58) | NC score | 0.034933 (rank : 65) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9QXX8, Q9CV69 | Gene names | Nufip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear fragile X mental retardation-interacting protein 1 (Nuclear FMRP-interacting protein 1). | |||||
|
PO6F2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 59) | NC score | 0.022667 (rank : 91) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P78424, P78425, Q75ME8, Q86UM6, Q9UDS7 | Gene names | POU6F2, RPF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 2 (Retina-derived POU-domain factor 1) (RPF-1). | |||||
|
RTN4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 60) | NC score | 0.032191 (rank : 74) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NQC3, O94962, Q9BXG5, Q9H212, Q9H3I3, Q9UQ42, Q9Y293, Q9Y2Y7, Q9Y5U6 | Gene names | RTN4, KIAA0886, NOGO | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-4 (Neurite outgrowth inhibitor) (Nogo protein) (Foocen) (Neuroendocrine-specific protein) (NSP) (Neuroendocrine-specific protein C homolog) (RTN-x) (Reticulon-5). | |||||
|
ZC3H6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 61) | NC score | 0.024084 (rank : 89) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P61129 | Gene names | ZC3H6, KIAA2035, ZC3HDC6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
AMPH_HUMAN
|
||||||
| θ value | 3.0926 (rank : 62) | NC score | 0.025006 (rank : 87) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P49418, O43538, Q75MJ8, Q75MK5, Q75MM3, Q8N4G0 | Gene names | AMPH, AMPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Amphiphysin. | |||||
|
DTX1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 63) | NC score | 0.023462 (rank : 90) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61010, Q3TER5, Q8C2H2, Q9ER09 | Gene names | Dtx1 | |||
|
Domain Architecture |
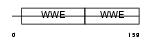
|
|||||
| Description | Protein deltex-1 (Deltex-1) (Deltex1) (mDTX1) (FXI-T1). | |||||
|
KCNC3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.017171 (rank : 117) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14003 | Gene names | KCNC3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
KCNC3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.014528 (rank : 131) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q63959, Q62088 | Gene names | Kcnc3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
MKL1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.039452 (rank : 46) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 67) | NC score | 0.033721 (rank : 68) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 3.0926 (rank : 68) | NC score | 0.070655 (rank : 16) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
RERE_HUMAN
|
||||||
| θ value | 3.0926 (rank : 69) | NC score | 0.040483 (rank : 44) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
RP3A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 70) | NC score | 0.015414 (rank : 122) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y2J0, Q96AE0 | Gene names | RPH3A, KIAA0985 | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
SENP3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 71) | NC score | 0.018046 (rank : 111) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H4L4, Q96PS4, Q9Y3W9 | Gene names | SENP3, SSP3, SUSP3 | |||
|
Domain Architecture |
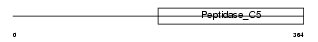
|
|||||
| Description | Sentrin-specific protease 3 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP3) (SUMO-1-specific protease 3). | |||||
|
3MG_HUMAN
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.026121 (rank : 85) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P29372, Q13770, Q15275, Q15961, Q96BZ6, Q96S33, Q9NNX5 | Gene names | MPG, AAG, ANPG, MID1 | |||
|
Domain Architecture |
|
|||||
| Description | DNA-3-methyladenine glycosylase (EC 3.2.2.21) (3-methyladenine DNA glycosidase) (ADPG) (3-alkyladenine DNA glycosylase) (N-methylpurine- DNA glycosylase). | |||||
|
AFAD_HUMAN
|
||||||
| θ value | 4.03905 (rank : 73) | NC score | 0.019456 (rank : 105) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P55196, O75087, O75088, O75089, Q5TIG6, Q9NSN7, Q9NU92 | Gene names | MLLT4, AF6 | |||
|
Domain Architecture |

|
|||||
| Description | Afadin (Protein AF-6). | |||||
|
CASC3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 74) | NC score | 0.044948 (rank : 42) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8K3W3, Q8K219, Q99NF0 | Gene names | Casc3, Mln51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC3 (Cancer susceptibility candidate gene 3 protein homolog) (Metastatic lymph node protein 51 homolog) (Protein MLN 51 homolog) (Protein barentsz) (Btz). | |||||
|
FGD1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 75) | NC score | 0.016601 (rank : 118) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 367 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P98174, Q8N4D9 | Gene names | FGD1, ZFYVE3 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
FMN2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | 0.054613 (rank : 27) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9NZ56, Q59GF6, Q5VU37, Q9NZ55 | Gene names | FMN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-2. | |||||
|
HXB3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.015023 (rank : 126) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P09026, P10285, Q4PJ06, Q61680 | Gene names | Hoxb3, Hox-2.7, Hoxb-3 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B3 (Hox-2.7) (Homeobox protein MH-23). | |||||
|
IQEC2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | 0.021376 (rank : 96) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5DU25 | Gene names | Iqsec2, Kiaa0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
JPH4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.016497 (rank : 119) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q80WT0, Q8BMI1 | Gene names | Jph4, Jphl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-4 (Junctophilin-like 1 protein). | |||||
|
MK07_HUMAN
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.008689 (rank : 148) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1087 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13164, Q16634 | Gene names | MAPK7, ERK4, ERK5, PRKM7 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase 7 (EC 2.7.11.24) (Extracellular signal-regulated kinase 5) (ERK-5) (ERK4) (BMK1 kinase). | |||||
|
MYO15_MOUSE
|
||||||
| θ value | 4.03905 (rank : 81) | NC score | 0.015566 (rank : 121) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9QZZ4, O70395, Q9QWL6 | Gene names | Myo15a, Myo15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
SETX_HUMAN
|
||||||
| θ value | 4.03905 (rank : 82) | NC score | 0.020591 (rank : 98) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7Z333, O75120, Q3KQX4, Q68DW5, Q6AZD7, Q7Z3J6, Q8WX33, Q9H9D1, Q9NVP9 | Gene names | SETX, ALS4, KIAA0625, SCAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable helicase senataxin (EC 3.6.1.-) (SEN1 homolog). | |||||
|
SMCA4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.018067 (rank : 110) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P51532, O95052, Q9HBD3 | Gene names | SMARCA4, BRG1, SNF2B, SNF2L4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L4 (EC 3.6.1.-) (ATP- dependent helicase SMARCA4) (SNF2-beta) (BRG-1 protein) (Mitotic growth and transcription activator) (Brahma protein homolog 1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4). | |||||
|
SMRC2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | 0.049594 (rank : 37) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6PDG5, Q6P6P3 | Gene names | Smarcc2, Baf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
SPR2F_MOUSE
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.033063 (rank : 71) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O70557 | Gene names | Sprr2f | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2F. | |||||
|
3BP2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.038139 (rank : 51) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q06649 | Gene names | Sh3bp2, 3bp2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
APLP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.014456 (rank : 132) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P51693, O00113, Q96A92 | Gene names | APLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 1 precursor (APLP) (APLP-1) [Contains: C30]. | |||||
|
FOG1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.008740 (rank : 147) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 981 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8IX07 | Gene names | ZFPM1, FOG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
GPTC8_HUMAN
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.018948 (rank : 108) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
MAPK2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.005003 (rank : 149) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P49137, Q8IYD6 | Gene names | MAPKAPK2 | |||
|
Domain Architecture |
|
|||||
| Description | MAP kinase-activated protein kinase 2 (EC 2.7.11.1) (MAPK-activated protein kinase 2) (MAPKAP kinase 2) (MAPKAPK-2) (MK2). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.065014 (rank : 21) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
PRP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.054006 (rank : 28) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PSL1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.036561 (rank : 56) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
SAPS1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.037852 (rank : 53) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
SC24C_HUMAN
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.046923 (rank : 40) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
SPR2E_MOUSE
|
||||||
| θ value | 5.27518 (rank : 96) | NC score | 0.035273 (rank : 63) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O70556 | Gene names | Sprr2e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2E. | |||||
|
TCGAP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 97) | NC score | 0.026772 (rank : 83) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q80YF9, Q6P7W6 | Gene names | Snx26, Tcgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
TRI66_HUMAN
|
||||||
| θ value | 5.27518 (rank : 98) | NC score | 0.038408 (rank : 50) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O15016, Q9BQQ4 | Gene names | TRIM66, KIAA0298 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
3BP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.034535 (rank : 66) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P78314, O00500, O15373, P78315 | Gene names | SH3BP2, 3BP2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
ARI1A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.040067 (rank : 45) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
CBX2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.033283 (rank : 70) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q14781, Q9BTB1 | Gene names | CBX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromobox protein homolog 2. | |||||
|
CJ047_HUMAN
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | 0.033689 (rank : 69) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q86WR7, Q5W0J9, Q5W0K0, Q5W0K1, Q5W0K2, Q6PJC8, Q8N317 | Gene names | C10orf47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf47. | |||||
|
GRIP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | 0.017532 (rank : 114) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9C0E4, Q8TEH9, Q9H7H3 | Gene names | GRIP2, KIAA1719 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor-interacting protein 2 (GRIP2 protein). | |||||
|
HLF_HUMAN
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | 0.020003 (rank : 102) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q16534 | Gene names | HLF | |||
|
Domain Architecture |

|
|||||
| Description | Hepatic leukemia factor. | |||||
|
HLF_MOUSE
|
||||||
| θ value | 6.88961 (rank : 105) | NC score | 0.020054 (rank : 101) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BW74, Q6PF83 | Gene names | Hlf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatic leukemia factor. | |||||
|
HRBL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 106) | NC score | 0.020478 (rank : 99) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q80WC7, Q8BKS5, Q99J67 | Gene names | Hrbl, Rabr | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
KBTB9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 107) | NC score | 0.010470 (rank : 143) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96CT2, Q8N388, Q96BF0, Q96PW7 | Gene names | KBTBD9, KIAA1921 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch repeat and BTB domain-containing protein 9. | |||||
|
MYO15_HUMAN
|
||||||
| θ value | 6.88961 (rank : 108) | NC score | 0.020292 (rank : 100) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
PTN23_HUMAN
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.018781 (rank : 109) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9H3S7, Q7KZF8, Q8N6Z5, Q9BSR5, Q9P257, Q9UG03, Q9UMZ4 | Gene names | PTPN23, KIAA1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His- domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14). | |||||
|
RBM22_HUMAN
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.022529 (rank : 92) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NW64, O95607 | Gene names | RBM22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-splicing factor RBM22 (RNA-binding motif protein 22). | |||||
|
RBM22_MOUSE
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.022529 (rank : 93) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BHS3, Q3TJB0, Q9CXA0 | Gene names | Rbm22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-splicing factor RBM22 (RNA-binding motif protein 22). | |||||
|
TSC2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 112) | NC score | 0.012548 (rank : 139) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61037, P97723, P97724, P97725, P97727, Q61007, Q61008, Q9WUF6 | Gene names | Tsc2 | |||
|
Domain Architecture |

|
|||||
| Description | Tuberin (Tuberous sclerosis 2 homolog protein). | |||||
|
UBIP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 113) | NC score | 0.009116 (rank : 145) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q811S7, Q3UPR3, Q3US11, Q60786, Q8C514 | Gene names | Ubp1, Cp2b, Nf2d9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Upstream-binding protein 1 (Nuclear factor 2d9) (NF2d9). | |||||
|
WASF2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.037413 (rank : 55) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BH43 | Gene names | Wasf2, Wave2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2). | |||||
|
WNK2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.014377 (rank : 133) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 116) | NC score | 0.048282 (rank : 38) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
AKAP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.014862 (rank : 129) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O08715, O08714, P97488 | Gene names | Akap1, Akap | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 1, mitochondrial precursor (Protein kinase A- anchoring protein 1) (PRKA1) (Dual specificity A-kinase-anchoring protein 1) (D-AKAP-1) (Spermatid A-kinase anchor protein) (S-AKAP). | |||||
|
ARHGB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.013261 (rank : 136) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O15085 | Gene names | ARHGEF11, KIAA0380 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 11 (PDZ-RhoGEF). | |||||
|
BCAR1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.028849 (rank : 81) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P56945 | Gene names | BCAR1, CAS, CRKAS | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
COHA1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 120) | NC score | 0.019862 (rank : 104) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q07563, Q99LK8 | Gene names | Col17a1, Bp180, Bpag2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVII) chain (Bullous pemphigoid antigen 2) (180 kDa bullous pemphigoid antigen 2). | |||||
|
DIAP2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 121) | NC score | 0.035085 (rank : 64) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O70566, Q8C2G8 | Gene names | Diaph2, Diap2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 2 (Diaphanous-related formin-2) (DRF2) (mDia3). | |||||
|
FACE1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 122) | NC score | 0.013096 (rank : 137) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75844, Q8NDZ8, Q9UBQ2 | Gene names | ZMPSTE24, FACE1, STE24 | |||
|
Domain Architecture |
|
|||||
| Description | CAAX prenyl protease 1 homolog (EC 3.4.24.84) (Prenyl protein-specific endoprotease 1) (Farnesylated proteins-converting enzyme 1) (FACE-1) (Zinc metalloproteinase Ste24 homolog). | |||||
|
FGFP3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | 0.019126 (rank : 107) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TAT2, Q8NBN0 | Gene names | FGFBP3, C10orf13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibroblast growth factor-binding protein 3 precursor (FGF-binding protein 3) (FGF-BP3) (FGFBP-3). | |||||
|
FOXC2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.004006 (rank : 151) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99958 | Gene names | FOXC2, FKHL14, MFH1 | |||
|
Domain Architecture |
|
|||||
| Description | Forkhead box protein C2 (Forkhead-related protein FKHL14) (Mesenchyme fork head protein 1) (MFH-1 protein) (Transcription factor FKH-14). | |||||
|
GLGB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.017563 (rank : 113) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q04446, Q96EN0 | Gene names | GBE1 | |||
|
Domain Architecture |
|
|||||
| Description | 1,4-alpha-glucan branching enzyme (EC 2.4.1.18) (Glycogen branching enzyme) (Brancher enzyme). | |||||
|
IRX5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 126) | NC score | 0.015015 (rank : 127) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P78411, P78416, Q7Z2E1 | Gene names | IRX5, IRX2A, IRXB2 | |||
|
Domain Architecture |
|
|||||
| Description | Iroquois-class homeodomain protein IRX-5 (Iroquois homeobox protein 5) (Homeodomain protein IRXB2) (IRX-2A). | |||||
|
K2027_MOUSE
|
||||||
| θ value | 8.99809 (rank : 127) | NC score | 0.013907 (rank : 135) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BWS5, Q69Z30 | Gene names | Kiaa2027 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA2027. | |||||
|
LARP4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.021179 (rank : 97) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BWW4, Q3UR69 | Gene names | Larp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | La-related protein 4 (La ribonucleoprotein domain family member 4). | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | 0.015217 (rank : 124) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
MNT_HUMAN
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | 0.030487 (rank : 78) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
NR1H2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.002381 (rank : 153) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60644 | Gene names | Nr1h2, Lxrb, Rip15, Unr, Unr2 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterols receptor LXR-beta (Liver X receptor beta) (Nuclear orphan receptor LXR-beta) (Ubiquitously-expressed nuclear receptor) (Retinoid X receptor-interacting protein No.15). | |||||
|
NUFP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | 0.036181 (rank : 58) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UHK0, Q8WVM5, Q96SG1 | Gene names | NUFIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear fragile X mental retardation-interacting protein 1 (Nuclear FMRP-interacting protein 1). | |||||
|
PCGF2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | 0.019906 (rank : 103) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P35227 | Gene names | PCGF2, MEL18, RNF110, ZNF144 | |||
|
Domain Architecture |
|
|||||
| Description | Polycomb group RING finger protein 2 (DNA-binding protein Mel-18) (RING finger protein 110) (Zinc finger protein 144). | |||||
|
PCGF2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 134) | NC score | 0.014683 (rank : 130) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P23798 | Gene names | Pcgf2, Mel-18, Mel18, Rnf110, Zfp144, Znf144 | |||
|
Domain Architecture |
|
|||||
| Description | Polycomb group RING finger protein 2 (DNA-binding protein Mel-18) (RING finger protein 110) (Zinc finger protein 144) (Zfp-144). | |||||
|
PHF1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 135) | NC score | 0.010916 (rank : 142) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z1B8, O54808 | Gene names | Phf1, Tctex-3, Tctex3 | |||
|
Domain Architecture |
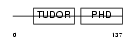
|
|||||
| Description | PHD finger protein 1 (Protein PHF1) (T-complex testis-expressed 3) (Polycomblike 1) (mPCl1). | |||||
|
PIGQ_HUMAN
|
||||||
| θ value | 8.99809 (rank : 136) | NC score | 0.015230 (rank : 123) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BRB3, O14927, Q96G00, Q96S22, Q9UJH4 | Gene names | PIGQ, GPI1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol N-acetylglucosaminyltransferase subunit Q (EC 2.4.1.198) (Phosphatidylinositol-glycan biosynthesis class Q protein) (PIG-Q) (N-acetylglucosamyl transferase component GPI1). | |||||
|
PLK3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 137) | NC score | 0.004132 (rank : 150) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 863 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q60806, Q60822, Q9R009 | Gene names | Plk3, Cnk, Fnk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PLK3 (EC 2.7.11.21) (Polo-like kinase 3) (PLK-3) (Cytokine-inducible serine/threonine-protein kinase) (FGF- inducible kinase). | |||||
|
PRD16_HUMAN
|
||||||
| θ value | 8.99809 (rank : 138) | NC score | -0.001747 (rank : 154) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 832 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9HAZ2, Q8WYJ9, Q9C0I8 | Gene names | PRDM16, KIAA1675, MEL1, PFM13 | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 16 (PR domain-containing protein 16) (Transcription factor MEL1). | |||||
|
PRRT1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 139) | NC score | 0.035622 (rank : 62) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99946, Q96DW3, Q96NQ8 | Gene names | PRRT1, C6orf31, NG5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 1. | |||||
|
PRRT1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 140) | NC score | 0.035885 (rank : 59) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O35449 | Gene names | Prrt1, Ng5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 1. | |||||
|
PTPRG_MOUSE
|
||||||
| θ value | 8.99809 (rank : 141) | NC score | 0.003917 (rank : 152) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q05909 | Gene names | Ptprg | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase gamma precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase gamma) (R-PTP-gamma). | |||||
|
PXK_MOUSE
|
||||||
| θ value | 8.99809 (rank : 142) | NC score | 0.014884 (rank : 128) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 320 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BX57, Q3TD60, Q3TEL7, Q3TZZ1, Q3U197, Q3U773, Q3UCS5, Q4FBH7, Q91WB6 | Gene names | Pxk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PX domain-containing protein kinase-like protein (Modulator of Na,K- ATPase) (MONaKA). | |||||
|
RSMB_MOUSE
|
||||||
| θ value | 8.99809 (rank : 143) | NC score | 0.019361 (rank : 106) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P27048, Q3UJT1 | Gene names | Snrpb | |||
|
Domain Architecture |
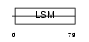
|
|||||
| Description | Small nuclear ribonucleoprotein-associated protein B (snRNP-B) (Sm protein B) (Sm-B) (SmB). | |||||
|
SFPQ_MOUSE
|
||||||
| θ value | 8.99809 (rank : 144) | NC score | 0.031748 (rank : 75) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
TERF2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 145) | NC score | 0.011918 (rank : 140) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35144 | Gene names | Terf2, Trf2 | |||
|
Domain Architecture |

|
|||||
| Description | Telomeric repeat-binding factor 2 (TTAGGG repeat-binding factor 2) (Telomeric DNA-binding protein). | |||||
|
TM119_MOUSE
|
||||||
| θ value | 8.99809 (rank : 146) | NC score | 0.017364 (rank : 115) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R138, Q8BP14 | Gene names | Tmem119 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 119 precursor. | |||||
|
WNK1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 147) | NC score | 0.009089 (rank : 146) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1158 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9H4A3, O15052, Q86WL5, Q8N673, Q9P1S9 | Gene names | WNK1, KDP, KIAA0344, PRKWNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1) (Kinase deficient protein). | |||||
|
YLPM1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 148) | NC score | 0.048259 (rank : 39) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
CF015_HUMAN
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.051037 (rank : 32) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6UXA7, Q5SQ81, Q86Z05, Q9UIG3 | Gene names | C6orf15, STG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf15 precursor (Protein STG). | |||||
|
CIC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.050405 (rank : 33) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
MLL3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.050355 (rank : 34) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.050247 (rank : 35) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
SON_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.077809 (rank : 13) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
SON_MOUSE
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.051798 (rank : 31) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
GR65_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 148 | |
| SwissProt Accessions | Q9BQQ3, Q8N272, Q96H42 | Gene names | GORASP1, GRASP65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 1 (Golgi reassembly-stacking protein of 65 kDa) (GRASP65) (Golgi peripheral membrane protein p65) (Golgi phosphoprotein 5) (GOLPH5). | |||||
|
GR65_MOUSE
|
||||||
| NC score | 0.957825 (rank : 2) | θ value | 1.77876e-181 (rank : 2) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91X51, Q9D3L9 | Gene names | Gorasp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 1 (Golgi reassembly-stacking protein of 65 kDa) (GRASP65) (Golgi peripheral membrane protein p65). | |||||
|
GORS2_HUMAN
|
||||||
| NC score | 0.904480 (rank : 3) | θ value | 1.54831e-84 (rank : 3) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H8Y8, Q96I74, Q96K84, Q9H946, Q9UFW4 | Gene names | GORASP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 2 (GRS2) (Golgi reassembly-stacking protein of 55 kDa) (GRASP55) (p59) (Golgi phosphoprotein 6) (GOLPH6). | |||||
|
GORS2_MOUSE
|
||||||
| NC score | 0.897445 (rank : 4) | θ value | 1.54831e-84 (rank : 4) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99JX3, Q3U9D2, Q8CCD0, Q91W68 | Gene names | Gorasp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 2 (GRS2) (Golgi reassembly-stacking protein of 55 kDa) (GRASP55). | |||||
|
PSMD9_MOUSE
|
||||||
| NC score | 0.227977 (rank : 5) | θ value | 0.00390308 (rank : 6) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CR00, Q3TG66 | Gene names | Psmd9 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 9 (26S proteasome regulatory subunit p27). | |||||
|
PSMD9_HUMAN
|
||||||
| NC score | 0.214607 (rank : 6) | θ value | 0.0113563 (rank : 7) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O00233, Q9BQ42 | Gene names | PSMD9 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 9 (26S proteasome regulatory subunit p27). | |||||
|
WASIP_HUMAN
|
||||||
| NC score | 0.101180 (rank : 7) | θ value | 0.47712 (rank : 26) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.097254 (rank : 8) | θ value | 0.000602161 (rank : 5) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
WASIP_MOUSE
|
||||||
| NC score | 0.087678 (rank : 9) | θ value | 0.813845 (rank : 30) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.082661 (rank : 10) | θ value | 0.0736092 (rank : 12) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
FMNL_HUMAN
|
||||||
| NC score | 0.080221 (rank : 11) | θ value | 0.0148317 (rank : 8) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O95466, Q6DKG5, Q6IBP3, Q86UH1, Q8N671, Q8TDH1, Q96H10 | Gene names | FMNL1, C17orf1, C17orf1B, FMNL | |||
|
Domain Architecture |

|
|||||
| Description | Formin-like protein 1 (Leukocyte formin) (CLL-associated antigen KW- 13). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.079546 (rank : 12) | θ value | 1.06291 (rank : 38) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.077809 (rank : 13) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.073164 (rank : 14) | θ value | 0.365318 (rank : 22) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
PDXL2_MOUSE
|
||||||
| NC score | 0.070774 (rank : 15) | θ value | 0.0736092 (rank : 11) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8CAE9, Q8CFW3 | Gene names | Podxl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Podocalyxin-like protein 2 precursor (Endoglycan). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.070655 (rank : 16) | θ value | 3.0926 (rank : 68) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
SF01_HUMAN
|
||||||
| NC score | 0.070199 (rank : 17) | θ value | 0.62314 (rank : 28) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q15637, Q14818, Q14819, Q15913, Q8IY00, Q92744, Q92745, Q969H7, Q9BW01, Q9UEI0 | Gene names | SF1, ZFM1, ZNF162 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 1 (Zinc finger protein 162) (Transcription factor ZFM1) (Zinc finger gene in MEN1 locus) (Mammalian branch point-binding protein mBBP) (BBP). | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.068793 (rank : 18) | θ value | 1.81305 (rank : 46) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
SMRC2_HUMAN
|
||||||
| NC score | 0.065895 (rank : 19) | θ value | 0.163984 (rank : 15) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8TAQ2, Q92923, Q96E12, Q96GY4 | Gene names | SMARCC2, BAF170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
ENAM_HUMAN
|
||||||
| NC score | 0.065498 (rank : 20) | θ value | 0.47712 (rank : 24) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NRM1, Q9H3D1 | Gene names | ENAM | |||
|
Domain Architecture |

|
|||||
| Description | Enamelin precursor. | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.065014 (rank : 21) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
CU118_HUMAN
|
||||||
| NC score | 0.063891 (rank : 22) | θ value | 1.06291 (rank : 32) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 2 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96PM8 | Gene names | C21orf118, PRED85 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B lymphocyte activation-related protein BC-1514. | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.061731 (rank : 23) | θ value | 2.36792 (rank : 50) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.056618 (rank : 24) | θ value | 1.38821 (rank : 44) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
FMN2_MOUSE
|
||||||
| NC score | 0.056583 (rank : 25) | θ value | 1.06291 (rank : 35) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9JL04 | Gene names | Fmn2 | |||
|
Domain Architecture |

|
|||||
| Description | Formin-2. | |||||
|
TAF6L_MOUSE
|
||||||
| NC score | 0.055239 (rank : 26) | θ value | 0.0961366 (rank : 13) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8R2K4, Q8C5T2 | Gene names | Taf6l, Paf65a | |||
|
Domain Architecture |

|
|||||
| Description | TAF6-like RNA polymerase II p300/CBP-associated factor-associated factor 65 kDa subunit 6L (PCAF-associated factor 65 alpha) (PAF65- alpha). | |||||
|
FMN2_HUMAN
|
||||||
| NC score | 0.054613 (rank : 27) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9NZ56, Q59GF6, Q5VU37, Q9NZ55 | Gene names | FMN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-2. | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.054006 (rank : 28) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
ENAH_MOUSE
|
||||||
| NC score | 0.052743 (rank : 29) | θ value | 1.38821 (rank : 42) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
NHS_HUMAN
|
||||||
| NC score | 0.052547 (rank : 30) | θ value | 1.06291 (rank : 39) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6T4R5, Q5J7Q0, Q5J7Q1, Q68DR5 | Gene names | NHS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nance-Horan syndrome protein (Congenital cataracts and dental anomalies protein). | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.051798 (rank : 31) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
CF015_HUMAN
|
||||||
| NC score | 0.051037 (rank : 32) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6UXA7, Q5SQ81, Q86Z05, Q9UIG3 | Gene names | C6orf15, STG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf15 precursor (Protein STG). | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.050405 (rank : 33) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.050355 (rank : 34) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.050247 (rank : 35) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
APXL_HUMAN
|
||||||
| NC score | 0.049729 (rank : 36) | θ value | 0.47712 (rank : 23) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13796 | Gene names | APXL | |||
|
Domain Architecture |

|
|||||
| Description | Apical-like protein (Protein APXL). | |||||
|
SMRC2_MOUSE
|
||||||
| NC score | 0.049594 (rank : 37) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6PDG5, Q6P6P3 | Gene names | Smarcc2, Baf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.048282 (rank : 38) | θ value | 6.88961 (rank : 116) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
YLPM1_MOUSE
|
||||||
| NC score | 0.048259 (rank : 39) | θ value | 8.99809 (rank : 148) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
SC24C_HUMAN
|
||||||
| NC score | 0.046923 (rank : 40) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.045913 (rank : 41) | θ value | 0.813845 (rank : 29) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
CASC3_MOUSE
|
||||||
| NC score | 0.044948 (rank : 42) | θ value | 4.03905 (rank : 74) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8K3W3, Q8K219, Q99NF0 | Gene names | Casc3, Mln51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC3 (Cancer susceptibility candidate gene 3 protein homolog) (Metastatic lymph node protein 51 homolog) (Protein MLN 51 homolog) (Protein barentsz) (Btz). | |||||
|
ALMS1_HUMAN
|
||||||
| NC score | 0.043464 (rank : 43) | θ value | 0.365318 (rank : 19) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8TCU4, Q53S05, Q580Q8, Q86VP9, Q9Y4G4 | Gene names | ALMS1, KIAA0328 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alstrom syndrome protein 1. | |||||
|
RERE_HUMAN
|
||||||
| NC score | 0.040483 (rank : 44) | θ value | 3.0926 (rank : 69) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
ARI1A_HUMAN
|
||||||
| NC score | 0.040067 (rank : 45) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
MKL1_MOUSE
|
||||||
| NC score | 0.039452 (rank : 46) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
MILK1_MOUSE
|
||||||
| NC score | 0.039387 (rank : 47) | θ value | 1.38821 (rank : 43) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
NF2L2_HUMAN
|
||||||
| NC score | 0.038794 (rank : 48) | θ value | 0.163984 (rank : 14) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q16236, Q96F71 | Gene names | NFE2L2, NRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 2 (NF-E2-related factor 2) (NFE2-related factor 2) (Nuclear factor, erythroid derived 2, like 2) (HEBP1). | |||||
|
ENAH_HUMAN
|
||||||
| NC score | 0.038731 (rank : 49) | θ value | 1.06291 (rank : 33) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8N8S7, Q502W5, Q5T5M7, Q5VTQ9, Q5VTR0, Q9NVF3, Q9UFB8 | Gene names | ENAH, MENA | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog. | |||||
|
TRI66_HUMAN
|
||||||
| NC score | 0.038408 (rank : 50) | θ value | 5.27518 (rank : 98) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O15016, Q9BQQ4 | Gene names | TRIM66, KIAA0298 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
3BP2_MOUSE
|
||||||
| NC score | 0.038139 (rank : 51) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q06649 | Gene names | Sh3bp2, 3bp2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
MTSS1_HUMAN
|
||||||
| NC score | 0.038127 (rank : 52) | θ value | 2.36792 (rank : 57) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O43312, Q8TCA2, Q96RX2 | Gene names | MTSS1, KIAA0429, MIM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein) (Metastasis suppressor YGL-1). | |||||
|
SAPS1_HUMAN
|
||||||
| NC score | 0.037852 (rank : 53) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
CEP41_HUMAN
|
||||||
| NC score | 0.037653 (rank : 54) | θ value | 1.06291 (rank : 31) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BYV8, Q7Z496, Q86TM1, Q8NFU8, Q9H6A3, Q9NPV3 | Gene names | TSGA14, CEP41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 41 kDa (Cep41 protein) (Testis-specific gene A14 protein). | |||||
|
WASF2_MOUSE
|
||||||
| NC score | 0.037413 (rank : 55) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BH43 | Gene names | Wasf2, Wave2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2). | |||||
|
PSL1_HUMAN
|
||||||
| NC score | 0.036561 (rank : 56) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
DIAP2_HUMAN
|
||||||
| NC score | 0.036469 (rank : 57) | θ value | 2.36792 (rank : 56) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O60879, O60878, Q8WX06, Q8WX48, Q9UJL2 | Gene names | DIAPH2, DIA | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 2 (Diaphanous-related formin-2) (DRF2). | |||||
|
NUFP1_HUMAN
|
||||||
| NC score | 0.036181 (rank : 58) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UHK0, Q8WVM5, Q96SG1 | Gene names | NUFIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear fragile X mental retardation-interacting protein 1 (Nuclear FMRP-interacting protein 1). | |||||
|
PRRT1_MOUSE
|
||||||
| NC score | 0.035885 (rank : 59) | θ value | 8.99809 (rank : 140) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O35449 | Gene names | Prrt1, Ng5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 1. | |||||
|
IRF3_HUMAN
|
||||||
| NC score | 0.035774 (rank : 60) | θ value | 0.365318 (rank : 20) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q14653 | Gene names | IRF3 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 3 (IRF-3). | |||||
|
CSKI1_MOUSE
|
||||||
| NC score | 0.035681 (rank : 61) | θ value | 2.36792 (rank : 53) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
PRRT1_HUMAN
|
||||||
| NC score | 0.035622 (rank : 62) | θ value | 8.99809 (rank : 139) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99946, Q96DW3, Q96NQ8 | Gene names | PRRT1, C6orf31, NG5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 1. | |||||
|
SPR2E_MOUSE
|
||||||
| NC score | 0.035273 (rank : 63) | θ value | 5.27518 (rank : 96) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O70556 | Gene names | Sprr2e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2E. | |||||
|
DIAP2_MOUSE
|
||||||
| NC score | 0.035085 (rank : 64) | θ value | 8.99809 (rank : 121) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O70566, Q8C2G8 | Gene names | Diaph2, Diap2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 2 (Diaphanous-related formin-2) (DRF2) (mDia3). | |||||
|
NUFP1_MOUSE
|
||||||
| NC score | 0.034933 (rank : 65) | θ value | 2.36792 (rank : 58) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9QXX8, Q9CV69 | Gene names | Nufip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear fragile X mental retardation-interacting protein 1 (Nuclear FMRP-interacting protein 1). | |||||
|
3BP2_HUMAN
|
||||||
| NC score | 0.034535 (rank : 66) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P78314, O00500, O15373, P78315 | Gene names | SH3BP2, 3BP2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
TCRG1_HUMAN
|
||||||
| NC score | 0.034226 (rank : 67) | θ value | 1.06291 (rank : 40) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.033721 (rank : 68) | θ value | 3.0926 (rank : 67) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
CJ047_HUMAN
|
||||||
| NC score | 0.033689 (rank : 69) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q86WR7, Q5W0J9, Q5W0K0, Q5W0K1, Q5W0K2, Q6PJC8, Q8N317 | Gene names | C10orf47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf47. | |||||
|
CBX2_HUMAN
|
||||||
| NC score | 0.033283 (rank : 70) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q14781, Q9BTB1 | Gene names | CBX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromobox protein homolog 2. | |||||
|
SPR2F_MOUSE
|
||||||
| NC score | 0.033063 (rank : 71) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O70557 | Gene names | Sprr2f | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2F. | |||||
|
TCRG1_MOUSE
|
||||||
| NC score | 0.033037 (rank : 72) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
CSKI1_HUMAN
|
||||||
| NC score | 0.032802 (rank : 73) | θ value | 2.36792 (rank : 52) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
RTN4_HUMAN
|
||||||
| NC score | 0.032191 (rank : 74) | θ value | 2.36792 (rank : 60) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NQC3, O94962, Q9BXG5, Q9H212, Q9H3I3, Q9UQ42, Q9Y293, Q9Y2Y7, Q9Y5U6 | Gene names | RTN4, KIAA0886, NOGO | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-4 (Neurite outgrowth inhibitor) (Nogo protein) (Foocen) (Neuroendocrine-specific protein) (NSP) (Neuroendocrine-specific protein C homolog) (RTN-x) (Reticulon-5). | |||||
|
SFPQ_MOUSE
|
||||||
| NC score | 0.031748 (rank : 75) | θ value | 8.99809 (rank : 144) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
CSKI2_MOUSE
|
||||||
| NC score | 0.031310 (rank : 76) | θ value | 2.36792 (rank : 55) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 812 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8VHK1, Q6ZPX1, Q7TS77 | Gene names | Caskin2, Kiaa1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
CSKI2_HUMAN
|
||||||
| NC score | 0.030502 (rank : 77) | θ value | 2.36792 (rank : 54) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8WXE0, Q7LG69, Q9ULT1 | Gene names | CASKIN2, KIAA1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
MNT_HUMAN
|
||||||
| NC score | 0.030487 (rank : 78) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
ZDHC8_HUMAN
|
||||||
| NC score | 0.030043 (rank : 79) | θ value | 1.81305 (rank : 49) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9ULC8, Q6ICL1, Q6ZNF5, Q7Z6L9 | Gene names | ZDHHC8, KIAA1292, ZDHHCL1, ZNF378 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC8 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 8) (DHHC-8) (Zinc finger protein 378). | |||||
|
ZDHC8_MOUSE
|
||||||
| NC score | 0.028901 (rank : 80) | θ value | 1.06291 (rank : 41) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5Y5T5, Q7TNF7 | Gene names | Zdhhc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable palmitoyltransferase ZDHHC8 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 8) (DHHC-8). | |||||
|
BCAR1_HUMAN
|
||||||
| NC score | 0.028849 (rank : 81) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P56945 | Gene names | BCAR1, CAS, CRKAS | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
OTU7B_HUMAN
|
||||||
| NC score | 0.028151 (rank : 82) | θ value | 1.81305 (rank : 47) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6GQQ9, Q8WWA7, Q9NQ53, Q9UFF4 | Gene names | OTUD7B, ZA20D1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | OTU domain-containing protein 7B (EC 3.-.-.-) (Zinc finger protein Cezanne) (Zinc finger A20 domain-containing protein 1) (Cellular zinc finger anti-NF-kappa B protein). | |||||
|
TCGAP_MOUSE
|
||||||
| NC score | 0.026772 (rank : 83) | θ value | 5.27518 (rank : 97) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q80YF9, Q6P7W6 | Gene names | Snx26, Tcgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
FCRLA_HUMAN
|
||||||
| NC score | 0.026650 (rank : 84) | θ value | 1.06291 (rank : 34) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7L513, Q5VXA1, Q5VXA2, Q5VXA3, Q5VXB0, Q8NEW4, Q8WXH3, Q96PC6, Q96PJ0, Q96PJ1, Q96PJ2, Q96PJ4, Q9BR57 | Gene names | FCRLA, FCRL, FCRL1, FCRLM1, FCRX, FREB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fc receptor-like and mucin-like 1 precursor (Fc receptor-like A) (Fc receptor homolog expressed in B-cells) (Fc receptor-related protein X) (FcRX) (Fc receptor-like protein). | |||||
|
3MG_HUMAN
|
||||||
| NC score | 0.026121 (rank : 85) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P29372, Q13770, Q15275, Q15961, Q96BZ6, Q96S33, Q9NNX5 | Gene names | MPG, AAG, ANPG, MID1 | |||
|
Domain Architecture |
|
|||||
| Description | DNA-3-methyladenine glycosylase (EC 3.2.2.21) (3-methyladenine DNA glycosidase) (ADPG) (3-alkyladenine DNA glycosylase) (N-methylpurine- DNA glycosylase). | |||||
|
K1849_MOUSE
|
||||||
| NC score | 0.026078 (rank : 86) | θ value | 1.06291 (rank : 37) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q69Z89, Q8BNL0, Q8C549, Q8CAL1 | Gene names | Kiaa1849 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1849. | |||||
|
AMPH_HUMAN
|
||||||
| NC score | 0.025006 (rank : 87) | θ value | 3.0926 (rank : 62) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P49418, O43538, Q75MJ8, Q75MK5, Q75MM3, Q8N4G0 | Gene names | AMPH, AMPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Amphiphysin. | |||||
|
PGCA_MOUSE
|
||||||
| NC score | 0.024445 (rank : 88) | θ value | 0.279714 (rank : 16) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
ZC3H6_HUMAN
|
||||||
| NC score | 0.024084 (rank : 89) | θ value | 2.36792 (rank : 61) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P61129 | Gene names | ZC3H6, KIAA2035, ZC3HDC6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
DTX1_MOUSE
|
||||||
| NC score | 0.023462 (rank : 90) | θ value | 3.0926 (rank : 63) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61010, Q3TER5, Q8C2H2, Q9ER09 | Gene names | Dtx1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein deltex-1 (Deltex-1) (Deltex1) (mDTX1) (FXI-T1). | |||||
|
PO6F2_HUMAN
|
||||||
| NC score | 0.022667 (rank : 91) | θ value | 2.36792 (rank : 59) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P78424, P78425, Q75ME8, Q86UM6, Q9UDS7 | Gene names | POU6F2, RPF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 2 (Retina-derived POU-domain factor 1) (RPF-1). | |||||
|
RBM22_HUMAN
|
||||||
| NC score | 0.022529 (rank : 92) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NW64, O95607 | Gene names | RBM22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-splicing factor RBM22 (RNA-binding motif protein 22). | |||||
|
RBM22_MOUSE
|
||||||
| NC score | 0.022529 (rank : 93) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BHS3, Q3TJB0, Q9CXA0 | Gene names | Rbm22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-splicing factor RBM22 (RNA-binding motif protein 22). | |||||
|
ABLM1_HUMAN
|
||||||
| NC score | 0.022340 (rank : 94) | θ value | 0.0736092 (rank : 10) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O14639, Q15039, Q68CQ9, Q9BUP1 | Gene names | ABLIM1, ABLIM, KIAA0059 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding LIM protein 1 (Actin-binding LIM protein family member 1) (abLIM-1) (Actin-binding double-zinc-finger protein) (LIMAB1) (Limatin). | |||||
|
HXB3_HUMAN
|
||||||
| NC score | 0.021676 (rank : 95) | θ value | 0.62314 (rank : 27) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P14651, O95615, P17484 | Gene names | HOXB3, HOX2G | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B3 (Hox-2G) (Hox-2.7). | |||||
|
IQEC2_MOUSE
|
||||||
| NC score | 0.021376 (rank : 96) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5DU25 | Gene names | Iqsec2, Kiaa0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
LARP4_MOUSE
|
||||||
| NC score | 0.021179 (rank : 97) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BWW4, Q3UR69 | Gene names | Larp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | La-related protein 4 (La ribonucleoprotein domain family member 4). | |||||
|
SETX_HUMAN
|
||||||
| NC score | 0.020591 (rank : 98) | θ value | 4.03905 (rank : 82) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7Z333, O75120, Q3KQX4, Q68DW5, Q6AZD7, Q7Z3J6, Q8WX33, Q9H9D1, Q9NVP9 | Gene names | SETX, ALS4, KIAA0625, SCAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable helicase senataxin (EC 3.6.1.-) (SEN1 homolog). | |||||
|
HRBL_MOUSE
|
||||||
| NC score | 0.020478 (rank : 99) | θ value | 6.88961 (rank : 106) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q80WC7, Q8BKS5, Q99J67 | Gene names | Hrbl, Rabr | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
MYO15_HUMAN
|
||||||
| NC score | 0.020292 (rank : 100) | θ value | 6.88961 (rank : 108) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
HLF_MOUSE
|
||||||
| NC score | 0.020054 (rank : 101) | θ value | 6.88961 (rank : 105) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BW74, Q6PF83 | Gene names | Hlf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatic leukemia factor. | |||||
|
HLF_HUMAN
|
||||||
| NC score | 0.020003 (rank : 102) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q16534 | Gene names | HLF | |||
|
Domain Architecture |

|
|||||
| Description | Hepatic leukemia factor. | |||||
|
PCGF2_HUMAN
|
||||||
| NC score | 0.019906 (rank : 103) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P35227 | Gene names | PCGF2, MEL18, RNF110, ZNF144 | |||
|
Domain Architecture |

|
|||||
| Description | Polycomb group RING finger protein 2 (DNA-binding protein Mel-18) (RING finger protein 110) (Zinc finger protein 144). | |||||
|
COHA1_MOUSE
|
||||||
| NC score | 0.019862 (rank : 104) | θ value | 8.99809 (rank : 120) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q07563, Q99LK8 | Gene names | Col17a1, Bp180, Bpag2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVII) chain (Bullous pemphigoid antigen 2) (180 kDa bullous pemphigoid antigen 2). | |||||
|
AFAD_HUMAN
|
||||||
| NC score | 0.019456 (rank : 105) | θ value | 4.03905 (rank : 73) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P55196, O75087, O75088, O75089, Q5TIG6, Q9NSN7, Q9NU92 | Gene names | MLLT4, AF6 | |||
|
Domain Architecture |

|
|||||
| Description | Afadin (Protein AF-6). | |||||
|
RSMB_MOUSE
|
||||||
| NC score | 0.019361 (rank : 106) | θ value | 8.99809 (rank : 143) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P27048, Q3UJT1 | Gene names | Snrpb | |||
|
Domain Architecture |

|
|||||
| Description | Small nuclear ribonucleoprotein-associated protein B (snRNP-B) (Sm protein B) (Sm-B) (SmB). | |||||
|
FGFP3_HUMAN
|
||||||
| NC score | 0.019126 (rank : 107) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TAT2, Q8NBN0 | Gene names | FGFBP3, C10orf13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibroblast growth factor-binding protein 3 precursor (FGF-binding protein 3) (FGF-BP3) (FGFBP-3). | |||||
|
GPTC8_HUMAN
|
||||||
| NC score | 0.018948 (rank : 108) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
PTN23_HUMAN
|
||||||
| NC score | 0.018781 (rank : 109) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9H3S7, Q7KZF8, Q8N6Z5, Q9BSR5, Q9P257, Q9UG03, Q9UMZ4 | Gene names | PTPN23, KIAA1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His- domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14). | |||||
|
SMCA4_HUMAN
|
||||||
| NC score | 0.018067 (rank : 110) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P51532, O95052, Q9HBD3 | Gene names | SMARCA4, BRG1, SNF2B, SNF2L4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L4 (EC 3.6.1.-) (ATP- dependent helicase SMARCA4) (SNF2-beta) (BRG-1 protein) (Mitotic growth and transcription activator) (Brahma protein homolog 1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4). | |||||
|
SENP3_HUMAN
|
||||||
| NC score | 0.018046 (rank : 111) | θ value | 3.0926 (rank : 71) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H4L4, Q96PS4, Q9Y3W9 | Gene names | SENP3, SSP3, SUSP3 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 3 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP3) (SUMO-1-specific protease 3). | |||||
|
CO8A2_MOUSE
|
||||||
| NC score | 0.017636 (rank : 112) | θ value | 2.36792 (rank : 51) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P25318, Q68ED0, Q6KAQ4, Q6P1C4 | Gene names | Col8a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-2(VIII) chain precursor (Endothelial collagen). | |||||
|
GLGB_HUMAN
|
||||||
| NC score | 0.017563 (rank : 113) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q04446, Q96EN0 | Gene names | GBE1 | |||
|
Domain Architecture |

|
|||||
| Description | 1,4-alpha-glucan branching enzyme (EC 2.4.1.18) (Glycogen branching enzyme) (Brancher enzyme). | |||||
|
GRIP2_HUMAN
|
||||||
| NC score | 0.017532 (rank : 114) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9C0E4, Q8TEH9, Q9H7H3 | Gene names | GRIP2, KIAA1719 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor-interacting protein 2 (GRIP2 protein). | |||||
|
TM119_MOUSE
|
||||||
| NC score | 0.017364 (rank : 115) | θ value | 8.99809 (rank : 146) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R138, Q8BP14 | Gene names | Tmem119 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 119 precursor. | |||||
|
FREM2_HUMAN
|
||||||
| NC score | 0.017256 (rank : 116) | θ value | 1.06291 (rank : 36) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5SZK8, Q4QQG1, Q5H9N8, Q5T6Q1, Q6N057, Q6ZSB4, Q7Z305, Q7Z341 | Gene names | FREM2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FRAS1-related extracellular matrix protein 2 precursor (ECM3 homolog). | |||||
|
KCNC3_HUMAN
|
||||||
| NC score | 0.017171 (rank : 117) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14003 | Gene names | KCNC3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
FGD1_HUMAN
|
||||||
| NC score | 0.016601 (rank : 118) | θ value | 4.03905 (rank : 75) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 367 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P98174, Q8N4D9 | Gene names | FGD1, ZFYVE3 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
JPH4_MOUSE
|
||||||
| NC score | 0.016497 (rank : 119) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q80WT0, Q8BMI1 | Gene names | Jph4, Jphl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-4 (Junctophilin-like 1 protein). | |||||
|
RX_HUMAN
|
||||||
| NC score | 0.016308 (rank : 120) | θ value | 0.47712 (rank : 25) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y2V3 | Gene names | RAX, RX | |||
|
Domain Architecture |

|
|||||
| Description | Retinal homeobox protein Rx (Retina and anterior neural fold homeobox protein). | |||||
|
MYO15_MOUSE
|
||||||
| NC score | 0.015566 (rank : 121) | θ value | 4.03905 (rank : 81) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9QZZ4, O70395, Q9QWL6 | Gene names | Myo15a, Myo15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
RP3A_HUMAN
|
||||||
| NC score | 0.015414 (rank : 122) | θ value | 3.0926 (rank : 70) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y2J0, Q96AE0 | Gene names | RPH3A, KIAA0985 | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
PIGQ_HUMAN
|
||||||
| NC score | 0.015230 (rank : 123) | θ value | 8.99809 (rank : 136) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BRB3, O14927, Q96G00, Q96S22, Q9UJH4 | Gene names | PIGQ, GPI1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol N-acetylglucosaminyltransferase subunit Q (EC 2.4.1.198) (Phosphatidylinositol-glycan biosynthesis class Q protein) (PIG-Q) (N-acetylglucosamyl transferase component GPI1). | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.015217 (rank : 124) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
KBTB9_MOUSE
|
||||||
| NC score | 0.015034 (rank : 125) | θ value | 0.365318 (rank : 21) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80T74 | Gene names | Kbtbd9, Kiaa1921 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch repeat and BTB domain-containing protein 9. | |||||
|
HXB3_MOUSE
|
||||||
| NC score | 0.015023 (rank : 126) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P09026, P10285, Q4PJ06, Q61680 | Gene names | Hoxb3, Hox-2.7, Hoxb-3 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B3 (Hox-2.7) (Homeobox protein MH-23). | |||||
|
IRX5_HUMAN
|
||||||
| NC score | 0.015015 (rank : 127) | θ value | 8.99809 (rank : 126) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P78411, P78416, Q7Z2E1 | Gene names | IRX5, IRX2A, IRXB2 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-5 (Iroquois homeobox protein 5) (Homeodomain protein IRXB2) (IRX-2A). | |||||
|
PXK_MOUSE
|
||||||
| NC score | 0.014884 (rank : 128) | θ value | 8.99809 (rank : 142) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 320 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BX57, Q3TD60, Q3TEL7, Q3TZZ1, Q3U197, Q3U773, Q3UCS5, Q4FBH7, Q91WB6 | Gene names | Pxk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PX domain-containing protein kinase-like protein (Modulator of Na,K- ATPase) (MONaKA). | |||||
|
AKAP1_MOUSE
|
||||||
| NC score | 0.014862 (rank : 129) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O08715, O08714, P97488 | Gene names | Akap1, Akap | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 1, mitochondrial precursor (Protein kinase A- anchoring protein 1) (PRKA1) (Dual specificity A-kinase-anchoring protein 1) (D-AKAP-1) (Spermatid A-kinase anchor protein) (S-AKAP). | |||||
|
PCGF2_MOUSE
|
||||||
| NC score | 0.014683 (rank : 130) | θ value | 8.99809 (rank : 134) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P23798 | Gene names | Pcgf2, Mel-18, Mel18, Rnf110, Zfp144, Znf144 | |||
|
Domain Architecture |

|
|||||
| Description | Polycomb group RING finger protein 2 (DNA-binding protein Mel-18) (RING finger protein 110) (Zinc finger protein 144) (Zfp-144). | |||||
|
KCNC3_MOUSE
|
||||||
| NC score | 0.014528 (rank : 131) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q63959, Q62088 | Gene names | Kcnc3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
APLP1_HUMAN
|
||||||
| NC score | 0.014456 (rank : 132) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P51693, O00113, Q96A92 | Gene names | APLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 1 precursor (APLP) (APLP-1) [Contains: C30]. | |||||
|
WNK2_HUMAN
|
||||||
| NC score | 0.014377 (rank : 133) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
SN1L2_MOUSE
|
||||||
| NC score | 0.013918 (rank : 134) | θ value | 0.0431538 (rank : 9) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8CFH6 | Gene names | Snf1lk2, Sik2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SNF1-like kinase 2 (EC 2.7.11.1) (Salt-inducible kinase 2). | |||||
|
K2027_MOUSE
|
||||||
| NC score | 0.013907 (rank : 135) | θ value | 8.99809 (rank : 127) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BWS5, Q69Z30 | Gene names | Kiaa2027 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA2027. | |||||
|
ARHGB_HUMAN
|
||||||
| NC score | 0.013261 (rank : 136) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O15085 | Gene names | ARHGEF11, KIAA0380 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 11 (PDZ-RhoGEF). | |||||
|
FACE1_HUMAN
|
||||||
| NC score | 0.013096 (rank : 137) | θ value | 8.99809 (rank : 122) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75844, Q8NDZ8, Q9UBQ2 | Gene names | ZMPSTE24, FACE1, STE24 | |||
|
Domain Architecture |

|
|||||
| Description | CAAX prenyl protease 1 homolog (EC 3.4.24.84) (Prenyl protein-specific endoprotease 1) (Farnesylated proteins-converting enzyme 1) (FACE-1) (Zinc metalloproteinase Ste24 homolog). | |||||
|
SN1L2_HUMAN
|
||||||
| NC score | 0.012823 (rank : 138) | θ value | 0.279714 (rank : 17) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H0K1, O94878, Q6AZE2, Q76N03, Q8NCV7, Q96CZ8 | Gene names | SNF1LK2, KIAA0781, QIK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SNF1-like kinase 2 (EC 2.7.11.1) (Qin- induced kinase). | |||||
|
TSC2_MOUSE
|
||||||
| NC score | 0.012548 (rank : 139) | θ value | 6.88961 (rank : 112) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61037, P97723, P97724, P97725, P97727, Q61007, Q61008, Q9WUF6 | Gene names | Tsc2 | |||
|
Domain Architecture |

|
|||||
| Description | Tuberin (Tuberous sclerosis 2 homolog protein). | |||||
|
TERF2_MOUSE
|
||||||
| NC score | 0.011918 (rank : 140) | θ value | 8.99809 (rank : 145) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35144 | Gene names | Terf2, Trf2 | |||
|
Domain Architecture |

|
|||||
| Description | Telomeric repeat-binding factor 2 (TTAGGG repeat-binding factor 2) (Telomeric DNA-binding protein). | |||||
|
TIE1_HUMAN
|
||||||
| NC score | 0.010984 (rank : 141) | θ value | 0.279714 (rank : 18) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P35590 | Gene names | TIE1, TIE | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase receptor Tie-1 precursor (EC 2.7.10.1). | |||||
|
PHF1_MOUSE
|
||||||
| NC score | 0.010916 (rank : 142) | θ value | 8.99809 (rank : 135) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z1B8, O54808 | Gene names | Phf1, Tctex-3, Tctex3 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 1 (Protein PHF1) (T-complex testis-expressed 3) (Polycomblike 1) (mPCl1). | |||||
|
KBTB9_HUMAN
|
||||||
| NC score | 0.010470 (rank : 143) | θ value | 6.88961 (rank : 107) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96CT2, Q8N388, Q96BF0, Q96PW7 | Gene names | KBTBD9, KIAA1921 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch repeat and BTB domain-containing protein 9. | |||||
|
TIE1_MOUSE
|
||||||
| NC score | 0.009786 (rank : 144) | θ value | 1.38821 (rank : 45) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1266 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q06806, Q811F4 | Gene names | Tie1, Tie, Tie-1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase receptor Tie-1 precursor (EC 2.7.10.1). | |||||
|
UBIP1_MOUSE
|
||||||
| NC score | 0.009116 (rank : 145) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q811S7, Q3UPR3, Q3US11, Q60786, Q8C514 | Gene names | Ubp1, Cp2b, Nf2d9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Upstream-binding protein 1 (Nuclear factor 2d9) (NF2d9). | |||||
|
WNK1_HUMAN
|
||||||
| NC score | 0.009089 (rank : 146) | θ value | 8.99809 (rank : 147) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1158 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9H4A3, O15052, Q86WL5, Q8N673, Q9P1S9 | Gene names | WNK1, KDP, KIAA0344, PRKWNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1) (Kinase deficient protein). | |||||
|
FOG1_HUMAN
|
||||||
| NC score | 0.008740 (rank : 147) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 981 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8IX07 | Gene names | ZFPM1, FOG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
MK07_HUMAN
|
||||||
| NC score | 0.008689 (rank : 148) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1087 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13164, Q16634 | Gene names | MAPK7, ERK4, ERK5, PRKM7 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase 7 (EC 2.7.11.24) (Extracellular signal-regulated kinase 5) (ERK-5) (ERK4) (BMK1 kinase). | |||||
|
MAPK2_HUMAN
|
||||||
| NC score | 0.005003 (rank : 149) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P49137, Q8IYD6 | Gene names | MAPKAPK2 | |||
|
Domain Architecture |

|
|||||
| Description | MAP kinase-activated protein kinase 2 (EC 2.7.11.1) (MAPK-activated protein kinase 2) (MAPKAP kinase 2) (MAPKAPK-2) (MK2). | |||||
|
PLK3_MOUSE
|
||||||
| NC score | 0.004132 (rank : 150) | θ value | 8.99809 (rank : 137) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 863 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q60806, Q60822, Q9R009 | Gene names | Plk3, Cnk, Fnk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PLK3 (EC 2.7.11.21) (Polo-like kinase 3) (PLK-3) (Cytokine-inducible serine/threonine-protein kinase) (FGF- inducible kinase). | |||||
|
FOXC2_HUMAN
|
||||||
| NC score | 0.004006 (rank : 151) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99958 | Gene names | FOXC2, FKHL14, MFH1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein C2 (Forkhead-related protein FKHL14) (Mesenchyme fork head protein 1) (MFH-1 protein) (Transcription factor FKH-14). | |||||
|
PTPRG_MOUSE
|
||||||
| NC score | 0.003917 (rank : 152) | θ value | 8.99809 (rank : 141) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q05909 | Gene names | Ptprg | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase gamma precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase gamma) (R-PTP-gamma). | |||||
|
NR1H2_MOUSE
|
||||||
| NC score | 0.002381 (rank : 153) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60644 | Gene names | Nr1h2, Lxrb, Rip15, Unr, Unr2 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterols receptor LXR-beta (Liver X receptor beta) (Nuclear orphan receptor LXR-beta) (Ubiquitously-expressed nuclear receptor) (Retinoid X receptor-interacting protein No.15). | |||||
|
PRD16_HUMAN
|
||||||
| NC score | -0.001747 (rank : 154) | θ value | 8.99809 (rank : 138) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 832 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9HAZ2, Q8WYJ9, Q9C0I8 | Gene names | PRDM16, KIAA1675, MEL1, PFM13 | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 16 (PR domain-containing protein 16) (Transcription factor MEL1). | |||||