Please be patient as the page loads
|
IRF3_HUMAN
|
||||||
| SwissProt Accessions | Q14653 | Gene names | IRF3 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 3 (IRF-3). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
IRF3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 129 | |
| SwissProt Accessions | Q14653 | Gene names | IRF3 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 3 (IRF-3). | |||||
|
IRF3_MOUSE
|
||||||
| θ value | 9.78303e-172 (rank : 2) | NC score | 0.980458 (rank : 2) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P70671 | Gene names | Irf3 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 3 (IRF-3). | |||||
|
IRF5_MOUSE
|
||||||
| θ value | 5.38352e-45 (rank : 3) | NC score | 0.900261 (rank : 4) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P56477 | Gene names | Irf5 | |||
|
Domain Architecture |
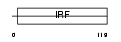
|
|||||
| Description | Interferon regulatory factor 5 (IRF-5). | |||||
|
IRF5_HUMAN
|
||||||
| θ value | 1.46607e-42 (rank : 4) | NC score | 0.899644 (rank : 5) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q13568, Q9BQF0 | Gene names | IRF5 | |||
|
Domain Architecture |
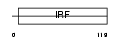
|
|||||
| Description | Interferon regulatory factor 5 (IRF-5). | |||||
|
IRF6_HUMAN
|
||||||
| θ value | 2.58786e-39 (rank : 5) | NC score | 0.891879 (rank : 6) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O14896 | Gene names | IRF6 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 6 (IRF-6). | |||||
|
IRF6_MOUSE
|
||||||
| θ value | 1.28434e-38 (rank : 6) | NC score | 0.891415 (rank : 7) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P97431 | Gene names | Irf6 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 6 (IRF-6). | |||||
|
IRF7_MOUSE
|
||||||
| θ value | 3.73686e-38 (rank : 7) | NC score | 0.900998 (rank : 3) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P70434 | Gene names | Irf7 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 7 (IRF-7). | |||||
|
IRF7_HUMAN
|
||||||
| θ value | 5.39604e-37 (rank : 8) | NC score | 0.877161 (rank : 8) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q92985, O00331, O00332, O00333, O75924 | Gene names | IRF7 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 7 (IRF-7). | |||||
|
IRF4_HUMAN
|
||||||
| θ value | 1.79631e-32 (rank : 9) | NC score | 0.870845 (rank : 9) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q15306, Q99660 | Gene names | IRF4, MUM1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 4 (IRF-4) (Lymphocyte-specific interferon regulatory factor) (LSIRF) (NF-EM5) (Multiple myeloma oncogene 1). | |||||
|
IRF4_MOUSE
|
||||||
| θ value | 1.16434e-31 (rank : 10) | NC score | 0.870277 (rank : 10) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q64287, Q60802 | Gene names | Irf4, Spip | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 4 (IRF-4) (Lymphocyte-specific interferon regulatory factor) (LSIRF) (NF-EM5) (PU.1 interaction partner) (Transcriptional activator PIP). | |||||
|
IRF8_HUMAN
|
||||||
| θ value | 1.08979e-29 (rank : 11) | NC score | 0.860701 (rank : 11) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q02556 | Gene names | IRF8, ICSBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 8 (IRF-8) (Interferon consensus sequence- binding protein) (ICSBP). | |||||
|
IRF8_MOUSE
|
||||||
| θ value | 3.50572e-28 (rank : 12) | NC score | 0.856896 (rank : 12) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P23611 | Gene names | Irf8, Icsbp, Icsbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 8 (IRF-8) (Interferon consensus sequence- binding protein) (ICSBP). | |||||
|
IRTF_HUMAN
|
||||||
| θ value | 1.42661e-21 (rank : 13) | NC score | 0.837651 (rank : 13) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q00978 | Gene names | ISGF3G, IRF9 | |||
|
Domain Architecture |
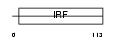
|
|||||
| Description | Transcriptional regulator ISGF3 subunit gamma (Interferon regulatory factor 9) (IRF-9) (IFN-alpha-responsive transcription factor subunit) (Interferon-stimulated gene factor 3 gamma) (ISGF3 p48 subunit) (ISGF- 3 gamma). | |||||
|
IRF2_MOUSE
|
||||||
| θ value | 4.02038e-16 (rank : 14) | NC score | 0.779761 (rank : 18) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P23906 | Gene names | Irf2 | |||
|
Domain Architecture |
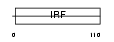
|
|||||
| Description | Interferon regulatory factor 2 (IRF-2). | |||||
|
IRF2_HUMAN
|
||||||
| θ value | 8.95645e-16 (rank : 15) | NC score | 0.782186 (rank : 17) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P14316, Q96B99 | Gene names | IRF2 | |||
|
Domain Architecture |
|
|||||
| Description | Interferon regulatory factor 2 (IRF-2). | |||||
|
IRF1_HUMAN
|
||||||
| θ value | 2.60593e-15 (rank : 16) | NC score | 0.786636 (rank : 15) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P10914, Q96GG7 | Gene names | IRF1 | |||
|
Domain Architecture |
|
|||||
| Description | Interferon regulatory factor 1 (IRF-1). | |||||
|
IRF1_MOUSE
|
||||||
| θ value | 3.40345e-15 (rank : 17) | NC score | 0.783154 (rank : 16) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P15314 | Gene names | Irf1 | |||
|
Domain Architecture |
|
|||||
| Description | Interferon regulatory factor 1 (IRF-1). | |||||
|
IRTF_MOUSE
|
||||||
| θ value | 3.76295e-14 (rank : 18) | NC score | 0.804459 (rank : 14) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q61179 | Gene names | Isgf3g | |||
|
Domain Architecture |
|
|||||
| Description | Transcriptional regulator ISGF3 subunit gamma (IFN-alpha-responsive transcription factor subunit) (Interferon-stimulated gene factor 3 gamma) (ISGF3 p48 subunit) (ISGF-3 gamma). | |||||
|
PTN23_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 19) | NC score | 0.029666 (rank : 30) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
PI5PA_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 20) | NC score | 0.051324 (rank : 19) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
PRDM2_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 21) | NC score | 0.014380 (rank : 76) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
MK07_HUMAN
|
||||||
| θ value | 0.163984 (rank : 22) | NC score | 0.007265 (rank : 110) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1087 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13164, Q16634 | Gene names | MAPK7, ERK4, ERK5, PRKM7 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase 7 (EC 2.7.11.24) (Extracellular signal-regulated kinase 5) (ERK-5) (ERK4) (BMK1 kinase). | |||||
|
MK07_MOUSE
|
||||||
| θ value | 0.163984 (rank : 23) | NC score | 0.006847 (rank : 113) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1054 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9WVS8 | Gene names | Mapk7, Erk5 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase 7 (EC 2.7.11.24) (Extracellular signal-regulated kinase 5) (ERK-5) (BMK1 kinase). | |||||
|
B4GN1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 24) | NC score | 0.033129 (rank : 29) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q00973 | Gene names | B4GALNT1, GALGT, SIAT2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4 N-acetylgalactosaminyltransferase 1 (EC 2.4.1.92) ((N- acetylneuraminyl)-galactosylglucosylceramide) (GM2/GD2 synthase) (GalNAc-T). | |||||
|
SN1L1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 25) | NC score | 0.002268 (rank : 127) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 911 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P57059, Q5R2V5, Q86YJ2 | Gene names | SNF1LK | |||
|
Domain Architecture |
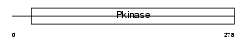
|
|||||
| Description | Serine/threonine-protein kinase SNF1-like kinase 1 (EC 2.7.11.1) (Serine/threonine-protein kinase SNF1LK). | |||||
|
GR65_HUMAN
|
||||||
| θ value | 0.365318 (rank : 26) | NC score | 0.035774 (rank : 25) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9BQQ3, Q8N272, Q96H42 | Gene names | GORASP1, GRASP65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 1 (Golgi reassembly-stacking protein of 65 kDa) (GRASP65) (Golgi peripheral membrane protein p65) (Golgi phosphoprotein 5) (GOLPH5). | |||||
|
WBP11_HUMAN
|
||||||
| θ value | 0.365318 (rank : 27) | NC score | 0.045634 (rank : 20) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
GP1BA_HUMAN
|
||||||
| θ value | 0.47712 (rank : 28) | NC score | 0.014311 (rank : 77) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P07359, Q14441, Q16469, Q8N1F3, Q8NG39, Q9HDC7, Q9UEK1, Q9UQS4 | Gene names | GP1BA | |||
|
Domain Architecture |

|
|||||
| Description | Platelet glycoprotein Ib alpha chain precursor (Glycoprotein Ibalpha) (GP-Ib alpha) (GPIbA) (GPIb-alpha) (Antigen CD42b-alpha) (CD42b antigen) [Contains: Glycocalicin]. | |||||
|
MOT8_MOUSE
|
||||||
| θ value | 0.47712 (rank : 29) | NC score | 0.024546 (rank : 39) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O70324, Q8K3S9 | Gene names | Slc16a2, Mct8, Xpct | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Monocarboxylate transporter 8 (MCT 8) (Solute carrier family 16 member 2) (X-linked PEST-containing transporter). | |||||
|
WBP11_MOUSE
|
||||||
| θ value | 0.47712 (rank : 30) | NC score | 0.044893 (rank : 21) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
FMN2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 31) | NC score | 0.033363 (rank : 28) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NZ56, Q59GF6, Q5VU37, Q9NZ55 | Gene names | FMN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-2. | |||||
|
MMP9_MOUSE
|
||||||
| θ value | 0.62314 (rank : 32) | NC score | 0.012554 (rank : 83) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P41245, Q06788, Q9DC02 | Gene names | Mmp9, Clg4b | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-9 precursor (EC 3.4.24.35) (MMP-9) (92 kDa type IV collagenase) (92 kDa gelatinase) (Gelatinase B) (GELB). | |||||
|
PI5PA_MOUSE
|
||||||
| θ value | 0.62314 (rank : 33) | NC score | 0.037798 (rank : 23) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P59644 | Gene names | Pib5pa | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
TAF3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 34) | NC score | 0.021330 (rank : 49) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
TM16H_HUMAN
|
||||||
| θ value | 0.62314 (rank : 35) | NC score | 0.025591 (rank : 37) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9HCE9 | Gene names | TMEM16H, KIAA1623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 16H. | |||||
|
CIC_HUMAN
|
||||||
| θ value | 0.813845 (rank : 36) | NC score | 0.034473 (rank : 27) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
ENAH_HUMAN
|
||||||
| θ value | 0.813845 (rank : 37) | NC score | 0.016437 (rank : 70) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8N8S7, Q502W5, Q5T5M7, Q5VTQ9, Q5VTR0, Q9NVF3, Q9UFB8 | Gene names | ENAH, MENA | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog. | |||||
|
FMN2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 38) | NC score | 0.035697 (rank : 26) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9JL04 | Gene names | Fmn2 | |||
|
Domain Architecture |

|
|||||
| Description | Formin-2. | |||||
|
KCNA5_HUMAN
|
||||||
| θ value | 0.813845 (rank : 39) | NC score | 0.010806 (rank : 95) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P22460 | Gene names | KCNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (HK2) (HPCN1). | |||||
|
ATF5_MOUSE
|
||||||
| θ value | 1.06291 (rank : 40) | NC score | 0.039468 (rank : 22) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O70191, Q58E51, Q99NH6 | Gene names | Atf5, Atfx, Nap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-5 (Activating transcription factor 5-alpha/beta) (Transcription factor ATFx) (Transcription factor-like protein ODA-10) (BZIP protein ATF7) (NRIF3- associated protein) (NAP1). | |||||
|
CDN1C_HUMAN
|
||||||
| θ value | 1.06291 (rank : 41) | NC score | 0.028092 (rank : 32) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P49918 | Gene names | CDKN1C, KIP2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
FMNL_HUMAN
|
||||||
| θ value | 1.38821 (rank : 42) | NC score | 0.027653 (rank : 33) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O95466, Q6DKG5, Q6IBP3, Q86UH1, Q8N671, Q8TDH1, Q96H10 | Gene names | FMNL1, C17orf1, C17orf1B, FMNL | |||
|
Domain Architecture |

|
|||||
| Description | Formin-like protein 1 (Leukocyte formin) (CLL-associated antigen KW- 13). | |||||
|
PELP1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 43) | NC score | 0.026258 (rank : 35) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8IZL8, O15450, Q5EGN3, Q6NTE6, Q96FT1, Q9BU60 | Gene names | PELP1, HMX3, MNAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3). | |||||
|
CIC_MOUSE
|
||||||
| θ value | 1.81305 (rank : 44) | NC score | 0.025661 (rank : 36) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
DIAP2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 45) | NC score | 0.024490 (rank : 40) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O70566, Q8C2G8 | Gene names | Diaph2, Diap2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 2 (Diaphanous-related formin-2) (DRF2) (mDia3). | |||||
|
IF4G1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 46) | NC score | 0.022612 (rank : 45) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
K1802_HUMAN
|
||||||
| θ value | 1.81305 (rank : 47) | NC score | 0.016657 (rank : 69) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
PHLA1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.025563 (rank : 38) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WV24, Q15184, Q2TAN2, Q9NZ17 | Gene names | PHLDA1, PHRIP, TDAG51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family A member 1 (T-cell death- associated gene 51 protein) (Apoptosis-associated nuclear protein) (Proline- and histidine-rich protein) (Proline- and glutamine-rich protein) (PQ-rich protein). | |||||
|
ZN358_HUMAN
|
||||||
| θ value | 1.81305 (rank : 49) | NC score | -0.001461 (rank : 129) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 840 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NW07, Q9BTM7 | Gene names | ZNF358 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 358. | |||||
|
ATBF1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 50) | NC score | 0.011489 (rank : 91) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
ATBF1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 51) | NC score | 0.011881 (rank : 87) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
CEND3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 52) | NC score | 0.014500 (rank : 75) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8WWN8 | Gene names | CENTD3, ARAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3). | |||||
|
DGKK_HUMAN
|
||||||
| θ value | 2.36792 (rank : 53) | NC score | 0.018052 (rank : 60) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5KSL6 | Gene names | DGKK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase kappa (EC 2.7.1.107) (Diglyceride kinase kappa) (DGK-kappa) (DAG kinase kappa) (142 kDa diacylglycerol kinase). | |||||
|
FOG1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 54) | NC score | 0.005037 (rank : 119) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 981 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8IX07 | Gene names | ZFPM1, FOG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
HD_MOUSE
|
||||||
| θ value | 2.36792 (rank : 55) | NC score | 0.023956 (rank : 41) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P42859 | Gene names | Hd, Hdh | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin (Huntington disease protein homolog) (HD protein). | |||||
|
JPH4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 56) | NC score | 0.012368 (rank : 84) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80WT0, Q8BMI1 | Gene names | Jph4, Jphl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-4 (Junctophilin-like 1 protein). | |||||
|
KCNC3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 57) | NC score | 0.009801 (rank : 100) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q63959, Q62088 | Gene names | Kcnc3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
ODP2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 58) | NC score | 0.019096 (rank : 56) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P10515, Q16783 | Gene names | DLAT, DLTA | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial precursor (EC 2.3.1.12) (Pyruvate dehydrogenase complex E2 subunit) (PDCE2) (E2) (Dihydrolipoamide S- acetyltransferase component of pyruvate dehydrogenase complex) (PDC- E2) (70 kDa mitochondrial autoantigen of primary biliary cirrhosis) (PBC) (M2 antigen complex 70 kDa subunit). | |||||
|
PELP1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 59) | NC score | 0.036945 (rank : 24) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
RX_HUMAN
|
||||||
| θ value | 2.36792 (rank : 60) | NC score | 0.005096 (rank : 118) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y2V3 | Gene names | RAX, RX | |||
|
Domain Architecture |

|
|||||
| Description | Retinal homeobox protein Rx (Retina and anterior neural fold homeobox protein). | |||||
|
ZPR1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 61) | NC score | 0.022852 (rank : 44) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75312, Q2TAA0 | Gene names | ZNF259, ZPR1 | |||
|
Domain Architecture |
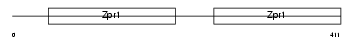
|
|||||
| Description | Zinc-finger protein ZPR1 (Zinc finger protein 259). | |||||
|
CBLB_HUMAN
|
||||||
| θ value | 3.0926 (rank : 62) | NC score | 0.014239 (rank : 78) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13191, Q13192, Q13193, Q3LIC0, Q8IVC5 | Gene names | CBLB, RNF56 | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase CBL-B (EC 6.3.2.-) (Signal transduction protein CBL-B) (SH3-binding protein CBL-B) (Casitas B-lineage lymphoma proto-oncogene b) (RING finger protein 56). | |||||
|
CEND3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 63) | NC score | 0.013446 (rank : 82) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R5G7, Q5DTN4, Q6NVF1, Q8R5G6 | Gene names | Centd3, Arap3, Drag1, Kiaa4097 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3) (Dual specificity Rho- and Arf-GTPase-activating protein 1). | |||||
|
DEN1A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.019210 (rank : 55) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8TEH3, Q5VWF0, Q8IVD6, Q9H796 | Gene names | DENND1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 1A. | |||||
|
GPNMB_HUMAN
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.021824 (rank : 47) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14956, Q6UVX1, Q8N1A1 | Gene names | GPNMB, HGFIN, NMB | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane glycoprotein NMB precursor (Transmembrane glycoprotein HGFIN). | |||||
|
PRR12_HUMAN
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.029661 (rank : 31) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
PTN23_HUMAN
|
||||||
| θ value | 3.0926 (rank : 67) | NC score | 0.019925 (rank : 52) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H3S7, Q7KZF8, Q8N6Z5, Q9BSR5, Q9P257, Q9UG03, Q9UMZ4 | Gene names | PTPN23, KIAA1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His- domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14). | |||||
|
RFTN1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 68) | NC score | 0.017093 (rank : 65) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6A0D4, Q3U654, Q3U8P6, Q80US1 | Gene names | Rftn1, Kiaa0084 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Raftlin (Raft-linking protein). | |||||
|
VMDL1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 69) | NC score | 0.011593 (rank : 89) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BGM5, Q8VCM0 | Gene names | Vmd2l1 | |||
|
Domain Architecture |

|
|||||
| Description | Bestrophin-2 (Vitelliform macular dystrophy 2-like protein 1). | |||||
|
CJ047_HUMAN
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.018410 (rank : 59) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86WR7, Q5W0J9, Q5W0K0, Q5W0K1, Q5W0K2, Q6PJC8, Q8N317 | Gene names | C10orf47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf47. | |||||
|
CNTP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.007306 (rank : 109) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P78357 | Gene names | CNTNAP1, CASPR, NRXN4 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein 1 precursor (Caspr) (Caspr1) (Neurexin 4) (Neurexin IV) (p190). | |||||
|
FBLI1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.006249 (rank : 117) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WUP2, Q5VVE0, Q5VVE1, Q8IX23, Q96T00, Q9UFD6 | Gene names | FBLIM1, FBLP1 | |||
|
Domain Architecture |
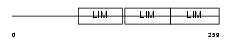
|
|||||
| Description | Filamin-binding LIM protein 1 (Filamin-binding LIM protein 1) (FBLP-1) (Mitogen-inducible 2-interacting protein) (MIG2-interacting protein) (Migfilin). | |||||
|
FOXJ3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 73) | NC score | 0.004751 (rank : 121) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UPW0, Q9NSS7 | Gene names | FOXJ3, KIAA1041 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein J3. | |||||
|
GNDS_MOUSE
|
||||||
| θ value | 4.03905 (rank : 74) | NC score | 0.010377 (rank : 97) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q03385 | Gene names | Ralgds, Rgds | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator (RalGEF) (RalGDS). | |||||
|
SMRC2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 75) | NC score | 0.017210 (rank : 64) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8TAQ2, Q92923, Q96E12, Q96GY4 | Gene names | SMARCC2, BAF170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
SMRC2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | 0.015765 (rank : 73) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6PDG5, Q6P6P3 | Gene names | Smarcc2, Baf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
SON_MOUSE
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.022072 (rank : 46) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
SYNJ1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | 0.020208 (rank : 50) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O43426, O43425, O94984 | Gene names | SYNJ1, KIAA0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
T53I2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.018603 (rank : 57) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IXH6, Q5JX64, Q8IYL5, Q9NU00 | Gene names | TP53INP2, C20orf110, PINH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor protein p53-inducible nuclear protein 2 (p53-inducible protein U) (PIG-U). | |||||
|
ANTR1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.017066 (rank : 66) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H6X2, Q96P02, Q9NVP3 | Gene names | ANTXR1, ATR, TEM8 | |||
|
Domain Architecture |
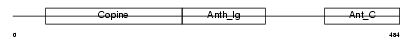
|
|||||
| Description | Anthrax toxin receptor 1 precursor (Tumor endothelial marker 8). | |||||
|
ANTR1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.016993 (rank : 68) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9CZ52 | Gene names | Antxr1, Atr, Tem8 | |||
|
Domain Architecture |
|
|||||
| Description | Anthrax toxin receptor 1 precursor (Tumor endothelial marker 8). | |||||
|
ATN1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.013793 (rank : 80) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
FARP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.003743 (rank : 123) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O94887, Q53QM5, Q8WU27, Q9UFE7 | Gene names | FARP2, KIAA0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
GGN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | 0.019708 (rank : 54) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86UU5, Q7RTU6, Q86UU4, Q8NAA1 | Gene names | GGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
GLSK_HUMAN
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.008327 (rank : 103) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O94925, Q9UL05, Q9UL06, Q9UL07, Q9UN40 | Gene names | GLS, GLS1, KIAA0838 | |||
|
Domain Architecture |

|
|||||
| Description | Glutaminase kidney isoform, mitochondrial precursor (EC 3.5.1.2) (GLS) (L-glutamine amidohydrolase) (K-glutaminase). | |||||
|
IRX3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.007050 (rank : 111) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P81067 | Gene names | Irx3, Irxb1 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-3 (Iroquois homeobox protein 3) (Homeodomain protein IRXB1). | |||||
|
K1949_MOUSE
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.016103 (rank : 71) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BQ30, Q3UDS6, Q8CEY9, Q8R3D8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1949 homolog. | |||||
|
PXK_MOUSE
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.011553 (rank : 90) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 320 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BX57, Q3TD60, Q3TEL7, Q3TZZ1, Q3U197, Q3U773, Q3UCS5, Q4FBH7, Q91WB6 | Gene names | Pxk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PX domain-containing protein kinase-like protein (Modulator of Na,K- ATPase) (MONaKA). | |||||
|
RB_MOUSE
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.011270 (rank : 94) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P13405 | Gene names | Rb1, Rb-1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoblastoma-associated protein (PP105) (RB). | |||||
|
SN_MOUSE
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.003321 (rank : 125) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62230, O55216, Q62228, Q62229 | Gene names | Sn, Sa | |||
|
Domain Architecture |

|
|||||
| Description | Sialoadhesin precursor (Sialic acid-binding Ig-like lectin-1) (Siglec- 1) (Sheep erythrocyte receptor) (SER) (CD169 antigen). | |||||
|
SPR2F_MOUSE
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.019913 (rank : 53) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O70557 | Gene names | Sprr2f | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2F. | |||||
|
TROAP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.023226 (rank : 43) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q12815 | Gene names | TROAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trophinin-associated protein (Tastin) (Trophinin-assisting protein). | |||||
|
TRRAP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.011420 (rank : 92) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y4A5, O75218, Q9Y631, Q9Y6H4 | Gene names | TRRAP, PAF400 | |||
|
Domain Architecture |

|
|||||
| Description | Transformation/transcription domain-associated protein (350/400 kDa PCAF-associated factor) (PAF350/400) (STAF40) (Tra1 homolog). | |||||
|
AFF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.007879 (rank : 106) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
ATF5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.026969 (rank : 34) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y2D1, Q9BSA1, Q9UNQ3 | Gene names | ATF5, ATFX | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-5 (Activating transcription factor 5) (Transcription factor ATFx). | |||||
|
CRSP7_HUMAN
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.018417 (rank : 58) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O95402 | Gene names | CRSP7, ARC70 | |||
|
Domain Architecture |

|
|||||
| Description | CRSP complex subunit 7 (Cofactor required for Sp1 transcriptional activation subunit 7) (Transcriptional coactivator CRSP70) (Activator- recruited cofactor 70 kDa component) (ARC70). | |||||
|
EAF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.015437 (rank : 74) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D4C5 | Gene names | Eaf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELL-associated factor 1. | |||||
|
FA13C_MOUSE
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.009852 (rank : 99) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBR2 | Gene names | Fam13c1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13C1. | |||||
|
GGA3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.007960 (rank : 104) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NZ52, Q15017, Q6IS16, Q9UJY3 | Gene names | GGA3, KIAA0154 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
HCN2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.005026 (rank : 120) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
HD_HUMAN
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.021624 (rank : 48) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P42858, Q9UQB7 | Gene names | HD, IT15 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin (Huntington disease protein) (HD protein). | |||||
|
KCNN3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | 0.006756 (rank : 114) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P58391 | Gene names | Kcnn3, Sk3 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 3 (SK3). | |||||
|
MYO15_MOUSE
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | 0.008887 (rank : 102) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9QZZ4, O70395, Q9QWL6 | Gene names | Myo15a, Myo15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | 0.012290 (rank : 86) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
SMCA4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 105) | NC score | 0.004118 (rank : 122) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P51532, O95052, Q9HBD3 | Gene names | SMARCA4, BRG1, SNF2B, SNF2L4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L4 (EC 3.6.1.-) (ATP- dependent helicase SMARCA4) (SNF2-beta) (BRG-1 protein) (Mitotic growth and transcription activator) (Brahma protein homolog 1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4). | |||||
|
SPR2D_MOUSE
|
||||||
| θ value | 6.88961 (rank : 106) | NC score | 0.012316 (rank : 85) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O70555 | Gene names | Sprr2d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2D. | |||||
|
SPR2E_MOUSE
|
||||||
| θ value | 6.88961 (rank : 107) | NC score | 0.017021 (rank : 67) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O70556 | Gene names | Sprr2e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2E. | |||||
|
WASF2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 108) | NC score | 0.010285 (rank : 98) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BH43 | Gene names | Wasf2, Wave2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2). | |||||
|
WNK2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.003698 (rank : 124) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
AUTS2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.015902 (rank : 72) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8WXX7, Q9Y4F2 | Gene names | AUTS2, KIAA0442 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autism susceptibility gene 2 protein. | |||||
|
CBLB_MOUSE
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.011720 (rank : 88) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q3TTA7 | Gene names | Cblb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase CBL-B (EC 6.3.2.-) (Signal transduction protein CBL-B) (SH3-binding protein CBL-B) (Casitas B-lineage lymphoma proto-oncogene b). | |||||
|
CO039_MOUSE
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.023581 (rank : 42) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q3TEI4, Q3TMF2, Q3U253, Q3UM75, Q9DA93 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39 homolog. | |||||
|
DIAP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.020181 (rank : 51) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O60610, Q9UC76 | Gene names | DIAPH1, DIAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1). | |||||
|
DLGP2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.006300 (rank : 115) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BJ42, Q6XBF3 | Gene names | Dlgap2, Dap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large-associated protein 2 (DAP-2) (SAP90/PSD-95-associated protein 2) (SAPAP2) (PSD-95/SAP90-binding protein 2). | |||||
|
EAF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.013866 (rank : 79) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96JC9, Q8IW10 | Gene names | EAF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELL-associated factor 1. | |||||
|
ENAH_MOUSE
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.017861 (rank : 61) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
FA44A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.007493 (rank : 108) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NFC6, Q6P0M8 | Gene names | FAM44A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM44A. | |||||
|
G3PT_MOUSE
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.008906 (rank : 101) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q64467, Q60650 | Gene names | Gapdhs, Gapd-s, Gapds | |||
|
Domain Architecture |
|
|||||
| Description | Glyceraldehyde-3-phosphate dehydrogenase, testis-specific (EC 1.2.1.12) (Spermatogenic cell-specific glyceraldehyde 3-phosphate dehydrogenase 2) (GAPDH-2). | |||||
|
GPNMB_MOUSE
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.017498 (rank : 62) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99P91, Q8BVV9, Q8BXL4, Q9QXA0 | Gene names | Gpnmb, Dchil, Nmb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane glycoprotein NMB precursor (Dendritic cell-associated transmembrane protein) (DC-HIL). | |||||
|
IQEC3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 120) | NC score | 0.006963 (rank : 112) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q3TES0, Q3UHP8, Q80TJ8 | Gene names | Iqsec3, Kiaa1110 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 3. | |||||
|
PEX6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 121) | NC score | 0.002991 (rank : 126) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99LC9, Q6YNQ9 | Gene names | Pex6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
PHLA1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 122) | NC score | 0.017483 (rank : 63) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62392, Q3TY48, Q3UG87 | Gene names | Phlda1, Tdag51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family A member 1 (T-cell death- associated gene 51 protein) (Proline- and glutamine-rich protein) (PQR protein). | |||||
|
PRRT1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | 0.013521 (rank : 81) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O35449 | Gene names | Prrt1, Ng5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 1. | |||||
|
RNPL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.006265 (rank : 116) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HAU8, Q96AC9, Q9H033, Q9NVD0 | Gene names | RNPEPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Arginyl aminopeptidase-like 1 (EC 3.4.11.-) (RNPEP-like protein). | |||||
|
RXRB_MOUSE
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.001159 (rank : 128) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P28704, P33243 | Gene names | Rxrb, Nr2b2 | |||
|
Domain Architecture |
|
|||||
| Description | Retinoic acid receptor RXR-beta (Retinoid X receptor beta) (MHC class I regulatory element-binding protein H-2RIIBP). | |||||
|
SET1A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 126) | NC score | 0.007953 (rank : 105) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
SPN90_MOUSE
|
||||||
| θ value | 8.99809 (rank : 127) | NC score | 0.007661 (rank : 107) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ESJ4, Q68G72 | Gene names | Nckipsd, Spin90, Wasbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3 adapter protein SPIN90 (NCK-interacting protein with SH3 domain) (SH3 protein interacting with Nck, 90 kDa) (VacA-interacting protein, 54 kDa) (VIP54) (N-WASP-interacting protein, 90kDa) (Wiskott-Aldrich syndrome protein-binding protein WISH) (N-WASP-binding protein). | |||||
|
UN119_HUMAN
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.011318 (rank : 93) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13432, O95126 | Gene names | UNC119, RG4 | |||
|
Domain Architecture |
|
|||||
| Description | Unc-119 protein homolog (Retinal protein 4) (HRG4). | |||||
|
WIRE_HUMAN
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | 0.010802 (rank : 96) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
IRF3_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 129 | |
| SwissProt Accessions | Q14653 | Gene names | IRF3 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 3 (IRF-3). | |||||
|
IRF3_MOUSE
|
||||||
| NC score | 0.980458 (rank : 2) | θ value | 9.78303e-172 (rank : 2) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P70671 | Gene names | Irf3 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 3 (IRF-3). | |||||
|
IRF7_MOUSE
|
||||||
| NC score | 0.900998 (rank : 3) | θ value | 3.73686e-38 (rank : 7) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P70434 | Gene names | Irf7 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 7 (IRF-7). | |||||
|
IRF5_MOUSE
|
||||||
| NC score | 0.900261 (rank : 4) | θ value | 5.38352e-45 (rank : 3) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P56477 | Gene names | Irf5 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 5 (IRF-5). | |||||
|
IRF5_HUMAN
|
||||||
| NC score | 0.899644 (rank : 5) | θ value | 1.46607e-42 (rank : 4) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q13568, Q9BQF0 | Gene names | IRF5 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 5 (IRF-5). | |||||
|
IRF6_HUMAN
|
||||||
| NC score | 0.891879 (rank : 6) | θ value | 2.58786e-39 (rank : 5) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O14896 | Gene names | IRF6 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 6 (IRF-6). | |||||
|
IRF6_MOUSE
|
||||||
| NC score | 0.891415 (rank : 7) | θ value | 1.28434e-38 (rank : 6) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P97431 | Gene names | Irf6 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 6 (IRF-6). | |||||
|
IRF7_HUMAN
|
||||||
| NC score | 0.877161 (rank : 8) | θ value | 5.39604e-37 (rank : 8) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q92985, O00331, O00332, O00333, O75924 | Gene names | IRF7 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 7 (IRF-7). | |||||
|
IRF4_HUMAN
|
||||||
| NC score | 0.870845 (rank : 9) | θ value | 1.79631e-32 (rank : 9) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q15306, Q99660 | Gene names | IRF4, MUM1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 4 (IRF-4) (Lymphocyte-specific interferon regulatory factor) (LSIRF) (NF-EM5) (Multiple myeloma oncogene 1). | |||||
|
IRF4_MOUSE
|
||||||
| NC score | 0.870277 (rank : 10) | θ value | 1.16434e-31 (rank : 10) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q64287, Q60802 | Gene names | Irf4, Spip | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 4 (IRF-4) (Lymphocyte-specific interferon regulatory factor) (LSIRF) (NF-EM5) (PU.1 interaction partner) (Transcriptional activator PIP). | |||||
|
IRF8_HUMAN
|
||||||
| NC score | 0.860701 (rank : 11) | θ value | 1.08979e-29 (rank : 11) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q02556 | Gene names | IRF8, ICSBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 8 (IRF-8) (Interferon consensus sequence- binding protein) (ICSBP). | |||||
|
IRF8_MOUSE
|
||||||
| NC score | 0.856896 (rank : 12) | θ value | 3.50572e-28 (rank : 12) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P23611 | Gene names | Irf8, Icsbp, Icsbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 8 (IRF-8) (Interferon consensus sequence- binding protein) (ICSBP). | |||||
|
IRTF_HUMAN
|
||||||
| NC score | 0.837651 (rank : 13) | θ value | 1.42661e-21 (rank : 13) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q00978 | Gene names | ISGF3G, IRF9 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ISGF3 subunit gamma (Interferon regulatory factor 9) (IRF-9) (IFN-alpha-responsive transcription factor subunit) (Interferon-stimulated gene factor 3 gamma) (ISGF3 p48 subunit) (ISGF- 3 gamma). | |||||
|
IRTF_MOUSE
|
||||||
| NC score | 0.804459 (rank : 14) | θ value | 3.76295e-14 (rank : 18) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q61179 | Gene names | Isgf3g | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ISGF3 subunit gamma (IFN-alpha-responsive transcription factor subunit) (Interferon-stimulated gene factor 3 gamma) (ISGF3 p48 subunit) (ISGF-3 gamma). | |||||
|
IRF1_HUMAN
|
||||||
| NC score | 0.786636 (rank : 15) | θ value | 2.60593e-15 (rank : 16) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P10914, Q96GG7 | Gene names | IRF1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 1 (IRF-1). | |||||
|
IRF1_MOUSE
|
||||||
| NC score | 0.783154 (rank : 16) | θ value | 3.40345e-15 (rank : 17) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P15314 | Gene names | Irf1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 1 (IRF-1). | |||||
|
IRF2_HUMAN
|
||||||
| NC score | 0.782186 (rank : 17) | θ value | 8.95645e-16 (rank : 15) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P14316, Q96B99 | Gene names | IRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 2 (IRF-2). | |||||
|
IRF2_MOUSE
|
||||||
| NC score | 0.779761 (rank : 18) | θ value | 4.02038e-16 (rank : 14) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P23906 | Gene names | Irf2 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 2 (IRF-2). | |||||
|
PI5PA_HUMAN
|
||||||
| NC score | 0.051324 (rank : 19) | θ value | 0.0563607 (rank : 20) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
WBP11_HUMAN
|
||||||
| NC score | 0.045634 (rank : 20) | θ value | 0.365318 (rank : 27) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
WBP11_MOUSE
|
||||||
| NC score | 0.044893 (rank : 21) | θ value | 0.47712 (rank : 30) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
ATF5_MOUSE
|
||||||
| NC score | 0.039468 (rank : 22) | θ value | 1.06291 (rank : 40) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O70191, Q58E51, Q99NH6 | Gene names | Atf5, Atfx, Nap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-5 (Activating transcription factor 5-alpha/beta) (Transcription factor ATFx) (Transcription factor-like protein ODA-10) (BZIP protein ATF7) (NRIF3- associated protein) (NAP1). | |||||
|
PI5PA_MOUSE
|
||||||
| NC score | 0.037798 (rank : 23) | θ value | 0.62314 (rank : 33) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P59644 | Gene names | Pib5pa | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
PELP1_MOUSE
|
||||||
| NC score | 0.036945 (rank : 24) | θ value | 2.36792 (rank : 59) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
GR65_HUMAN
|
||||||
| NC score | 0.035774 (rank : 25) | θ value | 0.365318 (rank : 26) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9BQQ3, Q8N272, Q96H42 | Gene names | GORASP1, GRASP65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 1 (Golgi reassembly-stacking protein of 65 kDa) (GRASP65) (Golgi peripheral membrane protein p65) (Golgi phosphoprotein 5) (GOLPH5). | |||||
|
FMN2_MOUSE
|
||||||
| NC score | 0.035697 (rank : 26) | θ value | 0.813845 (rank : 38) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9JL04 | Gene names | Fmn2 | |||
|
Domain Architecture |

|
|||||
| Description | Formin-2. | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.034473 (rank : 27) | θ value | 0.813845 (rank : 36) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
FMN2_HUMAN
|
||||||
| NC score | 0.033363 (rank : 28) | θ value | 0.62314 (rank : 31) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NZ56, Q59GF6, Q5VU37, Q9NZ55 | Gene names | FMN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-2. | |||||
|
B4GN1_HUMAN
|
||||||
| NC score | 0.033129 (rank : 29) | θ value | 0.21417 (rank : 24) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q00973 | Gene names | B4GALNT1, GALGT, SIAT2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4 N-acetylgalactosaminyltransferase 1 (EC 2.4.1.92) ((N- acetylneuraminyl)-galactosylglucosylceramide) (GM2/GD2 synthase) (GalNAc-T). | |||||
|
PTN23_MOUSE
|
||||||
| NC score | 0.029666 (rank : 30) | θ value | 0.0252991 (rank : 19) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.029661 (rank : 31) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
CDN1C_HUMAN
|
||||||
| NC score | 0.028092 (rank : 32) | θ value | 1.06291 (rank : 41) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P49918 | Gene names | CDKN1C, KIP2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
FMNL_HUMAN
|
||||||
| NC score | 0.027653 (rank : 33) | θ value | 1.38821 (rank : 42) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O95466, Q6DKG5, Q6IBP3, Q86UH1, Q8N671, Q8TDH1, Q96H10 | Gene names | FMNL1, C17orf1, C17orf1B, FMNL | |||
|
Domain Architecture |

|
|||||
| Description | Formin-like protein 1 (Leukocyte formin) (CLL-associated antigen KW- 13). | |||||
|
ATF5_HUMAN
|
||||||
| NC score | 0.026969 (rank : 34) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y2D1, Q9BSA1, Q9UNQ3 | Gene names | ATF5, ATFX | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-5 (Activating transcription factor 5) (Transcription factor ATFx). | |||||
|
PELP1_HUMAN
|
||||||
| NC score | 0.026258 (rank : 35) | θ value | 1.38821 (rank : 43) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8IZL8, O15450, Q5EGN3, Q6NTE6, Q96FT1, Q9BU60 | Gene names | PELP1, HMX3, MNAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3). | |||||
|
CIC_MOUSE
|
||||||
| NC score | 0.025661 (rank : 36) | θ value | 1.81305 (rank : 44) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
TM16H_HUMAN
|
||||||
| NC score | 0.025591 (rank : 37) | θ value | 0.62314 (rank : 35) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9HCE9 | Gene names | TMEM16H, KIAA1623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 16H. | |||||
|
PHLA1_HUMAN
|
||||||
| NC score | 0.025563 (rank : 38) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WV24, Q15184, Q2TAN2, Q9NZ17 | Gene names | PHLDA1, PHRIP, TDAG51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family A member 1 (T-cell death- associated gene 51 protein) (Apoptosis-associated nuclear protein) (Proline- and histidine-rich protein) (Proline- and glutamine-rich protein) (PQ-rich protein). | |||||
|
MOT8_MOUSE
|
||||||
| NC score | 0.024546 (rank : 39) | θ value | 0.47712 (rank : 29) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O70324, Q8K3S9 | Gene names | Slc16a2, Mct8, Xpct | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Monocarboxylate transporter 8 (MCT 8) (Solute carrier family 16 member 2) (X-linked PEST-containing transporter). | |||||
|
DIAP2_MOUSE
|
||||||
| NC score | 0.024490 (rank : 40) | θ value | 1.81305 (rank : 45) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O70566, Q8C2G8 | Gene names | Diaph2, Diap2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 2 (Diaphanous-related formin-2) (DRF2) (mDia3). | |||||
|
HD_MOUSE
|
||||||
| NC score | 0.023956 (rank : 41) | θ value | 2.36792 (rank : 55) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P42859 | Gene names | Hd, Hdh | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin (Huntington disease protein homolog) (HD protein). | |||||
|
CO039_MOUSE
|
||||||
| NC score | 0.023581 (rank : 42) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q3TEI4, Q3TMF2, Q3U253, Q3UM75, Q9DA93 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39 homolog. | |||||
|
TROAP_HUMAN
|
||||||
| NC score | 0.023226 (rank : 43) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q12815 | Gene names | TROAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trophinin-associated protein (Tastin) (Trophinin-assisting protein). | |||||
|
ZPR1_HUMAN
|
||||||
| NC score | 0.022852 (rank : 44) | θ value | 2.36792 (rank : 61) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75312, Q2TAA0 | Gene names | ZNF259, ZPR1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein ZPR1 (Zinc finger protein 259). | |||||
|
IF4G1_MOUSE
|
||||||
| NC score | 0.022612 (rank : 45) | θ value | 1.81305 (rank : 46) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.022072 (rank : 46) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
GPNMB_HUMAN
|
||||||
| NC score | 0.021824 (rank : 47) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14956, Q6UVX1, Q8N1A1 | Gene names | GPNMB, HGFIN, NMB | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane glycoprotein NMB precursor (Transmembrane glycoprotein HGFIN). | |||||
|
HD_HUMAN
|
||||||
| NC score | 0.021624 (rank : 48) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P42858, Q9UQB7 | Gene names | HD, IT15 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin (Huntington disease protein) (HD protein). | |||||
|
TAF3_HUMAN
|
||||||
| NC score | 0.021330 (rank : 49) | θ value | 0.62314 (rank : 34) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
SYNJ1_HUMAN
|
||||||
| NC score | 0.020208 (rank : 50) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O43426, O43425, O94984 | Gene names | SYNJ1, KIAA0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
DIAP1_HUMAN
|
||||||
| NC score | 0.020181 (rank : 51) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O60610, Q9UC76 | Gene names | DIAPH1, DIAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1). | |||||
|
PTN23_HUMAN
|
||||||
| NC score | 0.019925 (rank : 52) | θ value | 3.0926 (rank : 67) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H3S7, Q7KZF8, Q8N6Z5, Q9BSR5, Q9P257, Q9UG03, Q9UMZ4 | Gene names | PTPN23, KIAA1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His- domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14). | |||||
|
SPR2F_MOUSE
|
||||||
| NC score | 0.019913 (rank : 53) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O70557 | Gene names | Sprr2f | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2F. | |||||
|
GGN_HUMAN
|
||||||
| NC score | 0.019708 (rank : 54) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86UU5, Q7RTU6, Q86UU4, Q8NAA1 | Gene names | GGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
DEN1A_HUMAN
|
||||||
| NC score | 0.019210 (rank : 55) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8TEH3, Q5VWF0, Q8IVD6, Q9H796 | Gene names | DENND1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 1A. | |||||
|
ODP2_HUMAN
|
||||||
| NC score | 0.019096 (rank : 56) | θ value | 2.36792 (rank : 58) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P10515, Q16783 | Gene names | DLAT, DLTA | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial precursor (EC 2.3.1.12) (Pyruvate dehydrogenase complex E2 subunit) (PDCE2) (E2) (Dihydrolipoamide S- acetyltransferase component of pyruvate dehydrogenase complex) (PDC- E2) (70 kDa mitochondrial autoantigen of primary biliary cirrhosis) (PBC) (M2 antigen complex 70 kDa subunit). | |||||
|
T53I2_HUMAN
|
||||||
| NC score | 0.018603 (rank : 57) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IXH6, Q5JX64, Q8IYL5, Q9NU00 | Gene names | TP53INP2, C20orf110, PINH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor protein p53-inducible nuclear protein 2 (p53-inducible protein U) (PIG-U). | |||||
|
CRSP7_HUMAN
|
||||||
| NC score | 0.018417 (rank : 58) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O95402 | Gene names | CRSP7, ARC70 | |||
|
Domain Architecture |

|
|||||
| Description | CRSP complex subunit 7 (Cofactor required for Sp1 transcriptional activation subunit 7) (Transcriptional coactivator CRSP70) (Activator- recruited cofactor 70 kDa component) (ARC70). | |||||
|
CJ047_HUMAN
|
||||||
| NC score | 0.018410 (rank : 59) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86WR7, Q5W0J9, Q5W0K0, Q5W0K1, Q5W0K2, Q6PJC8, Q8N317 | Gene names | C10orf47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf47. | |||||
|
DGKK_HUMAN
|
||||||
| NC score | 0.018052 (rank : 60) | θ value | 2.36792 (rank : 53) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5KSL6 | Gene names | DGKK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase kappa (EC 2.7.1.107) (Diglyceride kinase kappa) (DGK-kappa) (DAG kinase kappa) (142 kDa diacylglycerol kinase). | |||||
|
ENAH_MOUSE
|
||||||
| NC score | 0.017861 (rank : 61) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
GPNMB_MOUSE
|
||||||
| NC score | 0.017498 (rank : 62) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99P91, Q8BVV9, Q8BXL4, Q9QXA0 | Gene names | Gpnmb, Dchil, Nmb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane glycoprotein NMB precursor (Dendritic cell-associated transmembrane protein) (DC-HIL). | |||||
|
PHLA1_MOUSE
|
||||||
| NC score | 0.017483 (rank : 63) | θ value | 8.99809 (rank : 122) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62392, Q3TY48, Q3UG87 | Gene names | Phlda1, Tdag51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family A member 1 (T-cell death- associated gene 51 protein) (Proline- and glutamine-rich protein) (PQR protein). | |||||
|
SMRC2_HUMAN
|
||||||
| NC score | 0.017210 (rank : 64) | θ value | 4.03905 (rank : 75) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8TAQ2, Q92923, Q96E12, Q96GY4 | Gene names | SMARCC2, BAF170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
RFTN1_MOUSE
|
||||||
| NC score | 0.017093 (rank : 65) | θ value | 3.0926 (rank : 68) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6A0D4, Q3U654, Q3U8P6, Q80US1 | Gene names | Rftn1, Kiaa0084 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Raftlin (Raft-linking protein). | |||||
|
ANTR1_HUMAN
|
||||||
| NC score | 0.017066 (rank : 66) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H6X2, Q96P02, Q9NVP3 | Gene names | ANTXR1, ATR, TEM8 | |||
|
Domain Architecture |

|
|||||
| Description | Anthrax toxin receptor 1 precursor (Tumor endothelial marker 8). | |||||
|
SPR2E_MOUSE
|
||||||
| NC score | 0.017021 (rank : 67) | θ value | 6.88961 (rank : 107) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O70556 | Gene names | Sprr2e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2E. | |||||
|
ANTR1_MOUSE
|
||||||
| NC score | 0.016993 (rank : 68) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9CZ52 | Gene names | Antxr1, Atr, Tem8 | |||
|
Domain Architecture |

|
|||||
| Description | Anthrax toxin receptor 1 precursor (Tumor endothelial marker 8). | |||||
|
K1802_HUMAN
|
||||||
| NC score | 0.016657 (rank : 69) | θ value | 1.81305 (rank : 47) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
ENAH_HUMAN
|
||||||
| NC score | 0.016437 (rank : 70) | θ value | 0.813845 (rank : 37) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8N8S7, Q502W5, Q5T5M7, Q5VTQ9, Q5VTR0, Q9NVF3, Q9UFB8 | Gene names | ENAH, MENA | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog. | |||||
|
K1949_MOUSE
|
||||||
| NC score | 0.016103 (rank : 71) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BQ30, Q3UDS6, Q8CEY9, Q8R3D8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1949 homolog. | |||||
|
AUTS2_HUMAN
|
||||||
| NC score | 0.015902 (rank : 72) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8WXX7, Q9Y4F2 | Gene names | AUTS2, KIAA0442 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autism susceptibility gene 2 protein. | |||||
|
SMRC2_MOUSE
|
||||||
| NC score | 0.015765 (rank : 73) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6PDG5, Q6P6P3 | Gene names | Smarcc2, Baf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
EAF1_MOUSE
|
||||||
| NC score | 0.015437 (rank : 74) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D4C5 | Gene names | Eaf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELL-associated factor 1. | |||||
|
CEND3_HUMAN
|
||||||
| NC score | 0.014500 (rank : 75) | θ value | 2.36792 (rank : 52) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8WWN8 | Gene names | CENTD3, ARAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3). | |||||
|
PRDM2_HUMAN
|
||||||
| NC score | 0.014380 (rank : 76) | θ value | 0.0563607 (rank : 21) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
GP1BA_HUMAN
|
||||||
| NC score | 0.014311 (rank : 77) | θ value | 0.47712 (rank : 28) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P07359, Q14441, Q16469, Q8N1F3, Q8NG39, Q9HDC7, Q9UEK1, Q9UQS4 | Gene names | GP1BA | |||
|
Domain Architecture |

|
|||||
| Description | Platelet glycoprotein Ib alpha chain precursor (Glycoprotein Ibalpha) (GP-Ib alpha) (GPIbA) (GPIb-alpha) (Antigen CD42b-alpha) (CD42b antigen) [Contains: Glycocalicin]. | |||||
|
CBLB_HUMAN
|
||||||
| NC score | 0.014239 (rank : 78) | θ value | 3.0926 (rank : 62) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13191, Q13192, Q13193, Q3LIC0, Q8IVC5 | Gene names | CBLB, RNF56 | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase CBL-B (EC 6.3.2.-) (Signal transduction protein CBL-B) (SH3-binding protein CBL-B) (Casitas B-lineage lymphoma proto-oncogene b) (RING finger protein 56). | |||||
|
EAF1_HUMAN
|
||||||
| NC score | 0.013866 (rank : 79) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96JC9, Q8IW10 | Gene names | EAF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELL-associated factor 1. | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.013793 (rank : 80) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
PRRT1_MOUSE
|
||||||
| NC score | 0.013521 (rank : 81) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O35449 | Gene names | Prrt1, Ng5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 1. | |||||
|
CEND3_MOUSE
|
||||||
| NC score | 0.013446 (rank : 82) | θ value | 3.0926 (rank : 63) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R5G7, Q5DTN4, Q6NVF1, Q8R5G6 | Gene names | Centd3, Arap3, Drag1, Kiaa4097 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3) (Dual specificity Rho- and Arf-GTPase-activating protein 1). | |||||
|
MMP9_MOUSE
|
||||||
| NC score | 0.012554 (rank : 83) | θ value | 0.62314 (rank : 32) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P41245, Q06788, Q9DC02 | Gene names | Mmp9, Clg4b | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-9 precursor (EC 3.4.24.35) (MMP-9) (92 kDa type IV collagenase) (92 kDa gelatinase) (Gelatinase B) (GELB). | |||||
|
JPH4_MOUSE
|
||||||
| NC score | 0.012368 (rank : 84) | θ value | 2.36792 (rank : 56) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80WT0, Q8BMI1 | Gene names | Jph4, Jphl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-4 (Junctophilin-like 1 protein). | |||||
|
SPR2D_MOUSE
|
||||||
| NC score | 0.012316 (rank : 85) | θ value | 6.88961 (rank : 106) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O70555 | Gene names | Sprr2d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2D. | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.012290 (rank : 86) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
ATBF1_MOUSE
|
||||||
| NC score | 0.011881 (rank : 87) | θ value | 2.36792 (rank : 51) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
CBLB_MOUSE
|
||||||
| NC score | 0.011720 (rank : 88) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q3TTA7 | Gene names | Cblb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase CBL-B (EC 6.3.2.-) (Signal transduction protein CBL-B) (SH3-binding protein CBL-B) (Casitas B-lineage lymphoma proto-oncogene b). | |||||
|
VMDL1_MOUSE
|
||||||
| NC score | 0.011593 (rank : 89) | θ value | 3.0926 (rank : 69) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BGM5, Q8VCM0 | Gene names | Vmd2l1 | |||
|
Domain Architecture |

|
|||||
| Description | Bestrophin-2 (Vitelliform macular dystrophy 2-like protein 1). | |||||
|
PXK_MOUSE
|
||||||
| NC score | 0.011553 (rank : 90) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 320 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BX57, Q3TD60, Q3TEL7, Q3TZZ1, Q3U197, Q3U773, Q3UCS5, Q4FBH7, Q91WB6 | Gene names | Pxk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PX domain-containing protein kinase-like protein (Modulator of Na,K- ATPase) (MONaKA). | |||||
|
ATBF1_HUMAN
|
||||||
| NC score | 0.011489 (rank : 91) | θ value | 2.36792 (rank : 50) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
TRRAP_HUMAN
|
||||||
| NC score | 0.011420 (rank : 92) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y4A5, O75218, Q9Y631, Q9Y6H4 | Gene names | TRRAP, PAF400 | |||
|
Domain Architecture |

|
|||||
| Description | Transformation/transcription domain-associated protein (350/400 kDa PCAF-associated factor) (PAF350/400) (STAF40) (Tra1 homolog). | |||||
|
UN119_HUMAN
|
||||||
| NC score | 0.011318 (rank : 93) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13432, O95126 | Gene names | UNC119, RG4 | |||
|
Domain Architecture |

|
|||||
| Description | Unc-119 protein homolog (Retinal protein 4) (HRG4). | |||||
|
RB_MOUSE
|
||||||
| NC score | 0.011270 (rank : 94) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P13405 | Gene names | Rb1, Rb-1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoblastoma-associated protein (PP105) (RB). | |||||
|
KCNA5_HUMAN
|
||||||
| NC score | 0.010806 (rank : 95) | θ value | 0.813845 (rank : 39) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P22460 | Gene names | KCNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (HK2) (HPCN1). | |||||
|
WIRE_HUMAN
|
||||||
| NC score | 0.010802 (rank : 96) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
GNDS_MOUSE
|
||||||
| NC score | 0.010377 (rank : 97) | θ value | 4.03905 (rank : 74) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q03385 | Gene names | Ralgds, Rgds | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator (RalGEF) (RalGDS). | |||||
|
WASF2_MOUSE
|
||||||
| NC score | 0.010285 (rank : 98) | θ value | 6.88961 (rank : 108) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BH43 | Gene names | Wasf2, Wave2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2). | |||||
|
FA13C_MOUSE
|
||||||
| NC score | 0.009852 (rank : 99) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBR2 | Gene names | Fam13c1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13C1. | |||||
|
KCNC3_MOUSE
|
||||||
| NC score | 0.009801 (rank : 100) | θ value | 2.36792 (rank : 57) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q63959, Q62088 | Gene names | Kcnc3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
G3PT_MOUSE
|
||||||
| NC score | 0.008906 (rank : 101) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q64467, Q60650 | Gene names | Gapdhs, Gapd-s, Gapds | |||
|
Domain Architecture |

|
|||||
| Description | Glyceraldehyde-3-phosphate dehydrogenase, testis-specific (EC 1.2.1.12) (Spermatogenic cell-specific glyceraldehyde 3-phosphate dehydrogenase 2) (GAPDH-2). | |||||
|
MYO15_MOUSE
|
||||||
| NC score | 0.008887 (rank : 102) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9QZZ4, O70395, Q9QWL6 | Gene names | Myo15a, Myo15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
GLSK_HUMAN
|
||||||
| NC score | 0.008327 (rank : 103) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O94925, Q9UL05, Q9UL06, Q9UL07, Q9UN40 | Gene names | GLS, GLS1, KIAA0838 | |||
|
Domain Architecture |

|
|||||
| Description | Glutaminase kidney isoform, mitochondrial precursor (EC 3.5.1.2) (GLS) (L-glutamine amidohydrolase) (K-glutaminase). | |||||
|
GGA3_HUMAN
|
||||||
| NC score | 0.007960 (rank : 104) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NZ52, Q15017, Q6IS16, Q9UJY3 | Gene names | GGA3, KIAA0154 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.007953 (rank : 105) | θ value | 8.99809 (rank : 126) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
AFF1_MOUSE
|
||||||
| NC score | 0.007879 (rank : 106) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
SPN90_MOUSE
|
||||||
| NC score | 0.007661 (rank : 107) | θ value | 8.99809 (rank : 127) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ESJ4, Q68G72 | Gene names | Nckipsd, Spin90, Wasbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3 adapter protein SPIN90 (NCK-interacting protein with SH3 domain) (SH3 protein interacting with Nck, 90 kDa) (VacA-interacting protein, 54 kDa) (VIP54) (N-WASP-interacting protein, 90kDa) (Wiskott-Aldrich syndrome protein-binding protein WISH) (N-WASP-binding protein). | |||||
|
FA44A_HUMAN
|
||||||
| NC score | 0.007493 (rank : 108) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NFC6, Q6P0M8 | Gene names | FAM44A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM44A. | |||||
|
CNTP1_HUMAN
|
||||||
| NC score | 0.007306 (rank : 109) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P78357 | Gene names | CNTNAP1, CASPR, NRXN4 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein 1 precursor (Caspr) (Caspr1) (Neurexin 4) (Neurexin IV) (p190). | |||||
|
MK07_HUMAN
|
||||||
| NC score | 0.007265 (rank : 110) | θ value | 0.163984 (rank : 22) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1087 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13164, Q16634 | Gene names | MAPK7, ERK4, ERK5, PRKM7 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase 7 (EC 2.7.11.24) (Extracellular signal-regulated kinase 5) (ERK-5) (ERK4) (BMK1 kinase). | |||||
|
IRX3_MOUSE
|
||||||
| NC score | 0.007050 (rank : 111) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P81067 | Gene names | Irx3, Irxb1 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-3 (Iroquois homeobox protein 3) (Homeodomain protein IRXB1). | |||||
|
IQEC3_MOUSE
|
||||||
| NC score | 0.006963 (rank : 112) | θ value | 8.99809 (rank : 120) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q3TES0, Q3UHP8, Q80TJ8 | Gene names | Iqsec3, Kiaa1110 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 3. | |||||
|
MK07_MOUSE
|
||||||
| NC score | 0.006847 (rank : 113) | θ value | 0.163984 (rank : 23) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1054 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9WVS8 | Gene names | Mapk7, Erk5 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase 7 (EC 2.7.11.24) (Extracellular signal-regulated kinase 5) (ERK-5) (BMK1 kinase). | |||||
|
KCNN3_MOUSE
|
||||||
| NC score | 0.006756 (rank : 114) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P58391 | Gene names | Kcnn3, Sk3 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 3 (SK3). | |||||
|
DLGP2_MOUSE
|
||||||
| NC score | 0.006300 (rank : 115) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BJ42, Q6XBF3 | Gene names | Dlgap2, Dap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large-associated protein 2 (DAP-2) (SAP90/PSD-95-associated protein 2) (SAPAP2) (PSD-95/SAP90-binding protein 2). | |||||
|
RNPL1_HUMAN
|
||||||
| NC score | 0.006265 (rank : 116) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HAU8, Q96AC9, Q9H033, Q9NVD0 | Gene names | RNPEPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Arginyl aminopeptidase-like 1 (EC 3.4.11.-) (RNPEP-like protein). | |||||
|
FBLI1_HUMAN
|
||||||
| NC score | 0.006249 (rank : 117) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WUP2, Q5VVE0, Q5VVE1, Q8IX23, Q96T00, Q9UFD6 | Gene names | FBLIM1, FBLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-binding LIM protein 1 (Filamin-binding LIM protein 1) (FBLP-1) (Mitogen-inducible 2-interacting protein) (MIG2-interacting protein) (Migfilin). | |||||
|
RX_HUMAN
|
||||||
| NC score | 0.005096 (rank : 118) | θ value | 2.36792 (rank : 60) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y2V3 | Gene names | RAX, RX | |||
|
Domain Architecture |

|
|||||
| Description | Retinal homeobox protein Rx (Retina and anterior neural fold homeobox protein). | |||||
|
FOG1_HUMAN
|
||||||
| NC score | 0.005037 (rank : 119) | θ value | 2.36792 (rank : 54) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 981 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8IX07 | Gene names | ZFPM1, FOG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
HCN2_HUMAN
|
||||||
| NC score | 0.005026 (rank : 120) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
FOXJ3_HUMAN
|
||||||
| NC score | 0.004751 (rank : 121) | θ value | 4.03905 (rank : 73) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UPW0, Q9NSS7 | Gene names | FOXJ3, KIAA1041 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein J3. | |||||
|
SMCA4_HUMAN
|
||||||
| NC score | 0.004118 (rank : 122) | θ value | 6.88961 (rank : 105) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P51532, O95052, Q9HBD3 | Gene names | SMARCA4, BRG1, SNF2B, SNF2L4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L4 (EC 3.6.1.-) (ATP- dependent helicase SMARCA4) (SNF2-beta) (BRG-1 protein) (Mitotic growth and transcription activator) (Brahma protein homolog 1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4). | |||||
|
FARP2_HUMAN
|
||||||
| NC score | 0.003743 (rank : 123) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O94887, Q53QM5, Q8WU27, Q9UFE7 | Gene names | FARP2, KIAA0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
WNK2_HUMAN
|
||||||
| NC score | 0.003698 (rank : 124) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
SN_MOUSE
|
||||||
| NC score | 0.003321 (rank : 125) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62230, O55216, Q62228, Q62229 | Gene names | Sn, Sa | |||
|
Domain Architecture |

|
|||||
| Description | Sialoadhesin precursor (Sialic acid-binding Ig-like lectin-1) (Siglec- 1) (Sheep erythrocyte receptor) (SER) (CD169 antigen). | |||||
|
PEX6_MOUSE
|
||||||
| NC score | 0.002991 (rank : 126) | θ value | 8.99809 (rank : 121) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99LC9, Q6YNQ9 | Gene names | Pex6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
SN1L1_HUMAN
|
||||||
| NC score | 0.002268 (rank : 127) | θ value | 0.279714 (rank : 25) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 911 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P57059, Q5R2V5, Q86YJ2 | Gene names | SNF1LK | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase SNF1-like kinase 1 (EC 2.7.11.1) (Serine/threonine-protein kinase SNF1LK). | |||||
|
RXRB_MOUSE
|
||||||
| NC score | 0.001159 (rank : 128) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P28704, P33243 | Gene names | Rxrb, Nr2b2 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor RXR-beta (Retinoid X receptor beta) (MHC class I regulatory element-binding protein H-2RIIBP). | |||||
|
ZN358_HUMAN
|
||||||
| NC score | -0.001461 (rank : 129) | θ value | 1.81305 (rank : 49) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 840 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NW07, Q9BTM7 | Gene names | ZNF358 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 358. | |||||